Key Points
The retroviral family is described, and an overview of the retroviral life cycle, the genetic organization and a brief description of the process of translation initiation in Eucaryotes is provided.
The role of viral RNA structures on translation initiation is discussed.
IRES elements characterized in HIV-1, HIV-2 and SIV, and those which are located both within the 5′-UTR and the Gag coding region, are responsible for the synthesis of the N-truncated shorter Gag isoforms.
Short upstream reading frames regulate translation initiation in avian simple retroviruses such as Rous sarcoma virus (RSV).
RNA translational enhancing elements that have been described in Mason-Pfizer monkey virus (M-PMV) and the avian spleen necrosis virus (SNV) are discussed.
The interaction between the TAR RNA structure and the Tat protein also has a significant regulatory role in translation that is mainly exerted through the PKR pathway.
The interaction of Rev with the RNA responsive element was shown to enhance Gag production by a mechanism that is not due to an increase of genomic RNA export, but is rather due to an enhanced recruitment of this genomic RNA into polyribosomes.
The viral protease of a large number of retroviruses including MLV, HIV-1, HIV-2 and SIV can induce the cleavage of the initiation factor eIF4GI, which is an essential component of the host translational apparatus. Such a proteolytic event was shown to strongly inhibit ribosomal scanning.
Retroviruses are a unique family of RNA viruses that depend on the translational machinery of the host cell for protein synthesis. Here, the mechanisms used by these viruses to ensure efficient protein synthesis within a highly competitive cellular environment are reviewed.
Abstract
All replication-competent retroviruses contain three main reading frames, gag, pol and env, which are used for the synthesis of structural proteins, enzymes and envelope proteins respectively. Complex retroviruses, such as lentiviruses, also code for regulatory and accessory proteins that have essential roles in viral replication. The concerted expression of these genes ensures the efficient polypeptide production required for the assembly and release of new infectious progeny virions. Retroviral protein synthesis takes place in the cytoplasm and depends exclusively on the translational machinery of the host infected cell. Therefore, not surprisingly, retroviruses have developed RNA structures and strategies to promote robust and efficient expression of viral proteins in a competitive cellular environment.
Main
Retroviruses are a unique family of RNA viruses that use virally encoded reverse transcriptase (RT) to replicate genomic RNA through a full-length viral DNA intermediate. These viruses have been shown to be present in the genomes of many vertebrates, including fish, rodents, birds, cats, ungulants, non-human primates and humans. Infection by retroviruses causes a wide variety of pathologies, most commonly cancers, such as leukaemias, sarcomas and mammary carcinomas, but retroviral infection can also cause immunodeficiencies, anaemias, arthritis and pneumonia1. The retroviridae are classified into seven different genera that are named α through to ε retroviruses, as well as the lentiviruses and spumaviruses (see the International Committee on Taxonomy of Viruses database). Historically, retroviruses have been the source of many key discoveries in biology during the twentieth century, including cell transformation, viral and cellular oncogenes, RT and viral transduction, which paved the way to cDNA cloning and design of retroviral vectors for gene therapy1.
This Review will focus on the mechanisms used by retroviruses to ensure the correct viral protein synthesis within the cytoplasm of the host infected cell with particular emphasis on α- and γ-retroviruses (whose prototypes are the avian leukosis virus (ALV) and murine leukaemia virus (MLV), respectively) and primate lentiviruses (HIV-1, HIV-2 and simian immunodeficiency virus (SIV)).
An overview of the retroviral life cycle. Retroviruses are enveloped RNA viruses that encapsidate two copies of the same capped and polyadenylated (positive sense) RNA molecule that ranges from 8,000 to 11,000 nucleotides in size. In the viral particle, the two RNA copies are linked by their 5′-end to form a dimeric RNA structure, and are coated by 1,500–2,000 molecules of nucleocapsid protein2. These viruses are characterized by their unique property to reverse transcribe their RNA genome into double-stranded (ds) DNA and to integrate this dsDNA into the nucleus of the host cell (to become the proviral DNA)3. All retroviruses have a similar genetic organization consisting of the gag, pol and env genes, which are translated as polyprotein precursors and are later processed to yield internal structural proteins, enzymes and the envelope glycoproteins, respectively. In addition, lentiviruses code for regulatory genes that are required, among other functions, to coordinate viral gene expression at different levels, such as transcription, nuclear export and translational regulation.
The retroviral life cycle begins with the interaction of the infectious viral particle with the appropriate membrane receptors and co-receptors (Fig. 1a). This triggers membrane fusion and nucleocapsid entry into the cytoplasm of the cell (Fig.1b). At this stage, the genomic RNA is reverse-transcribed into dsDNA by the virally encoded RT enzyme. Full-length viral DNA, in the form of a preintegration complex, is then imported to the nucleus where it integrates into the host chromosome (Fig. 1c). The viral long terminal repeats (LTRs), flanking the provirus, allow the synthesis of the viral genomic RNA by the cellular RNA polymerase II. As it is being synthesized, the primary transcription product interacts with the cellular RNA processing machinery and, similar to a typical cellular pre-mRNA, is spliced, exported to the cytoplasm, and translated by the host protein-synthesis machinery (Fig. 1d). A proportion of the pre-mRNA subverts typical RNA processing and interacts with viral and/or cellular nucleocytoplasmic shuttle proteins that activate nuclear export, despite the lack of intron removal4. In the cytoplasm, translation of the spliced, and incompletely–spliced, viral mRNAs depends exclusively on the host-cell translational machinery5 (Fig. 1e). The unspliced, genome-length RNA serves yet another function, as it is packaged into assembling virions as genomic RNA. In this respect, some simple retroviruses, such as MLV, contain two populations of unspliced viral RNAs that segregate either as mRNA for translation or as genomic RNA for encapsidation6,7. Complex retroviruses, however, present a single RNA population that functions interchangeably as both mRNA and genomic RNA8,9. After synthesis, the viral structural proteins are targeted to the plasma membrane where assembly takes place. For HIV-1 this process is associated with cholesterol-rich membrane microdomains known as rafts10,11,12,13 (Fig. 1f). However, with some retroviruses, such as mouse mammary tumour virus (MMTV), Mason-Pfizer monkey virus (M-PMV) and spumaviruses, Gag-containing polyproteins assemble within the cytoplasm, forming stable particles that are then transported to the plasma membrane where the envelope is acquired during the budding process5. Interestingly, certain defective endogenous retroviruses bud exclusively from the endoplasmic reticulum (ER)5.
Figure 1. An overview of the retroviral life cycle.
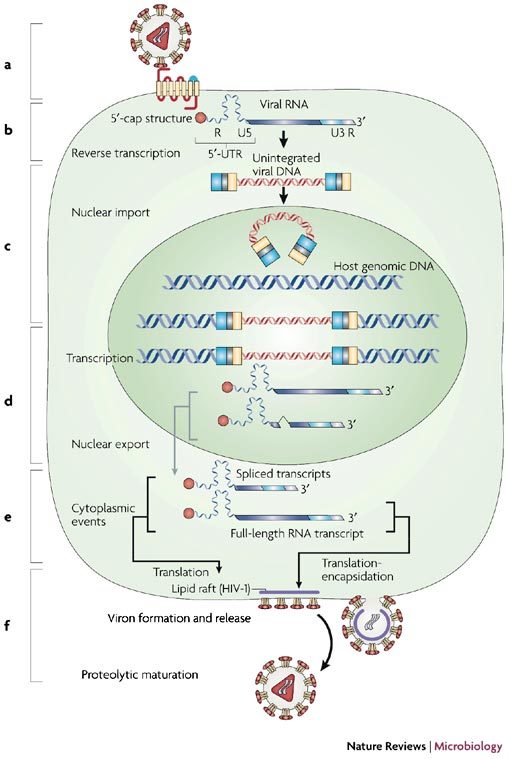
The life cycle of retroviruses can be broadly divided into six essential steps, which are shown schematically in the figure: interaction, adsorption and entry into the cell (a); synthesis of reverse transcribed DNA (b); integration (c); transcription, splicing and nuclear export (d); translation and encapsidation (e); and viral assembly and budding (f).
Genetic organization of the retroviral genomic RNA. The proviral DNA consists of three main open reading frames (ORFs) flanked by two LTRs that are involved in the integration and transcription processes2,3. Transcription of proviral DNA generates genomic full length retroviral mRNA that functions as genomic and messenger RNA, and possesses well-defined functional domains, including the R region, which corresponds to the +1 site of transcription and the site of capping, the 5′-untranslated region (5′-UTR) or leader, the viral ORFs, the 3′-UTR and the 3′-poly (A) tail. The 5′-UTR of the genomic RNA contains cis-acting RNA structures that are crucially involved in various aspects of the viral life cycle14,15,16,17,18 (Fig. 2). These structures include: first, the transactivation response element (TAR; only found in lentiviruses), which transactivates viral transcription and modulates viral mRNA translation (see below); second, the poly adenylation stem loop, the structure responsible for the addition of the 3′-poly (A) tail; third, the primer binding site (PBS), which is complementary to the 3′ portion of the cellular tRNAlys3 and functions as the recruitment point for the initiation of reverse transcription; fourth, the major splice donor (SD), which is the most upstream splice site used in the generation of viral subgenomic RNAs; fifth, the dimerization initiation site (DIS), which is involved in the establishment of the kissing-loop complex between two genomic RNA molecules2; and finally, the core packaging signals, necessary elements required for the selection and direction of the genomic RNA to the viral assembly machinery.
Figure 2. Genetic organization of the 5′-untranslated region (UTR) of the retroviral genomic RNA: the example of SIVmac.
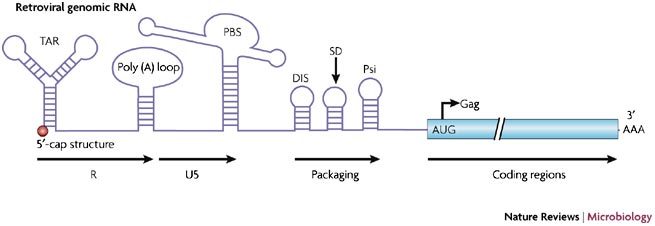
The 5′-UTR (or leader) of the simian immunodeficiency virus (SIV) genomic RNA contains numerous cis-acting sequences, such as the TAR stem-loop, the poly (A) loop, the primer-binding site (PBS), the dimerization initiation site (DIS), the core packaging signals (Psi) and the major splice donor (SD). The R-U5, packaging and coding regions are indicated in the figure.
General mechanism of retroviral protein synthesis. Similar to all cellular mRNAs, retroviral mRNA is synthesized by RNA polymerase II, therefore it exhibits all the characteristics associated with eukaryotic mRNA, including a 5′-cap structure, a 5′-UTR, a 3′-UTR and a 3′-poly (A) tail4. Following nuclear export, structural proteins and enzymes are synthesized from the unspliced mRNA (Gag and Gag-Pol) on free ribosomes, whereas the precursor envelope protein is expressed from spliced viral mRNA (simple retroviruses) or incompletely spliced viral mRNA (complex retroviruses) on membrane-bound ribosomes5.
The overall expression of Gag and Pol proteins is regulated by a subtle mechanism. In all retroviruses, the gag gene is the first reading frame encoded on the genome and is located upstream of pol; as such, ribosomes synthesize the Gag protein until they encounter a stop codon. At this point, the strategy differs among different members of the retrovirus family. In γ-retroviruses, the stop codon is misread and there is a termination suppression process that results in the translation of the downstream pol gene. This mechanism occurs in Moloney MLV (MoMLV) by the suppression of an amber termination codon (UAG), occasionally decoded by a glutamine tRNA19,20. The second mechanism, used by most retroviruses, is a frameshift mechanism whereby the ribosomes slip backward one nucleotide just before they encounter the stop codon of the gag gene21,22. This mechanism is dependent on two specific features characteristic of the viral mRNA: a heptanucleotide 'slippery' sequence21 located upstream of an RNA pseudoknot23. Recently, the molecular mechanism underlying '-1 frameshifting' was elegantly solved by cryoelectron microscopic imaging of ribosomes engaged in the coronavirus RNA pseudoknot. This analysis revealed that the 80S ribosome interacts with the RNA pseudoknot creating a physical block at the mRNA entrance channel. This results in the deformation of the P-site transfer RNA and this structural change impedes ribosomal translocation24. As a result, ribosomes leave the gag reading frame and translate the pol gene. In HIV-1, the efficiency of such a mechanism is comparable to that of termination suppression and yields approximately 20 Gag proteins for each Gag-Pol molecule synthesized. Disruption of this ratio in HIV-1 reduces the infectivity of the virus by 250- to 1,000–fold, showing the importance of this evolutionary-conserved mechanism25.
The role of viral RNA structures on translation
IRES elements and simple retroviruses. MLV has been extensively studied and is considered the prototype for mammalian g retroviruses. Although MLV is classified as a simple retrovirus, it encodes an additional longer glycosylated isoform of the Gag polyprotein (GCSA or glycoGag). GCSA is a membrane-bound protein that undergoes processing but is not incorporated into virions. Instead, the protein is exposed on the extra-cellular side of the plasma membrane, where it displays potential mitogenic activity26,27. In the case of the Friend MLV (FMLV), GCSA is not necessary for virus replication in cell culture but is required for virus pathogenesis, that is, haemolytic anaemia and erythroleukaemia in mice27,28. The synthesis of GCSA displays some unique characteristics as translation is initiated at two CUG codons, 300 nucleotides upstream from the Gag AUG and in the same reading frame26. Both Gag and GCSA are produced by two distinct internal ribosome entry segments (IRES) elements29,30(Box 1) that overlap the MLV and FMLV packaging signal31, indicating that the translation of the full-length mRNA and its recruitment as genomic RNA into newly formed virions could be functionally linked. Though possible, several reports suggest that simple retroviruses generate two independent pools of RNA, each predestined to a different fate: translation or encapsidation6,7. Interestingly, IRES-mediated translation is not restricted to full-length mRNA in MLV, as a mechanism of internal initiation is also used for the production of the Env glycoprotein32.
Sequence analysis indicates that IRES elements are highly conserved among the murine Oncoretroviruses as they also occur in viruses that are distantly related to MLV, including the defective leukaemia virus, Harvey murine sarcoma virus (HaMSV)33, and in retro-transposons such as mouse VL30 (Ref. 34). HaMSV is a highly pathogenic virus that arose from multiple genetic recombinations between MoMLV, the VL30 rat retro-element and the c-Ras oncogene. As a result, the v-Ras protein is expressed by a potent IRES element, which is derived from the 5′-UTR of the VL30 element. In a manner similar to the MLV 5′-IRES element, this element also overlaps the HaMSV RNA dimerization and packaging signals33. Finally, it is noteworthy that a human MLV-like retrovirus (called XMRV) was recently shown to be associated with prostate cancers35. Furthermore, sequence analyses indicates that it is highly likely that the synthesis of XMRV Gag and Gag-Pol is directed by an IRES element closely resembling that of FMLV35.
IRES elements have also been found in avian retroviruses, including the reticuloendotheliosis virus type A (REV-A)36. REV-A has the genetic organization of a simple retrovirus, similar to that of MLV, yet this virus does not produce a glycosylated Gag isoform, but still exhibits an IRES activity in its 5′-UTR36. However, in contrast to MLV, FMLV and HaMSV, this minimal IRES activity does not overlap with the main determinants of RNA encapsidation36.
ALV is another example of an α-retrovirus that possesses an IRES element. The 5′-UTR of ALV is composed of 330 nucleotides and has many functions both in the early and later steps of virus replication. It is thought to fold into a stable secondary structure with four independent stem loops37. Two of these structured regions encode small ORFs (uORFs) that have been shown to be crucial for Gag synthesis and virus replication38,39. These two structured domains have also been shown to be part of an active IRES element directing the initiation of Gag synthesis40.
Finally, it should be noted that two independent IRES elements have been found within the genomic RNA of the endogenous retrovirus Gypsy from Drosophila melanogaster41. The first element corresponds to the 5′-UTR of the genomic RNA encoding env and is active both in the reticulocyte lysate system and in cultured cells. However, the second IRES can only drive translation with rabbit reticulocyte lysate and strongly represses translation in cells41.
Taken together, these data indicate that the presence of IRES elements is a mechanistic theme that is common to all simple retroviruses as they are found in avian, mammalian and insect viruses that are only distantly related to each other. This distribution across different species, together with the fact that they are often co-localized with viral packaging signals, indicates that IRES elements could have an important role in the regulation of viral replication.
IRES elements and lentiviruses. In primate lentiviruses, IRES elements have been located and characterized from in the 5′-UTR and in the coding region of the genomic RNA42,43,44,45,46. Initially, the insertion of the 5′-UTR of HIV-1 into bicistronic vectors revealed that translation was extremely weak, both in the rabbit reticulocyte lysate and in transfected COS and HeLa cells, leading to the conclusion that this structure did not exhibit IRES element activity47. In fact, their data show that the cis-acting replication elements that are present in the 5′-UTR structure are an important barrier for ribosomal scanning and strongly inhibit translation initiation47.
This assertion was later revisited and it was shown that the 5′-UTR from the closely related SIV (Mac) was capable of internal translation initiation both in vitro and in vivo46. Nevertheless, the bicistronic expression driven by the 5′-UTR of SIV was relatively weak when compared with encephalomyocarditis virus (EMCV) IRES-positive control. In addition, some cap-dependence was observed when monocistronic constructs were used in the study. Another study that focused on the 5′-UTR of HIV-1 did reveal the presence of a cell-cycle-regulated IRES element that is expressed preferentially during the G2/M phase, both in HeLa cells and in translational extracts derived from the same cells43. These conflicting results indicate that lentiviruses might use alternative initiation strategies throughout the viral replication cycle. This could be explained by the existence of alternative RNA structures of the 5′-leader (see below) that would be favourable for either ribosomal scanning or IRES-dependent translation. Therefore, the physiological status of the cell, and/or the expression of viral proteins, would favour one or the other alternative conformations. This would, in turn, determine whether ribosomal binding occurs at the 5′-end of the genomic RNA or at an internal position within the untranslated region of the RNA. Moreover, as pointed out recently by Yilmaz et al., these inconsistencies could also reflect a disadvantage of the bicistronic reporter system, which does not fully recapitulate the RNA structures that can be found in the genomic mRNA48. In agreement with this possibility, it should also be noted that the assessment of the role of lentiviral internal initiation has not yet been carried out in the context of a complete replicating provirus in CD4+ primary cells.
By using chemical and enzymatic probing techniques, it was recently shown that the 5′-UTR of HIV-1 can fold into two alternative conformations49. The first conformation exposes the TAR, 3′-poly (A) tail, PBS, DIS and SD structures (as defined above), and one or several packaging signals (Fig. 2). This conformation, termed BMH (branched multiple hairpins), exists in equilibrium with another, more stable conformation, in which the 3′-poly (A) tail, DIS and SD structures and the packaging signals are paired with each other through a long distance interaction (LDI). The transition from the LDI to the BMH conformation is promoted by the presence of magnesium ions, high RNA concentrations and, most relevant to the physiological situation, by the presence of the nucleocapsid, a lentiviral protein that exhibits RNA chaperone activity50,51,52. Even though it is highly attractive to hypothesize that these changes in conformation are sufficient to explain the molecular switch that would determine the usage of genomic RNA as either mRNA for translation or as genomic RNA for the generation of new virions, new evidence indicates that this might not be the case53.
Although the characterization of IRES elements in the 5′-UTR of primate lentiviruses has been controversial, there is greater consensus regarding the discovery of IRES elements in the Gag coding region. In fact, alternative N-terminal truncated isoforms of the Gag polyprotein precursor have been observed in cells infected with HIV-1, HIV-2 and SIV42,44,45. Further investigation revealed that these isoforms are synthesized by IRES-mediated translation in which the element is located entirely within the gag coding region. Therefore, alternative translation initiation gives rise to one shorter isoform of Gag in the case of both HIV-1 and SIV (named p40 and p43, respectively), whereas two isoforms are produced from the HIV-2 coding region (named p50 and p44) (Fig. 3). The loss of these alternative Gag isoforms by mutagenesis of the AUG codons has a significant impact on viral infection, suggesting an important role for these proteins in pathogenicity42,45. Interestingly, production of the truncated Gag isoforms of HIV-1, HIV-2 and SIV occurs exclusively by internal entry of the ribosomes to the Gag coding region42,44,45 (Fig. 3).
Figure 3. IRESs in Lentiviruses.
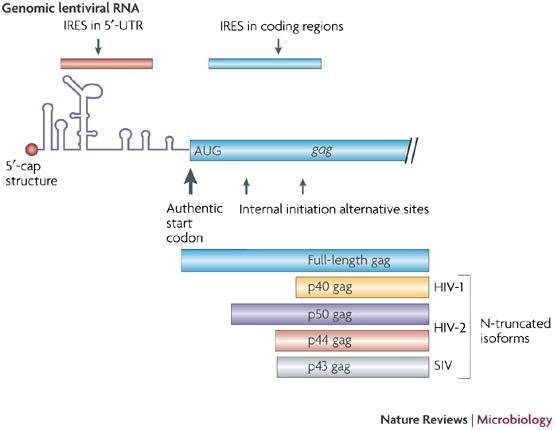
Schematic diagram of the position of the different IRESs within the lentiviral genomic RNAs. The N-truncated Gag isoforms that result from alternative translation initiation from the coding region are shown.
However, the most surprising and unexpected finding is that these IRES elements can also drive translation of the full-length polyprotein from the upstream authentic AUG codon42,44. This is particularly interesting as it implies that ribosomes can be recruited by determinants downstream of the first initiation codon, and gives rise to the possibility that the genomic RNA lacking the whole 5′-UTR sequence upstream of the AUG codon could be efficiently translated. Indeed, this property leads to the provocative suggestion that ribosomes can, in some contexts, scan backwards for a short distance. The presence of an IRES element in the coding region supports the idea that the gag gene can be translated independently despite the structural rearrangements and the various interactions that occur in the 5′-UTR. A structural model of the HIV-2 RNA coding region has been established by enzymatic and chemical analysis and is characterized by a collection of seven or eight hairpins (P1 to P8) and an LDI conformation termed P3 (Fig. 4). The region between the P8 and the 3′-end of the coding region is not conserved and is marginally structured44. Most of this structural model, derived for HIV-2, can be applied to the coding regions of HIV-1 and SIV, although no motifs of predictable function have emerged from a comparative analysis of the three structures. However, in several locations, all three structures have stretches of single-stranded purine nucleotides, a structural motif that is known to promote ribosome binding54. Although the precise molecular determinants required for ribosome recruitment are unknown, sucrose gradient analysis has indicated that three 43S, or 80S, ribosomal complexes can bind to each of the AUG initiation sites on the same HIV-2 mRNA molecule (Sargueil et al., unpublished data).
Figure 4. Schematic representation of the secondary structure adopted by the first 450 nucleotides of the HIV-2 Gag coding region.
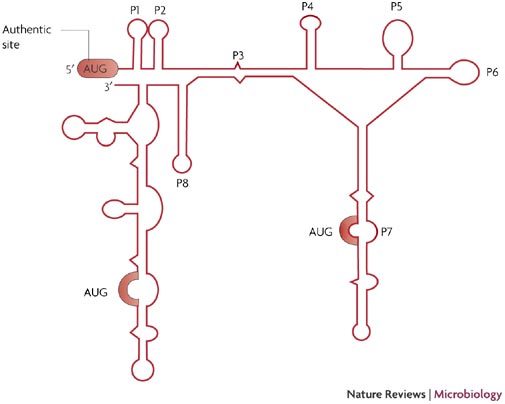
Secondary structure model deduced from chemical and enzymatic probing data (adapted from Ref. 44). Helices are numbered from P1 to P8, structures after P8 were not named and are not conserved in HIV-1 and simian immunodeficiency virus (SIV)mac. The P2 helix is also absent from HIV-1. The three initiation codons are highlighted in red.
The presence of an IRES element both in the 5′-UTR and in the coding region is likely to impact on the activity of each IRES element. In cells that were transfected with bicistronic constructs containing the HIV-1 IRES element in the 5′-UTR followed by segments of the gag coding region, Brasey et al. demonstrated that the IRES activity of the 5′-UTR was significantly inhibited when the structure was immediately upstream of gag coding region43. The converse is also true; interestingly, the HIV-2 coding region is less efficient as an IRES element when preceded by its 5′-UTR44. In both cases, this is likely to reflect an interaction between the 5′-UTR and the coding region, resulting in alternative folding of the two IRES elements. In agreement with this proposal, two LDI conformations have been predicted to occur in the 5′-UTR region of HIV-1, a prediction which can be extrapolated to HIV-2 and SIV. The first LDI is predicted to occur between the R-U5 region and the sequence in close proximity to the authentic AUG codon, resulting in the occlusion of the initiation site49,50,55,56. Site-directed mutagenesis analysis revealed that this interaction is crucial to regulate the conformational switch from BMH to LDI, but does not modulate translation efficiency53.
The interpretation of these experiments is hindered by the fact that translation could be initiated through different mechanisms. A second LDI conformation occurs between the 3′-poly (A) loop and a sequence separated by hundreds of nucleotides in the gag-coding region57. No functional information is available about this structure. In addition, neither of these LDI conformations have been detected either in vivo or from in vitro experiments in which the 5′-UTR and the coding region were probed together and/or independently44,58. Nevertheless, taking into account the dynamic nature of the RNA structure of HIV, it is conceivable that an interaction between the 5′-UTR and the gag-coding region could be one of several alternative conformations used by the genomic RNA. Therefore, the transition from one folding state to another might depend on the binding of one, or several, viral proteins to RNA signals located in the leader region. Consistent with this proposal is the recent finding that the Gag polyprotein was shown to modulate its own translation59 (Fig. 5). In the model described by Anderson and Lever, there is a bimodal effect of Gag in which low amounts of the protein initially stimulates translation. The resulting increasing local concentration of Gag then results in the oligomerization of the protein and coating of the 5′-UTR of HIV-1 (Ref. 59).
Figure 5. Translational control of lentiviral genomic RNAs.
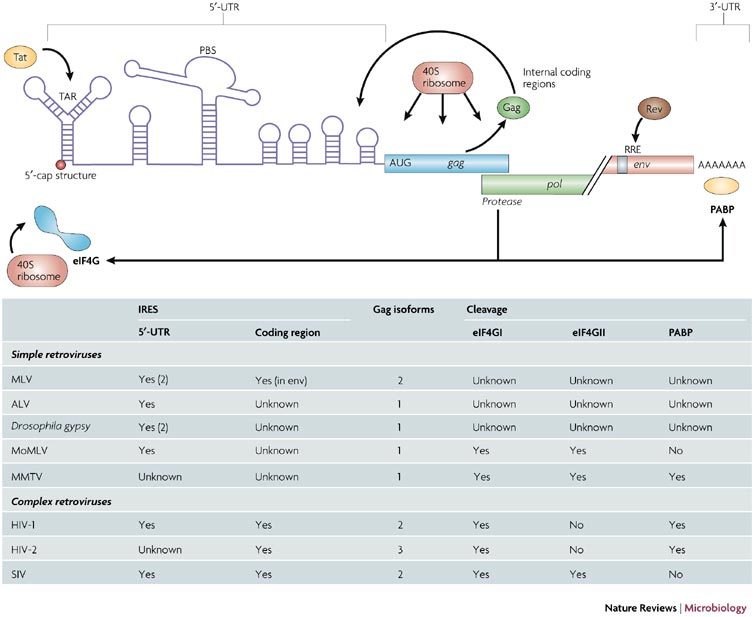
The figure summarizes the events that modulate and control the translation of the lentiviral genomic RNA. The table summarizes the location of the IRESs, the number of Gag isoforms and the effect of the protease on the translational apparatus for all the retroviruses described in the manuscript. ALV, avian leukosis virus; eIF4G, eukaryotic initiation factor (eIF) 4G (isoform I and II); MLV, murine leukaemia virus; MoMLV, Moloney MLV; MMTV, mouse mammary tumour virus; PABP, poly (A) binding protein; PBS, primer binding site; Rev, regulator of expression of virion proteins; RRE, Rev response element; SIV, simian immunodeficiency virus; TAR, transactivation response element.
Together, these results strongly indicate that the initiation of translation of the full-length Gag polyprotein can occur by three distinct mechanisms: first, ribosomal assembly, entry and scanning from the 5′-capped end of the genomic RNA; second, internal ribosome entry through the IRES element located within the 5′-UTR; third, internal entry of the ribosome through the IRES located in the coding region (Fig. 5). Future work will determine how the virus uses these alternative translation-initiation mechanisms during the course of infection.
Additional RNA structures involved in the initiation of translation. The genomic RNA of Rous sarcoma virus (RSV) possesses a long and structured 5′-UTR that contains three ORFs that are phylogenetically conserved in size and position60. In RSV, the packaging signal localizes to the third ORF and the integrity of this structure is crucial for viral replication. The introduction of point mutations to change the AUG initiation codons of the first and third reading frames leads to a dramatic reduction in RNA packaging (50- to 100-fold) indicating a functional link between translation and packaging39,61. A model for the packaging of RSV has emerged, in which the synthesis of Pr76gag represses translation from its cognate genomic RNA by binding to the packaging sequence62. This creates an autoregulatory mechanism that stops viral protein synthesis when the local concentration of p76gag to RSV genomic RNA reaches a ratio of about 2000:1 to allow packaging and viral assembly. However, studies have also mapped the minimal region for RSV packaging to a 160 nucleotide segment located downstream of the PBS that spans to the 5′-end of the gag coding sequence63,64. Therefore, the presence of this 160-nucleotide region (which contains the third ORF) on an RSV-based vector confers the ability to be encapsidated in trans; this is consistent with the use of RSV as a retroviral vector for gene-therapy purposes.
Another mechanism of translational enhancement mediated by an RNA structure was described in two retroviruses: M-PMV and the avian spleen necrosis virus (SNV). In these viruses, the region of genomic RNA between the 5′-LTR and the R-U5 domain was shown to enhance the translation of SNV genomic RNA and was subsequently termed the post-transcriptional control element (PCE). Interestingly, this effect was also demonstrated when this PCE was inserted upstream of both the main reading frame of the HIV-1 RNA and a Luciferase reporter gene65,66,67. Quantitative RNA analysis, together with ribosomal sedimentation profile analysis, has established that this translation enhancement is due to increased ribosome loading of the mRNA. RNA transfection assays, together with competition experiments, indicates that the R-U5 region interacts with nuclear proteins thereby creating a ribonucleoprotein complex that becomes actively translated in the cytoplasm68. Finally, it was recently shown that cellular RNA helicase A was involved in this process as it recognizes and binds specifically to the PCE in both the nucleus and the cytoplasm. This interaction probably induces structural changes at the level of RNA–RNA and/or RNA–protein that ultimately results in the enhanced translation of PCE-containing RNAs69.
Translational control by viral proteins
The TAR structure and the Tat protein. The TAR element is a stable 60-nucleotide stem-loop structure located at the 5′-extremities of all lentivirus transcripts that binds the virally encoded Tat protein with high affinity70,71,72. Efficient production of viral mRNA from integrated proviral DNA results in the coordinated action of TAR, the cellular transcriptional machinery and the viral Tat protein, which functions as an adaptor protein for cellular cofactors73. Initially it was shown that, owing to its location immediately downstream of the cap structure, TAR could inhibit protein synthesis by impairing ribosome binding to the 5′-end of the mRNA74. However, further experiments revealed that TAR is capable not only of modulating translation in cis but also in trans75. This phenomenon was shown to be related to the ability of TAR to induce phosphorylation of the α subunit of eukaryotic initiation factor (eIF)-2 by the activity of the interferon (IFN)-induced, dsRNA-dependent, serine/threonine protein kinase, PKR76,77. Phosphorylation of eIF2α blocks the recycling of eIF2B-GDP into eIF2-GTP77,78, which virtually shuts off protein synthesis. In this context, the TAR element has the intrinsic ability to activate both PKR and the 2–5A–synthetase — two enzymes associated with the IFN-mediated antiviral response76,77,79,80. Structural studies revealed that PKR binds TAR and undergoes a conformational change owing to PKR dimer formation on the TAR stem loop81,82.
To counteract PKR activation that is induced by TAR, HIV-1 has evolved the ability to evade the subsequent antiviral IFN response through the action of the viral protein Tat, which binds to PKR. Tat functions as a substrate homologue for PKR, competing with eIF2, and inhibiting the translational blockage induced by eIF2α phosphorylation76,79,80,83. Interestingly, the Tat–PKR interaction occurs directly and is not mediated by the presence of dsRNAs83,84. The action of Tat on PKR is not limited to the inhibition of its activation, as Tat can reverse the inhibition of translation that is induced by activated PKR84,85. Together, these findings lend support to a model whereby Tat functions as an inhibitor of PKR by competing with its natural substrate (eIF2) and by blocking the autophosphorylation of PKR.
However, the effect of Tat on protein synthesis goes beyond its ability to inhibit PKR activity; indeed, Tat can stimulate in vitro translation of a TAR-containing reporter gene independently of its effect on both PKR and the (2′–5′)-oligoadenylate synthetase activation pathways86(Fig. 5). Similar effects can be mediated by other TAR-binding proteins, including RNA helicase A, Staufen-1, and the La autoantigen87,88,89,90,91. Therefore, it seems that protein interaction with TAR can alleviate translational repression in cis by disrupting the stability of the TAR element. Interestingly, eIF2 can also interact with TAR92, allowing one to speculate that the interaction of both PKR and eIF2 with contiguous regions in TAR could facilitate the phosphorylation of eIF2 by PKR92. Finally, it should be noted that a cellular TAR-binding protein (TRBP) binds strongly to TAR, and transactivates the HIV-1 LTR synergistically with Tat function87. Moreover, similar to Tat, TRBP associates with PKR and inhibits its activity.
The RRE/ Rev interaction. The Rev (regulator of expression of virion proteins) protein is a 116 amino-acid protein that induces the transport of incompletely spliced lentiviral RNAs harbouring the cis-acting Rev response element (RRE) between the nucleus and the cytoplasm93,94,95,96. The complete RRE of HIV-1 consists of 351 nucleotides and is a highly ordered RNA stem-loop structure located in the env gene93,94,95,96,97. Initially, the Rev protein binds with high affinity at a site located at the apex of the central stem of the RRE, where it can recruit additional Rev molecules through cooperative protein–protein and protein–RNA interactions98,99,100,101,102. As a consequence, RRE-containing transcripts are compartmentalized differently, increasing their association with cytoskeleton polyribosomes103,104,105. Therefore, it is plausible that Rev not only helps in the transportation of viral RNA out of the nucleus but is also responsible for the localization of the RNA to polyribosomes, enhancing their use by the cellular translational machinery. In agreement with this suggestion, a number of reports have described a discordant relationship between the Rev-induced increase in cytoplasmic RRE-containing RNA levels and the dramatic increase reported for protein production from these same RNA transcripts. In one report, D'Agostino et al. (1992) used a reporter plasmid encoding the RRE-containing gag gene to show a 4- to 16-fold increase in the concentration of gag mRNA in the presence of Rev, a finding that cannot account for the more than an 800-fold increase in the production of the Gag protein105. A similar effect has been reported for the env mRNA, in which Rev expression results in a 1.6-fold increase in env mRNA transport, whereas translation from this mRNA was increased more than 100-fold106. Further studies confirmed the increased association of viral RNA with polyribosomes in response to the presence of the Rev protein104,105. In the absence of Rev or RRE, there is no association of viral RNA with polyribosomes and the RNA is found in 40S to 80S complexes104. Additional evidence to support a direct role for the Rev protein in mRNA translation was generated using a non-viral plasmid expression system capable of generating an intron-RRE-containing reporter RNA. This RNA lacks the viral nuclear retention sequences, thereby allowing the unspliced transcript to be exported from the nucleus to the cytoplasm. In this experimental setting, protein levels increased 3.5-fold in response to the viral Rev protein whereas the level of cytoplasmic mRNAs remained stable107.
Viral protease activity and the host-cell translational apparatus. In the retrovirus life cycle, different proteolytic events take place during the assembly of infectious virions (Fig. 1). The virally encoded aspartyl protease processes the Gag and Gag-Pol polyprotein precursors into mature proteins. The retroviral protease is a product derived from the pol gene and is one of the key targets in anti-HIV-1 therapies108,109. In addition to its function in the retroviral life cycle, this endopeptidase can also cleave certain cellular proteins, resulting in a cytotoxic effect110. One such protein is the initiation factor, eIF4GI, which is a scaffolding protein that has a key role in the assembly of the ribosomal pre-initiation complex111 (Box 2). The cleavage of eIF4GI was first observed in CD4+ cells infected with a laboratory strain of HIV-1, and this activity could be correlated with a global decrease in protein synthesis112. Expression of the HIV-1 protease (encoded on a plasmid) in COS-7 cells resulted in the proteolytic processing of eIF4GI and could be prevented by the addition of a specific HIV protease inhibitor112. Similar results were obtained by the addition of recombinant HIV-1 protease to HeLa-derived S10 extracts112 and to the rabbit reticulocyte system113. Sequence analysis from mass spectrometry revealed that the eIF4GI protein was cleaved twice between the eIF4E binding site and the first of the eIF4A binding sites (in fact, the two cleavage sites are only three amino acids apart). The protein was also cleaved further downstream in the region of the second eIF4A-binding domain112,113. The recombinant HIV-2 protease was also shown to be capable of cleaving eIF4GI at the same sites. However, in this case, the proteolytic activity occurred by a two-step reaction. First, a 100-kDa fragment (Ch-1 fragment) was generated as a result of proteolysis attack at the two sites located between the eIF4E- and eIF4A-binding sites. Second, at higher protease concentrations, the Ch-1 fragment was further digested into two 50-kDa C-terminal segments114. Interestingly, the Ch-1 fragment is only 40 amino acids shorter than the main cleavage product that results from the proteolysis of eIF4GI by the picornaviral protease, L/2A (Ref. 115). However, whereas the cleavage of eIF4GI by the picornaviral proteases results in a strong stimulation of virtually all IRES elements identified so far116,117,118, proteolysis mediated by the HIV-2 protease results in a strong inhibition of type I IRES-mediated translation with little or no effect on translation by type II IRES. This finding strongly indicates that the 40 amino-acid region between the L/2A and the HIV-2 protease cleavage sites (amino acids 641–681), which has RNA binding properties, has a role in ribosomal scanning114.
The cleavage of eIF4GI by retroviral proteases has been extended to those from HTLV-1, MMTV and MoMLV119(Fig. 5). Interestingly, with the exception of MoMLV, none of these proteases were able to cleave eIF4GII, the functional homologue of eIF4GI, despite the fact that eIF4GII harbours one of the consensus cleavage sites113,119.
Recently, the Poly (A) binding protein 1 (PABP1), which is required for mRNA circularization during translation initiation through its interaction with eIF4G and the mRNA poly (A) tail was also found to be a target for retroviral proteases120 (Fig. 5). This was shown both in the context of infected cells and in in vitro extracts derived from HeLa cells, and was demonstrated using proteases derived from MMTV, HIV-1 and HIV-2. However, the implications of PABP1 cleavage by retroviral proteases on the overall translation process deserves further investigation.
Following HIV-1 infection, protein synthesis by the infected cell is decreased; however, despite the fact that both eIF4GI and PABP1 are cleaved by the viral protease, translation still occurs in these cells112,120,121. This contrasts with the complete translational shut-off observed in host cells infected with picornaviruses, which is also a result of the cleavage of eIF4G (eIF4GI and eIF4GII) and PABP122. There are several explanations that could be used to explain these differences between picornaviral and retroviral infections: first, proteolysis of eIF4GI and PABP by retroviral proteases occurs at a much later time-point during the viral replication cycle; secondly, the HIV-1 protease is significantly less active on eIF4GI compared with the L picornaviral protease from the foot-and-mouth-disease virus (FMDV)123; third, eIF4GII is not cleaved by the retroviral proteases and therefore this protein could support translation in the absence of eIF4GI (as has been shown for picornaviral infection)124,125.
However, it is unknown how the translation of retroviral genomic RNA can benefit from these proteolytic events given that they occur at time when most, if not all, of the viral structural proteins and enzymes have been synthesized. The physiological role of eIF4GI cleavage by the retroviral proteases remains to be addressed in the context of viral infection. However, this might be very difficult to assess owing to the toxicity of the enzyme on the cellular apparatus110. Based on the in vitro data, it is tempting to speculate that the expression of low levels of the retroviral protease might sustain HIV translation at times when cellular protein synthesis is already severely affected112,114. However, at higher levels of protease, it is likely that the damages inflicted to the cell would result in polysomal dissociation and the inhibition of viral and cellular translation, which would favour viral packaging, assembly and the release of progeny virions110,113.
Perspectives and discussion
Over the past decade, retroviruses and lentiviruses have been the focus of intense research interest primarily as a result of the AIDS epidemic. Characterization of the IRES elements of several retroviruses has emphasized the importance of translation in viral replication. The potential use of several mechanisms, such as a combination of IRES elements located both in the 5′-UTR and the coding region, together with ribosomal entry at the 5′-capped end, suggests that viral gene expression is highly regulated at the translational level. An additional layer of complexity is supplied by the interplay of viral proteins that directly bind to the genomic RNA or that modify the host cell translational apparatus. The coordinated action of all these elements creates a selective translational advantage for viral genomic RNA, resulting in the recruitment of ribosomes and initiation factors in a competitive cellular environment (Fig. 5).
A second important issue relating to retroviral translation concerns its close relationship with the viral assembly process that leads to packaging of the genomic RNA into newly formed virions. It should be remembered that the full-length, unspliced, genomic retroviral RNA has a dual role in that it functions as a template for protein synthesis and ensures the flow of genetic information6. As a consequence, the same RNA molecules are either translated or packaged into newly formed retroviral particles (Fig. 6). However, the two mechanisms are believed to be mutually exclusive. Encapsidation of the unspliced RNA is dependent on the interaction between the Gag polyprotein and the RNA packaging motifs within the 5′-UTR. These Gag protein–RNA interactions result in a conformational change of the 5′-UTR as an increasing number of nascent Gag molecules progressively bind to the structure59. Therefore, it is conceivable that the use of multiple translational mechanisms could reflect the ability of the virus to adapt to these conformational changes in order to ensure continuous viral protein synthesis. As such, the early steps of infection might be characterized by the exclusive use of a 5′-cap-dependent mechanism as it is the most efficient way to rapidly build a stockpile of structural proteins and enzymes. At this stage, production of the Gag polyprotein could stimulate its own translation from the genomic RNA, as was recently proposed by Anderson and Lever59. Further progression in the viral cycle results in the production of high levels of Gag and Gag-Pol molecules that bind to the RNA packaging signals located within the 5′-UTR. This process results in the progressive occlusion of the 5′-UTR as the ribonucleoprotein scaffold is formed. Therefore, translation initiation switches to an 'all IRES'-based mechanism of ribosome entry at the AUGs that are located in the gag coding region. In the case of lentivirus replication, further support for this hypothesis comes from the fact that the Vpr protein probably blocks the cell cycle in a G2/M phase that is not favourable for cap-dependent translation initiation126,127. As such, this mechanism is a method to continue viral protein synthesis during the early steps of viral assembly. Finally, the accumulation of Gag and Gag-Pol in the cytoplasm of the infected cell ultimately triggers the maturation of the active protease that will, in turn, cleave the cellular translation initiation factor eIF4GI and PABP. This process results in a progressive inhibition of cellular and viral translation thereby promoting assembly and the release of new infectious particles.
Figure 6. Translation and encapsidation of the retroviral genomic RNA.
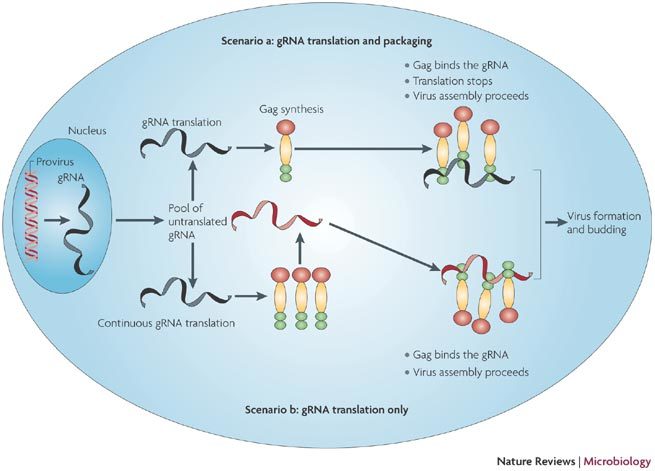
Transcription of the integrated provirus generates the genome length RNA (gRNA). A fraction of the gRNA is spliced to give the spliced viral RNAs (not shown here). The gRNA is exported to the cytoplasm where it is recruited by active ribosomes (scenarios a and b with gRNA in black) or eventually accumulates as a pool of non-translated RNA (gRNA in red). According to scenario a, the gRNA is translated to give Gag molecules and Gag-Pol (not shown here). Once Gag molecules accumulate, there is a switch from translation to packaging as Gag binds to the gRNA at the 5′-IRES-Psi signal. Virus formation and release by budding follow. In scenario b, the gRNA is continuously translated. Newly formed Gag molecules bind the gRNA from the non-translated pool (red gRNA). Virus formation follows.
Box 1 | The IRES mechanism.
Internal ribosomes entry segment (IRES) elements have been characterized within the genomic RNA of several retroviruses. IRES elements are defined RNA segments that allow translation initiation by direct ribosome binding to the 5′-untranslated region (UTR) of the mRNA128,129,130. Internal translation initiation was first demonstrated for the picornavirus family but has since emerged as a general mechanism for both viral and cellular RNAs131,132. The huge diversity of size, sequence and structure that has been reported for the 56 viral and 73 cellular IRES elements characterized so far indicates a polyphyletic origin133,134. Despite their differences, all IRES elements can be defined as an RNA domain that has the ability to recruit ribosomes in a cap-independent manner. This process occurs by either promoting direct contact between the mRNA and the small subunit of the ribosome, as is the case for the classical swine fever virus, hepatitis C virus and cricket paralysis virus135 or, more commonly, by the establishment of an RNA–protein–ribosome interaction through a set of initiation factors and/or by the use of specific IRES trans-acting factors129. Following their binding to the IRES sequence, the ribosomes can initiate at their landing site, as is the case in type II IRES (prototype encephalomyocarditis virus), or they can scan the 5′-UTR in a 5′ to 3′ direction until they locate the AUG initiation codon as in type I IRES (prototype poliovirus). Despite the relatively large number of IRES sequences that have now been identified, a structural 'prototype' for the internal initiation mechanism is still not available, which suggests that there might be not one, but several, RNA motifs that enable ribosomal entry. In agreement with this idea, there is a broad range of unrelated genes that use an IRES-mediated mechanism of translation initiation. From a functional standpoint, internal initiation allows the translation process to bypass the control mechanisms that specifically regulate cap-dependent translation, especially those involving modification of the eIF4E/4E-BP complex136. As a result, IRES-mediated translation occurs during many cellular stress conditions that are known to inhibit cap-dependent translation, including hypoxya137,138, irradiation139, apoptosis140, angiogenesis141, amino-acid starvation142, continuous heat shock143 and mitosis127.
Box 2 | Translation initiation in eukaryotes.
In the model for initiation (see figure), translation begins by the attachment of the 40S ribosomal subunit to the 5′-capped end of the transcript, followed by a linear scanning process, until it reaches an initiation codon. This process is mediated by a number of proteins called initiation factors that allow both efficient binding of the ribosome to the mRNA and migration to the initiation codon. Among these proteins, the eIF4F complex has a pivotal role144. It is composed of the eIF4E cap-binding protein, which drives the complex to the 5′-capped end of the eukaryotic mRNA, the eIF4G scaffolding protein and the eIF4A RNA helicase145. Two different isoforms of eIF4G (eIF4GI and eIF4GII) can connect eIF4E with the 43S ribosome complex, which is composed of the 40S ribosomal subunit associated with eIF3, eIF4A, eIF5, eIF1, eIF1A and eIF2-GTP bound to an initiator tRNA111 (see figure). The initiation factor eIF4G has been found to also interact with the poly (A) binding protein (PABP) which binds to the 3′-poly (A) tract of the eukaryotic mRNA, lending support to a closed-loop model for translation initiation146,147. After its attachment to the capped end of the mRNA, the 43S complex scans in a 5′ to 3′ direction until it reaches an initiation codon148. At this point, eIF5 promotes the dissociation of the preinitiation complex, GTP hydrolysis, and the 60S ribosomal subunit can assemble with the 40S to form the 80S ribosome.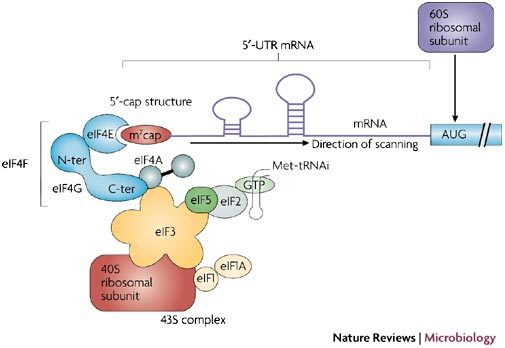
Acknowledgements
Work in the laboratory of T.O. is supported by grants from the Agence Nationale de la Recherche (ANR), the Agence Nationale de la Recherche centre le Sida (ANRS), SIDACTION and Action Concertee Incitative (ACI) 'jeune chercheur'. Work in the laboratory of J.L.D. is supported by grants from ANR, ANRS and Targeting Replication and Integration of HIV (TRIOH). Work in the laboratory of B.S. is supported by a grant from the ANR and ANRS. The work in the laboratory of M.L.L. associated to this manuscript is funded by the Fondo Nacional de Ciencia y Tecnología de Chile, and the Millennium Nucleus on Immunology and Immunotherapy. Collaborative efforts between France and Chile are partially supported by an ECOS-CONICYT grant to M.L.L., J.L.D. and T.O.
Glossary
- 5′-cap structure
This is a methylated guanosine (methylated at position 7) that is added as the first nucleotide of all RNA pol II transcripts by a 5′–5′ phosphodiester bond. In addition to its role in translation initiation, the presence of the cap structure protects the mRNA from exonucleases and has a role in splicing and mRNA export.
- RNA pseudoknot
An RNA motif formed by the pairing of a loop (apical or internal) with a sequence located outside of this loop. These common structures are formed in the presence of divalent ions and have peculiar functions.
- Reticulocyte lysate system
This is the most efficient in vitro translation system which is derived from immature red cells. As such, the system is devoid of any nuclear components but contains all the complete protein-synthesis machinery required to translate exogeneously added mRNAs.
- Bicistronic vectors
A construct that expresses two reading frames (cistrons) in tandem on the same transcriptional unit. The first cistron is translated by a normal cap-dependent mechanism, whereas the second cistron can only be expressed if an IRES structure is inserted upstream of its AUG initiation site. However, some proper controls must be undertaken in order to verify the integrity of the bicistronic RNA throughout the experiment.
- Ribosomal scanning
This term describes the linear progression of the eukaryotic ribosomal subunit along the 5′-UTR in a 5′ to 3′ direction from its initial binding to the mRNA and up to the recognition of the AUG initiation codon.
- R-U5 domain
The long terminal repeat (LTR) sequences are located at both the 5′ and 3′ termini of the retroviral genome. The LTRs can be broadly divided into three RNA sequences: U3, which contains promoter and enhancer regions for RNA synthesis; R (for repeat), which corresponds to the start (+1) of the genomic RNA and spans up to the end of the poly (A) loop; and U5, which starts from the end of the poly (A) loop, comprising the primer binding site and extends up to the packaging signal.
Biographies
Laurent Balvay obtained his Ph.D. in 1994 from the Pasteur Institut in Paris, where he worked on alternative splicing. Since 2005, he has recently joined the laboratory of T. Ohlmann to work on retroviral translation initiation.
Marcelo López-Lastra obtained his Ph.D. from the Université Claude Bernard, Lyon-1 (France), under the supervision of Jean-Luc Darlix. He then moved to McGill University (Canada), under the supervision of Nahum Sonenberg. He currently holds an Assistant professor position and is responsible for the Molecular Virology Laboratory at the Pontificia Universidad Católica de Chile.
Bruno Sargueil research has been dedicated to the study of the RNA structure–function relationship. Using enzymology and in vitro genetics he studied tertiary contacts in large and small ribozyme motifs. Recently he turned his interest to the role of RNA structure in translation initiation, focusing on lentiviral IRES.
Jean Luc Darlix obtained is Ph.D. in 1970 from the Pasteur Institut in Paris. From 1974–1985 he was assistant professor at Geneva University, Switzerland, where he investigated the retroviruses ASLV and MLV. From 1986–1990 he was Professor in microbiology at Toulouse University, investigating MLV and HIV-1. Since 1990, he has had the role of Director at INSERM and Founder of the INSERM -Human Virology Department at ENS Lyon (France).
Théophile Ohlmann earned his Ph.D. at the University of Sussex (UK) with V. Pain and S. Morley and completed a 2-year post doc with R. Jackson at Cambridge (UK). He then joined the Inserm and the laboratory of J.-L. Darlix in Lyon in 1998. He is now running a laboratory working on the translational control of Eucaryotic and viral RNAs at the Ecole Normale Supérieure de Lyon (France).
Related links
DATABASES
Entrez Genome
Entrez Genome Project
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
References
- 1.Telenitsky A, Goff SP. Retroviruses. 1997. pp. 121–160. [Google Scholar]
- 2.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral genomes: an inseperable pair. Nature Rev. Mircobiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 3.Vogt PK. Retroviruses. 1997. pp. 1–25. [Google Scholar]
- 4.Rabson AB, Graves BJ. Retroviruses. 1997. pp. 205–261. [Google Scholar]
- 5.Swanstrom R, Willis JW. Retroviruses. 1997. pp. 263–334. [Google Scholar]
- 6.Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin JG, Rosenak MJ. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl Acad. Sci. USA. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin SD, Allen JF, Lever AM. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye JF, Lever AM. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell. Mol. Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brugger B, et al. The HIV lipidome: a raft with an unusual composition. Proc. Natl Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morikawa Y. HIV capsid assembly. Curr. HIV Res. 2003;1:1–14. doi: 10.2174/1570162033352084. [DOI] [PubMed] [Google Scholar]
- 13.Saad JS, et al. From the cover: Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/S0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 15.Berkhout B, et al. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 2002;277:19967–19975. doi: 10.1074/jbc.M200950200. [DOI] [PubMed] [Google Scholar]
- 16.Corbin A, Darlix JL. Functions of the 5′ leader of murine leukemia virus genomic RNA in virion structure, viral replication and pathogenesis, and MLV-derived vectors. Biochimie. 1996;78:632–638. doi: 10.1016/S0300-9084(96)80009-5. [DOI] [PubMed] [Google Scholar]
- 17.Das AT, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klasens BI, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshinaka Y, Katoh I, Copeland TD, Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl Acad. Sci. USA. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honigman A, Wolf D, Yaish S, Falk H, Panet A. cis Acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology. 1991;183:313–319. doi: 10.1016/0042-6822(91)90144-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardon G, Varmus HE. Frameshift and intragenic suppressor mutations in a Rous sarcoma provirus suggest src encodes two proteins. Cell. 1983;32:871–879. doi: 10.1016/0092-8674(83)90072-7. [DOI] [PubMed] [Google Scholar]
- 23.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shehu-Xhilaga M, Crowe SM, Mak J. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prats AC, De Billy G, Wang P, Darlix JL. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J. Mol. Biol. 1989;205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 27.Portis JL, Fujisawa R, McAtee FJ. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV lacking expression of this protein. Virology. 1996;226:384–392. doi: 10.1006/viro.1996.0666. [DOI] [PubMed] [Google Scholar]
- 28.Corbin A, Prats AC, Darlix JL, Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J. Virol. 1994;68:3857–3867. doi: 10.1128/jvi.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berlioz C, Darlix JL. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vagner S, et al. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 31.Cepko CL, Roberts BE, Mulligan RC. Construction and applications of a highly transmissible murine retrovirus shuttle vector. Cell. 1984;37:1053–1062. doi: 10.1016/0092-8674(84)90440-9. [DOI] [PubMed] [Google Scholar]
- 32.Deffaud C, Darlix JL. Characterization of an internal ribosomal entry segment in the 5′ leader of murine leukemia virus env RNA. J. Virol. 2000;74:846–850. doi: 10.1128/JVI.74.2.846-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berlioz C, Torrent C, Darlix JL. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J. Virol. 1995;69:6400–6407. doi: 10.1128/jvi.69.10.6400-6407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Lastra M, Ulrici S, Gabus C, Darlix JL. Identification of an internal ribosome entry segment in the 5′ region of the mouse VL30 retrotransposon and its use in the development of retroviral vectors. J. Virol. 1999;73:8393–8402. doi: 10.1128/jvi.73.10.8393-8402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urisman A, et al. Identification of a novel γ retrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Lopez-Lastra M, Gabus C, Darlix JL. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 1997;8:1855–1865. doi: 10.1089/hum.1997.8.16-1855. [DOI] [PubMed] [Google Scholar]
- 37.Darlix JL, Zuker M, Spahr PF. Structure-function relationship of Rous sarcoma virus leader RNA. Nucleic Acids Res. 1982;10:5183–5196. doi: 10.1093/nar/10.17.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donze O, Damay P, Spahr PF. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donze O, Spahr PF. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deffaud C, Darlix JL. Rous sarcoma virus translation revisited: characterization of an internal ribosome entry segment in the 5′ leader of the genomic RNA. J. Virol. 2000;74:11581–11588. doi: 10.1128/JVI.74.24.11581-11588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronfort C, De Breyne S, Sandrin V, Darlix JL, Ohlmann T. Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement. Rna. 2004;10:504–515. doi: 10.1261/rna.5185604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buck CB, et al. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brasey A, et al. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbreteau CH, et al. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nature Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 45.Nicholson, M. G., Rue, S. M., Clements, J. E. & Barber, S. A. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology (2006). [DOI] [PubMed]
- 46.Ohlmann T, Lopez-Lastra M, Darlix JL. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000;275:11899–11906. doi: 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 47.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yilmaz A, Bolinger C, Boris-Lawrie K. Retrovirus translation initiation: Issues and hypotheses derived from study of HIV-1. Curr. HIV Res. 2006;4:131–139. doi: 10.2174/157016206776055039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 50.Dirac AM, Huthoff H, Kjems J, Berkhout B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002;30:2647–2655. doi: 10.1093/nar/gkf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. doi: 10.1021/bi025993n. [DOI] [PubMed] [Google Scholar]
- 53.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 54.Dorokhov YL, et al. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl Acad. Sci. USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanchy JM, Ivanovitch JD, Lodmell JS. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. Rna. 2003;9:1007–1018. doi: 10.1261/rna.5590603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 58.Paillart JC, et al. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 59.Anderson EC, Lever AM. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J. Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hackett PB, Dalton MW, Johnson DP, Petersen RB. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bieth E, Gabus C, Darlix JL. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonstegard TS, Hackett PB. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 63.Banks JD, Kealoha BO, Linial ML. An Mpsi-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J. Virol. 1999;73:8926–8933. doi: 10.1128/jvi.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Banks JD, Yeo A, Green K, Cepeda F, Linial ML. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Butsch M, Hull S, Wang Y, Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J. Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hull S, Boris-Lawrie K. RU5 of Mason-Pfizer monkey virus 5′ long terminal repeat enhances cytoplasmic expression of human immunodeficiency virus type 1 gag-pol and nonviral reporter RNA. J. Virol. 2002;76:10211–10218. doi: 10.1128/JVI.76.20.10211-10218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts TM, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J. Virol. 2000;74:8111–8118. doi: 10.1128/JVI.74.17.8111-8118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hull S, Boris-Lawrie K. Analysis of synergy between divergent simple retrovirus posttranscriptional control elements. Virology. 2003;317:146–154. doi: 10.1016/j.virol.2003.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartman TR, et al. RNA helicase A is necessary for translation of selected messenger RNAs. Nature Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 70.Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/S0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 71.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 72.Hauber J, Cullen BR. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J. Virol. 1988;62:673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 2005;3:61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 74.Parkin NT, et al. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edery I, Petryshyn R, Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 76.Maitra RK, et al. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 77.Kaufman RJ. Translational Control of Gene Expression. 2000. p. 1020. [Google Scholar]
- 78.Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 79.Schroder HC, Kelve M, Muller WE. The 2–5A system and HIV infection. Prog. Mol. Subcell. Biol. 1994;14:176–197. doi: 10.1007/978-3-642-78549-8_10. [DOI] [PubMed] [Google Scholar]
- 80.Silverman RH, Sengupta DN. Translational regulation by HIV leader RNA, TAT, and interferon-inducible enzymes. J. Exp. Pathol. 1990;5:69–77. [PubMed] [Google Scholar]
- 81.Carpick BW, et al. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 82.Cosentino GP, et al. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roy S, et al. Control of the interferon-induced 68-kilodalton protein kinase by the HIV-1 tat gene product. Science. 1990;247:1216–1219. doi: 10.1126/science.2180064. [DOI] [PubMed] [Google Scholar]
- 84.McMillan NA, et al. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 85.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 2000;373:361–367. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 86.SenGupta DN, Berkhout B, Gatignol A, Zhou AM, Silverman RH. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 88.Svitkin YV, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang YN, Kenan DJ, Keene JD, Gatignol A, Jeang KT. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fujii R, et al. A role of RNA helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 91.Dugre-Brisson S, et al. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ben-Asouli Y, Banai Y, Hauser H, Kaempfer R. Recognition of 5′-terminal TAR structure in human immunodeficiency virus-1 mRNA by eukaryotic translation initiation factor 2. Nucleic Acids Res. 2000;28:1011–1018. doi: 10.1093/nar/28.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hadzopoulou-Cladaras M, et al. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zapp ML, Green MR. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 95.Dillon PJ, Nelbock P, Perkins A, Rosen CA. Function of the human immunodeficiency virus types 1 and 2 Rev proteins is dependent on their ability to interact with a structured region present in env gene mRNA. J. Virol. 1990;64:4428–4437. doi: 10.1128/jvi.64.9.4428-4437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cochrane AW, Chen CH, Rosen CA. Specific interaction of the human immunodeficiency virus Rev protein with a structured region in the env mRNA. Proc. Natl Acad. Sci. USA. 1990;87:1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mann DA, et al. A molecular rheostat. Co-operative rev binding to stem I of the rev-response element modulates human immunodeficiency virus type-1 late gene expression. J. Mol. Biol. 1994;241:193–207. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 98.Tiley LS, Malim MH, Tewary HK, Stockley PG, Cullen BR. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc. Natl Acad. Sci. USA. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bartel DP, Zapp ML, Green MR, Szostak JW. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 100.Iwai S, Pritchard C, Mann DA, Karn J, Gait MJ. Recognition of the high affinity binding site in rev-response element RNA by the human immunodeficiency virus type-1 rev protein. Nucleic Acids Res. 1992;20:6465–6472. doi: 10.1093/nar/20.24.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kjems J, Calnan BJ, Frankel AD, Sharp PA. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992;11:1119–1129. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heaphy S, et al. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-Z. [DOI] [PubMed] [Google Scholar]
- 103.Kimura T, Hashimoto I, Nishikawa M, Fujisawa JI. A role for Rev in the association of HIV-1 gag mRNA with cytoskeletal b-actin and viral protein expression. Biochimie. 1996;78:1075–1080. doi: 10.1016/S0300-9084(97)86732-6. [DOI] [PubMed] [Google Scholar]
- 104.Arrigo SJ, Chen IS. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 105.D'Agostino DM, Felber BK, Harrison JE, Pavlakis GN. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell Biol. 1992;12:1375–1386. doi: 10.1128/MCB.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perales C, Carrasco L, Gonzalez ME. Regulation of HIV-1 env mRNA translation by Rev protein. Biochim. Biophys. Acta. 2005;1743:169–175. doi: 10.1016/j.bbamcr.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 107.Lawrence JB, Cochrane AW, Johnson CV, Perkins A, Rosen CA. The HIV-1 Rev protein: a model system for coupled RNA transport and translation. New Biol. 1991;3:1220–1232. [PubMed] [Google Scholar]
- 108.Gulick RM, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 109.Hammer SM, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 110.Blanco R, Carrasco L, Ventoso I. Cell killing by HIV-1 protease. J. Biol. Chem. 2003;278:1086–1093. doi: 10.1074/jbc.M205636200. [DOI] [PubMed] [Google Scholar]
- 111.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/S0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 112.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA. 2001;23:23. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ohlmann T, et al. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 114.Prevot D, et al. Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 2003;22:1909–1921. doi: 10.1093/emboj/cdg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 116.Ziegler E, Borman AM, Kirchweger R, Skern T, Kean KM. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ziegler E, et al. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology. 1995;213:549–557. doi: 10.1016/S0042-6822(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 118.Ohlmann T, Rau M, Pain VM, Morley SJ. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. doi: 10.1002/j.1460-2075.1996.tb00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alvarez E, Menendez-Arias L, Carrasco L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J. Virol. 2003;77:12392–12400. doi: 10.1128/JVI.77.23.12392-12400.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alvarez E, Castello A, Menendez-Arias L, Carrasco L. HIV protease cleaves poly (A)-binding protein. Biochem. J. 2006;396:219–226. doi: 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Butsch M, Boris-Lawrie K. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 2000;74:11531–11537. doi: 10.1128/JVI.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sarnow P. Translational control during virus infection. Virus Res. 2006;119:1. doi: 10.1016/j.virusres.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 123.Schlick P, Skern T. Eukaryotic initiation factor 4GI is a poor substrate for HIV-1 proteinase. FEBS Lett. 2002;529:337–340. doi: 10.1016/S0014-5793(02)03415-4. [DOI] [PubMed] [Google Scholar]
- 124.Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl Acad. Sci. USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Svitkin YV, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Planelles V, et al. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 1996;70:2516–2524. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sarnow P, Cevallos RC, Jan E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005;33:1479–1482. doi: 10.1042/BST0331479. [DOI] [PubMed] [Google Scholar]
- 129.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 130.Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/S0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- 131.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 132.Martinez-Salas E, Fernandez-Miragall O. Picornavirus IRES: structure function relationship. Curr. Pharm. Des. 2004;10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- 133.Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S. IRESdb: the internal ribosome entry site database. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mokrejs M, et al. IRESite: the database of experimentally verified IRES structures. Nucleic Acids Res. 2006;34:D125–D130. doi: 10.1093/nar/gkj081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 136.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 137.Stein I, et al. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/MCB.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/S0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 140.Stoneley M, et al. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 2000;20:1162–1169. doi: 10.1128/MCB.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Akiri G, et al. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17:227–236. doi: 10.1038/sj.onc.1202019. [DOI] [PubMed] [Google Scholar]
- 142.Fernandez J, et al. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 2001;276:12285–12291. doi: 10.1074/jbc.M009714200. [DOI] [PubMed] [Google Scholar]
- 143.Kim YK, Jang SK. Continuous heat shock enhances translational initiation directed by internal ribosomal entry site. Biochem. Biophys. Res. Commun. 2002;297:224–231. doi: 10.1016/S0006-291X(02)02154-X. [DOI] [PubMed] [Google Scholar]
- 144.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 145.Pestova TV, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tarun SZ, Jr, Sachs AB. Association of the yeast poly (A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. doi: 10.1002/j.1460-2075.1996.tb01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly (A)-binding protein and functions in poly (A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]


