Abstract
This report describes advances in the understanding of how microbes elicit and evade immune responses and the sensing of pathogens by host cells that leads to the activation and production of intra- and extracellular signaling molecules.
Subject terms: Immune evasion, Infection, Innate immunity, Signal transduction
On 16–18 February 2011, approximately 270 scientists met in Lorne, located 2 hours from Melbourne on the Great Ocean Road, Victoria, Australia, for the inaugural Lorne Infection and Immunity Conference. This multidisciplinary, international meeting was designed to stimulate collaborative research in infection and immunity by bringing together researchers specialized in the study of pathogen biology and host immune responses. The meeting built on the reputation of more than 20 years of Lorne annual conferences on proteins, cancer and genomes. Sessions covered diverse topics, including innate and adaptive immune mechanisms, microbial-host genomics and adaptation, microbial invasion, evasion and molecular pathogenesis, as well as structural biology of host and pathogen molecules and their effect on immunity. Here we summarize some of the newest ideas about infection and immunity research presented at the meeting that are shaping beliefs about how pathogens infect their hosts and how the host resists infection, as well as the translation of such ideas into the clinic.
Sensing and signaling
The human body has many innate receptors that probably evolved to sense microbial products such as nucleic acids and microbial cell-surface macromolecules from both pathogenic and commensal organisms. These receptors are now also known to sense endogenous molecules that may be present in an inappropriate time, place or form. For example, several classes of receptors sense pathogen nucleic acids but also respond to endogenous DNA that can arise from cell damage1. E. Latz (Bonn, Germany) opened the proceedings by introducing the nucleic acid–sensing receptors of the innate immune system and highlighted the importance of receptor-mediated trafficking of DNA into intracellular compartments for certain inflammatory responses.
The Toll-like receptors (TLRs) are known from structural studies to facilitate the assembly of large aggregates of adaptors such as MyD88, which form a scaffold for assembly of the subsequent components in TLR signaling. Indeed, the assembly of receptors and signaling molecules into large multimeric complexes is a common thread in the biology of signal transduction. The discovery and characterization of one such aggregate, called the 'Myddosome', has provided new insights into the mechanism and regulation of MyD88-dependent signal transduction by TLRs2,3. The Myddosome is a highly oligomeric signaling scaffold assembled sequentially from death domains of the receptor signaling adaptor MyD88 and protein kinases IRAK4 and IRAK1-IRAK2. It has a complex and variable stoichiometry and drives the formation of large signaling macro-complexes. The assembly process probably involves positive cooperation and takes place in lipid-rich membrane microdomains. These findings have important implications for the therapeutic targeting of TLR pathways in diseases as diverse as rheumatoid arthritis and neurodegeneration (N. Gay, Cambridge, UK). This suggests that effective therapies will need to target critical protein-protein interactions in the Myddosome, such as the Toll–interleukin 1 (IL-1) receptor domains at the heart of these assemblies, whose initial interactions are weak and are probably more readily disrupted. Signaling specificity is probably achieved by the localization of kinase and ubiquitin ligase substrates to the protein scaffold. Indeed, E3 ubiquitin ligases have key roles in regulating signaling in many biological processes, including the initiation, progression and maintenance of immune responses4. Using mice with targeted genes, M. Pellegrini (Melbourne, Australia) has characterized the critical role of a newly identified E3 ligase in maintaining the immune system in a quiescent state. The absence of this E3 ligase causes late embryonic death due to overwhelming immune activation. It seems to function in dendritic cells (DCs) to terminate spurious responses by the transcription factor NF-κB, and deficiency in this ligase results in enhanced and protracted transcriptional responses driven by the NF-κB subunit p65.
Other sensors of infection, belonging to the Nod-like receptor family of proteins, are reported to recognize pathogens as well as sterile inflammatory stimuli such as crystals via activation of the IL-1β-generating inflammasome5. A. Kupz (Melbourne, Australia) has identified and characterized a previously unknown in vivo pathway by which Salmonella enterica serovar Typhimurium (S. typhimurium) elicits cytokine production by distinct lymphocyte subsets involving inflammasome-mediated recognition of the bacteria by DCs. Although it has been known for some time that distinct cytokines are critical for the control of clinical and experimental infection with S. typhimurium, the exact regulation of their secretion during such infection has remained unclear. This inflammasome-mediated mechanism invokes intricate crosstalk between DCs and the responding lymphocyte subsets. The activation of this pathway has a substantial effect on the ability of infected mice to control the replication of S. typhimurium in vivo. In discussing another Gram-negative pathogen, Helicobacter pylori, M. Kaparakis (Melbourne, Australia) reported the use of bacterial outer membrane vesicles as a means of delivering bacterial products into the cytoplasm. The contents of outer membrane vesicles enter a compartment in which recognition by the cytosolic pattern-recognition receptor Nod1 occurs, leading to autophagy, inflammation and immune responses in the gut.
C. Mackay (Melbourne, Australia) showed how important the chemoattractant receptor GPR43 is for the normal regulation of inflammatory responses, particularly in the lower gut, in preventing inflammatory bowel disease. GPR43 is expressed by innate-type immune-response cells and binds short-chain fatty acids such as acetate, which are produced in the gut through bacterial fermentation of dietary fiber. Mackay proposed that diet, the makeup of the intestinal microbiota and bacterial production of acetate may account for changes in the incidence of asthma and autoimmune diseases in developed countries. Interestingly, acetate is emerging as an important factor produced by certain probiotic strains of bacteria6.
Microbial manipulation of host defense
Although the innate immune system has evolved to generate a rapid and effective inflammatory response to invading infectious organisms, it is no surprise that many microbial pathogens have evolved to avoid detection by this system and/or to deactivate the immune response. C. Roy (New Haven, Connecticut, USA) presented work on Legionella pneumophila, the causative agent of Legionnaire's disease, demonstrating that this opportunistic and accidental intracellular pathogen stimulates almost every pathway of the innate immune response as it infects and subsequently replicates in macrophages7. Indeed, L. pneumophila has proven a useful tool with which to study innate immune responses, particularly the NLRC4 inflammasome, which recognizes L. pneumophila flagellin8. L. pneumophila is an environmental organism whose natural hosts are unicellular eukaryotes, and this bacterium has thus not evolved to avoid detection by mammalian immune systems. In contrast, the closely related organism Coxiella burnetii, a cause of Q fever, is highly adapted to the host and evades detection by the immune system via mechanisms that are poorly appreciated. Both pathogens translocate virulence effector proteins into the host cell through the use of the Dot/Icm type IV secretion system, yet the effector protein repertoires of the two organisms are very different. The adaptation of C. burnetii to a mammalian host is evident in its ability to inhibit mammalian proapoptotic signaling. The C. burnetii Dot/Icm effector AnkG binds the host protein gC1qR (p32) and inhibits apoptosis in DCs, whereas L. pneumophila lacks this effector and induces rapid apoptosis in DCs in a p32-dependent manner9. Further work on the C. burnetii Dot/Icm effectors will undoubtedly identify additional mechanisms of host adaptation and microbial evasion of the immune response.
New information on host-cell invasion by the intracellular pathogen S. typhimurium presented by R. Teasdale (Brisbane, Australia) showed that there is still much to learn about the dynamic interaction between host and pathogen that enables S. typhimurium to form its intracellular niche. After invading the host, S. typhimurium stimulates a process similar to macropinocytosis to mediate its uptake into cells. By screening host-cell proteins involved in macropinocytosis through the use of small interfering RNA–mediated knockdown or pharmacological inhibition, researchers have discovered that phosphatidylinositol-(5)-kinase (PIKfyve) is essential for the replication of S. typhimurium inside cells10. The product of this kinase, phosphatidylinositol-(3,5)-bisphosphate, is required for the fusion of 'macropinosomes' (large vesicles filled with extracellular fluid that are formed through macropinocytosis) with late endosomes–lysosomes and proper maturation of the S. typhimurium–containing vacuole. Further screening has also identified a role for the SH3-PX-BAR–domain family of sorting nexins in macropinosome formation and the initial stages of S. typhimurium infection.
Antiviral immunity and viral evasion
Several presentations described the ongoing battle between clearance by the immune system and subversion of the immune response by viruses. New aspects of the host response were reported by M. Beard (Adelaide, Australia), who has used transcriptome analysis of liver infected with hepatitis C virus (HCV) to show that the interferon-stimulated gene product viperin and members of the interferon-inducible transmembrane family of proteins have distinct anti-HCV properties and act via inhibition of viral replication and entry, respectively, which thereby raises new possibilities for targeting chronic HCV infection. S. Lemon (Chapel Hill, North Carolina, USA) contrasted the ability of HCV to avoid immune responses and clearance with that of hepatitis A virus as a means of identifying and characterizing viral evasion strategies. Both viruses use a series of virus-produced proteases to target critical elements of the innate response machinery, yet surprisingly, it seems that hepatitis A virus is more evasive in a chimpanzee model of hepatitis infection, eliciting a minimal interferon response. These results suggest that although both viruses have evolved evasion strategies to target the same innate immune mechanisms, their survival and subsequent clearance rely on distinct processes11.
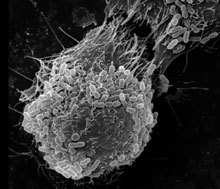
Attachment of enteropathogenic Escherichia coli to host cells infected for 3 hours and visualized by scanning electron microscopy. Photo courtesy of E. Hartland and Monash Micro Imaging.
Further exploring the host–hepatitis virus interaction, Z. Yuan (Shanghai, China) described the various strategies used by hepatitis B virus (HBV) to evade the host immune response. Liver biopsies of patients infected with HBV obtained before and after treatment have shown that HBV uses at least two mechanisms to avoid the innate immune response. First, impaired TLR-induced cytokine expression correlates with higher expression of HBV surface antigen and inhibition of the Jnk kinase pathway. Second, HBV polymerase inhibits the phosphorylation, dimerization and nuclear localization of the transcription factor IRF3 to dampen immune responses. Together these studies suggest HBV targets signaling by the innate immune response to evade detection by the immune system and establish chronic infection.
Reporting a twist on evasion of the immune system by hepatitis viruses, G. Ebert (Munich, Germany) harnessed the ability of 5′-triphosphorylated small interfering RNAs to trigger innate immune interferon responses. This research used small interfering RNAs designed to silence HBV replication in concert with more interferon production to suppress HBV replication in both cell-culture and transgenic HBV-infection models, which provides hope for modulation of the immune response to clear chronic infections.
New insight into immune regulation by 'old' molecules such as interferons is constantly being acquired. Progress in understanding the biology of the type III interferon IFN-λ (IL-28–IL-29) has led to considerable advances in the understanding of immunity. M. O'Keefe (Melbourne, Australia) discussed the cellular specificity of IFN-λ production and expression of the IFN-λ receptor by cells of the immune response. Her studies and those of others have shown that expression of this receptor is not ubiquitous, like that of receptors for other members of the interferon family, but instead is restricted to epithelial cells and, as described here, also plasmacytoid DCs (pDCs). In further contrast to other interferons, it seems that IFN-λ is produced by restricted subsets of DCs in response to specific ligands such as the synthetic RNA duplex poly(I:C), which makes IFN-λ a non-ubiquitous protein of the immune response. These subtle differences may account for the diverse immune functions now attributed to this newly identified cytokine.
A. Thompson (Melbourne, Australia) elaborated further on the role of IFN-λ in the pathogenesis of and therapeutic responses to HCV. Genome-wide association studies have identified polymorphisms in the region of IL28B (which encodes IFN-λ3) on chromosome 19 that correlate well with treatment outcome in patients infected with HCV genotype 1 who are treated with IFN-α (with covalently attached polyethylene glycol polymer chains) and ribavirin12. Patients carrying the 'good-response' variant have enhanced phase-one kinetics and two- to threefold higher rates of viral eradication. Furthermore, differences between patients of different ethnic background in allele frequency explain much of the recognized ethnic disparity in cure rates12. Moreover, IL28B polymorphisms have also been associated with spontaneous clearance of HCV infection13. The underlying biological mechanisms, however, remain unclear, which provides an important area for future research.
Emerging and re-emerging infections
Further evidence of the adaptation of microbes to their host or environment is apparent from genomic approaches to understanding Buruli ulcer, an emerging but neglected tropical disease. This is a disease of subcutaneous tissue that affects humans and other animals that is caused by infection with Mycobacterium ulcerans and for which there is no vaccine. T. Stinear (Melbourne, Australia) has used genomics to address some key research issues, including how M. ulcerans is transmitted to humans. Using multistrain comparative genomics, the group has shown that M. ulcerans evolved from the closely related Mycobacterium marinum by lateral gene transfer and genome reduction. Genes found in M. ulcerans but not in closely related mycobacteria are predicted to encode M. ulcerans–specific antigens, and recombinant proteins derived from these genes seem to elicit specific serum antibody responses in humans living in a region in which Buruli ulcer is endemic14. This suggests it may be possible to use serology to assess exposure to the bacterium.
K. Holt (Melbourne, Australia) discussed a study of the microevolution of Shigella sonnei, a common agent of endemic dysentery. Whole-genome sequencing and phylogenetic analysis of a collection of over 200 isolates from global sources has categorized the population into several distinct clusters, with most of the recent isolates restricted to a single cluster. Analysis of 91 isolates from Ho Chi Minh City has shown nearly all local cases can be attributed to the local expansion of a single clone. Similar rapid microbial adaptation is evident in sub-Saharan Africa, where a distinct sequence type of S. typhimurium has emerged as a frequent cause of invasive disease. M. Levine (Baltimore, Maryland, USA) reported that the high disease burden and case fatality caused by this strain has made vaccine development an urgent priority. This emergence was discovered through a comprehensive surveillance program and highlights the importance of monitoring infectious diseases in vulnerable populations.
Microbes also form an unusual interaction with hosts such as bats, which serve as the reservoirs for many emerging viruses, including paramyxoviruses (Hendra and Nipah viruses), coronaviruses (severe acute respiratory syndrome coronaviruses) and filoviruses (Ebola and Marburg viruses). Wild bat populations or experimentally infected bats, however, do not show any clinical signs of disease. Work by L. Wang (Geelong, Australia) is focused on understanding virus-bat interactions from the perspective of genomics and 'transcriptomics' (expression profiling). Considerable progress has also been made in establishing bat cell lines and in functional studies of key molecules in bat innate immunity, such as TLRs and the interferon system. Ultimately, these fundamental investigations may help to explain why bats seem to be asymptomatic carriers of some viruses that are deadly to humans.
Cells of the innate immune response
An effective immune response requires the ordered, sequential expression and secretion of inflammatory cytokines by the appropriate cells. J. Stow (Brisbane, Australia) has used live-cell imaging to show that cytokines traffic through recycling endosomes in macrophages, which allows the orchestrated release of multiple cytokines through the secretory and lysosomal pathways. These studies have demonstrated that recycling endosomes coordinate cytokine release while allowing macrophages to engulf and destroy invading pathogens. Macrophage activation and the induction of a robust immune response are critical for the generation of protective immunity to Mycobacterium tuberculosis. B. Saunders (Sydney, Australia) proposed that macrophages use a previously unknown communication strategy that involves the release of microparticles after stress induced by bacillus Calmette-Guérin. These microparticles contain M. tuberculosis antigens that are processed by antigen-presenting cells, thereby inducing antigen-specific T cell responses. Therefore, microparticles may represent a mode of immunological transfer that induces cell migration and the inflammatory response to infection with M. tuberculosis.

Surf beach at Lorne, Victoria, Australia. Photo courtesy of G. Jones.
The signal-transduction programs of core cells of the innate immune response, such as macrophages, are mediated by key transcription factors such as the IRF proteins. For example, IRF5 controls the differentiation of M1-type macrophages in both mice and humans15. P. Pitha (Baltimore, Maryland, USA) discussed the phenotype of IRF5-deficient mice, which includes B cell abnormalities and a propensity to develop autoimmunity. This finding underscores the role of key transcription factor–mediated genetic programs in the interface between the innate and adaptive immune systems. These factors mediate the innate responses of both mRNA and microRNA in cells of the innate immune system, such as macrophages, whose coordinated induction was presented by S. Forster (Melbourne, Australia).
Additional functions for another innate cell type, the pDC, were reported by D. Ang (Melbourne, Australia). Although pDCs have a recognized role as producers of type I interferon during viral infection, here pDCs were also shown to combat the intracellular bacterial pathogen L. pneumophila. Depletion of pDCs in mice infected with L. pneumophila exacerbates infection16, whereas supplementation of pDCs into the lungs ameliorates infection by diminishing the bacterial load. Notably, resistance to pulmonary L. pneumophila infection is independent of type I interferon signaling, which shows that pDCs control bacterial infection in the lungs by a mechanism distinct from antiviral pathways such as type I interferons. In summary, these presentations confirmed the idea that the presence of pathogen-associated stimulating molecules and their detection by cells of the innate immune system drive the signaling and production of cytokines, in a context-dependent manner, that shape the nature of the response that ensues.
Modulation of the adaptive response
Innate sculpting of adaptive immunity occurs through cytokines such as IL-10, a potent immunosuppressive cytokine with wide-ranging effects on T cells and B cells as well as cells of the innate immune response, such as macrophages and DCs. The tropical disease visceral leishmaniasis is characterized by splenomegaly, remodeling of the red and white pulp compartments of the spleen and local IL-10-mediated immunosuppression. P. Kaye (York, UK) presented data that have extended published work17 to show that the cellular control of splenic remodeling is highly compartment specific. Although inflammatory monocytes negative for matrix metalloproteinase 9 and positive for the neutrophil marker Ly6G (MMP9−Ly6G+), not MMP9+Ly6G+ neutrophils, are responsible for regulating vascular changes in red pulp, neither of these populations has a substantial role in regulating the destruction of follicular DCs or fibroblastic reticular cells in the white pulp. Through the use of a model in which mice were depleted of CD11c+ cells during chronic infection and then were reconstituted by adoptive transfer, this work has provided new insight into how CD11chi and CD11clo–int cells regulate the development of IL-10-producing T helper type 1 cells and immunopathology differently.
How IL-10 acts on a large number of genes in diverse cells is unclear at present. C. McCoy (Melbourne, Australia) and colleagues have built on published work18 showing that expression of the microRNA miR-155 is inhibited by IL-10. This microRNA has been linked to the modulation of inflammatory responses, inflammation-induced cancer and the pathogenesis of autoimmunity. This latest work suggests that through miR-155, IL-10 regulates a set of genes previously unknown to be regulated by this cytokine, which helps to explain its biological potency. This work also strengthens the case for the development of therapeutics based on miR-155 and surely other microRNAs that regulate immune responses.
Concluding remarks
The field of host-pathogen interactions has seen discoveries that extend far beyond greater knowledge of pathogen or immune-response factors. Multidisciplinary approaches to understanding the complex interaction between the pathogen and the immune response during infection have led to a heightened appreciation of the adaptation of pathogens to their host as well as greater insight into so-called 'sterile' inflammation, innate and adaptive immune control and autoimmunity. The first Lorne Infection and Immunity Conference was established to bring together aspects of infection, the pathogen and the immune response. Collaborations and ideas that result from discussions at these meetings are set to change the way researchers view infection, leading to improved basic knowledge and its translation into better treatment and prevention of infectious and inflammatory diseases. We look forward to further rapid developments in this field and the second Lorne Infection and Immunity Conference in February 2012.
Acknowledgements
We apologize to the many presenters whose work was not included here because of space constraints. The Lorne Infection and Immunity Conference was hosted by the Victorian Infection and Immunity Network (The University of Melbourne Infection and Immunity Domain, Monash University, Walter and Eliza Hall Institute, Victorian Infectious Disease Reference Laboratory, The Burnet Institute, The Commonwealth Scientific and Industrial Research Organisation) and was sponsored by Pfizer Centres of Therapeutic Innovation, Roche, CSL, Gilead Life Research and Bio-Rad.
Competing interests
The authors declare no competing financial interests.
References
- 1.Hornung V, Latz E. Nat. Rev. Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 2.Lin SC, Lo YC, Wu H. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maslowski KM, et al. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malynn BA, Ma A. Immunity. 2010;14:843–852. doi: 10.1016/j.immuni.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philpott DJ, Girardin SE. Curr. Opin. Immunol. 2010;22:428–434. doi: 10.1016/j.coi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Maslowski KM, Mackay CR. Nat. Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 7.Archer KA, Ader F, Kobayashi KS, Flavell RA, Roy CR. Infect. Immun. 2010;78:2477–2487. doi: 10.1128/IAI.00243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Case CL, Shin S, Roy CR. Infect. Immun. 2009;77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luhrmann A, Nogueira CV, Carey KL, Roy CR. Proc. Natl. Acad. Sci. USA. 2010;107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr MC, et al. EMBO J. 2010;29:1331–1347. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu L, Lemon SM. Semin. Liver Dis. 2010;30:319–332. doi: 10.1055/s-0030-1267534. [DOI] [PubMed] [Google Scholar]
- 12.Ge D, et al. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 13.Thomas DL, et al. Nature. 2009;461:798–802. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pidot SJ, et al. PLoS Negl. Trop. Dis. 2010;4:e872. doi: 10.1371/journal.pntd.0000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krausgruber T, et al. Nat. Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 16.Ang DK, et al. J. Immunol. 2010;184:5429–5433. doi: 10.4049/jimmunol.1000128. [DOI] [PubMed] [Google Scholar]
- 17.Dalton JE, et al. J. Clin. Invest. 2010;120:1204–1216. doi: 10.1172/JCI41281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy CE, et al. J. Biol. Chem. 2010;285:20492–20498. doi: 10.1074/jbc.M110.102111. [DOI] [PMC free article] [PubMed] [Google Scholar]


