Abstract
Structural biology is rapidly accumulating a wealth of detailed information about protein function, binding sites, RNA, large assemblies and molecular motions. These data are increasingly of interest to a broader community of life scientists, not just structural experts. Visualization is a primary means for accessing and using these data, yet visualization is also a stumbling block that prevents many life scientists from benefiting from three-dimensional structural data. In this review, we focus on key biological questions where visualizing three-dimensional structures can provide insight and describe available methods and tools.
Supplementary information
The online version of this article (doi:10.1038/nmeth.1427) contains supplementary material, which is available to authorized users.
Subject terms: Software
Main
Decades ago, when structural biology was still in its infancy, structures were rare and structural biologists often dedicated years of their life to studying just one structure at atomic detail. The first tools used for visualizing macromolecular structures were tools for specialists.
Today's situation is very different: the rate at which structures are solved has greatly increased, with over 60,000 high-resolution protein structures now available in the consolidated Worldwide Protein Data Bank (wwPDB)1. These data provide a wealth of detailed information that can yield significant insight into macromolecular function. To use this information most effectively, visualization tools were developed and are increasingly becoming everyday tools for biologists. For example, many biochemists regularly view protein structures to gain insight into protein function (Fig. 1). Chemists look at ligand-binding sites as part of drug design. Molecular biologists view RNA structures and complexes with proteins to gain insight into RNA signal and message processing. Some aspects of structure visualization remain mostly the domain of the specialist, such as molecular motion and large-scale molecular assemblies. Even in these intrinsically more complex fields, however, resources are beginning to enable bench biologists to visualize and use this information.
Figure 1. Visualizing a tyrosine kinase structure (PDB 1QCF)97.
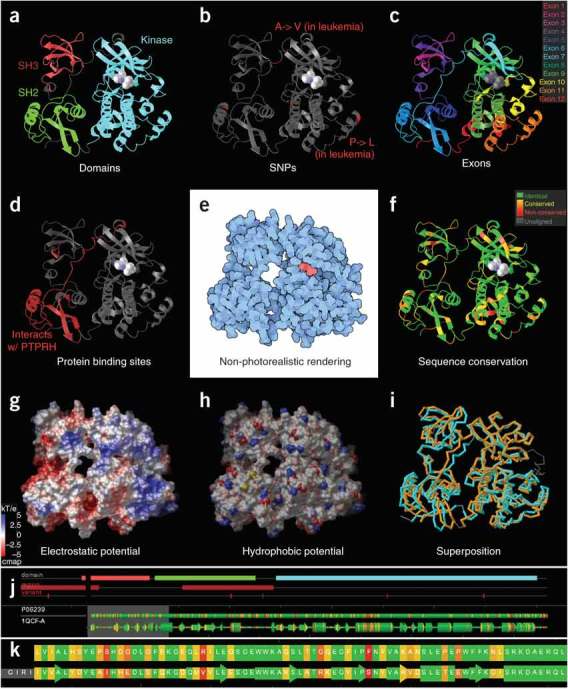
(a–d,f) A simple way to gain insight into function is to use ribbon representation colored by sequence features: for example, domains (a), SNPs (b), exons (c), protein binding sites (d) and sequence conservation (f). (e) An effective way to show overall shape is with nonphotorealistic rendering using flat colors and outlines. (g,h) Solvent-accessible surfaces are often used for displaying electrostatic (g) and hydrophobic potentials (h; hydrophilic in saturated colors and hydrophobic in white). (i) Superposition is commonly used to compare two or more related structures—for example, two distinct states of the same protein, or, as shown here, two separate proteins with similar structure (PDB 1QCF and 1FMK)98. (j,k) Increasingly many tools have an integrated, interactive sequence viewer, which helps users understand the relationship between sequence and three-dimensional structure. Images were made using SRS 3D7 (a–d,f,j,k), PMV25 (e,g,h) and RCSB PDB5 (i).
However, although structural information is now viewed and used by a large and diverse group of scientists, most of them are not prepared to spend months learning complex user interfaces or scripting languages. Even today, complex user interfaces in visualization tools are often a stumbling block, preventing many scientists from benefiting from structural data. Even structural experts have come to expect ease of use from molecular graphics tools, in addition to improved speed, features and capabilities.
In the past, molecular graphics tools were invariably stand-alone, designed to view one molecular system at once. Today's tools are increasingly internet aware, often integrated tightly with structure databases (Table 1), as well as with databases containing sequences and other features (for example, domains, single-nucleotide polymorphisms (SNPs), interactions).
Table 1.
Selected resources for finding and visualizing macromolecules
| Name | Cost | OS | Description | URL |
|---|---|---|---|---|
| Stand-alone | ||||
| Amira | $ | Win, Mac, Linux | Combines many different methods and scripting (EDM, MRI, optical) | http://www.amiravis.com/ |
| Cn3D17 | Free | Win, Mac, Linux | Integrated sequence alignment view; embeddable | http://tinyurl.com/Cn3D-NCBI/ |
| Chime | Free | Win | Widely used; structure editing; electrostatic maps; embeddable | http://tinyurl.com/chime-pro/ |
| Chimera16 | Free | Win, Mac, Linux | Popular; integrated sequence alignment viewer (EDM, MD) | http://www.cgl.ucsf.edu/chimera/ |
| DS Visualizer | Free | Win, Mac, Linux | Free version of Accelrys's powerful viewer/editor program | http://tinyurl.com/DSVisualizer/ |
| ICM-Browser | Free | Win, Mac, Linux | High quality images; integrates with sequence alignment viewer | http://tinyurl.com/icm-browser/ |
| IMOD109 | Free | Win, Mac, Linux | Tomogram alignment, display, segmentation (EDM, optical) | http://bio3d.colorado.edu/imod/ |
| Jmol | Free | Win, Mac, Linux | Widely used; embeddable | http://www.jmol.org/ |
| KiNG | Free | Win, Mac, Linux | Generic tool for creating 'kinemages' | http://tinyurl.com/KiNGapp/ |
| Mage6 | Free | Win, Mac, Linux | Generic tool for creating 'kinemages'; allows structure editing | http://tinyurl.com/kinemage/ |
| MOE | $ | Win, Mac, Linux | Integrated multifunctional suite; useful for drug design (MM) | http://www.chemcomp.com/ |
| Molscript23 | Free | Unix | Useful for preparing manuscript images | http://www.avatar.se/molscript/ |
| MolSurfer | Free | Win, Mac, Linux | Shows macromolecular interfaces, for example, by electrostatic potential | http://tinyurl.com/molsurfer/ |
| MOLMOL35 | Free | Win, Mac, Linux | Many features, particularly suited for NMR structures | http://tinyurl.com/molmol1/ |
| OpenAstexViewer18 | Free | Win, Mac, Linux | Embedded in many PDBe (see below) services | http://www.openastexviewer.net/ |
| ProSAT2 (ref. 31) | Free | Win, Mac, Linux | Displays sequence features on three-dimensional structure | http://tinyurl.com/ProSAT2/ |
| PMV25 | Free | Win, Mac, Linux | Dynamically extensible; multiple structures, large assemblies (MM) | http://tinyurl.com/PMV-MGL/ |
| PyMOL | Free | Win, Mac, Linux | Widely used; embeddable; high-quality images (EDM, MM) | http://www.pymol.org/ |
| RasMol110 | Free | Win, Mac, Linux | Widely used; fast; scripting | http://www.rasmol.org/ |
| Raster3D24 | Free | Win, Mac, Linux | High-quality, photorealistic rendering | http://tinyurl.com/raster3d/ |
| SPICE27 | Free | Win, Mac, Linux | Adds DAS features to three-dimensional structures | http://tinyurl.com/spice-browser/ |
| STRAP19 | Free | Win, Mac, Linux | Editor for structural alignments of proteins (HM) | http://tinyurl.com/STRAP1/ |
| Swiss-PdbViewer20 | Free | Win, Mac, Linux | Integrated sequence view (EDM, MM) | http://spdbv.vital-it.ch/ |
| SYBYL | $ | Win, Mac, Linux | Popular molecular modeling tool (MM) | http://tinyurl.com/triposSYBYL/ |
| VMD26* | Free | Win, Mac, Linux | Widely used; extensible, many add-ons (EDM, MD, MM, NMR) | http://tinyurl.com/VMD-viewer/ |
| WHAT IF42 | $ | Win, Mac, Linux | Powerful features; good support (EDM, HM, MM) | http://swift.cmbi.ru.nl/whatif/ |
| Yasara | Free | Win, Mac, Linux | Innovative 'virtual reality' graphical user-interface (EDM, MM, NMR) | http://www.yasara.org/ |
| Web-based | ||||
| CAME | Free | Assesses structure quality (ProSA-Web111); finds structural homologs | http://www.came.sbg.ac.at/ | |
| EMDB | Free | Central repository for electron microscopy density maps | http://emdatabank.org/ | |
| Entrez Structure | Free | Finds related structures for a sequence | http://tinyurl.com/entrez3d/ | |
| FirstGlance | Free | Useful for a first impression of a structure | http://firstglance.jmol.org/ | |
| JenaLib28 | Free | Displays sequence features on three-dimensional structure | http://tinyurl.com/JenaLib/ | |
| NDB68 | Free | Central repository for nucleic acid structures | http://ndbserver.rutgers.edu/ | |
| PDBe | Free | European branch of wwPDB (formerly MSD); many services | http://www.ebi.ac.uk/pdbe/ | |
| PDBsum29 | Free | Pictorial structural annotations | http://www.ebi.ac.uk/pdbsum/ | |
| PISA33 | Free | Predicts biologically relevant quaternary structure | http://tinyurl.com/piserver/ | |
| Relibase58,59 | Free/$ | Finds similar ligands and binding sites; free version has limits | http://tinyurl.com/relibase/ | |
| RSCB PDB5* | Free | US branch of wwPDB; has wide range of services | http://www.pdb.org/ | |
| PMP10 | Free | Consolidated portal for homology-modeled structures | http://tinyurl.com/ThePMP/ | |
| Proteopedia94 | Free | Community annotation of structures | http://www.proteopedia.org/ | |
| SRS 3D7 | Free | Finds related structures for a sequence; displays sequence features | http://SRS3D.org/ | |
| Swiss-Model11 | Free | Finds related structures for a sequence | http://swissmodel.expasy.org/ | |
| TraceSuite II | Free | Maps phylogenetic information onto structures, finds functional residues | http://tinyurl.com/TraceSuite/ |
The table shows only tools with outstanding features or strengths; more complete lists can found on Wikipedia (http://tinyurl.com/moleculargraphics/), at the World Index of Molecular Visualization Resources (http://www.molvisindex.org/), at the PDB (http://tinyurl.com/moleculargraphics-pdb/) and at http://molviz.org/. *Our recommendations. Free means the tool is free for academic use; $ means there is a cost. OS, operating system: Win, Microsoft Windows; Mac, Macintosh OS X. Tools running on Linux usually also run on other versions of Unix. EDM, electron density maps; HM, homology modeling; MD, molecular dynamics; MM, molecular modeling and molecular orbital visualization; MRI, magnetic resonance imaging; NMR, nuclear magnetic resonance; optical, optical microscopy.
Today, we are spoiled for choice when it comes to molecular graphics tools for viewing proteins and other macromolecular structures. Indeed, the sheer range of available tools can be overwhelming. Many molecular graphics tools have been developed to address diverse requirements, as documented in recent reviews2,3,4 and in several web resources maintaining lists of such tools (see footnote to Table 1). Most of these tools have a large set of features in common, including standard representations (ribbon, space-filling, ball-and-stick and so on) and coloring schemes (element-based coloring of atoms, coloring by secondary structure and so on). It is beyond the scope of this review to comprehensively compare all of these tools; instead, we focus on key biological questions for which visualizing structures can provide insight, and we highlight practical methods and tools with outstanding features that are particularly suited to addressing these questions.
Protein structures
Finding three-dimensional structures. For a biochemist looking to use three-dimensional structures to gain insight into the functions of a particular protein, the typical first step is a search for relevant structures. This task is considerably simplified by the remarkable degree to which all experimentally determined protein three-dimensional structures are consolidated into a single data repository, the Worldwide Protein Data Bank (wwPDB)1. Three primary distribution sites (RSCB PDB5, PDB Europe and PDB Japan; Table 1) provide access to the same underlying data bank, each with a wide range of integrated visualization and analysis tools. In addition, the PDB is mirrored at many other sites, some of which provide innovative visualization tools tailored to make specific questions easier to answer (Table 1). Most of these sites offer, embedded directly in their web-pages, one or more molecular graphics tools (for example, Jmol, PyMol, KiNG and Mage6). Increasingly, the process of finding and visualizing structures is becoming one seamless step for most users.
Finding structures from sequence. Several websites (for example, RCSB PDB5) allow the user to find structures using a sequence identifier or BLAST search (Table 1). Entrez Structure and SRS 3D7 allow the sequence to be aligned to any related three-dimensional structure (Fig. 1f). So far, experimental three-dimensional structures have been determined for less than 1% of all known proteins (based on direct links from PDB to protein sequences in UniProt8). However, for around 42–48% of all proteins, at least part of their sequence is considered significantly similar to a PDB entry, so that some structural information can be inferred9,10. Several websites (for example, Swiss-Model11) provide comparative models for such cases12,13. Each service uses slightly varying cut-off criteria for defining 'significant sequence similarity' (for example, in some cases depending on the length of aligned regions), but generally >40% sequence identity to a PDB structure is considered sufficiently good to create a high-quality comparative model structure10. These comparative models can be accessed at a single consolidated website, the Protein Model Portal (PMP)10. The original PDB templates also include information on experimental conditions, ligands and cofactors, which can be relevant in deciding to use or discard a comparative model.
For sequences where no template PDB structure can be found by the above resources, it may be possible to calculate a structure using so-called ab initio methods14. However, in spite of progress15, ab initio methods still require much improvement14 and we recommend they be used with caution.
Getting a first impression. To gain an initial overview of a protein structure, it is often useful to choose a representation that hides side chain atoms; ribbon-like representations do that well and also convey information about secondary structure (Fig. 1a–d). Ligand molecules are best displayed in space-filling or ball-and-stick atom representations. Many of the websites in Table 1 provide such a view (for example, FirstGlance, among others), some by default. Typically, each protein chain is colored differently, thus giving a quick insight into the number of molecules present in the PDB entry. To highlight overall shape and form, nonphotorealistic rendering can be very effective (Fig. 1e), especially with images for presentation and publication.
Some molecular graphics tools (for example, Chimera16, Cn3D17, OpenAstexViewer18, SRS 3D7, STRAP19 and Swiss-PdbViewer20) offer an integrated view of both the amino acid sequence and the three-dimensional structure, and further enable interaction between these two views (Fig. 1j). For example, clicking on a residue in the sequence view causes the corresponding residue to be highlighted and selected in the three-dimensional view, and vice versa. This feature can significantly help a scientist in understanding and using three-dimensional structures. For example, by viewing the location of key residues or sequence motifs, a scientist can assess whether they are likely to be accessible for posttranslational modification, such as phosphorylation21. Some viewers (for example, STRAP19) go one step further, showing structure integrated with a multiple sequence alignment viewer—a feature we anticipate will continue to become available for other viewers22.
For publication and presentations, some viewers can create impressive, ray-traced images (for example, Amira, Chimera16 ICM-Browser, Molscript23 plus Raster3D24, PMV25, PyMOL, VMD26).
The majority of PDB structures are derived from X-ray crystallography (Box 1, Fig. 2), about 13% from NMR spectroscopy (Box 2, Fig. 3) and less than 1% from electron microscopy (Box 3). These three experimental methods often require specific considerations and visualization methods (discussed in each display box).
Figure 2. Caution for beginners: symmetry in crystal structures.
PDB entries often do not have explicit three-dimensional coordinates for all parts of symmetric oligomers. (a,b) For example, in PDB 2C2A107, coordinates are given for only one monomer (a), although the biologically active state is a homodimer (b). (a–e) Usually this information is given in 'REMARK 350', however we recommend using PISA33, which automatically constructs a range of assemblies that occur in the crystal and predicts which of these is most biologically relevant. In this case, PISA gives the asymmetric unit (a), three dimer forms (b,c,d) and the unit cell (e). Increasingly, sites such as RCSB PDB5 provide the biologically relevant assembly precalculated with PISA. Image of PISA output made using VMD26.
Figure 3. Visualization of an NMR ensemble for SH3 (ref. 108).
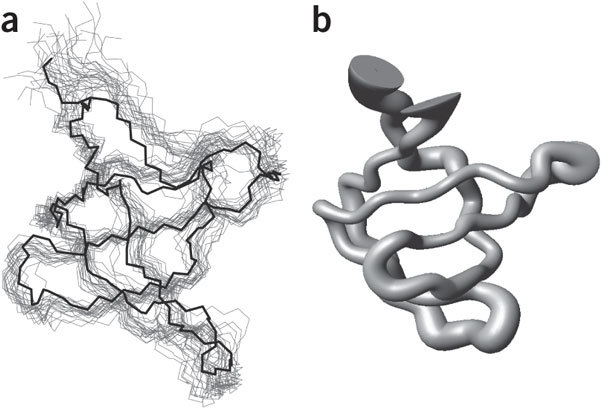
(a,b) NMR structures are typically deposited in the PDB as an ensemble of superimposed structures (a), with the spread of the ensemble giving an indication of precision, but not of accuracy. The 'sausage' representation (b) gives an informative summary of an ensemble by adjusting the width of the tube to match to the width of the ensemble. Images made using MOLMOL35 (a) and VMD26 (b).
Viewing sequence features on three-dimensional structures. A very straightforward way to use three-dimensional structures to gain insight into function is by coloring based on features such as domains, SNPs, exon boundaries, secondary structure and so forth. (Fig. 1a–d,f). The ability to easily see where sequence features are located in the three-dimensional structure can be of substantial practical value to bench biochemists and molecular biologists. For example, the spatial location of residues within the structure and the proximity to solvent can help in designing primers and mutation experiments. The ability to show such views for a wide range of features is a particular strength of SRS 3D7 and SPICE27 and is also facilitated by JenaLib28, PDBsum29 and Entrez Structure. Viewers such as STRAP30 that provide easy access to multiple sequence alignment information mapped onto three-dimensional structures can help locate key conserved residues. ProSAT2 (ref. 31) can display SNPs and also predict their effects, allowing a scientist to gauge the potential impact of a SNP on the protein structure.
Protein-protein binding sites. Typically, as part of its biological role, a protein will bind to several other proteins through comparatively large but flat binding surfaces. In fact, a large percentage of PDB entries contain not just a single protein chain but several. In some cases, this means identical subunits assembled together; in other cases, it means a complex of several different protein chains. The arrangement of subunits, and of the interface residues that form the subunit-subunit contacts, is often of biological significance. Several websites specialize in finding and visualizing subunit-subunit interface residues32. In PDBsum29 the interacting residues, and the types of their interaction across the interface, are shown schematically. MolSurfer (Table 1) provides a range of methods that help users explore macromolecular interfaces.
For symmetric assemblies (dimers, trimers and so on), the PDB entry of an X-ray crystal structure will often have explicit three-dimensional coordinates for only one monomer. To construct the coordinates for all subunits in the biologically relevant assembly, we recommend PISA33 (see Box 1, Fig. 2).
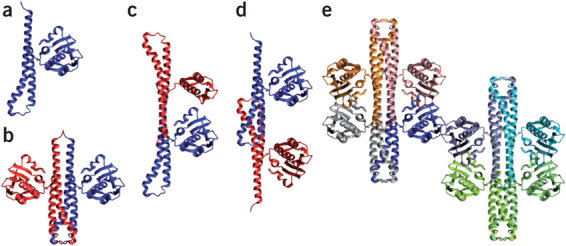
Comparing related structures. It is often informative to visualize two related structures superimposed—for example, two states of the same molecule, or two proteins with homologous sequences, or two structural homologs found by structural comparison tools34. Many molecular graphics tools offer automatic superposition as a standard feature (for example, MOLMOL35, MOE, PyMOL or VMD26). These tools allow the researcher to specify a portion of the molecule to be superimposed. The results are highly dependent on the regions chosen for the superposition. Typically, the researcher identifies a more-or-less rigid core of the molecule and superimposes this region using a subset of the atoms (typically the α-carbons or the backbone atoms). But many other combinations are possible for addressing specific questions (Figs. 1i and 4d–f). For difficult cases—for example, low sequence similarity or large regions that cannot be aligned in sequence—it is best to use more robust, dedicated superimposition tools (for example, STAMP36, STRAP19 or THESEUS37).
Figure 4. Visualizing ligand-binding sites.
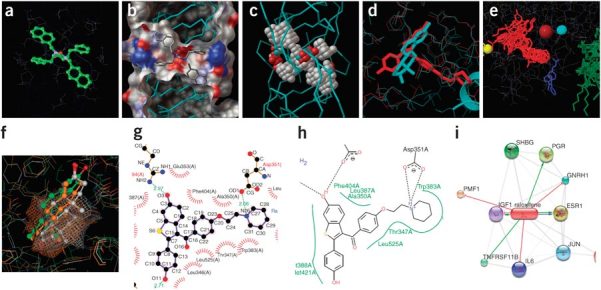
(a) A useful initial view is to show ligands and binding site residues in ball-and-stick and wire-frame representations, respectively. Here, an inhibitor is shown bound to HIV protease (PDB 1HVR99). (b) Visualizing the same binding site using a molecular surface colored by atom type reveals the catalytic oxygen atoms (center, red). (c) Here, AutoLigand44 has been used to find regions that might bind a ligand-sized molecule. (d) Two structures of the same protein (estrogen receptor) superimposed using Relibase58,59, one with estrogen (blue, PDB 1QKU)100, a second with an antagonist (red, PDB 1ERR)101, give insight into the antagonist mechanism. (e) All 74 structures of human estrogen receptor compared using PDBsum, showing estrogen (red) and cofactors (green). (f) Comparing binding sites of related structures can give insight into drug specificity. Image shows estrogen receptor (green), progesterone receptor (gray) and androgen receptor (orange). (g,h) Simplified two-dimensional schematics can be useful for visualizing binding site interactions, such as hydrogen bonds (dashed lines), unbonded contacts ('eyelashes', g) and hydrophobic interactions (green curves, h). (i) To study drug specificity, interaction networks can be used to show all proteins known to interact with a drug. Images made using SRS 3D7 (a), PMV25 (b,c), OpenAstexViewer18 (d), Jmol (e), MOE (f), LIGPLOT65 (g), PoseView66 (h) and STITCH63 (i).
Molecular surfaces and electrostatic potentials. Many tools can generate molecular surfaces, most commonly the so-called Connolly surface38, which is derived by rolling a sphere the radius of a water molecule around the atomic van der Waals surface of the molecule. This surface, also known as the solvent-excluded surface, can be used as a canvas to map a wide variety of properties such as residue conservation scores, hydrophobicity (Fig. 1h), depth-cue information (Fig. 1e), mean-force potentials39 and electrostatics (Fig. 1g). Such colored surfaces (sometimes called texture mappings) can give insight into molecular interactions and conformational changes, for example, by highlighting surface regions with complementary shape and charge. The molecular surface can also be used to estimate the energetics of molecular interactions, including the entropic cost of desolvation, by calculating the area buried from solvent upon binding of other molecules40.
Although many program can generate a surface, the program MSMS41 is widely used as it provides a good estimate of molecular surface area and volume, and the most relevant molecular geometry when analyzing molecular interactions and interfaces.
Box 1: X-ray crystal structures.
About 86% of PDB entries are derived from X-ray crystallography. Each X-ray structure has a resolution value, that is, a measure of the crystal order. The expected error in the three-dimensional atomic coordinates is correlated to, but much smaller than, the resolution. An average resolution is about 2.5 Å, and 1.2 Å is very high quality112. At low resolution (≥4 Å), there is a significant chance of errors in the structure. Each atom also has a B-factor, a parameter correlated with molecular motion, and many molecular graphics tools can use this parameter to color the molecule. However, the correlation with motion is only partial, as other effects contribute to the B-factor113. Regions with very high B-factor (>>80) should generally be treated as of unknown structure. The goodness of fit of the structure to the X-ray crystal data is indicated by the R-value and free R-value114, where values of around 20% are considered to indicate a good structure. The difference between the R-value and the free R-value should be low. The goodness of fit can be examined at the Electron Density Server115. It can be useful to also check further independent measures of structure accuracy, and many are available directly on the web page for each structure at the RCSB PDB site. The PDBe site also has several useful services for assessing accuracy, particularly PDBsum29. A very useful, and independent, accuracy estimate is also available from ProSA-Web111, which can show residue-specific quality scores mapped on the three-dimensional structure (Table 1). When viewing crystal structures, it is important to be aware that PDB entries only give explicit coordinates of the 'asymmetric subunit' of the crystal (that is, the smallest portion of a crystal needed to produce the unit cell of the crystal) which often is only part of the full biologically relevant assembly (Fig. 2).
A primary limitation of X-ray crystallography is the need to crystallize the macromolecule; this is especially difficult for membrane proteins, for proteins with natively disordered regions and for transient complexes. To aid crystallization, macromolecules are often tampered with, for example, by mutating surface residues and truncating segments of sequence; in addition, modifications present in vivo are often missing in X-ray crystal structures. To help with structure calculation, macromolecules are often crystallized with seleno-methionine, heavy metal ions, or other impurities (sometimes even other proteins) that are not present in vivo and that may sometimes distort the structure.
X-ray crystal structures are calculated by fitting an atomic model of the molecule into an electron-density map (EDM) or isosurface and can be visualized by tools such as COOT116, FRODO117 and O118. Highly mobile parts of the molecule often are missing in the EDM and are usually removed from the molecular model, resulting in missing atoms or residues. Current methods for structure determination are dominated by the concept of a rigid structure, although improved methods have very recently been proposed119 that account for molecular motion and produce structural ensembles similar to NMR structures (Box 2).
Box 2: NMR structures.
About 13% of PDB entries are derived from NMR spectroscopy. NMR structures are usually deposited in the PDB as an ensemble of 10–50 structures (Fig. 3), providing a visual representation of precision. The ensemble precision derives not from the dynamics of the molecule in solution but from the lack of data to describe the structure fully. The method used for structure calculation also affects the ensemble precision, and recently a significantly improved method has been developed that ensures that the ensemble precision more truly reflects the data120. Often, a single 'minimized average' structure, or a representative structure, is also provided in the PDB. The ensemble precision is often measured as the root-mean-square (r.m.s.) deviation to the average structure, where a value of about 2.0 Å is typical121; more precise ensembles with lower r.m.s. deviation may indicate an overfitting of data, rather than high quality. An ensemble r.m.s. deviation of ≥5 Å generally indicates a low quality structure, although values this low may occur, for example, in a structure comprising two well ordered domains connected by a flexible linker region. There is still no standardized measure for assessing the goodness of fit of structures to NMR data, although it is common to report the r.m.s. deviation of violations of distance and other constraints. As with X-ray structures, it can be useful to check independent measures of accuracy (for example, available at RCSB PDB, PDBe and via CAME/ProSA-Web111).
Compared with X-ray crystallography, NMR has the advantage that it is not necessary to crystallize a molecule; instead, NMR usually studies biomolecules in solution, hence arguably in a more natural state. NMR structures also lack the heavy metal contaminants of many X-ray structures. NMR can also dynamically track specific reactions in living cells122 and can more easily study weak associations as well as disordered protein states123. However, NMR has the disadvantage that it imposes an upper limit on the size of the molecule studied: the largest molecule solved by NMR so far is 82 kDa (ref. 124), and most NMR structures are 25–30 kDa or less. Another disadvantage is that, for the same molecular system, NMR usually produces less precise structures than does X-ray crystallography. As with X-ray crystallography, studying structures by NMR often requires tampering with the target macromolecule. Frequently, truncated segments, often single domains, are studied by NMR, rather than full-length proteins. In addition, mutations of surface residues are often introduced to avoid aggregation. Finally, post-translational modifications that are present in vivo are often missing in NMR structures. NMR structures are calculated automatically from constraints derived from the NMR spectra, primarily constraints on interatomic distances125. However, manual checks are sometimes needed during structure determination, and several molecular graphics tools (for example, MOLMOL35) offer the possibility of easily displaying NMR data, such as distance constraints, directly on the structure. In contrast to X-ray crystal structure calculation, it is customary to retain residues with very little or no data, with the result that some regions of the molecule can be very divergent in the ensemble. The display of ensembles requires prior superposition of the structures. Some graphics packages offer automatic superposition (VMD26, MOLMOL35), but for more difficult cases, with highly divergent regions, dedicated programs should be used; for example, THESEUS35.
Box 3: Macromolecular structures from electron microscopy.
The Electron Microscopy Database (EMDB, http://emdatabank.org/) now has nearly 700 entries, mostly three-dimensional electron density maps (EDMs) of macromolecular and cellular structures. For about 250 of these entries, an atomic model has been calculated, usually by fitting an X-ray crystal structure (for example, Fig. 5b): these atomic-detail models may be deposited in linked entries in the PDB, where they account for about 0.4% of PDB entries.
A significant advantage of electron microscopy structure determination126,127,128 is that it can be used to study a wide range of sample types, from large, ordered assemblies such as helical arrays to isolated complexes (single particles) and irregular objects such as cells or subcellular components. The upper size limit is mainly the sample thickness (up to a few hundred nanometers). Compared with X-ray and NMR structures, electron microscopy is almost always at lower resolution. Three-dimensional maps are viewed by choosing an appropriate density threshold value, normally one that gives a surface enclosing the correct molecular volume and displaying the isosurface (for example, with Chimera16). EMDB provides two map viewers, of which we recommend OpenAstexViewer18 because it displays surfaces well and lets the user change threshold levels.
Transmission electron microscopy images are projections, and three-dimensional structure determination involves the collection and merging of different projections (views) of the object. The main task is usually the determination of the relative positions and orientations for the set of views. Atomic structures of components in larger assemblies that have been determined by crystallography can be docked into the electron microscopy map. Often there are conformational changes between a structure in a crystal lattice and in solution. Flexible fitting makes it possible to account for changes, such as hinge rotations, in fitting. In the most favorable cases, the resolution of macromolecular electron microscopy structures can reach about 3 Å, although for cell or tissue sections, radiation damage limits the resolution to >>30–40 Å. At this high resolution end, the conformation of protein and nucleic acid backbones and bulky side chains can be determined directly from the electron microscopy density129 with the same tools as in X-ray crystallography.
However, there are uncertainties in determining the resolution and in validation for noncrystalline samples. When atomic-detail structures are known that correspond to any part of the electron microscopy structure, docking of the atomic coordinates into the map provides an independent test of reliability. Two density maps can be compared by maximizing their cross-correlation (for example, using Chimera), and the comparison can be visualized using semitransparent and solid or wire mesh surface displays that can be overlaid. For tomographic reconstruction and for three-dimensional reconstruction of sections, IMOD109 is commonly used.
Ligand binding sites
Interactions between macromolecules and small molecules often occur in buried active sites; these may be catalytic active sites, allosteric sites, or sites that may either disrupt or stabilize protein-protein interactions. The PDB at present contains over 37,000 binding sites involving about 10,000 different types of ligand molecules. A range of methods are available to characterize and visualize these sites, depending on the questions asked by the end user.
Annotation and highlighting. For gaining an initial insight into the atomic interactions in the binding site, a useful representation is to display ligands using a ball-and-stick representation and to display only backbone atoms of the protein or nucleic acid, except for those residues in direct contact with ligands (Fig. 4a). Many molecular graphics tools have been developed to support working with small molecules (for example, DS Visualizer, MOE, PMV25, PyMOL, STRAP19, Swiss-PdbViewer20, SYBYL, VMD26, WHAT IF42, Yasara; Table 1) Almost any can implement such views, and those with scripting capabilities can often be programmed to recreate this view on demand.
In addition, many PDB entries or related files (for example, UniProt) have annotations indicating which residues form the binding site. It can be instructive to display these annotations directly on three-dimensional structures, and many molecular graphics tools enable such displays (for example, JenaLib28, PDBsum29, ProSAT2 (ref. 31), SRS 3D7 and Ligand Explorer in the RCSB PDB).
Surface-based approaches. Structural details of binding sites are widely used in rational drug design, usually to generate ideas for classes of compounds for screening43. A common question is to ask what kinds of small molecules may bind to a given binding site. Many molecular graphics viewers allow the surface to be colored by local properties, such as hydrogen bonding ability, hydrophobicity or electrostatics, to allow exploration of chemical complementarily (Fig. 1g,h). The local curvature of the surface may also be used to evaluate steric complementarily.
Volume-based approaches. An alternative approach is to analyze the space around the target molecule, highlighting regions that may form strong interactions with small molecules. Some tools (for example, AutoLigand44) allow probe atoms, such as carbon atoms or oxygen atoms, to be scanned through the entire space and the interaction energies of the probes with the molecule to be evaluated. The resultant three-dimensional data sets are then rendered to show the areas of most favorable interaction45. More recently, atomic probes have been used to create maps of the atomic affinity. These may be rendered using isocontours, text-mapped clipping planes or volume rendering (Fig. 4b,c). Many researchers are now analyzing these volume data sets to identify and visualize ligand-sized regions of maximal affinity44.
Sequence-profile approaches. Another approach to identify ligand binding sites uses multiple sequence alignments mapped onto three-dimensional structures46. This approach is based on the observation that binding site residues tend to be more conserved than other positions, so it can be particularly useful when little is known about a protein. Even for well studied proteins, however, these methods sometimes find binding sites not previously noticed. Some examples of such services are TraceSuite47, ETV48 and others48,49,50.
Multiple ligands. A three-dimensional structure gives a snapshot of a single state; however, in some cases, several different structures of the same protein exist with different ligands. We can use this information to help explore the range of conformations available to the system. For example, such comparisons can highlight interactions common to all known binding partners, which may help to guide the search for further possible binding partners51,52,53. For such comparisons, it can be useful to try different sets of atoms for superposition—for example, the ligand alone, or all atoms involved in the binding site. Each of these superimpositions can highlight different aspects of the conformational differences.
Often, it is of interest to compare structures with multiple ligands obtained by means of docking tools (for example, FlexX54, AutoDock55). To preselect promising compounds, computational chemists can scan large libraries of drug-like molecules and dock 'hits' into the binding site of the protein target56. Subsequently, the docked structures can be inspected visually to find ways of enhancing the predicted strength of binding57. Some docking tools now provide graphical interfaces (for example, FlexV and AutoDockTools) for the preparation of the input structures and the analysis of the results. These tools allow the comparison of interaction geometries of different ligands with the same protein.
Two useful resources for comparing multiple ligand structures are Relibase58,59 and Superligands60, which both contain information about all ligands in the PDB and take special care to ensure the assignment of chemically correct atom and bond types. Both resources allow searching by identifiers as well as chemical substructure searches and similarity searches; Relibase also offers keyword searches and sequence similarity searches. The structures can be displayed in two or in three dimensions in embedded viewers. When exploring a specific protein, it is especially useful to search for similar complexes; Relibase lists similar proteins with their respective ligands, which can subsequently be superimposed and displayed in the embedded OpenAstexViewer18 (Fig. 4d and Supplementary Fig. 1). The extended functionalities of Relibase+ (which requires a paid license) give an analysis of the differences in the superimposed structures (protein movements and ligand overlap).
PDBsum can also help visualize multiple ligands binding to the same protein by superimposing the protein's different structural models in the PDB and identifying any 'ligand clusters'; that is, sites where the ligands from the different structures overlap (Fig. 4e).
Multiple proteins and ligands. Finding features that are specific to a given target adds another level of complexity when studying protein-ligand interactions. To identify features determining selectivity, it is useful to compare the target binding site with binding sites of similar proteins. The “similar binding site” as well as the “similar ligand” search of Relibase can help to identify and compare similar protein complexes. Here, again, the Relibase+ comparison table is especially useful for detecting differences in the protein binding sites—mutations, insertions and residue movements. MOE provides a similar facility to help compare multiple proteins bound to multiple ligands (Fig. 4f).
Structural visualization can be useful for predicting side effects and 'off-label' uses of known drugs by comparing the target binding site to other known protein structures61,62. Some graphic tools support this: for example, Relibase+ offers a search for “similar cavities,” where the protein comparison is based on physico-chemical properties rather than residues, hence finding remote similarities not evident from sequence similarity.
Structural visualization can also help in developing more selective drugs. Although promising, such approaches remain speculative, and their success will be fundamentally limited, as the PDB contains only a small fraction of all binding site geometries. A complementary approach is to use the much larger set of known protein-drug interactions where no three-dimensional structure is available. For example, STITCH63 can be used to show a network featuring all proteins known to interact with a given drug, based on a wide range of experimental databases, including the PDB (Fig. 4i). In the future, we anticipate that such approaches will be improved, and that PDB data will be increasingly incorporated into network visualization methods64.
Schematic illustrations. For presentations and printouts, it can be useful to highlight key interactions in the binding site using simplified schematic illustrations produced by tools such as LIGPLOT65, PoseView66 and Ligand:Protein Interaction Diagrams67 (part of MOE). These illustrations show the ligand and interacting protein side chains 'flattened' in a plane, and indicating relevant hydrogen bonds, covalent bonds, unbonded contacts and water-mediated hydrogen bonds (Fig. 4g,h). For comparing different complexes, LIGPLOT65 and MOE allow the user to generate a series of plots for related proteins binding the same or different ligands. Equivalent components of each plot are plotted in the same relative location, thus highlighting residues and interactions present in some of the structures but missing in others.
RNA structures
Over 4,000 nucleic acid three-dimensional structures are on deposit in the Nucleic Acid Databank (NDB68), mostly RNA structures, either determined experimentally or by ab initio prediction. NDB is also synchronized with the PDB1, and RNA structures account at present for nearly 8% of PDB entries. Many standard aspects of visualizing three-dimensional structures of RNA can be performed completely adequately by molecular graphics tools designed for proteins, such as PyMOL and Swiss-PdbViewer20 (Table 1).
Knowing the secondary structure of an RNA molecule often gives significant insight into its function, much more so than for protein secondary structure. RNA secondary structure can be derived either from multiple sequence alignments or from thermodynamic predictions, although the process requires specialized features and capabilities not available in most tools for visualizing protein alignments or structures. Multiple sequence alignment is particularly important in RNA research; alignments can be used to find covariations between nucleotide positions, which are then taken as evidence for a contact between the two nucleotide positions, and these contacts in turn define secondary structure (Fig. 5).
Figure 5. Visualization of RNA structure in one, two and three dimensions.
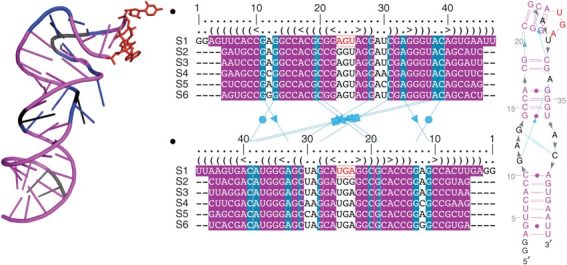
Viewing multiple sequence alignment simultaneously with two-and three-dimensional representations greatly helps in assigning two-dimensional structure and understanding function. This process is aided by synchronizing colors in all three views. The RNA structure shown is from SARS virus102, and the image was made using S2S Assemble69 with PyMOL.
Because of these special-purpose requirements, the RNA community has developed their own specialized visualization tools (Supplementary Table 1) for viewing RNA secondary structure. Some of these RNA tools (for example, S2S Assemble69) provide an integrated environment for interactively visualizing multiple sequence alignments, intramolecular contacts and RNA three-dimensional structures (Fig. 5). The most useful tools provide the option to manually edit the two-dimensional contacts, allowing not only reorientations of elements but also deletion and addition of nucleotides or a whole element, such as a helix.
At present, two of the main challenges in RNA visualization are as follows: first, RNA often adopts multiple structures depending on experimental conditions, and none of the available tools can deal with this properly. Second, RNA in vivo usually occurs in complex with proteins, however the RNA-specific tools cannot yet manage such complexes. RNA researchers can use standard molecular graphics tools to view such complexes, but of course this means losing RNA-specific features and capabilities.
Molecular motion
Biomacromolecules are dynamic entities, and motion is usually essential to function70. Visualizing dynamic molecular processes is often key toward understanding these processes. Recently, several visualization tools have become available that allow quick and easy exploration of dynamic transitions between two known states of a molecule. For example, the Yale Morph Server71 (http://molmovdb.org/) provides morphed animations of potential plausible pathways between two structures; Moviemaker72 (http://tinyurl.com/moviemaker-v1/) is a web server that permits the user to generate simple animations of a variety of types of protein motion. These tools provide very approximate, often simply schematic, descriptions of the molecular motions.
To explore large-amplitude, low-frequency motions, such as protein domain flexing, methods based on normal mode analysis and elastic network models provide a computationally efficient approach73. There are now several websites, for example, NOMAD-ref74 and ANM75, where even a novice user can enter a PDB file, compute normal modes, and visualize and analyze the results.
At a slightly higher level of complexity, several programs allow users to generate conformational ensembles and trajectories using constraint-based methods. Such programs include tCONCOORD76 and FIRST/FRODA77. One application of these methods is to identify segmental flexibility in proteins. The researcher identifies rigid domains in the protein connected by flexible tethers, then defines the geometry of the hinge or shear motions that occur as the proteins change conformation78. The Database of Macromolecular Movements71 provides a service for analyzing hinge motion in proteins. Other websites enable molecular motions to be analyzed by means of hierarchical, multiresolution flexibility trees79.
More realistic and detailed studies of motion require molecular dynamics simulations, which typically simulate 10–100 ns of motion in ∼1-fs time-steps. Unfortunately, such calculations are generally too CPU-intensive to be provided as a free service; hence, users usually need to calculate their own trajectories. For a first look at molecular dynamics simulations, DSMM80 (http://tinyurl.com/dsmm-eml/) is a site that collects movies showing molecular dynamics simulations. Generally, molecular dynamics simulations are recorded as trajectory files that can be played back in a range of molecular graphics tools that support molecular dynamics (Table 1). There is as yet no unified resource to deposit or access trajectory files, although there are several initiatives in this direction—for example, the MoDEL Molecular Dynamics Extended Library (http://mmb.pcb.ub.es/model/). A related project, called Dynameomics81 (http://www.dynameomics.org/), provides online interactive views of simulations of 30 proteins and plans to extend this to all known protein folds. Such services are still very new, and we can expect significant advances in the next few years.
Of the molecular graphics tools with molecular dynamics support, VMD26 is probably the most widely used. It can display 'movies', analyze properties such as atomic fluctuations and allows flexible integration with other computational tools and with the user's own scripts. Although VMD is popular, many other molecular graphics tools support molecular dynamics trajectories, and each tool often has unique features that may be useful for particular projects (Table 1).
In general, visualization of molecular dynamics trajectories remains challenging owing to intrinsic complexity, such as the large number of atoms involved and the many orders of magnitude in time relevant for biological processes. The most straightforward visualization is to superimpose several molecular dynamics snapshots (Fig. 6a). While often useful, this method has obvious limits. Overall motion can be viewed using 'sausage-like' representations (Fig. 3b); however, often dimension-reduction methods are needed58. An increasing number of such methods are being developed for visualization of specialized cases—for example, transient cavities (Fig. 6b) and molecular diffusion (Fig. 6c–e).
Figure 6. Visualizations of molecular motion.
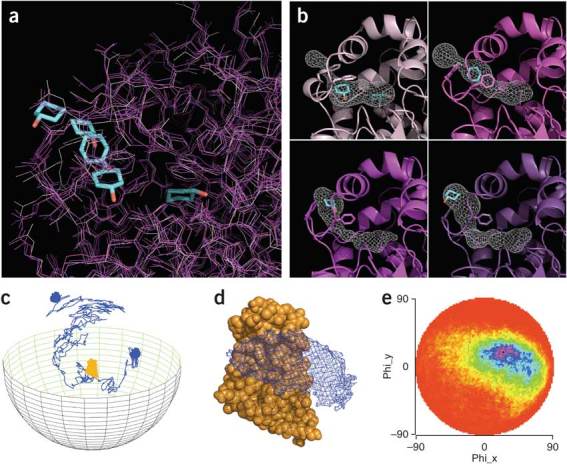
(a) Four snapshots from a molecular dynamics simulation visualized (darker protein coloring indicating later snapshots). A ligand is shown moving from its initial position buried in an active site (right) to the protein exterior (left). (b) Same four snapshots using a simplified representation highlighting residues undergoing conformational changes as the ligand escapes. The contoured surface (generated with CAVER103) shows changes to the transient tunnel used by the ligand. (c–e) Visualization of protein-protein diffusion simulations made using SDA (http://tinyurl.com/SDA-EML/). (c) Representative trajectory of a protein (blue) diffusing around a second, target protein (orange). (d) Isocontours (blue) show the region most occupied by the diffusing protein during thousands of trajectories. Target protein, orange. (e) Two-dimensional map of occupancy versus protein-protein center-to-center distance; blue, the most occupied region104.
Large macromolecular assemblies
X-ray crystallography is being used to solve the structures of larger and more complex systems, and there is now considerable overlap in the size range of structures from X-ray crystallography and from electron microscopy (Box 3). It is common to see electron microscopy isosurfaces into which atomic-detail X-ray structures have been fitted. Meanwhile, electron microscopy continues to produce higher-resolution density maps of large assemblies and of single particles, such as viruses or other isolated complexes (Box 3), in addition to tomograms of higher-order, unique structures such as cell sections or isolated organelles82.
These data on large-scale assemblies that integrate data from X-ray crystallography (Box 1), NMR spectroscopy (Box 2), electron microscopy (Box 3) and even light microscopy82,83,84 pose many new challenges for visualization. Many of these data are not at atomic detail, so other representations must be used. In addition, the systems can be very large, and there are often issues with computational and graphics performance. There is a need for high-performance, interactive visualization of such large assemblies, and across very different distance scales, although some tools, such as Amira (Visage Imaging) and PMV25, were designed with such challenges in mind.
At present, researchers typically use a hierarchical approach to visualizing large macromolecular assemblies. For portions for which atomic information is available, atomic representations may be used, and then abstracted to simpler, surface-based representations. These surfaces may then be integrated with density sections or volumes from the lower-resolution methods (for example, electron microscopy tomography). This approach scales nicely from the level of atoms to the level of cells, allowing the use of simpler, more abstracted representations of the individual components as one moves to large systems, such as intracellular components (Fig. 7a) or even whole-cell visualization (Fig. 7b), and to multiscale movies85.
Figure 7. Two examples of multiscale, hierarchical visualization.
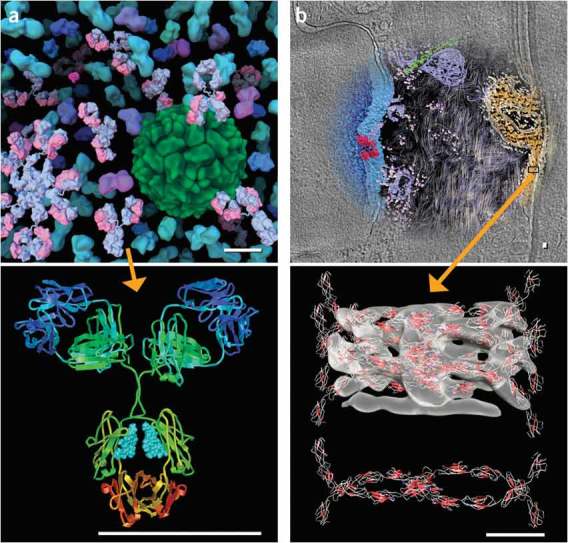
(a) An atomic structure of an antibody (bottom) was used to create a smoothed surface as part of a more complex scene of blood serum (top). Images made with AVS (http://www.avs.com/) and PMV25. (b) Top, a 2.4-nm electron tomogram slice of a human skin section showing part of the nuclear envelope (blue), cytoplasm (black background) and a desmosome (orange) at the boundary of the two cells. Using sub-tomogram averaging, the interaction of cadherin proteins can be resolved105, and they were used to calculate isosurfaces (below) into which the atomic-detail structure of C-cadherins106 has been fitted. Images created using MATLAB and Amira. Scale bars, 10 nm.
Visualization hardware
Most of this review has focused exclusively on software developments, tacitly assuming that computer and display hardware are adequate for all visualization tasks we require. In the early days of molecular graphics tools, hardware limitations were a key issue; display systems were often very expensive, and they relied on nonstandard hardware. Significant effort in software development was directed toward ameliorating hardware limitations. Today, although most molecular graphics tools run comfortably on standard desktop computers, many hardware issues remain, particularly for the more complex visualization tasks, such as the study of molecular motion and of large assemblies.
Stereo capabilities can greatly enhance molecular graphics and, although available for many years on expensive and specialized systems, stereo is only just now becoming available for desktop LCD screens.
For particularly large assemblies, computational speed is often still an issue—here, it is important to use a top-of-the-line graphics card, and also to use molecular graphics tools that can take advantage of hardware acceleration. Fortunately, most tools can, the principal exceptions being RasMol and Chime.
Immersive virtual reality. Visualizing large, complex and multi-scale macromolecular assemblies, especially combined with molecular motion, is not only challenging computationally, but ultimately may require display systems significantly better than current computer monitors can provide. Immersive virtual reality is very promising, enabling the user to virtually enter a microscopic world, flying through and interactively manipulating macromolecules. Experimental immersive environments—for example, CAVE86—have been in development for over 20 years, and concepts from this research have been used to enhance the user experience of several molecular graphics tools (for example, Yasara; Table 1). But such techniques have yet to find widespread use for molecular visualization—partially because of the still high cost and cumbersome nature of such systems, but perhaps also because the sense of immersion is not critical for interaction with the molecular world.
Today, however, some of the hardware components for virtual reality are becoming affordable and practical, such as head-mounted displays with head tracking, and a variety of haptic devices (mechanical input devices that are touch sensitive), such as the Wii controller, as well as devices such as wired gloves that can provide force feedback. These improvements are largely driven by the gaming market and are expected to continue rapidly. For most molecular graphics tools, minimal modifications should be required to allow them to work with such hardware, and some tools have been built with such support already in mind (for example, VMD26 and SRS 3D7). However fully exploiting the promise of virtual reality will require substantial further software development, particularly to the user interface layer.
Physical models. Today, molecular visualization relies almost exclusively on computer-generated images. Although physical wooden and wire molecular models played a critical early role in structural chemistry and biology, the advent of three-dimensional interactive computer graphics in the 1970's provided new and much improved utility in macromolecular structure determination and analysis. However, a more recent technology, computer autofabrication, or 'solid printing', initially developed for industrial rapid prototyping, is now being used to produce physical molecular models. Such models bring back the properties of real-object perception and manipulation that were lost when the model resided only in the computer. Over the past decade, the variety of such printers has increased steadily as the entry price has dropped to below $10,000 and printing services have sprouted to fill this new niche. Because accurate and complex tangible models can be produced automatically as computer 'printouts' of molecular geometrical representations, the barrier to custom production has disappeared. Physical models with functional parts have been autofabricated with analog physical constraints, affinities and/or structural behavior of the molecular system87 (Fig. 8).
Figure 8. Tangible models in research.
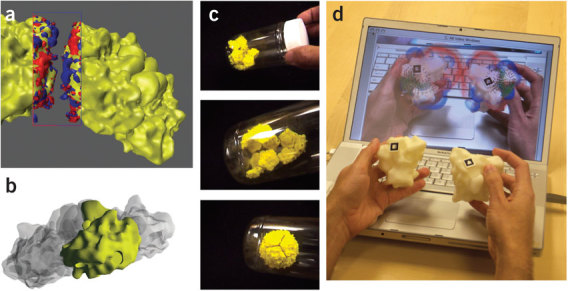
Tangible models were used to explore the modes of self-assembly of viral capsids88. (a) The electrostatic and charge complementarity is displayed using isosurfaces for the protein and electrostatic potential. (b) Affordances for placement of magnets were designed into the protein surface using constructive solid geometry methods. (c) Physical models were built and fitted with magnets. Twelve pentameric subunits then self-assemble when shaken for several minutes in a tube. Images created with PMV25. (d) An augmented-reality interface used to study molecular interactions of the enzyme superoxide dismutase. An inexpensive video camera (not in the picture) views the models, and embedded markers on the surface (small black squares) are used to determine the orientation of the model from the video image. Volume-rendered electrostatic potentials and small animated arrows for the electrostatic field vectors are then overlapped onto the video image, following the video image as the user manipulates the model.
Such models have begun to be used for structural research. As persistent objects they are convenient, accessible and naturally manipulable. They can be used as springboards to ideas and hypotheses88. Such characteristics also make physical models useful in multidisciplinary collaborations, helping structural experts communicate better with other colleagues. In addition, physical models lend themselves to teaching89. We are in the early stages of learning how to best use physical models in structural biology education and research, perhaps comparable to where computer graphics was in the 1970s. This is an ongoing area of research90,91,92.
Future perspectives
Methods for visualizing molecular structures are very mature. In the near future, we can expect more effective computational approaches for representing, analyzing and synthesizing ever-more-complex molecular systems. Increased collaboration with the graphic design community will also lead to the development of more effective and intelligible rendering approaches. However, we expect that most of the advances in molecular visualization will come in the areas of computer interfaces, user interaction and new ways to represent and visualize nonspatial information. These changes will help structures reach an even broader audience. Navigating a synthesis of structural data with image data82 and genomic22,93 and biological network information64 will require new methods that combine spatial and dynamic representations with statistical and high-dimensional abstract relationships. We also anticipate that collaborative community editing of structure-related data sources (for example, Proteopedia94) will change how scientists relate to structural data, and to each other. The fields of information visualization and visual analytics have developed over the past decade to address problems in making such complex data intelligible and navigable95,96.
Some of the drawbacks of immersive virtual reality may be overcome by the emerging technology of augmented reality (Fig. 8d), which provides inexpensive and accessible ways to interact in intuitive and perceptually rich ways with our computational models. Whatever direction new technologies will take us, the roles of macromolecular visualization in understanding, gaining insight and developing ideas will remain the same.
Note: Supplementary information is available on the Nature Methods website.
Supplementary information
Supplementary Figure 1 and Supplementary Table 1 (PDF 729 kb)
Acknowledgements
Thanks to M. Berynskyy and L. Biedermannova for assistance with Figure 6. This work was partly supported by the European Union Framework Programme 6 grant 'TAMAHUD' (LSHC-CT-2007-037472). R.C.W. gratefully acknowledges the support of the Klaus Tschira Foundation.
Accession codes
Accessions
Protein Data Bank
Competing interests
The authors declare no competing financial interests.
References
- 1.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 2.Goddard TD, Ferrin TE. Visualization software for molecular assemblies. Curr. Opin. Struct. Biol. 2007;17:587–595. doi: 10.1016/j.sbi.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tate J. Molecular visualization. Methods Biochem. Anal. 2003;44:135–158. [PubMed] [Google Scholar]
- 4.Olson AJ, Pique ME. Visualizing the future of molecular graphics. SAR QSAR Environ. Res. 1998;8:233–247. doi: 10.1080/10629369808039142. [DOI] [PubMed] [Google Scholar]
- 5.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richardson DC, Richardson JS. The kinemage: a tool for scientific communication. Protein Sci. 1992;1:3–9. doi: 10.1002/pro.5560010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Donoghue SI, Meyer JEW, Schafferhans A, Fries K. The SRS 3D module: integrating sequence, structure, and annotation data. Bioinformatics. 2004;20:2476–2478. doi: 10.1093/bioinformatics/bth260. [DOI] [PubMed] [Google Scholar]
- 8.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schafferhans A, Meyer JEW, O'Donoghue SI. The PSSH database of alignments between protein sequences and tertiary structures. Nucleic Acids Res. 2003;31:494–498. doi: 10.1093/nar/gkg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold K, et al. The Protein Model Portal. J. Struct. Funct. Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieger E, Nabuurs SB, Vriend G. Homology modeling. Methods Biochem. Anal. 2003;44:509–523. doi: 10.1002/0471721204.ch25. [DOI] [PubMed] [Google Scholar]
- 13.Cozzetto D, et al. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77(Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-David M, et al. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77(Suppl 9):50–65. doi: 10.1002/prot.22591. [DOI] [PubMed] [Google Scholar]
- 15.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–1871. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 16.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 18.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 19.Gille C, Frommel C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- 20.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 21.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35(Database issue):D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Procter JB, et al. Visualization of multiple alignments, phylogenies and gene family evolution. Nat. Methods. 2010;7:S16–S25. doi: 10.1038/nmeth.1434. [DOI] [PubMed] [Google Scholar]
- 23.Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 24.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 25.Sanner MF. A component-based software environment for visualizing large macromolecular assemblies. Structure. 2005;13:447–462. doi: 10.1016/j.str.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Prlic A, Down TA, Hubbard TJ. Adding some SPICE to DAS. Bioinformatics. 2005;21(Suppl. 2):ii40–ii41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huehne, R. & Suehnel, J. The Jena Library of Biological Macromolecules – JenaLib. Preprint at 〈http://precedings.nature.com/documents/3114/version/1/〉 (2009).
- 29.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gille C. Structural interpretation of mutations and SNPs using STRAP-NT. Protein Sci. 2006;15:208–210. doi: 10.1110/ps.051882006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabdoulline RR, Ulbrich S, Richter S, Wade RC. ProSAT2–protein structure annotation server. Nucleic Acids Res. 2006;34(Web Server issue):W79–W83. doi: 10.1093/nar/gkl216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bordner AJ, Gorin AA. Comprehensive inventory of protein complexes in the Protein Data Bank from consistent classification of interfaces. BMC Bioinformatics. 2008;9:234. doi: 10.1186/1471-2105-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 37.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171–2172. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 39.Sippl MJ. Boltzmann's principle, knowledge-based mean fields, and protein folding. An approach to the computational determination of protein structures. J. Comput. Aided Mol. Des. 1993;7:473–501. doi: 10.1007/BF02337562. [DOI] [PubMed] [Google Scholar]
- 40.Wesson L, Eisenberg D. Atomic solvation parameters applied to molecular dynamics of proteins in solution. Protein Sci. 1992;1:227–235. doi: 10.1002/pro.5560010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanner MF, Olson AJ, Spehner J-C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3<305::AID-BIP4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 42.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 43.Hunter WN. Structure-based ligand design and the promise held for antiprotozoan drug discovery. J. Biol. Chem. 2009;284:11749–11753. doi: 10.1074/jbc.R800072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris R, Olson AJ, Goodsell DS. Automated prediction of ligand-binding sites in proteins. Proteins. 2008;70:1506–1517. doi: 10.1002/prot.21645. [DOI] [PubMed] [Google Scholar]
- 45.Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 46.Campbell SJ, Gold ND, Jackson RM, Westhead DR. Ligand binding: functional site location, similarity and docking. Curr. Opin. Struct. Biol. 2003;13:389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 47.Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 48.Morgan DH, Kristensen DM, Mittelman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33(Web Server issue):W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinoshita K, Murakami Y, Nakamura H. eF-seek: prediction of the functional sites of proteins by searching for similar electrostatic potential and molecular surface shape. Nucleic Acids Res. 2007;35(Web Server issue):W398–W402. doi: 10.1093/nar/gkm351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolber, G. & Kosara, R. Pharmacophores from macromolecular complexes with LigandScout. in Pharmacophores and Pharmacophore Searches (ed. Langer, T. & Hoffmann, R.D.) vol. 32, 131–150 (Wiley-VCH, Weinheim, Germany, 2006).
- 52.Vulpetti A, Pevarello P. An analysis of the binding modes of ATP-competitive CDK2 inhibitors as revealed by X-ray structures of protein-inhibitor complexes. Curr. Med. Chem. Anticancer Agents. 2005;5:561–573. doi: 10.2174/1568011054866928. [DOI] [PubMed] [Google Scholar]
- 53.Zou J, et al. Towards more accurate pharmacophore modeling: Multicomplex-based comprehensive pharmacophore map and most-frequent-feature pharmacophore model of CDK2. J. Mol. Graph. Model. 2008;27:430–438. doi: 10.1016/j.jmgm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 55.Morris GM, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoete V, Grosdidier A, Michielin O. Docking, virtual high throughput screening and in silico fragment-based drug design. J. Cell. Mol. Med. 2009;13:238–248. doi: 10.1111/j.1582-4934.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karkola S, Alho-Richmond S, Wahala K. Pharmacophore modelling of 17β-HSD1 enzyme based on active inhibitors and enzyme structure. Mol. Cell. Endocrinol. 2009;301:225–228. doi: 10.1016/j.mce.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 58.Hendlich M. Databases for protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1178–1182. doi: 10.1107/s0907444998007124. [DOI] [PubMed] [Google Scholar]
- 59.Gunther J, Bergner A, Hendlich M, Klebe G. Utilising structural knowledge in drug design strategies: applications using Relibase. J. Mol. Biol. 2003;326:621–636. doi: 10.1016/s0022-2836(02)01409-2. [DOI] [PubMed] [Google Scholar]
- 60.Michalsky E, Dunkel M, Goede A, Preissner R. SuperLigands—a database of ligand structures derived from the Protein Data Bank. BMC Bioinformatics. 2005;6:122. doi: 10.1186/1471-2105-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie L, Li J, Bourne PE. Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLOS Comput. Biol. 2009;5:e1000387. doi: 10.1371/journal.pcbi.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinnings SL, et al. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLOS Comput. Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gehlenborg N, et al. Visualization of omics data for systems biology. Nat. Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 65.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 66.Stierand K, Maass PC, Rarey M. Molecular complexes at a glance: automated generation of two-dimensional complex diagrams. Bioinformatics. 2006;22:1710–1716. doi: 10.1093/bioinformatics/btl150. [DOI] [PubMed] [Google Scholar]
- 67.Clark AM, Labute P. 2D depiction of protein-ligand complexes. J. Chem. Inf. Model. 2007;47:1933–1944. doi: 10.1021/ci7001473. [DOI] [PubMed] [Google Scholar]
- 68.Berman HM, et al. The nucleic acid database: a comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 70.Ringe D, Petsko GA. The 'glass transition' in protein dynamics: what it is, why it occurs, and how to exploit it. Biophys. Chem. 2003;105:667–680. doi: 10.1016/s0301-4622(03)00096-6. [DOI] [PubMed] [Google Scholar]
- 71.Flores S, et al. The Database of Macromolecular Motions: new features added at the decade mark. Nucleic Acids Res. 2006;34(Database issue):D296–D301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maiti R, Van Domselaar GH, Wishart DS. MovieMaker: a web server for rapid rendering of protein motions and interactions. Nucleic Acids Res. 2005;33(Web Server issue):W358–W362. doi: 10.1093/nar/gki485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chennubhotla C, Rader AJ, Yang LW, Bahar I. Elastic network models for understanding biomolecular machinery: from enzymes to supramolecular assemblies. Phys. Biol. 2005;2:S173–S180. doi: 10.1088/1478-3975/2/4/S12. [DOI] [PubMed] [Google Scholar]
- 74.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34(Web Server issue):W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eyal E, Yang LW, Bahar I. Anisotropic network model: systematic evaluation and a new web interface. Bioinformatics. 2006;22:2619–2627. doi: 10.1093/bioinformatics/btl448. [DOI] [PubMed] [Google Scholar]
- 76.Seeliger D, De Groot BL. tCONCOORD-GUI: visually supported conformational sampling of bioactive molecules. J. Comput. Chem. 2009;30:1160–1166. doi: 10.1002/jcc.21127. [DOI] [PubMed] [Google Scholar]
- 77.Thorpe MF, Lei M, Rader AJ, Jacobs DJ, Kuhn LA. Protein flexibility and dynamics using constraint theory. J. Mol. Graph. Model. 2001;19:60–69. doi: 10.1016/s1093-3263(00)00122-4. [DOI] [PubMed] [Google Scholar]
- 78.Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y, Stoffler D, Sanner M. Hierarchical and multi-resolution representation of protein flexibility. Bioinformatics. 2006;22:2768–2774. doi: 10.1093/bioinformatics/btl481. [DOI] [PubMed] [Google Scholar]
- 80.Finocchiaro G, Wang T, Hoffmann R, Gonzalez A, Wade RC. DSMM: a database of simulated molecular motions. Nucleic Acids Res. 2003;31:456–457. doi: 10.1093/nar/gkg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kehl C, Simms AM, Toofanny RD, Daggett V. Dynameomics: a multi-dimensional analysis-optimized database for dynamic protein data. Protein Eng. Des. Sel. 2008;21:379–386. doi: 10.1093/protein/gzn015. [DOI] [PubMed] [Google Scholar]
- 82.Walter T, et al. Visualization of image data from cells to organisms. Nat. Methods. 2010;7:S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodsell DS. Visual methods from atoms to cells. Structure. 2005;13:347–354. doi: 10.1016/j.str.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 84.Goodsell DS. Making the step from chemistry to biology and back. Nat. Chem. Biol. 2007;3:681–684. doi: 10.1038/nchembio1107-681. [DOI] [PubMed] [Google Scholar]
- 85.McGill G. Molecular movies. coming to a lecture near you. Cell. 2008;133:1127–1132. doi: 10.1016/j.cell.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 86.Cruz-Neira C, Sandin DJ, DeFanti TA, Kenyon RV, Hart JC. The CAVE: audio visual experience automatic virtual environment. Commun. ACM. 1992;35:64–72. [Google Scholar]
- 87.Gillet A, Sanner MF, Stoffler D, Olson AJ. Tangible interfaces for structural molecular biology. Structure. 2005;13:483–491. doi: 10.1016/j.str.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Olson AJ, Hu YH, Keinan E. Chemical mimicry of viral capsid self-assembly. Proc. Natl. Acad. Sci. USA. 2007;104:20731–20736. doi: 10.1073/pnas.0709489104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Herman T, et al. Tactile teaching: exploring protein structure/function using physical models. Biochem. Mol. Biol. Educ. 2006;34:247–254. doi: 10.1002/bmb.2006.494034042649. [DOI] [PubMed] [Google Scholar]
- 90.Creem SH, Proffitt DR. Grasping objects by their handles: a necessary interaction between cognition and action. J. Exp. Psychol. Hum. Percept. Perform. 2001;27:218–228. doi: 10.1037//0096-1523.27.1.218. [DOI] [PubMed] [Google Scholar]
- 91.Kozma R. The material features of multiple representations and their cognitive and social affordances for science understanding. Learning and Instruction. 2003;13:205–226. [Google Scholar]
- 92.Zhang J, Patel VL. Distributed cognition, representation, and affordance. Pragmatics & Cognition. 2006;14:333–341. [Google Scholar]
- 93.Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T. Visualizing genomes: techniques and challenges. Nat. Methods. 2009;7:S5–S15. doi: 10.1038/nmeth.1422. [DOI] [PubMed] [Google Scholar]
- 94.Hodis E, et al. Proteopedia—a scientific 'wiki' bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol. 2008;9:R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cook K, Earnshaw R, Stasko J. Discovering the unexpected. IEEE Comput. Graph. Appl. 2007;27:15–19. doi: 10.1109/mcg.2007.126. [DOI] [PubMed] [Google Scholar]
- 96.Kerren, A., Stasko, J.T., Fekete, J.D. & North, C. Information Visualization (Springer, New York, 2008).
- 97.Schindler T, et al. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 98.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 99.Lam PY, et al. Rational design of potent, bioavailable, nonpeptide cyclic ureas as HIV protease inhibitors. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- 100.Gangloff M, et al. Crystal structure of a mutant hERα ligand-binding domain reveals key structural features for the mechanism of partial agonism. J. Biol. Chem. 2001;276:15059–15065. doi: 10.1074/jbc.M009870200. [DOI] [PubMed] [Google Scholar]
- 101.Brzozowski AM, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 102.Robertson MP, et al. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3:e5. doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petrek M, et al. CAVER: a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Spaar A, Dammer C, Gabdoulline RR, Wade RC, Helms V. Diffusional encounter of barnase and barstar. Biophys. J. 2006;90:1913–1924. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- 106.Boggon TJ, et al. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- 107.Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mal TK, Matthews SJ, Kovacs H, Campbell ID, Boyd J. Some NMR experiments and a structure determination employing a {15N,2H} enriched protein. J. Biomol. NMR. 1998;12:259–276. doi: 10.1023/a:1008238009056. [DOI] [PubMed] [Google Scholar]
- 109.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 110.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 111.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rhodes, G. Crystallography Made Crystal Clear: A Guide for Users of Macromolecular Models 3rd edn. (Academic Press, 2006).
- 113.Glykos NM. On the application of molecular-dynamics simulations to validate thermal parameters and to optimize TLS-group selection for macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2007;63:705–713. doi: 10.1107/S0907444907014928. [DOI] [PubMed] [Google Scholar]
- 114.Brünger AT. The free R factor: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 115.Kleywegt GJ, et al. The Uppsala electron-density server. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 116.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 117.Jones TA. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 118.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 119.Levin EJ, Kondrashov DA, Wesenberg GE, Phillips GN., Jr. Ensemble refinement of protein crystal structures: validation and application. Structure. 2007;15:1040–1052. doi: 10.1016/j.str.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–306. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- 121.Nederveen AJ, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 122.Selenko P, Wagner G. Looking into live cells with in-cell NMR spectroscopy. J. Struct. Biol. 2007;158:244–253. doi: 10.1016/j.jsb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 123.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tugarinov V, Choy W-Y, Orekhov VY, Kay LE. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc. Natl. Acad. Sci. USA. 2005;102:622–627. doi: 10.1073/pnas.0407792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Markwick PR, Malliavin T, Nilges M. Structural biology by NMR: structure, dynamics, and interactions. PLOS Comput. Biol. 2008;4:e1000168. doi: 10.1371/journal.pcbi.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang L, Sigworth FJ. Cryo-EM and single particles. Physiology (Bethesda) 2006;21:13–18. doi: 10.1152/physiol.00045.2005. [DOI] [PubMed] [Google Scholar]
- 127.Frank, J. Three-dimensional Electron Microscopy of Macromolecular Assemblies 2nd edn. (Oxford University Press, 2006).
- 128.Frank, J. ed. Electron Tomography 2nd edn. (Springer, 2006).
- 129.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 and Supplementary Table 1 (PDF 729 kb)


