Key Points
Autophagy has a role in degrading intracellular bacteria, parasites and viruses.
Intracellular pathogens have strategies to subvert or evade the autophagy pathway.
Autophagy has a role in innate immune signalling by delivering viral nucleic acids to endosomal Toll-like receptors.
Autophagy is active in antigen-presenting cells and is involved in MHC class II presentation of certain endogenous antigens.
Immune signalling molecules that are important in the control of viruses, parasites and intracellular bacteria activate autophagy.
Autophagy is involved in regulating T-cell homeostasis.
Autophagy genes are strongly linked to susceptibility to Crohn's disease, a form of inflammatory bowel disease.
Autophagy — a cellular process for recycling, remodelling or disposing of unwanted cytoplasmic constituents — is emerging as an important pathway in innate and adaptive immunity. This Review describes its role in pathogen defence, antigen processing and presentation, T-cell homeostasis and disease.
Abstract
Cells digest portions of their interiors in a process known as autophagy to recycle nutrients, remodel and dispose of unwanted cytoplasmic constituents. This ancient pathway, conserved from yeast to humans, is now emerging as a central player in the immunological control of bacterial, parasitic and viral infections. The process of autophagy may degrade intracellular pathogens, deliver endogenous antigens to MHC-class-II-loading compartments, direct viral nucleic acids to Toll-like receptors and regulate T-cell homeostasis. This Review describes the mechanisms of autophagy and highlights recent advances relevant to the role of autophagy in innate and adaptive immunity.
Main
Our armamentarium for fighting intracellular pathogens includes multiple facets of the innate and adaptive immune response. In the past few decades, we have witnessed an explosion in our understanding of the mechanisms underlying antigen presentation, immune recognition of infected cells, cellular sensing of pathogens, and signalling pathways that induce antimicrobial states in infected cells. Surprisingly, however, microbiologists and immunologists have long overlooked one of the greatest challenges that the immune system faces in dealing with intracellular pathogens — that is, the problem of how to dispose of a microorganism without disposing of the entire infected cell. A seemingly simple strategy to tackle this challenge is now beginning to unveil itself. Mammalian cells use an evolutionarily conserved lysosomal degradation pathway known as autophagy to selectively dispose of intracellular pathogens.
Autophagy is a fundamental cellular homeostatic process that enables cells to clean up, in a regulated manner, portions of their own cytoplasm and degrade their constituents1. This primordial function is preserved in all eukaryotic organisms, from yeast to humans. During autophagy, an isolation membrane wraps around portions of the cytoplasm to form a double-membrane organelle known as the autophagosome. The engulfed cytoplasmic material in an autophagosome is degraded after fusion of the autophagosome with late endosomes or lysosomes (Fig. 1).
Figure 1. Cellular events in autophagy.

The cellular events during digestion of self constituents or intracellular pathogens follow three distinct stages: initiation (formation of the phagophore), elongation (growth and closure) and maturation of a double membrane autophagosome into an autolysosome. a | Autophagy sequesters and removes cellular constituents from the cytosol, including surplus or damaged organelles from the cytosol. b | Autophagy can eliminate bacteria (free in the cytosol or inside a phagosome), viruses and protozoan parasites in a manner similar to the elimination of self constituents.
The autophagy pathway has many physiological roles, and is often used to remove damaged or surplus organelles. It is also used by cells to turn over long-lived proteins and other macromolecules, either to supply nutrients for essential anabolic needs under conditions of nutrient deprivation or growth factor withdrawal, or to rid cells of potentially toxic aggregate-prone proteins2. The broad spectrum of autophagy functions is intricately linked to a wide range of health and disease states3,4. For example, autophagy is involved in the control of development, tissue homeostasis and the lifespan of an organism; the suppression of tumour development; and the prevention of neurodegeneration. Aberrant regulation of autophagy has been mechanistically linked to cancer, Huntington's disease, Parkinson's disease, myodegeneration and cardiomyopathy (Box 1).
Autophagy also functions in diverse aspects of immunity5,6,7. Although the term autophagy means 'to digest oneself', it is now clear that the autophagy pathway also eliminates intracellular pathogens, including viruses, parasites and bacteria (a process sometimes referred to as xenophagy)7,8,9,10. The autophagic sequestration of viral components can also fuel MHC class II presentation of endogenous antigens11 and the production of type I interferons (IFNs) in response to Toll-like receptor 7 (TLR7) signalling12. Furthermore, autophagy may directly affect T-cell homeostasis13,14. More speculatively, autophagy may have a role in the prevention of autoimmunity and inflammatory disorders15,16,17,18,19,20. Here, we provide an introduction to the molecular mechanisms of autophagy, discuss the major recent advances in autophagy and immunity, and highlight important unanswered questions in the field.
Autophagy: a lysosomal degradation pathway
There are several morphologically and functionally distinct forms of autophagy, including macroautophagy (herein referred to as autophagy), microautophagy, chaperone-mediated autophagy and others that involve the selective degradation of specific organelles (such as peroxisomes, mitochondria and the endoplasmic reticulum). During the initiation of autophagy, a damaged organelle or a portion of cytosol is sequestered in a structure known as the isolation membrane or phagophore (Fig. 1a). The phagophore then becomes enlarged during the elongation stages by the addition of new membrane — the origin of which is still unclear. The phagophore seals to form an autophagosome, an organelle that is distinguished from the conventional phagosome by the presence of a double delimiting membrane (two lipid bilayers) and intra-lumenal cytoplasmic content. During maturation, autophagosomes fuse with lysosomes to form autolysosomes in which the captured material is degraded. The capture of intracellular pathogens is thought to follow a similar path (Fig. 1b), although the sequestration of microorganisms during autophagy has not been studied as extensively as that of cellular contents.
Most cells undergo some level of autophagy while adjusting their biomass, removing protein aggregates or eliminating damaged organelles (such as mitochondria). The classical signalling pathways that regulate autophagy have been reviewed extensively elsewhere21,22, whereas signals of particular relevance to immunity are discussed later. Key players in autophagy regulation are the serine/threonine kinase mammalian target of rapamycin (mTOR; also known as FRAP1) and the class I and class III phosphoinositide 3-kinases (PI3Ks). Two well-characterized stimuli that induce autophagy are amino-acid starvation and growth-factor withdrawal2. In response to growth-factor stimulation, class I PI3Ks generate phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3) on the plasma membrane by phosphorylating phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) and in turn PtdIns(3,4,5)P3 activates mTOR, thereby repressing autophagy. The class III PI3K VPS34 (also known as PIK3C3) generates phosphatidylinositol-3-phosphate (PtdIns3P) by phosphorylating PtdIns on endomembranes and acts at several steps along the signalling pathway associated with autophagy (Fig. 2). When amino acids are plentiful, VPS34 contributes to mTOR activation, thereby repressing autophagy23,24. By contrast, the initiation stages of autophagy depend on VPS34 in a complex with the autophagy-associated protein beclin 1 (also known as ATG6)25. The beclin-1–VPS34 autophagy complex can be activated by the beclin-1-interacting partners, UVRAG (UV radiation resistance associated gene)26 and AMBRA1 (activating molecule in beclin-1-regulated autophagy)27, and inhibited by another beclin-1-interacting partner, BCL-2 (B-cell lymphoma 2)28. Pharmacological induction of autophagy can be achieved using rapamycin, a drug that inhibits mTOR activity29, whereas pharmacological inhibition of autophagy can be achieved using 3-methyladenine, a drug that inhibits class III PI3K activity30. It is noteworthy, however, that as mTOR and class III PI3Ks have pleiotropic cellular functions, both of these drugs are potent, but not specific, regulators of autophagy. Knockdown of expression of autophagy-related genes by small interfering RNA (siRNA) is therefore an indispensable tool for selectively probing the role of autophagy in defined biological processes.
Figure 2. Molecular events in autophagy.
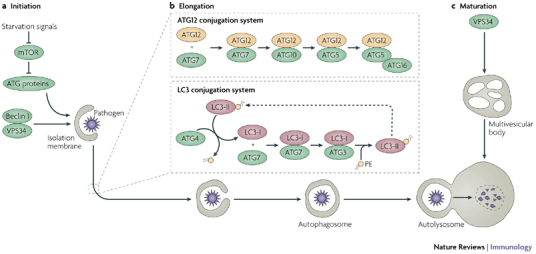
Autophagy is regulated by a set of autophagy-related proteins (ATG proteins). In the absence of amino acids or in response to other stimuli, ATG1 and a complex of the class III PI3K (phosphoinositide 3-kinase) VPS34 and beclin 1 lead to the activation of downstream ATG factors that are involved in the initiation (a), elongation (b) and maturation (c) of autophagy. a | In amino-acid-rich conditions, VPS34 contributes to mTOR (mammalian target of rapamycin) activation and inhibition of ATG1 and autophagy. The sources of membrane for autophagosome initiation and elongation may include those containing the only known membrane integral ATG protein ATG9, redistributing between a resting location to autophagosomes in an ATG1- and PI3K-dependent manner. ATG9 redistribution may depend on ATG18, which binds phosphatidylinositol-3-phosphate (PtdIns3P). b | The elongation and shape of the autophagosome are controlled by two protein (and lipid) conjugation systems, similar to the ubiquitylation systems: the ATG12 and LC3 (also known as ATG8)–phosphatidylethanolamine (PE) conjugation pathways, which include E1-activating and E2-conjugating enzymes. ATG12 is initially conjugated to ATG7 (an E1-activating enzyme) and then is transferred to the E2-like conjugating enzyme ATG10. This intermediate presents ATG12 for conjugation to an ATG5 lysine residue. The ATG5–ATG12 conjugate, stabilized non-covalently by ATG16, triggers oligomerization on the outside membrane of the growing autophagosome, and enhances LC3 carboxy-terminal lipidation through the LC3 conjugation system. Upon autophagosome closure, ATG5–ATG12–ATG16 and LC3 (delipidated by ATG4) are recycled. c | LC3 associated with the lumenal membrane remains trapped in the autophagosome and is degraded during maturation into the autolysosome, which involves fusion of autophagosomes with late endosomes, including endosomal multivesicular bodies and lysosomal organelles, and dissolution of the internal membrane. VPS34 has a role in the formation of late endosomal multivesicular bodies and lysosomal organelles contributing to the maturation stages of autophagy.
The execution of autophagy is mediated by evolutionarily conserved proteins known as the autophagy-related (ATG) proteins31. The molecular mechanisms by which this group of proteins mediates autophagy has been the subject of recent reviews3,32,33,34 and is depicted schematically in Fig. 2. Autophagosomal membrane formation and expansion is facilitated by two specialized protein conjugation systems (Fig. 2), the ATG8 (known as LC3 in mammals) system and the ATG12 system. This results in the carboxy-terminal conjugation of LC3 to the lipid phosphatidylethanolamine (PE) and the localization of lipidated LC3 (LC3-II; also known as ATG8-PE) to autophagic membranes. Membrane-associated LC3-II has become the most universally used marker for the detection of membranes that are undergoing autophagy. Interestingly, LC3 interacts with p62 (sequestosome-1; also known as SQSTM1), a protein that recognizes polyubiquitylated protein aggregates (that are too big to be disposed of by the proteasome) and may deliver them to autophagosomes for degradation35. Although only studied so far in the context of targeting polyubiquitylated protein aggregates, an interesting question is whether LC3 may interact with p62 or other proteins to target microorganisms for degradation or antigens for presentation through the MHC class II pathway.
Autophagy in bacterial and parasitic defence
The first indication that intracellular bacteria may be degraded by an autophagy-like pathway emerged more than two decades ago in morphological studies of polymorphonuclear cells infected with Rickettsia conorii36. However, before the discovery of the components of the autophagic machinery, this was difficult to prove. There was a lack of markers to unequivocally identify autophagosomes; it was difficult to follow the dynamic fate of intracellular bacteria; and it was difficult to determine the significance of bacterial association with autophagosomal membranes in host defence. Indeed, several studies proposed that autophagy was a 'microorganism-friendly process' that supported the intracellular survival of certain pathogens37. However, with new tools available to label autophagosomes and to inactivate the autophagy pathway in infected cells, we now know that autophagy is an important host mechanism for the removal of intracellular bacteria and protozoans, in keeping with its primary function as a cytoplasmic clean-up process38 (Fig. 3). In parallel, many pathogens have evolved strategies to protect themselves against autophagy or to harness components of the autophagy pathway for their own benefit, although, in general, the molecular details of such strategies are not well defined37.
Figure 3. Autophagy eliminates intracellular microorganisms.
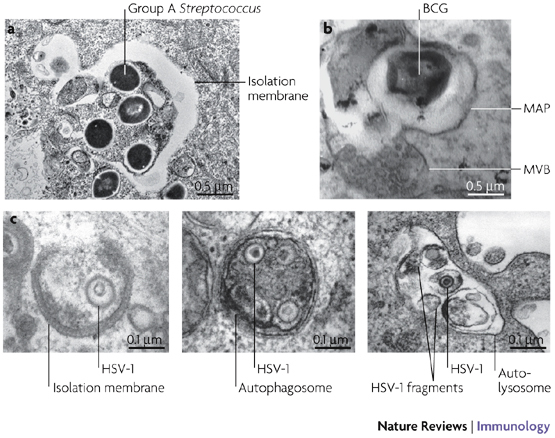
a | Group A Streptococci captured within an autophagosome. Image kindly provided by Tamotsu Yoshimori, Osaka University, Japan. b | Mycobacterium bovis bacillus Calmette–Guérin (BCG) present in a mycobacterial autophagosome (MAP) that is fusing with a multivesicular body (MVB). Image reproduced with permission from Ref. 42 © (2004) Cell Press. c | Herpes simplex virus type I (HSV-1) virion(s) in the process of being surrounded by an isolation membrane (left panel), engulfed inside an autophagosome (middle panel) or degraded inside an autolysosome (right panel). Image reproduced with permission from Ref. 77 © (2006) Landes Bioscience.
One of the initial indications that autophagy may have a role in immunity against intracellular bacteria was provided by observations regarding the role of PtdIns3P in innate immunity. It was known that PtdIns3P participated in phagolysosomal biogenesis and microbial clearance upon macrophage phagocytosis of microorganisms39,40. This link prompted investigations into a potential role of increased PtdIns3P production41 and, by extension, autophagy42 — as PtdIns3P is involved in the initiation of autophagy downstream of mTOR — in eliminating certain intracellular pathogens, such as Mycobacterium tuberculosis, that block normal phagolysosome biogenesis. Gutierrez et al. showed that the mycobacterial-imposed block in phagolysosomal maturation can be overcome by activating cellular autophagy, either through starvation or inhibition of mTOR42. Nearly simultaneously, other studies determined that autophagy can capture intracellular bacteria that lyse the phagosome and escape into the cytosol (such as Shigella spp.)43 or extracellular bacteria that manage to invade the host cytoplasm (such as Group A Streptococcus)44. Other studies have confirmed these initial findings and extended the list of intracellular bacteria and parasites targeted by autophagy to include Listeria monocytogenes, Salmonella enterica, Francisella tularensis and Toxoplasma gondii8,9,45,46,47,48,49.
There are some aspects of autophagy that may be particular to the control of intracellular bacteria. First, the size of autophagosomes that engulf intracellular bacteria tends to be considerably larger than 'typical' autophagosomes (that is, those clearing up cytoplasmic constituents)44, raising the possibility that these autophagosomes may have a different biogenesis than typical autophagosomes. Nevertheless, there are similarities in size between LC3-positive structures that clear large protein aggregates50 and the LC3-positive bacteria-containing autophagosomes44. This suggests that the formation of these large autophagosomes, if distinct from classical autophagy, is not reserved exclusively for microorganisms.
Another area of debate is whether autophagosomes can only target intracellular bacteria that reside in the cytosol or whether they can sequester pathogens that reside in membranous or intravacuolar compartments42,44,48,51. As autophagy is designed to engulf membranous organelles4,38, the autophagic enclosure of a pathogen within a membranous vacuole, such as a phagosome, should not represent an obstacle to autophagic elimination. This question has been answered experimentally using the parasite T. gondii. In T. gondii-infected cells, two different but equally potent pathways of autophagic elimination of the pathogen operate: one that disrupts the parasitophorous vacuole that harbours the protozoan9 and the other that does not require disruption of the parasitophorous vacuole8. Thus, encasement of the pathogen in a vacuole does not seem to represent a physical barrier to autophagic capture and elimination. It has been proposed that damage to the vacuolar membrane containing the organism may precede its autophagic sequestration9, but it is not yet known whether such damage or a molecular modification of the target membrane leads to autophagic uptake.
Another pertinent question is how pathogens that are free in the cytosol are recognized by the autophagic machinery. One possibility is that microbial proteins may be marked for autophagy by modifications, such as ubiquitylation, that are already known to modify bacterial products52, or by other yet to be identified molecular tags. Alternatively (or in addition), pattern-recognition receptors, such as TLRs, NOD-like receptors and RIG-I (retinoic-acid-inducible gene I)-like helicases, may recognize pathogen-associated molecular patterns (PAMPs) to stimulate autophagy. A related possibility is that exposure of certain epitopes at the surface of a microorganism may be involved in microbial targeting to autophagosomes. For example, exposure of a Shigella epitope that is normally unexposed (due to masking by another Shigella-encoded protein) leads to autophagic bacterial capture in cells infected with mutants lacking the epitope-masking protein43. Another possible signal that links microbial presence and autophagy could be the generation of reactive oxygen intermediates, as these are often associated with pathogen recognition by host cells and are also known to induce autophagy53,54. Future research into the mechanisms underlying the induction of autophagy by microorganisms and their targeting to autophagosomes is likely not only to uncover the specifics of how the autophagic machinery recognizes foreign material in the cytoplasm but also may shed light on how the autophagic machinery detects the cell's own damaged organelles, aggregated proteins and other cytoplasmic targets for lysosomal degradation.
Autophagy in viral defence
The sequestration of intracellular pathogens during autophagy is not limited to bacteria and parasites. Autophagy can also capture virions that are newly assembled inside their host cells. In neurons and fibroblasts infected with herpes simplex virus — a DNA virus that replicates in the nucleus — viral nucleocapsids are engulfed by autophagosomes as they egress out of the nucleus into the cytoplasm55 (Fig. 3). It is not yet known whether autophagy also targets viruses during cell entry, but this seems probable based on extrapolations from findings in bacterial systems. If true, this could explain why the ratio of virus particles per cell often needs to be very high for cells to become productively infected by viruses. The possibilities for how viruses are targeted to autophagosomes are conceptually similar to those discussed previously for intracellular pathogens in general. However, the specific molecules involved are likely to differ given the unique pathogen-specific receptors and PAMPs that recognize viral versus bacterial components.
Similar to studies in bacteria, several investigators have also proposed that autophagy may be a 'virus-friendly' pathway, as it can provide viruses with a source of intracellular membrane that serves as a scaffold for viral RNA replication complexes, an event that is necessary for efficient cytoplasmic replication of certain viruses37. So, similar to bacteria, viruses may have also found ways to harness the autophagic machinery for their own replicative benefit. The best-studied examples are the mammalian picornavirus poliovirus and the murine coronavirus mouse hepatitis virus56,57. In infections with either of these viruses, knockdown of expression of autophagy-related genes reduces viral yields. Interestingly, however, a Drosophila melanogaster picornavirus also replicates in association with double-membrane vacuoles (which are morphologically similar to the 'autophagy-like' double-membrane vacuoles associated with mammalian picornavirus replication) but autophagy genes are not required for its normal replication58. This suggests that the requirement for the autophagic machinery in the generation of double-membrane vacuoles that support picornavirus replication may be cell-type specific or restricted to certain phylogenetic hosts. Furthermore, the replication of vaccinia virus, a DNA virus that replicates in the cytoplasm in association with double-membrane vacuoles, also does not require the autophagic machinery59. So, it is unclear to what extent viruses exploit the autophagic machinery to obtain membrane anchors for their cytoplasmic replication. Even in circumstances in which components of the autophagic machinery are necessary for viral replication, it is unknown whether the viruses are truly subverting the host autophagic pathway or merely using a partially overlapping pathway that is involved in membrane trafficking and rearrangement.
Although there is perhaps less direct in vitro evidence for viruses, compared with bacteria, that autophagy functions in pathogen elimination, studies with viruses have provided the first in vivo evidence for a role of autophagy in immunity. In two different phylogenetic kingdoms, genetic manipulation of autophagy-related genes has been shown to have striking effects on viral diseases. In plants, RNAi-mediated silencing of several different autophagy-related genes increases local replication of tobacco mosaic virus and results in the uncontrolled spread of programmed cell death beyond infected plant cells60. In mice, enforced neuronal expression of the autophagy-associated protein beclin 1 reduces alphavirus replication and alphavirus-induced neuronal apoptosis and protects mice from lethal virus-induced encephalitis61. Together, these studies suggest that autophagy functions in antiviral immunity in vivo not only by restricting viral replication but also by restricting pathogen-induced cell death.
The mechanisms underlying these protective functions of autophagy are not yet defined. In principle, autophagy may function in the direct elimination of viruses (as shown in vitro), in the breakdown of host factors required for viral replication or the inhibition of innate immune signalling, and in the promotion of cell survival either by maintaining bioenergetics in virally infected cells or by removing toxic self or viral components. As discussed in more detail later, autophagy may also promote adaptive immunity by the endogenous presentation of certain viral antigens through the MHC class II pathway. Moreover, the role of autophagy in innate antiviral immunity may not be confined to direct pathogen elimination. Lee et al. recently found that the autophagic machinery can deliver viral nucleic acids to endosomal TLRs in plasmacytoid dendritic cells in vitro, resulting in type I IFN production12. Perhaps autophagy has a similar role in type I IFN production during viral infections in vivo and in other cell types infected with viruses.
Another major line of evidence that autophagy is important in antiviral immunity in vivo is the recent discovery that, to be pathogenic, viruses may need to successfully counter autophagy. An essential herpes simplex virus neurovirulence protein, ICP34.5, confers pathogenicity by binding to beclin 1 and by antagonizing the host autophagy response62. Although this is the first example of a viral virulence factor directly targeting the autophagic machinery to elicit disease, it seems probable that viral evasion of autophagy will prove to be a more general strategy that viruses use to evade host antiviral defence. Other viruses encode proteins that inhibit the autophagy function of beclin 1, including BCL-2-like proteins encoded by the oncogenic gammaherpesviruses28. In addition, numerous viruses inhibit the PKR (IFN-inducible double-stranded-RNA-dependent protein kinase) antiviral signalling pathway that is required for the induction of autophagy in virally infected cells or activate the autophagy-inhibitory class I PI3K–AKT–mTOR signalling pathway63. The multiplicity of mechanisms that diverse viruses have to turn off autophagy highlights a probable fundamental role for autophagy in antiviral immunity.
Autophagy and viral recognition by plasmacytoid dendritic cells. Recent evidence indicates that autophagy functions in delivering viral nucleic acids to the innate immune system12. A subset of receptors of the TLR family sense viral nucleic acids in the lumen of endosomes64. However, viral nucleic acids are most often released directly into the cytoplasm after fusion of a viral envelope with the endosomal membrane or penetration of cellular membranes by a viral capsid. This poses a topological challenge for the efficient detection of viral nucleic acids by endosomal TLRs. This challenge may be met, at least in part, by using the autophagic pathway for the delivery of cytoplasmic viral nucleic acids to endosomal TLRs, which would then lead to the induction of type-I-IFN-dependent innate immune responses (Fig. 4). Recently, Lee et al. demonstrated that, for two different single-stranded RNA viral infections (Sendai virus and vesicular stomatitis virus), robust IFN production by mouse plasmacytoid dendritic cells required live, not UV-inactivated, virus infection, TLR7 expression and the autophagy gene Atg5(Ref. 12).
Figure 4. Functions of autophagy in innate and adaptive immunity during infection with intracellular pathogens.

a | Intracellular pathogens (bacteria, parasites and viruses) that are either free inside the cytosol, inside phagosomes or inside pathogen-containing vacuoles are surrounded by isolation membranes, engulfed into autophagosomes, which fuse with lysosomes, and then degraded inside autolysosomes. b | Viral nucleic acids are transferred by autophagy from the cytoplasm to intracellular compartments containing Toll-like receptor 7 (TLR7), which signals the induction of type I interferon (IFN) production. c | Viral antigens (and potentially other endogenously synthesized microbial antigens and self antigens) are engulfed into autophagosomes that fuse with MHC-class-II-containing late endosomes (MIICs), and then loaded onto MHC class II molecules for presentation to CD4+ T cells. Cytosolic antigens that contain a KFERQ recognition motif may also be directly imported into MIICs by chaperone-mediated autophagy. CLIP, class II-associated invariant chain peptide.
So, similar to the use of autophagy for endogenous antigen presentation in adaptive immunity (described later), autophagy may be used for the delivery of endogenous viral nucleic acids to their cognate innate immunity detectors. The precise details of how the autophagy machinery senses viral RNA and targets it to endosomal TLR7 remain to be elucidated. Moreover, it is still unclear how the nucleic acids of DNA viruses, and even other RNA viruses, are targeted to endosomal TLRs in plasmacytoid dendritic cells, as viral replication is not required for type I IFN production in plasmacytoid dendritic cells infected with herpesviruses (DNA viruses) or influenza virus (an RNA virus)12,65. Another unexplored question is whether there is any molecular interplay between the autophagy pathway and type I IFN signalling mediated by cytoplasmic RNA helicases that function as receptors for cytoplasmic double-stranded RNA produced during viral infection.
Autophagy and antigen presentation
The role of autophagy in immunity is not limited to the direct elimination of intracellular pathogens or stimulation of type I IFN production. At least in certain contexts, autophagy promotes MHC class II presentation of cytosolic antigens66,67,68,69,70, thereby connecting autophagy with adaptive immunity. Brazil et al. demonstrated that the inhibition of autophagy with 3-methyladenine abrogated MHC class II presentation of endogenously synthesized C5 protein66. Nimmerjahn et al. reported that 3-methyladenine decreased MHC class II presentation of an endogenously expressed bacterial peptide67, and Dorfel et al. reported similar results using a tumour-associated antigen68. Dengjel et al. noted that changes in MHC class II presentation of peptides derived from intracellular proteins may occur upon amino-acid starvation69. Paludan et al. showed that pharmacological and genetic inhibition of autophagy decreases efficient MHC class II presentation of an endogenously synthesized viral protein (Epstein–Barr virus nuclear antigen 1 (EBNA1))70. Together, these initial studies led to the generally accepted idea that the autophagy pathway allows the transfer of cytosolic antigens to late endosomal or lysosomal compartments (Fig. 4), in contrast to the processing of exogenous antigens captured through endocytosis or phagocytosis in antigen-presenting cells.
The autophagic delivery of cytosolic antigens to endosomes and/or lysosomes represents an attractive model for explaining why the MHC class II peptidome contains many peptides of cytosolic or nuclear origin that cannot be delivered to MHC class II compartments by the classical exogenous route. However, it is not yet clear how universal a role autophagy has in the delivery of self and foreign cytosolic and nuclear antigens. Interestingly, high levels of autophagy activity are observed in the thymic epithelial cells of newborn mice that transgenically express a fluorescently tagged autophagy marker (green fluorescent protein (GFP)–LC3)71, suggesting that autophagy may indeed enable thymic epithelial cells to present self antigens to lymphocytes during positive and negative selection. Yet, the role of autophagy in MHC class II presentation of self antigens has not yet been directly examined. Moreover, whereas EBNA1 is processed by autophagy, two other Epstein–Barr-virus-encoded nuclear antigens, EBNA2 and EBNA3C, are preferentially processed by intracellular transfer of antigenic moieties and endocytic uptake from the culture media72, indicating that autophagy may only be used for MHC class II presentation of certain endogenously produced viral antigens. A fascinating question is why, even in the same cell, some microbial antigens, but not others, gain access to the MHC class II antigen-presentation pathway by autophagy.
As some of the earlier studies that showed a role for autophagy in MHC class II presentation of endogenous antigens were performed in conditions in which autophagy was upregulated by cell starvation, another important question has been whether cytosolic antigens are delivered to MHC class II molecules by autophagy under normal conditions. Recently, Schmid et al. observed constitutive autophagosome formation in MHC-class-II-positive cells, including B cells, dendritic cells and epithelial cells73. In these cells, at least one half of all autophagosomes intersect or fuse with MHC-class-II-loading compartments. This trafficking pathway may be highly relevant for antigen presentation, as the targeting of an influenza virus matrix protein (MP) to autophagosomes by fusion with the autophagosomal protein LC3 led to a 20-fold enhancement of MHC class II presentation to MP-specific CD4+ T-cell clones. This has exciting implications for vaccine development; targeting proteins for autophagic delivery to MHC-class-II-loading compartments may be an effective means to improve T helper (TH)-cell responses. However, the contribution of autophagic delivery of viral antigens to adaptive immunity during natural infections has not yet been explored.
The 'individualized' form of autophagy termed chaperone-mediated autophagy also has a role in endogenous MHC class II presentation74. Chaperone-mediated autophagy imports individual cytosolic proteins containing specific pentapeptide recognition motifs into the lysosome via a particular isoform of lysosome-associated membrane protein 2 (LAMP2a) and an accessory chaperone, the heat-shock protein HSC70. Importantly, targeting to the chaperone-mediated autophagy pathway is intrinsic to a large fraction of self proteins, as the targeting signal (KFERQ) is present in roughly 30% of all cytosolic proteins75. Although the relative contribution of autophagy and chaperone-mediated autophagy in endogenous MHC class II antigen presentation is not yet known, Zhou et al. have shown that chaperone-mediated autophagy may regulate MHC class II presentation of several cytoplasmic antigens76. Overexpression of LAMP2a or HSC70 increases cytoplasmic self antigen presentation, and diminished HSC70 expression reduces MHC-class-II-restricted T-cell responses to these antigens.
Now that it is known that autophagic pathways may have a role in MHC class II presentation of endogenous antigen, many important new questions arise. How important are these pathways for adaptive immunity to intracellular pathogens? What is the relationship between autophagic elimination of intracellular pathogens and MHC class II presentation of microbial antigens? Does autophagic degradation of pathogens provide a source of antigens for loading into MHC class II compartments and/or does the autophagic machinery independently capture newly synthesized microbial peptides? How are microbial (and self) antigens targeted for autophagic delivery to MHC-class-II-loading compartments? Beyond immunity against infection, what is the broader significance of autophagic antigen processing and presentation in an MHC-class-II-dependent manner? It will be interesting to unravel the role of this pathway not only in immunity against infection, but also in cancer immunology, central and peripheral tolerance, autoimmunity and transplant rejection.
Autophagy regulation by immune signals
The relationship between autophagy and immunity is bidirectional. Not only does autophagy, at least in certain contexts, enhance innate and adaptive immune responses, but in parallel, cytokines, receptors and ligands involved in innate and adaptive immunity enhance autophagy. Immune signalling molecules that have been shown to positively regulate autophagy in some contexts include PKR77, IFNγ (and its downstream effector immunity-related GTPases)9,42,45,78,79, tumour-necrosis factor (TNF)53,80,81, and the CD40–CD40L (CD40 ligand) interaction8. By contrast, autophagy is negatively regulated by the TH2-type cytokines, interleukin-4 (IL-4) and IL-13, although this has been shown so far only in a non-immune cell line82,83,84.
In general, there is a correlation between activation of autophagy by immune mediators and the control of infection with intracellular pathogens. The PKR signalling pathway is an important arm of the innate defence pathway against viruses85 and is required for virus-induced autophagy55,77. Cell-mediated immunity can induce autophagy through CD40–CD40L stimulation and protect target cells against the vacuolar parasite T. gondii8. IFNγ and TNF are crucial for protection against infection by mycobacteria and other pathogens that replicate in macrophages, and are potent inducers of autophagy in both macrophages and other cell types9,42,45,53,78,79,80,81. It is interesting to note the contrasting roles that the TH1-type cytokines IFNγ and TNF, and the TH2-type cytokines IL-4 and IL-13 (Ref. 81) may have on the regulation of autophagy. Perhaps, TH1-cell responses activate autophagy, thereby affording protection against intracellular microorganisms, whereas TH2-cell responses dampen the autophagic response, thus, potentially explaining the negative role that the TH2-cell response has in the control of intracellular pathogens86.
p47 GTPase-mediated regulation of autophagy. Recent advances have been made in identifying the molecular mechanisms that underlie the antimicrobial action of IFNγ-induced autophagy. The mouse genome contains 23 different immunity-related GTPases that are responsive to IFNγ and that have been long known to have a role in defence against a wide range of intracellular pathogens87. However, until recently, the mechanisms by which these immunity-related GTPases control intracellular pathogens have remained unclear, as has the question of whether human immunity-related GTPases have a similar role in defence against intracellular pathogens. This has been partially resolved by the recent discoveries that both the mouse and human p47 GTPase family member immunity-related GTPase family, M (IRGM; also known as LRG47) are required for IFNγ-induced autophagy and antimycobacterial activity in macrophages42,45, and that mouse IRGM is also required for antiparasitic activity associated with macrophage autophagy9.
Although the expression of human IRGM is not regulated by IFNγ, cells must be stimulated with this cytokine, or other physiological or pharmacological inducers of autophagy, for IRGM to exert its action. The exact mechanisms by which IRGM promotes autophagy are not known, but they may depend on direct or indirect interactions with organelles, regulators or effectors of the autophagic pathway. Alternatively, in view of the putative function of IRGM as a dynamin-like membrane remodelling protein, autophagy induction may be indirectly promoted by IRGM-induced changes to the parasitic vacuole membrane.
Autophagy and T-cell homeostasis
Autophagy has a central role in life and death decisions of numerous cell types across diverse phyla, functioning both as a pro-survival mechanism during nutrient deprivation and other forms of cell stress and as a cell-death mechanism in other contexts, such as in cells defective in apoptosis and in cells with very high levels of autophagy88,89. Recent studies indicate that this homeostatic role of autophagy extends to T cells.
Pools of mature T cells in the periphery are subject to tight regulation that must balance naive T-cell flux following thymic selection with effector T-cell proliferation, cell death and differentiation. A role for autophagy in T-cell survival and proliferation has been shown in vivo using lethally irradiated mice repopulated with haematopoietic cells from fetal livers of Atg5−/− mice13. CD4+ and CD8+ T cells from Atg5−/− mice fail to undergo efficient proliferation after T-cell receptor stimulation. Moreover, Atg5−/− T cells develop normally in the recipient thymus, but fail to repopulate the periphery due to overwhelming cell death. One interpretation of this finding is that T cells, on exit from the thymus, become exposed to nutritional stress owing to limitations in trophic factor support (such as IL-7) and require autophagy to sustain them during this period. At present, it is not known how autophagy affects immunological memory, but based on its role in the maintenance of other long-lived cells, such as neurons, the prediction is that autophagy may also have a role in maintaining memory T cells.
In contrast to its function as a T-cell survival process, excessive autophagy has been linked to the cell death of effector T cells under conditions that model normal homeostasis. Li et al. found that TH2 cells become more resistant to cell death induced by growth-factor withdrawal when autophagy is blocked using pharmacological or genetic methods14. This cell death process may be exploited by viruses such as HIV, as the HIV envelope glycoprotein has been shown to induce autophagic cell death by binding to CXC-chemokine receptor 4 (CXCR4) in uninfected bystander CD4+ T cells90. However, in CD4+ T cells, the natural ligand of CXCR4, CXC-chemokine ligand 12 (CXCL12; also known as SDF1α), induces lymphocyte activation and homing rather than cell death, indicating divergent outcomes in T-cell physiology in response to engagement of the same cellular chemokine receptor by a viral glycoprotein versus its endogenous ligand.
Future studies are needed to determine the role of this phenomenon in CD4+ T-cell depletion in patients with AIDS and to better define the factors that regulate whether autophagy has a pro-survival or pro-death role in lymphocytes. Given the extensive molecular interplay between autophagy and apoptosis88, it is not surprising that autophagy might have a dual role in T-cell homeostasis, executing both life and death decisions. Furthermore, there is no reason to think that the homeostatic role of autophagy will be confined to T cells; as research in the field progresses, we are likely to witness the unfolding of crucial roles for autophagy in maintaining not only homeostasis but also proper differentiation and function of other populations of immune cells.
Autophagy in inflammation and autoimmunity
An unexpected link between autophagy and the removal of apoptotic cell corpses has recently been reported15, which raises some intriguing possibilities about a role for autophagy in the prevention of inflammation and autoimmunity. Qu et al. found that autophagy provides apoptotic cells with signals to ensure their clearance during programmed cell death15. It is well established that the rapid removal of apoptotic cell corpses is crucial for the prevention of tissue inflammation91, and indeed, autophagy-deficient Atg5−/− embryos have increased inflammation in tissues that have impaired clearance of apoptotic cells15. Moreover, it is now believed that defective clearance of apoptotic cells overcomes tolerance to self antigens and leads to autoimmune diseases such as systemic lupus erythematosus (SLE)92,93. So, it is also possible that defective autophagy may contribute to the pathogenesis of SLE or other autoimmune diseases.
Of great interest, a strong genetic link has recently been uncovered between autophagy and Crohn's disease, a chronic inflammatory disease of the intestine. Several recent genome-wide scans have identified a strong association between a non-synonymous single-nucleotide polymorphism (SNP) in the autophagy gene ATG16L1 (T300A variant) and susceptibility to Crohn's disease16,17,18,19. In addition, the gene encoding the autophagy-stimulatory GTPase IRGM has been identified as a susceptibility gene for Crohn's disease20. Together, these studies are suggestive of a role for autophagy dysregulation in the pathogenesis of Crohn's disease. This hypothesis will be strengthened if the ATG16L1 variant (T300A) associated with Crohn's disease is found to lead to defective autophagy function.
It is not yet known how autophagy might be mechanistically linked to susceptibility to Crohn's disease. The pathogenic mechanisms of Crohn's disease are poorly understood but are speculated to involve a dysregulated immune response to commensal gut bacteria and possibly defects in mucosal barrier function or bacterial clearance94,95. It is therefore possible that defects in autophagy lead to altered clearance of and/or altered immune responses to commensal gut bacteria. Given the possible role of autophagy in peripheral tolerance, another speculation is that, in the setting of decreased autophagy, tolerance induction might fail and produce gut-reactive immune responses. As the intestine is a site of constant epithelial-cell shedding owing to apoptosis and regeneration, defective autophagy might also contribute to the pathogenesis of this inflammatory disorder by interfering with apoptotic cell clearance. Studies in targeted mutant mice with conditional deletions or mutations of autophagy genes should help to elucidate the pathogenetic mechanisms of Crohn's disease.
Conclusion
Autophagy probably originated to degrade cellular constituents, recycle nutrients and maintain cellular survival during starvation. Yet perhaps, with confrontations between primitive eukaryotes such as amoebae and bacteria, this ancient lysosomal degradation pathway evolved and became exquisitely adapted to orchestrate a multipronged defence against intracellular pathogens. The autophagy pathway degrades intracellular pathogens, and delivers microbial genetic material and antigens to the necessary cellular compartments for activation of innate and adaptive immunity. In addition to its role in defence against pathogens, it also is involved in immune-cell homeostasis and potentially, in preventing inflammation and autoimmunity. The journey forward — in deciphering how autophagy executes these and, similarly, other not yet identified immune functions — will be an exciting challenge for immunologists.
Note added in proof
While this manuscript was in press a new link was reported between pattern-recognition receptors of innate immunity and stimulation of autophagy. Xu et al. found that the Gram-negative bacterial lipopolysaccharide induces autophagy in macrophages through a TLR4 signalling pathway96.
Box 1 | Autophagy in health and disease.
The autophagy pathway has numerous adaptive functions in eukaryotic organisms. In cellular starvation settings, autophagy functions to preserve cellular bioenergetics by providing metabolic substrates (obtained through bulk cytoplasmic degradation), which maintains macromolecular synthesis and ATP production. Another important function of autophagy that probably underlies its protective role against diverse pathologies is its ability to perform 'routine housecleaning' and also to clean-up toxic or damaged cytoplasmic constituents, a process that may have more selectivity than autophagy induced by cell starvation. This function of autophagy contributes to its protection against neurodegenerative disease; it has a basal role in preventing the abnormal accumulation of ubiquitylated protein aggregates and it specifically degrades toxic aggregate-prone mutant polyglutamine expansion proteins. The degradation of damaged mitochondria and other organelles also may underlie the anti-ageing effects and the tumour suppressor effects of autophagy, by helping to reduce genotoxic stress and to prevent DNA damage and genomic instability. In parallel to cleaning-up endogenous cellular constituents, autophagy cleans up intracellular microorganisms, thereby protecting against disease caused by intracellular pathogens. Autophagy also can selectively deliver microbial genetic material and antigens to the innate and adaptive immune systems. Some of these effects on immune regulation and bacterial clearance may explain the recently uncovered genetic linkage between autophagy genes and susceptibility to Crohn's disease.
Given the diverse functions of autophagy in health and disease, there is now considerable interest in targeting the autophagy pathway in the treatment of different diseases, including cancer, neurodegenerative diseases, heart diseases, ageing, infectious diseases and Crohn's disease. However, for some of these diseases, such as cancer and heart disease, there is intense debate as to whether autophagy should be turned on or turned off, with no clear-cut data to resolve the debate. For other conditions, such as ageing and neurodegenerative diseases, presently available data suggest that autophagy augmentation is likely to be beneficial. In the case of infectious diseases, it is also likely that, at least in most cases, autophagy induction will foster increased innate and adaptive immunity. Nonetheless, the possibility that certain microorganisms may fare better in the setting of increased autophagy remains.
Acknowledgements
The original work from the authors' laboratories was supported by the National Institutes of Health, USA (V.D. and B.L.) and the Ellison Medical Foundation (B.L.).
Glossary
- Lysosomal degradation
The digestion of macromolecules in lysosomal organelles, which are the terminal organelles of degradative pathways, such as phagosomal or endosomal and autophagy pathways.
- Autophagy
Any process involving degradative delivery of a portion of the cytoplasm to the lysosome that does not involve direct transport through the endocytic or vacuolar protein sorting pathways.
- Xenophagy
The selective degradation of microorganisms (such as bacteria, fungi, parasites or viruses) through an autophagy-related mechanism.
- Macroautophagy
(Also known as autophagy). The largely non-specific autophagic sequestration of cytoplasm into a double- or multiple-membrane-delimited compartment (an autophagosome) of non-lysosomal origin. Note that certain proteins, organelles and pathogens may be selectively degraded via macroautophagy.
- Microautophagy
The uptake and degradation of cytoplasm by invagination of the lysosomal membrane.
- Chaperone-mediated autophagy
The import and degradation of soluble cytosolic proteins by chaperone-dependent, direct translocation across the lysosomal membrane.
- Small interfering RNA
(siRNA). Synthetic RNA molecules of 19–23 nucleotides that are used to 'knockdown' (that is, silence the expression of) a specific gene. This is known as RNA interference (RNAi) and is mediated by the sequence-specific degradation of mRNA.
- Proteasome
A giant multicatalytic protease resident to the cytosol and the nucleus.
- Peptidome
The repertoire of peptides that is presented by antigen-presenting molecules.
- Positive selection
The process in the thymus that selects thymocytes expressing T-cell receptors that have the ability to interact with self MHC molecules.
- Negative selection
The process in the thymus that eliminates T cells that express T-cell receptors with high affinity for self antigens.
- T helper 1 cell
(TH1 cell). The term used for a CD4+ T cell that has differentiated into a cell that produces the cytokines interferon-γ, lymphotoxin-α and tumour-necrosis factor, and supports cell-mediated immunity.
- T helper 2 cell
(TH2 cell). The term used for a CD4+ T cell that has differentiated into a cell that produces interleukin-4 (IL-4), IL-5 and IL1-3, supports humoral immunity and downregulates TH1-cell responses.
- p47 GTPase family
A group of 47–48-kDa proteins that are produced in response to interferons (IFNs) and that are involved in resistance to intracellular protozoa, bacteria and viruses. Members of this family include IFNγ-induced GTPase (IGTP), immunity-related GTPase family, M (IRGM; also known as LRG47) and T-cell-specific GTPase (TGTP).
Biographies
Beth Levine is the Chief of Infectious Diseases at the University of Texas Southwestern Medical Center, Dallas, USA. She received her M.D. from Cornell University Medical College, New York, USA, and undertook postdoctoral training in viral pathogenesis at Johns Hopkins University School of Medicine, Baltimore, Maryland, USA. Her research focuses on the role of beclin 1 and other autophagy proteins in host defence against viral infections, in tumour suppression and in the regulation of cell death.
Vojo Deretic is the Chair of Molecular Genetics and Microbiology at the University of New Mexico Health Sciences Center, Albuquerque, USA. He received his Ph.D. in molecular biology from the University of Belgrade, Serbia, and undertook postdoctoral training in molecular bacteriology at the University of Illinois at Chicago, USA. His research focuses on cystic fibrosis, phagolysosomal biogenesis, autophagy and the intracellular trafficking of Mycobacterium tuberculosis and HIV in macrophages.
Related links
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
Contributor Information
Beth Levine, Email: beth.levine@utsouthwestern.edu.
Vojo Deretic, Email: vderetic@salud.unm.edu.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nature Rev. Mol. Cell. Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deretic V. Autophagy in innate and adaptive immunity. Trends. Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Schmid D, Dengjel J, Schoor O, Stevanovic S, Munz C. Autophagy in innate and adaptive immunity against intracellular pathogens. J. Mol. Med. 2006;84:1–9. doi: 10.1007/s00109-005-0014-4. [DOI] [PubMed] [Google Scholar]
- 7.Deretic V. Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 2006;18:375–382. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amano A, Nakagawa I, Yoshimori T. Autophagy in innate immunity against intracellular bacteria. J. Biochem. 2006;140:161–166. doi: 10.1093/jb/mvj162. [DOI] [PubMed] [Google Scholar]
- 11.Menéndez-Benito V, Neefjes J. Autophagy in MHC class II presentation: sampling from within. Immunity. 2007;26:1–3. doi: 10.1016/j.immuni.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cell. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 13.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J. Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 15.Qu X, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 16.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 17.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prescott NJ, et al. A nonsynonymous SNP in ATG16L predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 19.The Wellcome. Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nature Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell. Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Meijer AJ, Codogno P. Signalling and autophagy regulation in health and disease. Mol. Aspects. Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 24.Nobukuni T, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl Acad. Sci. USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin–phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin 1-binding protein UVRAG. Nature Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 27.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 28.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 32.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nature Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 33.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl. 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell. Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat. Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- 37.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nature Rev. Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B. Eating oneself and uninvited guests; autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell. Biol. 2001;154:631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergne I, Chua J, Deretic V. Mycobacterium tuberculosis phagosome maturation arrest: selective targeting of PI3P-dependent membrane trafficking. Traffic. 2003;4:600–606. doi: 10.1034/j.1600-0854.2003.00120.x. [DOI] [PubMed] [Google Scholar]
- 41.Vergne I, et al. Mycobacterium tuberculosis phagosome maturation arrest: mycobacterial phosphatidylinositol analog phosphatidylinositol mannoside stimulates early endosomal fusion. Mol. Biol. Cell. 2004;15:751–760. doi: 10.1091/mbc.E03-05-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 45.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 46.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 47.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 48.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 49.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl Acad. Sci. USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J. Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa M, Sasakawa C. Intracellular survival of Shigella. Cell. Microbiol. 2006;8:177–184. doi: 10.1111/j.1462-5822.2005.00652.x. [DOI] [PubMed] [Google Scholar]
- 52.Balachandran P, et al. The ubiquitin ligase Cbl-b limits Pseudomonas aeruginosa exotoxin T-mediated virulence. J. Clin. Invest. 2007;117:419–427. doi: 10.1172/JCI28792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djavaheri-Mergny M, et al. NF-κB activation represses tumor necrosis factor-α-induced autophagy. J. Biol. Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 54.Scherz-Shouvai R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talloczy Z, Virgin HW, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 56.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cherry S, et al. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006;2:e102. doi: 10.1371/journal.ppat.0020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, et al. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2:91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Liang XH, et al. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Levine B. Autophagy in Immunity and Infection: a novel immune effector. 2006. pp. 227–241. [Google Scholar]
- 64.Kawai T, Akira S. Innate immune recognition of viral infection. Nature Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 65.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur. J. Immunol. 1997;27:1506–1514. doi: 10.1002/eji.1830270629. [DOI] [PubMed] [Google Scholar]
- 67.Nimmerjahn F, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 68.Dorfel D, et al. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 69.Dengjel J, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl Acad. Sci. USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paludan C, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 71.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor GS, et al. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J. Immunol. 2006;177:3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 73.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- 75.Cuervo AM. Autophagy: many paths to the same end. Mol. Cell. Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- 76.Zhou D, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Talloczy Z, et al. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J. Cell. Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pyo JO, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 80.Jia G, Cheng G, Gangahar DM, Agrawal DK. Insulin-like growth factor-1 and TNF-α regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol. Cell Biol. 2006;84:448–454. doi: 10.1111/j.1440-1711.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 81.Subauste C, Andrade R, Wessendarp M. CD40–TRAF6 and autophagy-dependent anti-microbial activity in macrophages. Autophagy. 2007;3:245–248. doi: 10.4161/auto.3717. [DOI] [PubMed] [Google Scholar]
- 82.Wright K, Ward SG, Kolios G, Westwick J. Activation of phosphatidylinositol 3-kinase by interleukin-13. An inhibitory signal for inducible nitric-oxide synthase expression in epithelial cell line HT-29. J. Biol. Chem. 1997;272:12626–12633. doi: 10.1074/jbc.272.19.12626. [DOI] [PubMed] [Google Scholar]
- 83.Arico S, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 84.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 85.Gale M, Jr, Katze MG. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 86.Harris, J. et al. Autophagy is an effector of Th1–Th2 polarization: Th2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity (in the press). [DOI] [PubMed]
- 87.Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nature Rev. Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 88.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshimori T. Autophagy: paying Charon's toll. Cell. 2007;128:833–836. doi: 10.1016/j.cell.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 90.Espert L, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 2006;116:2161–2172. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maderna P, Godson C. Phagocytosis of apoptotic cells and the resolution of inflammation. Biochim. Biophys. Acta. 2003;1639:141–151. doi: 10.1016/j.bbadis.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 92.Grossmayer GE, et al. Removal of dying cells and systemic lupus erythematosus. Mod. Rheumatol. 2005;15:383–390. doi: 10.1007/s10165-005-0430-x. [DOI] [PubMed] [Google Scholar]
- 93.Gaipl US, et al. Inefficient clearance of dying cells and autoreactivity. Curr. Top. Microbiol. Immunol. 2006;305:161–176. doi: 10.1007/3-540-29714-6_8. [DOI] [PubMed] [Google Scholar]
- 94.Sartor RS. Mechanisms of disease pathogenesis: pathogenesis of Crohn's disease and ulcerative colitis. Nature Clin. Pract. Gastroenterol. Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 95.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 96.Xu Y, et al. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


