The use of pathogen profiles in the management of disease could integrate typing and epidemiological data to enable the early detection of hospital infections and real-time global epidemiological surveillance of pathogens. This article outlines approaches to the translation of pathogen genotyping and microbial genomics into formats that are suitable for communicable disease management, surveillance and control.
Supplementary information
The online version of this article (doi:10.1038/nrmicro1656) contains supplementary material, which is available to authorized users.
Abstract
The usefulness of rapid pathogen genotyping is widely recognized, but its effective interpretation and application requires integration into clinical and public health decision-making. How can pathogen genotyping data best be translated to inform disease management and surveillance? Pathogen profiling integrates microbial genomics data into communicable disease control by consolidating phenotypic identity-based methods with DNA microarrays, proteomics, metabolomics and sequence-based typing. Sharing data on pathogen profiles should facilitate our understanding of transmission patterns and the dynamics of epidemics.
Supplementary information
The online version of this article (doi:10.1038/nrmicro1656) contains supplementary material, which is available to authorized users.
Main
The accurate classification of pathogens with epidemic potential can optimize communicable disease control and reduce associated costs1,2. Recognition of the usefulness of rapid genotyping for this purpose has led to a call for closer interplay between epidemiological surveillance and disease-management strategies3. The application and interpretation of genetic typing in clinical and epidemiological studies requires not only an understanding of the typing techniques involved, but also efficient integration of the results into clinical and public health decision-making4,5.
Clinical genomics and bioinformatics have been dominated by eukaryotic paradigms in which genomic rearrangements typically denote dysfunction. However, prokaryotic genomes, particularly those of bacteria, have a mosaic structure and can vary significantly, even within a species; it remains unclear, therefore, how microbial genomic data should be processed so that they are easy to interpret, accessible and easy to share. There is a growing mismatch between the volume of microbial genome data available and the ability to automate its systematic analysis and interpretation6,7. In this Perspective we outline selected approaches to the translation of pathogen genotyping and microbial genomics into formats that can be incorporated into communicable disease management, surveillance and control. Further, we introduce the concept of pathogen profiling as a tool for disease management in public health.
Moving beyond the phenotype
Pathogen profiles. Analysing the dynamics of infections that have epidemic potential relies on the accurate demarcation and identification of individual strains or epidemic clones, together with the identification of specific virulence factors and other validated markers. Together, this information can be consolidated into a pathogen profile, which comprises information derived from traditional phenotype-based methods, such as bacterial culture identification (often based on biochemical properties and antibiotic resistance), and other information, such as that derived from nucleic-acid-based techniques. Nucleic-acid-based techniques include various high-throughput epidemiological typing methods that have the capacity to simultaneously identify and analyse multiple selected regions within a given pathogen genome and are relatively new to mainstream clinical microbiology8,9.
The argument that a species-based description of pathogens has inherent limitations is not new. Many bacterial species contain different strains that are associated with distinct clinical features and epidemiology, and which cannot be distinguished by traditional means4,10. Strains of the same species can vary by as much as 35% in either the complement or number of unique genes present and sometimes have significant variation within individual genes. For example, the sizes of the Escherichia coli and Salmonella enterica chromosomes can vary by more than 1 Mb and 300 kb, respectively11, and most bacterial species are a mosaic of different subpopulations. In many bacteria the characteristics that determine pathogenicity for hosts are encoded on mobile genetic elements that are transferred between strains at different rates. Organizing bacterial strains into clonal complexes rather than traditional species groupings is therefore often more relevant to clinicians and is better suited to epidemiological analyses. For example, the diversity of hundreds of distinct Campylobacter jejuni strains, as defined by multilocus sequence typing (MLST), is represented by 17 clonal complexes, six of which comprise more than 60% of the strains isolated from human campylobacteriosis12.
The heterogeneity of pathogens, hosts and the environment means that no single characteristic can adequately reflect the clinical and epidemiological complexity of infection or reliably predict the outcome(s). The systematic construction of pathogen profiles from a combination of genomic or other 'omic' markers in a manner that enables data to be integrated and shared, is essential for successful surveillance and disease management13. Consider, for example, an infection that is potentially caused by several different strains of the same species, each of which has different sets of virulence factors that can be distinguished by genotyping. If the optimal management strategies varied for infections caused by different subtypes, then rapid subtype identification would optimize disease management. For example, antibiotic resistant strains of Mycobacterium tuberculosis, detection of which indicates potential therapeutic failure, can be identified using genetic markers2,8. Similarly, evidence from the monitoring of HIV or hepatitis C virus (HCV) infections supports this approach14,15 (Boxes 1,2).
Profile attributes. A pathogen profile is a single, multivariate observation (or set of observations) that is composed of classes of specific attributes, for example, genome, transcriptome, proteome or metabolome data, which are designed to allow interrogation of existing (or future) databases (see Further information; Table 1), and integration with clinical observations and patient outcomes (Fig. 1). The profile can indicate the probability that a specific marker is associated with a clinically relevant phenotype, such as in vivo antimicrobial resistance or high transmissibility. This information would allow classification of strains into risk groups for either treatment failure or a propensity to cause outbreaks. It is often important to also capture quantitative information about a pathogen in vivo, for example, viral or bacterial loads and their units of measurement.
Table 1.
Classes of determinants for pathogen profiling
| Class of determinant | Data type | Uses | Data standards | Refs |
|---|---|---|---|---|
| Pathogen identification | Presence of pathogen, genus and species-specific gene | Confirmation of identity of a pathogen | SNOMED, LOINC | 13, 19, 20 |
| Virulence | Presence or absence of individual genes or mutants associated with virulence | Primary risk assessment or outcome prediction* | Clinical, bioinformatics, ontologies | 21 |
| Transmissibility | Presence or absence of individual genes associated with transmissibility | Secondary risk assessment or outcome prediction* | N/A | – |
| Antimicrobial resistance | Presence or absence of individual genes or mutations associated with resistant phenotype | Treatment response prediction | SNOMED, XML | 22 |
| Clonality | Genotypes and epidemiological data | Confirmation of epidemiological links or generation of hypotheses about relationships in the absence of epidemiological data‡; Tracking geographical and temporal spread of pathogens of public health importance | PIML, RDF Microarray & Gene Expression Markup Language | 23,24 |
| Clinical information | Patient's demographics and location, laboratory number | Unique identifier, temporal and geo-positioning | HL-7, UMLS | 25,70 |
| *Identifying risk factors for recent infection or rapidly progressive disease. | ||||
| ‡Identifying an outbreak in what appears to be sporadic cases of infection. LOINC, Logical Observation Identifier Names and Codes (Regenstrief Institute); N/A, not available; PIML, Pathogen Information Markup Language; SNOMED, Systematised Nomenclature of Medicine (College of American Pathologists); UMLS, United Medical Language System. | ||||
Figure 1. Interaction of the different 'omes' in a microbial cell.

Each 'ome' is a complex function of the other 'omes', and the amount of integration increases from the bottom to the top.
In contrast to traditional subtyping, which is based on phenotypic characteristics such as serotype, biotype, phage type or antimicrobial susceptibility, genetic profiling describes the phenotypic potential in the nucleic acid sequence. Genotyping systems that are based on comparison of sizes and numbers of different DNA fragments separated by gel electrophoresis — pulse field gel electrophoresis (PFGE), or nucleic acid amplification-based typing methods such as restriction fragment length polymorphism (RFLP) or random amplified polymorphic DNA chain reaction (RAPD) — have been less reliable than direct sequence-based methods, due to a lack of precision and reproducibility16. Sequence-based typing and RAPD, plasmid fingerprinting or PFGE can be viewed as examples of direct and indirect methods of assessing nucleic acid sequence, respectively. All of these methods provide both strain typing and phylogenetic data2,17,18 that can be processed using sequence alignment and clustering techniques and are amenable to standardization and database cataloguing. The derived information often correlates well with clinically relevant phenotypic characteristics, such as virulence19,20,21. Typing systems that use markers with specific or binary values, including MLST, are more reproducible and are therefore more appropriate for pathogen profiling19,20. Such typing systems enable classification of pathogens that are relevant to the investigation of chains of infection transmission and are useful tools for studies of global epidemiology18. Detailed descriptions of molecular typing techniques that are used for epidemiology studies can be found elsewhere20,21,22.
Selection of attributes. The choice of attributes used to construct a profile depends on the clonality of the species, the function, diversity and rates of change of chosen genes, and their clinical or public health relevance. As a rule, microbial profiles should include key molecular markers that are potentially associated with specific patient outcomes or risk factors, and antimicrobial resistance markers. Profiles of different types of viruses and bacteria can differ significantly as there is no unique or common template or genotyping method that can capture all of the attributes required to describe all types of microorganisms. Some genome profiling techniques are based on conserved genes — genes that are associated with metabolism or other 'housekeeping' functions — whereas others target variable genes that are often associated with virulence20. Virulence determinants are frequently present on transferable genetic material, such as plasmids, pathogenicity islands and bacteriophages, with genetic histories and dynamics distinct from those of the conserved genes of the host bacterial population.
The specific disease and the type of control measures influence both the clinical relevance and discriminatory power of the typing system that is used for profiling and the level of statistical significance that is required to identify clustering23. Microbial genotyping alone might not always be the correct classification method as outbreaks are occasionally caused by several different agents, rather than a single, virulent clone; for example, sewage contamination of water or food could cause an outbreak of diarrhoea. Therefore a combination of genomic and phenotypic microbial characteristics and comparison of genotypic clusters with those identified by epidemiological investigations, is important in outbreak investigations. Using a combination of methods can enhance the discriminatory power and precision of microbial profiling24 and might be required to define genotypes that are composed of conserved and variable portions of the genome, but would increase the cost and the complexity of data interpretation and sharing.
The task of defining which information to include in the pathogen profile is non-trivial and is becoming even more complex as the number and scope of molecular typing methods increases and are linked with treatment and public health decisions25. The nature of clinical reports of antimicrobial resistance illustrates this problem26. Currently, clinical microbiologists usually report the pathogen name and antibiotic susceptibilities, but few, if any, other details. In future, routine reports could include predictive prognostic markers such as a calculated post-test probability based on the pre-test information. For example, interpretative reports of antiretroviral susceptibility testing might include information about mutations and cumulative sensitivity scores to rank the likely efficacy of individual drugs and combinations27.
A pathogen profile is a synthesis of different markers and clinical end-points that can be extracted from medical charts and that characterize an individual patient's clinical and public health outcomes. The profile can be heuristic, when only a single genetic marker is associated with a specific patient outcome, however greater insight can be achieved when attributes from different levels of the biological hierarchy (that is, gene detection, gene expression, metabolite profiles and so on) corroborate and complement each other. Large-scale genotyping generates valuable information that can be translated into databases to search for strain-specific epidemiological markers or to construct an evolutionary history of strains for a particular epidemiological catchment area. This objective becomes greatly simplified if the genomic data are categorized, archived and electronically portable so as to facilitate access, retrieval and comparisons. The task of designing, capturing and correlating pathogen profiles can be assisted by the development of a standards-based representation of attributes and pathogen-specific ontologies.
The medical and cost benefits of highly integrated, comprehensive disease-control programmes that include routine microbial genotyping have been demonstrated28,29, yet incorporating multiple data sources remains a technical challenge16. The need for models that define data elements in communicable disease informatics, and the relationships between them, have been identified30,31. Microbial profiles provide data models with discrete elements amenable for standardization. Figure 2 illustrates such a data model by demonstrating the relationships between meticillin-resistant Staphylococcus aureus (MRSA) as a concept (object) and the determinants of its pathogen profile. However, the vocabulary of profiling data (the words or individual components), syntax (the 'sentence' structure) and messaging protocols are yet to be developed. Healthcare vocabularies such as the UMLS (United Medical Language System, National Library of Medicine), LOINC (Logical Observation Identifier Names and Codes, Regenstrief Institute) and SNOMED (Systematised Nomenclature of Medicine, College of American Pathologists)32,33 provide integration mechanisms for high-level terms used in medical charts (for example, tuberculosis) with the relatively low-level terms used in the clinical laboratory (for example, Mycobacterium tuberculosis Beijing Family spoligotype).
Figure 2. Relationships between MRSA as a concept (object) and determinants of the pathogen profile.
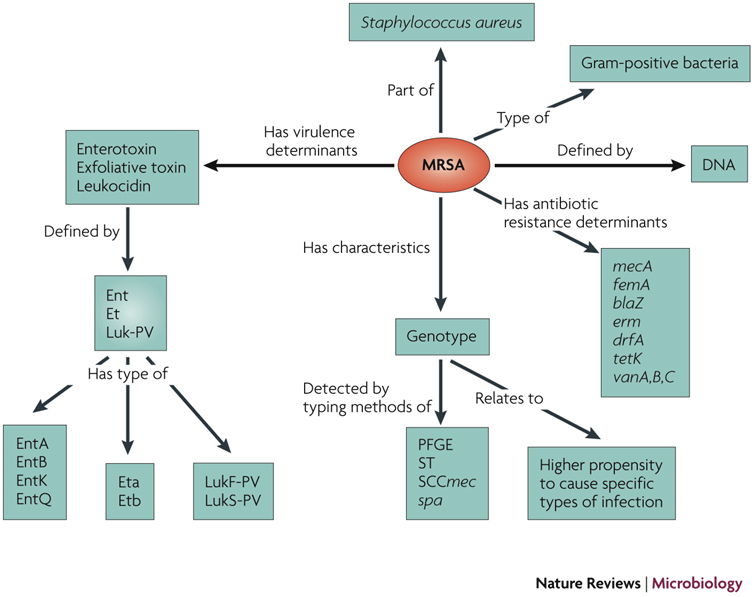
This data model defines major classes of attributes for an MRSA profile (for example, genotyping methods, virulence factors and clinical outcomes) and relationships between them. blaZ, β-lactamase gene; drfA, trimethoprim resistance gene; Ent, enterotoxin; erm, macrolide resistance gene; Et, exfoliative toxin; femA, gene encoding a cytoplasmic protein necessary for the expression of meticillin resistance; Luk-PV, Panton-Valentine leukocidin; mecA, gene encoding PBP2a, the low-binding-affinity penicillin-binding protein that mediates meticillin-resistance; MRSA, meticillin-resistant Staphylococcus aureus; SCCmec, Staphylococcus cassette chromosome; spa, staphylococcal protein A gene type; ST, sequence type; tetK, tetracycline resistance gene; tst, staphylococcal toxic shock toxin gene; vanA, vanB, vanC, vancomycin resistance genes.
Successful initiatives that have focused on common interchange standards in genomics and proteomics, such as minimum information about a microarray experiment (MIAME), minimum information requested in the annotation of biochemical models (MIRIAM)34 and minimum information to describe a proteomic experiment (MIAPE)35,36, should be informative in the push to integrate databases in the management of disease. These projects have introduced formats to enable the unambiguous interpretation of results and aim to ensure that experimental results in genomics, proteomics and metabolomics are deposited in public databases before publication, as has already been long established for nucleotide sequences. The Pathogen Information Markup Language (PIML) has also been recently introduced to enhance the interoperability of microbiology datasets for pathogens with epidemic potential31 by capturing the data elements that describe determinants of pathogen profiles.
Matching profiles. Once a profile has been constructed for a strain, it can be matched with those of others or with existing datasets using similarity measures and clustering techniques (see Supplementary information S1 (box) for a list of microbial databases). Sequence similarity or genotype matching of microorganisms implies a common lineage rather than a unique identity, in contrast to eukaryotic DNA matching. Different distance functions for phylogenetic assessments and clustering algorithms have been applied to reveal or compare microbial patterns in bacterial or viral fingerprints (for example, Euclidian distance or Pearson correlation, index of diversity, approximate matching heuristics and information theoretic similarity measures)37,38. For example, Simpson's index of diversity estimates the probability that two unrelated strains will be placed into two different typing groups38. The closer this numerical index is to 0 the higher the chance that two microbial profiles match.
Alternatively, the level of reported similarity between sequences, which can indicate biological relationships, can be measured as E values (expert value) which range from 0 (100% identity), or close to 0, to larger numbers which indicate lower similarity. The relatedness of isolates can be visualized using dendrograms that are based on unweighted pair group methods with arithmetic means (UPGMA) for small numbers of isolates or clustering, for example using eBURST, for larger datasets39. The eBURST algorithm, which was developed for the interpretation of MLST results, first identifies mutually exclusive groups of related genotypes in the population, then identifies the group's founding genotype, predicts the descent — from the founder — of other genotypes, and shows the output as a radial diagram, centred on the predicted founding genotype. The computational power required and the confidence limits used depend on the number of markers and their diversity within and among species, and the number of representative samples. Computational pattern matching and validation techniques have received little attention in the biomedical literature so far40,41.
Uses of pathogen profiling
Knowledge discovery from databases. Although the number and range of data relevant to microbial profiles have increased, they do not characterize the entire phenotype of a pathogen in an environmental or experimental context. Linking systematically annotated profiles with clinical and research databases can identify previously unrecognized associations between phenotype, genotype, environment and host responses and, potentially, the specific genes that govern them42. Functionally linked genes or proteins have been identified by examining connections between them, using computational methods like the Rosetta Stone43,44, Phylogenetic Profile45 or Operon46. Networks, created by relationships among phenotype, disease expression, environment and experimental context and associated genes with differential expression, could provide new insights into microbial interactions and pathogenesis47,48,49. This approach has been fruitful in metagenomics50 and information management systems designed to assist with genotyping or functional genomics are now being developed51,52. For example, in silico analyses that combine molecular phylogeny and targeted sequencing have identified possible target genes for antimalarial treatment53 and predicted candidate antigens for vaccine development (reverse vaccinology54).
A great deal of data that are relevant to microbial profiling already exist. Public electronic bacterial typing databases such as MLSTNet, PulseNet, the BioPortal and SPOTCLUST, among others, use web-based formats that allow universal access and matching of bacterial or viral isolates to each other and to those represented in databases. More recently, structured polymorphism databases have been built, yet data sharing and integration remain difficult, due to the lack of common structures47,55. Several hundred public domain molecular biology databases are currently online but few contain raw data. Most represent the efforts of individuals to organize, annotate and interpret data from other sources. These databases are highly valued and are increasingly expected to replace paper publication as the medium of communication46. Some are classification databases (for example, the Staphylococcus aureus spa typing tool or the SPOTCLUST database for Mycobacterium tuberculosis genotyping). Critical factors that distinguish the best databases include networks of subscribers willing to share data, the availability of statistical algorithms to analyse these data and the quality of the curation process.
MLST and PulseNet are good examples of advanced databases. At the core of the MLST concept is the provision of freely accessible nucleotide-sequence databases, which function as a common dictionary to enable direct comparison of bacterial isolates without requiring the physical exchange of cultures. In this sense they provide the basis of a common language for bacterial typing45. In contrast to archival databases such as GenBank, MLST databases are curated for accuracy. To overcome some limitations of the first MLST stand-alone web sites, a new network-based database (MLSTdB-Net) has been implemented with more than 30 MLST schemes, for different bacterial species. It is hosted at 33 websites to ensure greater computational power and better analytical performance. Some of the MLST websites allow researchers to run and curate their own schemes remotely. The PulseNet system, which is based on PFGE patterns, is the most developed system for the characterization of bacterial isolates with a fingerprinting approach. It is one of the few networks that integrate epidemiological and typing data over wide geographical regions45,50.
Antimicrobial therapy optimization. The great diversity of mutational patterns contributing to antimicrobial resistance complicates the choice of optimal therapies. A range of bioinformatics tools, which are designed to predict drug resistance or response to therapy from genotype, have been developed to provide clinicial support. These tools use either a statistical approach, in which the inferred model and prediction are treated as regression problems, or machine learning algorithms, in which the model is treated as a classification problem17. A statistical learning approach to ranking of therapeutic choices often relies on a direct correlation between baseline microbial profile, the therapeutic decision and response to treatment, for example, expected reduction in viral load resulting from anti-HIV combination therapy (Box 1). Several susceptibility scores have been used for combination antiretroviral therapy that take into account specific resistance mutations and add up the activities of individual drugs in the regimen27,56,57. Computer-assisted therapy is an attractive way to reduce the complexity of prescribing antimicrobial combinations. It highlights the need for databases that can be widely shared, and that allow correlation of quality-controlled data from genotypic resistance assays and treatment regimens with short- and long-term clinical outcomes. Differences in antimicrobial sensitivities reflect variation in amino-acid composition of resistant microorganisms, but simply counting mutations is not enough to detect most functional differences, which affect treatment outcomes. The data links between laboratory and clinical databases will unlock the full utility of microbial profiles.
Efficiency in outbreak investigation and disease monitoring. The genetic signatures of pathogens enrich the accuracy and predictive power of laboratory experiments2,3. Microbial typing can confirm or refute putative epidemiological links among and between cases and potential environmental sources, and therefore might trigger public health investigations. Alternatively, typing studies can demonstrate that putative clusters are unrelated and so rule out the need for further action. However, the usefulness of pathogen profiling goes beyond specific questions related to the investigation of possible outbreaks. It can also be used for disease monitoring, by identifying transmission and associations between microbial types and clinical outcomes41. Molecular profiling can assist in the assessment of the reproductive number (R0) of an infectious organism during epidemics, in making infection control policies more organism-specific41 and in predicting clinical outcomes. For example, multiple isolates of the same pathogen that have indistinguishable profiles, which are highly clustered in time and space, would suggest an outbreak and trigger an epidemiological investigation supplemented by a social network analysis of patients involved. This could potentially identify a 'superspreader' — an individual who is responsible for 80% of transmission events58. Evidence suggests that, for some infections such as severe acute respiratory syndrome (SARS) that have epidemic potential, public health control strategies that are focused on 'superspreaders' would be three times more effective than the random interventions currently used58.
Molecular typing also facilitates the detection of chains and patterns of infection transmission and the construction of epidemic trees3. For example, by distinguishing tuberculosis (TB) due to recent infection from reactivation, typing allows the assessment of current rates of active transmission in a community and hence guides appropriate control efforts. Molecular typing has led to a reassessment of the role of casual contacts in the transmission of TB59. Specifically, a two-stage TB contact tracing strategy, based on clustering of genetically related M. tuberculosis isolates, can improve the identification of epidemiological links and prevent more cases of secondary infections in low prevalence settings, and therefore augment traditional contact tracing59,60. This capacity of pathogen profiling is especially important as changes in contact patterns often underlie the re-emergence of disease.
Early warning for population health and infection control. A particularly exciting prospect is the integration of typing databases with epidemiological information, potentially producing global real-time epidemiological surveillance of pathogens that have epidemic potential61,62. There is increasing evidence of the value of rapid molecular profiling in assisting outbreak detection and hospital infection control26,28,29,63. For example, rapid outbreak detection by routine MRSA spa typing is a potential alternative to traditional approaches to hospital-acquired infection control28,63. In a prospective study, automated clonal alerts, which were based on real-time spa typing of hospital MRSA isolates and temporal-scan test statistics, were 100% and 95.2% sensitive and specific, respectively, in identifying outbreaks and were more sensitive and timely than routine surveillance by infection control nurses63.
In such an 'on-line' surveillance system, novel and previously characterized strains can be compared, grouped by cluster analysis and depicted as dendrogram or multidimensional graphs to simplify the presentation of complex time–space relationships. Spatial surveillance, using emerging geographical information systems, will enhance the ability to measure the extent and variables of an outbreak in space and time and the power to detect localized events64. The output from these systems ultimately needs to be integrated into clinical and diagnostic processes. Real-time data sharing, especially of genotypes of microbial isolates from different animal species as well as humans (for zoonotic infections) and from different jurisdictions or countries, could enhance rapid response using input and action triggers provided by multiple diagnostic, veterinary and public health laboratories and other partner organizations.
Concluding remarks
In this Opinion we have identified some of the major steps that are needed to generate and translate accessible genomic information about pathogens of clinical and public health importance. The synergistic use of high-throughput molecular testing, with advanced machine-learning approaches, has already redefined several traditional classifications of cancer65. A similar approach has started to affect communicable disease control. The concept of pathogen profiling described here provides a framework for data integration and sharing to ensure that the flood of data from new molecular technologies will be used effectively in public health surveillance and disease management.
We argue that diagnostic pathogen profiling will help to predict patient outcomes and identify markers that can be used for early diagnosis and to predict and monitor treatment responses. Pathogen profiling to identify individual genetic variation, along with a detailed knowledge of polymorphisms, will allow tailored interventions, a process commonly referred to as 'personalized medicine'. The potential value of pathogen profiles can be shown by, for example, the use of HIV and HCV genotyping to direct the choice of antiviral therapy, or specific genetic signatures in cancer tissue or host immune responses to predict outcomes27,31,57.
There are, however, many challenges in producing useful pathogen profiles. The methods used to generate input data and standards for sharing data are still evolving. A shift of emphasis towards integrative data analysis and sharing is difficult, but might prove to be the key to the successful translation and integration of laboratory diagnostics into improving clinical and public health outcomes in medicine.
Box 1 | HIV case study.
HIV is a complex retrovirus characterized by extensive genetic variability. On the basis of phylogenetic analyses, multiple circulating HIV-1 group M genetic subtypes and recombinant forms have been recognized. Inter-subtype diversity is relevant to the development of antiretroviral drug resistance, diagnostic tests and rates of virus transmission and disease progression that influence the dynamics of the HIV pandemic56,66. For example, subtype C has lower replicative efficiency than subtype B but is associated with a greater propensity for transmission in utero and higher levels of shedding from the genital tract than subtypes A or D.
Interpretation of genotypic data (see table) must account for both the number of mutations that contribute to resistance and the various patterns of mutations. Different algorithms, which use public or commercial databases to correlate genotypes collected from patients before and after antiviral therapy with corresponding phenotypic susceptibilities15,56,57, have been developed for bioinformatics-assisted antiretroviral therapy. They produce cumulative susceptibility scores that are increasingly recognized for their clinical value. Susceptibility scores range from 0 (Stanford, intermediate or low-level resistance; ANRS*, resistant) to 1 (Stanford, potential low-level resistance or susceptible; ANRS, susceptible); the sum of drugs' individual scores provides the genetic susceptibility score of the antiretroviral regimen or genotypic inhibitory quotient (the ratio of drug concentration to the number of target mutations)27,57.
In the table, examples of specific mutations (indicating the site and effect of a mutation) encoding drug resistance include: inhibitors of nucleoside reverse transcriptase (Zidovudin – K70R; T215Y or F; M41L; D67N; L210W and Lamivudine – M184V or I); inhibitors of nucleotide reverse transcriptase (Fenofovir – K65R); inhibitors of non-nucleoside reverse transcriptase (Nevirapine – K103N; V106A; V108I; Y181C or I;G190A); and protease inhibitors (Indinavir – V32I; V82A or T or F; I84V; L90M).
In the table entry for data sources, examples of specific rule-based algorithms developed for the interpretation of anti-retroviral susceptibility from genotypic data are shown. These algorithms are periodically updated as new mutations in the HIV genome are linked with resistance and as new treatments become available. Currently more than 20 interpretation systems are available. *ANRS, Agence Nationale de Recherches sur le Siaa; ‡CREST, can resistance testing enhance selection of therapy (study).
Box 2 | Hepatitis C virus case study.
Hepatitis C virus (HCV) is classified into six major genotypes, numbered 1 to 6, which can vary in nucleotide sequence by as much as 30% and occupy unique geographical niches. Chronic HCV infection is responsible for inflammation of the liver, and ∼20% of patients progress to liver cirrhosis with an increased risk for the development of hepatocellular carcinoma67. The complications of HCV infection can be prevented by antiviral therapy. The combination of pegylated interferon-α (IFN-α) and ribavirin has become the standard treatment for chronic HCV infection. HCV profiles (see figure) are important clinically because they most accurately predict the chance of an antiviral response, dictate the duration of therapy, the dosage of ribavirin and determine the virological monitoring procedures67,68. The amino-acid variability of HCV proteins reflects different sensitivities to IFN-α-based therapy and a range of mutations in genes encoding protein kinase receptors and the IFN-α sensitivity-determining region associated with HCV protease and polymerase inhibitors have been identified69. HCV genotypes 1 and 4 are more resistant to antivirals and are cleared from infected cells more slowly. Current algorithms for the use of HCV profiles in the treatment of chronic hepatitis C are shown in the figure.
Supplementary information
Public Microbial Databases on the web (PDF 73 kb)
Acknowledgements
The authors wish to thank Enrico Coiera and Dominic Dwyer for helpful comments. Funding from the National Health & Medical Research Council (grants 358351, 457472 and 457122) and from a Capacity Building Infrastructure Grant from the New South Wales Department of Health is acknowledged.
Biographies
Vitali Sintchenko is a Staff Specialist Microbiologist at the Centre for Infectious Diseases and Microbiology–Public Health at Westmead Hospital, a Senior Lecturer with the Western Clinical School, The University of Sydney, and a Visiting Fellow to the Centre for Health Informatics, University of New South Wales, Australia. His research interests are in communicable disease informatics, molecular epidemiology of pathogens with epidemic potential and outbreak pattern discovery. He has over 50 peer-reviewed publications and has received research fellowships from the National Health & Medical Research Council and the National Institute of Clinical Studies.
Associate Professor Jonathan Iredell is based at the Centre for Infectious Diseases and Microbiology at Westmead Hospital, where he has worked as a Staff Specialist in Intensive Care, Infectious Diseases and Microbiology. Relevant interests include the pathogenesis of Gram-negative bacteria and the epidemiology of mobile antibiotic resistance genes, with particular reference to the management of infection in the critically ill. Research in his laboratory is supported by the National Health & Medical Research Council and the Australia and New Zealand Intensive Care Foundation.
Lyn Gilbert is Director of the Centre for Infectious Diseases and Microbiology Laboratory at Westmead Hospital, Sydney, which includes a large diagnostic microbiology service and New South Wales public health microbiology reference laboratory. Her clinical and research interests are in the diagnosis, surveillance, prevention and molecular epidemiology of communicable diseases, especially those with significant public health impact. In particular, she supports the development of informatics tools to interpret complex laboratory data in context and provide appropriate decision support for clinical and public health action. She has a particular interest in public health ethics, especially relating to healthcare associated infections.
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
References
- 1.Burke MD. Laboratory medicine in the 21st century. Am. J. Clin. Pathol. 2001;114:841–846. doi: 10.1309/TH8P-1CAL-9K3G-VFTM. [DOI] [PubMed] [Google Scholar]
- 2.Fey PD, Rupp ME. Molecular epidemiology in the public health and hospital environment. Clin. Lab. Med. 2003;23:885–901. doi: 10.1016/S0272-2712(03)00100-8. [DOI] [PubMed] [Google Scholar]
- 3.Matthews L, Woolhouse M. New approaches to quantifying the spread of infection. Nature Rev. Microbiol. 2005;3:529–536. doi: 10.1038/nrmicro1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansmann U. Genomic profiling: Interplay between clinical epidemiology, bioinformatics and biostatistics. Methods Inf. Med. 2005;44:454–460. doi: 10.1055/s-0038-1633982. [DOI] [PubMed] [Google Scholar]
- 5.Sintchenko V, Iredell J, Gilbert GL. Culture independent PCR in diagnostic bacteriology: expectations and reality (is it time to replace the Petri dish with PCR?) Pathology. 1999;31:436–439. doi: 10.1080/003130299104909. [DOI] [PubMed] [Google Scholar]
- 6.Kasturi J, Acharya R. Clustering of diverse genomic data using information fusion. Bioinformatics. 2005;21:423–429. doi: 10.1093/bioinformatics/bti186. [DOI] [PubMed] [Google Scholar]
- 7.Budowle B, et al. Genetic analysis and attribution of microbial forensic evidence. Crit. Rev. Microbiol. 2005;31:233–254. doi: 10.1080/10408410500304082. [DOI] [PubMed] [Google Scholar]
- 8.Campbell CJ, Ghazal P. Molecular signatures for diagnosis of infection: application of microarray technology. J. Appl. Microbiol. 2004;96:18–23. doi: 10.1046/j.1365-2672.2003.02112.x. [DOI] [PubMed] [Google Scholar]
- 9.Wilson WJ, et al. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Nucleic Acids Res. 2004;32:1848–1856. doi: 10.1093/nar/gkh329. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinidis K, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkhill J, Thomson NR. Microbial genomes. 2004. pp. 269–290. [Google Scholar]
- 12.Dingle KE, et al. Molecular characterization of Campylobacter jejuni clones: a rational basis for epidemiological investigations. Emerg. Infect. Dis. 2002;8:949–955. doi: 10.3201/eid0809.02-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rotz LD, Hughes JM. Advances in detecting and responding to treats from bioterrorism and emerging infectious disease. Nature Med. 2004;10:S130–S136. doi: 10.1038/nm1152. [DOI] [PubMed] [Google Scholar]
- 14.Brun-Vezinet F, et al. Clinically validated genotype analysis: guiding principles and statistical concerns. Antivir. Therapy. 2004;9:465–478. [PubMed] [Google Scholar]
- 15.Liu TF, Shafer RW. Web resources for HIV type I genotypic-resistance test interpretation. Clin. Infect. Dis. 2006;42:1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagen RM, et al. Development of real-time PCR assay for rapid identification of methicillin-resistant Staphylococcus aureus from clinical samples. Intern. J. Med. Microbiol. 2005;295:77–86. doi: 10.1016/j.ijmm.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. doi: 10.1016/S0966-842X(99)01609-1. [DOI] [PubMed] [Google Scholar]
- 18.Urwin R, Maiden MCJ. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 2003;11:479–487. doi: 10.1016/j.tim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Blanc DS. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infect. Genet. Evol. 2004;4:193–197. doi: 10.1016/j.meegid.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Tenover FC, et al. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections. Infect. Control Hosp. Epidemiol. 1997;18:426–439. doi: 10.2307/30141252. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, et al. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 2006;19:512–530. doi: 10.1128/CMR.00025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struelens MJ. Members of the european study group on epidemiological markers. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 23.Wartenberg D. Investigating disease clusters: why, when and how? J. R. Statist. Soc. A. 2001;164:13–22. doi: 10.1111/1467-985X.00181. [DOI] [Google Scholar]
- 24.van Deutekom H, et al. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 2005;43:4473–4479. doi: 10.1128/JCM.43.9.4473-4479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchevsky AM, Wick MR. Evidence-based medicine, medical decision analysis, and pathology. Hum. Pathol. 2004;35:1179–1188. doi: 10.1016/j.humpath.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Kuperman GJ, et al. Improving response to critical laboratory results with automation. J. Am. Med. Inform. Assoc. 1999;6:512–522. doi: 10.1136/jamia.1999.0060512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengauer T, Sing T. Bioinformatics-assisted anti-HIV therapy. Nature Rev. Microbiol. 2006;4:790–797. doi: 10.1038/nrmicro1477. [DOI] [PubMed] [Google Scholar]
- 28.Hacek DM, et al. Computer-assisted surveillance for detecting clonal outbreaks of nosocomial infection. J. Clin. Microbiol. 2004;42:1170–1175. doi: 10.1128/JCM.42.3.1170-1175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hacek DM, et al. Medical and economic benefit of a comprehensive infection control program that includes routine determination of microbial clonality. Am. J. Clin. Pathol. 1999;111:647–654. doi: 10.1093/ajcp/111.5.647. [DOI] [PubMed] [Google Scholar]
- 30.Huang SH, Triche T, Jong AY. Infectomics: Genomics and proteomics of microbial infections. Funct. Integr. Genomic. 2002;1:331–344. doi: 10.1007/s10142-002-0048-4. [DOI] [PubMed] [Google Scholar]
- 31.He Y, et al. PIML: the pathogen information markup language. Bioinformatics. 2005;21:116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]
- 32.McDonald CJ, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin. Chem. 2003;49:624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 33.Wurtz R, Cameron BJ. Electronic laboratory reporting for the infectious diseases physician and clinical microbiologist. Clin. Infect. Dis. 2005;40:1638–1643. doi: 10.1086/429904. [DOI] [PubMed] [Google Scholar]
- 34.Le Novere N, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nature Biotech. 2005;23:1509–1515. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 35.Orchard S, et al. Common interchange standards for proteomics data: public availability of tools and schema. Proteomics. 2004;4:490–491. doi: 10.1002/pmic.200300694. [DOI] [PubMed] [Google Scholar]
- 36.Louie B, Mork P, Martin F, Halevy A, Tarczy-Hornoch P. Data integration and genomic medicine. J. Biomed. Inform. 2007;40:5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feil EJ, Enright MC. Analyses of clonality and the evolution of bacterial pathogens. Curr. Opin. Microbiol. 2004;7:308–313. doi: 10.1016/j.mib.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Handl J, Knowles J, Kell DB. Computational cluster validation in post-genomic data analysis. Bioinformatics. 2005;21:3201–3212. doi: 10.1093/bioinformatics/bti517. [DOI] [PubMed] [Google Scholar]
- 41.Wallinga J, Edminds WJ, Kretzschmar M. Perspective: human contact patterns and the spread of airborne infectious diseases. Trends Microbiol. 1999;7:372–377. doi: 10.1016/S0966-842X(99)01546-2. [DOI] [PubMed] [Google Scholar]
- 42.Werner T, Nelson J. Joining high-throughput technology with in silico modelling advances genome-wide screening towards targeted discovery. Brief Funct. Genom. Proteom. 2006;5:32–36. doi: 10.1093/bfgp/ell010. [DOI] [PubMed] [Google Scholar]
- 43.Marcotte EM, et al. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 44.Rachman H, et al. Mycobacterium tuberculosis gene expression profiling within the context of protein networks. Microb. Infect. 2006;8:747–757. doi: 10.1016/j.micinf.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Maiden MC. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 2006;60:561–588. doi: 10.1146/annurev.micro.59.030804.121325. [DOI] [PubMed] [Google Scholar]
- 46.Lisacek F, Cohen-Boulakia S, Appel RD. Proteome bioinformatics II. Bioinformatics for comparative proteomics. Proteomics. 2006;6:5445–5466. doi: 10.1002/pmic.200600275. [DOI] [PubMed] [Google Scholar]
- 47.Achard F, Vaysseix G, Barillot E. XML, bioinformatics and data integration. Bioinformatics. 2001;17:115–125. doi: 10.1093/bioinformatics/17.2.115. [DOI] [PubMed] [Google Scholar]
- 48.Pelegrini M, et al. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol. Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 50.Saminathan B, et al. PulseNet: The molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 2001;7:382–389. doi: 10.3201/eid0703.017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donofrio NM, et al. PACLIMS: a component LIM system for high throughput functional genomic analysis. BMC Bioinformatics. 2005;6:94. doi: 10.1186/1471-2105-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao L-J, Li M-X, Guo Y-F, Xu F-H, Li J-L, Deng H-W. SNPP: automating large scale SNP genotype data management. Bioinformatics. 2005;21:266–268. doi: 10.1093/bioinformatics/bth486. [DOI] [PubMed] [Google Scholar]
- 53.Birkholtz L-M, et al. Integration and mining of malaria molecular, functional and pharmacological data: how far are we from a chemogenomic knowledge space? Malaria J. 2006;5:110. doi: 10.1186/1475-2875-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol. 2000;3:445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 55.Boguski MS, McIntosh MW. Biomedical informatics for proteomics. Nature. 2003;422:233–237. doi: 10.1038/nature01515. [DOI] [PubMed] [Google Scholar]
- 56.DeGruttola V, et al. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir. Ther. 2000;5:41–48. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 57.De Luca A, et al. Variable prediction of antiretroviral treatment outcome by different systems for interpreting genotypic human immunodeficiency virus type 1 drug resistance. J. Infect. Dis. 2003;187:1934–1943. doi: 10.1086/375355. [DOI] [PubMed] [Google Scholar]
- 58.Lloyd-Smith JO, et al. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malik ANJ, Godfrey-Faussett P. Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect. Dis. 2005;5:174–183. doi: 10.1016/S1473-3099(05)01310-1. [DOI] [PubMed] [Google Scholar]
- 60.Sintchenko V, Gilbert GL. Utility of genotyping of Mycobacterium tuberculosis in the contact investigation: a decision analysis. Tuberculosis. 2007;87:176–184. doi: 10.1016/j.tube.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 61.Gardner SP. Ontologies and semantic data integration. Drug Disc. Today:Biosilico. 2005;10:1001–1007. doi: 10.1016/S1359-6446(05)03504-X. [DOI] [PubMed] [Google Scholar]
- 62.Ecker DJ, et al. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl Acad. Sci. USA. 2005;102:8012–8017. doi: 10.1073/pnas.0409920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mellmann A, et al. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PloS Medicine. 2006;3:e3. doi: 10.1371/journal.pmed.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gierl L, Schmidt R. Geomedical warning system against epidemic. Int. J. Hyg. Environ. Health. 2005;208:287–297. doi: 10.1016/j.ijheh.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 65.King HC, Sinha AA. Gene expression profile analysis by DNA microarrays: promise and pitfalls. J. Am. Med. Assoc. 2001;286:2280–2288. doi: 10.1001/jama.286.18.2280. [DOI] [PubMed] [Google Scholar]
- 66.Geretti A. HIV-1 subtypes: epidemiology and significance for HIV management. Curr. Opin. Infect. Dis. 2006;19:1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 67.Berman JJ. Pathology data integration with eXtensible Markup Language. Hum. Pathol. 2005;36:139–145. doi: 10.1016/j.humpath.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 68.Pawlotsky J-M. Therapy of hepatitis C: from empiricism to eradication. Hepatol. 2006;43:S207–S220. doi: 10.1002/hep.21064. [DOI] [PubMed] [Google Scholar]
- 69.Scott JD, Gretch DR. Molecular diagnostics of hepatitis C virus infection: a systematic review. J. Am. Med. Assoc. 2007;297:724–732. doi: 10.1001/jama.297.7.724. [DOI] [PubMed] [Google Scholar]
- 70.Wohnsland A, Hofmann WP, Sarrazin C. Viral determinants of resistance to treatment in patients with hepatitis C. Clin. Microbiol. Rev. 2007;20:23–38. doi: 10.1128/CMR.00010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Public Microbial Databases on the web (PDF 73 kb)


