Abstract
The ability to attach different functional moieties to a molecular building block1,2 could lead to applications in nanoelectronics3, nanophotonics4, intelligent sensing5 and drug delivery6,7. The building unit needs to be both multivalent and anisotropic, and although many anisotropic building blocks have been created1,8,9,10,11,12, these have not been universally applicable. Recently, DNA has been used to generate various nanostructures13,14,15,16,17 or hybrid systems18,19,20,21,22,23,24,25, and as a generic building block for various applications26,27,28,29,30. Here, we report the creation of anisotropic, branched and crosslinkable building blocks (ABC monomers) from which multifunctional nanoarchitectures have been assembled. In particular, we demonstrate a target-driven polymerization process in which polymers are generated only in the presence of a specific DNA molecule, leading to highly sensitive pathogen detection. Using this monomer system, we have also designed a biocompatible nanovector that delivers both drugs and tracers simultaneously. Our approach provides a general yet versatile route towards the creation of a range of multifunctional nanoarchitectures.
Supplementary information
The online version of this article (doi:10.1038/nnano.2009.93) contains supplementary material, which is available to authorized users.
Multifunctional nanostructures have been created from DNA-based anisotropic, branched and crosslinkable building blocks — ABC monomers. Using these monomers, a target-driven polymerization process is demonstrated where polymers are generated only in the presence of a specific DNA molecule, leading to highly sensitive pathogen detection. The nanoarchitectures can also be used to deliver drugs to cells.
Supplementary information
The online version of this article (doi:10.1038/nnano.2009.93) contains supplementary material, which is available to authorized users.
Main
In this work we have developed a modular (‘plug-and-play’) approach to constructing a multifunctional nanoarchitecture from an ABC monomer building block. To create the ABC monomer, branched, Y-shaped DNA (Y-DNA) conjugated with different moieties served as the modular donor and X-shaped DNA (X-DNA) was used as the core acceptor molecule (Fig. 1). Anisotropy was achieved by designing unique DNA sequences at the end of each branch of the core X-DNA. Because of the freedom in synthesizing any desired DNA sequence at any position, the diversity of anisotropy (from different sequences) is practically unlimited. Multifunctionality was realized by the specific connection of different donor Y-DNA onto the same acceptor X-DNA. Single-stranded bridge DNA with a sequence complementary to both acceptor and donor sequences was used to connect the Y-DNA to the X-DNA. To simplify the notation, we named each unique end-sequence as West, North and East based on the branch orientation (designated W, N and E for Y-DNA and W′, N′ and E′ for X-DNA, with subscript i used sometimes to distinguish different sequences; Fig. 1). The bridge DNAs were notated correspondingly (w, n and e in lower-case letters).
Figure 1. Schematic of the assembly of an ABC monomer and target-driven photo-polymerization.
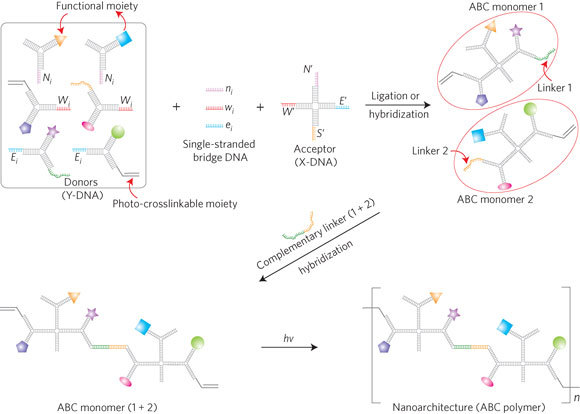
Multiple moieties are conjugated on Y-DNA donors. The functionalized Y-DNAs are then connected to corresponding end-sequences of acceptor X-DNA to form the ABC monomer. The bridge DNA glues the donor and acceptor together. To perform photo-polymerization, ABC monomers 1 and 2 are linked together through hybridization with a complementary linker DNA. These ABC monomers (1 + 2) are then photo-polymerized.
Because of the built-in modularity, the type of moiety that can be linked to the core is virtually unrestricted. To build up a multifunctional polymer from ABC monomers, we designed different modules separately including DNA capture probes, fluorescent dyes, quantum dots and gold nanoparticles (AuNPs, not shown in this paper; for DNA sequences see Supplementary Tables S1, S2). In addition, by incorporating a polyethylene glycol monoacrylate (PEGA) moiety onto the Y-DNA (see Supplementary Figs S1–S3), we further introduced a new capability for photo-crosslinking (for sequences, see Supplementary Table S3). The resulting ABC monomers, now with multiple and varying functional groups, can thus be photo-polymerized using the built-in photoreactive groups.
To demonstrate the precision of our approach, we created ABC monomers that carried different fluorescence dyes at a predetermined ratio within a single monomer (for scheme, see Supplementary Fig. S4a). The monomers were characterized using gel electrophoresis (Fig. 2a, b; see also Supplementary Fig. S4b), which revealed that the fluorescence colours of the ABC monomers corresponded to the various combinations of donor Y-DNAs. Without adding the bridge DNA, only unconnected green donor Y-DNAs were shown (Fig. 2b, lane 1). Unreacted acceptor X-DNAs, which carried no fluorescence dyes, were dark and undetectable but could be seen upon staining with a DNA-specific dye (ethidium bromide; see Supplementary Fig. S4b). After the w bridge DNA was added to the reaction, a single green band with retarded electrophoretic mobility could be observed (Fig. 2b, lane 2), suggesting that (i) the green fluorescence dye was exclusively attached to the West branch of the X-DNA (X–W′) and (ii) the product was monodispersed. Similar results were obtained with fluorescence dye attached to the East and North branches of the X-DNA after adding e and n bridge DNA (Fig. 2b, lanes 3 and 4, respectively). To link two different configurations of fluorescence dyes simultaneously onto two different branches of a single X-DNA, we added two bridge DNAs simultaneously (Fig. 2b, lanes 5–7). By adding all three bridge DNAs, all three different donor Y-DNAs were anisotropically linked onto the X-DNA simultaneously (Fig. 2b, lane 8) in a controlled fashion. This approach is robust and efficient; the estimated yield of ABC monomers is close to 90%, as estimated by densitometry.
Figure 2. ABC monomers with precisely positioned fluorescence dyes and nanoparticles.
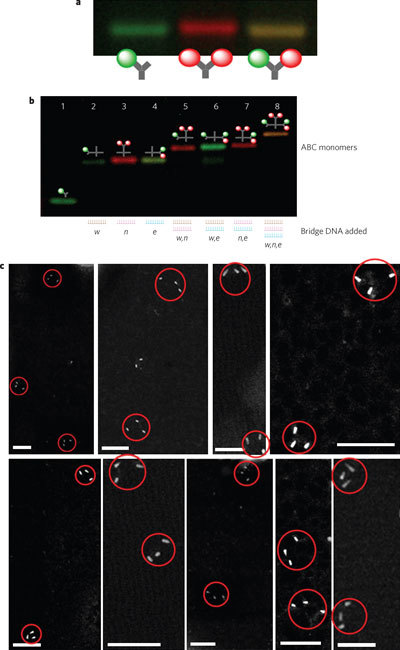
a, Gel electrophoresis image of three donor Y-DNAs conjugated with one green fluorescence dye (1G), two red fluorescence dyes (2R), and one green and one red fluorescence dye (1G1R). b, Gel electrophoresis image of ABC monomers with different configurations. The resulting ABC monomers show different colour bands determined by the donor Y-DNAs, which are connected to the X-DNA by a specific bridge DNA. Unrelated donors, if any, are shown at the bottom, because they cannot be connected to acceptor X-DNA without the presence of a corresponding bridge DNA (lane 1). Lanes 5, 6 and 7 represent w + n (1G + 2R), w + e (1G + 1G1R) and n + e (2R + 1G1R), respectively. c, Annular dark-field STEM image of ABC monomers with three quantum dots. Scale bars, 100 nm. Although DNA molecules themselves were not visible in STEM, the distance between two quantum dots should be close to the length of the DNA between them, which was ∼41 nm (considering a rigid DNA model with 0.34 nm per base pair; for detailed calculations see Supplementary Fig. S7a). The measured distance based on STEM annular dark-field images was 44.8 ± 7.3 nm.
To characterize the ABC monomer at the individual molecule level, we generated donor Y-DNAs tethered with two different colours of quantum dots to be linked onto X-DNA (see Supplementary Figs S5, S6a). Quantum dots were selected because they were individually detectable by means of scanning transmission electron microscopy (STEM). In addition, quantum dots were also visible at both the bulk and solution scales through their intrinsic fluorescence (see Supplementary Fig. S6b, c). As seen in Fig. 2c, the three quantum dots within one ABC monomer were obviously tethered together and were at the expected distance from one another (see Supplementary Fig. S7). These images reveal that different multi-moieties were individually and anisotropically placed accurately within one ABC monomer.
To synthesize multifunctional nanoarchitectures from an ABC monomer we designed each ABC monomer to have two quantum dots with three different colour configurations (2G, 1G1R or 2R; see Supplementary Fig. S8), one photo-responsive PEGA moiety, and one single-stranded oligonucleotide probe that was complementary to a specific pathogen DNA such as SARS coronavirus, Ebola virus or Bacillus anthracis (this unique DNA is termed the ‘capture probe’; see 1A and 1B in Fig. 3a and Supplementary Table S3). Consequently, an ABC dimer formed only in the presence of a targeted pathogen DNA because the pathogen DNA now served as a complementary linker DNA to link the two ABC monomers 1A and 1B together (see 2 in Fig. 3a). Following a short UV illumination (10 min), photo-crosslinking occurred with the dimers but not with the monomers (Fig. 3a, species 3). In other words, we have successfully developed a ‘target-driven’ polymerization where polymers (diblock co-polymers in this case) can only be synthesized in the presence of a specific target DNA. To the best of our knowledge, this is the first time that a polymerization process has been designed to be dependent on the presence of a pathogen DNA. This novel polymerization approach can be used to generate detectable polymers for pathogen sensing.
Figure 3. Photo-polymerization and characterization of ABC monomers.
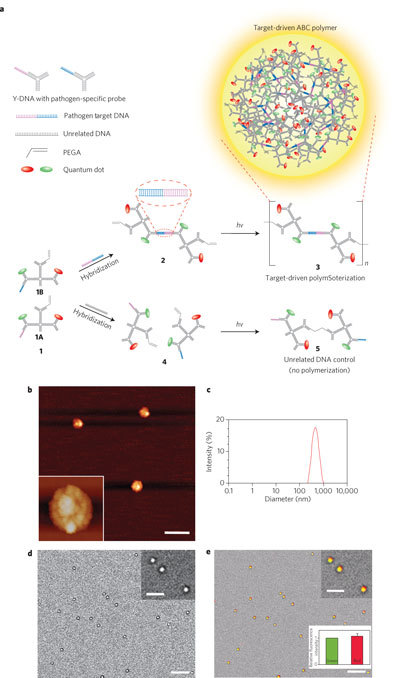
a, Schematic of target-driven polymerization. b, Atomic force microscopy (AFM) image of polymeric spheres. The inset shows a higher magnification image. Scale bar, 1 µm. c, Size distribution of polymeric spheres obtained using dynamic light scattering measurements. d, Bright-field optical microscopy images after photo-crosslinking. e, Overlay image obtained using both bright-field and fluorescence microscopy. Error bars represent standard deviations from three replicates. Scale bar, 5 µm (inset, 2 µm).
We used atomic force microscopy (AFM) to image the morphology of the pathogen-target-driven polymer. The image in Fig. 3b shows that the polymerized ABC monomers were nearly spherical. Dynamic light scattering (DLS) measurement revealed that the average diameter was 410 ± 70 nm (Fig. 3c). Interestingly, the polymerization efficiency was very high, as shown by the lack of a monomer peak post-polymerization (see Supplementary Fig. S9a). In addition, the diameters of the resulting polymeric spheres can be further controlled by varying the concentrations of the starting ABC monomer (see Supplementary Fig. S9b). Because each monomer contained two specific quantum dots with a predetermined ratio (1G1R), the formation of the target-driven polymers was further evaluated by both bright-field and epi-fluorescence optical microscopy (Fig. 3d, e). The overlay of the bright-field and fluorescence images in Fig. 3e confirms again that the polymeric spheres were generated from ABC monomers due to the unique 1G1R fluorescence ratio.
As well as being able to link several hundred quantum dots together and thus effectively amplify signals from a single target-binding event, each target-driven polymer also contains a unique fluorescence code with a specific ratio of green and red; this feature makes it possible to detect multiple targets simultaneously. Here, three different pathogen DNAs (SARS, Ebola and Anthrax) were used as target DNAs (for sequences see Supplementary Table S3). We first added an ‘unknown’ DNA, and after target-driven photo-polymerization the resultant spheres had a colour ratio of 1G1R (Fig. 4a). By referring to the pre-assigned fluorescence codes (see Supplementary Table S4), this result indicated that the ‘unknown’ DNA was the SARS DNA. Similarly, Anthrax and Ebola DNA were detected with 1G3R and 4G0R, respectively (Fig. 4b, c). The detection was highly specific: in the presence of an unrelated DNA, no polymerized spheres were observed (Fig. 4d) because polymerization could not occur with only mono-PEGA–ABC monomers. Interestingly, we found that the concentrations of polymeric spheres (that is, the number per unit area obtained from microscopy images) were linearly proportional to the log of concentrations of target DNA (Fig. 4e). Thus we can easily detect the presence and approximate concentration of the pathogen DNA by simply counting the polymeric spheres under the microscope. The estimated detection limit was 100 fM under current conditions (Fig. 4e). This method also allows for a wide dynamic range (four orders of magnitude) of detection.
Figure 4. Detection of pathogen DNA by means of target-driven polymerization of ABC monomers.
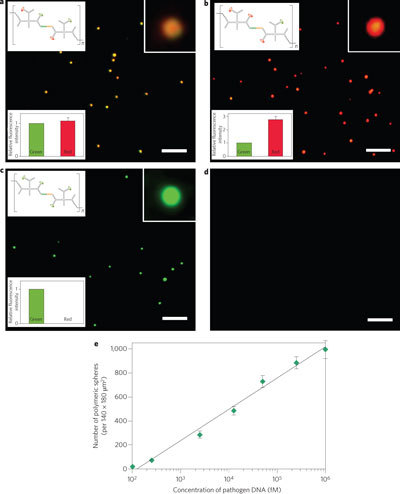
a–c, Fluorescence microscopic images of target-driven polymers with pathogen DNAs including SARS, Bacillus anthracis and Ebola, respectively. Orange (1G1R) polymeric spheres were generated from ABC monomers that were dimerized by SARS DNA (a). 1G3R and 4G coded polymeric spheres were produced by Bacillus anthracis (b) and Ebola pathogen (c) DNA, respectively. d, By incubating 1G1R ABC monomers with an unrelated pathogen DNA, no polymerized DNA materials were observed. Scale bars, 5 µm. e, Relationship between the number of polymerized spheres per 140 × 180 µm2 and the concentration of pathogen DNA (for determining the sensitivity of detection through target-driven polymerization). The coefficient of variation (CV) of the assay ranged from 5.6 to 10.4%, depending on the concentration of the target pathogen DNA (for more details see Supplementary Table S5). Error bars represent standard deviations from three replicates.
In addition to detecting pathogen DNA, the ABC polymeric spheres can also serve as multi-drug delivery vectors due to their intrinsic multivalency and anisotropicity. The built-in DNA scaffolds also provide an ideal interface for nucleic acid-based drugs (see Supplementary Fig. S10). Polymerized spheres were delivered to cells with both model drugs (oligonucleotide (ODN) and siRNA) and tracers (quantum dots). Microscopy images revealed that similarly sized spheres were internalized by HeLa cells (Fig. 5a–c). The cellular uptake was attributed to endocytosis, because no fluorescence was observed inside the cells when endocytosis was inhibited (see Supplementary Fig. S11a,b). These results suggest that the cells can internalize these multi-drug carriers without any additional treatment with a transfection reagent. To further understand the uptake mechanisms, we evaluated the cellular uptake in the presence of various endocytosis-specific inhibitors. Internalization of polymeric spheres was severely reduced in cells treated with inhibitor cytochalasin B, suggesting that actin-dependent endocytosis such as phagocytosis and macropinocytosis was the main mechanism (see Supplementary Fig. S12).
Figure 5. Microscopic images of a HeLa cell treated with polymeric spheres at 37 °C overnight and cytotoxicity studies.
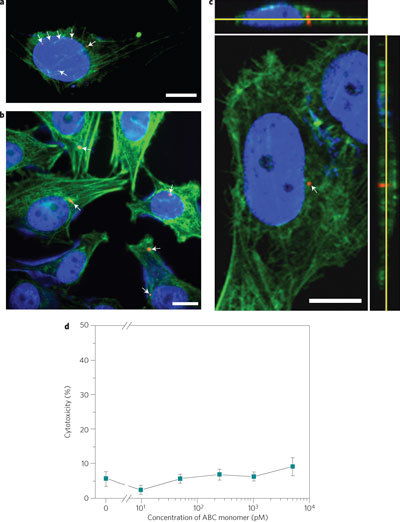
a,b, Fluorescence microscopy images (blue, nuclei; green, actin; red, polymeric spheres). Scale bars, 10 µm. c, Confocal microscopy image with resliced regions of interest. The resliced image shows that polymeric spheres are taken up by HeLa cells. d, Cytotoxicity studies of polymeric spheres. The assay shows cytotoxicity after 36-h exposures of cells to 0.01, 0.05, 0.25, 1 and 5 nM ABC monomers. Error bars represent standard deviations from three replicates.
It is noteworthy that these ABC polymers exhibited little cytotoxicity. After a 36-h treatment with ABC monomers, HeLa cells showed more than 90% cell viability (or less than 10% cytotoxicity was observed; Fig. 5d). Using the conditions reported here, we were able to obtain a delivery efficiency of 25% (data not shown). Although further investigations are needed to optimize delivery efficiency, we have demonstrated that our ABC polymers provide a general route to carry different moieties within one entity for both delivery and tracing.
In summary, we have created anisotropic, branched and crosslinkable monomers that are DNA-based and can be precisely manipulated. Using these ABC monomers, we have also developed a general approach to generate multifunctional nanoarchitectures that cannot be achieved with currently available building blocks. In particular, we have demonstrated for the first time a target-driven polymerization that can be used to rapidly amplify signals to detect pathogens with high specificity and sensitivity. In addition, we have shown that these nanoarchitectures can be used as multi-drug carriers that are readily internalized by cells without any other chemical reagents. It is important to note that our ABC monomer system is designed to be modular. The built-in ‘plug-and-play’ feature coupled with its ‘mix-and-match’ flexibility makes the ABC monomer a versatile platform technology—any other functional groups, both organic and inorganic, that can be conjugated with DNA (or RNA) can be incorporated into the final nanoarchitectures. Thus, we anticipate that our ABC monomers will lead to many possibilities for creating more novel nanostructures and nanomaterials with multiple functionalities.
Methods
Conjugation of PEGA onto Y-DNA
PEGA (0.5 µM, 3,400 Da) was added into a solution (water) containing 5′ amine-modified Y-DNA (0.2 µM). The reaction was carried out overnight at room temperature. Non-reacted amine-modified Y-DNA and PEGA were removed by HPLC equipped with a photodiode array detector (Waters).
Preparation of the ABC monomer and dimer
ABC monomers were synthesized by incubating equimolar quantities of donor Y-DNA and corresponding bridge DNA with X-DNA at 30 °C for 1 h. ABC dimer was assembled by mixing equimolar quantities of each ABC monomer with pathogen DNA in a solution containing 15 mM MgCl2 and 10 mM Tris buffer (pH 8). The mixture was incubated at 30 °C for 30 min.
Formation of the ABC polymer by photo-polymerization
ABC dimers were photo-polymerized with 265 nm UV light (8 mW cm−2) in an aqueous solution of 5 wt% photoinitiator Irgacure (Ciba Geigy) using a UV crosslinker (Spectronics Corporation, XL-1000) for 10 min.
Atomic force microscopy imaging
A 5 µl sample was placed onto the surface of mica (Ted Pella) that was already functionalized by being placed into a 0.1% (v/v) aminopropyltriethoxysilane (APTES, Aldrich) water solution for 15 min, and the sample was allowed to adsorb to the mica surface for ∼20 min. The mica was then rinsed in Milli-Q water. Tapping-mode AFM images were taken in air using a Dimension 3100 AFM (Digital Instruments).
Microscopy of the nanoarchitectures
A 10 µl sample was placed onto the microscope glass and then covered by cover glass (22 × 22 mm2). The DNA nanoarchitectures were observed with a microscope (Olympus IX70) equipped with a ×100 (high-magnification) oil-immersion objective. Images were acquired with MetaMorph image acquisition software. All observations were performed at room temperature.
Cell culture
HeLa cells were cultured at 37 °C with 5% CO2 in Dulbecco's Minimum Essential Medium (DMEM, Mediatech) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 2% penicillin/streptomycin (P/S, Mediatech).
Fluorescence labelling and imaging of cell
Cells (3 × 104) were cultured in each well on Lab-Tek chamber slides (8 wells, Permanox slide, Nunc) for one day. Cells were further cultured in the presence of 20 µl polymeric spheres (2.9 pM) overnight at 37 °C or 4 °C, or 10 min at 37 °C. Cells were washed three times with PBS, then fixed with 4% paraformaldehyde. Actin filaments and nuclei were stained with Alexa Fluor488 phalloidin (Invitrogen) and DAPI (4′,6-diamidino-2-phenylindole) with an antifade reagent (Invitrogen) according to the supplier's protocol. Cross-sectional images of the cell were obtained by confocal microscopy (Zeiss LSM 510 Meta Confocal Microscope).
Cell cytotoxicity evaluation
The cytotoxicity of the polymer spheres (with ABC monomer from 10 pM to 5 nM) was determined after a 36-h incubation with HeLa cells (1 × 104 cell) by measuring the release of lactate dehydrogenase (LDH) from damaged membranes of the cells using CytoTox-One homogeneous membrane integrity assay (Promega).
Supplementary information
Supplementary information (PDF 1392 kb)
Acknowledgements
This work was partially supported by the US Department of Agriculture (USDA) National Research Initiatives (NRI) and the US National Science Foundation (NSF) CAREER award and performed in part at the Nanobiotechnology Center and Cornell Center for Material Research, which are supported by the NSF, Cornell University and industrial affiliates. We thank E. J. Rice, J. C. March, J. S. Kahn, S. Tan and M. Campolongo for proofreading this manuscript.
Author Contributions
J.B.L., Y.H.R. and D.L. conceived and designed the experiments. J.B.L., Y.H.R. and H.F. performed the experiments. J.B.L., Y.H.R., H.F. and D.L. analysed the data. J.B.L., Y.H.R., S.H.U., W.C., P.K., J.J.C. and D.A.M. contributed materials and analysis tools. J.B.L., Y.H.R. and D.L. co-wrote the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Glotzer SC, Solomon MJ. Anisotropy of building blocks and their assembly into complex structures. Nature Mater. 2007;6:557–562. doi: 10.1038/nmat1949. [DOI] [PubMed] [Google Scholar]
- 2.Grill L, et al. Nanoarchitectures by covalent assembly of molecular building blocks. Nature Nanotech. 2007;2:687–691. doi: 10.1038/nnano.2007.346. [DOI] [PubMed] [Google Scholar]
- 3.Javey A, Nam S, Friedman RS, Yan H, Lieber CM. Layer-by-layer assembly of nanowires for three-dimensional, multifunctional electronics. Nano Lett. 2007;7:773–777. doi: 10.1021/nl063056l. [DOI] [PubMed] [Google Scholar]
- 4.Slocik JM, Tam F, Halas NJ, Naik RR. Peptide-assembled optically responsive nanoparticle complexes. Nano Lett. 2007;7:1054–1058. doi: 10.1021/nl070267x. [DOI] [PubMed] [Google Scholar]
- 5.Pregibon DC, Toner M, Doyle PS. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315:1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 6.Torchilin VP. Multifunctional nanocarriers. Adv. Drug Del. Rev. 2006;58:1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Salem AK, Searson PC, Leong KW. Multifunctional nanorods for gene delivery. Nature Mater. 2003;2:668–671. doi: 10.1038/nmat974. [DOI] [PubMed] [Google Scholar]
- 8.Alivisatos P. The use of nanocrystals in biological detection. Nature Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 9.Busby M, Kerschbaumer H, Calzaferri G, De Cola L. Orthogonally bifunctional fluorescent zeolite-L microcrystals. Adv. Mater. 2008;20:1614–1618. doi: 10.1002/adma.200702354. [DOI] [Google Scholar]
- 10.Popovic Z, Otter M, Calzaferri G, De Cola L. Self-assembling living systems with functional nanomaterials. Angew Chem. Int. Ed. 2007;46:6188–6191. doi: 10.1002/anie.200701019. [DOI] [PubMed] [Google Scholar]
- 11.Lee CC, et al. A single dose of doxorubicin-functionalized bow-tie dendrimer cures mice bearing C-26 colon carcinomas. Proc. Natl Acad. Sci. USA. 2006;103:16649–16654. doi: 10.1073/pnas.0607705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CC, Mackay JA, Frechet JM, Szoka FC. Designing dendrimers for biological applications. Nature Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 13.Goodman RP, et al. Reconfigurable, braced, three-dimensional DNA nanostructures. Nature Nanotech. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 14.Venkataraman S, Dirks RM, Rothemund PW, Winfree E, Pierce NA. An autonomous polymerization motor powered by DNA hybridization. Nature Nanotech. 2007;2:490–494. doi: 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- 15.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 16.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 17.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Rosi NL, Wang Y, Huo F, Mirkin CA. Asymmetric functionalization of gold nanoparticles with oligonucleotides. J. Am. Chem. Soc. 2006;128:9286–9287. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu A, et al. Discrete nanostructures of quantum dots/Au with DNA. J. Am. Chem. Soc. 2004;126:10832–10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 20.Feldkamp U, Niemeyer CM. Rational design of DNA nanoarchitectures. Angew Chem. Int. Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J, et al. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C, Liu Y, Yan H. Self-assembled combinatorial encoding nanoarrays for multiplexed biosensing. Nano Lett. 2007;7:507–512. doi: 10.1021/nl062998n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SH, et al. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005;5:729–733. doi: 10.1021/nl050175c. [DOI] [PubMed] [Google Scholar]
- 24.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand–protein binding. Nature Nanotech. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claridge SA, Liang HW, Basu SR, Frechet JM, Alivisatos AP. Isolation of discrete nanoparticle–DNA conjugates for plasmonic applications. Nano Lett. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, et al. Controlled assembly of dendrimer-like DNA. Nature Mater. 2004;3:38–42. doi: 10.1038/nmat1045. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Cu YT, Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nature Biotechnol. 2005;23:885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Um SH, Lee J, Kwon S, Li Y, Luo D. Dendrimer-like DNA (DL-DNA) based fluorescence nanobarcodes. Nature Protoc. 2006;1:995–1000. doi: 10.1038/nprot.2006.141. [DOI] [PubMed] [Google Scholar]
- 29.Um SH, et al. Enzyme-catalysed assembly of DNA hydrogel. Nature Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 30.Cheng W, Park N, Walter MT, Hartman MR, Luo D. Nanopatterning self-assembled nanoparticle superlattices by molding microdroplets. Nature Nanotech. 2008;3:682–690. doi: 10.1038/nnano.2008.279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information (PDF 1392 kb)


