Abstract
Hepatitis C virus (HCV) nonstructural protein 4B (NS4B) is a multi-transmembrane protein, but little is known about how NS4B contributes to HCV replication and tumorigenesis. Its C-terminal domain (CTD) has been shown to associate with intracellular membrane, and we have previously shown that NS4B CTD contains a class I PDZ-binding motif (PBM). Here, we demonstrated that NS4B PBM interacts with the PDZ-containing tumor suppressor protein, Scribble, using immunofluorescence and co-immunoprecipitation assays, and this interaction requires at least three contiguous PDZ domains of Scribble. In addition, NS4B PBM specifically induced Scribble degradation by activating the proteasome-ubiquitin pathway. Similar Scribble degradation was also observed in HCV-infected cells, suggesting NS4B could work in the context of HCV. Finally, NS4B PBM mutants showed reduced colony formation capacity compared with its wild-type counterpart, indicating that NS4B PBM plays important roles in NS4B-mediated cell transformation. Altogether, we provide a mechanism by which NS4B induces cell transformation through its PBM, which specifically interacts with the PDZ domains of Scribble and targets Scribble for degradation.
Keywords: HCV, NS4B, PBM, Ubiquitin, Scribble, Tumorigenesis
Introduction
Hepatitis C virus (HCV) is an enveloped positive single-stranded RNA virus that causes hepatitis C and HCV-related liver diseases including hepatocellular carcinoma (HCC) [1]. Up until now, there is no effective HCV vaccine available and hepatitis C is becoming a serious global health problem that affects 170 to 200 million people worldwide. The current standard of care is the combined use of nucleoside analog ribavirin and pegylated interferon-α; however, the efficacy of this treatment is limited and depends largely on the viral genotypes [2–4].
The HCV genome encodes a polyprotein of about 3010 amino acids, which is cleaved into functional proteins including the mature structural capsid protein (C), envelope proteins (E1 and E2), ion channel (p7), and six nonstructural proteins (NS2-NS5B) [5]. Although the precise mechanism underlying HCV replication process remains largely unclear, most of the HCV nonstructural (NS) proteins are known to be involved in membranous web (MW) and double-membrane vesicles (DMVs) formed by protein–protein and protein–lipid interactions, which is the site of the viral RNA synthesis [6–8]. One of the NS proteins that are indispensable for viral replication is HCV NS4B, which is a hydrophobic protein highly conserved in Flaviviridae family [9]. NS4B expression causes alteration of the endoplasmic reticulum (ER) membrane and formation of MW, which could provide a platform for HCV viral replication [8, 10]. Our previous work has shown that NS4B expression activates unfolded protein response (UPR), ER overload response, and NF-κB pathway in human hepatic cells, which could contribute to HCV replication and pathogenesis [11–13].
HCV NS4B is a 27-KDa protein consisting of an N-terminal domain (aa 1 to 69), C-terminal domain (aa 191 to 261), and a central transmembrane domain (aa 70 to 190). Topology structure studies of NS4B showed that its N-terminal domain comprises two amphipathic α-helices (AH1 and AH2) with several functional properties. Its central domain consists of four transmembrane domains (TMs) and a nucleotide-binding Walker A motif, mediating HCV viral replication and replication-related focus formation [9]. The C-terminal domain has been suggested to be the major membrane-binding domain, but recent studies showed that it is involved in protein–protein and protein–RNA interactions [14, 15]. In addition, the predicted helices and motifs of the C-terminal domain are involved in HCV replication [16].
We have previously analyzed the interaction motifs within NS4B using basic ELM software and found that its C-terminus contains a PDZ-binding motif (PBM) [9], which could bind cellular PDZ-containing proteins such as the membrane protein, Scribble. Scribble is a PDZ-containing protein and has been well known for its roles in cellular polarity [17] and apoptosis [18]. The functions of Scribble have been shown to be modulated by viral proteins [19]. Human T-lymphotropic virus type 1 (HTLV-1) Tax protein affects the localization of Scribble protein and inhibits its activity to attenuate T cell receptor (TCR)-induced activation of nuclear factor of activated T cells (NFAT). Tick-borne encephalitis virus (TBEV) NS5 protein interacts with Scribble to inhibit interferon-mediated JAK-STAT signaling. In addition, high-risk human papillomavirus (HPV) E6 protein binds Scribble protein to cause its degradation through proteasome-mediated proteolysis [19].
HCV NS4B has been reported to transform NIH3T3 cells, but little is known about the underlying mechanism [14]. Here, we showed that NS4B PBM is a key motif that enables NS4B to interact with Scribble protein, and at least three PDZ domains of Scribble are required for this interaction. Furthermore, this interaction leads to scribble degradation through the proteasome-ubiquitin pathway. We also observed this proteasome-dependent Scribble degradation in HCV-infected Huh7.5.1 cells, suggesting that NS4B has some roles for HCV-mediated Scribble degradation. In addition, NS4B PBM confers the ability of NS4B to transform cells. Taken together, our study revealed a novel mechanism by which NS4B contributes to HCV tumorigenesis by targeting tumor suppressor protein, Scribble, for degradation.
Materials and methods
Cells and plasmids
293T, HepG2, Huh7.5.1, and HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10 % fetal bovine serum (FBS) and cultured at 37 °C in a 5 % CO2 incubator. The replicon Con-1 was kindly provided by Prof. Ralf Bartenschlager (University of Heidelberg, Germany). To construct NS4B PBM mutants, the last four amino acids of NS4B were deleted or site-direct-mutated as alanine (AAAA) with appropriate sets of primers. The full-length cDNA of NS4B gene was designed as NS4BWT, the NS4B with last four amino acids deleted as NS4BD, and the NS4B with last four amino acids mutated as NS4BM. These NS4B sequences were inserted into pEGFPC1 and pFLAG-CMV2 plasmids (Invitrogen, USA) to construct pEGFPC1-NS4B, pEGFPC1-NS4BD, and pEGFPC1-NS4BM and pFLAG-CMV2-NS4B, pFLAG-CMV2-NS4BD, and pFLAG-CMV2-NS4BM, respectively. The plasmid pcDNA/Flag-Scribble (kindly provided by Prof. Rice, Baylor College of Medicine, USA) was used as template to amplify all the fragments of Scribble and cloned into indicated mammalian expression vectors. All the constructs were verified by DNA sequencing. The plasmids were transfected into cells at approximately 80 % confluence by Lipofectamine 2000 (Invitrogen, Karlsruhe, USA) in 150-μl DMEM medium without 10 % FBS, according to manufacturers’ instructions.
Immunofluorescence analysis
At 24 h post-transfection, cells were washed with phosphate-buffered saline (PBS) and fixed in 4 % formaldehyde for 15 min at room temperature. Cells were permeabilized in 0.1 % Triton X-100 in PBS, blocked with 3 % FBS in PBS, and then incubated with primary antibody overnight at 4 °C followed by incubation with secondary antibody conjugated to red fluorochromes. The cell nuclei were stained with 1 μg/ml dihydrochloride (Beyotime, China). Cover slips were mounted upside down on slides with suitable volume of ProLong Gold Antifade Reagent (Invitrogen, USA). The cells were examined using an Olympus IX81 fluorescence microscope equipped with the appropriate filter sets and Olympus FluoView Ver.2.0a Viewer software. The antibodies used were anti-Scribble (Santa Cruz Biotechnology, USA), anti-Goat TRITC labeled IgG antibody (KPL, USA).
Co-immunoprecipitation
At 48 h post-transfection, 293T cells were washed in ice-cold PBS and lysed on ice with lysis buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 % Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM NaVO4, 1 μg/ml Leupeptin, 1 mM PMSF). Cell-free lysate was mixed with anti-FLAG agarose (Abmart, China), washed three times with lysis buffer, and boiled in 40 μl of 1× SDS-PAGE loading buffer.
Western blotting analysis
Proteins were separated on SDS-PAGE gel and transferred into PVDF membrane. Membrane was blocked with 5 % nonfat milk and incubated with primary antibodies overnight at 4 °C and secondary antibodies (KPL Company) in 2.5 % nonfat milk at room temperature for 2 h. Proteins were visualized by SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific).
JFH1 production and infection
Plasmid pJFH1 that contains the full-length HCV genotype 2a JFH1 strain cDNA was kindly provided by Professor Wakita (National Institute of Infectious Diseases, Tokyo, Japan). The infectious JFH1 HCV 2a virus was generated, and virus titers were quantified by RT-PCR as previously described [20]. Huh7.5.1 was infected by JFH1 at TCID50 of 100. Cells were harvested at the indicated time, and Scribble expression was analyzed by qRT-PCR and Western blot.
Colony formation assay
At 24 h post-transfection, trypsinized cells were suspended in DMEM medium and subsequently 0.3 % low-melting-point agarose overlaid onto a solidified layer of DMEM medium and 0.6 % agarose in 20-mm plates (1000 cells/plate). Two weeks later, the colonies were stained with 0.005 % crystal violet (Sigma, USA). The colony-forming units (CFU) with more than 100 cells were counted under a light microscope.
Results
HCV NS4B interacts with Scribble
We have previously reported that HCV NS4B contains a PBM domain within its C-terminal domain, which enables it to interact with PDZ domain-containing proteins [9]. As Scribble protein contains four PDZ domains and interacts with the PBM of some viral proteins [19], we hypothesized that HCV NS4B PBM domain could interact with the PDZ domain of Scribble. To test this hypothesis, we co-transfected pEGFP-NS4B and pFLAG-CMV2-Scribble expression vectors into 293T cells and analyzed their co-localization by immunofluorescence. HCV NS4B localized throughout the cytoplasm and has a perinuclear staining (Fig. 1a, second panel), consistent with previous reports about NS4B localization [21, 22]. Most importantly, we found that NS4B co-localized with Scribble (Fig. 1a, second and third panels), indicating an interaction between these two proteins. We also observed the co-localization between NS4B and Scribble in other cells lines including HeLa (Fig. 1a, fourth panel) and HepG2 cells (Fig. 1a, fifth panel), indicating that HCV NS4B interacts with Scribble protein irrespective of cell lines.
Fig. 1.
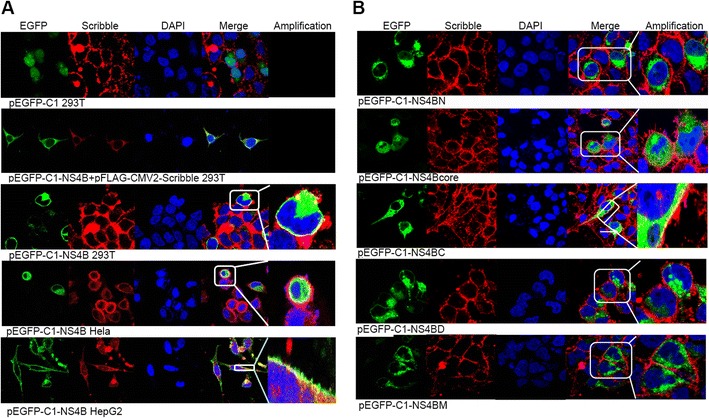
HCV NS4B co-localized with Scribble protein. Cells were fixed for immunofluorescence analysis at 24 h post-transfection. Cell nuclei were stained by DAPI (in blue). The co-localization between NS4B and Scribble was shown in the merged images (fourth columns)
To identify which NS4B domain(s) interacts with Scribble, we constructed vectors that express EGFP-fused NS4B N-terminal domain (EGFP-NS4BN), central domain (EGFP-NS4BCore), and C-terminal domain (EGFP-NS4BC) and transfected them into 293T cells. We found that NS4B N-terminal domain distributes around the nucleus but not nearby the membrane (Fig. 1b, first panel). NS4B central domain displays small punctate or dot-like structures throughout the cells (Fig. 1b, second panel). However, neither NS4B N-terminal domain nor central domain co-localized with Scribble. Only NS4B C-terminal domain showed co-localization with Scribble (Fig. 1b, third panel), indicating that NS4B C-terminal domain is required for NS4B–Scribble interaction.
HCV NS4B interacts with Scribble via its PBM
As PBM resides in NS4B C-terminal domain, to further explore whether NS4B PBM mediates the NS4B–Scribble interaction, we mutated NS4B PBM either by deleting all PBM residues (EGFP-NS4BD) or changing all PBM residues to alanine (EGFP-NS4BM) and transfected these two mutants into 293T cells. We then analyzed the interaction between NS4B and endogenous Scribble protein by immunofluorescence assay. As shown in Fig. 1b, EGFP-NS4BD and EGFP-NS4BM proteins were expressed throughout the cytoplasm, almost the same as EGFP-NS4B, indicating that NS4B PBM domain does not affect its cellular localization. However, there is no overlay area of endogenous Scribble with either EGFP-NS4BD or EGFP-NS4BM (Fig. 1b, fourth and fifth panels), indicating that NS4B PBM domain is required for NS4B–Scribble interaction.
We further analyzed NS4B–Scribble interaction by co-immunoprecipitation (Co-IP). We transfected 293T or HepG2 cells with plasmids expressing FLAG-tagged NS4B (Flag-NS4B) or its PBM mutants (Flag-NS4BD or Flag-NS4BM). NS4B was immunoprecipitated with anti-FLAG resin. As expected, Scribble protein was precipitated with wild-type NS4B but not NS4B PBM mutants (Fig. 2, compare lane 6 to lanes 7 and 8), confirming that NS4B PBM is required for NS4B to interact with Scribble.
Fig. 2.
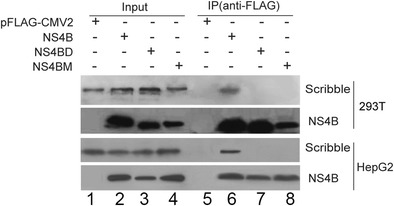
HCV NS4B PBM is required to bind Scribble. Cell extract was incubated with anti-FLAG resin, and the co-immunoprecipitated Scribble was analyzed by Western blot with anti-Scribble antibody
The PDZ domains of Scribble are required for its interaction with NS4B PBM
Scribble consists of three primary domains: leucine-rich repeats (LRRs), LAP-specific domain (LAPSD), and four contiguous PDZ domains (Fig. 3a). Both LRRs and PDZ domains are protein–protein interaction modules. To determine whether HCV NS4B interacts with PDZ domains of Scribble, we transfected the plasmid expressing Myc-tagged four Scribble PDZ domains into 293T cells and examined its interaction with NS4B by Co-IP assay. As shown in Fig. 3b, NS4B binds the four PDZ fragment of Scribble, indicating that the other two domains, LRR and LAPSD, are not required for NS4B–Scribble interaction. To identify which PDZ domains are required for this interaction, we constructed a series of plasmids expressing Myc-tagged Scribble PDZ domains and examined their interactions with NS4B by Co-IP assay. When PDZ1, PDZ2, PDZ3, or PDZ4 was expressed alone, they were not able to interact with NS4B (Fig. 3c–f). Expression of two tandem PDZ domains (PDZ1-2, PDZ2-3, PDZ3-4) of Scribble also failed to interact with NS4B (Fig. 3g–i). Only Scribble that contains the first three (PDZ1-3) or the last three PDZ domains (PDZ2-4) interacts with NS4B (Fig. 3j, k), indicating that the interaction between HCV NS4B PBM and Scribble involves at least three contiguous PDZ domains of Scribble.
Fig. 3.
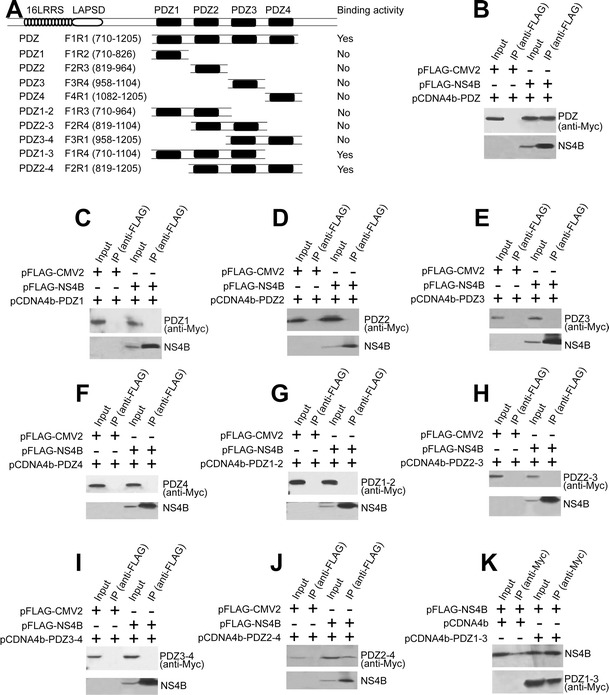
Three adjacent PDZ domains of Scribble are required to bind HCV NS4B. a Diagram showing the structure of Scribble protein. The Myc-tagged PDZ domains of Scribble used in the Co-IP assay were indicated, and the binding activities of Scribble PDZ domains for HCV NS4B were summarized. b–j 293T cells were transfected with pFLAG-CMV2 or pFLAG-CMV2-NS4B and Myc-tagged Scribble PDZ (b), PDZ1 (c), PDZ2 (d), PDZ3 (e), PDZ4 (f), PDZ1-2 (g), PDZ2-3 (h), PDZ3-4 (i), and PDZ2-4 (j). Cell lysates were mixed with anti-FLAG resin, and the precipitates were analyzed by Western blot with indicated antibodies. k 293T cells were co-transfected with pFLAG-CMV2-NS4B, pCDNA4b, or Myc-tagged Scribble PDZ1-3. Cell lysates were mixed with anti-Myc resin, and the precipitates were analyzed by Western blot with indicated antibodies
HCV NS4B induces the degradation of Scribble
As HCV NS4B interacts with Scribble, we next investigated the effects of NS4B on Scribble. Viral proteins have been shown to regulate Scribble either by altering its localization or triggering Scribble degradation [19]. From Fig. 1a, we found that NS4B does not affect the localization of Scribble. Hence, we explored whether NS4B affects the protein levels of Scribble. We transfected different amounts of NS4B expression plasmids into 293T and HepG2 cells and then examined the protein levels of Scribble. Increased expression of NS4B significantly reduced Scribble protein levels in both 293T (Fig. 4a) and HepG2 cells (Fig. 4b). In NS4B-transfected 293T and HepG2 cells, NS4B expression was higher at 48-h post-transfection compared with 24-h post-transfection; however, Scribble protein levels were lower at 48-h post-transfection compared with 24-h post-transfection, revealing a negative correlation between NS4B expression and Scribble protein level. We also examined the impact of NS4B on the other two PDZ-containing proteins, lethal giant larvae2 (Lgl2) and protease-activated receptor-3 (Par3). Our data showed that their intracellular protein levels were not altered by NS4B expression (Fig. 4b–d, f; the Par3 protein data was not shown). These results indicate that NS4B specifically induced Scribble degradation.
Fig. 4.
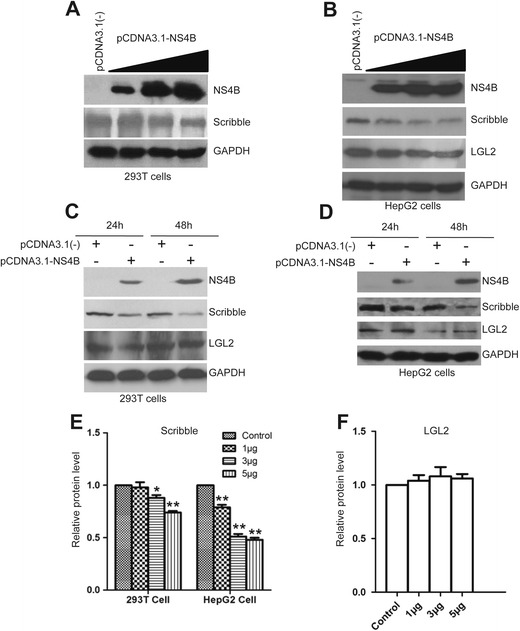
HCV NS4B induced Scribble degradation. a, b 293T (a) and HepG2 (b) cells were transfected with 1, 3, and 5 μg pCDNA3.1-NS4B expression plasmid. Twenty-four hours post-transfection, cell lysates were prepared and analyzed by Western blot with indicated antibodies. pCDNA3.1(−) plasmid was used as a negative control. c, d 293T (c) and HepG2 (d) cells were transfected with 1 μg of pCDNA3.1-NS4B expression plasmid. Twenty-four and 48 h post-transfection, the protein levels of NS4B, Scribble, and LGL2 were analyzed by Western blot. e, f Relative protein levels of Scribble (e) and LGL2 (f) were quantitated with standard deviation (SD). GAPDH levels were used to normalize these two proteins. *P < 0.05; **P < 0.01
HCV NS4B activates the proteasome-ubiquitin pathway to degrade Scribble
To understand how HCV NS4B triggered Scribble degradation, we examined the involvement of the proteasome-ubiquitin pathway. First, we examined the effects of NS4B on the protein levels of ubiquitin-specific peptidase 14 (USP14), a cytoplasmic protein that belongs to the ubiquitin-specific processing family of deubiquitinating enzymes [23, 24]. As shown in Fig. 5a, expression of HCV NS4B led to reduced USP14 protein levels in both 293T cells and HepG2 cells especially at 48-h post-transfection, indicating that HCV NS4B activates the proteasome-ubiquitin pathway.
Fig. 5.
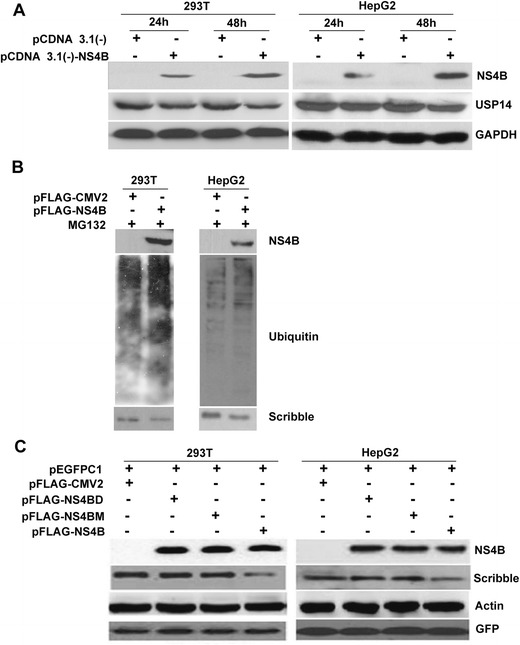
HCV NS4B PBM induced Scribble degradation via the proteasome. a 293T and HepG2 cells were transfected with 1 μg pCDNA3.1(−) or pCDNA3.1-NS4B. Twenty-four and 48 h post-transfection, the protein levels of NS4B, USP14, and GAPDH were analyzed by Western blot. b 293T cells and HepG2 cells were transfected with pFLAG-CMV2 or pFLAG-NS4B, and proteasome inhibitor was added to the cells 5 h before harvesting cells. The ubiquitination levels of Scribble were analyzed by Western blot using anti-ubiquitin antibody. c 293T and HepG2 cells were transfected with full-length NS4B, NS4BD, NS4BM, pFLAG-CMV2, and pEFGPC1. Twenty-four hours post-transfection, cell lysates were prepared and analyzed by Western blot with indicated antibodies
To examine whether the proteasome pathway is involved in the Scribble protein degradation, we performed a series of ubiquitination assays. Cells were transfected with plasmids expressing full-length Flag-tagged NS4B and then treated cells with the proteasome inhibitor, MG132. Our data showed that NS4B induced higher ubiquitination of Scribble than empty plasmids in both 293T cells and HepG2 cells (Fig. 5b), suggesting that NS4B induced Scribble degradation via the proteasome-ubiquitin pathway.
We also transfected NS4B PBM mutants (Flag-NS4BD and Flag-NS4BM) into 293T and HepG2 cells and found that the protein levels of Scribble were not significantly reduced by these two NS4B mutants when compared with the full-length NS4B (Fig. 5c). As a control, the expression of endogenous β-actin and exogenous GFP was not altered (Fig. 5c), indicating that the specific degradation of Scribble requires its interaction with NS4B PBM.
The effect of HCV infection on Scribble expression levels
To analyze whether NS4B triggers Scribble degradation in the context of HCV, we investigated the effect of HCV JFH1 infection on Scribble expression. Huh7.5.1 cells were infected with HCV JFH1, and the expression of Scribble was analyzed by qRT-PCR and Western blotting. Our results showed that Scribble mRNA was not significantly changed by HCV infection (Fig. 6a). However, Scribble protein was decreased in HCV-infected Huh7.5.1 cells compared with mock-infected cells (Fig. 6b), indicating that HCV infection resulted in Scribble degradation. Altogether, these results implied that NS4B works in the context of HCV to trigger Scribble degradation.
Fig. 6.
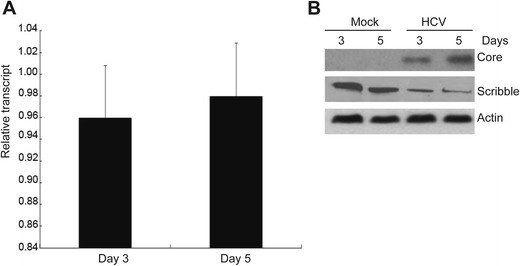
HCV infection induced Scribble protein degradation. Huh7.5.1 cells were infected by HCV JFH1 and harvested at day 3 and day 5. a Total mRNA was extracted, and RT-PCR was performed using primers for Scribble and β-actin. Data were presented as mean ± SE (n = 3, P>0.05). b Cells lysates were subjected to Western blot analysis using antibodies against HCV Core protein, Scribble, and β-actin
The NS4B PBM promotes the colony formation of hepatoma cells by degrading Scribble
As Scribble is a tumor suppressor protein and HCV NS4B PBM targets Scribble for degradation, it is highly possible that NS4B PBM may promote cell transformation by targeting Scribble. To test this hypothesis, we transfected HepG2 cells with NS4B-expressing plasmids. Our results showed that NS4B induced more colonies (increased anchorage-independent growth) than empty vector-transfected cells (Fig. 7), consistent with its reported roles in transformation [14]. Moreover, transfection of full-length NS4B significantly yielded more colonies than its PBM mutants, NS4BD or NS4BM (Fig. 7). These data suggest that NS4B promotes cell transformation primarily by its PBM domain-mediated Scribble degradation.
Fig. 7.
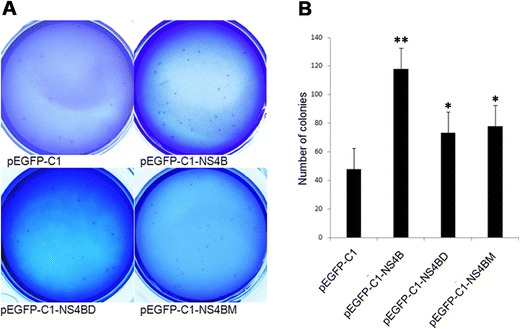
NS4B PBM facilitated HepG2 cells transformation. Soft-agar colony formation assay for HepG2 cells transfected with pEGFP-C1 or pEGFP-C1-NS4B or pEGFP-C1-NS4B or pEGFP-C1-NS4BM. Quantification of foci was counted. Values are means ± SD (n = 3). *P < 0.01,**P < 0.05
Discussion
Cell-derived membrane is the key platform for viral replication and assembly. The interaction between HCV proteins and host cellular proteins plays important roles in HCV life cycle and pathogenesis [25]. HCV NS4B is a highly hydrophobic protein and considered to be the least characterized HCV protein [26]. The major function of NS4B is to induce intracellular host membrane alteration and provide a platform for virus replication [8, 9, 27]. HCV NS4B has also been shown to cause cell transformation [14]. However, it remains to be determined about the underlying mechanism. In this study, we provided a plausible mechanism that NS4B induces cell transformation by degrading the tumor-suppressor protein, Scribble, through the interaction between NS4B PBM and Scribble PDZ domains.
Our study provides the first evidence that HCV NS4B interacts with the tumor suppressor protein Scribble via its PBM. Several virus proteins have been shown to contain PBM that can bind PDZ domain(s). These proteins include human adenovirus E4-ORF1, hepatitis B virus (HBV) core protein, influenza A virus NS1 protein, HPV E6 protein, HTLV1 Tax protein, TBEV NS5 protein, Dengue virus (DV) NS5 protein, Rabies G protein, SARS E protein, RhPV E7 protein, and CRPV LE6 and SE6 proteins (reviewed in [19]). Among these proteins, influenza A virus NS1 protein, HPV E6 protein, HTLV1 Tax protein, and TBEV NS5 protein have been shown to interact with Scribble. Here, we provide evidence that HCV NS4B is a novel PBM-containing protein that interacts with Scribble.
The PDZ domain is a protein–protein interaction module that is widespread throughout evolution. Many cellular PDZ proteins are targets for viral proteins including Dlg1, Dlg4, MAGl-1, MAGl-2, MAGl-3, Scribble, PDLIM2, MUPP-1, PATJ, ZO-1, ZO-2, TIP-1, TIP-2/GIPC, Erbin, pro-IL-16, ß1-syntrophin, PDZD8, PTPN4, and PALS1 [19]. In this study, we found that HCV NS4B specifically interacts with Scribble. Moreover, we found that at least three contiguous PDZ domains of Scribble are required for this interaction (Fig. 3). This phenomenon is quite different from the other reported virus proteins (HPV E6, HTLV-1 Tax, and TBEV NS5) that target the cellular PDZ protein [19]. For example, only PDZ domains 4 and 5 of PATJ are required to bind HPV E6 [19]. It is highly likely that the interaction between HCV NS4B and Scribble requires the three contiguous Scribble PDZ domains to form a favorable conformation and cooperative binding among these PDZ domains.
Different viruses employ various strategies to modulate Scribble to facilitate their replication and/or pathogenesis. Some viral proteins alter Scribble localization. The influenza A virus NS1 and HTLV-1 Tax proteins interact with Scribble and result in mislocalization of Scribble into cytoplasmic puncta [19]. The influenza A virus NS1-mediated relocalization of Scribble was thought to inhibit the pro-apoptotic function of Scribble. However, we did not observe any location change of Scribble in NS4B expression cells (Fig. 1a). Other viral proteins regulate Scribble protein level. The HPV E6 binds Scribble and results in its degradation through the proteasome pathway [28–30]. Similar to HPV E6, we found that HCV NS4B specifically induced Scribble protein degradation with little to no effects on the other two PDZ-containing proteins, Lgl2 and Par3. This is the first report regarding the role of HCV NS proteins in regulating Scribble functions by affecting its protein levels. Moreover, HCV NS4B PBM induced Scribble degradation through the ubiquitin-proteasome pathway, indicating that HCV NS4B utilized the same strategy with HPV E6 in regulating Scribble. The TBEV NS5 protein directly interacts with Scribble and causes the inhibition of the JAK/STAT pathway by an unclear mechanism [19]. It is unknown whether HCV NS4B affects the JAK/STAT pathway.
The NS4B PBM–Scribble PDZ interaction is important for colony formation of transfected cells, indicating that NS4B PBM contributes to HCV pathogenesis and cellular transformation by inducing Scribble degradation. Interestingly, colony formation assay also showed that in the cells expressing the NS4B PBM mutants, more colonies were formed compared to empty vector-transfected cells, suggesting that there are additional unknown mechanisms whereby NS4B promotes transformed cell growth in addition to PBM. As NS4B nucleotide-binding motif (NBM) has been reported to induce tumor transformation [14], it is highly likely that both NBM and PBM confer NS4B the potential to promote tumor transformation.
Taken together, we identified an important motif, PBM, within NS4B and characterized its functions in facilitating tumor cell transformation through binding the PDZ domains of tumor suppressor, Scribble, and inducing Scribble degradation via the proteasome-ubiquitin pathway. Our findings reveal a novel mechanism by which NS4B contributes to HCV pathogenesis and tumorigenesis.
Acknowledgments
This work was supported by Scientific Research Funding for Returned Scholars of Ministry of Education (no. 2012940) and by Natural Science Foundation of Guangdong Province (no. 2014A030313726). We thank Prof. Ralf Bartenschlager, University of Heidelberg, Germany, for providing the subreplicon Con-1 and Prof. Andrew Rice for providing the plasmid pcDNA/Flag-Scribble.
Abbreviations
- HCV
Hepatitis C virus
- HCC
Hepatocellular carcinoma
- NS4B
Nonstructural protein 4B
- PBM
PDZ-binding motif
- CTD
C-terminal domain
- NS
Nonstructural
- MW
Membranous web
- DMVs
Double-membrane vesicles
- TMs
Transmembrane domains
- HTLV-1
Human T-lymphotropic virus type 1
- TCR
T cell receptor
- NFAT
Nuclear factor of activated T cells
- HPV
Human papillomavirus
- TBEV
Tick-borne encephalitis virus
- LRRs
Leucine-rich repeats
- LAPSD
LAP-specific domain
- TCID50
Tissue culture infective dose 50
- CFU
Colony-forming units
- Lgl2
Lethal giant larvae2
- Par3
Protease-activated receptor-3
- USP14
Ubiquitin-specific peptidase 14
- NBM
Nucleotide-binding motif
- HBV
Hepatitis B virus
- DV
Dengue virus
Compliance with ethical standards
Conflicts of interest
None
Footnotes
Bo Hu and Shanshan Li contributed equally to this work.
Contributor Information
Xianzhang Huang, Email: huangxianzhanggd@126.com.
Yi Zheng, Email: zhengyi1205@126.com.
References
- 1.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 2.Halliday J, Klenerman P, Barnes E. Vaccination for hepatitis C virus: closing in on an evasive target. Expert Rev Vaccines. 2011;10:659–672. doi: 10.1586/erv.11.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendt A, Adhoute X, Castellani P, et al. Chronic hepatitis C: future treatment. J Clin Pharmacol. 2014;6:1–17. doi: 10.2147/CPAA.S30338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrova M, Imbert I. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-5414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gosert R, Egger D, Lohmann V, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Yu X, Guo Y, Kong L. Interaction networks of hepatitis C virus NS4B: implications for antiviral therapy. Cell Microbiol. 2012;14:994–1002. doi: 10.1111/j.1462-5822.2012.01773.x. [DOI] [PubMed] [Google Scholar]
- 10.Egger D, Wolk B, Gosert R, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Ye L, Yu X, et al. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappa B activation. Virology. 2009;391:257–264. doi: 10.1016/j.virol.2009.06.039. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Gao B, Ye L, et al. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol. 2005;43:529–536. [PubMed] [Google Scholar]
- 13.Kong L, Li S, Huang M, et al. The roles of endoplasmic reticulum overload response induced by HCV and NS4B protein in human hepatocyte viability and virus replication. PLoS One. 2015;10(4):e0123190. doi: 10.1371/journal.pone.0123190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einav S, Sklan EH, Moon HM, et al. The nucleotide binding motif of hepatitis C virus NS4B can mediate cellular transformation and tumor formation without Ha-ras co-transfection. Hepotology. 2008;47:827–835. doi: 10.1002/hep.22108. [DOI] [PubMed] [Google Scholar]
- 15.Jones DM, Patel AH, Targett-Adams P, McLauchlan J. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol. 2009;83:2163–2177. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dvory-Sobol H, Pang PS, Glenn JS. The future of HCV therapy: NS4B as an antiviral target. Viruses. 2010;2:2481–2492. doi: 10.3390/v2112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Assemat E, Bazellieres E, Pallesi-Pocachard E, Le-Bivic A, Massey-Harroche D. Polarity complex proteins. Biochim Biophys Acta. 1778;2008:614–630. doi: 10.1016/j.bbamem.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 18.Zhan L, Rosenberg A, Bergami KC, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javier RT, Rice AP. Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J Virol. 2011;85:11544–11556. doi: 10.1128/JVI.05410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 21.Hugle T, Fehrmann F, Bieck E, et al. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 22.Lundin M, Monne M, Widell A, Von-Heijne G, Persson MA. Topology of the membrane- associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee BH, Lee MJ, Park S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome- associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10:R110–003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Hage N, Luo G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol. 2003;84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 26.Liefhebber JM, Brandt BW, Broer R, Spaan WJ, van Leeuwen HC. Hepatitis C virus NS4B carboxy terminal domain is a membrane binding domain. Virol J. 2009;6:62. doi: 10.1186/1743-422X-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol. 2013;11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massimi P, Shai A, Lambert P, Banks L. HPV E6 degradation of p53 and PDZ containing substrates in an E6AP null background. Oncogene. 2008;27:1800–1804. doi: 10.1038/sj.onc.1210810. [DOI] [PubMed] [Google Scholar]
- 29.Latorre IJ, Roh MH, Frese KK, Weiss RS, Margolis B, Javier RT. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J Cell Sci. 2005;118:4283–4293. doi: 10.1242/jcs.02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storrs CH, Silverstein SJ. PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6. J Virol. 2007;81:4080–4090. doi: 10.1128/JVI.02545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


