The presence of nucleic acids in the cytosol alerts the cell to viral infection or damaged self. The oligoadenylate synthase (OAS) proteins and cyclic GMP–AMP synthase (cGAS) are enzymes that detect this danger and promote antiviral immunity. Recent structural studies reveal that these enzymes have a common mechanism of action and probably the same evolutionary origin.
Supplementary information
The online version of this article (doi:10.1038/nri3719) contains supplementary material, which is available to authorized users.
Subject terms: Innate immunity
Abstract
Recent discoveries in the field of innate immunity have highlighted the existence of a family of nucleic acid-sensing proteins that have similar structural and functional properties. These include the well-known oligoadenylate synthase (OAS) family proteins and the recently identified OAS homologue cyclic GMP–AMP (cGAMP) synthase (cGAS). The OAS proteins and cGAS are template-independent nucleotidyltransferases that, once activated by double-stranded nucleic acids in the cytosol, produce unique classes of 2′–5′-linked second messenger molecules, which — through distinct mechanisms — have crucial antiviral functions. 2′–5′-linked oligoadenylates limit viral propagation through the activation of the enzyme RNase L, which degrades host and viral RNA, and 2′–5′-linked cGAMP activates downstream signalling pathways to induce de novo antiviral gene expression. In this Progress article, we describe the striking functional and structural similarities between OAS proteins and cGAS, and highlight their roles in antiviral immunity.
Supplementary information
The online version of this article (doi:10.1038/nri3719) contains supplementary material, which is available to authorized users.
Main
To recognize potentially harmful signals, the innate immune system has evolved a conserved set of receptors — known as pattern recognition receptors (PRRs) — that can detect microbial pathogens through the presence of microorganism-associated molecular patterns (MAMPs). Certain PRRs sense MAMPs that are truly (structurally) non-self, whereas others sense the non-self origin of molecules by their presence in compartments that are normally devoid of them1. Viral infection can be sensed by the recognition of virus-derived nucleic acids through structural features that are not found in self nucleic acids. For example, unmodified, fully base-paired 5′-triphosphorylated RNAs activate the cytosolic RNA helicase retinoic acid-inducible gene I (RIG-I), whereas long double-stranded RNAs (dsRNAs) are detected by melanoma differentiation-associated gene 5 (MDA5). By contrast, Toll-like receptors (TLRs) can sense RNA and DNA molecules of both exogenous and endogenous origin, although they show preferences for microbial nucleic acids. In this regard, the mislocalization of nucleic acids in the endolysosomal compartment acts as a signal of non-self origin (as reviewed in Ref. 2).
In this Progress article, we describe the recent studies that have revealed the existence of a new family of cytosolic nucleic acid-sensing proteins that includes the well-known dsRNA-sensing 2′–5′-oligoadenylate synthase (OAS) proteins and the DNA sensor cyclic GMP–AMP (cGAMP) synthase (cGAS). cGAS functions in a classical PRR pathway that monitors the cytosol for the presence of DNA, and that triggers type I interferon (IFN) production and antiviral gene expression through activation of stimulator of IFN genes (STING) (Fig. 1). By contrast, OAS proteins function as nucleic acid sensors in a more immediate antiviral restriction pathway by impeding translation3. Despite these different functionalities, a remarkable finding from recent studies is that OAS1 and cGAS share closely related structural and enzymatic features — they possess the same structural fold, they are activated by a similar double-stranded nucleic acid-induced structural switch and they form a nucleotide second messenger that contains an unusual 2′–5′ phosphodiester linkage (Fig. 1).
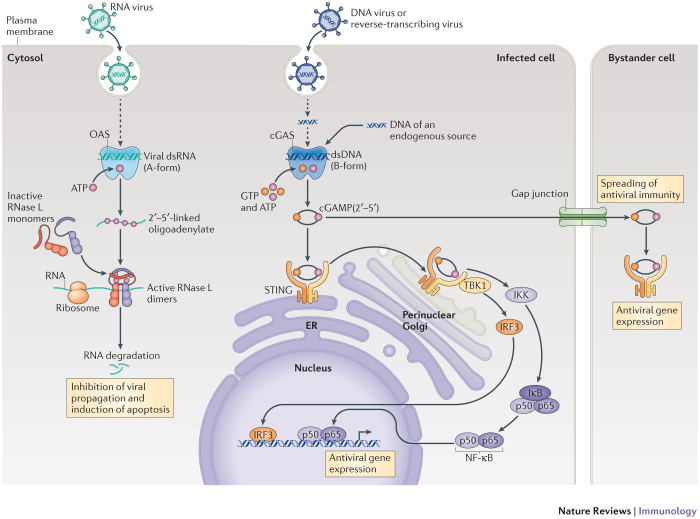
Figure 1. Simplified schematic comparison of the OAS1–RNase L and cGAS–STING axes in innate immune signalling and antiviral defence.
Upon double-stranded RNA (dsRNA) binding, oligoadenylate synthase (OAS) enzymes undergo a conformational switch, which results in their catalytic activity — that is, the synthesis of 2′–5′-linked oligoadenylates using ATP as a substrate. 2′–5′-linked oligoadenylates subsequently act as second messenger molecules by activating the latent endoribonuclease RNase L in the cytoplasm. RNase L then forms a crossed dimer and degrades RNA that is of both cellular and viral origin, leading to the inhibition of viral propagation. On the other hand, cyclic GMP–AMP (cGAMP) synthase (cGAS) is activated by cytosolic B-form dsDNA to synthesize the non-canonical cyclic dinucleotide (CDN) cGAMP(2′–5′) as its second messenger molecule (using the substrates ATP and GTP). cGAMP(2′–5′) binds to and activates the endoplasmic reticulum (ER)-resident receptor stimulator of interferon genes (STING), which subsequently translocates to a perinuclear Golgi compartment where it obtains its signalling-competent state. This results in the activation of transcription factors that initiate antiviral and pro-inflammatory gene expression. At the same time, cGAMP(2′–5′) can also diffuse through gap junctions to initiate antiviral activity in bystander cells. In addition to its role in sensing the endogenous second messenger molecule cGAMP(2′–5′), STING responds to exogenous CDNs that are derived from prokaryotes (not shown). IκB, inhibitor of NF-κB; IKK, IκB kinase complex; IRF3, interferon-regulatory factor 3; NF-κB, nuclear factor-κB; TBK1, TANK-binding kinase 1.
The OAS–RNase L system
The human OAS family. The human OAS family consists of four IFN-regulated genes — namely, OAS1, OAS2, OAS3 and OASL (which encodes OAS-like protein)4,5. The OAS1, OAS2 and OAS3 proteins are all active enzymes that can generate 2′–5′-linked oligoadenylates, whereas OASL is devoid of 2′–5′-linked oligoadenylate synthase activity. Nevertheless, OASL (and Oasl2 in mice) has potent antiviral activity6, which can be ascribed to its positive regulatory role in RIG-I signalling7. By contrast, mouse Oasl1 has been shown to negatively regulate antiviral immunity by inhibiting the translation of IFN-regulatory factor 7 (IRF7)8.
2′–5′-linked oligoadenylates are second messengers that activate RNase L. The OAS proteins sense viral dsRNA and synthesize 2′–5′-linked oligoadenylates, which are second messengers that activate RNase L (Fig. 1). All three human OAS isoforms can be activated by dsRNA in vitro, although the precise in vivo activators are unknown. The 2′–5′-linked oligoadenylates bind to RNase L, which dimerizes and degrades cellular and viral RNA9,10. The structural mechanisms of RNase L activation by 2′–5′-linked oligoadenylates and its dimer formation have recently been described11,12. The full activation of the OAS system in virally infected cells leads to the inhibition of protein synthesis and induces apoptosis, and therefore interferes with the production of new viruses13.
Activation of the OAS–RNase L system limits the replication of many different viruses, in particular, positive-strand viruses — such as picornaviruses, flaviviruses and alphaviruses3,14,15,16 — which is in line with the notion that these viruses display large amounts of dsRNA during their life cycle (as reviewed in Ref. 15). Indirect evidence for the importance of the OAS–RNase L system in restricting viral propagation is provided by the existence of virus-encoded inhibitors of this pathway. For example, the coronavirus mouse hepatitis virus encodes non-structural protein 2 (NS2), which can degrade 2′–5′-linked oligoadenylates17. Similarly, several picornaviruses have mechanisms that antagonize RNase L18,19,20. Although the OAS–RNase L pathway seems to inhibit a wide variety of viruses, this inhibition is rarely complete and is often redundant with other pathways in vitro. Therefore, the OAS–RNase L pathway probably cooperates with other translational inhibitors that are induced during viral infection in vivo to efficiently prevent the production of viral proteins in infected cells.
The cGAS–STING axis
Intracellular DNA sensing. In 2000, TLR9 was identified as the first bona fide DNA-sensing PRR21. TLR9, which is localized in endolysosomal compartments, detects the presence of DNA with unmethylated CpG-containing motifs22. However, TLR9-deficient cells still mount antiviral immune responses following cytosolic DNA challenge23,24,25,26. During the search for the underlying cytosolic DNA sensor (or sensors), it was noted that AT-rich dsDNA is transcribed by RNA polymerase III to form an immunostimulatory RNA that triggers the RIG-I pathway27,28. However, most DNA molecules — including those that are derived from microorganisms — do not initiate RNA polymerase III-dependent transcription, which suggested the existence of at least one additional sensing pathway.
cGAS functions upstream of STING. While carrying out functional cDNA screens to characterize new IFN-inducing molecules, several groups identified the endoplasmic reticulum (ER)-resident protein STING, which turned out to be a crucial factor for the sensing of cytosolic DNA29,30,31. STING-deficient cells or animals showed a severely impaired antiviral immune response following cytosolic DNA delivery (with the notable exception of AT-rich DNA) or DNA virus infection29,32. In addition, STING was shown to be a direct receptor for prokaryotic cyclic dinucleotides (CDNs)33 (Box 1). The crystal structure of cyclic di-GMP (c-di-GMP) bound to the carboxy-terminal domain of STING demonstrated that a preformed STING dimer provides a highly complementary V-shaped binding pocket for CDNs34,35,36,37,38. The dual role of STING as a direct receptor for CDNs and an indirect DNA sensor remained puzzling until the recent discovery of the cytoplasmic nucleotidyltransferase cGAS and its product cGAMP39,40. Indeed, cGAS proved to be the elusive cytoplasmic DNA receptor that functions upstream of STING. Upon DNA binding, cGAS catalyses the synthesis of cGAMP, which, in turn, binds to and activates STING. This exciting finding identified a role for a CDN-dependent signalling process in metazoans (Fig. 1). Surprisingly, the cGAS-derived cGAMP molecule was found to have a mixed phosphodiester linkage. Unlike known prokaryotic CDNs, cGAMP contains a 2′–5′ and a 3′–5′ phosphodiester linkage between its two ring-forming nucleotides, thus constituting >Gp(2′–5′)Ap(3′–5′)> or cGAMP(2′–5′)41,42,43. This finding extended the existing family of 2′–5′-linked antiviral biomolecules — which now encompasses both 2′–5′-linked CDNs, as well as 2′–5′-linked oligoadenylates — and supported a functional relationship between cGAS and the OAS system. Of note, human STING is more responsive to cGAMP(2′–5′) than to cGAMP(3′–5′), which also translates into greater antiviral activity of the 2′–5′ phosphodiester-containing CDNs43,44. This might be due to a greater affinity of human STING for cGAMP(2′–5′)44, although this model has been questioned45.
cGAS in antimicrobial immunity. Studies using cGAS-deficient cells and animals have highlighted the pivotal and non-redundant role of the cGAS–STING axis in detecting cytosolic DNA. cGAS-deficient cells show a markedly compromised antiviral immune response following challenge with various synthetic DNA molecules, DNA viruses or reverse-transcribing viruses46,47. Although the natural cGAS-stimulatory DNA species have not yet been studied in the context of microbial infection, dsDNA species that originate from the viral genome or from viral replication intermediates are candidates. DNA that is derived from bacteria might also be sensed by cGAS. Several bacterial species that replicate inside or outside the cytoplasm have been shown to trigger cytokine production in a STING-dependent manner32,48,49. However, given the dual role of STING as a sensor for prokaryotic CDNs and endogenous cGAMP, it remains to be determined whether these microbial pathogens are indeed sensed by cGAS. As some of these bacteria can trigger the activation of absent in melanoma 2 (AIM2) — which is a bona fide cytosolic DNA sensor — a cytosolic cGAS ligand should, in principle, be available50,51,52.
cGAS as a sensor for endogenous DNA. cGAS seems to function as a general dsDNA sensor without a preference for microbial DNA. This suggests that the cGAS–STING pathway might erroneously sense endogenous DNA species that have gained access to the cytosol. In this regard, it is noteworthy that several inflammatory disorders have been described in which the failure to keep the cytoplasm clear of PRR-stimulatory nucleic acids forms the mechanistic basis of disease (as reviewed in Refs 53, 54). For example, in the context of the deficiency or decreased activity of the 3′ repair exonuclease TREX1, endogenous DNA species that presumably originate from reverse-transcribed cDNA elements accumulate in the cytoplasm55. This, in turn, triggers a cell-autonomous antiviral immune response that is associated with the production of type I IFNs and can result in severe autoimmune disease pathology in affected patients or the respective mouse model. Knocking out the genes encoding STING or cGAS completely abrogates the spontaneous induction of antiviral gene expression in TREX1-deficient cells, and TREX1-deficient mice can be rescued from lethal autoimmunity by deleting the gene that encodes STING56,57. In light of these findings, it is likely that the cGAS–STING axis is also involved in other inflammatory conditions, in which endogenous DNA erroneously gains access to the cytoplasm.
Somewhat unexpectedly, cGAS was also shown to contribute to the innate control of the positive-strand RNA virus West Nile virus58. In this case, it seems that cGAS is not directly involved in sensing the virus itself but instead contributes to a tonic type I IFN response that is required to facilitate the primary response to the virus. This low but constitutive type I IFN response could be triggered by endogenous DNA species that are sensed by cGAS under steady-state conditions, during which they are present at levels that are below the threshold required for triggering disease pathology. At the same time, in the context of cell damage that is inflicted by viral propagation or cell stress, it is possible that endogenous DNA species act as ligands for cGAS. In line with this hypothesis, it is interesting to note that several reports have ascribed the adjuvanticity of cell damage to the release and sensing of endogenous DNA species59,60.
Box 1: Cyclic dinucleotides in prokaryotes and metazoans.
3′–5′,3′–5′-linked cyclic dinucleotides (CDNs) are common second messengers in bacteria. The first described and best-studied CDN is cyclic di-GMP (c-di-GMP)80. C-di-GMP is produced from GTP precursors by enzymes with GGDEF domains. From its original identification as an allosteric regulator of bacterial cellulose synthase more than 25 years ago, it is now known that the intracellular and local extracellular concentration of c-di-GMPs regulates numerous processes in bacteria. It also became evident that c-di-GMP is immunostimulatory81,82 and can inhibit cancer cell growth83, which suggested that this class of molecules has therapeutic potential. More recently, synthases for c-di-AMP and cyclic GMP–AMP (cGAMP) have been reported, expanding the range of prokaryotic CDNs. Key features of the currently known CDNs are listed below.
C-di-GMP: >Gp(3′–5′)Ap(3′–5′)>
• Produced by the GGDEF domain and hydrolysed by EAL and HD-GYP domains, which are found in bacterial proteins
• Allosterically regulates a diverse class of receptors and riboswitches
• Controls complex processes such as biofilm formation, virulence, cell morphology, cell cycle and differentiation
C-di-AMP: >Ap(3′–5′)Ap(3′–5′)>
• Identified as a product of a bacterial DNA damage sensor (known as DisA)67
• Produced by DAC domains, which are found in proteins in both bacteria and archaea
• Regulates cell wall metabolism, osmotic stress response and sporulation
cGAMP(3′–5′): >Gp(3′–5′)Ap(3′–5′)>
• Identified as the product of a dinucleotide cyclase in Vibrio cholerae (known as DncV)66
• DncV downregulates V. cholerae chemotaxis, is required for intestinal colonization and is implicated in virulence
cGAMP(2′–5′): >Gp(2′–5′)Ap(3′–5′)>
• Identified as a product of the metazoan cGAMP synthase (cGAS)
• Activates the type I interferon response via the stimulator of interferon genes (STING)
• Can diffuse through gap junctions to activate bystander cells
Mechanism of cGAS and OAS activation
OAS proteins and cGAS are structurally related nucleotidyltransferases. Structural studies have shown that OAS proteins and cGAS have a highly similar fold41,61,62,63 (Fig. 2). OAS proteins belong to the class of template-independent polymerases that includes poly(A) polymerase, for example64,65. Enzymes of this family have a common two-lobed catalytic core (Fig. 2a) and they transfer a 'donor' nucleotide triphosphate to the 2′-OH or 3′-OH of an 'acceptor' nucleotide. In the case of OAS1, the acceptor is the 2′-OH of ATP and subsequently of 2′–5′-linked oligoadenylates. In general, the products of these enzymes are linear nucleotides (Fig. 3). However, cGAS generates CDNs rather than linear nucleotides by carrying out a second nucleotidyl transfer reaction39,40,66. In the case of cGAS, the first reaction is the transfer of the donor ATP onto the 2′-OH of the acceptor GTP41,42,43. The resulting pppG(2′–5′)pA is cyclized by an additional link between the α-phosphate of GTP and the 3′-OH of ATP. This two-step mechanism is distinct from that of the broadly distributed bacterial diadenylate or diguanylate cyclases (which have DAC and GGDEF domains, respectively) that are dimers or oligomers, in which each active site binds one ATP or GTP molecule67,68. Here, two opposing active sites carry out two transferase reactions in parallel to form c-di-AMP or c-di-GMP. Although OAS1 and cGAS seem to be closely related in evolutionary terms, no phylogenetic connection is apparent between cGAS and bacterial diadenylate or diguanylate cyclases. Of note, Vibrio cholerae contains a cGAMP(3′–5′) synthase that has sequence homology to OAS proteins66. It will be interesting to clarify the catalytic mechanism of this bacterial enzyme and the potential evolutionary connections to cGAS and OAS proteins.
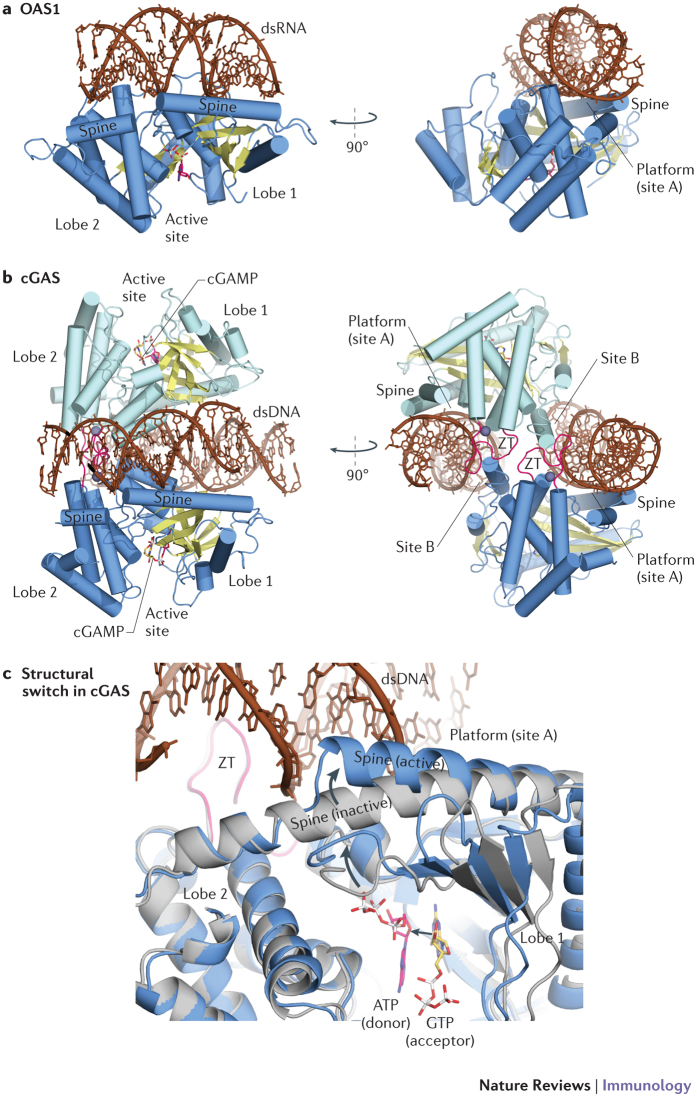
Figure 2. Structures of OAS1 and cGAS.
a | Two views of 2′–5′-oligoadenylate synthase 1 (OAS1) in complex with double-stranded RNA (dsRNA). b | Two views of the cyclic GMP–AMP (cGAMP) synthase (cGAS) dimer (light and dark blue) in complex with dsDNA. c | Close up comparison of cGAS in the apo form (grey) and DNA-bound form (blue). DNA binds to the platform at site A and induces a structural switch in the spine helix (indicated by arrows) and active site loops, thereby facilitating ATP and GTP binding and catalysis. ZT, zinc thumb.
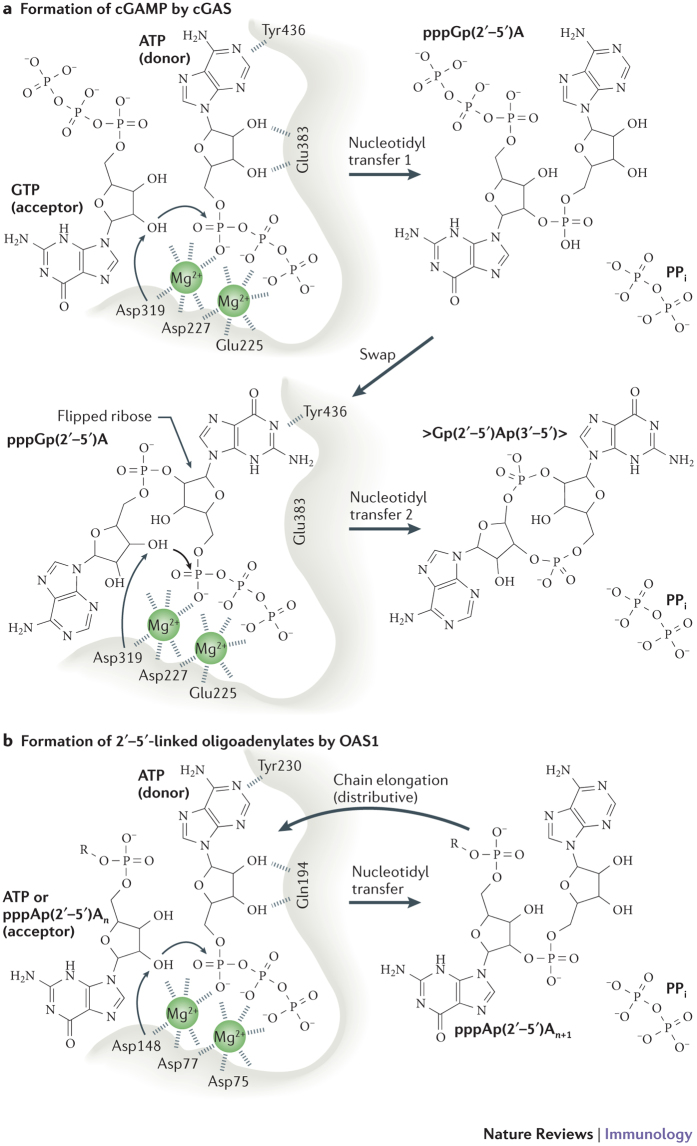
Figure 3. Unified mechanism of nucleotidyl transfer by cGAS and OAS1 proteins.
a | The mechanism for two-step nucleotidyl transfer in the formation of cyclic GMP–AMP (cGAMP) by cGAMP synthase (cGAS). Selected catalytically important residues, as well as two magnesium ions, are indicated. In particular, Glu225 and Asp227 bind the two magnesium ions that are crucial to orient and activate the donor triphosphate moiety. Asp319 polarizes the attacking 2′-OH, and Tyr436 and Glu383 bind base and ribose, respectively. After the first catalytic step, the pppGp(2′–5′)A intermediate needs to dissociate and rebind in reverse order (swap) for the second catalytic step. Human cGAS numbering is used for the amino acid residues shown. b | Nucleotidyl transfer 1 by oligoadenylate synthase 1 (OAS1) is similar to that of cGAS. Instead of a nucleotide swap, as observed in cGAS, the product of nucleotidyl transfer 1 rebinds to the acceptor site only. In the beginning, the acceptor is ATP (in ATP acceptor, 'R' denotes diphosphate group), whereas in subsequent steps, during chain elongation, the acceptor is pppAp(2′–5′)A and longer 2′–5′-linked oligoadenylates ('R' denotes pppAp(2′–5′)A and longer chains). Human OAS1 numbering is used for the amino acid residues shown.
Nucleic acids allosterically activate OAS proteins and cGAS by a structural switch. In the absence of DNA, cGAS is monomeric in vitro and contains a partially unstructured active site that is not properly aligned for binding donor and acceptor nucleotides41,61,62,63 (Fig. 2). Although single nucleotide triphosphates or the cGAMP product can be bound to the cGAS active site in the absence of DNA41,61,69, they do not seem to induce a fully folded and nucleotidyltransferase-competent conformation.
dsDNA binds to cGAS in a sequence-independent manner to a highly positively charged 'platform' on the opposite site of the enzyme with respect to the nucleotidyltransferase active site41,61 (Fig. 2b). A similar surface area of OAS1 binds to dsRNA70 (Fig. 2a), which suggests a common overall activation mechanism. The binding of B-form DNA to cGAS and of A-form RNA to human OAS1 induces a structural switch in the long 'spine' helix that spans both lobes of the nucleotidyltransferase fold in cGAS and OAS1 (Fig. 2c). This structural change modulates the conformation of active site loops for productive binding of GTP and ATP substrates, as well as active site Mg2+ ions. The active conformation is further stabilized in cGAS by dimer formation, in which two dsDNA moieties are sandwiched between two cGAS protomers69,71 (Fig. 2b).
The specificity of cGAS for DNA is, in part, provided by a 'zinc thumb' motif, which is not found in OAS1 and which is important for proper DNA binding and DNA-induced cGAS dimer formation69,71. Besides the zinc thumb, crucial cGAS dimer interactions are mediated by the binding of each dsDNA moiety to the 'platform' (site 'A') of one protomer and a secondary DNA-binding site 'B' on the other protomer. The cooperative binding of DNA to both sites A and B has been shown to be crucial for cGAS activation and stabilizes the conformationally active dimer structure69,71. Currently, there is no evidence that OAS1 forms a similar dimer structure, which suggests that dimer formation is a unique feature of cGAS and may help to generate specificity for DNA rather than RNA.
Although cGAS can bind dsRNA in vitro, this does not lead to its activation61. This could be because only B-form nucleic acids can form appropriate contacts with the platform, the zinc thumb and the secondary binding site in the cGAS dimer. By contrast, the OAS1 platform binds A-form dsRNA70 and single-stranded RNA (ssRNA) but only dsRNA triggers formation of the active conformation. Thus, it seems that OAS proteins and cGAS use double-stranded nucleic acid topology to distinguish between RNA and DNA.
Catalytic mechanism of OAS1 and cGAS. After the DNA-induced structural switch, cGAS can bind ATP and GTP to the active site. Crystal structures of porcine cGAS61 with transferase-trapping active site mutations defined the catalytic step that leads to the formation of the linear intermediate pppGp(2′–5′)A. GTP and ATP bind to the acceptor and donor pockets, respectively. The ATP donor nucleotide and its triphosphate moiety are positioned in such a way that the 2′-OH of GTP can attack the α-phosphate of ATP. Structural studies using wild-type cGAS protein have revealed the subsequent steps of catalysis that occur in the cyclization reaction41. It seems that, before the second catalytic step, the linear dinucleotide intermediate (pppG(2′–5′)pA) needs to rebind in the reversed order — that is, GTP now occupies the donor pocket and 2′–5′-linked AMP occupies the acceptor pocket. In this setting, the 3′-OH of AMP attacks the α-phosphate of GTP, which is in keeping with the universal nucleotidyltransferase mechanism72,73. The proposed reaction mode would result in a cGAMP product with guanine in the donor site and adenine in the acceptor site, which is indeed observed in a crystal structure71.
Although cGAS is very specific for GTP as the acceptor, biochemical experiments have shown that, to some extent, it can tolerate GTP instead of ATP as a donor41,43. Similarly, OAS enzymes selectively use nucleotides with an adenine base and a free 2′-OH as the acceptor substrate, whereas any triphosphate nucleotide can be used as a donor74. It therefore seems that the triphosphate moiety in pppGp(2′–5′)A overrules the donor and acceptor specificities. Further work is necessary to decipher the timescale and nucleotide specificities of the different reaction states. On the basis of the available structures, it is possible to formulate unified activation and catalytic mechanisms for both cGAS and OAS1 (Fig. 3).
An interesting open question is why cGAS forms CDNs, whereas OAS1 forms linear oligomeric chains. One possibility is that OAS1 simply suppresses the swapped binding of a pppA(2′–5′)pA intermediate. Moreover, we need to understand why dsDNA or dsRNA needs to be ≥50 base pairs in length to trigger efficient antiviral immunity through OAS1 and cGAS in vivo27,75,76. According to the crystal structures and biochemical analyses, the binding site of DNA on cGAS or dsRNA on OAS1 is much shorter and, in vitro, cGAS is partially activated by dsDNA that is 16 base pairs long and almost fully activated by dsDNA that is 20 base pairs long71. This length-dependence in vivo cannot be explained by the structure of the cGAS dimer, which binds two shorter DNA molecules side-by-side, rather than one long DNA molecule. Thus, further work is necessary to understand the precise mode by which these two related sensors operate.
Concluding remarks
We now have a general understanding of how viral nucleic acids are sensed by innate immune receptors and which signalling cascades are triggered by these sensors to initiate antiviral immunity. Most recently, a family of evolutionarily, structurally and functionally related nucleotidyltransferases have been defined that sense cytosolic viral RNA and DNA (Box 2). Despite having obtained a good insight into this family of innate sensors, there are several key aspects that are poorly understood and require further clarification.
In particular, the precise ligands for OAS1 and cGAS enzymes need to be determined in vivo. In the case of cGAS, it will be interesting to identify the nature of the endogenous ligands that are sensed in the context of sterile inflammatory conditions or, presumably, in the course of cell damage. In this regard, interaction studies coupled with next generation sequencing could reveal physiological ligands. Moreover, if it can be proved that cGAS cannot distinguish between self and non-self, what happens during cell division when the nuclear envelope breaks down and cGAS is exposed to nuclear DNA?
Further work is also required with respect to the regulation of the cGAS–STING axis, as its signalling output can initiate a self-perpetuating inflammatory response. The systems could be regulated at the level of ligand (DNA or RNA) availability, the activity of the primary sensor (cGAS or OAS proteins), the level of the second messenger (cGAMP or 2′–5′-linked oligoadenylates) or its secondary sensor (STING or RNase L). Regulating the intracellular levels of the second messenger molecule itself seems to be a crucial point of regulation. 2′–5′-linked oligoadenylates were found to undergo rapid degradation when incubated with cellular extracts from resting or IFN-stimulated cells77,78,79. To this end, a 5′ exonuclease and a 2′–5′-specific phosphodiesterase activity had been described, yet the exact nature of the respective enzymes remains controversial. It is possible that enzymes with similar, if not overlapping, functions are present that prevent the accumulation of cGAMP in the cytosol. Moreover, by analogy to other PRR systems, microorganism-encoded virulence factors constitute another promising reservoir to search for negative regulators of cGAMP, and of the cGAS and OAS protein pathways in general.
Box 2: A family of nucleic acid-sensing nucleotidyltransferases.
On the basis of recent structural and biochemical work, shared features of oligoadenylate synthase (OAS) proteins and cyclic GMP–AMP synthase (cGAS) are summarized below.
• OAS and cGAS share a common fold and probably the same evolutionary origin.
• OAS and cGAS bind to double-stranded nucleic acid ligands in a similar manner, which triggers an activating conformational change.
• OAS and cGAS both generate 2′–5′-linked phosphodiesters at the starting point of a signalling cascade.
OAS enzymes and cGAS belong to a much larger family of proteins that have a common nucleotidyltransferase structure84,85. The OAS proteins constitute a well-defined subfamily within this superfamily86,87. Human cGAS (also known as MB21D1) has been placed into the MAB21 family of proteins, which includes MAB21 domain-containing protein 2 (MB21D2), MAB21-like protein 1 (MAB21L1), MAB21L2 and MAB21L3. However, cGAS differs from other vertebrate MAB21 proteins in two important ways. The 'zinc thumb' motif is only present in cGAS and the remaining members of the vertebrate MAB21 family members lack some of the conserved active site residues that are required for catalysis. Interestingly, both insects and chordates have a potential cGAS homologue and MAB21 family member that seems to be catalytically active but does not contain the zinc thumb. It is plausible that OAS proteins and MAB21 proteins diverged earlier in evolution, and that cGAS evolved by the insertion of a zinc thumb motif into a pre-existing catalytically active MAB21-like ancestral protein. To clarify the evolutionary relationships, it will be important to reveal the functional mechanisms of other MAB21 proteins.
Acknowledgements
V.H. is supported by grants from the German Research Foundation (SFB704 and SFB670) and the European Research Council (AIM2 Inflammasome). V.H. and A.A. are members of the Cluster of Excellence ImmunoSensation. R.H. is supported by the Danish Cancer Society and the Danish council for independent research. K.-P.H. is supported by the Bavarian Research Network for Molecular Biosystems (BioSysNet; Bavarian Government), the US National Institutes of Health (grant U19U19AI083025), the Center for Integrated Protein Science Munich (CIPSM) Cluster of Excellence and by a European Research Council Advanced Grant (ATMMACHINE).
Glossary
- A-form RNA
Double-stranded RNA (dsRNA) molecules usually assemble into A-form helices within the cell. A-form helices are right-handed with 11 base pairs per helical turn and the bases are not completely perpendicular to the helical axis. In A-form dsRNA, the major groove is deep and narrow.
- B-form DNA
This is the most commonly found conformation of double-stranded DNA (dsDNA) in nature. Its double helix is right-handed with 10.5 base pairs per helical turn, whereby the bases are oriented perpendicular to the helical axis. B-form dsDNA has a wide major groove and a narrow minor groove.
Biographies
Veit Hornung is the Director of the Institute of Molecular Medicine at the University Hospital, University of Bonn, Germany. His research focuses on the mechanisms of non-self recognition by the innate immune system and its functional consequences. Current research in his laboratory focuses on two main topics — nucleic acid recognition leading to antiviral immunity, and the activation and regulation of inflammasome pathways. Veit Hornung's laboratory homepage
Rune Hartmann received his Ph.D. at Aarhus University, Denmark, and carried out postdoctoral training in the laboratory of Vivien Yee at the Cleveland Clinic Foundation, Ohio, USA. During his postdoctoral studies, he solved the structure of an oligoadenylate synthase (OAS) enzyme, providing the first structural insight into the catalysis of 2′–5′-linked oligonucleotides. He then established his own group, continuing to work on the OAS family of proteins and the type III interferons. Rune Hartmann's laboratory homepage
Andrea Ablasser completed her medical studies at the Ludwig-Maximilians University in Munich, Germany, in 2008. She then joined the laboratory of Veit Hornung at the University of Bonn, Germany, where she has been working on intracellular DNA sensing mechanisms. In 2014, she became an assistant professor at the École Polytechnique Fédérale de Lausanne (EPFL), Switzerland, where she is currently setting up her own laboratory. Andrea Ablasser's homepage
Karl-Peter Hopfner is a professor of biochemistry at the Gene Center of the Ludwig-Maximilians University in Munich, Germany. His laboratory uses integrative structural biology and biochemistry methods to analyse how cells detect and signal the presence of damaged and foreign nucleic acids. Karl-Peter Hopfner's laboratory homepage
PowerPoint slides
Competing interests
The authors declare no competing financial interests.
Contributor Information
Veit Hornung, Email: veit.hornung@uni-bonn.de.
Karl-Peter Hopfner, Email: hopfner@genzentrum.lmu.de.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristiansen H, et al. Extracellular 2′-5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J. Virol. 2010;84:11898–11904. doi: 10.1128/JVI.01003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 5.Melchjorsen J, et al. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interferon Cytokine Res. 2009;29:199–207. doi: 10.1089/jir.2008.0050. [DOI] [PubMed] [Google Scholar]
- 6.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, et al. Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor. Immunity. 2014;40:936–948. doi: 10.1016/j.immuni.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nature Immunol. 2013;14:346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- 9.Dong B, Silverman RH. A bipartite model of 2-5A-dependent RNase L. J. Biol. Chem. 1997;272:22236–22242. doi: 10.1074/jbc.272.35.22236. [DOI] [PubMed] [Google Scholar]
- 10.Clemens MJ, Williams BR. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 11.Han Y, et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science. 2014;343:1244–1248. doi: 10.1126/science.1249845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H, et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol. Cell. 2014;53:221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castelli JC, et al. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 1998;5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 14.Zhou A, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brehin AC, et al. The large form of human 2′,5′-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology. 2009;384:216–222. doi: 10.1016/j.virol.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, et al. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc. Natl Acad. Sci. USA. 2013;110:13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L, et al. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorgeloos F, Jha BK, Silverman RH, Michiels T. Evasion of antiviral innate immunity by Theiler's virus L* protein through direct inhibition of RNase L. PLoS Pathog. 2013;9:e1003474. doi: 10.1371/journal.ppat.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han JQ, et al. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 2007;81:5561–5572. doi: 10.1128/JVI.01857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 22.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 23.Fox BA, et al. Lipofection indirectly increases expression of endogenous major histocompatibility complex class I molecules on tumor cells. Cancer Gene Ther. 1998;5:307–312. [PubMed] [Google Scholar]
- 24.Park JH, et al. Up-regulation of the expression of major histocompatibility complex class I antigens by plasmid DNA transfection in non-hematopoietic cells. FEBS Lett. 1998;436:55–60. doi: 10.1016/s0014-5793(98)01097-7. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda K, et al. Endosomal translocation of vertebrate DNA activates dendritic cells via TLR9-dependent and -independent pathways. J. Immunol. 2005;174:6129–6136. doi: 10.4049/jimmunol.174.10.6129. [DOI] [PubMed] [Google Scholar]
- 26.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nature Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 27.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nature Struct. Mol. Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 35.Ouyang S, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang G, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nature Struct. Mol. Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 37.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature Struct. Mol. Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Q, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao P, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao D, et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer JD, et al. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 2010;40:1545–1551. doi: 10.1002/eji.201040425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nature Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nature Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann. NY Acad. Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 54.Ablasser A, Hertrich C, Wassermann R, Hornung V. Nucleic acid driven sterile inflammation. Clin. Immunol. 2013;147:207–215. doi: 10.1016/j.clim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ablasser A, et al. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 58.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marichal T, et al. DNA released from dying host cells mediates aluminum adjuvant activity. Nature Med. 2011;17:996–1002. doi: 10.1038/nm.2403. [DOI] [PubMed] [Google Scholar]
- 60.Wegmann F, et al. Polyethyleneimine is a potent mucosal adjuvant for viral glycoprotein antigens. Nature Biotech. 2012;30:883–888. doi: 10.1038/nbt.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Civril F, et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato K, et al. Structural and functional analyses of DNA-sensing and immune activation by human cGAS. PloS One. 2013;8:e76983. doi: 10.1371/journal.pone.0076983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 65.Xiong Y, Steitz TA. A story with a good ending: tRNA 3′-end maturation by CCA-adding enzymes. Curr. Opin. Struct. Biol. 2006;16:12–17. doi: 10.1016/j.sbi.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 68.Chan C, et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl Acad. Sci. USA. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl Acad. Sci. USA. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li X, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 73.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 74.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desai SY, Sen GC. Effects of varying lengths of double-stranded RNA on binding and activation of 2′-5′-oligoadenylate synthetase. J. Interferon Cytokine Res. 1997;17:531–536. doi: 10.1089/jir.1997.17.531. [DOI] [PubMed] [Google Scholar]
- 76.Abe T, et al. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams BR, Kerr IM, Gilbert CS, White CN, Ball LA. Synthesis and breakdown of pppA2′p5′A2′p5′A and transient inhibition of protein synthesis in extracts from interferon-treated and control cells. Eur. J. Biochem. 1978;92:455–462. doi: 10.1111/j.1432-1033.1978.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 78.Schmidt A, et al. An interferon-induced phosphodiesterase degrading (2′-5′) oligoisoadenylate and the C-C-A terminus of tRNA. Proc. Natl Acad. Sci. USA. 1979;76:4788–4792. doi: 10.1073/pnas.76.10.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Minks MA, Benvin S, Maroney PA, Baglioni C. Metabolic stability of 2′ 5′oligo (A) and activity of 2′ 5′oligo (A)-dependent endonuclease in extracts of interferon-treated and control HeLa cells. Nucleic Acids Res. 1979;6:767–780. doi: 10.1093/nar/6.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 81.Karaolis DK, et al. Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- 82.Karaolis DK, et al. Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infect. Immun. 2007;75:4942–4950. doi: 10.1128/IAI.01762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem. J. 1995;311:921–927. doi: 10.1042/bj3110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 85.Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pari M, et al. Enzymatically active 2′,5′-oligoadenylate synthetases are widely distributed among Metazoa, including protostome lineage. Biochimie. 2014;97:200–209. doi: 10.1016/j.biochi.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 87.Torralba S, Sojat J, Hartmann R. 2′-5′ Oligoadenylate synthetase shares active site architecture with the archaeal CCA-adding enzyme. Cell. Mol. Life Sci. 2008;65:2613–2620. doi: 10.1007/s00018-008-8164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]