Abstract
Body size plays a key role in production, health, selection, and environmental adaptation of animals, but the genetic basis of body size variation is not clearly understood. Here, we conducted genome-wide association studies (GWAS) of 15 body size traits using autosomal single nucleotide polymorphisms (SNPs) derived from whole-genome sequences of 31 Brahman cattle and 131 Yunling cattle and identified 20 significant loci, which implicated 18 candidate genes. For ischium width, the most significant SNP was assigned to LCORL, a famous gene controlling body size. For chest width, the most significant SNP was located upstream of BMP5, a secreted ligand of transformation growth factor-beta superfamily of proteins involved in bone and cartilage development. Subsequently, we detected selective sweeps in Brahman cattle using integrated Haplotype Score, composite likelihood ratio, and nucleotide diversity. The results showed CNTNAP5 locus associated with hip cross height and LIMCH1 locus associated with forehead size were in selective signals, which were consistent with higher hip cross height and higher forehead size in Brahman cattle compared with Yunling cattle. Our findings provide genetic insights into variation and selection of body size using GWAS and selective signals and will accelerate future efforts aimed at cattle improvement.
Keywords: body size, Brahman cattle, genome-wide association study, selective sweep, Yunling cattle
Introduction
Body size consists of a series of complex quantitative traits, such as body weight, stature, and chest circumference. Body size is an important indicator owing to its correlation with productive, fitness, and adaptive traits. For example, the genetic correlation between stature and gestation length was high (0.49) in Holstein cows (Pozveh et al., 2009). Interestingly, numerous studies have shown that larger stature individuals have a shorter longevity (Zavadilová et al., 2009; Plassais et al., 2019). In addition, Tibetan cattle living in Tibetan Plateau is the shortest in 53 indigenous breeds according to the book about genetic resources of bovines in China (Zhang, 2011), indicating the importance of body size for survival in extreme environments. Therefore, understanding the genetic basis of interindividual variation in body size might not only provide basic material for animal production and health but also help us identify the mechanism of environmental adaptation.
Over the past decades, many genome-wide association studies (GWAS) or mappings of quantitative traits loci (QTL) for body size have identified over 100 loci in the cattle genome. An excellent work identified eight clustered candidate quantitative trait nucleotides for bovine stature at PLAG1 locus (Karim et al., 2011). Another famous QTL containing LCORL gene was found to be associated with body weights (birth, carcass, direct weaning, mature, and yearling weights), calving ease direct, and weaning weight maternal in nine U.S. cattle breeds (Saatchi et al., 2014). Recently, based on 25.4 million imputed whole-genome sequence variants, a meta-analysis of GWAS showed that 163 leading variants explained 13.8% of the phenotypic variance in stature (Bouwman et al., 2018), suggesting that numerous genes of very small effect control body size. The similar findings were also observed in humans (Wood et al., 2014).
In contrast, variants of strong impact could explain greater than 90% of body size variation in domestic dog (Plassais et al., 2019). However, the analyses were based on the difference between breeds, rather than within breeds, suggesting that genomic variants affecting body size were selective targets during breed formation. In fact, the conclusion might be also suitable for domestic cattle. It has been reported that the shoulder height in extinct aurochs was larger than those in its domesticated descendent (Karim et al., 2011). Furthermore, selection for stature has been demonstrated in 5 Bos taurus breeds for LCORL locus and in 10 B. taurus breeds for PLAG1 locus (Bouwman et al., 2018). However, this phenomenon was not explored in Bos indicus.
Yunling cattle is a crossbreed that consists of one-half Brahman cattle, one-fourth Murray Grey cattle, and one-fourth Yunnan indigenous cattle, thus it is an ideal material for exploring the genetic mechanism of complex traits (e.g., body size) in domestic cattle. To investigate the genetic background of body size in Brahman cattle and Yunling cattle, we performed a GWAS for 15 body size traits using autosomal single nucleotide polymorphisms (SNPs) derived from whole-genome sequence. Additionally, to further investigate the relationship between genes identified in GWAS mapping studies and genes with selective signals, we applied the integrated Haplotype Score (iHS) and the composite likelihood ratio (CLR) to identify the selective sweep regions in Brahman cattle. Our results should provide basic materials for those interested in further understanding the genetic mechanism of body size traits and will facilitate the improvement of these traits through genomic selection.
Material and Methods
Care and use of animals
Ethics approval for all animal experiments was granted by the Institutional Animal Care and Use Committee of Northwest A&F University following the recommendation of the Regulations for the Administration of Affairs Concerning Experimental Animals of China.
Phenotype measurements
All animals in the experiment were originally from the core breeding farm of Yunnan Academy of Grassland and Animal Science in China. The experimental animals used were multiparous cows. Our samples consisted of 36 pen-feeding individuals (5 Brahman cattle and 31 Yunling cattle) and 126 free-grazing individuals (26 Brahman cattle and 100 Yunling cattle). The pen-feeding individuals were fed a total mixed ration (TMR), which consisted of 65% grass silage and 35% concentrate on a dry matter basis. The free-grazing individuals ate grass in the meadow from June to November every year and were properly fed above TMR from December to May every year.
Each individual was encouraged to a squeeze crush to measure body size traits and collect ear tissue. Firstly, 15 body size traits were measured using a measuring tape and measuring stick. Secondly, the ear tissue was collected using ear punch and then stored 75% ethanol to extract genomic DNA. The genomic DNA was isolated using the standard phenol/chloroform protocol (Green and Sambrook, 2012). We checked the effect of breed on body size using the general linear model of R program with the consideration of feeding pattern.
Genetic analysis
The genomic DNA was transported to the Novogene Bioinformatics Institute, (Beijing, China) to sequence genome. Paired-end libraries with an insert size of 350 bp were constructed and sequenced using Illumina Novaseq 6000 platform. A total of ~17 billion clean reads were generated and then aligned to the B. taurus reference genome sequence ARS-UCD1.2 using Burrows-Wheeler Aligner - Maximal Exact Match (Li and Durbin, 2009) with default parameters. Potential duplicate reads were removed using Picard tools (“REMOVE_DUPLICATES=true”). The average alignment rate and coverage were 99.55% and 5.61, respectively (Supplementary Table S1). Initial variant site identification was performed using Genome analysis toolkit 3.8 (GATK) (Nekrutenko and Taylor, 2012) (“HaplotypeCaller”, “GenotypeGVCFs,” and “SelectVariants” modules). Subsequently, to exclude false positives during SNP calling, high-quality SNPs were calculated using GATK with following options (“VariantFiltration” module): QualByDepth < 2.0, FisherStrand > 60.0, RMSMappingQuality < 40.0, MQrankSum < −12.5, ReadPosRankSum < −8.0, StrandOddsRatio > 3.0, read depth < 303 (1/3-fold of the total sequencing depth of all the individuals) and read depth > 2,727 (3-fold of the total sequencing depth of all the individuals). A total of ~41 M SNPs were detected using the abovementioned criteria. For GWAS analysis, we also filtered the SNPs using VCFtools (Danecek et al., 2011) with the following parameters: minor allele frequency < 0.05 and missing rate > 0.1, leaving 18,060,230 autosomal SNPs. We also used Beagle to infer haplotype and impute missing alleles (Browning and Browning, 2007) to further carry out GWAS and calculate iHS. In addition, we used smartPCA of EIGENSOFT v5.0 package (Patterson et al., 2006) to estimate the eigenvectors to adjust the population structure in GWAS.
Subsequently, we performed a univariate GWAS by applying a mixed linear model to reveal the potential associations between body size traits and genomic variants using the software GEMMA (Zhou and Stephens, 2012). The statistical model was as follows:
where y denotes the phenotypic values of body size traits, W refers to a covariate matrix (fixed effect: principal component 1 and feeding regime), α denotes a vector of corresponding effects, X denotes the marker genotypes, β refers to the effects of corresponding markers, K refers to the kinship matrix, and μ refers to the effects of corresponding kinship, and ε is a vector of random residuals. The significant and suggestive threshold were set at Pwald = 5 × 10–8 and Pwald = 1 × 10–6, respectively (Bouwman et al., 2018; Schaid et al., 2018). In our model, the principal component 1 and kinship matrix were used to eliminate the fixed effect of genetic ancestry and the random effects of relatedness between individuals, respectively (Price et al., 2010).
After completing GWAS, we used the following strategy to narrow down our findings to obtain corresponding candidate genes. Firstly, the linkage disequilibrium (LD) correlation (r2) between the associated (suggestive and significant) SNPs was calculated using PLINK (Purcell et al., 2007) with the parameters (--r2 --ld-window-kb 1000 --ld-window 99999). Borders of the associated loci were defined according to the LD correlation (r2 > 0.6). To reduce the false positives, the loci with the number of associated SNPs <3 were excluded. Secondly, we focused on the SNPs (leading SNPs) with the smallest Pwald values in the associated loci for body size traits. Finally, we performed functional annotation for suggestive SNPs associated with body size traits using ANNOVAR (Wang et al., 2010) according to the B. taurus reference genome ARS-UCD1.2.
Genome scans for selection in Brahman cattle were performed using the following strategy. Firstly, we used VCFtools (Danecek et al., 2011) to extract the SNPs of 31 Brahman cattle and then calculated the iHS using selscan (Szpiech and Hernandez, 2014). The output results for each SNP were then normalized over all chromosomes using the norm module of selscan (--winsize 40000). This resulted in 62,183 windows across all autosomes. Secondly, based on the empirical frequency spectrum with an allele frequency file combined across all autosomes, we performed the CLR test using SweepFinder2 (DeGiorgio et al., 2016). The grid size was set as 40,000. This resulted in 62,185 local CLR values. Finally, we also calculated nucleotide diversity using VCFtools (--window-pi 40000 --window-pi-step 20000). After excluding the windows <10 SNPs, 124,306 windows were retained. The value in the top 1% of empirical distribution in each algorithm was designated as threshold.
Data availability statement
Sequences are available from GenBank with the BioProject accession number PRJNA555741.
Results
Phenotypic variation in Brahman cattle and Yunling cattle
We measured 15 body size traits in 31 Brahman cattle and 131 Yunling cattle (Table 1). These traits comprised withers height, hip cross height, body length, chest circumference, abdominal circumference, cannon circumference, chest width, chest depth, hip circumference, hip width, ischium width, head length, forehead size, rump length, and body weight. The coefficient of variation ranged from 3.78% to 9.83% (median: 6.75%). We also detected the effect of breed on body size using general linear model with consideration of feeding regime. The results showed that the withers height, hip cross height, hip circumference, forehead size, and cannon circumference in Yunling cattle were significantly lower than those in Brahman cattle (P < 0.05) (Table 2).
Table 1.
Descriptive statistics of 15 body size traits
| Traits | Maximum | Minimum | Mean | SD | CV, % | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|
| Withers height | 143.0 | 118.0 | 129.2 | 4.89 | 3.78 | 0.19 | 2.82 |
| Hip cross height | 152.0 | 121.0 | 133.3 | 5.24 | 3.93 | 0.29 | 3.04 |
| Body length | 176.0 | 126.0 | 156.3 | 8.43 | 5.39 | −0.28 | 3.44 |
| Chest circumference | 250.0 | 177.0 | 197.7 | 10.04 | 5.08 | 1.80 | 10.54 |
| Abdominal circumference | 265.0 | 200.0 | 229.4 | 11.43 | 4.98 | 0.05 | 2.88 |
| Cannon circumference | 23.00 | 12.00 | 18.61 | 1.52 | 8.16 | −0.09 | 4.84 |
| Chest width | 68.00 | 39.00 | 49.02 | 4.60 | 9.38 | 0.52 | 4.05 |
| Chest depth | 88.00 | 47.00 | 68.62 | 5.98 | 8.71 | 0.34 | 4.38 |
| Hip circumference | 136.0 | 95.0 | 112.1 | 7.57 | 6.75 | 0.40 | 3.11 |
| Hip width | 70.00 | 46.00 | 57.96 | 4.90 | 8.45 | 0.19 | 2.64 |
| Ischium width | 28.00 | 18.00 | 22.46 | 2.03 | 9.04 | 0.13 | 2.45 |
| Head length | 57.00 | 40.00 | 48.62 | 2.43 | 5.00 | 0.04 | 3.97 |
| Forehead size | 26.00 | 20.00 | 22.53 | 1.21 | 5.37 | 0.31 | 3.32 |
| Rump length | 65.00 | 43.00 | 50.93 | 3.60 | 7.07 | 0.77 | 4.23 |
| Body weight | 725.0 | 451.5 | 574.7 | 56.47 | 9.83 | 0.27 | 2.50 |
Table 2.
Difference in body size traits between in 31 Brahman cattle and 131 Yunling cattle (least square mean ± standard error)
| Traits | Brahman cattle | Yunling cattle | P-value |
|---|---|---|---|
| Withers height | 132 ± 0.909 | 127 ± 0.473 | 3.37 × 10–4 |
| Hip cross height | 137 ± 0.972 | 133 ± 0.506 | 2.18 × 10–4 |
| Body length | 155 ± 1.607 | 156 ± 0.848 | 0.499 |
| Chest circumference | 200 ± 1.893 | 199 ± 0.987 | 0.543 |
| Abdominal circumference | 229 ± 2.21 | 230 ± 1.15 | 0.647 |
| Cannon circumference | 20.0 ± 0.264 | 18.6 ± 0.139 | 2.29 × 10–6 |
| Chest width | 49.2 ± 0.864 | 49.9 ± 0.450 | 0.451 |
| Chest depth | 68.6 ± 1.148 | 68.5 ± 0.607 | 0.975 |
| Hip circumference | 117 ± 1.413 | 112 ± 0.736 | 4.40 × 10–3 |
| Hip width | 58.2 ± 0.939 | 58.2 ± 0.496 | 0.999 |
| Ischium width | 23.2 ± 0.385 | 22.4 ± 0.203 | 0.058 |
| Head length | 48.4 ± 0.456 | 48.3 ± 0.240 | 0.699 |
| Forehead size | 23.1 ± 0.225 | 22.4 ± 0.119 | 2.08 × 10–3 |
| Rump length | 52.3 ± 0.656 | 51.5 ± 0.346 | 0.224 |
| Body weight | 597 ± 11.25 | 576 ± 6.34 | 0.076 |
GWAS for 15 body size traits
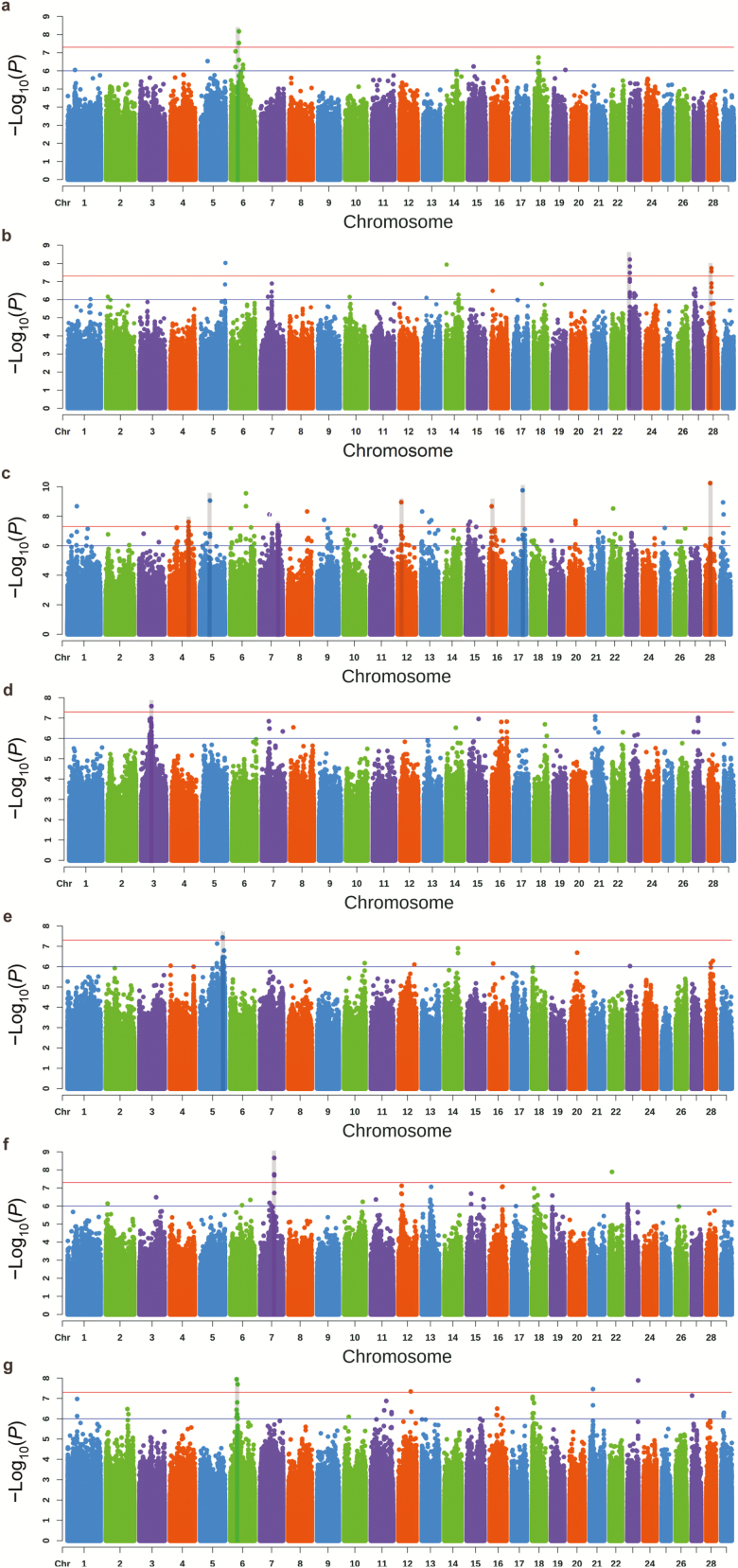
The entire set of GWAS results was presented in Supplementary Table S2. In total, 780 suggestive SNPs (Pwald < 1 × 10–6) and 61 significant SNPs (Pwald < 5 × 10–8) were detected for 15 body size traits. After calculating the LD among suggestive and significant SNPs, a total of 56 suggestively associated loci (Pwald < 1 × 10–6) were remained across 12 out of 15 body size traits (Supplementary Table S3). Among these loci, 20 loci were significant (Pwald < 5 × 10–8) in nine traits (Table 3). There were no suggestively associated loci for bodyweight, abdominal circumference, and hip circumference.
Table 3.
A descriptive summary of GWAS for 15 body size traits (Pwald < 5 × 10–8)
| Associated loci | Leading variants | MAF | −Log10Pwald | Traits | Nearest gene |
|---|---|---|---|---|---|
| 2:74232049-76428806 | 2:76123001 | 0.293 | 8.27 | HCH | CNTNAP5 |
| 2:77234797-79549419 | 2:77705883 | 0.259 | 7.36 | HCH | CNTNAP5 |
| 3:48619698-49685573 | 3:48903215 | 0.454 | 7.59 | CAC | F3 |
| 4:61097399-61109497 | 4:61097399 | 0.194 | 7.48 | HCH | EEPD1 |
| 4:90719918-91208754 | 4:91135749 | 0.068 | 7.61 | CC | GRM8 |
| 5:49425124-49524195 | 5:49522901 | 0.105 | 9.06 | CC | C5H12orf66 |
| 5:108168319-108851195 | 5:108451308 | 0.404 | 7.44 | BL | CACNA1C |
| 6:25768970-31681813 | 6:26182890 | 0.136 | 8.68 | FS | RAP1GDS1 |
| 6:30080542-30099052 | 6:30087434 | 0.090 | 7.94 | CD | PDLIM5 |
| 6:38272742-38308265 | 6:38272802 | 0.148 | 8.18 | IW | LCORL |
| 6:60373051-60432904 | 6:60391823 | 0.080 | 7.46 | FS | LIMCH1 |
| 7:68161294-68306145 | 7:68164002 | 0.201 | 8.66 | RL | HAVCR1 |
| 7:87107538-87300437 | 7:87131231 | 0.380 | 7.38 | CC | CCNH |
| 11:29374698-29493941 | 11:29374698 | 0.065 | 7.35 | FS | MCFD2 |
| 12:17259287-17279977 | 12:17259287 | 0.090 | 8.95 | CC | HTR2A |
| 15:11813883-12824779 | 15:11813883 | 0.315 | 7.42 | CC | Gene desert |
| 17:56698365-56747811 | 17:56698365 | 0.065 | 9.76 | CC | SUDS3 |
| 23:4387170-4419935 | 23:4419914 | 0.049 | 8.01 | CW | BMP5 |
| 28:15174368-15620173 | 28:15620173 | 0.213 | 7.58 | CW | CCDC6 |
| 28:24097580-24111037 | 28:24107275 | 0.080 | 10.24 | CC | CTNNA3 |
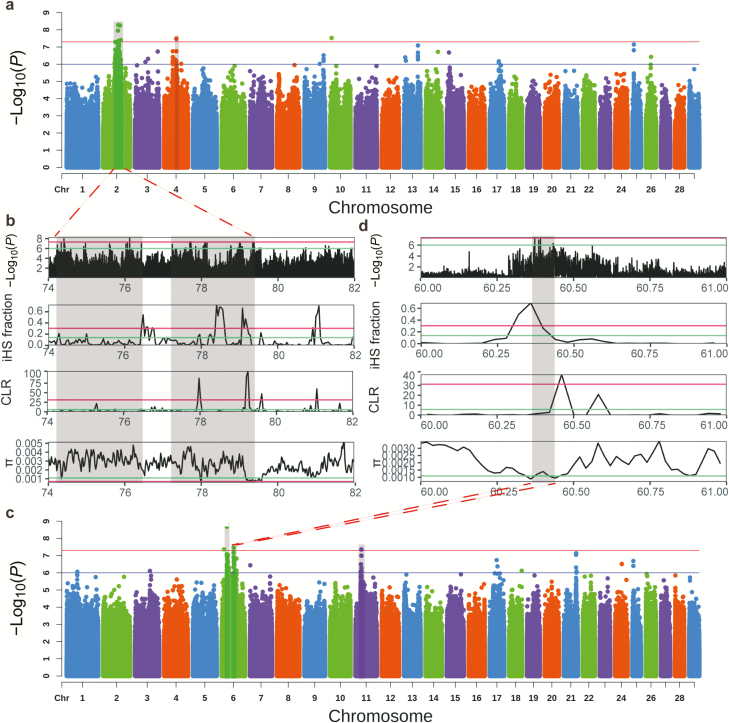
For ischium width, there was only one significantly associated locus, which was observed on Bos taurus autosome 6 (BTA6) and was assigned to LCORL (Figure 1a). For chest width, there were two significantly associated loci, including BMP5 and CCDC6 (Figure 1b). The most significant locus was observed on BTA23 and was located ~50 kb upstream of BMP5. Another associated locus was observed on BTA28 and was located ~50 kb downstream of CCDC6. For chest circumference, seven significantly associated loci were detected on BTA4, BTA5, BTA7, BTA12, BTA15, BTA17, and BTA28, respectively (Figure 1c). The most significant locus was located in the intron of CTNNA3. For cannon circumference, only one significant locus was detected, located on BTA3 and assigned to F3 (Figure 1d). For hip cross height, two most significant loci were observed on BTA2 and were assigned to CNTNAP5 (Figure 2a). Another significant locus was found on BTA4 and was located in the intron of EEPD1. For forehead size, two most significant loci were observed on BTA6 and were assigned to RAP1GDS1 and LIMCH1, respectively (Figure 2c). Another significant locus was observed on BTA11 and assigned to MCFD2. The remaining significant loci were observed on BTA5, BTA7, BTA6, and were associated with body length, rump length, and chest width, and their candidate genes were assigned to CACNA1C, HAVCR1, and PDLIM5, respectively (Figure 1e, f, and g).
Figure 1.
Manhattan plots for ischium width (a), chest width (b), chest circumference (c), cannon circumference (d), body length (e), rump length (f), and chest depth (g). Red line and blue line indicate the significant threshold and suggestive threshold, respectively. The shading of each rectangle shows a significant association locus for body size.
Figure 2.
Manhattan plots for hip cross height (a), forehead size (c), and selective signals on chromosome 2 (chr2:74 to 76 Mb) (b), and on chromosome 6 (chr6:60 to 61 Mb) (d) in Brahman cattle. In Manhattan plots, red line and blue line indicate the significant threshold and suggestive threshold, respectively. In selective signals, red line and blue line indicate top 1% and top 5% threshold in whole genome, respectively. The shading of each rectangle shows a significant association locus for body size. The selective signals were validated by iHS, CLR, and π (nucleotide diversity).
Signals of selection on candidate genes
To identify whether there were selective signatures in Brahman cattle overlapped with aforementioned GWAS signals for body size traits, we calculated iHS, CLR, and nucleotide diversity (π) in Brahman cattle. The entire selective signal results were presented in Supplementary Tables S4–S6. We found CNTNAP5 locus identified from GWAS for hip cross height fell into the 1% tail of iHS score and CLR statistic (Figure 2b). Moreover, LIMCH1 locus identified from GWAS for forehead size showed extreme iHS value and a local reduction in nucleotide diversity (Figure 2d). In suggestive loci, DDX18 locus and TMEFF2 locus (Supplementary Table S3) identified from GWAS for hip cross height appeared in the 1% tail of iHS and CLR in Brahman cattle (Supplementary Tables S4 and S5). In addition, PTBP2 gene, a candidate gene for cannon circumference (Supplementary Table S3), also showed a strong CLR signal (Supplementary Table S5) and a local reduction in nucleotide diversity (Supplementary Table S6).
Discussion
By performing a GWAS using autosomal SNPs derived from the whole-genome sequence in Brahman cattle and its crossbred (Yunling cattle), we identified 56 suggestively associated loci (including 20 significant loci) for 15 body size traits, supporting the complex genetic architecture for cattle body size. These 56 loci implicated 54 candidate genes, including LCORL, BMP5, CNTNAP5, F3, EEPD1, GRM8, C5H12orf66, CACNA1C, RAP1GDS1, PDLIM5, LIMCH1, HAVCR1, CCNH, MCFD2, HTR2A, SUDS3, CCDC6, and CTNNA3. Although our sample size is smaller, the genotype–phenotype interactions with large effects and higher frequency could be detected. Future GWAS mapping with larger sizes might result in the identification of additional variants with small effect and lower frequency. Moreover, although further functional validation experiments could allow us to consolidate our studies, some associated genes (e.g., LCORL, BMP5, CNTNAP5, and LIMCH1) have biologically plausible links to the body size traits.
For ischium width, the most significant SNP was located downstream of LCORL, a famous gene that controlled body size. LCORL encodes a transcription factor that may function in spermatogenesis. In humans, a previous study has demonstrated that polymorphisms in LCORL gene were associated with skeletal size measurements, including hip axis length and spine length (Soranzo et al., 2009). Moreover, many studies have pointed to the correlation of LCORL locus with body size measurements and growth traits in domestic cattle, such as stature (Bouwman et al., 2018), body weights (Saatchi et al., 2014), weaning weight (Weng et al., 2016), average daily gain (Lindholm-Perry et al., 2011). In fact, this correlation also exists in other organisms, such as standard breed height in domestic dog (Plassais et al., 2019), withers height in horse (Boyko et al., 2014), and body weight in sheep (Al-Mamun et al., 2015). Therefore, we concluded that LCORL is a strong candidate gene underpinning the body size of cattle.
For chest width, the most significant SNP was located upstream of BMP5. The gene encodes a secreted ligand of transformation growth factor-beta superfamily of proteins, which plays a role in bone and cartilage development. Previous models of null mutations in the mouse BMP5 gene showed defect in the formation and growth of cartilages (Green and Green, 1942; Green, 1958). It has been reported that a germline mutation at BMP5 locus affected the formation of multiple skeletal features, including misshapen xiphisternum and missing ribs (Ho et al., 2008). These results imply that BMP5 might participate in the body size of cattle through the effect of skeletal formation.
For hip cross height, the most significant locus was observed on BTA2 and contained only the gene CNTNAP5. CNTNAP5 belongs to the neurexin family, members of which play a key role in the vertebrate nervous system as cell adhesion molecules and receptors. In goat, a GWAS of body size traits reported that CNTNAP5 locus was associated with bicoastal diameter (Rahmatalla et al., 2018). Although CNTNAP5 locus was scarcely found to be associated with height, partial deletion of the neurexin 1 (NRXN1), a homologous gene of CNTNAP5, showed short stature in humans (Bermudez-Wagner et al., 2013). Moreover, we also observed CNTNAP5 locus was in selective signals in Brahman cattle (extreme iHS score and CLR statistic, and a reduction in nucleotide diversity). In addition, hip cross height in Brahman cattle was significantly higher than those in Yunling cattle. From above results, we speculated that CNTNAP5 locus might contribute to higher hip cross height in Brahman cattle compared with Yunling cattle.
For forehead size, the second-ranked significant locus was located in the intron of LIMCH1, an actin stress fibers-associated protein which is a paralogous protein with C-terminal LIM domains (Zhang et al., 2019). The mutation of LIM domain showed defect in forebrain and midbrain tissue in mouse, suggesting that LIM domain is essential for head development (Cheah et al., 2000). An experiment in a targeted deletion of LIM1, a homologous gene of LIMCH1, demonstrated that LIM1-null mice lacked anterior head structures (Shawlot and Behringer, 1995). Moreover, we also observed LIMCH1 locus was in selective signals in Brahman cattle (extreme iHS score and a local reduction in nucleotide diversity). In addition, forehead size in Brahman cattle was significantly higher than those in Yunling cattle. Therefore, we concluded that LIMCH1 locus is a putative region underlying higher forehead size in Brahman cattle compared with Yunling cattle.
Conclusion
Using GWAS for body size traits with autosomal SNPs derived from the whole-genome sequence in Brahman cattle and Yunling cattle, we identified 56 suggestively associated loci (including 20 significant loci), which implicated 54 candidate genes. The identification of QTL for body size will provide genomic targets for genetic improvement of body size in domestic cattle. The revelation of some strong candidate genes (e.g., LCORL and BMP5) will help us understand the genetic mechanism and identify causal variants underlying body size variation. Of particular interest is the implication of selective sweep for body size (e.g., CNTNAP5 and LIMCH1), which will advance our understanding of complex demographic history of Brahman cattle.
Supplementary Material
Acknowledgments
This work was supported by the Program of National Beef Cattle and Yak Industrial Technology system (CARS-37), the Program of Yunling Scholar, the Young and Middle-aged Academic Technology Leader Backup Talent Cultivation Program in Yunnan Province, China (2018HB045), and Yunnan Provincial Major S&T Project (2019ZG007 and 2019ZG011).
Glossary
Abbreviations
- BL
body length
- CAC
cannon circumference
- CC
chest circumference
- CD
chest depth
- CLR
composite likelihood ratio
- CW
chest width
- FS
forehead size
- GATK
genome analysis toolkit
- GWAS
genome-wide association studies
- HCH
hip cross height
- iHS
integrated haplotype score
- IW
ischium width
- LD
linkage disequilibrium
- QTL
quantitative traits loci
- RL
rump length
- TMR
total mixed ration
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Al-Mamun H. A., Kwan P., Clark S. A., Ferdosi M. H., Tellam R., and Gondro C.. . 2015. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 47:66. doi: 10.1186/s12711-015-0142-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Wagner K., Jeng L. J., Slavotinek A. M., and Sanford E. F.. . 2013. 2p16.3 microdeletion with partial deletion of the neurexin-1 gene in a female with developmental delays, short stature, and a congenital diaphragmatic hernia. Clin. Dysmorphol. 22:22–24. doi: 10.1097/MCD.0b013e32835b8df2 [DOI] [PubMed] [Google Scholar]
- Bouwman A. C., Daetwyler H. D., Chamberlain A. J., Ponce C. H., Sargolzaei M., Schenkel F. S., Sahana G., Govignon-Gion A., Boitard S., Dolezal M., . et al. 2018. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 50:362–367. doi: 10.1038/s41588-018-0056-5 [DOI] [PubMed] [Google Scholar]
- Boyko A. R., Brooks S. A., Behan-Braman A., Castelhano M., Corey E., Oliveira K. C., Swinburne J. E., Todhunter R. J., Zhang Z., Ainsworth D. M., . et al. 2014. Genomic analysis establishes correlation between growth and laryngeal neuropathy in Thoroughbreds. BMC Genomics. 15:259. doi: 10.1186/1471-2164-15-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., and Browning B. L.. . 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81:1084–1097. doi: 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah S. S., Kwan K. M., and Behringer R. R.. . 2000. Requirement of LIM domains for LIM1 function in mouse head development. Genesis 27:12–21. doi: [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., . et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGiorgio M., Huber C. D., Hubisz M. J., Hellmann I., and Nielsen R.. . 2016. SweepFinder2: increased sensitivity, robustness and flexibility. Bioinformatics 32:1895–1897. doi: 10.1093/bioinformatics/btw051 [DOI] [PubMed] [Google Scholar]
- Green M. C. 1958. Effects of the short ear gene in the mouse on cartilage formation in healing bone fractures. J. Exp. Zool. 137:75–88. doi: 10.1002/jez.1401370105 [DOI] [PubMed] [Google Scholar]
- Green E. L., and Green M. C.. . 1942. The development of three manifestations of the short ear gene in the mouse. J. Morphol. 70(1):1–19. doi: 10.1002/jmor.1050700102 [DOI] [Google Scholar]
- Green M. R., and Sambrook J.. . 2012. Molecular cloning: a laboratory manual. 4th ed. New York (NY):Cold Spring Harbor Laboratory Press. [Google Scholar]
- Ho A. M., Marker P. C., Peng H., Quintero A. J., Kingsley D. M., and Huard J.. . 2008. Dominant negative BMP5 mutation reveals key role of BMPs in skeletal response to mechanical stimulation. BMC Dev. Biol. 8:35. doi: 10.1186/1471-213X-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim L., Takeda H., Lin L., Druet T., Arias J. A., Baurain D., Cambisano N., Davis S. R., Farnir F., Grisart B., . et al. 2011. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43:405–413. doi: 10.1038/ng.814 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. . 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm-Perry A. K., Sexten A. K., Kuehn L. A., Smith T. P., King D. A., Shackelford S. D., Wheeler T. L., Ferrell C. L., Jenkins T. G., Snelling W. M., . et al. 2011. Association, effects and validation of polymorphisms within the NCAPG – LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet. 12:103. doi: 10.1186/1471-2156-12-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A., and Taylor J.. . 2012. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet. 13:667–672. doi: 10.1038/nrg3305 [DOI] [PubMed] [Google Scholar]
- Patterson N., Price A. L., and Reich D.. . 2006. Population structure and eigenanalysis. Plos Genet. 2:e190. doi: 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassais J., Kim J., Davis B. W., Karyadi D. M., Hogan A. N., Harris A. C., Decker B., Parker H. G., and Ostrander E. A.. . 2019. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun. 10:1489. doi: 10.1038/s41467-019-09373-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozveh S. T., Shadparvar A. A., Shahrbabak M. M., and Taromsari M. D.. . 2009. Genetic analysis of reproduction traits and their relationship with conformation traits in Holstein cows. Livest. Sci. 125(1):84–87. doi: 10.1016/j.livsci.2009.02.015 [DOI] [Google Scholar]
- Price A. L., Zaitlen N. A., Reich D., and Patterson N.. . 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11:459–463. doi: 10.1038/nrg2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., . et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmatalla S. A., Arends D., Reissmann M., Wimmers K., Reyer H., and Brockmann G. A.. . 2018. Genome-wide association study of body morphological traits in Sudanese goats. Anim. Genet. 49:478–482. doi: 10.1111/age.12686 [DOI] [PubMed] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., and Garrick D. J.. . 2014. Large-effect pleiotropic or closely linked QTL segregate within and across ten US cattle breeds. BMC Genomics. 15(1):442. doi: 10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid D. J., Chen W., and Larson N. B.. . 2018. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19:491–504. doi: 10.1038/s41576-018-0016-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawlot W., and Behringer R. R.. . 1995. Requirement for Lim1 in head-organizer function. Nature 374:425–430. doi: 10.1038/374425a0 [DOI] [PubMed] [Google Scholar]
- Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J. B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., . et al. 2009. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. Plos Genet. 5:e1000445. doi: 10.1371/journal.pgen.1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech Z. A., and Hernandez R. D.. . 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31:2824–2827. doi: 10.1093/molbev/msu211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., and Hakonarson H.. . 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16):e164–e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Z., Su H., Saatchi M., Lee J., Thomas M. G., Dunkelberger J. R., and Garrick D. J.. . 2016. Genome-wide association study of growth and body composition traits in Brangus beef cattle. Livest. Sci. 183:4–11. doi: 10.1016/j.livsci.2015.11.011 [DOI] [Google Scholar]
- Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H., Gustafsson S., Chu A. Y., Estrada K., Luan J., Kutalik Z., . et al. ; Electronic Medical Records and Genomics (eMEMERGEGE) Consortium; MIGen Consortium; PAGEGE Consortium; LifeLines Cohort Study. 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46:1173–1186. doi: 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadilová L., Němcová E., Štípková M., and Bouška J.. . 2009. Relationships between longevity and conformation traits in Czech Fleckvieh cows. Czech J. Anim. Sci. 54(9):387–394. doi: 10.17221/1685-cjas [DOI] [Google Scholar]
- Zhang Y. 2011. Animal genetic resources in China – bovines (in Chinese). Beijing (China): China Agriculture Press. [Google Scholar]
- Zhang Y., Zhang Y., and Xu H.. . 2019. LIMCH1 suppress the growth of lung cancer by interacting with HUWE1 to sustain p53 stability. Gene 712:143963. doi: 10.1016/j.gene.2019.143963 [DOI] [PubMed] [Google Scholar]
- Zhou X., and Stephens M.. . 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44:821–824. doi: 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences are available from GenBank with the BioProject accession number PRJNA555741.