ADP-ribosylation catalyzed by PARPs and sirtuins is an important post-translation modification. Macrodomain proteins MacroD1 and D2 are now shown to preferentially bind mono-ADP-ribosylated proteins and to act as proximal ADP-ribosylhydrolases. The crystal structure of the MacroD2–ADPr complex suggests a catalytic mechanism for the reaction.
Supplementary information
The online version of this article (doi:10.1038/nsmb.2523) contains supplementary material, which is available to authorized users.
Subject terms: Hydrolases, Post-translational modifications, Structural biology
Abstract
ADP-ribosylation is a reversible post-translational modification with wide-ranging biological functions in all kingdoms of life. A variety of enzymes use NAD+ to transfer either single or multiple ADP-ribose (ADPr) moieties onto distinct amino acid substrates, often in response to DNA damage or other stresses. Poly-ADPr-glycohydrolase readily reverses poly-ADP-ribosylation induced by the DNA-damage sensor PARP1 and other enzymes, but it does not remove the most proximal ADPr linked to the target amino acid. Searches for enzymes capable of fully reversing cellular mono-ADP-ribosylation back to the unmodified state have proved elusive, which leaves a gap in the understanding of this modification. Here, we identify a family of macrodomain enzymes present in viruses, yeast and animals that reverse cellular ADP-ribosylation by acting on mono-ADP-ribosylated substrates. Our discoveries establish the complete reversibility of PARP-catalyzed cellular ADP-ribosylation as a regulatory modification.
Supplementary information
The online version of this article (doi:10.1038/nsmb.2523) contains supplementary material, which is available to authorized users.
Main
ADP-ribosylation is a highly dynamic post-translational modification carried out by a variety of enzymes that share the substrate NAD+ and transfer single or multiple ADPr moieties onto target proteins1,2,3,4,5,6,7,8, often in response to cellular stresses including DNA damage, infection and neuronal signaling. Specific PARP family members (ARTD enzymes), including PARP10, PARP14 and PARP16 as well as certain sirtuins (hSirT4 and hSirT6), bacterial toxins and ecto-mARTs (ARTC enzymes), transfer only a single ADPr unit onto amino acid side chains1,3,6,9,10,11,12. Other ADP-ribosyltransferases can attach the initial ADPr moiety onto an acceptor residue and then add additional ADPr units to the 2′-OH group of the covalently linked ADPr moiety, thus acting as genuine oligo- or poly-ADPr polymerases, the most notable example of which is PARP1, an enzyme involved in DNA-damage signaling and the control of chromatin structure.
ADP-ribosylhydrolase 3 (ARH3) and poly-ADPr-glycohydrolase (PARG) are two enzymes known to efficiently hydrolyze poly-ADPr (PAR)3,6,9,10,11,12,13,14. They have been reported to also act on the most protein-proximal ADPr moiety6, but their activities have not been rigorously tested. More recently, human PARG has been specifically tested on mono-ADP-ribosylated PARP1 E988Q substrate and has shown essentially no activity toward the terminal ADPr14. Two additional enzymatic activities have been reported that can catalyze the removal of the protein-proximal ADPr unit from acceptor proteins. The first is ADP-ribosylhydrolase 1 (ARH1), which specifically hydrolyzes ADP-ribosyl-arginine linkages15. The second is an unidentified ADP-ribosyl lyase activity that was first described in biochemical extracts from rat liver13, which is thought to function on ADP-ribosylated glutamate or lysine residues, releasing a deoxy form of ADPr, but that has not been biochemically identified to date. Indeed, searches for enzymes capable of fully reversing cellular mono-ADP-ribosylation back to the unmodified 'ground state' have proved elusive, despite the textbook assumption of full reversibility for PARP-mediated ADP-ribosylation.
Enzymes that reverse cellular mono-ADP-ribosylation should bind mono-ADP-ribosylated proteins, remove ADPr and generate an unmodified protein that can be post-translationally remodified by ADP-ribosyltransferases. Macrodomain proteins are good candidates because distinct macrodomains bind ADP-ribosylated proteins2,3,4,5,6,7,8,16,17,18 and hydrolyze the metabolites ADP-ribose-1′′-phosphate and O-acetyl-ADPr (OAADPr, the product of sirtuin deacetylases)19,20,21,22, and X-ray structures of PARG enzymes from various organisms reveal homology to macrodomains14,23,24.
To identify enzymes that reverse cellular mono-ADP-ribosylation, we biochemically screened human macrodomains for catalytic activity toward the reversal of PARP1- and PARP10-mediated ADP-ribosylation. Here, we show that the macrodomain proteins MacroD1 and MacroD2 act as hydrolases on the terminal, protein-proximal ADPr ester linkage, releasing ADPr and an unmodified amino acid product that is readily available for the next round of ADP-ribosylation. Using crystallography, site-directed mutagenesis and molecular dynamics simulations of the enzyme–substrate complex, we probe the catalytic mechanism of the enzyme and use the derived structure-and-function model to predict and validate protein-proximal ADP-ribosyl-hydrolase function in a family of active macrodomain enzymes across genomes from viruses to animals.
Our findings establish PARP-catalyzed cellular ADP-ribosylation as a genuinely fully reversible regulatory post-translational modification. We anticipate that the identification of these terminal ADP-ribosyl-hydrolases will improve understanding of disease-relevant ADP-ribosylation networks and help establish new therapeutic approaches.
Results
MacroD2 interacts directly with weakly modified PARP1
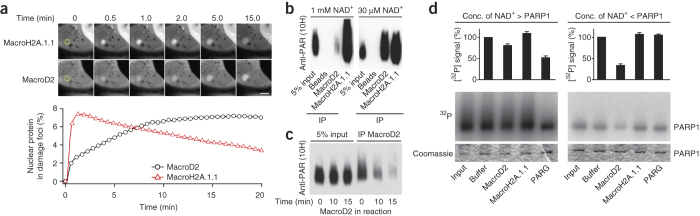
During our search for proteins that could modify the targets of cellular ADP-ribosylation, human MacroD2 caught our attention because a fragment containing its macrodomain (termed MacroD2 from here on) did not bind PAR25 yet bound ADPr and was recruited to DNA lesions (Fig. 1a and Supplementary Fig. 1). Treatment with PARP1 inhibitor AG14361 (30 μM) or mutation of MacroD2 ADPr-binding pocket (G188E) abolished recruitment (Supplementary Fig. 2a,b), consistent with a conserved role of its ADPr pocket and a PARP1-dependent mechanism. In contrast to the PAR-binding MacroH2A.1.1 macrodomain18,26, where more DNA damage only increased the amount of recruited protein but not its kinetics, MacroD2 recruitment acquired bimodal features, with a rapid phase followed by a slower phase, thus causing the maximum intensity of recruitment at the DNA lesions to shift to later time points concordantly with increasing DNA damage (Supplementary Fig. 2c,d). These observations suggested that MacroD2 binding sites become available through two distinct mechanisms. The first phase of recruitment is probably the result of the initial DNA damage–induced mono-ADP-ribosylation, as it depended on PARP inhibition (Supplementary Fig. 2a). The second phase, in turn, may represent MacroD2's binding to mono-ADP-ribosylated species generated by the degradation of PAR through PARG activity. Consistently, short interfering RNA–mediated PARG depletion, which increased PAR levels (Supplementary Fig. 3a), slowed the second MacroD2 recruitment phase without affecting the first phase of recruitment or the recruitment profile of the MacroH2A.1.1 macrodomain (Supplementary Fig. 3b–d).
Figure 1. Macrodomain proteins interact with and demodify weakly ADP-ribosylated PARP1.
(a) Recruitment of tagged mEGFP-MacroD2 and mCherry-MacroH2A.1.1 macrodomains to sites of laser-induced DNA damage. The focus of laser microirradiation is indicated with a yellow circle. Scale bar, 10 μm. (b) Anti-PAR (10H) western blot of poly-ADP-ribosylated PARP1 immunoprecipitates (IP). MacroD2 and MacroH2A.1.1 immunoprecipitation of highly (1 mM NAD+) or weakly (30 μM NAD+) poly-ADP-ribosylated PARP1. (c) Immunoprecipitation and anti-PAR (10H) western blot of PARP1 automodification reactions co-incubated with MacroD2 for the indicated times. (d) Autoradiography and quantification of PARP1 demodification reactions. PARP1 automodified with molar excess (left) or substoichiometric amounts (right) of NAD+ spiked with [32P]NAD+, subjected to demodification by MacroD2, MacroH2A.1.1 or PARG are shown. The residual radioactivity is normalized to buffer control. Error bars, s.e.m. (n = 3). Conc, concentration.
The human genome contains a second MacroD2-related protein known as MacroD1, which localizes to mitochondria and has related identified biochemical functions20,25. Indeed, MacroD1 lacking its mitochondrial targeting sequence (residues 1–85) responded to DNA damage as did MacroD2, showing similar delayed recruitment at higher DNA-damage levels (Supplementary Fig. 2c,d). These results indicate that MacroD1 may also preferentially bind mono-ADP-ribosylated proteins in living cells.
Further, although peptide mass fingerprinting (data not shown) and western blots showed that wild-type MacroD2, but not the ADPr binding–deficient G188E mutant, pulled down PARP1 from extracts (Supplementary Fig. 4a), MacroD2 efficiently interacted with recombinant PARP1 only when PARP1 was incubated with low NAD+ concentrations (Fig. 1b), which generated primarily mono-ADP-ribosylated PARP1. In contrast, high NAD+ levels resulted in poly-ADP-ribosylation but did not lead to efficient MacroD2 binding, whereas MacroH2A.1.1 bound efficiently under these conditions. MacroD2 thus prefers weakly modified (mono-ADP-ribosylated) substrates, consistent with its selective affinity for ADPr over PAR.
MacroD2 suppresses PARP1-mediated mono-ADP-ribosylation
When MacroD2 was co-incubated with PARP1 during the NAD+-dependent automodification, it did not pull down PARP1 (Fig. 1c). In fact, MacroD2 pulled down PARP1 only when it was incubated at 4 °C and added after automodification. In contrast, incubation with automodified PARP1 at 37 °C disrupted MacroD2-PARP1 interactions (Supplementary Fig. 4b). Because MacroD2 remained folded up to 50 °C (Supplementary Fig. 4c), MacroD2 may thus directly affect PARP1 auto-ADP-ribosylation.
MacroD2 and its paralog MacroD1 deacetylate OAADPr20. Because glutamate-linked, proximal ADPr units in mono-ADP-ribosylated PARP1 are chemically related to OAADPr and because MacroD2 preferred binding to weakly modified PARP1 (Fig. 1b), we tested whether it cleaved ADP-ribosylated PARP1. MacroD2 incubation with substoichiometrically [32P]NAD+-modified PARP1 showed efficient [32P]ADPr removal (Fig. 1d), whereas PARG failed to remove ADPr. In contrast, PARG removed most radioactivity from poly-ADP-ribosylated PARP1 (Fig. 1d), and MacroD2 showed little activity. MacroH2A.1.1, consistently with its nonenzymatic roles, removed no ADPr under either condition. Thus, MacroD2 acts on weakly modified mono-ADP-ribosylated PARP1, whereas PARG acts on highly modified poly-ADP-ribosylated PARP1. MacroD2 appears to be an enzyme that reverses PARP1-mediated mono-ADP-ribosylation.
If MacroD2 reverses the first step of PARP1 activity, that is, mono-ADP-ribosylation, it should interfere with PAR synthesis. Indeed, under substoichiometric NAD+ concentrations MacroD2 limited [32P]ADPr incorporation on PARP1, regardless of when it was added (Supplementary Fig. 4d). In contrast, when high NAD+ concentrations were used, MacroD2 only reduced total ADPr when it was present in the reaction from the outset (Supplementary Fig. 4d). To exclude the possibility that MacroD2 would affect PARP1 ADP-ribosylation by hydrolyzing NAD+, we assayed NAD+ levels upon incubation in the presence of MacroD2 or MacroH2A.1.1. Neither MacroD2 nor MacroH2A.1.1 affected NAD+ levels (Supplementary Fig. 4e). Thus, MacroD2 suppresses PARP1-mediated mono-ADP-ribosylation but cannot modify poly-ADP–ribosylated sites.
ADPr is released from mono-ADP-ribosylated substrates
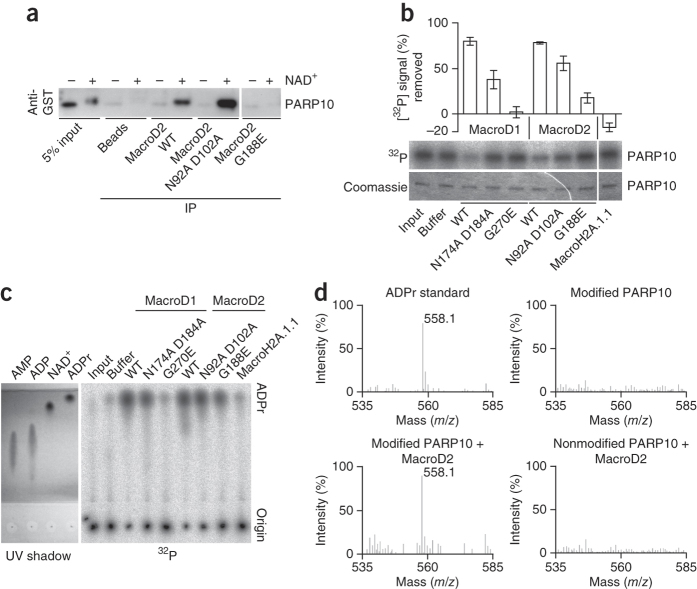
To test whether MacroD2 and MacroD1 modify genuinely mono-ADP-ribosylated proteins, we tested MacroD2 with PARP1 E988K, a chain elongation–defective mutant27, and PARP10, which adds single ADPr units onto glutamate residues6. Wild-type MacroD2, but not an ADPr binding–deficient mutant, pulled down ADP-ribosyl-PARP10 (Fig. 2a). Further, MacroD2 and MacroD1 strongly reduced both PARP1 E988K and PARP10 automodification (Fig. 2b and Supplementary Fig. 4f). In contrast, MacroH2A.1.1, which like MacroD2 bound ADPr and pulled down modified PARP10 (Supplementary Fig. 4g), showed no activity (Fig. 2b). Further, mutation of MacroD1 or MacroD2 residues with a role in OAADPr hydrolysis also reduced the activity toward ADP-ribosylated PARP10 (Fig. 2b).
Figure 2. Macrodomain proteins reversibly remove ADPr from ADP-ribosylated PARP10.
(a) Immunoprecipitates of GST-PARP10 catalytic domain and anti-GST western blot. Samples are of unmodified (−) or ADP-ribosylated (+) GST-PARP10 pulled down with MacroD2 wild type (WT), the O-acetyl-ADP-ribose hydrolysis–deficient double mutant N92A D102A and the ADPr binding–deficient mutant G188E. (b) Autoradiography and quantification of GST-PARP10 catalytic-domain demodification reactions. Samples are demodification reactions containing the indicated macrodomain proteins. The removed radioactive signal is normalized to the buffer control. Error bars, s.e.m. (n = 3). (c) UV shadowing and autoradiography of thin-layer chromatograph. Samples from b alongside with AMP, ADP, NAD+ and ADPr standards separated by thin-layer chromatography and visualized by UV shadowing or 32P autoradiography are shown. (d) MALDI-MS analysis of PARP10 demodification reactions. Analysis of ADPr standard (top left), modified PARP10 (top right), modified PARP10 together with MacroD2 (bottom left) or nonmodified PARP10 together with MacroD2 (bottom right) samples. The 558.1 m/z peak corresponds to ADPr.
To identify the product of MacroD2 activity, we used thin-layer chromatography and MS. The radioactive signal from the demodification reaction was consistent with ADPr (Fig. 2c), and the single MS peak (558.1 m/z) matched ADPr (Fig. 2d). We detected no ADPr upon MacroD2 incubation with unmodified PARP10, nor for modified PARP10 alone. MacroD2 thus hydrolyzes ADPr from mono-ADP-ribosylated PARP10.
The release of ADPr from PARP10 suggested that MacroD2 reverses mono-ADP-ribosylation to generate a protein that can be readily re-ADP-ribosylated. To test the reversibility of PARP10- and MacroD2-catalyzed reactions, we modified PARP10 with cold NAD+, incubated PARP10 with or without MacroD2 and then reincubated PARP10 with [32P]NAD+. As predicted, modified PARP10 incubated with MacroD2 incorporated radioactivity once MacroD2 was removed, compared to modified PARP10 incubated with buffer only, for which only a low amount of radioactivity was incorporated (Supplementary Fig. 5a). Further, repeating the MacroD2 incubation removed [32P]ADPr. Thus, MacroD2 generated a product that could be readily remodified by PARP10, which establishes the identity of the enzyme that makes cellular ADP-ribosylation a reversible post-translational modification.
To test the function of MacroD2 as a genuine enzyme, we tested hallmarks of enzyme-mediated catalysis. We found that the ADPr product inhibited MacroD2-mediated PARP10 demodification (Supplementary Fig. 5b). Hydrolysis depended on temperature, enzyme concentration and incubation time and was efficient at high pH (Supplementary Fig. 5c–f), as would be expected for base-mediated hydrolysis. Consistent with MacroD2's specificity for glutamate-ADPr linkages, arginine-ADPr hydrolase (that is, ARH1) showed no activity on automodified PARP10, whereas MacroD2 showed no activity on automodified cholera toxin (Supplementary Fig. 5g,h). Furthermore, neither MacroD1 nor MacroD2 removed ketamine-linked ADPr from a lysine-containing peptide, which was glycated nonenzymatically by ADPr28 (Supplementary Fig. 5i), thus indicating specificity for the hydrolysis of ADPr-glutamate conjugates. Thus, MacroD1 and MacroD2 are genuine enzymes that reverse PARP-mediated mono-ADP-ribosylation.
The MacroD2–ADPr complex and cleavage of 1′′-OH–linked ADPr
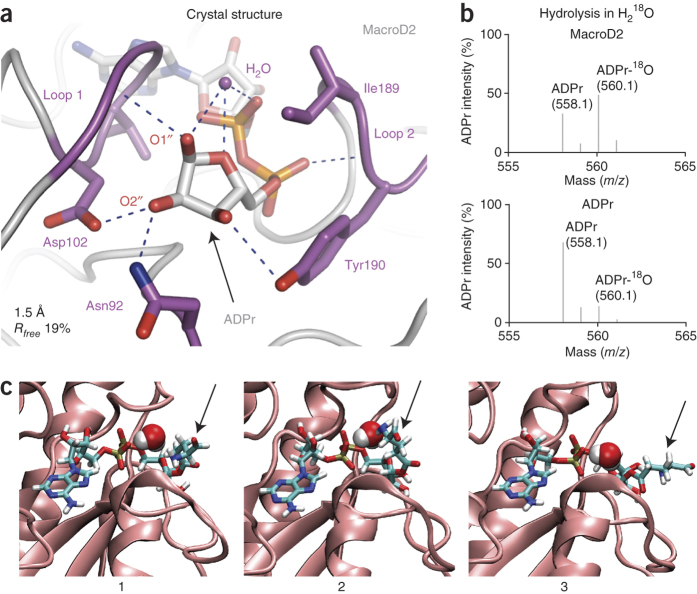
To gain insight into the substrate binding and catalytic mechanism, we solved the structure of the MacroD2–ADPr complex to 1.5-Å resolution (Fig. 3a, Supplementary Fig. 6 and Table 1). Like other macrodomains, MacroD2 binds ADPr in a deep cleft, but the distal ribose unit lined by two glycine-rich loops (loop 1, 97-GGGGV-101 and loop 2, 188-GIYG-191; Fig. 3a) is especially tightly coordinated. Besides forming van der Waals contacts mediated by Ile189 and Tyr190, it hydrogen-bonds several residues. Notably, a water molecule sits in the cleft between loop 1 and 2, held in place by hydrogen bonds between the distal ribose and the neighboring ADPr α-phosphate (Fig. 3a). This tight interaction network establishes high ADPr affinity (Supplementary Fig. 7a) and maintains the distal ribose in an orientation in which the 1′′- and 3′′-OH groups point toward solvent while the 2′′-OH is shielded, forming hydrogen bonds to Asn92 and Asn102. The 1′′-OH is positioned to accommodate mono-ADP-ribosyl-protein substrates.
Figure 3. MacroD2 hydrolyzes mono-ADP-ribosylated PARP10 at the C1′′ atom of ADPr.
(a) Close-up view of the 1.5-Å-resolution X-ray structure of the MacroD2 macrodomain in complex with ADPr (gray), focusing on the distal ribose unit of ADPr. Residues in the vicinity of the distal ribose are shown in stick representation (purple). Hydrogen bonds between protein and ligand are indicated by dashed lines (blue). The positioned H2O, coordinated by hydrogen bonds to the 1′′-OH, 5′′-O and α-phosphate, is shown (purple sphere). The regions forming the two sides of the distal ribose binding cleft are labeled loop 1 and 2. (b) ESI-MS analysis of PARP10 demodification reaction in the presence of H218O. Samples are PARP10 incubated with MacroD2 (top) or ADPr under the same reaction conditions (bottom) in the presence of 66% H218O. The 560.1 m/z peak indicates the 18O atom containing ADPr. (c) Snapshots from a molecular dynamics simulation of MacroD2 in complex with an ADPr-glutamate ester. The MacroD2 binding region is indicated as cartoon (pink), and the bound ADPr ester is shown as a stick model with the glutamate residue (arrow) adopting various conformations and orientations (1–3). A water molecule adopts the placement close to the presumed catalytically active water and is represented as van der Waals surface.
Table 1.
Data collection and refinement statistics
| MacroD2–ADPr | |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 40.71, 49.88, 66.97 |
| α, β, γ (°) | 69.82, 72.35, 86.08 |
| Resolution (Å) | 1.47 (1.55–1.47)a |
| R merge | 0.045 (0.416) |
| I / σI | 10.4 (2.0) |
| Completeness (%) | 90.9 (87.8) |
| Redundancy | 2.0 (2.1) |
| Refinement | |
| Resolution (Å) | 1.55 |
| No. reflections | 62,450 |
| Rwork / Rfree | 0.159 / 0.187 |
| No. atoms | |
| Protein | 3,474 |
| Ligand/ion | 74 |
| Water | 531 |
| B factors | |
| Protein | 20.78 |
| Ligand/ion | 11.45 |
| Water | 35.33 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.213 |
aValues in parentheses are for highest-resolution shell.
To identify which distal-ribose OH group mediates the covalent linkage between ADPr and target protein, we performed de-ADP-ribosylation reactions with H218O. If ADPr is protein linked through its 1′′-OH, 18O would be incorporated into the hydrolyzed ADPr. In the case of 2′′- or 3′′-OH linkage, 18O would be incorporated into the released peptide29. In fact, 18O incorporated into ADPr (Fig. 3b). Consistent with this, molecular dynamics simulations confirmed that a 1′′-OH–linked glutamate-ADPr ester stably binds MacroD2 and that the interloop position remains occupied by a rapidly exchanging water molecule (Fig. 3c). In contrast, molecular dynamics simulations with lysine-ADPr ketamine or di-ADPr indicated that neither of them can stably interact with MacroD2 (Supplementary Fig. 8), which supports our biochemical results. We surmise that an activated H2O performs a nucleophilic attack on the C1′′ atom of the distal ribose.
A catalytic mechanism for terminal ADP-ribosyl hydrolases
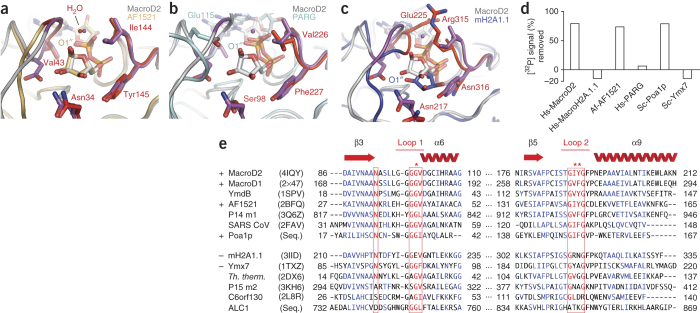
The orientation of the distal ribose in the MacroD2–ADPr complex is notably similar to that of other ADPr ligand–bound macrodomains (Fig. 4a–c). Archaeoglobus fulgidus AF1521 (Fig. 4a), human PARP14 macro1 (data not shown), viral macrodomains (data not shown) and the macrodomain-related Thermomonospora curvata PARG (Fig. 4b) are isostructural. In AF1521, all key residues and even the positioned water in the cleft between loop 1 and loop 2 are conserved. In the case of PARG, only loop 2 is isostructural and conserved (Fig. 4b). The other side of the ADPr cleft contains an insertion, leaving room for the catalytic Glu115, which nucleophilically attacks the C1′′ atom, thus explaining the catalytic differences between MacroD2 and PARG14 (Fig. 4d). In contrast, the MacroH2A.1.1 macrodomain, showing no activity (Figs. 2b and 4d), has a distinct ribose conformation with bulky side chains (Glu225, Arg315) on either side of the distal ribose that close the gap over the bound ADPr (Fig. 4c). In addition, Tyr190 in loop 2 is replaced by Asn316, which hydrogen-bonds the 3′′-OH. These changes result in a buried and rotated position of the distal ribose with respect to MacroD2–ADPr, consistent with a binding role.
Figure 4. The mode of ADPr coordination in the macrodomain pocket determines mono-ADP-ribosyl hydrolase function and identifies evolutionarily conserved enzymes.
(a–c) Superposition of MacroD2 with A. fulgidus AF1521 (PDB 2BFQ) (a), T. curvata PARG (PDB 3SIG), with the catalytic residue Glu115 unique to PARG labeled (b) or MacroH2A.1.1 (PDB 3IID) (c). Isostructural residues and conserved water molecules are highlighted in red. The positions of the 1′′-OH groups of ADPr are indicated. (d) Quantification of macrodomain proteins' demodification activity on [32P]ADP-ribosylated PARP10. Samples include either MacroD2, MacroH2A.1.1, AF1521, PARG, Poa1p or Ymx7 macrodomains. Prefix indicates organism: Af, A. fulgidus; Hs, human, Sc, Saccharomyces cerevisiae. 32P removal is normalized to buffer control. (e) Structure-guided sequence alignment of selected macrodomains. Two sequence regions around loop 1 and 2 are shown. Secondary structure elements are taken from the MacroD2 structure. Signature motif residues important for the coordination of the ADPr distal ribose and indicative for hydrolysis activity are highlighted in red. Plus indicates confirmed hydrolysis activity; minus indicates no activity; asterisks highlight the residues most important for catalytic activity.
Comparison of the MacroD2–ADPr complex with the ligand-free structure of the catalytically active homolog MacroD1 showed a structural rearrangement of loop 2 (amino acids 188–191 in MacroD2) upon ADPr binding20. In MacroD1, the loop is in an open conformation but is closed in the structure of the MacroD2–ADPr complex, where it stacks tightly against the distal ribose (a 13-Å movement of Tyr190 in MacroD2 compared to the corresponding Phe272 in MacroD1; Supplementary Fig. 6). The similarities in structure are consistent with a highly related biochemical function.
Further, the structural similarity between human MacroD2 and archaeal AF1521 prompted us to test whether AF1521 shows catalysis. Indeed, AF1521 efficiently cleaved protein-ADPr linkages, whereas PARG and MacroH2A.1.1 were inactive (Fig. 4d). This suggested that the conformation of the distal ribose in MacroD2 is ideal for ester hydrolysis at the 1′′-OH and that residues Asn92, Gly99, Gly100, Val101, Ile188 and Tyr189—isostructural in AF1521—are important for mono-ADP-ribosyl-hydrolase activity, thus allowing us to propose a catalytic mechanism.
Asn92 and Asp102 of MacroD2 have been suggested to be catalytic for OAADPr deacetylation20. We found that MacroD2 N92A D102A and the corresponding MacroD1 mutations N174A D184A retain strong activity toward PARP10 (Fig. 2b), which suggests rather indirect roles in de-ADP-ribosylation. Consistent with this, Asp102 is not conserved in AF1521 (Fig. 4a,e). Likewise, Asn92, which is also conserved in noncatalytic macrodomains, helps to coordinate the distal ribose (Fig. 4c,e). The only other hydrophilic group in the vicinity of the distal ribose is Tyr190, which hydrogen-bonds the 3′′-OH group and mediates van der Waals contacts with the distal ribose. However, Tyr190 is exchanged to phenylalanine in the catalytically active MacroD1, which argues against catalytic roles. The absence of other candidate residues leads us to tentatively propose a substrate-assisted mechanism, in which a coordinated water molecule ideally positioned between α-phosphate and distal ribose would be catalytic (Fig. 3a). We propose that the ADPr α-phosphate activates this water molecule, allowing it to make a nucleophilic attack on the ADPr C1′′ atom30 (Supplementary Fig. 7b).
Evolutionary conservation of mono-ADP-ribosylase activity
To validate our suggested mechanism, we created a mutant that displaces the catalytic water and influences the conformation of the distal ribose by binding ADPr as does the inert MacroH2A.1.1. Starting from a MacroD2 G100E scaffold (no catalytic activity, no ADPr binding), we introduced mutations that reestablish ADPr affinity but not catalysis. The ADPr-binding G100E Y190N and G100E I189R Y190N mutants exhibited no detectable de-ADP-ribosylation activity, despite their interaction with modified PARP10 (Supplementary Fig. 9a–d). This indicated that the orientation of the distal ribose, the distance between Gly100 and Ile189 and the positioned water are crucial for catalysis.
The discovery of the mono-ADP-ribosyl-hydrolase function in MacroD1, MacroD2 and archaeal AF1521 allowed us to identify hydrolases across genomes. Structure-based alignments showed that the above identified signature residues required for catalytic activity in MacroD2, MacroD1 and AF1521 (Gly100 in loop 1, Ile189 and Tyr190 in loop 2 of MacroD2) are conserved in a number of macrodomain proteins, including viral SARS-CoV Nsp3, Escherichia coli YmdB, yeast Poa1p and human PARP14 (Fig. 4e), identifying these proteins as potential mono-ADP-ribosyl hydrolases. We validated hydrolase activity in AF1521 and yeast Poa1p (Fig. 4d). Consistent with our predictions, a yeast macrodomain not matching the signature (Ymx7) was not catalytically active (Fig. 4d). Biochemical analysis thus revealed conserved 1′′-O-linked mono-ADP-ribosyl-hydrolase activity in an evolutionarily distant family of macrodomain proteins (Supplementary Fig. 9e).
Discussion
Our biochemical, structural and modeling analyses identify a conserved family of macrodomains that reverse and antagonize cellular glutamate-linked mono-ADP-ribosylation as mediated by PARP1 and PARP10 and are inactive toward lysine- and arginine-linked mono-ADP-ribosylation. We suggest that the hydrolytic activities of macrodomain-like proteins on OAADPr and mono-ADP-ribosylated PARP1 and PARP10 are overlapping functions. OAADPr maintains an equilibrium between acetylated 2′′- and 3′′-OH, yet its acetyl group can also isomerize to the 1′′-OH27, so hydrolysis may occur at the 1′′-ester linkage. ADP-ribosylated glutamates are linked through the 1′′-OH, and our assays with H218O showed that MacroD2 hydrolyzes 1′′-linked ADPr. Further, the ADPr-1′′-phosphate hydrolase activity described in yPoa1p19 but also present in MacroD1, MacroD2 and other macrodomains25,31 correlates very well with our reported mono-ADP-ribosyl-hydrolase activity. Notably, a mutational analysis of SARS-CoV Nsp3 classified residues important for phosphatase activity that overlap our signature motif for protein-ADPr hydrolysis31. Further, the SARS-CoV Nsp3–ADPr complex is isostructural for distal ribose coordination with MacroD2–ADPr. Signature motif–containing macrodomain proteins thus act as 1′′-ester hydrolases.
MacroD2 recruitment to DNA-damage sites in vivo shows a role for this enzyme in PARP1-mediated functions at DNA lesions. In particular, MacroD2's reversal of ADP-ribosylation on weakly automodified PARP1 in vitro (Figs. 1 and 2c) suggests that MacroD2 could suppress PARP1 activation by removing mono-ADP-ribosylated PARP1 species until a certain threshold for full PARP1 activation is achieved, for example upon DNA damage. Inhibitors targeting macrodomain-like proteins most probably alter PARP1 signaling and could be therapeutically useful for cancer treatment. Further, there are many other ADP-ribosylating enzymes in humans, including PARPs and sirtuins, as well as nonenzymatic ADP-ribosylation mechanisms. Here we reported what is, to our knowledge, the first set of conserved enzymes capable of removing terminal, protein-proximal ADPr units from PARP-modified mono-ADP-ribosylated proteins. Although detailed mechanistic analysis awaits the generation of highly defined mono-ADP-ribosylated substrates, our discovery of enzymes that fully reverse this regulatory modification will allow systematic dissection of ADP-ribosylation signaling.
Methods
Reagents.
Human PARP1 protein (ALX-201-063-C020), PARP1 E988K mutant (ALX-201-254-C010), PARG protein (ALX-202-045-UC01), ARH1 (ALX-201-290-C010) and PJ-34 inhibitor (ALX-270-289) were bought from Enzo Life Sciences. Buffer chemicals, including ADP-ribose (A0752), anti-V5 affinity gel (A7345), H218O (32987) and cholera toxin A subunit (C8180) were all from Sigma.
Plasmids.
Mammalian expression constructs of wild-type MacroH2A.1.1 macrodomain and MacroD2 macrodomain were described previously32. The mammalian expression construct of wild-type MacroD1 (aa 86–325) was generated by cloning of the PCR-amplified corresponding sequence into the pmEGFP-C1 vector by using BglII and EcoRI restriction sites. The mutant G188E was generated by using QuikChange site-directed mutagenesis (Stratagene) according to the manufacturer's protocol. All constructs were sequence verified.
Bacterial expression constructs were as follows: GST-PARP10 catalytic domain (aa 818–1025) was a gift from B. Lüscher33; GST-AF1521 was described previously34; pGEX-2TKN-Poa1p (full-length S. cerevisiae YBR022W ORF), pETM11-His6-Ymx7 (full-length S. cerevisiae YMR087W ORF), pETM-CN vector-based His6-TEV-V5 tagged human MacroH2A.1.1 (aa 162–372), MacroD1 macrodomain (aa 91–325) and MacroD2 macrodomain (aa 7–243) constructs were generated in house from cDNA sequences. Corresponding point mutants were generated through the QuikChange site-directed mutagenesis (Stratagene) protocol.
Cell culture, transfection and siRNA treatment.
Human U2OS cell lines stably expressing fluorescent protein–tagged MacroD2 macrodomain or MacroH2A.1.1 macrodomain were grown in DMEM (Sigma) containing 10% (v/v) FBS (Gibco), 1 mM sodium pyruvate (Sigma), 2 mM L-glutamine (Sigma), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin (Sigma) and 200 μg ml−1 G418 (Gibco). For transient transfections, Xfect (Clontech) was used according to the manufacturer's recommendations. For siRNA-mediated PARG depletion, we used reverse transfection according to a published protocol35. Lab-Tek chambered coverslip glass was coated with s16158 Silencer Select siRNA oligos (Ambion), and the cells were grown on the coated coverslip for 24 h.
Live-cell imaging, pulsed-laser microirradiation and image analysis.
Imaging was performed on a Zeiss AxioObserver Z1 confocal spinning-disk microscope equipped with an AxioCam HRm CCD camera (Zeiss) through a Zeiss Plan/Apo 63×/1.4 oil-immersion objective lens. For laser microirradiation, we used a 355-nm-wavelength diode-pumped solid-state pulsed laser (DPSL-355/14, Rapp OptoElectronics). DNA damage was induced by focusing a circle with a diameter of 5 μm on the nucleus. We have defined as low laser energy the amount of microirradiation resulting in the recruitment of 2% of nuclear MacroH2A.1.1 at the sites of microirradiation and, consequently, high laser power as eight-fold increased energy compared to the former, which results in recruitment of about 6% of the nuclear MacroH2A.1.1 macrodomain to the site of microirradiation. The microirradiated cells were imaged every 3 s for 5–20 min. Cells were plated in borosilicate 8-well Lab-Tek chambered cover glasses (ThermoScientific), and Hoechst 33342 (Sigma) was added to the culture medium at least 30 min before microirradiation at 200 ng ml−1 final concentration. When indicated, the PARP inhibitor AG14361 (Selleckchem) was added to the culture medium 30 min before microirradiation at 30 μM final concentration. During the experiments, cells were kept at 37 °C in a CO2-independent imaging medium (Gibco) supplemented with 10% (v/v) FBS (Gibco), 1 mM sodium pyruvate (Sigma) and 2 mM L-glutamine (Sigma), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin (Sigma).
Time-lapse images were registered and analyzed with Fiji (http://fiji.sc/wiki/index.php/Fiji), and the MultiStackReg. Igor Pro (WaveMetrics) Fiji plug-in was used for analyzing and plotting the data. To quantify protein recruitment following microirradiation, data were background subtracted, corrected to premicroirradiation, adjusted for changes in nuclear matrix intensity and corrected for fluorescence loss according to the formula R(t) = ((I(t)−Iback(t))−((I(t0)−Iback(t0))*((T(t)−Iback(t))−(I(t)−Iback(t)))/((T(t0)−Iback(t0))−(I(t0)−Iback(t0))))/(T(t)−Iback(t))×100, where R is percentage of recruitment, I is intensity acquired along the laser path region, Iback is the background region outside the cell of interest, and T is the total fluorescence within the nucleus.
In vivo poly-ADP-ribosylation assay.
To validate the effect of siRNA-mediated PARG depletion, cells were washed in PBS (Sigma) after live-cell imaging and treated with 1.2 mM H2O2 (Sigma) and fixed at different time points (10 min and 30 min). Subsequently, cells were fixed in prechilled 70–30% (v/v) methanol/acetone for 10 min at −20 °C, washed in PBS and blocked in 5% (w/v) milk in 0.05% (v/v) PBS-Tween20 at room temperature. The primary antibody, anti-poly-ADPr mouse monoclonal 10H antibody (ascites), 1:1,000, was used overnight at 4 °C. AlexaFluor647-conjugated donkey anti-mouse IgG secondary antibody (Molecular Probes cat. no. A31571) was used at 1:500 dilution. DNA was stained by using Hoechst 33342 (Sigma). Images of fixed cells were taken on a Zeiss AxioObserver Z1 confocal spinning-disk microscope and analyzed with CellProfiler 2.0 (http://www.cellprofiler.org/). To quantify poly-ADPr signal intensities, images were background subtracted, and far-red integrated intensities were calculated in the nuclei of cells positively expressing MacroD2 macrodomain. For the CellProfiler pipeline, nuclei were segmented by Hoechst staining, the segmented nuclei were filtered for MacroD2 expression on the basis of GFP fluorescence, and PAR levels were quantified as with the AlexaFluor647 integrated fluorescence intensities. Additional information is provided in the Supplementary Note.
Protein expression and purification.
Macrodomains were expressed in E. coli Rosetta(DE3)pLysS cells at 18 °C for 18 h after 200 μM IPTG induction. Cell pellets were snap frozen with liquid nitrogen and stored at −80 °C. For histidine-tagged-protein purification, the thawed pellet was resuspended in lysis/wash buffer (50 mM Tris-Cl, pH 7.9, 0.5 M NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol and protease inhibitor cocktail (Roche)). Lysates were sonicated for 3 × 30 s at medium setting (Branson ) until the lysate was not viscous and centrifuged for 45 min at ~45,000g at 4 °C. The supernatant was incubated for 1h with Ni-NTA resin (Macherey Nagel), washed five times with 45 ml wash buffer and eluted with wash buffer containing 500 mM imidazole. Where appropriate, the histidine tag was cleaved with TEV protease overnight at 4 °C. The proteins were dialyzed overnight in storage buffer (25 mM Tris-Cl, pH 7.9, 0.25 M NaCl, 1 mM DTT), concentrated and subjected to gel-filtration chromatography with a Superdex S75 16/60 column (GE Healthcare) using storage buffer as eluent. Peak fractions confirmed by SDS-PAGE and Coomassie staining were pooled and concentrated with 10,000 MWCO concentrators (Amicon). Concentrations were determined by absorbance measurements at 280 nm wavelength by using calculated molar extinction coefficients.
For GST-tagged macrodomain protein purification, the procedure was as above, with the exception of lysis buffer composition (50 mM Tris-Cl, pH 7.9, 0.1 mM EDTA, 0.5 M NaCl, 10% glycerol, 1 mM DTT and protease inhibitors) and glutathione Sepharose, with 20 mM reduced-glutathione elution (GE Healthcare).
GST-fused PARP10 catalytic domain was expressed and purified essentially as previously described33, with the exception that the lysis/wash buffer for protein purification was 50 mM HEPES, pH 7.2, 500 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT and included protease inhibitors. The PARP10 protein was incubated with glutathione Sepharose, washed with wash buffer and eluted with 20 mM reduced glutathione in wash buffer. The fractions were dialyzed in storage buffer (25 mM HEPES, pH 7.2, 300 mM KCl, 10% glycerol, 1 mM DTT), concentrated to 1 mg/ml. The concentration was determined by absorbance measurements.
Crystallization and data collection.
MacroD2 macrodomain was purified to homogeneity in 25 mM Tris, pH 7.5, 100 mM NaCl, 1 mM DTT and concentrated to 5 mg/ml in the presence of 6 mM ADPr. Crystals of the macrodomain–ADPr complex (Table 1) were grown at room temperature by vapor diffusion from hanging drops composed of equal volumes (1 + 1 μl) of protein solution and crystallization buffer (18% (w/v) polyethylene glycol 3350, 0.1 M HEPES, pH 7.5, and 0.1 M Mg formate) suspended over 1.0 ml of the latter as reservoir solution. Needle-shaped crystals of up to 500 × 50 × 50 μm were transferred into reservoir solution containing 20% (v/v) glycerol and flash frozen in liquid nitrogen. A 1.5-Å resolution data set at 100 K was recorded at beamline ID-29 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Processing and scaling were carried out with XDS36 and Scala37.
Structure solution and refinement.
The structure of the complex was solved with the molecular replacement method as implemented in PHASER (University of Cambridge38), by using the previously determined human MacroD1 macrodomain as a search model39 (PDB 2X47). The structure was finalized in alternating cycles of manual model correction in COOT40 and restrained TLS refinement in Phenix41 (Table 1) . The structure has excellent stereochemistry with 100% in allowed and 98.9% in favored regions of the Ramachandran plot, as implemented in MolProbity42. Structural visualization was done with PyMol (http://www.pymol.org/).
Accession codes.
Atomic coordinates and structure factors for the reported crystal structure has been deposited in the Protein Data Bank under accession number 4IQY.
Supplementary information
Supplementary Figures 1–9 and Supplementary Note (PDF 23089 kb)
Acknowledgements
We thank P. Heckmeyer for technical assistance (Ludwig Maximilians University of Munich, Munich, Germany), B. Lüscher (Rheinisch-Westfälische Technische Hochschule Aachen University, Aachen, Germany) for PARP10 plasmids, P.O. Hassa and M. Hottiger (University of Zurich, Zurich, Switzerland) for PARP1 protein and antibody, A. Scholz and the European Molecular Biology Laboratory crystallization facility, staff at the European Synchrotron Radiation Facility beamline ID29 for assistance during data collection, I. Forné and A. Imhof of the Ludwig-Maximilian-University of Munich's ZfP facility for mass spectrometry, S. Uebel and E. Weyher-Stingl from the biochemistry core facility of Max Planck Institute of Biochemistry for access to the isothermal titration calorimeter and ESI-MS mass spectrometer, A. Bolzer and Carl Zeiss MicroImaging GmbH for assistance with spinning-disk confocal microscopy, as well as A. Eberharter and C. Laverty for comments on the manuscript. A.G.L. and coworkers are funded by EMBL, Ludwig Maximilians University of Munich, EU FP7 Marie Curie Initial Training Network “Nucleosome4D”, the Deutsche Forschungsgemeinschaft (LA 2489/1-1) and the Center for Integrated Protein Science Munich.
Author Contributions
G.J. conducted pulldown, ADP-ribosylation and MS assays; G.J. and V.R. conducted isothermal titration calorimetry assays; M.H. conducted crystallographic work; G.J. and M.H. purified proteins; B.G. and G.T. conducted live-cell imaging; M.Z. conducted molecular dynamics simulations; G.J., M.H., G.T. and A.G.L. contributed to the design of the study; G.T. and A.G.L. supervised the work; G.J., M.H., G.T. and A.G.L. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Accession codes
Primary accessions
Protein Data Bank
Referenced accessions
Protein Data Bank
Competing interests
The authors declare no competing financial interests.
Footnotes
Gytis Jankevicius and Markus Hassler: These authors contributed equally to this work.
Contributor Information
Gyula Timinszky, Email: gyula.timinszky@med.uni-muenchen.de.
Andreas G Ladurner, Email: andreas.ladurner@med.uni-muenchen.de.
References
- 1.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 4.Kawaichi M, Oka J, Ueda K, Hayaishi O. A new method for oligo (ADP-ribose) fractionation according to chain length. Biochem. Biophys. Res. Commun. 1981;101:672–679. doi: 10.1016/0006-291X(81)91311-5. [DOI] [PubMed] [Google Scholar]
- 5.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010;38:6350–6362. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Z, Gao P, Liu H-W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009;131:14258–14260. doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 9.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogehase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 10.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. doi: 10.1042/bj3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber V, Dantzer F, Amé J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 12.Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies Karyopherin-β1. PLoS ONE. 2012;7:e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka J, Ueda K, Hayaishi O, Komura H, Nakanishi K. ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J. Biol. Chem. 1984;259:986–995. [PubMed] [Google Scholar]
- 14.Slade D, et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss J, Oppenheimer NJ, West RE, Stanley SJ. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1986;25:5408–5414. doi: 10.1021/bi00367a010. [DOI] [PubMed] [Google Scholar]
- 16.Dani N, et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Front. Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 18.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 19.Martzen MR, et al. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 20.Chen D, et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson FC, et al. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 2011;286:35955–35965. doi: 10.1074/jbc.M111.276238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 23.Dunstan MS, et al. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012;3:878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- 24.Kim I-K, et al. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat. Struct. Mol. Biol. 2012;19:653–656. doi: 10.1038/nsmb.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuvonen M, Ahola T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 2009;385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karras GI, et al. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolli V, O'Farrell M, Ménissier-de Murcia J, de Murcia G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry. 1997;36:12147–12154. doi: 10.1021/bi971055p. [DOI] [PubMed] [Google Scholar]
- 28.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J. Biol. Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- 29.Kasamatsu A, et al. Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J. Biol. Chem. 2011;286:21110–21117. doi: 10.1074/jbc.M111.237636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dall'Acqua W, Carter P. Substrate-assisted catalysis: molecular basis and biological significance. Protein Sci. 2000;9:1–9. doi: 10.1110/ps.9.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egloff MP, et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 33.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Dani N, et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erfle H, et al. Reverse transfection on cell arrays for high content screening microscopy. Nat. Protoc. 2007;2:392–399. doi: 10.1038/nprot.2006.483. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen D, et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–9 and Supplementary Note (PDF 23089 kb)