Abstract
Molecular machines orchestrate the translocation and entry of pathogens through host cell membranes, in addition to the uptake and release of molecules during endocytosis and exocytosis. Viral cell entry requires a family of glycoproteins, and the structural organization and function of these viral glycoproteins are similar to the SNARE proteins, which are known to be involved in intracellular vesicle fusion, endocytosis and exocytosis. Here, we propose that a family of bacterial membrane proteins that are responsible for cell-mediated adherence and entry resembles the structural architecture of both viral fusion proteins and eukaryotic SNAREs and might therefore share similar, but distinct, mechanisms of cell membrane translocation. Furthermore, we propose that the recurrence of these molecular machines across species indicates that these architectural motifs were evolutionarily selected because they provided the best solution to ensure the survival of pathogens within a particular environment.
Main
The eukaryotic plasma membrane has evolved to control and regulate the entry of fluids, solutes and particles into cells from the extracellular environment and to regulate the export of intracellular components stored in vesicles. This is achieved by endocytosis and exocytosis. In parallel, microorganisms such as viruses and bacteria have developed mechanisms to enter host cells to facilitate their replication, transport and transmission. One of the recent breakthroughs in cell biology has been the understanding that molecular machines that share similarities both in structural domains and functional mechanisms mediate both viral entry and cellular exocytosis events.
The fusion between cell membranes during vesicle trafficking and exocytosis (Fig. 1) is mediated by a conserved set of tetrameric proteins, collectively known as SNAREs1,2,3,4 (see Glossary). Similarly, the entry of enveloped viruses into host cells is mediated by a family of glycoproteins that usually fold as homotrimers and that mediate the fusion between two cell membranes (Fig. 1). Also, non-enveloped viruses follow a separate pathway of cell entry5,6, which seems to be mediated by proteins that have a similar topology.
Figure 1. Schematic representation of intracellular vesicle exocytosis, and viral and bacterial cell entry.

a | Attachment of intracellular vesicles to the plasma membrane is mediated by vesicle and target SNAREs (vSNAREs and tSNAREs, respectively), viral fusion proteins and bacterial invasins. b | Assembly and conformational changes of surface proteins result in the formation of a coiled-coil metastable structure, which induces tethering, followed by intimate adherence of vesicles and pathogens to the plasma membrane. In the case of HIV, the fusion protein undergoes a conformational change that results in insertion of the fusion peptide (shown in black) into the plasma membrane. Influenza virus entry is pH dependent — the virus is internalized and the acidification of the endosome results in a conformational change of haemagglutinin. This exposes the fusion peptide, which then inserts into the vesicular membrane, causing fusion of the viral and endosomal membrane. c | Energy derived from the conformational changes in b promote either fusion of cellular membranes (as seen for both vesicles and viruses) or invasion of host cells by bacteria. SNARE, soluble NSF (N-ethylmaleimide-sensitive fusion protein) accessory protein (SNAP) receptor.
Compared with the above-mentioned viral and eukaryotic fusion events, relatively little is known about the molecular mechanisms that allow bacteria access to the internal milieu of host cells (Fig. 1). What is known is that this process is mediated by a family of surface proteins that are known as bacterial INVASINS and, as in viral entry, bacterial entry requires actin polymerization. Following host cell-receptor binding, invasins trigger the phosphorylation and dephosphorylation of cytoskeleton effector molecules and scaffolding proteins, which results in bacterial internalization6,7,8,9,10 (endocytosis). However, it is not known whether invasins play a further part in bacterial pathogenesis. Evidence from this report indicates that a family of bacterial cell-entry proteins shares the modular organization of SNARE heterotetramers and viral homotrimeric spike proteins (Fig. 2). It is intriguing to speculate that, because of their conserved modular architecture — which is defined by a membrane anchor, a central COILED-COIL motif and a receptor-binding domain (RBD) (Fig. 2a) — viral fusion proteins, bacterial invasins and SNAREs might share similar mechanisms of action. The absence of amino-acid homology coupled to the analogous distribution of domains indicates a common mechanism that might add selective advantages to the pathogen, which would ensure survival in a particular environment. Furthermore, this indicates that different organisms might have evolved common solutions when challenged with the task of penetrating cellular barriers.
Figure 2. Architecture of eukaryotic membrane and viral fusion proteins compared with bacterial invasins.
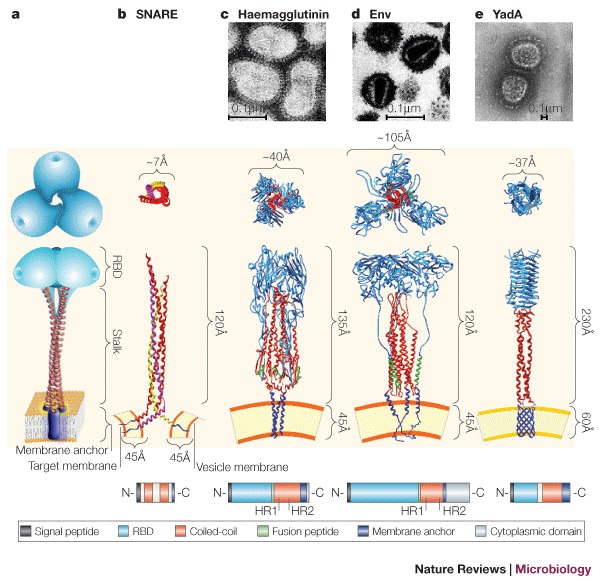
a | Schematic representation of the common trimeric structure consisting of a membrane anchor (purple), a central coiled-coil motif (stalk, red) and a globular receptor-binding domain (RBD, cyan). b | Partial three-dimensional structure of the coiled-coil domains of the synaptic SNARE complex. Synaptobrevin 2 (also known as VAMP2) (yellow) is anchored to the vesicle membrane. Syntaxin (pink) and SNAP25 (red) are anchored to the target membrane. These proteins are anchored to the cytoplasmic membrane by a hydrophobic α-helical C- terminal domain, which allows the protein to extend 120Å from the cell membrane. c | Structural representation of the influenza haemagglutinin (HA) trimer with the fusion peptide (green). The stalk is in its metastable pre-fusogenic state. The globular receptor-binding subunit HA1 is shown in blue, and the triple-stranded, coiled-coil fibrous region, known as HA2, is shown in red. d | The HIV envelope protein (Env) is composed of two non-covalently associated trimeric gp120 (red) and gp41 (blue) subunits. Here, gp120 is artificially fused to gp41. The fusion peptide, which is located in gp41, is depicted in green. In their monomeric form, both fusion proteins — haemagglutinin and Env — span the viral envelope with a single transmembrane α-helix near the C-terminus. e | Structural model of the trimeric Yersinia spp. YadA molecule anchored to the bacterial outer membrane through a β-barrel (blue). The anchor domain is represented by a porin-like outer membrane protein. Top–down views of the heterotrimeric SNARE complex, homotrimeric viral and bacterial fusion proteins (invasins), are shown. Structures are not to scale. The structures used to build the models have been obtained from the PDB protein databank (see the Online links box). Short domains for which coordinates were not available were built manually on the basis of secondary structure predictions. The anchor domain was modelled on the integral outer membrane protein (OmpA) from Escherichia coli (strain 1BXW), whereas the coiled-coil stalk was modelled on the structure of chicken cartilage trix protein (1AQ5). Transmission electron microscopy (TEM) of negatively stained influenza virus (c), HIV (d) and Yersinia spp. (e) are shown. The bottom panel is a schematic representation of the protein sequence architecture of the corresponding proteins. HR1 and HR2 are experimentally determined heptad repeats for haemagglutinin and Env. White boxes represent sequence portions with an undefined role. SNARE, soluble NSF (N-ethylmaleimide-sensitive fusion protein) accessory protein (SNAP) receptor. Panel c TEM courtesy of Linda M. Stannard, University of Cape Town, South Africa. Panel c (structural image) reproduced, with permission, from Ref. 98 © (2002) Elsevier Science. Panel d TEM courtesy of H. Gelderblom, Koch Institute, Berlin. Panel e TEM reproduced, with permission, from Ref. 60 © (2000) European Molecular Biology Organization.
Viruses and vesicles
Vesicles — the SNARE family. In 1992, a seminal discovery showed that proteolytic cleavage of a membrane fusion protein, known as VAMP1, by the tetanus toxin was sufficient to inhibit the secretion of neurotransmitters from synaptic junctions and cause the deadly tetanus disease11. This finding led to the understanding that intracellular fusion during exocytosis was carried out by specialized proteins, SNAREs. The SNARE superfamily comprises more than 35 eukaryotic membrane fusion proteins. To mediate fusion, three SNAREs from the target membrane (tSNAREs) recognize one vSNARE (vesicle SNARE) from the vesicle membrane12,13, and the crystal structures of short coiled-coil fragments indicate that this interaction results in the formation of a supercoil complex, which consists of a heterotetramer of four parallel α-helices2,14,15 (Fig. 2b). This complex assembles through the hydrophobic and ionic core-packing interactions that are formed between vSNAREs and tSNAREs. The free energy that is derived from the formation of the four-helix bundle and a subsequent conformational change promotes tethering of the two opposing membranes to mediate subsequent fusion events2,3,4,12,14,15. Such fusion events mediate either exocytosis of secretory vesicles or endosomal trafficking (Fig. 1); and they involve additional factors, such as the exocyst multi-subunit protein docking complex, microtubules and the recruitment of tethering proteins through Rab GTPases (such as Rab3A)15,16 that specify the fusion of transport vesicles with a particular target membrane.
Viral fusion. Conceptually, all enveloped viruses require a similar membrane fusion event to deliver their genomes into the host cell cytoplasm for the replication and transmission of new virions17. The entry of many enveloped viruses is mediated by glycoproteins (called 'spikes'), which are expressed as trimers on the external surface of the viral particle, where they form a halo-like appearance (Fig. 2c,d). These viral fusion proteins have several common features: first, the presence of a generally hydrophobic FUSION PEPTIDE that participates in the first phase of membrane fusion; second, an energetically metastable region that is characterized by coiled-coil HEPTAD REPEATS (HR1 and HR2), which form higher order oligomers on the viral membrane; and last, all possess a C-terminal membrane anchor domain17 (Fig. 2c,d).
On binding with a receptor, many viral fusion proteins undergo a conformational change that is required to expose the fusion peptide, which anchors the host cell membrane. Although fusion peptides of different viruses are variable in structure, hydrophobic residues are important for coiled-coil formation, whereas the ability of these peptides to form trimers is crucial for fusogenic activity18,19,20. It has been hypothesized that the energy released from a conformational change is important to expose and deliver the fusion peptide to the target membrane, successively driving the formation of the fusion pore and eventually joining the membranes14,19,21,22.
Viral fusion proteins are classified as class I and class II fusion proteins according to the location of the fusion peptide and the orientation of the glycoprotein relative to the viral envelope22,23,24. Class I fusion proteins are expressed by human pathogens such as the influenza virus and HIV and form trimeric 'spikes' in their native metastable conformation at the surface of the infectious virions25. Their post-fusion conformation is a hairpin-like structure in which both the fusion peptide (proximal to the N-terminus) and the membrane anchor (distal to the N-terminus) are juxtaposed at the same end of a stable protein 'rod' or 'spike' (Fig. 1).
The influenza virus haemagglutinin (HA) is considered the prototype of class I fusion proteins. The fusion that is mediated by haemagglutinin is a pH-dependent process. Haemagglutinin is a trimeric glycoprotein that projects 135Å from the target membrane26 (Fig. 2c). Each monomer is composed of a globular receptor-binding subunit HA1 (shown in blue in Fig. 2), and a triple-stranded coiled-coil fibrous region, known as HA2 (shown in red in Fig. 2). Data from the crystal structure of haemagglutinin before and after the pH-induced conformational change allowed an understanding of the mechanism of viral fusion with the host cell27. In this multistep process, haemagglutinin binds surface-exposed cell receptors (Fig. 1). The virus is then internalized, and the subsequent acidification of the endosome causes a conformational change of haemagglutinin (irreversible in most cases), and triggers the fusion of the viral and endosomal membrane28,29.
Similar to haemagglutinin of the influenza virus, class I viral fusion proteins from HIV, Visna virus, and SARS-CoV are characterized by a central domain, which during fusion, adopts a trimer of 'hairpins' folds, which leads to the assembly of a six-helix, coiled-coil bundle25,30,31,32,33. However, in these cases, the fusion event is not dependent on a pH change.
Specifically, during cell entry of HIV, insertion of the HIV envelope protein (Env) fusion peptide into the host cell membrane is followed by the formation of a complete coiled-coil structure, which involves the folding of the HR1 heptad repeat of the stalk over the second heptad repeat HR2, and assembly of the six-helix bundle21. The free energy that is released by this conformational change is required to draw the viral and host membranes into close proximity until fusion occurs30 (Fig. 1). The formation of this coiled-coil structure is essential for the fusogenic function of Env and for the infection of CD4+ T cells34,35,36. Inhibiting its formation by blocking the HR1 domain — for example, with a synthetic peptide such as Enfuvirtide, which mimics the function of HR2 — impedes the fusion of HIV37.
By contrast, class II fusion protein heterodimers contain an internal fusion peptide and, when exposed to low pH, these fusion proteins reversibly dissociate from their partner to form irreversible homotrimers, which in turn mediate membrane fusion by a mechanism similar to the one described above38,39. Class II fusion proteins are found in flaviviruses such as dengue virus29, hepatitis C virus (HCV), tick-borne encephalitis (TBE) virus39, and alphaviruses such as the Semliki Forest virus40.
Non-enveloped viruses, such as picornaviruses and rotaviruses, lack a lipid bilayer membrane and therefore cannot achieve cell entry through membrane fusion41. Despite years of study, the mechanism that allows this class of viruses to cross a membrane remains poorly understood. However, it seems that these viruses might enter cells using proteins that have a similar topology to those used by enveloped viruses. Recently, work by Dormitzer and colleagues reported the structural characterization of a domain of the membrane penetration protein — the VP4 spike of rotavirus5. VP4 haemagglutinin is a dimer that is present on the external surface of the virion42,43. Following trypsin cleavage of the VP4 spike, two virion-associated fragments, VP8 and VP5, are generated. The VP5 fragments cluster to form a homotrimer that is composed of three globular domains that are folded over an α-helical triple coiled-coil stalk, which is reminiscent of the structure of spike proteins from enveloped viruses5. A possible mechanism of cell entry is proposed in which the interaction of the rotavirus membrane penetration protein with the host cell receptor promotes a conformational change that leads to the formation of the observed trimer and to the release of the VP8 domain. The hydrophobic fusion domain of VP5 is exposed and might be used to breach or permeabilize the host cell membrane, thereby allowing the virion to penetrate into the cytoplasm. This mechanism of cell entry, although not well understood, recalls that described for enveloped viruses. Also, such a membrane-fusion-independent method of host cell penetration might represent a possible link between the cell entry mechanisms and molecular machines that are used by viruses and bacteria.
Membrane fusion and bacteriophages. In addition to the fusion of enveloped viruses with their host, membrane-fusion events in prokaryotes have also been described. In vitro assays coupled with transmission electron microscopy provide evidence that the segmented double-stranded RNA enveloped bacteriophage φ6 expresses a trimeric structure that is involved in the fusion with the outer membrane of its host, the plant pathogen Pseudomonas syringae44,45. An additional lipid-containing, double-stranded DNA bacteriophage, PRD1, known to infect Gram-negative pathogens such as Escherichia coli, Salmonella spp. and Pseudomonas spp., has been proposed to fuse with the bacterial membrane on infection46,47 (S. Butcher, personal communication).
What about bacteria?
Whereas bacteria do not require access to the host cell cytoplasm for replication, many pathogens induce their own uptake to gain access to an immunologically protected environment that is optimal for survival and replication. Bacterial entry is a complex process that involves direct adherence to host cell receptors or indirect adherence through extracellular matrix ligands, followed by host–pathogen signalling events. Once internalized, some pathogens remain within vacuoles (Salmonella spp., Yersinia spp., Streptococcus spp., Neisseria spp.)48,49,50,51, whereas others escape to infect neighbouring cells (Shigella spp., Listeria spp.)52,53.
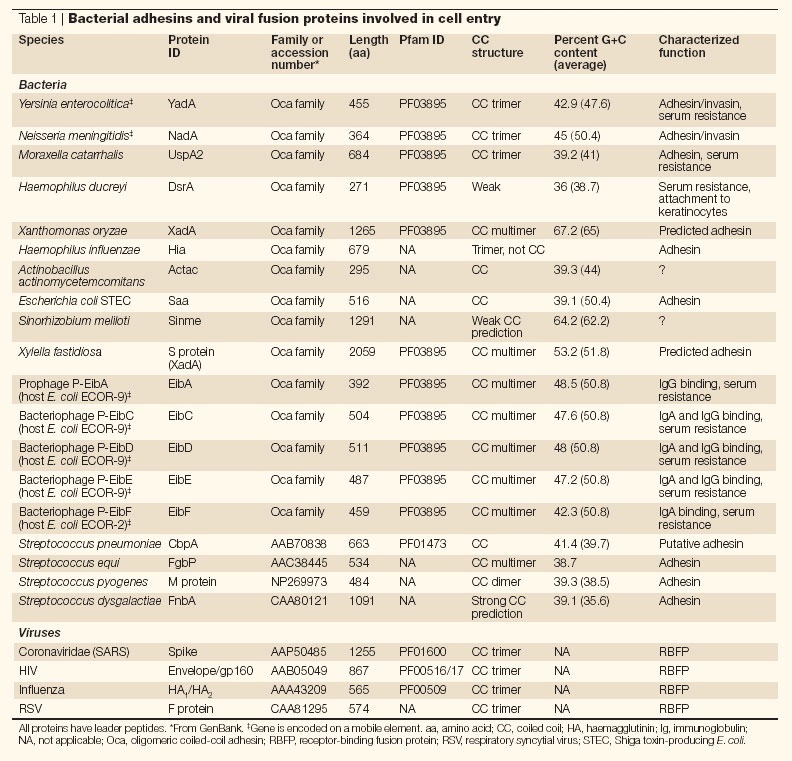
Proteins that mediate the entry of bacteria into non-phagocytic host cells are known as invasins, the prototype of which is the Yersinia spp. invasin protein (InvA), which was discovered in 1987 (Ref. 54). More recently, other invasins such as the Yersinia spp. YadA and Neisseria meningitidis NadA have been described55,56,57,58,59. YadA and NadA, together with the ubiquitous surface proteins UspA1 and UspA2 of Moraxella catarrhalis, represent the prototypes of the Oca (oligomeric coiled-coil adhesin) family of putative non-fimbrial adhesins60 (Table 1). Genome sequence analysis and structural prediction algorithms have identified more than 20 novel members of this family in Gram-negative bacteria, as well as in plant pathogens60,61, all of which share several structural and functional features. Oca family proteins are grouped by homologies found in the C-terminal anchor domain, but how did diverse bacterial species acquire these cell entry machines?
Table 1.
Bacterial adhesins and viral fusion proteins involved in cell entry
As suggested by the anomalous GC composition of the oca genes, and by the conserved nature of their C-terminal domains, we propose that these domains might have originated by horizontal transfer. A possible origin could be envisaged in the ubiquitous siphoviridae P-Eib prophages, which encode a family of four genes (eib-A, -C, -D and -E), the protein products of which confer immunoglobulin-binding activity to the ECOR group of E. coli strains62. The Eib (E. coli immunoglobulin binding) proteins form high-molecular-weight oligomers. They show the same YadA-like tripartite organization (Fig. 2e) centred on a core coiled-coil motif with a C-terminal membrane anchor that shares sequence identity with the corresponding domains of UspA2 (60.3%), YadA (56.7%) and NadA (58.6%). One hypothesis is that eib genes were transferred among E. coli strains by phage vectors. A similar mechanism of prophage-driven transfer might have been involved in the propagation of these surface molecules (or of their anchor domains) to different recipient bacterial species. Support for this idea comes from the fact that some of the genes that code for proteins of the Oca family, such as YadA and NadA, are carried on mobile genetic elements (Table 1).
In addition to theories of evolutionary origins, we provide further evidence that bacterial invasins have a role in cell entry that is similar to that mediated by viral fusion proteins and SNAREs.
Structural similarities. Like most class I, class II and non-enveloped viral spike proteins, YadA and NadA are expressed as homotrimers on the surface of Yersinia spp. and N. meningitidis, respectively, and they participate in the binding to and uptake of the bacterium into host cells. Despite the lack of complete three-dimensional structures for proteins of the Oca family, much has been learned about the topology of these proteins through ultramicroscopy and primary and secondary structure prediction.
Similar to viral spike proteins, both YadA and UspA appear as distinct 'lollipop'-shaped structures, forming a halo-like surface projection on the outer membrane of the bacteria in electron micrographs (Fig. 2e). Whereas the shape of the projections seems similar, the rod-like segments are three times longer in UspA compared with YadA, which extends approximately 230Å from the bacterial cell surface60. This structure is reminiscent of the spike proteins that are expressed by many enveloped viruses (Fig. 2c,d).
Furthermore, sequence analysis of YadA, NadA and the UspA proteins has helped to define the molecular architecture of this class of molecules, which is remarkably similar to that of many viral envelope proteins. According to these predictions, three main domains can be envisaged for proteins of the Oca family: a C-terminal outer membrane anchor domain; a rod-like intermediate segment formed mainly by extended right- or left-handed coiled-coil segments, which are implicated in the formation of higher-order oligomers; and an N-terminal globular head region that is involved in binding to host cells and the extracellular matrix63 (Fig. 2a,e).
Recently, the crystal structure of the collagen-binding domain of YadA was solved (Fig. 2e), revealing a novel, nine-coiled, left-handed parallel β-roll64, a structure that is commonly found in fibrous proteins65. In Fig. 2e, the X-ray-determined structure of the YadA head domain has been arbitrarily fused to structural models of the stalk and membrane anchor regions, which have been derived from THREADING ANALYSES.
Like YadA, NadA forms stable, high-molecular-weight oligomers on the surface of N. meningitidis58 and, when expressed in E. coli, is exported to the outer membrane where it assembles into trimers. The trimeric conformation of NadA has been confirmed experimentally by light scattering analysis (S. Savino, personal communication). Detailed analysis of the NadA primary and secondary structure profiles, and comparison with coiled-coil regions of viral glycoproteins has allowed the prediction of the two putative heptad repeat domains HR1 and HR2 within the stalk region. In the absence of three-dimensional data, the presence of a coiled-coil structure for NadA was partially confirmed by CIRCULAR DICHROISM (CD) SPECTRA ANALYSIS, which shows a prominent α-helical content for this protein (S. Savino, personal communication). Although it is not clear whether these predicted domains have a function, preliminary analysis shows that purified NadA undergoes temperature- and pH-inducible conformational changes that are visible in SDS-PAGE, indicative of a metastable structure similar to that of viral spikes and SNAREs (S. Savino, personal communication).
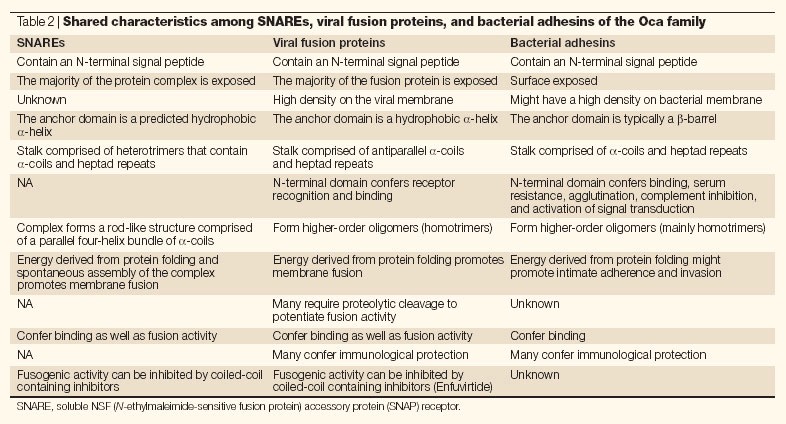
Functional similarities. Besides common structural features, the Oca family proteins and viral fusion proteins also share functional characteristics, such as binding to host cell structures and mediating immunological protection (Table 2). It has been reported that, similar to InvA, YadA-promoted cell entry occurs through the interaction with β1-integrins66. Whereas the interaction between Yersinia spp. InvA and β1-integrins is direct67, YadA interaction with host cells is mediated indirectly by a bridging mechanism involving extracellular matrix components, such as collagen and fibronectin68,69,70.
Table 2.
Shared characteristics among SNAREs, viral fusion proteins, and bacterial adhesins of the Oca family
Several important reports have linked YadA to bacterial invasion. YadA is expressed in both Yersinia enterocolitica and Yersinia pseudotuberculosis, but not in Yersinia pestis71. As with many virulence factors, YadA is located on the 70-kD plasmid (pYV). Unlike the β-immunoglobulin-like Yersinia spp. InvA, YadA is a homotrimer that is induced during exponential growth in minimal media at 37°C (Ref. 72). In vitro, YadA binds collagen, laminin, cellular fibronectin, intestinal submucosa and hydrophobic surfaces70. Single amino-acid mutations in the N-terminal receptor-binding domain resulted in the abrogation of YadA binding to extracellular matrix proteins in vitro, which led to a marked reduction in virulence in an animal model of infection63. Eitel and Dersch argue that YadA belongs to a secondary uptake pathway that might complement InvA-mediated cell entry when synthesis of InvA is repressed72. Earlier reports showed that yadA genes that were encoded on the pYV plasmid promoted internalization of Yersinia invA mutants, but with a lower frequency of occurrence55,57,71. More recent studies provide evidence that E. coli strains that express YadA under the pBAD-inducible arabinose promoter, in gentamicin protection assays, had the same ability to adhere to and enter cell monolayers as E. coli that expressed InvA72.
Recombinant NadA mediates binding and uptake of E. coli into Chang epithelial cells59. It also has strong immunogenic properties.
Little information is available on the biological function of the UspA proteins. Nevertheless, they seem to share central functional properties with YadA and NadA. All these proteins form high-molecular-weight oligomers, have a role in cell attachment, mediate serum resistance and are good immunogens.
Gram-positive bacteria and cell entry. Gram-positive organisms have also evolved strategies that facilitate host cell adherence and entry. Similar to SNAREs and viral membrane fusion proteins, a group of Gram-positive bacterial adhesins are known to undergo a conformational change on receptor binding, which brings the molecule from a disordered to an ordered state, therefore reinforcing binding and facilitating subsequent cell invasion73,74. Proteins that mediate these functions also share a fibril-like elongated architecture, usually composed of coiled coils. Examples of such proteins include the well characterized dimeric M protein of Streptococcus pyogenes75, the fibronectin binding protein (FnbA) of Streptococcus dysgalactiae, the fibrinogen binding protein (FgbP) of Streptoccoccus equi76,77, and the choline binding protein (CbpA) of Streptococcus pneumoniae78. The M protein of S. pyogenes is a fibrillar molecule that binds to fibrinogen and albumin and promotes bacterial adhesion to host cells75. Interestingly, FnbA and structurally similar proteins that are produced by Staphylococcus aureus and S. pyogenes mediate bacterial cell entry by binding to integrins through a fibronectin-based bridging mechanism79. This mechanism involves a conformation change that is known to trigger the recruitment of effector molecules, such as focal adhesion kinase (FAK), to the entry site.
Intracellular signalling cascades
Both bacterial and viral pathogens use the cell and its effector molecules to induce intracellular uptake. Pathogens not only depend on the host cell machinery for their internalization, but also for trafficking within the cytoplasm and for the ability to find sites that allow replication and transmission. Endosomal trafficking and exocytosis of secretory vesicles use molecular mechanisms and signalling pathways that are subverted by bacteria and viruses for their own purposes.
Exocytosis and endocytic events involve docking factors such as the exocyst protein complex and Rab GTPases, such as Rab3A (Fig. 3a). Vesicle trafficking also involves the activation of intracellular cytoskeleton effector molecules such as phosphatidylinositol 3-kinase (PI3K) and other small GTPases such as Cdc42. During the final stages of exocytosis, F-actin forms a cortical network under the plasma membrane of a cell. It has been shown that exocytotic vesicles are transported along microtubules to the plasma membrane, but are not secreted before the cortical actin network opens locally to form the exit pore. Also, PI3K activates phosphatidylinositol 4,5-bisphosphate (PIP2), which has been implicated in the regulation of the actin cytoskeleton and vesicle trafficking. PIP2 stimulates de novo actin polymerization by activating a pathway that comprises the Wiskott–Aldrich syndrome protein (WASP) and the actin-related protein complex ARP2/3 (Fig. 3). Other studies show that actin polymerizes from cholesterol-sphingolipid-rich membrane microdomains called 'rafts', in a tyrosine phosphorylation-dependent manner80.
Figure 3. Intracellular signalling events.

The signalling cascades and intracellular pathways that are used by viruses and bacteria to induce host cell uptake converge on those used in vesicle trafficking. A common theme is the activation of GTPases and the subsequent cytoskeletal changes that aid in the movement of vesicles or in the uptake of pathogens. a | In SNARE-mediated vesicle trafficking and exocytosis, intracellular factors such as the exocyst protein complex and the small GTPase Rab3A are important in the docking of the vesicle to the plasma membrane. Vesicle trafficking also involves the activation of intracellular cytoskeleton effector molecules such as phosphatidylinositol 3-kinase (PI3K) and other small GTPases such as Cdc42. F-actin nucleation mediated by N-WASP (neural Wiskott–Aldrich syndrome protein) and ARP2/3, and microtubule elongation are crucial in trafficking events. Receptor-mediated attachment of viruses such as influenza virus (b) and bacteria such as Yersinia spp. (c) also leads to the signalling and activation of GTPases (such as dynamin, Ras, Rac1 and Cdc42), as well as actin polymerization through the ARP2/3 complex (mediated by N-WASP in bacteria, and N-WASPand intersectin in viruses). PI3K is activated by the binding of certain viruses and bacteria to cell surface receptors, and is also implicated in actin polymerization. AP2, adaptor protein-2; CAS, Crk-associated substrate; CAT2, cationic amino-acid transporter; DAG, diacylglycerol; FAK, focal adhesion kinase; GRB2, growth factor receptor-bound protein-2; MAPK, mitogen-activated protein kinase; PIP2, phosphatidylinositol-4,5-bisphosphate; PI4P, phosphatidylinositol 4-phosphate; PKC, protein kinase C; SH2, Src-homology domain-2; SNARE, soluble NSF (N-ethylmaleimide-sensitive fusion protein) accessory protein (SNAP) receptor.
Productive viral and bacterial infections are also frequently associated with profound changes of the host cytoskeleton. Such changes are often mediated through phosphorylation-regulated signalling cascades. Similar to SNARE-mediated intracellular trafficking, some viral uptake mechanisms involve a tightly controlled interplay of intracellular molecules such as clathrin — an effector protein that interacts with intracellular transport effector molecules, such as Esp15 (Ref. 81), amphiphysin and the AP2 adaptor proteins82. For example, during cell entry, the influenza virus might be taken up by a clathrin-dependent or clathrin-independent endocytic route, following the initial interaction of viral haemagglutinin with its sialic-acid receptor17,83. Also, in vitro data indicate that the GTPase dynamin, which is known to be involved in the release of endosomes from the plasma membrane, is also essential in the early events of influenza virus endocytosis84,85,86 (Fig. 3b).
The role of actin in the endocytosis of viruses is not clear, although a role for actin in viral exocytosis and budding has been described28,87. Microtubules might also be involved in these processes, as seen in the trancytosis of HIV in vitro22,88. Furthermore, HIV is known to activate a FAK pYK2 kinase and to signal to the mitogen-activated protein kinase (MAPK) pathway88, which is important in the uptake and intracellular transport of the virus (Fig. 3b). Signalling is a common theme in cell entry, as internalization of the adenovirus through ligand binding to its co-receptor integrin activates both protein kinase C and PI3K89, as well as subsequent effector molecules that are involved in vesicular trafficking (Rab5) and cytoskeletal organization (Rac1, Cdc42 and dynamin). Interestingly, pre-treatment of epithelial cells with cytochalasin D, a product that disrupts actin fibres, causes a dose-dependent inhibition of adenovirus internalization89. These results indicate that assembly of the actin cytoskeleton plays a key part in viral endocytosis, and therefore resembles the general mechanism of invasion that is used by many bacterial pathogens.
Bacterial internalization is accompanied by changes in the phosphorylation status of cytoskeleton effector molecules and scaffolding proteins6,7,8,9,10 (endocytosis). However, so far, there is no data on the signalling events that occur in YadA-mediated cell entry. It is known that, following tight adhesion of Yersinia spp. to the host cellular surface, InvA-mediated bacterial uptake seems to involve FAK125, Src, PI3K, the small GTPase Rac1 and the ARP2/3 complex90,91 (Fig. 3c). Similarly, recent data support the hypothesis that YadA-mediated bacterial invasion is dependent on protein phosphorylation events, as the addition of tyrosine kinase inhibitors strongly impairs Yersinia spp. cell entry through YadA72,92. InvA- and YadA-promoted cell entry occurs through the interaction with the β1-integrins66 and the interaction of YadA with the β1-integrins is mediated through collagen and fibronectin68,69,70. On the basis of in vitro experiments that describe collagen signal transduction, one might infer that YadA-mediated cell entry is dependent on cell signalling — collagen IV and laminin, but not fibronectin, are known to stimulate tyrosine phosphorylation of intracellular FAK and other signalling molecules. Notably, one of the molecules involved in the InvA signalling pathway, PI3K, is implicated in actin polymerization and can also interact with FAK125, which itself associates with β1-integrins93,94. This implicates modulation of the actin cytoskeleton in YadA-mediated cell entry.
In conclusion, both InvA-promoted and YadA-promoted cell entry occur through a 'zipper-like' mechanism; that is, a high affinity-binding event that takes advantage of phosphorylation-regulated signalling pathways and that requires the action of phosphokinases and actin polymerization. Recent experimental data from the NadA protein indicates that, similar to YadA, NadA-mediated invasion is actin-dependent59. These results indicate that many of the signalling cascades described in pathogen uptake and eukaryotic vesicle trafficking converge on the machinery involved in host cell reorganization, providing new theories on the evolution of pathogen survival strategies.
Conclusions and implications
In this report, we provide evidence that a large family of bacterial surface proteins has an overall modular organization that resembles membrane fusion proteins such as viral spikes and SNAREs. Two bacterial membrane proteins, YadA and NadA, have specifically been shown to mediate bacterial entry into host cells, a function that is analogous to that of viral spike proteins. The data available on the mechanism of invasion promoted by YadA and NadA, compared with what is known about viral uptake, allow some speculation on a possible common route of cell entry that is shared by viruses and bacteria.
As bacterial entry does not involve fusion between bacterial and host plasma membranes, we do not yet have an explanation for all the similarities observed. However, recent data indicates that, even in viruses, these molecular machines are used for common cell entry mechanisms and not solely for membrane fusion events — non-enveloped rotaviruses do not fuse but instead disrupt the host cell membrane, thereby allowing the virion access to the cytoplasm5. The mechanism of cell entry that is exploited by this class of viruses might represent the missing link between viral and bacterial cell-entry mechanisms and might help to explain the possible mechanism of invasion that is promoted by coiled-coil bacterial invasins.
The most probable and supported hypothesis is that, after mediating adhesion, NadA and YadA undergo a conformational change and bring the bacterial and host membranes into tight contact (intimate adhesion). This process would initiate the bacterial–host signalling events that induce actin polymerization, formation of membrane protrusions around the bacteria, and ultimately result in bacterial uptake by endocytosis. A less likely possibility is that these bacterial invasins function as real fusion proteins by binding the membrane protrusions that surround the bacteria, bringing them together until they fuse and engulf the bacteria into the endosome.
The discovery of a large family of bacterial proteins with structural and potentially functional similarities to viral spikes and SNAREs indicates that pathogens use a recurrent theme to cross membranes. These common machineries might confer a selective advantage to the pathogen and provide a significant contribution to pathogen fitness.
Other examples of similar viral and bacterial cell-entry structures have recently been described. The first is IncA of Chlamydia trachomatis, a coiled-coil protein that has been shown to assemble into tetramers and to interact homotypically to promote a vesicle fusion mechanism that is similar to that of eukaryotic SNAREs95. The second example is the structural resemblance between the major coat protein of the bacteriophage PRD1 and the human adenovirus Hexon. Although the two proteins have different amino-acid sequences, they show an identical topology and are organized in a double-barrel trimer that contains two eight-stranded JELLY-ROLL MOTIFS96. We have no doubt that evolutionary pressure has selected machineries with similar architectural domains to perform similar functions in bacteria, viruses and eukaryotic cells, but the absence of any primary sequence similarity leaves open the question of whether this is the result of convergent or divergent evolution. In the first case, a similar architecture would be a common solution for molecules with different origins. In the second case, the morphology that is necessary for function would be the only remaining feature from an ancestral molecule and, as suggested by the LINNAEAN APPROACH, “morphology might be the real link to phylogeny”97.
The main limits to our hypothesis relate to the lack of structural information on Oca bacterial invasins, and the dearth of experimental data on the mechanistic processes that lead to bacterial cell entry. However, structural prediction on YadA and NadA and recent experimental results on NadA support a close resemblance between class I and non-enveloped viral spike proteins and Oca molecules. The fact that class II viral fusion proteins are functionally related to class I viral fusion proteins, despite the lack of structural similarities, leads us to speculate that microorganisms have found various solutions to the problem of host cell entry. Hopefully, this Opinion article will stimulate further experimentation and will help to unveil more bacterial examples of cell entry machineries.
It is fascinating to think that we might be able to exploit these perfect molecular machineries to develop novel antimicrobial drugs such as the HIV fusion inhibitor, or for the development of novel nanomachines that would use programmed fusion between membrane-bound vesicles for drug delivery and for in vivo targeting of cancer cells. Preliminary data also indicates that functional domains of bacterial and viral entry proteins can be exchanged to obtain chimeric proteins with novel properties. Future studies to resolve the crystal structure, identify structural rearrangements involved in conformational changes, as well as to understand the intracellular signalling events mediated by the Oca family of proteins will be crucial in elucidating the speculative function of this set of proteins.
Acknowledgements
The authors would like to thank S. Savino, B. Capecchi, J. Adu-Bobie, M. Pizza and B. Aricò for sharing unpublished data on the NadA protein, crucial to support our hypotheses. They are also grateful to M. Scarselli for help in structural model reconstruction of the discussed proteins, and to K. Stadler and I. Ferlenghi for useful discussion and advice. A special acknowledgment goes to G. Corsi for artwork and to C. Mallia for assistance in manuscript preparation.
Glossary
- CIRCULAR DICHROISM SPECTRUM ANALYSIS
A widely used technique for obtaining information about protein structure and conformation. It is a sensitive and reliable tool to study the structure and stability of proteins.
- COILED COIL
An important protein–protein interaction motif often used to control oligomerization, characterized by a conserved heptad repeat. Coiled coils consist of helices that wrap around one another with a superhelical twist.
- FUSION PEPTIDE
A sequence of 20–30 mainly hydrophobic residues (Ala and Gly) found at the N-terminus of the stalk exposed after a conformational change of the viral fusion protein. Insertion of the peptide into the host cell membrane leads to cell fusion events.
- HEPTAD REPEAT
Seven residue patterns denoted (abcdefg)n in which the a and d residues (core positions) are generally hydrophobic. As there are 3.6 residues to each turn of the α-helix, these a and d residues form a hydrophobic seam, which, as each heptad is slightly under two turns, slowly twists around a helix. Heptad repeats are characteristic of certain proteins such as the intermediate filament proteins.
- INVASIN
Any bacterial surface protein that provokes endocytic uptake by host cells. Adhesins, on the other hand, promote binding to cell surface receptors, but do not elicit uptake by the host cell.
- JELLY-ROLL MOTIF
These motifs are found in proteins in which the primary fold contains only β-antiparallel strands. Like other elements of super-secondary structure involving the β-strand (for example, the β–α–α–β unit) the known structure forms a right-handed superhelix.
- LINNAEAN APPROACH
The first formal classification scheme was created by Carolus Linnaeus and relied on classification of species according to hierarchical structure, from most general to most similar. However, this scheme ignores their evolutionary history, and important aspects of the origin of those similarities and differences are overlooked.
- SNAREs
Soluble NSF (N-ethylmaleimide-sensitive fusion protein) accessory protein (SNAP) receptor). These proteins contain a heptad repeat of 60–90 residues that participate in coiled-coil formation. The family of SNARE proteins are involved in intracellular fusion events.
- THREADING ANALYSIS
This method uses computer modelling to obtain structural information based on the amino-acid sequence of an uncharacterized protein structure. Threading analysis is mostly used to detect remote homologues that can not be detected by standard sequence alignment.
Biographies
Michèle A. Barocchi received her Ph.D from the University of California, Berkeley, in the Infectious Diseases and Immunity Programme, studying molecular host–pathogen interactions. She is currently a postdoctoral fellow in the Department of Cellular Microbiology and Bioinformatics at Chiron Vaccines in Siena, Italy.
Vega Masignani got her Ph.D in biotechnology with a thesis on the computational approach to the development of a novel Neisseria meningitidis protein-based vaccine. At present, she works in the Cellular Microbiology and Bioinformatics Unit of Chiron Vaccines, doing research in the field of computer analysis applied to microbial pathogenesis.
Rino Rappuoli is Head of Research at Chiron Vaccines and his area of expertise includes bacterial toxins and vaccines and bacterial pathogenesis. His pioneering studies in infectious diseases have brought to market vaccines against diphtheria, pertussis, hepatitis A and meningococcus C.
Related links
DATABASES
Swiss-Prot
FURTHER INFORMATION
Competing interests
The authors are employed by Chiron Vaccines
References
- 1.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2. 4Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 2.Antonin W, Fasshauer D, Becker S, Jahn R, Schneider TR. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nature Struct. Biol. 2002;9:107–111. doi: 10.1038/nsb746. [DOI] [PubMed] [Google Scholar]
- 3.Chen YA, Scheller RH. Snare-mediated membrane fusion. Nature Rev. Mol. Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 4.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- 5.Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isberg RR, Leong JM. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacteria penetration into mammalian cells. Cell. 1990;60:861–871. doi: 10.1016/0092-8674(90)90099-Z. [DOI] [PubMed] [Google Scholar]
- 7.Van Nhieu GT, Krukonis ES, Reszka AA, Horwitz AF, Isberg RR. Mutations in the cytoplasmic domain of the integrin β1 chain indicate a role for endocytosis factors in bacterial internalization. J. Biol. Chem. 1996;271:7665–7672. doi: 10.1074/jbc.271.13.7665. [DOI] [PubMed] [Google Scholar]
- 8.Bruce-Staskal PJ, Weidow CL, Gibson JJ, Bouton AH. Cas, Fak and Pyk2 function in diverse signaling cascades to promote Yersinia uptake. J. Cell Sci. 2002;115:2689–2700. doi: 10.1242/jcs.115.13.2689. [DOI] [PubMed] [Google Scholar]
- 9.Weidow CL, Black DS, Bliska JB, Bouton AH. CAS/Crk signaling mediates uptake of Yersinia into human epithelial cells. Cell. Microbiol. 2000;2:549–560. doi: 10.1046/j.1462-5822.2000.00079.x. [DOI] [PubMed] [Google Scholar]
- 10.Boyle EC, Finlay BB. Bacterial pathogenesis: exploiting cellular adherence. Curr. Opin. Cell Biol. 2003;15:633–639. doi: 10.1016/S0955-0674(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 11.Schiavo G, et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 12.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/S0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 13.Hu C, et al. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 14.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 15.Harbury PAB. Springs and zippers: coiled coils in SNARE-mediated membrane fusion. Curr. Biol. 1998;6:1487–1491. doi: 10.1016/s0969-2126(98)00147-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Sugita S, Sudhof TC. The RIM/NIM familyof neuronal C2 domain proteins. Interactions with Rab3 and a newclass of Src homology 3 domain proteins. J. Biol. Chem. 2000;275:20033–20044. doi: 10.1074/jbc.M909008199. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus–cell and cell–cell fusion. Ann. Rev. Cell. Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 18.Adam B, Lins L, Stroobant V, Thomas A, Brasseur R. Distribution of hydrophobic residues is crucial for the fusogenic properties of the Ebola virus GP2 fusion peptide. J. Virol. 2004;78:2131–2136. doi: 10.1128/JVI.78.4.2131-2136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta. 2003;1614:116–121. doi: 10.1016/S0005-2736(03)00169-X. [DOI] [PubMed] [Google Scholar]
- 20.Lau WL, Ege DS, Lear JD, Hammer DA, DeGrado WF. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skehel JJ, Wiley DC. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/S0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov DS. Virus entry: molecular mechanisms and biochemical applications. Nature Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poranen MM, Daugelavicius R, Bamford DH. Common principles in viral entry. Annu. Rev. Microbiol. 2002;56:521–538. doi: 10.1146/annurev.micro.56.012302.160643. [DOI] [PubMed] [Google Scholar]
- 24.Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2004;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nature Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 26.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 27.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-W. [DOI] [PubMed] [Google Scholar]
- 28.Pelkmans L, Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/S0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 29.Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl Acad. Sci. USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marti DN, Bjelic S, Lu M, Bosshard HR, Jelesarov I. Fast folding of the HIV-1 and SIV gp41 six-helix bundles. J. Mol. Biol. 2004;336:1–8. doi: 10.1016/j.jmb.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 31.Malashkevich VN, Singh M, Kim PS. The trimer-of-hairpins motif in membrane fusion: Visna virus. Proc. Natl Acad. Sci. USA. 2001;98:8502–8506. doi: 10.1073/pnas.151254798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripet B, et al. Structural characterization of the SARS coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sackett K, Shai Y. How structure correlates to function for membrane associated HIV-1 gp41 constructs corresponding to the N-terminal half of the ectodomain. J. Mol. Biol. 2003;333:47–58. doi: 10.1016/j.jmb.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Sackett K, Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochem. 2002;41:4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Wang S, Hoxie JA, LaBranche CC, Lu M. Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus–CPmac envelope glycoprotein complex. J. Biol. Chem. 2002;277:12891–12900. doi: 10.1074/jbc.M110315200. [DOI] [PubMed] [Google Scholar]
- 37.Matthews T, et al. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nature Rev. Drug. Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 38.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the Dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 39.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 40.Gibbons DL, et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature. 2004;427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 41.Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw AL, et al. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeager M, Berriman JA, Baker TS, Bellamy AR. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bamford DH, Romantschuk M, Somerharju PJ. Membrane fusion in prokaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romantschuk M, Olkkonen VM, Bamford DH. The nucleocapsid of bacteriophage φ6 penetrates the host cytoplasmic membrane. EMBO J. 1988;7:1821–1829. doi: 10.1002/j.1460-2075.1988.tb03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L, Benson SD, Butcher SJ, Bamford DH, Burnett RM. The receptor binding protein P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure. 2003;11:309–322. doi: 10.1016/S0969-2126(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 47.Grahn AM, Daugelavicius R, Bamford DH. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 2002;46:1199–1209. doi: 10.1046/j.1365-2958.2002.03250.x. [DOI] [PubMed] [Google Scholar]
- 48.Finlay BB, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying salmonella entry into epithelial cells. J. Cell Sci. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 49.Small PL, Isberg RR, Falkow S. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect. Immun. 1987;55:1674–1679. doi: 10.1128/iai.55.7.1674-1679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dombek PE, et al. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol. Microbiol. 1999;31:859–870. doi: 10.1046/j.1365-2958.1999.01223.x. [DOI] [PubMed] [Google Scholar]
- 51.McGee ZA, Stephens DS, Hoffman LH, Schlech WF, 3rd, Horn RG. Mechanisms of mucosal invasion by pathogenic Neisseria. Rev. Infect. Dis. 1983;4:S708–S714. doi: 10.1093/clinids/5.Supplement_4.S708. [DOI] [PubMed] [Google Scholar]
- 52.Sansonetti PJ, Ryter A, Clerc P, Maurelli AT, Mounier J. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gedde MM, Higgins DE, Tilney LG, Portnoy DA. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 2000;68:999–1003. doi: 10.1128/IAI.68.2.999-1003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isberg RI, Voorhis DL, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 55.Bliska JB, Copass MC, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect. Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Nhieu GT, Krukonis ES, Reszka AA, Horwitz AF, Isberg RR. Mutations in the cytoplasmic domain of the integrin β1 chain indicate a role for endocytosis factors in bacterial internalization. J. Biol. Chem. 1996;271:7665–7672. doi: 10.1074/jbc.271.13.7665. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Isberg RR. Cellular internalization in the absence of invasin expression is promoted by the Yersinia pseudotuberculosis yadA product. Infect. Immun. 1993;61:3907–3913. doi: 10.1128/iai.61.9.3907-3913.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comanducci M, et al. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 2002;195:1445–1454. doi: 10.1084/jem.20020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capecchi B, et al. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 2005;55:687–698. doi: 10.1111/j.1365-2958.2004.04423.x. [DOI] [PubMed] [Google Scholar]
- 60.Hoiczyk E, Roggenkamp A, Reichenbecher M, Lupas A, Heesemann J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 2000;19:5989–5999. doi: 10.1093/emboj/19.22.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang P, et al. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl Acad. Sci. USA. 2004;101:13630–13635. doi: 10.1073/pnas.0405284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandt CH, Hill CW. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun. 2000;68:2205–2214. doi: 10.1128/IAI.68.4.2205-2214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roggenkamp A, et al. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 2003;185:3735–3744. doi: 10.1128/JB.185.13.3735-3744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nummelin H, et al. The Yersinia adhesin YadA collagen-binding domain sructure is a novel left-handed parallel β-roll. EMBO J. 2004;23:701–711. doi: 10.1038/sj.emboj.7600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soding J, Lupas AN. More than the sum of their parts: on the evolution of proteins from peptides. BioEssays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 66.Bliska JB, Copass MC, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect. Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isberg RR, Leong JM. Multiple β1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60:861–871. doi: 10.1016/0092-8674(90)90099-Z. [DOI] [PubMed] [Google Scholar]
- 68.El Tahir YE, Kuusela P, Skurnik M. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG-S motifs in the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 2000;37:192–206. doi: 10.1046/j.1365-2958.2000.01992.x. [DOI] [PubMed] [Google Scholar]
- 69.Schulze-Koops H, et al. Outer membrane protein YadA of enteropathogenic yersiniae mediates specific binding to cellular but not plasma fibronectin. Infect. Immun. 1993;61:2513–2519. doi: 10.1128/iai.61.6.2513-2519.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flugel A, et al. Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and-2, and nidogen/entactin. J. Biol. Chem. 1994;269:29732–29738. [PubMed] [Google Scholar]
- 71.Rosqvist R, Skurnik M, Wolf-Watz H. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature. 1988;334:522–525. doi: 10.1038/334522a0. [DOI] [PubMed] [Google Scholar]
- 72.Eitel J, Dersch P. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect. Immun. 2002;70:4880–4891. doi: 10.1128/IAI.70.9.4880-4891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.House-Pompeo K, Xu Y, Joh D, Speziale P, Hook M. Conformational changes in the fibronectin binding MSCRAMMs are induced by ligand binding. J. Biol. Chem. 1996;271:1379–1384. doi: 10.1074/jbc.271.3.1379. [DOI] [PubMed] [Google Scholar]
- 74.Schwarz-Linek U, Höök M, Potts JR. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 2004;52:631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 75.Frick IG, Schmidtchen A, Sjöbring U. Interactions between M proteins of Streptococcus pyogenes and glycosaminoglycans promote bacterial adhesion to host cells. Eur. J. Biochem. 2003;270:2303–2311. doi: 10.1046/j.1432-1033.2003.03600.x. [DOI] [PubMed] [Google Scholar]
- 76.Meehan M, Kelly SM, Price NC, Owen P. The C-terminal portion of the fibrinogen-binding protein of Streptococcus equi subsp. equi contains extensive-helical coiled-coil structure and contributes to thermal stability. FEMS Microbiol. Lett. 2002;206:81–86. doi: 10.1111/j.1574-6968.2002.tb10990.x. [DOI] [PubMed] [Google Scholar]
- 77.Meehan M, Nowlan P, Owen P. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiol. 1998;144:993–1003. doi: 10.1099/00221287-144-4-993. [DOI] [PubMed] [Google Scholar]
- 78.Rosenow C, et al. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 1997;25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 79.Sinha B, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1999;2:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 80.Rozelle AL, et al. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP–Arp2/3. Curr. Biol. 2000;10:311–320. doi: 10.1016/S0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- 81.Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchhausen T. Esp15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J. Biol. Chem. 1996;271:28727–28730. doi: 10.1074/jbc.271.46.28727. [DOI] [PubMed] [Google Scholar]
- 82.Marsh M, McMahon HT. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 83.Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J. Gen. Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 84.Roy AMM, Parker JS, Parrish CR, Whittaker GR. Early stages of influenza virus entry into Mv-1 lung cells: involvement of dynamin. Virology. 2000;267:17–28. doi: 10.1006/viro.1999.0109. [DOI] [PubMed] [Google Scholar]
- 85.Greber UF. Signalling in viral entry. Cell. Mol. Life Sci. 2002;59:608–626. doi: 10.1007/s00018-002-8453-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ludwig S, Planz O, Pleschka S, Wolff T. Influenza-virus-induced signaling cascades: targets for antiviral therapy. Trends Mol. Med. 2003;9:46–52. doi: 10.1016/S1471-4914(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 87.Cudmore S, Reckmann I, Way M. Viral manipulations of the actin cytoskeleton. Trends Microbiol. 1997;5:142–148. doi: 10.1016/S0966-842X(97)01011-1. [DOI] [PubMed] [Google Scholar]
- 88.Davis CB, et al. Signal transduction due to HIV-1 envelope interactions with chemokine receptors CXCR4 or CCR5. J. Exp. Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li E, Stupack D, Bokoch GM, Nemerow GR. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 1998;72:8806–8812. doi: 10.1128/jvi.72.11.8806-8812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alrutz MA, Isberg RR. Involvement of focal adhesion kinase in invasin-mediated uptake. Proc. Natl Acad. Sci. USA. 1998;95:13658–13663. doi: 10.1073/pnas.95.23.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Isberg RR, Hamburger Z, Dersch P. Signaling and invasin-promoted uptake via integrin receptors. Microbes Infect. 2000;2:793–801. doi: 10.1016/S1286-4579(00)90364-2. [DOI] [PubMed] [Google Scholar]
- 92.El Tahir Y, Skurnik M. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 2001;291:209–218. doi: 10.1078/1438-4221-00119. [DOI] [PubMed] [Google Scholar]
- 93.Schlaepfer DD, Broome MA, Hunter T. Fibronectin-stimulated signaling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol. Cell. Biol. 1997;17:1702–1713. doi: 10.1128/MCB.17.3.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sieg DJ, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nature Cell Biol. 2000;2:249–256. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 95.Delevoye C, Nilges M, Dautry-Varsat A, Subtil A. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 2004;279:46896–46906. doi: 10.1074/jbc.M407227200. [DOI] [PubMed] [Google Scholar]
- 96.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/S0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 97.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 98.Skehel JJ, Wiley DC. Influenza haemagglutinin. Vaccine. 2002;20:S51–S54. doi: 10.1016/S0264-410X(02)00131-7. [DOI] [PubMed] [Google Scholar]


