Abstract
A stapled α-helix peptide library was designed and constructed using a chemically modified phage display system for screening stapled-peptide ligands against target proteins. The α-helix peptide library, with two cysteine residues on the opposite side of the randomized face, was modified with a rigid hydrocarbon staple linker on a phage. The stapled α-helix peptide phage library was screened against galectin-3 (Gal-3), a cancer-related galactose-binding protein. The obtained stapled peptides showed a high binding affinity (Kd = 0.45 μM) despite being nonsugar ligands. The stapled modification played important roles in stabilizing the α-helical structure that contributed to the high binding affinity to Gal-3. In addition, the best stapled peptide ligands showed specific binding to Gal-3 among various carbohydrate-binding proteins. Thus, the designed α-helix peptide phage library with a constrained structure by the staple linker will advance the discovery of peptide ligands with improved specificity and affinity.
Introduction
An α-helical peptide is one of the important components in protein–protein interactions (PPIs) and protein functions within cells. There are many helical peptide-mediated biological interactions, such as the tumor protein p53/hDM2 interaction,1 HIV infection (membrane fusion protein),2 estrogen receptor/coactivator proteins,3 and the B-cell lymphoma 2 (Bcl-2) family.4,5 α-Helix peptides have a great potential in the regulation of PPIs related to various diseases; however, α-helical peptides alone do not have a stable conformation due to the lack of factors to stabilize their folding. To overcome this limitation, stapled peptides have been developed, in which a pair of amino acids on the same helix face are linked together via covalent bonds between side chains6,7 through several methods, including ring-closing metathesis,8 lactamization,9 cycloadditions,10,11 and thioether formation.12 Stapled peptides have an improved α-helical structure,7 which is associated with increased binding affinity13,14 and specificity,13,14 cell-penetrating property,15 and extended half-life in serum.7,16 In addition, stapling and further modification of peptides can pharmacokinetically stabilize peptide to make them long acting and enhance the oral bioavailability.17 Although various stapled peptide ligands have been developed as PPI inhibitors based on natural α-helix peptide scaffolds,13,14,16,18−20 the stapling strategy is limited to fragment mimetics of natural proteins. The de novo design of PPI regulators is still limited.
Recently, structure-designed peptide phage libraries have been developed to discover de novo peptide ligands for target proteins.21−24 Diverse peptides with stable secondary structures can produce ligands with good selectivity and binding affinity to target proteins. In addition, selective chemical modification methods of phage-displayed peptides have attracted further attention in various fields, including chemical biology and medicinal chemistry, because these methods can extend the functionalities of peptide libraries by the addition of artificial functional groups. A pioneering work in the field of chemically modified peptide phage library was the development of a bicyclic peptide library and the screening of peptides to inhibit a disease-related protease.25 Structurally constrained cyclic peptide libraries are useful for obtaining specific ligands to target proteins.26,27 In addition, conjugating a sugar molecule or an enzyme inhibitor to a phage-displayed library produced target-directed peptide libraries and enabled the discovery of low-micromolar peptide ligands for each target protein.28 Alternatively, the peptide phage library was modified with a bait fragment that can specifically bind to the desired target location to screen covalent binder peptides, which has a benefit of high selectivity and less off-target binding.29
Previously, we reported a chemically modified phage-display system for screening monosaccharide-modified peptide (glycopeptide) ligands to a carbohydrate-binding protein.23,30,31 Selective glycopeptides with high binding affinity to concanavalin A, a mannose-binding protein, were successfully identified from structure-constrained peptide libraries designed based on α-helix and β-loop structures.30−33 These studies revealed that not only target directivity by glyco-modification but also a stable secondary structure of peptides is important for the interaction with the protein. However, it is difficult to strongly stabilize a secondary structure of short peptides less than 20 amino acids. The stabilizing method of short peptides will benefit the ligand screening for target proteins.
Galectin-3 (Gal-3) is a galactose-binding protein and is expressed in the cytoplasm and nucleus and secreted from cells.34,35 Gal-3 has a carbohydrate-binding domain at the C-terminal side, which binds to N-acetyllactosamine (Kd = 0.2 mM)36−38 and lactose (Kd = 1 mM),36−38 through hydrogen bonding with amino acids (His158, Arg162, Asn174, and Glu184). The elevated expression level of Gal-3 is the evidence of its ability to enhance tumor cell adhesion to common extracellular matrix proteins,39 to increase lung carcinoma metastasis,40 and to help cancer cells to resist from apoptosis.41−44 Therefore, Gal-3 is an important target for cancer treatment and diagnosis.
Many types of antagonists that bind to the carbohydrate-binding domain of Gal-3 have been reported. The anti-Gal-3 antibody can reduce the occurrence of metastatic lung colonies. The complex polysaccharide ligands, GCS-100,35,45 GR-MD02, and GM-CT-01,46 induce apoptosis in myeloma cells.47 Peptide ligands, identified in a phage library for Gal-3, blocked the adhesion of human breast carcinoma cells to endothelial cells.48 However, potential ligands are still being researched.
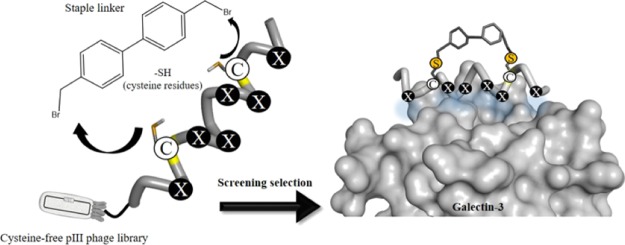
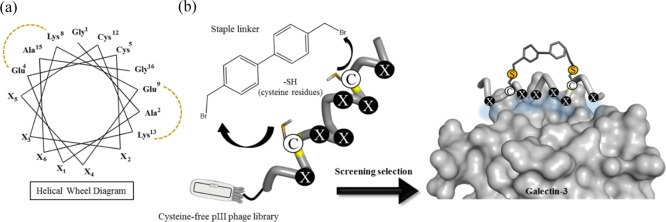
Based on these contexts, in this study, we constructed a stapled α-helix peptide library by combining a chemical modification with a phage display method to produce a diverse structure-constrained peptide pool for ligand screening (Figure 1a). Gal-3 was selected as a target protein because it has a cleft-like structure around its galactose-binding site to which an α-helical peptide can bind (Figure 1b). An α-helix peptide library with six randomized sites was displayed on the filamentous phage fd with the cysteine-free gene-3 protein (pIII).25,49 The library was designed to contain two fixed cysteine residues as modification sites located on the opposite face of a randomized region. A biphenyl (BP) derivative was used as a hydrocarbon staple linker with a rigid structure to enhance the α-helical structure of the peptides. Before affinity-based screening (biopanning), the peptide phage library was chemically modified with a BP linker via two thioether bonds of cysteine residues (Figure 1b).
Figure 1.
(a) Illustration of the helical wheel diagram showing the positions of two cysteines (Cys5 and Cys12) and random amino acid residues (X1–X6) in the designed stapled α-helix peptide library (16-mer peptide, GAX1ECX2X3KEX4X4CKX6AG, Xn = randomized positions). (b) Illustration of the staple modification of the peptide library. The BP linker was reacted with two cysteines via thioether bonds. A designed stapled peptide is expected to have a stabilized α-helical structure and bind to Gal-3 at the galactose-binding site with a cleft-like structure.
Results and Discussion
Construction of the Stapled Peptide Phage Design
An α-helical peptide library was designed as 16-mer peptides, GAX1ECX2X3KEX4X5CKX6AG (Xn = any of the 20 natural amino acids), with six randomized positions on the same face of the helix. The sequence of the peptide library was designed to have an α-helical behavior by containing alanine residues (Ala2, Ala15) due to its α-helix-stabilizing properties,50 two pairs of glutamate and lysine residues (Glu4-Lys8 and Glu9-Lys13) to maintain the helical structure by an intramolecular salt bridge formation (Figure 1a), and glycine residues at both ends of the peptide to stabilize the backbone hydrogen bonds. Two cysteine residues were arranged at the 5th and 12th (i and i + 7) positions as the modification sites that were placed on the opposite side of the randomized region (Figure 1a). 4,4-Bis(bromomethyl) biphenyl (BP) was used as a cysteine-reactive hydrocarbon staple linker because a BP scaffold is not only suitable to crosslink the two cysteine residues at the i and i + 7 positions, but also efficient to promote the cell permeability of peptides due to its hydrophobic property.51 As a result, the BP-stapled peptides will have a stabilized helical structure that can display six randomized amino acids on the same face of the helix. Before the preparation of a peptide phage library, a model peptide (mentioned as a control peptide later) with six alanine residues at the randomized positions (H-GAAECAAKEAACKAAG-NH2) was tested for the staple modification by a BP linker (Supporting Information). The model peptide gave the stapled product within 1 h without any side reaction (Figure S1). Although the model peptide did not show an α-helical structure, the stapling reaction of the model peptide almost completed within 1 h, indicating that any sequences, including less helical peptides displayed on the phage, could give the stapled product.
The non-cys fd bacteriophage was used to achieve the selective staple modification of desired peptides displayed on the phage. The phage library displaying the designed α-helix peptides was produced by the genetic manipulation of a phagemid vector. The obtained peptide phage library covered the theoretical diversity (6.4 × 107) of peptides with six randomized amino acids. Then, the peptide phage library was amplified and modified with a BP staple linker to produce the stapled peptide phage library before the screening process.
Affinity Screening
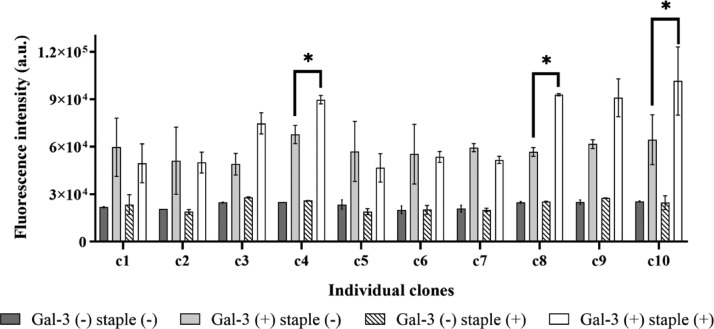
The peptide phage library was reduced with TCEP to avoid disulfide bonds. After reduction, the BP staple linker was reacted with a phage solution ([BP linker] = 10 μM, [phage] = 100 nM) at 42 °C for 1 h. The BP-modified peptide phage was purified and collected by a standard phage precipitation protocol and resuspended in HBS [10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 150 mM NaCl, pH 7.0]. The modification of peptides on phages in previous studies confirmed that the modification condition is sufficient.30,31 In addition, the number of phages used for affinity screening was more than 1.0 × 1010 (Table S1), and the theoretical diversity of the phage library was 6.4 × 107, meaning that more than 150 copies for each sequence were included in the library. Therefore, all kinds of peptide sequences could be modified with a BP linker. Accordingly, the BP-stapled peptide phage was screened against biotinylated Gal-3. Phages binding to Gal-3 were captured by streptavidin-magnetic beads. After weakly bound phages were washed out, the remaining phages were selectively eluted using an excess amount of lactose as a competitive binder to Gal-3 to obtain peptide ligands that bind to the galactose-binding site of Gal-3. To concentrate the phages bound to Gal-3, the amount of Gal-3 was decreased from 1.0 μg (first and second rounds) to 0.5 μg in the third and fourth rounds of biopanning. The recovery yield of the phage pool in each round was monitored by phage titration (Table S1 and Figure S2). The yield increased from the first to second round, indicating that Gal-3-bound phages increased. In the third round, the yield decreased due to less Gal-3; however, the yield increased again in the fourth round, indicating the enrichment of Gal-3-bound phages. The screening efficiency was also checked by a phage ELISA using all phage pools (Figure S3). In the case of the phage library before screening, phages displaying non-stapled peptides showed fluorescent intensity similar to the phage displaying stapled peptides, indicating that the phage library contains many phages that do not specifically bind to Gal-3. The phage pools with a staple modification after the first and second biopanning clearly showed higher fluorescent intensity than those without staple modification. The fourth-round phage pool showed the most significant difference in fluorescent intensity with and without staple modification, suggesting that phages displaying stapled peptides binding to Gal-3 were successfully obtained. After the cloning and DNA sequencing of individual clones from the fourth-round phage pool, 27 phage clones were identified (Table S2). Although there was no consensus sequence, most peptides contained polar amino acids (Gln, Asn, Ser, and Thr) that could form hydrogen bonds to the binding site of Gal-3. Twenty-six clones were classified based on their total net charge from −2 to +2 at pH 7.0. The clone with an extra cysteine residue in the peptide sequence was ignored because this peptide might form an undesired cross-linked structure. There were many peptide sequences in the category of +1 and neutral net charge. Then, 10 different representative clones were selected from each category (Table 1), and a phage ELISA using individual clones with and without staple modification was performed to verify the ability of the phages to bind Gal-3 (Figure 2). Among the representative clones, five phage clones (c3, c4, c8, c9, and c10) showed increased fluorescent intensity. In particular, these five clones displaying a stapled peptide exhibited higher fluorescent intensity than non-stapled phages did; in contrast, other phage clones did not show a significant difference despite the presence or absence of staple modification. These results suggest that several stapled peptides that bind to Gal-3 were successfully screened and that the staple modification contributed to their binding ability. Among these five clones, c4, c8, and c10 were selected for further validation experiments. The c3 and c9 clones were removed because they have the same net charge as c4 and c10, respectively, and showed lower fluorescent intensity than those clones.
Table 1. Sequences of the Individual Clones for Phage-Based ELISAa.
| clone | sequence (GA-X1-EC-X2X3-KE-X4X5-CK-X6-AG) | net charge |
|---|---|---|
| c1 | YVDEGG | –2 |
| c2 | TDDHNQ | –1 |
| c3 | FYSQMP | 0 |
| c4 | QVYQSS | 0 |
| c5 | GQVPRS | +1 |
| c6 | AQVFSH | +1 |
| c7 | TQMTPH | +1 |
| c8 | QPPPAK | +1 |
| c9 | KPHTTQ | +2 |
| c10 | PKRYEK | +2 |
The underlined amino acids mean X1–X6 of random positions.
Figure 2.
Phage-based ELISA results. The gray solid and black strip bars indicate the fluorescence intensity of non-stapled and stapled phage clones to the microplate without Gal-3 (Gal-3(−)), respectively. The light gray solid and white solid bars indicate the fluorescence intensity of non-stapled and stapled phage clones to the Gal-3-immobilized microplate (Gal-3(+)), respectively. For all samples, n = 3; the error bars represent the standard deviation. *p < 0.05.
Structural Analysis of Peptides
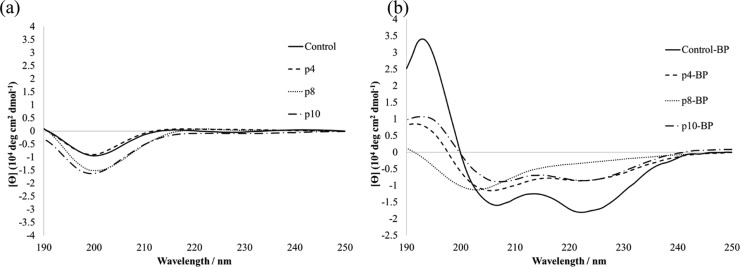
To investigate the detailed characteristics of displayed peptides, the three peptides from the c4, c8, and c10 phage clones were chemically synthesized as p4, p8, and p10, respectively (Table 2). These peptides were modified with the BP linker to produce the stapled peptides, p4-BP, p8-BP, and p10-BP. In addition, a peptide with six alanine residues at randomized positions was synthesized as a control peptide (control) and modified with the BP linker to produce control-BP. The secondary structure of all peptides was evaluated by circular dichroism spectrometry (Figure 3). All non-stapled peptides showed a negative maximum at 200 nm, suggesting a random coil structure (Figure 3a). On the other hand, p4-BP, p10-BP, and control-BP exhibited typical spectra for α-helical structures with double-negative maxima at 208 and 222 nm (Figure 3b). The p8-BP did not show a significant change after BP modification. Since p8-BP and p10-BP contain proline residues, their α-helical structure might be distorted.52 In the case of p10-BP containing one proline residue, the BP linker can maintain the α-helical structure, because this proline residue is placed outside of a stapled region. In the case of control-BP, the alanine has a helix-forming propensity and a small side chain structure that can avoid steric hindrance between side chains and the BP linker. As a result, control-BP exhibited the highest α-helical content (60%), followed by p4-BP (32%) and p10-BP (30%) (Table 2). This result clearly suggests that the BP linker can enhance the α-helical structure of these peptides.
Table 2. Sequences, Helical Contents, and Dissociation Constants of Peptides to Gal-3c.
| peptide | sequence | helicity | Kd (μM) |
|---|---|---|---|
| p4-BP | H-GAQECVYKEQSCKSAG-NH2 | 32% | |
| p4-flu-BP | H-GAQECVYKEQSCKSAG-GGGK-(flu)-NH2 | 0.45 ± 0.18 | |
| (3.75 ± 2.22)a | |||
| p8-BP | H-GAQECPPKEPACKKAG-NH2 | 14% | |
| p8-flu-BP | H-GAQECPPKEPACKKAG-GGGK-(flu)-NH2 | nd | |
| p10-BP | H-GAPECKRKEYECKKAG-NH2 | 30% | |
| p10-flu-BP | H-GAPECKRKEYECKKAG-GGGK-(flu)-NH2 | 1.16 ± 0.42 | |
| (4.74 ± 3.53)a | |||
| control-BP | H-GAAECAAKEAACKAAG-NH2 | 60% | |
| control-flu-BP | H-GAAECAAKEAACKAAG-GGGK-(flu)-NH2 | nd | |
| p4 | H-GAQECVYKEQSCKSAG-NH2 | nd | |
| p4-flu | H-GAQECVYKEQSCKSAG-GGGK-(flu)-NH2 | nd | |
| p8 | H-GAQECPPKEPACKKAG-NH2 | nd | |
| p8-flu | H-GAQECPPKEPACKKAG-GGGK-(flu)-NH2 | nd | |
| p10 | H-GAPECKRKEYECKKAG-NH2 | 5.7% | |
| p10-flu | H-GAPECKRKEYECKKAG-GGGK-(flu)-NH2 | nd | |
| control | H-GAAECAAKEAACKAAG-NH2 | 3.4% | |
| control-flu | H-GAAECAAKEAACKAAG-GGGK-(flu)-NH2 | nd | |
| lactose | 1 mMb |
Figure 3.
Circular dichroism spectra of the peptides. (a) Non-stapled peptides. (b) Stapled peptides. All peptides (50 μM) were measured at 25 °C in 10 mM Tris-HCl buffer (pH 7.0).
Binding Affinity of Peptides
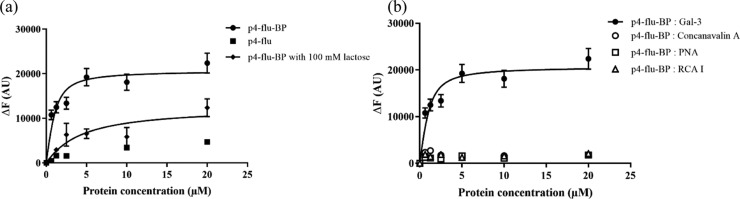
To evaluate the binding affinity of the screened peptides (p4, p8, and p10) and the control peptide to Gal-3, 5(6)-carboxyfluorescein-labeled peptides (p4-flu, p8-flu, p10-flu, and control-flu) were chemically synthesized. Fluorescein was introduced into the side chain of a lysine residue at the C-terminal of peptides via a triglycine spacer (Table 2). These labeled peptides were also modified with the BP linker to produce stapled-fluorescent peptides (p4-flu-BP, p8-flu-BP, p10-flu-BP, and control-flu-BP). The binding affinity of the peptides to Gal-3 was determined by fluorescence titration experiments (Figures 4a and S4–S6). Fluorescein-labeled peptides were titrated with Gal-3, and the dissociation constants (Kd) were estimated by analyzing the fluorescent intensity change with a nonlinear least-square curve-fitting method based on the 1:1 stoichiometry model (Table 2). The Kd value of p4-flu-BP was determined to be 0.45 μM, which was 2000-fold lower than that of lactose (1 mM). This result suggests that a high binding affinity ligand was successfully screened from the stapled α-helix peptide phage library. The p4-flu peptide without staple modification did not show significant fluorescent changes, indicating that staple modification is important for high binding affinity. In addition, the apparent Kd value of p4-flu-BP was increased to 3.75 μM in the presence of excess lactose (100 mM) compared with the absence of lactose (0.45 μM), which was 8.3-fold higher than that of p4-flu-BP alone (Figure 4a, Table 2). This result suggests that p4-flu-BP bound to the galactose-binding site of Gal-3. Similarly, p10-flu-BP exhibited good binding affinity (Kd = 1.16 μM) to Gal-3 in a competitive manner to lactose (Kd = 4.74 μM in the presence of 100 mM lactose), and p10-flu without staple modification did not show any Gal-3 binding (Figure S5, Table 2). These results suggest that p10-BP also competitively binds to the galactose-binding site of Gal-3 and that staple modification plays an important role in high binding affinity. On the other hand, the binding affinity of p8-flu and p8-flu-BP peptides could not be evaluated because the fluorescent intensity change was not well related to the Gal-3 concentration (Figure S4). Since the excess amount of lactose did not significantly affect the change in the fluorescent intensity of p8-flu-BP, the p8-BP peptide might interact with Gal-3 in a nonspecific manner. The Kd values of the control-flu and control-flu-BP peptides were also not estimated for the same reason (Figure S6). As a result, selected amino acids of p4 and p10 peptides are important components for the specific binding to the carbohydrate-binding domain of Gal-3. However, the different driving forces between p4-flu-BP and p10-flu-BP in binding to Gal-3 should be considered because p4-flu-BP contains non-charged amino acids (glutamine and serine), while p10-flu-BP contains charged amino acids (lysine, arginine, and glutamic acid). The Gal-3 protein contains some hydrophilic amino acids, such as Asn160 and Asn174, that interact with lactose via hydrogen bonding (Figure S7a), implying that the non-charged amino acids of the p4 peptide partly mimic the sugar unit. In contrast, Gal-3 has charged amino acids, such as Glu165, Glu184, Arg144, His158, and Arg162, around the galactose-binding site that could make electrostatic interactions with the p10 peptide. The tyrosine residue is common between two peptides, which might make a hydrophobic interaction with Trp181 located at the galactose-binding site of Gal-3 (Figure S7b). The valine residue in p4-flu-BP seems to also contribute to hydrophobic interactions. Additional hydrophobic interactions might contribute to the improved Gal-3 binding of the p4-BP peptide compared with that of the p10-BP peptide.
Figure 4.
Fluorescence titration experiments: (a) p4-flu-BP ( ●), p4-flu ( ■), p4-flu-BP with 100 mM lactose (⧫) to Gal-3. (b) p4-flu-BP to Gal-3 (●), p4-flu-BP to concanavalin A ( ○), p4-flu-BP to PNA (□), and p4-flu-BP to RCA I (△). The curves represent a nonlinear least-squares curve fitting based on a 1:1 stoichiometry model.
Other carbohydrate-binding proteins, such as concanavalin A (Con A, a mannose-binding protein), peanut agglutinin (PNA, a galactose-binding protein), and Ricinus communis agglutinin I (RCA I, a galactose-binding protein), were used to evaluate the binding specificity of the p4-flu-BP peptide to Gal-3 (Figure 4b). The results of the fluorescence titration experiment showed no significant fluorescent change in p4-flu-BP in the presence of Con A, PNA, or RCA I. This result clearly suggests that the p4-BP peptide specifically binds to Gal-3. In particular, p4-BP could discriminate Gal-3 from PNA and RCA I although these three proteins have the same galactose-binding function, meaning that the p4-BP peptide not only mimics the galactose moiety but also selectively recognizes the surface cleft-like structure of Gal-3.
Conclusions
We successfully constructed a stapled α-helical peptide phage library and identified Gal-3-binding stapled peptides with good binding affinity and selectivity. The designed peptides displayed on the pIII coat protein of the phage library were modified with a rigid hydrocarbon staple linker through thioether bonds via two cysteine residues. After affinity-based screening, several phage clones with binding affinity to Gal-3 were identified. The secondary structure of the isolated peptides was stabilized as an α-helix structure by stapling with a rigid BP linker. The stapled peptides, p4-flu-BP and p10-flu-BP, showed high binding affinity to the target protein, Gal-3; the binding affinity of these peptides was 1000-fold better than that of lactose, a simple carbohydrate. These stapled peptides bound to the galactose-binding site with a cleft-like structure and the best peptide p4-flu-BP showed specific binding only to Gal-3. The stapled peptide ligand could be conjugated with nanoparticles for drastic improvement in the binding affinity to Gal-3, because we previously demonstrated that glycopeptide ligands displayed on gold nanoparticles exhibited high binding affinity by a multivalent effect.33 In addition, different kinds of staple linkers will be examined for further improvement in the quality of the peptide library, including the availability for other PPIs in the future. Therefore, the structure-based design of a stapled α-helix peptide library could be utilized for the discovery of peptide ligands with high specificity and improved binding affinity toward the target protein.
Experimental Section
Construction of the Peptide Phage Library
A detailed procedure is described in the Supporting Information Briefly, the phagemid vector (fdg3p0ss21-library) encoding α-helix peptides with six random amino acids (GAX1ECX2X3KEX4X5CKX6AG; Xn = any of the 20 natural amino acids) was constructed according to a previously reported method.25,32,33,49 The PCR primer sequences are shown in Table S3. NNK codon (N = A, T, C, G; K = G, T nucleotide) was used for the randomized positions to encode all 20 amino acids eliminating possible stop codons. A 16-mer peptide phage library was generated by the electroporation of the fdg3p0ss21-library phagemid vector into TG1 Escherichia coli cells. The diversity of the library was estimated to be approximately 5.92 × 109 colonies that can cover the theoretically possible peptides (206 = 6.4 × 107). Before affinity screening (biopanning), glycerol stocks of library-transformed TG1 E. coli cells were cultured in a 2YT medium supplemented with chloramphenicol (30 μg/mL) to prepare the phage pool. The amplified phage was collected by a standard phage precipitation protocol.53,54
Preparation of the Stapled Peptide Phage Library by Chemical Modification
The peptides displayed on the phages were chemically modified with the BP staple linker to prepare a stapled α-helix peptide phage library. The disulfide bond was reduced by incubating phage (100 μL, 100 nM) with excess TCEP (1.0 mM) in a degassed reaction buffer [20 mM NH4HCO3, 5 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0] at 42 °C for 1 h. The reduced phage was collected by the standard phage precipitation protocol and resuspended in degassed reaction buffer. The BP staple linker dissolved in acetonitrile (1.0 mM) was added to the phage solution ([BP linker] = 10 μM, [phage] = 100 nM), and the mixture was incubated at 42 °C for 1 h. Finally, the BP-modified phage was purified and collected by the standard phage precipitation protocol and resuspended in HBS (10 mM HEPES, 150 mM NaCl, pH 7.0).
Expression of Gal-3 Protein and Biotin Modification
The pGEX-4T-2 bacterial vector with the cDNA of human galectin-3 (LGAL3, Bioresource Center, RIKEN)55 was transformed into E. coli BL21-Codonplus (DE3)-RIPL competent cells (Agilent Technologies). The expression of GST-tagged Gal-3 was conducted in a 2YT medium with antibiotics, protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (1 mM), and the cells were incubated at 37 °C for 3 h. After the induction period, the bacterial pellet was collected by centrifugation and lysed by ultrasonication in the presence of protease inhibitors. After centrifugation, the supernatant was applied to a Glutathione-Sepharose 4B chromatography column (GE Healthcare) for affinity purification of the GST-Gal-3 protein. The GST-Gal-3 protein was eluted with an elution buffer (50 mM Tris, 10 mM reduced glutathione, pH 8.0) according to the manufacturer’s instructions. The GST tag was removed from the GST-Gal-3 protein by the thrombin treatment. The cleaved Gal-3 was purified by the lactose gel affinity column chromatography (EY Laboratories). The bound Gal-3 was eluted with 100 mM lactose (phosphate buffered saline, pH 7.3), and the buffer was exchanged for HBS by ultracentrifugation with an Amicon centrifugal filter (10 kDa, MWCO) at 4 °C and 7500g for 15 min.
Purified Gal-3 was biotinylated by using biotin-AC5-OSu (10 equiv) (Dojindo) at 37 °C for 1 h. The excess biotin-AC5-OSu was removed by ultracentrifugation in the same manner. Biotinylation was confirmed using streptavidin-immobilized magnetic beads (Dynabeads).
Affinity Screening (Biopanning)
The BP-modified phage solution (100 nM in 100 μL HBS) was incubated with biotinylated Gal-3 (1 μg) at room temperature for 1 h. The phage bound to Gal-3 was captured with streptavidin-immobilized magnetic beads and blocked by 1% bovine serum albumin (BSA) by incubating at room temperature for 15 min with gentle rotation (5 rpm). After the magnetic beads were washed 5 times with HBS-T (HBS with 0.1% v/v Tween 20), the bound phages were competitively eluted 2 times with excess lactose (200 mM in HBS). The eluted phages were infected with log-phase E. coli TG1 cells at 37 °C for 90 min, and infected TG1 cells were cultured overnight for the amplification of the eluted phage. The amplified phage was used for the next screening round. The second screening was conducted in the same manner. Third and fourth rounds of biopanning were conducted using reduced amounts of biotinylated Gal-3, 0.5 μg for both rounds. The selection efficiency was evaluated based on the percentage yield of the bound phage (output/input, %).56 After the fourth round, 27 colonies were selected for the preparation of phage clones. Peptide sequences were identified by a DNA sequence analysis from isolated phage clones.
Phage ELISA
A phage-based enzyme-linked immunosorbent assay (ELISA) was conducted to evaluate the binding of phages to Gal-3. A 96-well microplate (Corning, Costar 3690) was coated with Gal-3 (100 μL, 1.0 μg per well) at 4 °C overnight. After 3 washes with HBS, each well was blocked with 1% BSA (w/v) in HBS at 4 °C for 1 h. The phage solution (1.0 nM, 50 μL) in HBS was added to the Gal-3-immobilized well and incubated at room temperature for 1 h. The well was washed with HBS 3 times, and an anti-M13 phage antibody labeled with horseradish peroxidase (Santa Cruz Biotechnology) was added to each well (70 μL, dilution of 1:500 in 1% (w/v) BSA/HBS) and incubated at room temperature for 1 h. After the well was washed with HBS 3 times, the substrate for the fluorogenic enzyme reaction (QuantaBlu Fluorogenic Peroxidase Substrate kit, Pierce) was added. The fluorescence (λex = 390 nm, λem = 475 nm) was measured with a microplate fluorometer (Twinkle LB 970, Berthold Technologies). All phage pools that underwent biopanning and the 10 selected phage clones after the fourth round biopanning were used for phage ELISA with and without staple modification. Statistical significance was accepted at p < 0.05.
Peptide Synthesis
A detailed procedure for the peptide synthesis is described in the Supporting Information. The introduction of 5(6)-carboxyfluorescein into the side chain of the C-terminal lysine was conducted on the solid support. The purified peptides were dissolved (100 μM) in a degassed reaction buffer (20 mM NH4HCO3, 5 mM EDTA, pH 8.0). Then, the BP linker in acetonitrile (10 mM) was added to the peptide solution (final concentration was 1 mM). After the modification reaction was conducted at 42 °C for 1 h, the stapled peptides were purified by RP-HPLC and identified by ESI–MS.
Circular Dichroism Study
The circular dichroism spectra of peptides were recorded on a JASCO J-1100 spectropolarimeter using a quartz cell with a 0.1 cm path length at 25 °C. Stapled peptides were dissolved in Tris-HCl buffer (10 mM, pH 7.0) to prepare a 50 μM solution. Non-stapled peptides were dissolved in Tris-HCl buffer containing TCEP (1.0 mM). The α-helical contents of peptides were calculated by a reported method.57
Evaluation of the Binding Affinity of Peptides
Fluorescein-labeled peptides were used for a titration experiment with Gal-3 and other carbohydrate-binding proteins (ConA, PNA, and RCA I). The stock solution (100 μM) of fluorescein-labeled peptides was prepared with HBS and diluted (10 μM) into protein solutions with various concentrations from 0.625 to 20 μM in HBS. The mixture (100 μL) was incubated at room temperature for 25 min in a test tube with gentle rotation (5 rpm). After incubation, the mixture was transferred into a 96-well plate (Corning, Costar 3915) and fluorescence intensity was measured with a plate reader (PerkinElmer, 1420 Multilabel Counter ARVO MX) (λex = 485 nm, λem = 535 nm). The measurement was conducted 3 times for reproducibility. The fluorescent intensity change was analyzed by a Graphpad Prism software using a nonlinear least-square curve-fitting method based on a 1:1 stoichiometry model to determine the dissociation constants of peptides to proteins. In the cases of p4-flu-BP and p10-flu-BP, the same measurement was conducted in the presence of 100 mM lactose as a competitor.
Acknowledgments
This work was supported in part by KAKENHI, MEXT, Japan. The authors thank the Biomaterials Analysis Division, Tokyo Institute of Technology, for the DNA sequence analysis. The CD spectra measurement was supported by the Open Research Facilities for Life Science and Technology.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03461.
HPLC trace of the staple modification of the model peptide, titer check of biopanning, phage ELISA of each biopanning phage pool, peptide sequences of identified phage clones, fluorescent titration results of peptides with Gal-3, and the structural figure of the Gal-3-binding site (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kussie P. H.; Gorina S.; Marechal V.; Elenbaas B.; Moreau J.; Levine A. J.; Pavletich N. P. Structure of the MDM2 Oncoprotein Bound to the p53 Tumor Suppressor Transactivation Domain. Science 1996, 274, 948–953. 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Chan D. C.; Fass D.; Berger J. M.; Kim P. S. Core Structure of gp41 from the HIV Envelope Glycoprotein. Cell 1997, 89, 263–273. 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Bruning J. B.; Parent A. A.; Gil G.; Zhao M.; Nowak J.; Pace M. C.; Smith C. L.; Afonine P. V.; Adams P. D.; Katzenellenbogen J. A.; Nettles K. W. Coupling of receptor conformation and ligand orientation determine graded activity. Nat. Chem. Biol. 2010, 6, 837–843. 10.1038/nchembio.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar P. E.; Lee E. F.; van Delft M. F.; Day C. L.; Smith B. J.; Huang D. C. S.; Fairlie W. D.; Hinds M. G.; Colman P. M. Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 6217–6222. 10.1073/pnas.0701297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Dai S.; Zhu Y.; Marrack P.; Kappler J. W. The Structure of a Bcl-xL/Bim Fragment Complex. Immunity 2003, 19, 341–352. 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- Tsomaia N. Peptide therapeutics: Targeting the undruggable space. Eur. J. Med. Chem. 2015, 94, 459–470. 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Schafmeister C. E.; Po J.; Verdine G. L. An All-Hydrocarbon Cross-Linking System for Enhancing the Helicity and Metabolic Stability of Peptides. J. Am. Chem. Soc. 2000, 122, 5891–5892. 10.1021/ja000563a. [DOI] [Google Scholar]
- Blackwell H. E.; Grubbs R. H. Highly Efficient Synthesis of Covalently Cross-Linked Peptide Helices by Ring-Closing Metathesis. Angew. Chem., Int. Ed. 1998, 37, 3281–3284. . [DOI] [PubMed] [Google Scholar]
- Felix A. M.; Heimer E. P.; Wang C.-T.; Lambros T. J.; Fournier A.; Mowles T. F.; Maines S.; Campbell R. M.; Wegrzynski B. B.; Toome V.; Fry D.; Madison V. S. Synthesis, biological activity and conformational analysis of cyclic GRF analogs. Int. J. Pept. Protein Res. 2009, 32, 441–454. 10.1111/j.1399-3011.1988.tb01375.x. [DOI] [PubMed] [Google Scholar]
- Moses J. E.; Moorhouse A. D. Correction: The growing applications of click chemistry. Chem. Soc. Rev. 2016, 45, 6888. 10.1039/c6cs90108e. [DOI] [PubMed] [Google Scholar]
- Scrima M.; Le Chevalier-Isaad A.; Rovero P.; Papini A. M.; Chorev M.; D’Ursi A. M. CuI-Catalyzed Azide-Alkyne Intramolecular i-to-(i+4) Side-Chain-to-Side-Chain Cyclization Promotes the Formation of Helix-Like Secondary Structures. Eur. J. Org. Chem. 2010, 2010, 446–457. 10.1002/ejoc.200901157. [DOI] [Google Scholar]
- Brunel F. M.; Dawson P. E. Synthesis of constrained helical peptides by thioether ligation: application to analogs of gp41. Chem. Commun. 2005, 2552. 10.1039/b419015g. [DOI] [PubMed] [Google Scholar]
- Brown C. J.; Quah S. T.; Jong J.; Goh A. M.; Chiam P. C.; Khoo K. H.; Choong M. L.; Lee M. A.; Yurlova L.; Zolghadr K.; Joseph T. L.; Verma C. S.; Lane D. P. Stapled Peptides with Improved Potency and Specificity That Activate p53. ACS Chem. Biol. 2012, 8, 506–512. 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- Chang Y. S.; Graves B.; Guerlavais V.; Tovar C.; Packman K.; To K.-H.; Olson K. A.; Kesavan K.; Gangurde P.; Mukherjee A.; Baker T.; Darlak K.; Elkin C.; Filipovic Z.; Qureshi F. Z.; Cai H.; Berry P.; Feyfant E.; Shi X. E.; Horstick J.; Annis D. A.; Manning A. M.; Fotouhi N.; Nash H.; Vassilev L. T.; Sawyer T. K. Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E3445 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thean D.; Ebo J. S.; Luxton T.; Lee X. E. C.; Yuen T. Y.; Ferrer F. J.; Johannes C. W.; Lane D. P.; Brown C. J. Enhancing Specific Disruption of Intracellular Protein Complexes by Hydrocarbon Stapled Peptides Using Lipid Based Delivery. Sci. Rep. 2017, 7, 1763. 10.1038/s41598-017-01712-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky L. D.; Kung A. L.; Escher I.; et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 2004, 305, 1466–1470. 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear S.; Amso Z.; Shen W. Engineering PEG-fatty acid stapled, long-acting peptide agonists for G protein-coupled receptors. Methods Enzymol. 2019, 622, 183–200. 10.1016/bs.mie.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Baek S.; Kutchukian P. S.; Verdine G. L.; Huber R.; Holak T. A.; Ki Won L.; Popowicz G. M. Structure of the Stapled p53 Peptide Bound to Mdm2. J. Am. Chem. Soc. 2012, 134, 103–106. 10.2210/pdb3v3b/pdb. [DOI] [PubMed] [Google Scholar]
- Bird G. H.; Irimia A.; Ofek G.; Kwong P. D.; Wilson I. A.; Walensky L. D. Stapled HIV-1 peptides recapitulate antigenic structures and engage broadly neutralizing antibodies. Nat. Struct. Mol. Biol. 2014, 21, 1058–1067. 10.1038/nsmb.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. L.; Fire E.; Keating A. E.; Walensky L. D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 2010, 6, 595–601. 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii I.; Takaoka Y.; Suzuki K.; Tanaka T. A conformationally purified α-helical peptide library. Tetrahedron Lett. 2001, 42, 3323–3325. 10.1016/s0040-4039(01)00382-3. [DOI] [Google Scholar]
- Suzuki N.; Fujii I. Optimazation of the loop length for folding of a helix-loop-helix peptide. Tetrahedron Lett. 1999, 40, 6013–6017. 10.1016/s0040-4039(99)01095-3. [DOI] [Google Scholar]
- Sawada T.; Ishiguro K.; Takahashi T.; Mihara H. A novel β-loop scaffold of phage-displayed peptides for highly specific affinities. Mol. BioSyst. 2011, 7, 2558. 10.1039/c1mb05085k. [DOI] [PubMed] [Google Scholar]
- Tsutsumi H.; Nakano K.; Mihara H. Dihydrofolate reductase inhibitory peptides screened from a structured designed β-loop peptide library displayed on phage. Mol. BioSyst. 2015, 11, 2713–2716. 10.1039/c5mb00316d. [DOI] [PubMed] [Google Scholar]
- Heinis C.; Rutherford T.; Freund S.; Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 2009, 5, 502–507. 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- Rebollo I. R.; Heinis C. Phage selection of bicyclic peptides. Methods 2013, 60, 46–54. 10.1016/j.ymeth.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Fukunaga K.; Hatanaka T.; Ito Y.; Minami M.; Taki M. Construction of a crown ether-like supramolecular library by conjugation of genetically-encoded peptide linkers displayed on bacteriophage T7. Chem. Commun. 2014, 50, 3921–3923. 10.1039/c4cc00811a. [DOI] [PubMed] [Google Scholar]
- Ng S.; Bennett N. J.; Schulze J.; Gao N.; Rademacher C.; Derda R. Genetically-encoded fragment-based discovery of glycopeptide ligands for DC-SIGN. Bioorg. Med. Chem. 2018, 26, 5368–5377. 10.1016/j.bmc.2018.08.036. [DOI] [PubMed] [Google Scholar]
- Uematsu S.; Tabuchi Y.; Ito Y.; Taki M. Combinatorially Screened Peptide as Targeted Covalent Binder: Alteration of Bait-Conjugated Peptide to Reactive Modifier. Bioconjugate Chem. 2018, 29, 1866–1871. 10.1021/acs.bioconjchem.8b00301. [DOI] [PubMed] [Google Scholar]
- Chang I. V.; Tsutsumi H.; Mihara H. Screening for concanavalin A binders from a mannose-modified α-helix peptide phage library. Mol. BioSyst. 2017, 13, 2222–2225. 10.1039/c7mb00495h. [DOI] [PubMed] [Google Scholar]
- Arai K.; Tsutsumi H.; Mihara H. A monosaccharide-modified peptide phage library for screening of ligands to carbohydrate-binding proteins. Bioorg. Med. Chem. Lett. 2013, 23, 4940–4943. 10.1016/j.bmcl.2013.06.059. [DOI] [PubMed] [Google Scholar]
- Usui K.; Ojima T.; Tomizaki K.-Y.; Mihara H. A Designed Glycopeptide Array for Characterization of Sugar-Binding Proteins Toward a Glycopeptide Chip Technology. NanoBiotechnology 2005, 1, 191–200. 10.1385/nbt:1:2:191. [DOI] [Google Scholar]
- Tsutsumi H.; Ohkusa H.; Park H.; Takahashi T.; Yuasa H.; Mihara H. Gold nanoparticles conjugated with monosaccharide-modified peptide for lectin detection. Bioorg. Med. Chem. Lett. 2012, 22, 6825–6827. 10.1016/j.bmcl.2012.09.051. [DOI] [PubMed] [Google Scholar]
- Nangia-Makker P.; Hogan V.; Raz A. Galectin-3 and cancer stemness. Glycobiology 2018, 28, 172–181. 10.1093/glycob/cwy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streetly M. J.; Maharaj L.; Joel S.; Schey S. A.; Gribben J. G.; Cotter F. E. GCS-100, a novel galectin-3 antagonist, modulates MCL-1, NOXA, and cell cycle to induce myeloma cell death. Blood 2010, 115, 3939–3948. 10.1182/blood-2009-10-251660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumic J.; Dabelic S.; Flögel M. Galectin-3: An open-ended story. Biochim. Biophys. Acta 2006, 1760, 616–635. 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Seetharaman J.; Kanigsberg A.; Slaaby R.; Leffler H.; Barondes S. H.; Rini J. M. X-Ray Crystal Structure Of The Human Galectin-3 Carbohydrate Recognition Domain (Crd) At 2.1 Angstrom Resolution. J. Biol. Chem. 1998, 273, 13047–13052. 10.2210/pdb1a3k/pdb. [DOI] [PubMed] [Google Scholar]
- Ahmed H.; Alsadek D. M. M. Galectin-3 as a Potential Target to Prevent Cancer Metastasis. Clin. Med. Insights: Oncol. 2015, 9, 113. 10.4137/cmo.s29462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochieng J.; Warfield P.; Green-Jarvis B.; Fentie I. Galectin-3 regulates the adhesive interaction between breast carcinoma cells and elastin. J. Cell. Biochem. 1999, 75, 505–514. . [DOI] [PubMed] [Google Scholar]
- Raz A.; Zhu D.; Hogan V.; Shah N.; Raz T.; Karkash R.; Pazerini G.; Carmi P. Evidence for the role of 34-kDa galactoside-binding lectin in transformation and metastasis. Int. J. Cancer 1990, 46, 871–877. 10.1002/ijc.2910460520. [DOI] [PubMed] [Google Scholar]
- Akahani S.; Nangia-Makker P.; Inohara H.; Kim H.-R. C.; Raz A. Galectin-3: A Novel Antiapoptotic Molecule with A Functional BH1 (NWGR) Domain of Bcl-2 Family. Cancer Res. 1997, 57, 5272–5276. [PubMed] [Google Scholar]
- Matarrese P.; Fusco O.; Tinari N.; Natoli C.; Liu F.-T.; Semeraro M. L.; Malorni W.; Iacobelli S. Galectin-3 overexpression protects from apoptosis by improving cell adhesion properties. Int. J. Cancer 2000, 85, 545–554. . [DOI] [PubMed] [Google Scholar]
- Yu F.; Finley R. L.; Raz A.; Kim H.-R. C. Galectin-3 Translocates to the Perinuclear Membranes and Inhibits CytochromecRelease from the Mitochondria. J. Biol. Chem. 2002, 277, 15819–15827. 10.1074/jbc.m200154200. [DOI] [PubMed] [Google Scholar]
- Zou J. Peptides specific to the galectin-3 carbohydrate recognition domain inhibit metastasis-associated cancer cell adhesion. Carcinogenesis 2004, 26, 309–318. 10.1093/carcin/bgh329. [DOI] [PubMed] [Google Scholar]
- Ruvolo P. P.; Ruvolo V. R.; Benton C. B.; Alrawi A.; Burks J. K.; Schober W.; Rolke J.; Tidmarsh G. Jr; Hail N.; Eric Davis R.; Andreeff M. Combination of galectin inhibitor GCS-100 and BH3 mimetics eliminates both p53 wild type and p53 null AML cells. Biochim. Biophys. Acta 2016, 1863, 562–571. 10.1016/j.bbamcr.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Zhou L.; Liao W.; Chen H.; Du Z.; Shao C.; Wang P.; Ding K. HH1-1, a novel Galectin-3 inhibitor, exerts anti-pancreatic cancer activity by blocking Galectin-3/EGFR/AKT/FOXO3 signaling pathway. Carbohydr. Polym. 2019, 204, 111–123. 10.1016/j.carbpol.2018.10.008. [DOI] [PubMed] [Google Scholar]
- Chauhan D.; Li G.; Podar K.; Hideshima T.; Neri P.; He D.; Mitsiades N.; Richardson P.; Chang Y.; Schindler J.; Carver B.; Anderson K. C. A Novel Carbohydrate-Based Therapeutic GCS-100 Overcomes Bortezomib Resistance and Enhances Dexamethasone-Induced Apoptosis in Multiple Myeloma Cells. Cancer Res. 2005, 65, 8350–8358. 10.1158/0008-5472.can-05-0163. [DOI] [PubMed] [Google Scholar]
- Zou J.; Glinsky V. V.; Landon L. A.; Matthews L.; Deutscher S. L. Peptides specific to the galectin-3 carbohydrate recognition domain inhibit metastasis-associated cancer cell adhesion. Carcinogenesis 2004, 26, 309–318. 10.1093/carcin/bgh329. [DOI] [PubMed] [Google Scholar]
- Kather I.; Bippes C. A.; Schmid F. X. A Stable Disulfide-free Gene-3-protein of Phage fd Generated by In vitro Evolution. J. Mol. Biol. 2005, 354, 666–678. 10.1016/j.jmb.2005.09.086. [DOI] [PubMed] [Google Scholar]
- López-Llano J.; Campos L. A.; Sancho J. α-helix stabilization by alanine relative to glycine: Roles of polar and apolar solvent exposures and of backbone entropy. Proteins: Struct., Funct., Bioinf. 2006, 64, 769–778. 10.1002/prot.21041. [DOI] [PubMed] [Google Scholar]
- Muppidi A.; Wang Z.; Li X.; Chen J.; Lin Q. Achieving cell penetration with distance-matching cysteine cross-linkers: a facile route to cell-permeable peptide dual inhibitors of Mdm2/Mdmx. Chem. Commun. 2011, 47, 9396. 10.1039/c1cc13320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. C.; Goto N. K.; Williams K. A.; Deber C. M. Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 6676–6681. 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M. A.. In Phage Display: A Laboratory Manual; Barbas C. F. III, Burton D. R., Scott J. K., Silverman G. J., Eds.; Analytical Biochemistry; CSHL Press, 2001; Vol. 294 ( (2), ), p 194 [Google Scholar]
- Kay B. K.; Winter J.; MacCafferty J.. Phage Display of Peptides and Proteins: A Laboratory Manual; Academic Press: San Diego, 1996. [Google Scholar]
- Matsushita N.; Nishi N.; Seki M.; Matsumoto R.; Kuwabara I.; Liu F.-T.; Hata Y.; Nakamura T.; Hirashima M. Requirement of Divalent Galactoside-binding Activity of Ecalectin/Galectin-9 for Eosinophil Chemoattraction. J. Biol. Chem. 2000, 275, 8355–8360. 10.1074/jbc.275.12.8355. [DOI] [PubMed] [Google Scholar]
- Benhar I.; Azriel R.; Nahary L.; Shaky S.; Berdichevsky Y.; Tamarkin A.; Wels W. Highly efficient selection of phage antibodies mediated by display of antigen as Lpp-OmpA′ fusions on live bacteria. J. Mol. Biol. 2000, 301, 893–904. 10.1006/jmbi.2000.4021. [DOI] [PubMed] [Google Scholar]
- Luo P.; Baldwin R. L. Mechanism of Helix Induction by Trifluoroethanol: A Framework for Extrapolating the Helix-Forming Properties of Peptides from Trifluoroethanol/Water Mixtures Back to Water. Biochemistry 1997, 36, 8413–8421. 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.