Abstract
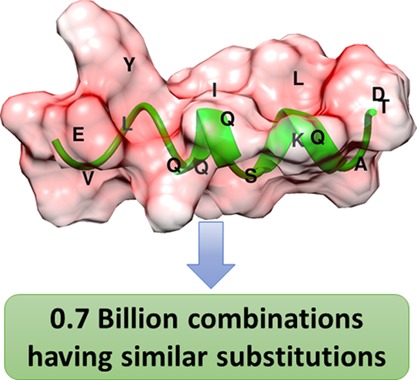
Peptides are used as reagents both for basic research and diagnostic purposes. Therefore, there is a need for novel methods for the design of peptide molecules with a particular specific physiochemical profile. The properties of the peptides are governed by the nature of amino acids constituting the peptide. There is a lack of a web server or tools which could predict all the possible combinations of the peptides generated because of the combinations of amino acids based on the physiochemical properties. We have developed a peptide combination generator (PepCoGen), a web server for generating all the possible combinations of peptides by varying the amino acids having similar physiochemical properties at a particular position. It also predicts other properties of the peptides including molecular weight, charge, solubility, hydrophobic plot, and isoelectric point, and random three-dimensional models for each generated combination.
Introduction
In the past decade, peptides have gained diverse applications in both medicine and biotechnology; moreover, the therapeutic peptide research has experienced revolution because of commercial reasons. Peptide sequences are constituents of bigger proteins, where they assist in molecular recognition and various biological functions by acting as precursors for proteins,1 alkaloids,2 hormones,3 anti-malarial agents,4 antimicrobial agents,5−7 anti-oxidants,8 anti-neurodegenerative,9 immunogens,10 and many more.11 Inhibition of protein–protein interactions by peptides and thereby therapeutic intervention of various disease-causing pathways have been the key for peptide drug design.12 Moreover, owing to their high specificity, safety, tolerability, efficacy, and excellent pharmacological profile, peptides seem to be ideal drug leads.13 There are more than 60 FDA-approved peptide drugs in the market, and more than 150 peptide drugs are in clinical trials and more than 500 therapeutic peptides in preclinical development.14,15 Numerous peptides including insulin, calcitonin, oxytocin, and vasopressin have been used as therapeutic peptides.13,15 Similarly, peptides from snake venom have been used for therapeutic purposes.16 There has been an increase in the development of combinatorial methods for discovery of membrane-active peptides17 and cell-permeable bicyclic peptides.18 Similarly, there has also been an increase in computational methods for the prediction of therapeutic peptides,11,19,20 peptide structure,21,22 peptide docking,23 and peptide binding.24 Peptide libraries are routinely used as a powerful tool in biological research.25 They provide a quick and economical solution for a wide range of bioactivity screening purposes and identifying bioactive peptides. Dedicated libraries are ideal for “T-cell epitope mapping and searching”. Overlapping linear peptide libraries provide a rapid route to mapping of the linear or complex epitope of the antibody. Peptide libraries are also used as enzyme substrates, ligands, or inhibitors in biochemical studies. The 20 amino acids can be differentiated into six broad groups, namely, hydrophobic (aliphatic), aromatic, basic, acidic, unique, and polar uncharged based on their physiochemical properties26−28 (Table S1). Currently, there is no standalone tool or web server, which could generate all the possible combination of the peptides by varying or mutating the amino acids with other residues having similar properties at a particular position in the peptide sequence. To predict all such combinations for the peptide, we have developed a user-friendly web server, which can be freely accessed at https://www.bicfri.in/pepcogen/. The front end of the web server is illustrated in Figure 1.
Figure 1.
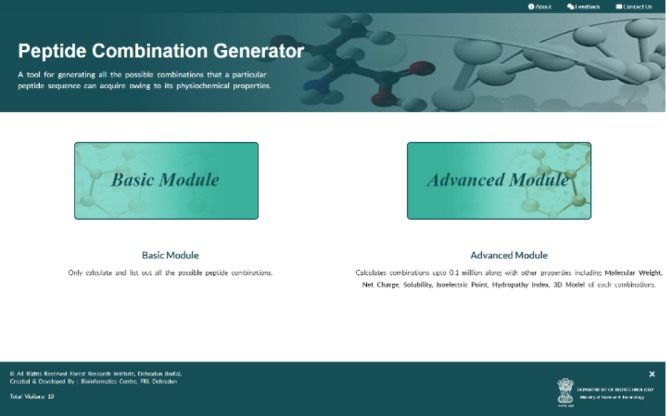
Image indicates the main web server front page, which can be used to access both basic and advanced modules of the web server.
Results and Discussion
The basic module only calculates the total combinations along with the peptide sequence, while the advanced modules calculate the physiochemical properties for all the combinations as well. Peptides of length 4 to 21 residues have been explored, as mostly the wet lab synthesis of peptides has been focused on developing peptides of these lengths. The peptide length can be increased, but the computational cost of calculating all the physiochemical properties was found to be high for such peptides. Therefore, we restricted the length of peptides to this range. In both basic and advanced modules, after initially selecting the sequence of the peptide fragment ranging from 4 to 21 amino acids (see Figures S1–S3), a drop-down menu will pop up asking the user to select the desired amino acids having the selected physiochemical property for each position in the peptide (Figure S4). After submitting the request on the front end, all the possible combinations of the peptide sequences based on the selected properties are calculated for both the modules (Figure S5). For example, for calculating the number of combinations for the peptide having sequence length 6 and having the possibility of acidic amino acids for positions 1 and 2, basic amino acids for positions 3 and 4, and hydrophobic amino acids for positions 5 and 6, the total possible combinations generated were 900, which were displayed on the browser window of the client end. For the advanced module (which is limited for generating combinations of up to 0.1 million), all the combinations are listed along with the calculated physiochemical properties including molecular weight, isoelectric point, charge, solubility, and hydropathy index, and a random 3D model (Figure S6). For instance, for calculating the combinations for the peptide with length 5, having the possibility of hydrophobic residues at all positions, we generated 3125 combinations (Figure S7). The hydropathy index is displaced in a new window along with its average value (Figure S8). The models can be generated, by clicking on the model icon in the corresponding tab, in a separate window using the JSmol library (Figure S9). However, the 3D models were generated accurately for the peptides having a length of up to 8 amino acids. Methods described previously were found to be more accurate22,23 and can be used for generating models using the string format generated for each combination.
Conclusions
We introduce a user-friendly web server tool, peptide combination generator (PepCoGen), which is the first freeware to assist the researchers in creating all the possible combinations of the peptide sequence by the variation of amino acids having similar physiochemical properties. The peptide combinations generated for a particular peptide sequence can be used by the researchers in making crucial decisions for selecting peptide sequences. The decision for selecting the peptides could be based on their molecular weight, charge, isoelectric point, and hydropathy index. Furthermore, by conserving the critical residues, all the possible combinations based on the physiochemical profile can be used to design analogues for sequence optimizations. Custom peptide libraries can be shortlisted, which have widespread applications in identifying critical amino acid residues, designing analogues, identifications of more potent peptide-based drugs, and many more. The web server could be used for the design of virtual peptide library databases, which further have numerous applications.
Computational Methods
The web server “PepCoGen” is organized as the REST architecture, with the front end of the server for the user interface and the back end for the algorithm scripts. The front end was designed using HTML, CSS, JQUERY, AJAX, and BOOTSTRAP, while the back end scripts were written in PHP. The physiochemical properties of the peptides including molecular mass, charge, solubility, and isoelectric point (at pH 7) were calculated using scripts 1, 2, 3, and 4, respectively, described in the Supporting Information (scripts s1, s2, s3, and s4). The hydropathy index was calculated using script 5, described in the Supporting Information (script s5). The models were generated using JSmol plugin.29 PepCoGen was implemented in PHP language, and the complete workflow has been described in Figure 2. PepCoGen is an easy-to-use tool to generate combinations for peptide lengths ranging from 4 to 21 residues and has widespread practical applications. The tool has been divided into basic and advanced modules. The basic module only calculates and reports all the combinations in the screen. The advanced module can calculate combinations up to 0.1 million peptides, along with the other features including molecular weight, charge, solubility, isoelectric point, and hydropathy index, and generate a random 3D model for each peptide. For calculating the isoelectric point, computationally optimized dissociation constant (pKa) values of amino acids were used.30 For calculating the hydropathy index values, the Hoops and Woods method was used as the values calculated could be useful in developing synthetic peptide immunogens and for understanding the relationship of the peptide sequence and its interaction with macromolecules.31 Models for each peptide were built using JSmol plugin. For the advanced module, the results can be viewed in the screen in batches of 10, 25, 50, and 100 per page. The results can also be downloaded in the excel sheet and pdf format.
Figure 2.
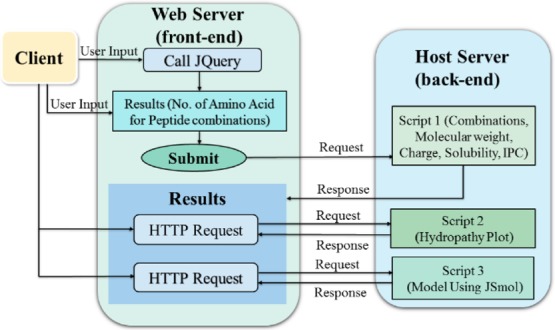
Description of the workflow implemented in web.
Acknowledgments
The authors are grateful to both FRI, Dehradun, and Department of Biotechnology, India.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b03848.
Classification table of various amino acids based on their physiochemical properties, screenshots of the different modules and how to use them for inferring results, and different scripts used for calculating various physiochemical properties (PDF)
This study was supported by the Department of Biotechnology, Government of India.
The authors declare no competing financial interest.
Supplementary Material
References
- Trimaille T.; Mabrouk K.; Monnier V.; Charles L.; Bertin D.; Gigmes D. SG1-Functionalized peptides as precursors for polymer-peptide conjugates: A straightforward Approach. Macromolecules 2010, 43, 4864–4870. 10.1021/ma100598d. [DOI] [Google Scholar]
- Giacomelli S. R.; Maldaner G.; Gonzaga W. A.; Garcia C. M.; da Silva U. F.; Dalcol I. I.; Morel A. F. Cyclic peptide alkaloids from the bark of Discaria americana. Phytochemistry 2004, 65, 933–937. 10.1016/j.phytochem.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Hutchinson J. A.; Burholt S.; Hamley I. W. Peptide hormones and lipopeptides: from self-assembly to therapeutic applications. J. Pept. Sci. 2017, 23, 82–94. 10.1002/psc.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundez C.; Sellanes D.; Serra G. Synthesis of Cyclic Peptides as Potential Anti-Malarials. ACS Comb. Sci. 2018, 20, 212–219. 10.1021/acscombsci.7b00154. [DOI] [PubMed] [Google Scholar]
- Sierra J. M.; Fusté E.; Rabanal F.; Vinuesa T.; Viñas M. An overview of antimicrobial peptides and the latest advances in their development. Expert Opin. Biol. Ther. 2017, 17, 663–676. 10.1080/14712598.2017.1315402. [DOI] [PubMed] [Google Scholar]
- Juretić D.; Vukicević D.; Ilić N.; Antcheva N.; Tossi A. Computational design of highly selective antimicrobial peptides. J. Chem. Inf. Model. 2009, 49, 2873–2882. 10.1021/ci900327a. [DOI] [PubMed] [Google Scholar]
- Vishnepolsky B.; Gabrielian A.; Rosenthal A.; Hurt D. E.; Tartakovsky M.; Managadze G.; Grigolava M.; Makhatadze G. I.; Pirtskhalava M. Predictive Model of Linear Antimicrobial Peptides Active against Gram-Negative Bacteria. J. Chem. Inf. Model. 2018, 58, 1141–1151. 10.1021/acs.jcim.8b00118. [DOI] [PubMed] [Google Scholar]
- Kobayashi K.; Maehata Y.; Okada Y.; Kusubata M.; Hattori S.; Tanaka K.; Miyamoto C.; Yoshino F.; Yoshida A.; Tokutomi F.; Wada-Takahashi S.; Komatsu T.; Otsuka T.; Takahashi S.-S.; Lee M.-C. -I. Medical-grade collagen peptide in injectables provides antioxidant protection. Pharm. Dev. Technol. 2015, 20, 219–226. 10.3109/10837450.2013.860547. [DOI] [PubMed] [Google Scholar]
- Goyal D.; Shuaib S.; Mann S.; Goyal B. Rationally Designed Peptides and Peptidomimetics as Inhibitors of Amyloid-β (Aβ) Aggregation: Potential Therapeutics of Alzheimer’s Disease. ACS Comb. Sci. 2017, 19, 55–80. 10.1021/acscombsci.6b00116. [DOI] [PubMed] [Google Scholar]
- Lee B.-S.; Huang J.-S.; Jayathilaka L. P.; Lee J.; Gupta S. Antibody Production with Synthetic Peptides. Methods Mol. Biol. 2016, 1474, 25–47. 10.1007/978-1-4939-6352-2_2. [DOI] [PubMed] [Google Scholar]
- Cui W.; Wei Z.; Chen Q.; Cheng Y.; Geng L.; Zhang J.; Chen J.; Hou T.; Ji M. Structure-based design of peptides against G3BP with cytotoxicity on tumor cells. J. Chem. Inf. Model. 2010, 50, 380–387. 10.1021/ci900404p. [DOI] [PubMed] [Google Scholar]
- Chang Y. S.; Graves B.; Guerlavais V.; Tovar C.; Packman K.; To K. H.; Olson K. A.; Kesavan K.; Gangurde P.; Mukherjee A.; Baker T.; Darlak K.; Elkin C.; Filipovic Z.; Qureshi F. Z.; Cai H.; Berry P.; Feyfant E.; Shi X. E.; Horstick J.; Annis D. A.; Manning A. M.; Fotouhi N.; Nash H.; Vassilev L. T.; Sawyer T. K. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, E3445–E3454. 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosgerau K.; Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discovery Today 2015, 20, 122–128. 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Kaspar A. A.; Reichert J. M. Future directions for peptide therapeutics development. Drug Discovery Today 2013, 18, 807–817. 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Carney R. P.; Thillier Y.; Kiss Z.; Sahabi A.; Heleno Campos J. C.; Knudson A.; Liu R.; Olivos D.; Saunders M.; Tian L.; Lam K. S. Combinatorial Library Screening with Liposomes for Discovery of Membrane Active Peptides. ACS Comb. Sci. 2017, 19, 299–307. 10.1021/acscombsci.6b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros E.; Gonçalves R. M.; Cardoso M. H.; Santos N. C.; Franco O. L.; Cândido E. S. Snake Venom Cathelicidins as Natural Antimicrobial Peptides. Front. Pharmacol. 2019, 10, 1415. 10.3389/fphar.2019.01415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh T. B.; Upadhyaya P.; Qian Z.; Pei D. Discovery of a Direct Ras Inhibitor by Screening a Combinatorial Library of Cell-Permeable Bicyclic Peptides. ACS Comb. Sci. 2016, 18, 75–85. 10.1021/acscombsci.5b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J. L.; Dunn M. K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Freire J. M.; Almeida Dias S.; Flores L.; Veiga A. S.; Castanho M. A. R. B. Mining viral proteins for antimicrobial and cell-penetrating drug delivery peptides. Bioinformatics 2015, 31, 2252–2256. 10.1093/bioinformatics/btv131. [DOI] [PubMed] [Google Scholar]
- Sharma T.; Siddiqi M. I. In silico identification and design of potent peptide inhibitors against PDZ-3 domain of Postsynaptic Density Protein (PSD-95). J. Biomol. Struct. Dyn. 2019, 37, 1241–1253. 10.1080/07391102.2018.1454851. [DOI] [PubMed] [Google Scholar]
- Singh S.; Singh H.; Tuknait A.; Chaudhary K.; Singh B.; Kumaran S.; Raghava G. P. PEPstrMOD: structure prediction of peptides containing natural, non-natural and modified residues. Biol. Direct 2015, 10, 73. 10.1186/s13062-015-0103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamiable A.; Thevenet P.; Tufféry P. A critical assessment of hidden markov model sub-optimal sampling strategies applied to the generation of peptide 3D models. J. Comput. Chem. 2016, 37, 2006–2016. 10.1002/jcc.24422. [DOI] [PubMed] [Google Scholar]
- Zhou P.; Li B.; Yan Y.; Jin B.; Wang L.; Huang S.-Y. Hierarchical Flexible Peptide Docking by Conformer Generation and Ensemble Docking of Peptides. J. Chem. Inf. Model. 2018, 58, 1292–1302. 10.1021/acs.jcim.8b00142. [DOI] [PubMed] [Google Scholar]
- Yordanov V.; Dimitrov I.; Doytchinova I. Proteochemometrics-Based Prediction of Peptide Binding to HLA-DP Proteins. J. Chem. Inf. Model. 2018, 58, 297–304. 10.1021/acs.jcim.7b00026. [DOI] [PubMed] [Google Scholar]
- Marasco D.; Perretta G.; Sabatella M.; Ruvo M. Past and future perspectives of synthetic peptide libraries. Curr. Protein Pept. Sci. 2008, 9, 447–467. 10.2174/138920308785915209. [DOI] [PubMed] [Google Scholar]
- Amino Acid Reference, Properties of Common Amino Acids. https://www.sigmaaldrich.com/lifescience/metabolomics.html (accessed October 15, 2018).
- Lide D. R.Handbook of Chemistry and Physics, 72nd ed.; CRC Press: Boca Raton, FL, 1991. [Google Scholar]
- Nelson D. L.; Cox M. M.. Lehninger Principles of Biochemistry, 5th ed.; W. H. Freeman: New York, 2008. [Google Scholar]
- Hanson R. M.; Lu X.-J. DSSR-enhanced visualization of nucleic acid structures in Jmol. Nucleic Acids Res. 2017, 45, W528–W533. 10.1093/nar/gkx365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski L. P. IPC - Isoelectric Point Calculator. Biol. Direct 2016, 11, 55. 10.1186/s13062-016-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P. Use of hydrophilicity plotting procedures to identify protein antigenic segments and other interaction sites. Methods Enzymol. 1989, 178, 571–585. 10.1016/0076-6879(89)78040-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


