Abstract
Objective
Lack of specific marker-sets prohibits definition and functional distinction of cellular subtypes in the intervertebral disc (IVD), such as those from the annulus fibrosus (AF) and the nucleus pulposus (NP).
Design
We recently generated immortalized cell lines from human NP and AF tissues; these comprise a set of functionally distinct clonal subtypes. Whole transcriptome analyses were performed of 12 phenotypically distinct clonal cell lines (4× NP-Responder, 4× NP-nonResponder, 2× AF-Sheet forming, and 2× AF-nonSheet forming). Data sets were filtered for membrane-associated marker genes and compared to literature.
Results
Comparison of our immortal cell lines to published primary NP, AF, and articular chondrocytes (AC) transcriptome datasets revealed preservation of AF and NP phenotypes. NP-specific membrane-associated genes were defined by comparison to AF cells in both the primary dataset (46 genes) and immortal cell-lines (161 genes). Definition of AF-specific membrane-associated genes yielded 125 primary AF cell and 92 immortal cell-line markers. Overlap between primary and immortal NP cells yielded high-confidence NP-specific marker genes for NP-R (CLDN11, TMEFF2, CA12, ANXA2, CD44) and NP-nR (EFNA1, NETO2, SLC2A1). Overlap between AF and immortal AF subtypes yielded specific markers for AF-S (COLEC12, LPAR1) and AF-nS (CHIC1).
Conclusions
The current study provides a reference platform for preclinical evaluation of novel membrane-associated cell type–specific markers in the IVD. Future research will focus on their biological relevance for IVD function in development, homeostasis, and degenerate conditions.
Keywords: intervertebral disc, cell lines, nucleus pulposus, annulus fibrosus, biomarkers, membranome
Introduction
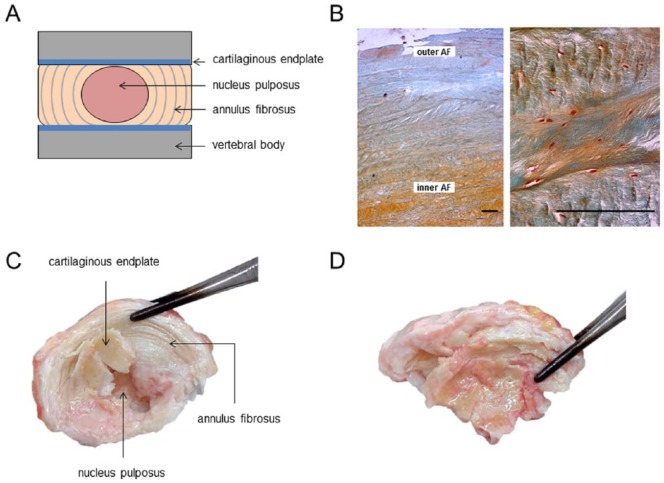
Low back pain has an extensive socioeconomic impact.1,2 Degeneration of the intervertebral disc is strongly associated with low back pain and disc herniation.3 Severe disc degeneration is estimated to occur in 10% of the general population at 50 years of age, and up to 60% in 70-year-old individuals.4 The intervertebral disc (IVD) degenerates more rapidly than other and surrounding musculoskeletal tissues.5 In particular, lumbar IVDs are susceptible to damage and injuries, underscoring important unique anatomical/mechanical features that contribute to the etiology of disc degeneration. IVD physiology is unique in that it is subject to a number of specific stressors. It is the most hypoxic tissue naturally occurring in the human body. Its nutrient supply to the nucleus pulposus (NP) and the surrounding annulus fibrosus (AF) occurs exclusively through the cartilaginous endplates (CEP; Fig. 1A and B ) and vessels surrounding the AF.5,6 In addition, the IVD is subject to constant heavy loading, predominantly by muscle pressure.7 The osmolarity of the NP is higher compared to articular cartilage (AC), thereby retaining tissue-associated water to maintain disc height.8 The AF confines the NP to its central position within the IVD; this mediates vertical pressure absorbance within the spine. In addition, the AF endows the spinal column with a degree of flexibility.
Figure 1.
Intervertebral disc morphology and degenerative disc disease. (A) Schematic representation of an intervertebral disc. The IVD comprises 3 distinctive tissues: a central gelatinous nucleus pulposus (NP), surrounded by the lamellar annulus fibrosus (AF). The AF consists of multiple, concentrically arranged lamellae in which cells and ECM components interact to provide function and structure. In the AF, two distinct zones are distinguished primarily based on their collagen/proteoglycan content: the inner and outer AF. The NP and AF are flanked at each side by cartilaginous endplates (CEP), which mediate the contact between the IVD body and vertebra. (B) Sections of a healthy AF (15-year-old male donor) stained with Safranin O and Fast green. The outer AF predominantly contains collagens while the inner AF contains proteoglycans. The characteristic lamellar structure is clearly visible; bar represents 200 µm. (C) A nondegenerate L1/L2 intervertebral disc of 63-year-old male donor. The lamellar annulus fibrosus is clearly recognizable. The central nucleus pulposus displays no aberrant morphology; the cartilaginous endplate is relatively thin. (D) The L4/L5 intervertebral disc of the same donor. Degenerative changes cause apparent loss of lamellar structure in the AF. The NP is no longer distinguishable from the AF and additional ossification is found near the CEP (For interpretation of the references to colours in this figure legend, refer to the online version of this article).
Degenerative disc disease (DDD) is characterized by reduced proteoglycan content, loss of structural integrity, and height of the disc and CEP erosion ( Fig. 1C and D ). The CEPs are thought to be the weak link in the context of mechanical compression of the IVD. Loss of CEP integrity allows the NP to bulge into the vertebral body; this loss of structural integrity within the IVD in turn results in overloading of the AF. Consequential wear and tear may lead to AF rupture and NP protrusion (disc herniation). The exact nature of the processes that leads to irreversible disc degeneration are poorly understood.6 A number of cellular changes have been observed in the degenerate IVD: a decrease in cell numbers, the appearance of senescent cells, and cell cluster formation.9,10 Concomitant production of inflammatory mediators and catabolic enzymes lead to a decrease in proteoglycan content in the NP and loss of structural integrity of the AF.11,12 Finally, posttranslational modifications of ECM molecules13-15 and alterations in nutrient supply have been reported to correlate with DDD.16,17 Incomplete understanding of the underlying biology of functionally distinct cell populations in the IVD, however, hampers the identification of signaling pathways and factors important for IVD development, homeostasis, and disease.
We recently generated immortal clonal NP and AF cell lines: we identified multiple recurrent, functionally distinct cellular NP and AF phenotypes from independent young (13-15 years old) donors.18-20 While these findings provided vital proof-of-concept that multiple relevant cellular functions can be distinguished in the NP and AF, a clear understanding of the contribution of these cells to IVD biology in vivo awaits further elucidation. Heterogeneity of cells in the NP was first described in the early 1980s.21,22 Importantly, differences in cell phenotypes and cellular composition in current human IVD literature are likely to contribute to large variation in reported NP markers in various species (Suppl. Table S1).23-32 In addition, variations in age, gender, or clinical stage of DDD inevitably introduce substantial variation in human datasets, which limits understanding of these markers in healthy human disc biology.33-35 The availability of single cell–derived clonal NP and AF subpopulations provides an obvious advantage in the context of defining cellular heterogeneity in the IVD. Morphologically and functionally distinctive cell types provide an excellent starting point for gene expression profile-based, comparative phenotypic analysis. Biological specificity of cell types within diverse tissues is determined by specialized proteins, such as membrane-associated proteins, which have structural roles, determine morphology, and movement and mediate signaling from the microenvironment (e.g., cytokines, growth factors) and/or interaction with other cells and ECM components. A comprehensive analysis of molecules expressed at the cell surface of the different IVD cell types and a detailed understanding of underlying molecular signaling networks is expected to help identify specific cellular subtypes and to define cellular responses to their environment in development and disease. Definition of cancer cell membranomes have yielded novel druggable targets and biomarkers for tumor staging,36-38 and provided important proof-of-principle that crucial advances can be made in translational research by using selective approaches to identify unique features of cells. To obtain an unbiased molecular description of our immortal NP and AF cellular phenotypes, we adapted this approach in the current study by combining unbiased comparative whole-transcriptomic analysis with a selective filtering strategy specifically tailored toward identification of novel membrane-associated markers for IVD cell subtypes. The herein outlined membranomics approach using our unique AF and NP models aims to generate a reference platform for comprehensive preclinical evaluation of novel cell type-specific markers in the IVD.
Methods
Human IVD Tissue
Human IVD tissue from young idiopathic scoliosis patients (age 8-15 years) was obtained as surplus material during scoliosis correction surgery.18,20 Degenerate and nondegenerate lumbar IVD tissue was obtained post-mortem from a 63-year-old donor.19 Acquisition and use of all human materials was approved by the MUMC-Medical Ethical Review Committee (Approval ID 08-4-021, July 11, 2012).18-20,26
Cell Isolation, Culturing, and RNA Extraction
The isolation of primary NP and AF cells, the immortalization procedures, and the establishment and characterization of immortal NP and AF cell lines have been described in detail elsewhere.18,20 Briefly, 12 immortal cell clones derived from one male donor (D5, 13 years old, male spina bifida patient) were used for RNA isolation and comparative transcriptome analysis. Culturing procedures are described in detail in the Supplemental Methods section. RNA isolation, quality assessment, and real-time polymerase chain reaction (PCR) analysis were essentially carried out as published.18-20 Procedures are described in detail in the Supplemental Methods section.
Whole Transcriptome Expression Datasets
Raw datasets from published whole transcriptome expression measurements (Affymetrix U133 Plus 2.0 array) of primary human AF (3), NP (3), and AC (3) cell donors (age range: 46-60 years; n = 9) were derived from a previous study.26 Processing of RNA samples, labeling, hybridization of 12 Illimuna humanHT-12v4 arrays, data extraction, and analyses are described in detail in the Supplemental Methods section. Briefly, an arbitrary cutoff of fold change >2.0 up or down and an average expression above 100 or higher (mean fluorescence intensity) in the cell type of interest was used to define and select expressed genes. Modified t tests were used to evaluate statistically significant differences between groups (Benjamini Hochberg FDR adjusted P value <0.05 was considered significant).
Results and Discussion
High-Throughput Expression Phenotyping of Novel Immortal NP and AF Clones
We previously defined functionally distinct immortal NP and AF cell lines based on their (in)ability to respond to established chondrocyte-differentiation conditions (cf. (non)Responder NP clones: NP-R and NP-nR18,20 and based on their (in)ability to process procollagens in vitro (non) Sheet-forming AF clones: AF-S and AF-nS20). Four representative clones for both the NP-R and NP-nR cellular phenotypes were chosen for large scale expression analyses, in addition to 2 AF-S and 2 AF-nS clones (Suppl. Fig. S1A). We first evaluated expression of previously reported marker-sets for NP-R cells (FOXF1, CA12,18 NP-nR (CD24), and AF cells (SFRP2, COL12A120) prior to whole array expression analyses. This initial marker analysis confirmed that FOXF1 and CA12 discriminated between NP-R, NP-nR, and AF cell clones (Suppl. Fig. S1B). Expression levels of COL12A1 and SFRP2 were higher in AF and NP-nR cell clones compared to NP-R (Suppl. Fig. S1B). NP-nR and AF cell clones showed similar levels of SFRP2 and COL12A1 mRNA. CD24 levels, however, were significantly higher in NP-nR compared to NP-R or AF cell clones. This initial combined distinctive marker expression profiling of clonal IVD cell subtypes provided a solid basis for comparative genome-wide transcriptomic analysis.
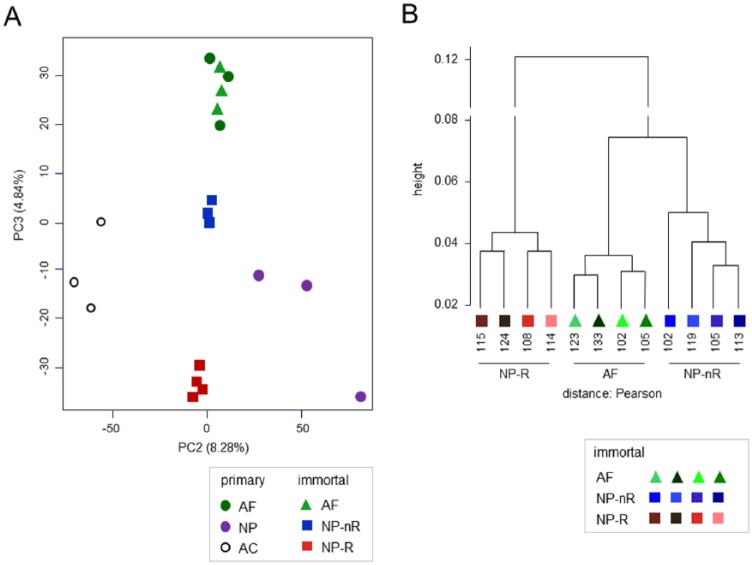
To determine the degree of transcriptome preservation of our immortalized AF and NP cell clones, we compared our immortal array dataset with published array datasets of primary isolates, comparing 3 donor-matched AC, AF, and NP primary cell isolates.26 Principal component analysis (PCA) on the normalized datasets produced a high-resolution clustering overview. The first component strongly separated the 2 datasets, which were generated on different array platforms (see Supplemental Methods section). The second and third components clearly separated AC cells from all IVD cells, in line with their divergent tissues of origin ( Fig. 2A ). Notably, primary AF and immortal clonal AF cell lines, all from unrelated donors, were presented as a distinctive cluster based on gene expression profiling. Although primary NP isolates showed considerable variation (3 independent donors), the immortal NP-R and NP-nR clones all clustered close to the primary NP isolates. Hierarchical clustering of immortal clones resulted in the anticipated separation of the PCA clusters ( Fig. 2B ; Suppl. Fig. S2). In line with their earlier observed overlap in marker expression of SFRP2 and COL12A1, NP-nR clones clustered closer to the AF clones than the NP-R group. Thus, PCA analyses indicated significantly distinctive gene expression profiles among immortal clonal IVD cellular subtypes and supported considerable preservation of primary AF and NP transcriptomic phenotype.
Figure 2.
Principle component analysis reveals overlap between primary and immortalized NP and AF clones. Transcriptome comparison between immortal cell clone and primary cell datasets generated on Affymetrix and Illumina platforms (first component), respectively. PCA analysis was preceded by ranking genes (descending expression value; see Supplemental Methods for detailed description). (A) PCA plot of the second and third components of primary AC, NP, and AF samples from 3 independent healthy donors (circles) and immortalized NP-Responder (n = 4), NP non-Responder (n = 3), and AF (n = 3) cell clones from a single donor (squares); these clones have been described in previous reports.18,20 The first component separated the dataset on platform (primary cells: Affymetrix platform, immortalized cell lines: Illumina platform). (B) Dendrogram showing the hierarchical clustering of immortalized NP-Responder (n = 4), NP non-Responder (n = 4), and AF (n = 4) cell clones; clustering was based on Pearson correlation. The NP-R, AF, NP-nR clusters clearly differ. NP-nR clone 102 fell outside the NP-nR cluster (Suppl. Fig. S2). As this effect persisted in subsequent analyses, this clone was omitted from further analysis. Analyses were performed and figures generated using the open source scripting language R (version 2.13.0) and R packages of Bioconductor 2.8.
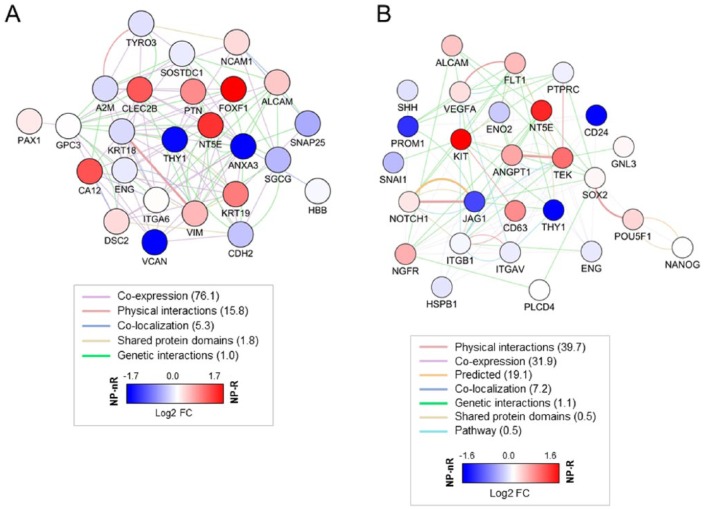
To obtain insight into genetic network activity underlying the preservation of immortal phenotypes in the NP, we first generated a list of putative NP, AC, and AF markers in rat, canine, and human IVDs. Rodents, in contrast to humans, retain notochordal cells (NC) throughout their life time39; for this reason published NP markers in, for example, rat studies, include NC markers. These putative markers were based on whole transcriptome expression studies performed on freshly isolated cells.23-26 We identified a total of 37 unique markers that showed differential expression between NP and AF, NP and AC, or NP and NC in one or more of these studies (Suppl. Table S1). Twenty-five positive NP markers (showing no overlap with AF- or AC-specific genes) were selected to build a literature-based NP gene-network ( Fig. 3A ; Suppl. Table S2); this network was then used to visualize clone-specific differences in gene expression between NP-R and NP-nR clones. NP-R cells showed relatively higher expression of CA12, CLEC2B, DSC2, KRT19, FOXF1, and PTN, while NP-nR cells expressed A2M, SNAP25, SOSTDC1, and VCAN ( Fig. 3A ). The differential expression of FOXF1, CA12, KRT19, and PTN is consistent with our previous qPCR-based analyses.18 A2M, CLEC2B, DSC2, SNAP25, and SOSTDC1 potentially provide additional functional definition of NP cell subtypes. The NP-nR specific Alpha 2 Macroglobullin (A2M) is a general protease inhibitor that functionally interacts with important cytokines like bFGF, PDGF, NGF, IL1β, and IL6 that are thought to be involved in DDD.40-42 The C-type lectin domain family 2, member B (CLEC2B) prevents blood vessel formation by endothelial cells and may fulfill a similar function in the NP.43 Desmocollin 2 (DSC2) is an ECM glycoprotein and component of the rat NP matrix.23 Synaptosomal-associated protein 25 (SNAP25) is involved in intracellular membrane trafficking and has been extensively studied in the context of brain function.44 Genes with known functions in the brain, such as axon guidance, have been reported before as NP markers.45,46 Finally, Sclerostin domain-containing protein 1 (SOSTDC1) is a BMP antagonist. SOSTDC1 deficiency was recently shown to promote fracture healing through expansion of periosteal mesenchymal stem cells.47
Figure 3.
Differential expression of published NP markers among NP cellular subtypes. (A) Selected marker genes (NP cell markers; Suppl. Tables S1 and S2) were used as input for GeneMANIA network building using Cytoscape visualization software. Gene expression differences between NP-R (red) and NP-nR (blue) clones are depicted as log2-based fold change (FC) in colored circles. Colored lines in the network represent established biological connections between markers; parentheses: percentage representation. The network is strongly interconnected based on co-expression and genetic interactions studies. Of note: established NP markers are not equally expressed by isolated cellular subpopulations derived from the NP. (B) Progenitor-associated gene expression in NP-Responder clones. Selected genes (NP progenitor markers; Table 1 , Suppl. Table S2) were used as input for network building using GeneMANIA and Cytoscape visualization software. Gene expression differences between NP-R (red) and NP-nR (blue) clones are depicted as log2-based FC in colored circles. Colored lines in the network represent established biological connections between markers; parentheses: percentage representation (For interpretation of the references to colours in this figure legend, refer to the online version of this article).
To visualize relative gene activity in this set of 25 markers in primary NP cells, the published transcriptome dataset26 was plotted within the same gene network. This approach revealed that NP markers KRT18, FoxF1, and VCAN were relatively highly expressed in primary NP cells (Suppl. Fig. S3A). Comparative gene network analysis thus reveals a number of striking differences in activity profiles of previously proposed general NP markers between immortal NP cellular subtypes and primary isolates, and emphasizes the importance of functional definition of cellular heterogeneity in the NP.
Progenitor Cells in the Mature NP
Progenitor or stem cells in the mature IVD were first isolated and studied in 1995 and have since been studied in both NP and AF of various species.48-54 Progenitor cells were first isolated from the IVD by fluorescence-activated cell sorting (FACS) using a discriminatory candidate cell surface marker-based approach.55 NP progenitor cells were isolated by FACS from mouse, human, and recently cow intervertebral discs using a combination of membrane-associated Cluster of Differentiation 24 (CD24), Ganglioside G2 (GD2), and/or receptor tyrosine kinase, epithelial specific (Tie2) cell surface markers.56,57 A summary of these reported findings shows little overlap in the markers deployed to identify progenitor cells in the mature IVD ( Table 1 ).
Table 1.
Overview of Literature Reports on Cell Heterogeneity and Progenitor Populations in the Intervertebral Disc.
| Tissue Origin (Ref) | Reference | Isolation method | Markers |
|---|---|---|---|
| Humana; NP, AF | 54 | Flow cytometry | CD105, CD166, CD63, CD90, CD73 (NT5E), P75, CD133 |
| Human; NP | 48 | IHC | HSP27, HSP72 |
| Lapine, ovine, humana; NP | 49 | IHC | Notch1, Jag1, Delta4, Stro1, C-kit (CD117), CD105, CD166 |
| Human; AF | 50 | Flow cytometry | CD24, CD49, CD51, NSE |
| Human; CEP | 52 | CFU assay; agarose | CD133, CD105, CD90, CD73 (NT5E), Stro1, CD166, CD44 |
| Canine; NP | 53 | CFU assay | Sox2, Oct3/4, Nanog, CD133 (mRNA only) |
| Murine, human; NP | 55 | CFU assay; agarose | CD24, Tie2, GD2, CD44, (CD49f, CD56, CD73 (NT5E), CD90, CD105, CD166)b; (CD271, Flt1)c |
| Lapine; AF | 51 | CFU assay; fibroblast | CD29, CD44, CD166, Oct-4, Gnl3, SSEA-4 |
Various markers have been used to isolate multipotent cell populations from IVD tissues (as indicated in the corresponding column). Typical MSC markers (CD73, CD90, and CD105) are often used in addition to CD24 and CD44. We measured gene and protein expression of genes printed in bold in immortal cell clones of a single donor (NP and AF).
Degenerate tissue.
Expressed on all mouse and human NP cells.
Correlated to progenitor phenotype.
In our immortal clonal NP cell lines, distinctive gene signatures were identified that correlated with morphological characteristics: wave-like NP-Responder (NP-R) clones expressed high levels of FoxF1 and CA12, whereas cobble stone-shaped NP-non-Responder (NP-nR) clones expressed high levels of membrane-associated CD24.18 NP-R clones exhibited spontaneous spheroid-forming capacity on specific culture substrates (Aggrecan [ACAN], Matrigel). This stemness-associated feature has previously been utilized to identify progenitor cells in multiple systems.58,59 Hence, we hypothesized that the immortal NP-R clones harbor a progenitor-like phenotype; in contrast, our analyses suggested NP-nR clones maintain a more differentiated phenotype.18-20
To obtain additional support for this hypothesis, we focused on the “cell heterogeneity and progenitor” genes listed in Table 1 and used these to generate a progenitor gene-network representing the NP progenitor cell phenotype. This network was then used to visualize relative expression differences between NP-R and NP-nR clones ( Fig. 3B ). Significantly higher expression of ANGPT1, CD44, CD63, FLT1 (VEGFR1), KIT, NT5E (CD73), and TEK was observed in NP-R clones. FLT1 (VEGFR1) and ANGPT1 are known as important signaling molecules mediating survival of NP cells.56 In the same study TEK (Tie-2) was identified as an NP progenitor marker. The expression of these markers in NP-R clones strongly supports the progenitor-like nature of this cell type.
NP-nR clones were characterized by increased CD24, JAG1, PROM1 (CD133), and THY1 (CD90) expression. Primary NP cell isolates (mixed cell population) had high expression of CD24, ENO2, ITGB1 (CD29), POU5F1 (OCT3/4), and VEGFA, whereas TEK1 and ANGPT1 were expressed at relatively low levels (Suppl. Fig. S3B). Of note, the expression level of progenitor cell marker genes in the primary NP cell dataset was expected to be low, as progenitor cells are not likely to represent a predominant phenotype in the mature human IVD.
Although we evaluated a significant proportion of these cell markers, it is currently not clear how the 2 distinctive cellular NP phenotypes, NP-R and NP-nR, can be positioned on an imaginary differentiation scheme that was proposed based on membrane expression of CD24, CD44, Tie2, and GD2 ( Table 2 ).56 Our findings fit this model to certain extent: our progenitor cells (NP-R) are indeed CD24 negative (CD24−), whereas the more differentiated NP-nR cells are CD24+. None of our immortal clones were Tie2+ however, which was proposed as a hallmark of NP progenitor cells. Although NP-R clones were initially negative for GD2, GD2-expression could be induced using chondrogenic differentiation stimuli.18 In summary, NP-R clones displayed a gene signature compatible with NP progenitor characteristics.
Table 2.
Positioning of Immortal NP-Phenotypes in NP-Based Progenitor/Differentiation Model.
| Marker | Progenitor |
Differentiation |
Immortal Clones |
||||
|---|---|---|---|---|---|---|---|
| Dormant | Activated | 1 | 2 | 3 | NP-R | NP-nR | |
| Tie2 | + | + | − | − | − | − | − |
| GD2 | − | + | + | + | − | − | + |
| CD24 | − | − | − | + | + | − | + |
| CD44 | +/− | + | + | + | + | + | + |
| CD271 | + | + | +/− | − | − | ND | ND |
| FLT1 | + | + | +/− | − | − | ND | ND |
A recently postulated progenitor/differentiation model based on expression (+) or absence (−) of markers55 is represented in the columns marked Progenitor and Differentiation. The right most columns show the measurements we performed on our immortal NP cell clones. ND = analysis not done.
Novel Cell Surface Markers in the NP
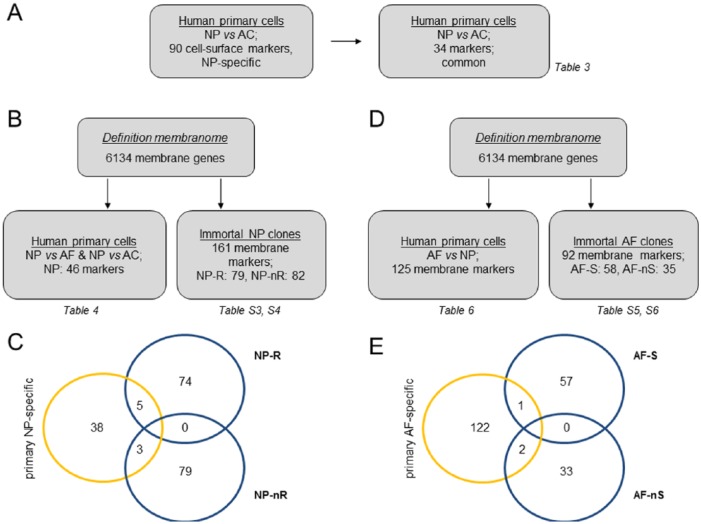
A previous study identified membrane-associated genes specific for the NP compared with AC, by using an in silico screening of differentially expressed genes for the presence of transmembrane domains.26 To test whether these results could be reproduced in an independent study, we compared 90 previously identified membrane-associated genes27 with primary NP/AC dataset26 ( Fig. 4A ). Thirty-four overlapping NP membrane-associated marker genes (NP/AC) were identified between these 2 studies; the expression of these markers was at least 2-fold higher in primary NP cells in comparison to AC ( Table 3 ). CA12 remained the top hit (3 probes in the top 10; relative expression level: >20-fold). In addition expression of ABCG1, SLC27A2, TMEM71, FLRT2, CLDN11, and DNER was more than 10-fold higher in primary NP isolates. As these genes may define functional differences between NP cells and ACs, their potential involvement in IVD biology is briefly discussed below. ATP binding cassette subfamily G member 1 (ABCG1) is expressed in articular cartilage and downregulated in osteoarthritis and has a regulatory function in cholesterol and phospholipid transport.60 Solute carrier family 27 member2 (SLC27A2) is a fatty acid transport protein. TMEM71 function is unknown; it is regulated by the candidate tumor suppressor gene inhibitor of growth 2 (ING2).61 Fibronectin-like domain-containing leucine-rich transmembrane protein (FLRT2) interacts with fibronectin and has distinct expression patterns in mouse development that are linked to FGF signaling.62,63 Claudin 11 (CLDN11) is a tight junction protein originally identified in oligodendrocytes and is known to be expressed in a small cellular subpopulation in the human IVD.64,65 Delta- and notch-like epidermal growth factor-related receptor (DNER) plays an important role in brain development.66 Taken together, 34 membrane-associated marker genes that show a relatively higher transcriptional activity in NP compared to AC were identified in 2 independent datasets, further emphasizing the unique phenotype of NP cells. With the exception of CA12 these markers have not been studied in the IVD.
Figure 4.
Identification of membrane-associated NP marker genes. Comparison strategy of whole transcriptome datasets of published primary IVD cellular isolates and clonal cell lines. (A) Marker genes significantly more highly expressed in primary NP compared to AC from 2 independent published datasets26,27 were compared. (B) Definition of primary NP-specific membrane marker genes compared to AF and AC26 and definition of NP cell clone (R/nR) specific marker genes.18 (C) Subsequently both primary and clonal NP membrane marker gene sets were compared to each other. (D) Definition of primary AF-specific membrane marker genes compared to NP and definition of AF cell clone (S/nS) specific marker genes. (E) Subsequent comparison of primary and clonal AF membrane marker genes.
Table 3.
Common Human NP and AC Markers.
| Gene | IDa | AC Meanb | NP Meanb | FCc |
|---|---|---|---|---|
| ABCG1 | 9619 | 20.3 | 739.7 | 36.4 |
| AGPAT9 | 84803 | 675.2 | 2611.6 | 3.9 |
| C1orf9 | 51430 | 1166 | 3352.1 | 2.9 |
| CA12 | 771 | 71.3 | 2726.9 | 38.2 |
| CDH19 | 28513 | 761.7 | 2760.6 | 3.6 |
| CDS1 | 1040 | 76.9 | 249.5 | 3.2 |
| CLDN11 | 5010 | 19.3 | 204.2 | 10.6 |
| CLEC2B | 9976 | 53.4 | 303.3 | 5.7 |
| DNER | 92737 | 365.6 | 3671.4 | 10 |
| FAM62B | 57488 | 78.5 | 196.5 | 2.5 |
| FKBP11 | 51303 | 1102.8 | 2438.5 | 2.2 |
| FLRT2 | 23768 | 21.1 | 240.5 | 11.4 |
| FNDC3B | 64778 | 385.9 | 1598.9 | 4.1 |
| HS6ST3 | 266722 | 54.2 | 494.7 | 9.1 |
| INSIG1 | 3638 | 885.9 | 2010.5 | 2.3 |
| JPH3 | 57338 | 50.3 | 252.9 | 5 |
| KCNS3 | 3790 | 61.2 | 175.8 | 2.9 |
| MFAP3L | 9848 | 49.2 | 128.8 | 2.6 |
| MXRA7 | 439921 | 238.6 | 844.3 | 3.5 |
| NETO2 | 81831 | 49.3 | 323 | 6.6 |
| ODZ3 | 55714 | 36.3 | 217.6 | 6 |
| ORAI3 | 93129 | 431.5 | 2239.2 | 5.2 |
| PPAPDC1B | 84513 | 774.6 | 2823.4 | 3.6 |
| REEP1 | 65055 | 42.2 | 129.8 | 3.1 |
| SEL1L | 6400 | 190.6 | 1166.5 | 6.1 |
| SGCG | 6445 | 24.3 | 75.5 | 3.1 |
| SLC27A2 | 11001 | 19.2 | 350 | 18.3 |
| SLC44A1 | 23446 | 86.6 | 477.2 | 5.5 |
| SLC7A1 | 6541 | 99.9 | 237 | 2.4 |
| TMEM27 | 57393 | 19.3 | 44.2 | 2.3 |
| TMEM71 | 137835 | 29.5 | 336.6 | 11.4 |
| TSPAN31 | 6302 | 574 | 1177.6 | 2.1 |
| WFS1 | 7466 | 484.1 | 1093.4 | 2.3 |
| WNT11 | 7481 | 179.9 | 406.2 | 2.3 |
A comparison between 90 transmembrane cell surface markers27 and primary tissue-specific markers26 extracted from human micro-array data (NP vs. AC).
ENTREZ Gene ID.
Adjusted P value <0.05, fold change >2.0, and expression in NP >100 arbitrary units; see Methods section for details.
Fold change (NP/AC).
Comparative Membranome Analysis of primary NP, AF, and AC Isolates
To identify additional NP- and AF-specific cell surface markers, we first analyzed the primary NP, AF, and AC cell datasets26 and filtered for differentially expressed genes (FC > 2.0; adjusted P value <0.05; see Methods section for additional expression criteria). This yielded 898 genes of which expression was significantly higher in NP versus AF and 226 genes that were more highly expressed in NP versus AC. From these 2 gene lists 143 NP-specific markers (significantly higher in NP compared to AF and AC) were extracted. Using published meta-analyses,36 6134 membrane-associated proteins were defined. Of the 143 NP-specific markers, 46 were found to be cell membrane-associated ( Fig. 4B and C ). This marker set definition in NP and AF cell-lines showed overlap with our previous analyses in published primary NP cells and AC datasets (compare Table 3 with Table 4 ): CA12, CLDN11, Heparan Sulfate 6-0-sulfotransferase 3 (HS6ST3), Neuropilin- and tolloid-like 2 (NETO2), Calcium Release-Activated Calcium Modulator 3 (ORAI3, TMEM142C), and Choline transporter-like protein 1 (SLC44A1) ( Table 4 ; bold print). The latter 4 genes have thus far not been studied in the musculoskeletal system and appear highly specific for the NP in 2 independent human IVD cell studies.
Table 4.
NP-Specific Membrane-Associated Markers Compared to AF and AC in Primary Cell Expression Datasets.
| Gene | IDa | AC Meanb | AF Meanb | NP Meanb | FCc | FCd |
|---|---|---|---|---|---|---|
| ANKRD10 | 55608 | 687.4 | 909.1 | 2267.0 | 3.3 | 2.5 |
| ANXA2 | 302 | 18.5 | 23.5 | 380.8 | 20.6 | 16.2 |
| ATP6V0E1 | 8992 | 35.4 | 37.7 | 124.4 | 3.5 | 3.3 |
| C11orf1 | 64776 | 50.3 | 48.5 | 178.3 | 3.5 | 3.7 |
| C9orf91 | 203197 | 68.0 | 75.3 | 276.3 | 4.1 | 3.7 |
| CA12 | 771 | 71.3 | 134.4 | 2726.9 | 38.2 | 20.3 |
| CLDN11 | 5010 | 19.3 | 47.2 | 204.2 | 10.6 | 4.3 |
| CLIP4 | 79745 | 98.1 | 63.9 | 207.2 | 2.1 | 3.2 |
| DKFZp451A211 | 400169 | 505.2 | 989.2 | 2828.0 | 5.6 | 2.9 |
| DUSP6 | 1848 | 533.6 | 680.5 | 2172.2 | 4.1 | 3.2 |
| DYRK2 | 8445 | 238.8 | 247.3 | 746.3 | 3.1 | 3.0 |
| EFNA1 | 1942 | 909.3 | 2144.1 | 6752.4 | 7.4 | 3.1 |
| F3 | 2152 | 68.8 | 55.4 | 263.1 | 3.8 | 4.8 |
| FAM134A | 79137 | 173.5 | 188.9 | 408.8 | 2.4 | 2.2 |
| FGFR2 | 2263 | 25.9 | 26.5 | 109.7 | 4.2 | 4.1 |
| FLRT1 | 23769 | 79.5 | 90.6 | 243.3 | 3.1 | 2.7 |
| FUS | 2521 | 43.2 | 41.3 | 105.2 | 2.4 | 2.5 |
| GPR153 | 387509 | 319.2 | 310.1 | 686.7 | 2.2 | 2.2 |
| GRB10 | 2887 | 179.9 | 154.4 | 651.0 | 3.6 | 4.2 |
| HS6ST3 | 266722 | 54.2 | 183.9 | 494.7 | 9.1 | 2.7 |
| KHDRBS3 | 10656 | 164.8 | 289.8 | 1349.9 | 8.2 | 4.7 |
| KIAA1217 | 56243 | 39.9 | 184.3 | 529.1 | 13.2 | 2.9 |
| LDLR | 3949 | 140.3 | 148.1 | 339.9 | 2.4 | 2.3 |
| LRRFIP1 | 9208 | 398.6 | 499.9 | 1150.3 | 2.9 | 2.3 |
| NCAM1 | 4684 | 49.2 | 43.7 | 116.8 | 2.4 | 2.7 |
| NETO2 | 81831 | 27.7 | 48.6 | 122.4 | 4.4 | 2.5 |
| NNMT | 4837 | 89.5 | 52.7 | 263.0 | 2.9 | 5.0 |
| ORAI3 | 93129 | 431.5 | 1058.3 | 2239.2 | 5.2 | 2.1 |
| PGK1 | 5230 | 39.1 | 54.9 | 471.6 | 12.1 | 8.6 |
| PSMB4 | 5692 | 1084.0 | 800.1 | 2256.8 | 2.1 | 2.8 |
| RP1-21O18.1 | 23254 | 198.8 | 153.9 | 422.4 | 2.1 | 2.7 |
| RPL38 | 6169 | 1114.4 | 863.3 | 6333.9 | 5.7 | 7.3 |
| RPLP2 | 6181 | 234.9 | 194.1 | 739.6 | 3.1 | 3.8 |
| RRAD | 6236 | 234.2 | 247.8 | 745.4 | 3.2 | 3.0 |
| SLC16A3 | 9123 | 33.5 | 42.1 | 169.9 | 5.1 | 4.0 |
| SLC2A1 | 6513 | 353.9 | 851.8 | 2506.6 | 7.1 | 2.9 |
| SLC2A3 | 6515 | 410.9 | 437.5 | 1330.5 | 3.2 | 3.0 |
| SLC44A1 | 23446 | 86.6 | 208.1 | 477.2 | 5.5 | 2.3 |
| SLC6A6 | 6533 | 264.9 | 185.9 | 569.8 | 2.2 | 3.1 |
| STX3 | 6809 | 356.4 | 327.7 | 746.2 | 2.1 | 2.3 |
| TMEFF2 | 23671 | 271.5 | 321.8 | 2501.6 | 9.2 | 7.8 |
| TMEM200A | 114801 | 61.7 | 53.1 | 183.0 | 3.0 | 3.4 |
| TNS1 | 7145 | 2142.0 | 3537.6 | 7840.3 | 3.7 | 2.2 |
| TRPM8 | 79054 | 31.6 | 65.0 | 269.2 | 8.5 | 4.1 |
| ZNF395 | 55893 | 1956.3 | 4013.4 | 9607.8 | 4.9 | 2.4 |
| ZNF791 | 163049 | 711.1 | 1397.7 | 3029.9 | 4.3 | 2.2 |
ENTREZ Gene ID.
Differentially expressed genes were determined by adjusted P value <0.05, fold change >2.0, and expression in NP >100 (arbitrary units; see Methods section for details).26 Marker genes overlapping with those in Table 4 are printed in bold.
Fold change (NP/AC).
Fold change (NP/AF).
NP Clone-Specific Membrane-Associated Markers
In parallel, we identified unique membrane-associated markers for the NP clonal subtypes. The immortal cell clone dataset was filtered using a published membranome dataset.36 Of 161 differentially expressed membrane-associated genes, 79 were more highly expressed in the NP-R and 82 in the NP-nR ( Fig. 4B ; Suppl. Tables S3 and S4). These genes and their possible relevance for the NP cell phenotype are discussed below.
NP Responder-Specific Membrane-Associated Markers
Among NP-R-specific membrane markers, 3 genes showed a larger than 10-fold change: (1) plasticity related gene 1 (LPPR4), (2) transient receptor potential cation channel, subfamily V, member 2 (TRPV2), and (3) protocadherin 10 (PCDH10). LPPR4 is known for its role in brain plasticity67 and has not been studied in the context of the IVD or articular cartilage. TRPV2 is known to be expressed in chondrogenic high-density cultures of limb bud cells in chickens and mice and was recently associated with genetic skeletal disorders.68,69 PCDH10 is a negative regulator of the WNT pathway.70 Platelet-Derived Growth Factor Receptor Alpha (PDGFRA) has a fold change of 8.2 (NP-R/NP-nR) and is required for the formation of chondrogenic mesoderm and interacts with CAV1, which inhibits its signaling.71,72 Interestingly, the NP-marker PAX1 is a transcriptional regulator of PDGFRA.73 The PDGFRA ligand PDGF partakes in a complex autocrine feedback with its receptor and modulates the effect of TGFβ-stimulation.74 In addition, the PDGFR ligand PDGF-BB was recently shown to delay disc degeneration in a rabbit model.75 In contrast, PDGF was shown to potentiate the chondrocyte’s response to IL1β in rheumatoid arthritis.76 Thus, differences in a cell’s ability to activate the PAX1-PDGFRA-PDGF signaling axis may be relevant to local inflammatory responses and contribute to disc degeneration. CLDN11 (fold change 6.9 NP-R/nR) expression was recently inversely correlated with SDC2 expression.77 Of note: NP-nR clones showed a higher expression of SDC2 than CLDN11 expressing NP-R clones. RGM (repulsive guidance molecule) domain family member B (RGMB) was restricted to NP-R clones (fold change 5.5). RGMa and RGMb are both highly expressed in the developing chick notochord.78 RGMb functions as a co-receptor for BMP and acts as an antagonist of noggin, which in turn antagonizes BMP-signaling and has an important role in the embryonic development of the NP.79 In the NP, RGM expression is thought to prevent nerve in-growth in the IVD.45,80 As qPCR analysis independently confirmed the micro-array-based differences (Suppl. Fig. 4A), these markers are likely to have potential with regards to progenitor isolation from the NP.
NP nonResponder-Specific Membrane-Associated Markers
Among the 82 NP-nR specific membrane genes, protein tyrosine phosphatase, receptor type, F (PTPRF), tumor necrosis factor (ligand) superfamily, member 7 (CD70), chromosome 13 open reading frame 13 (SHISA2), G protein-coupled receptor, family C, group 5, member C (GPRC5C), chemokine orphan receptor 1 (CXCR7), and prostaglandin F2 receptor negative regulator (PTGFRN) all have more than 10-fold higher expression in NP-nR clones than in NP-R clones. Strikingly, multiple genes could be traced to the WNT signaling pathway. PTPRF shares an extracellular domain with NCAM and controls beta-catenin signaling.81 NCAM was previously suggested as a NP marker.23,30 SHISA2 modulates both WNT and FGF signaling in the developing chicken embryo82; in Xenopus it promotes somatic maturation of precursor cells.83 Low-density lipoprotein Receptor-related Protein, 5 (LRP5; fold change 5) is part of the canonical WNT signaling. Induced expression of LRP5 mediates cartilage destruction in ostheoarthritis.84,85 PTGFRN is involved in cell migration by selective interaction with Collagen type I and Fibronectin. This effect is modulated by the expression of the tetraspanins CD9 and CD81.86 Differential expression of these genes in NP-nR (vs. NP-R and AF cells) was confirmed by qPCR (Suppl. Fig. 4B).
Markers for NP Cellular Heterogeneity In Vivo
To begin to verify the potential usefulness of the clone-specific marker sets to distinguish between NP cell subtypes, we lastly compared the set of 46 primary NP cell-specific membrane-associated genes to NP-R and NP-nR-specific membrane-associated markers ( Fig. 4C ). A subset of 8 genes was identified both as primary NP-specific cell surface markers and as being differentially expressed between immortalized NP clonal cell types. NP-R cells were marked by CLDN11, TMEFF2, CA12, ANXA2, CD44 expression; NP-nR by EFNA1, NETO2, and SLC2A1 ( Table 5 ). TransMembrane protein with EGF-like and two Follistatin-like domains 2 (TMEFF2) regulates non-canonical BMP4/activin signaling and is involved in PI3K and RAS/ERK1/2 signaling.87 Annexin A2 (ANXA2) regulates membrane organization and trafficking including endo- and exocytosis.88 CD44 is the receptor for hyaluronan, known to be expressed by notochordal and mature NP cells.56,89 Finally, Solute carrier family 2 member 1 (SLC2A1; also known as GLUT1) is a known NP marker90 and was expressed at substantially higher levels in NP-nR clones. Importantly, expression of SLC2A1 is reported to be elevated in the context of human disc degeneration.91
Table 5.
Differentially Expressed NP-Specific Membrane-Associated Markers in NP-R and NP-nR Cell Clones.
| Gene | IDa | FCb | P c | Gene Description |
|---|---|---|---|---|
| CLDN11 | 5010 | 6.90 | 0.011 | Claudin 11 (oligodendrocyte transmembrane protein) |
| TMEFF2 | 23671 | 5.46 | 0.005 | Transmembrane protein with EGF-like and 2 follistatin-like domains 2 |
| CA12 | 771 | 2.72 | 0.025 | Carbonic anhydrase XII |
| ANXA2 | 302 | 2.51 | 0.009 | Annexin A2 |
| CD44 | 960 | 2.16 | 0.042 | CD44 antigen (homing function and Indian blood group system) |
| EFNA1 | 1942 | −3.24 | 0.003 | Ephrin-A1 |
| NETO2 | 81831 | −3.64 | 0.002 | Neuropilin (NRP) and tolloid (TLL)-like 2 |
| SLC2A1 | 6513 | −4.55 | 0.022 | Solute carrier family 2 (facilitated glucose transporter), member 1 |
ENTREZ Gene ID.
Fold change (NP-R/NP-nR).
Adjusted P value <0.05, fold change >2.0, and expression in NP >100 (arbitrary units; see Methods section for de tails).
Cellular Heterogeneity in the AF
Cellular heterogeneity in the inner (iAF) and outer AF (oAF) has been suggested based on morphological differences: oAF cells have an elongated appearance while iAF cells are more rounded and showed morphological similarities with NP cells. A recent study demonstrated differences in ECM expression after prolonged culturing of iAF and oAF cells on a polycarbonate urethane substrate: oAF cells expressed higher amounts of COL1A1, while iAF cells expressed more COL2A1, ACAN, and VCAN.91 COMP and Glypican 3 (GPC3) have been reported, although not consistently, as AF cell markers in vivo (Suppl. Table S1). GPC3 is a heparan sulfate proteoglycan involved in WNT, BMP and FGF signaling. Relevantly, GPC3 is an important inhibitor of SHH signaling92; as SHH is expressed in the embryonic notochord, GPC3 might be an important regulator of NP/AF boundary demarcation in IVD development. Secreted Frizzled Related Protein 2 (SFRP2) is an inhibitor of WNT signaling and was identified as an AF marker in primary bovine IVD cells,25 which we validated as an mRNA marker of human AF cells in vitro.20 Finally, differential expression of mitochondrial genes was suggested to differentiate between senescent and non-senescent cells in the AF.93,94 Overall, this limited gene list stresses the need for identification of additional AF-specific markers ( Table 6 ).
Table 6.
Expression of AF-Specific Membrane-Associated Markers Genes Compared to NP.
| Gene | Gene IDa | AF Meanb | NP Meanb | FCc |
|---|---|---|---|---|
| SLC40A1 | 30061 | 1241.8 | 148.2 | 8.4 |
| RNFT1 | 51136 | 271.2 | 38.0 | 7.1 |
| C18orf55 | 29090 | 369.5 | 66.6 | 5.5 |
| COLEC12 | 81035 | 257.7 | 47.9 | 5.4 |
| DAD1 | 1603 | 3141.1 | 584.2 | 5.4 |
| LPAR1 | 1902 | 1546.4 | 294.7 | 5.2 |
| IFITM2 | 10581 | 1411.7 | 297.8 | 4.7 |
| ALG10B | 144245 | 307.2 | 68.5 | 4.5 |
| IFITM1 | 8519 | 995.7 | 222.5 | 4.5 |
| THBD | 7056 | 388.0 | 88.4 | 4.4 |
| HEATR5A | 25938 | 286.0 | 66.2 | 4.3 |
| LRP6 | 4040 | 704.8 | 173.6 | 4.1 |
| SLC38A9 | 153129 | 249.9 | 62.3 | 4.0 |
| ATP11C | 286410 | 397.1 | 100.6 | 3.9 |
| FLRT2 | 23768 | 688.3 | 175.1 | 3.9 |
| GPR125 | 166647 | 374.9 | 96.5 | 3.9 |
| SLC30A5 | 64924 | 115.6 | 30.1 | 3.8 |
| GPR137B | 7107 | 232.0 | 61.0 | 3.8 |
| TMEM133 | 83935 | 331.6 | 88.7 | 3.7 |
| CD55 | 1604 | 3992.5 | 1074.7 | 3.7 |
| EFHA2 | 286097 | 149.8 | 41.6 | 3.6 |
| LRIG1 | 26018 | 1224.4 | 340.5 | 3.6 |
| SLC35A5 | 55032 | 486.7 | 135.8 | 3.6 |
| ANKRD46 | 157567 | 778.0 | 220.9 | 3.5 |
| CD58 | 965 | 237.3 | 68.8 | 3.4 |
| TMEM27 | 57393 | 151.3 | 44.2 | 3.4 |
| EMP3 | 2014 | 868.2 | 254.0 | 3.4 |
| CD59 | 966 | 333.2 | 97.9 | 3.4 |
| FAM69A | 388650 | 348.7 | 102.9 | 3.4 |
| TACSTD2 | 4070 | 2031.0 | 603.1 | 3.4 |
| C5orf28 | 64417 | 195.1 | 58.0 | 3.4 |
| SCNN1A | 6337 | 513.8 | 153.3 | 3.4 |
| SLC10A7 | 84068 | 388.9 | 116.1 | 3.4 |
| LRIG2 | 9860 | 524.8 | 158.6 | 3.3 |
| SMC3 | 9126 | 957.8 | 296.4 | 3.2 |
| ATP2C1 | 27032 | 122.2 | 38.4 | 3.2 |
| FLJ20160 | 54842 | 236.1 | 76.0 | 3.1 |
| LIFR | 3977 | 447.5 | 144.3 | 3.1 |
| FLJ11171 | 55783 | 216.9 | 72.3 | 3.0 |
| OMA1 | 115209 | 327.8 | 111.3 | 2.9 |
| C9orf123 | 90871 | 212.5 | 72.9 | 2.9 |
| ITM2C | 81618 | 1483.2 | 514.1 | 2.9 |
| ABHD2 | 11057 | 854.9 | 297.1 | 2.9 |
| EMP2 | 2013 | 999.4 | 350.1 | 2.9 |
| CD99 | 4267 | 606.0 | 213.8 | 2.8 |
| IL13RA1 | 3597 | 169.0 | 59.7 | 2.8 |
| C20orf199 | 441951 | 1288.8 | 466.2 | 2.8 |
| LOC440993 | 440993 | 276.4 | 100.4 | 2.8 |
| ADAM9 | 8754 | 229.3 | 85.2 | 2.7 |
| MGC24039 | 160518 | 131.7 | 49.0 | 2.7 |
| TNFSF10 | 8743 | 290.6 | 109.2 | 2.7 |
| PCNX | 22990 | 168.4 | 63.7 | 2.6 |
| TMEM200B | 399474 | 300.5 | 113.7 | 2.6 |
| ZMYM6 | 9204 | 220.5 | 83.9 | 2.6 |
| SEPSECS | 51091 | 131.7 | 50.6 | 2.6 |
| FAT4 | 79633 | 213.8 | 82.8 | 2.6 |
| LPAR4 | 2846 | 212.2 | 82.3 | 2.6 |
| LOC493869 | 493869 | 206.8 | 80.9 | 2.6 |
| AGTRAP | 57085 | 136.9 | 54.0 | 2.5 |
| CHIC1 | 53344 | 385.5 | 152.1 | 2.5 |
| CLCN3 | 1182 | 174.6 | 69.1 | 2.5 |
| STX7 | 8417 | 194.0 | 77.0 | 2.5 |
| SLC39A7 | 7922 | 782.0 | 312.4 | 2.5 |
| ACVR1 | 90 | 3337.6 | 1337.0 | 2.5 |
| DISP1 | 84976 | 178.9 | 73.0 | 2.4 |
| REEP1 | 65055 | 316.9 | 129.8 | 2.4 |
| SLC35B3 | 51000 | 1395.7 | 572.9 | 2.4 |
| SLC39A14 | 23516 | 344.9 | 142.4 | 2.4 |
| ROBO1 | 6091 | 606.3 | 251.1 | 2.4 |
| SLC41A2 | 84102 | 272.7 | 113.4 | 2.4 |
| FDFT1 | 2222 | 593.3 | 247.6 | 2.4 |
| MIA3 | 375056 | 362.0 | 151.4 | 2.4 |
| HIGD1A | 25994 | 2291.2 | 960.2 | 2.4 |
| ITGB1 | 3688 | 221.4 | 92.9 | 2.4 |
| SLC33A1 | 9197 | 1725.7 | 731.3 | 2.4 |
| UPK1B | 7348 | 160.2 | 68.0 | 2.4 |
| SLC16A3 | 9123 | 183.0 | 77.6 | 2.4 |
| FAS | 355 | 192.3 | 82.1 | 2.3 |
| C20orf30 | 29058 | 217.5 | 92.9 | 2.3 |
| SLC30A6 | 55676 | 365.7 | 156.2 | 2.3 |
| PTPLAD2 | 401494 | 108.2 | 46.3 | 2.3 |
| LRP12 | 29967 | 129.7 | 55.8 | 2.3 |
| STEAP2 | 261729 | 312.0 | 134.9 | 2.3 |
| FAM73A | 374986 | 365.2 | 158.3 | 2.3 |
| LAG3 | 3902 | 330.2 | 144.4 | 2.3 |
| KCNA1 | 3736 | 167.5 | 73.6 | 2.3 |
| ULBP2 | 80328 | 184.1 | 81.2 | 2.3 |
| ATL3 | 25923 | 1223.0 | 540.4 | 2.3 |
| ATP11B | 23200 | 236.3 | 104.9 | 2.3 |
| PROCR | 10544 | 568.0 | 252.3 | 2.3 |
| C7orf44 | 55744 | 237.1 | 105.6 | 2.2 |
| ASAM | 79827 | 195.6 | 87.3 | 2.2 |
| C14orf101 | 54916 | 230.7 | 103.0 | 2.2 |
| TSPAN4 | 7106 | 115.5 | 51.7 | 2.2 |
| SLC31A2 | 1318 | 545.7 | 246.3 | 2.2 |
| SACM1L | 22908 | 1277.7 | 577.6 | 2.2 |
| FZD2 | 2535 | 197.5 | 89.6 | 2.2 |
| TM2D2 | 83877 | 1033.3 | 469.6 | 2.2 |
| USMG5 | 84833 | 3457.8 | 1573.8 | 2.2 |
| MFSD8 | 256471 | 161.8 | 73.7 | 2.2 |
| JAM2 | 58494 | 318.1 | 144.9 | 2.2 |
| ARMCX5 | 64860 | 273.9 | 125.2 | 2.2 |
| AXL | 558 | 1232.8 | 566.4 | 2.2 |
| STX17 | 55014 | 421.2 | 194.1 | 2.2 |
| LNPEP | 4012 | 215.2 | 99.5 | 2.2 |
| DLEU2 | 8847 | 406.6 | 188.7 | 2.2 |
| LOC219854 | 219854 | 395.2 | 183.6 | 2.2 |
| C9orf105 | 401505 | 649.6 | 303.2 | 2.1 |
| GHITM | 27069 | 649.2 | 303.7 | 2.1 |
| SLC16A7 | 9194 | 237.6 | 112.6 | 2.1 |
| TMEM154 | 201799 | 172.1 | 81.9 | 2.1 |
| EGFR | 1956 | 253.3 | 121.0 | 2.1 |
| TMEM19 | 55266 | 113.9 | 55.0 | 2.1 |
| CAPRIN1 | 4076 | 308.4 | 149.0 | 2.1 |
| SEL1L | 6400 | 889.5 | 433.1 | 2.1 |
| IFNAR1 | 3454 | 197.7 | 96.8 | 2.0 |
| SPA17 | 53340 | 125.9 | 61.8 | 2.0 |
| GLIPR1 | 11010 | 850.1 | 417.6 | 2.0 |
| CNIH4 | 29097 | 2894.7 | 1424.1 | 2.0 |
| KCNK1 | 3775 | 582.0 | 287.8 | 2.0 |
| LOC203547 | 203547 | 1509.4 | 747.3 | 2.0 |
| IL17RD | 54756 | 149.7 | 74.3 | 2.0 |
| INTS7 | 25896 | 102.8 | 51.2 | 2.0 |
| DPY19L4 | 286148 | 283.4 | 141.6 | 2.0 |
| SLC13A4 | 26266 | 1083.4 | 541.6 | 2.0 |
ENTREZ Gene ID.
Adjusted P value <0.05, fold change >2.0, and expression: >100 (arbitrary units; see Methods section for details).
Fold change (AF/NP).
Novel AF Subclone-Specific Membrane-Associated Markers
To identify AF-specific membrane markers, we first aimed to define AF-specific genes by comparing primary cell isolates from AF and NP26; this analysis yielded 1161 genes that showed a significantly higher expression in AF than in NP. Application of the membranome-filter to this gene collection produced a subset of 125 AF-specific genes encoding membrane-associated proteins ( Fig. 4D , Table 7 ). We have limited discussing the potential relevance of these markers in regard to AF cell type–specific biology to a selection of marker genes. Of interest, we identified 2 receptors from the canonical WNT pathway (LRP4 and FZD2). LRP4 was recently shown to facilitate chondrogenic differentiation and articular cartilage homeostasis.95,96 In addition, expression of the ligand WNT16 was significantly higher in primary AF. WNT16 was shown to repress osteoclastogenesis and to protect articular cartilage in a murine model for osteoarthritis.97,98 Combined with our earlier observation that the SFRP2 is an AF marker in vitro, these results suggest that regulation of WNT signaling separates AF cells from NP cells. Of note, none of these factors have thus far been investigated in the IVD. WNT signaling is known to be involved in IVD development, decreases in the adult mouse IVD and is reactivated in old IVDs.
Table 7.
Differentially Expressed AF-Specific Membrane-Associated Markers in AF-S and AF-nS Cell Clones.
| Gene | Gene IDa | FCb | P c | Gene Description | FCd |
|---|---|---|---|---|---|
| CHIC1 | 53344 | 2.3 | 0.0017 | Cysteine-rich hydrophobic domain 1 | 2.5 |
| COLEC12 | 81035 | −2.3 | 0.0432 | collectin sub-family member 12 | 5.4 |
| LPAR1 | 1902 | −2.0 | 0.0232 | lysophosphatidic acid receptor 1 | 5.2 |
ENTREZ Gene ID.
Fold change (AF2/AF1).
Adjusted P value <0.05, fold change >2.0, and expression in AF >100 (arbitrary units; see Methods section for details).
Fold change (AF/NP).
To expose AF clone-specific markers, we next compared the 2 AF clone subtypes that were distinguished on their (in)ability to process procollagens in vitro: Sheet-forming (AF-S) and nonSheet-forming (AF-nS) AF clones.20 A total of 120 differentially expressed genes remained after membranome-filtering ( Fig. 4D , Suppl. Tables S5 and S6). CLDN11, identified as an NP-R marker gene (cf. Suppl. Fig. S2), has also been implicated as a polarity gene in human AF cells in situ.64 CLDN11 was preferentially expressed in AF-S clones. Of interest, AF-nS clones showed conspicuous Notch activity, as NOTCH3, JAG1, and downstream target gene HES1 were more highly expressed in AF-nS cells compared to AF-S cells (Suppl. Fig. S5C). Notch/Jagged/Hes1 activity was shown to be differentially regulated in NP an AF in response to hypoxia.99 In addition, polymorphisms in the NOTCH3 gene have been associated with lumbar disc herniation and Notch signaling has been implicated in degenerative disc disease.100,101 The exact biological implication of increased Notch-signaling in AF cells is currently unknown. It has been suggested that Notch is important for maintaining proliferative capacity of disc cells.99 Our data add to these observations and for the first time identifies a cellular AF sub-phenotype in which Notch signaling has a distinctive function.
Last, to determine the validity of these clone-specific markers, we compared primary AF cell specific genes to AF subtype-specific markers ( Fig. 4E ). This identified 3 genes that were expressed both in primary as well as immortal monoclonal AF cell extracts: Cysteine-rich hydrophobic domain 1 (CHIC1), Collectin sub-family member 12 (COLEC12), and Lysophosphatidic acid receptor 1 (LPAR1; Table 7 ). CHIC1 (also known as AKAP13) is an anchoring protein involved in Protein Kinase A signaling. In addition to its involvement in cardiac hypertrophy, it is known to regulate osmotic responses and integration of inflammatory signals in T lymphocytes.102,103 Little is known about COLEC12 in the context of IVD biology. LPAR1 was shown to play a role in synovial fibroblast activation during arthritis and plays a role in osteoclastogenesis.104,105 Taken together, our analyses identify a substantial set of novel AF membrane-associated markers. Subsets among these markers, such as WNT receptors LRP4, LRP5 (NP-nR and AF marker), FZD2, and additional genes described above, discriminate between our previously established immortal IVD cell types and may prove useful to functionally define cellular heterogeneity in the AF.
Summary and Perspectives
In this study, we present a meta-analysis of molecular markers in relation to cellular NP and AF phenotypes in the IVD. We here provide independent evidence of whole-transcriptome-based clustering of our novel immortal NP and AF cell lines with their respective IVD-derived primary isolates; in addition, PCA analysis clearly separates the NP clonal subtypes from each other and from AF cellular subpopulations. Importantly, the data accrued confirms cellular heterogeneity in NP and AF cell populations and supports the notion that cellular phenotypes are epigenetically preserved during the immortalization process.
Our membranome-based transcriptomics analysis aimed to identify novel cell surface markers for NP and AF cellular (sub)types. PCA analysis and validation of membrane and non–membrane-associated marker expression using qPCR analysis consistently reveal a closer similarity of NP-nR with AF clones than with NP-R clones. Besides being lineage specific, a cell’s transcriptomic phenotype is also determined through interaction with its microenvironment.106 Hence, it is conceivable that despite their dissimilar ontogeny, original NP-nR and AF cells were derived from areas with a relatively similar microenvironment in their respective tissues of origin.
The novel membrane markers identified in this study will be crucial for the isolation, systematic analysis, and functional characterization of NP cellular subtypes and their implication in the context of disc homeostasis and disease. Investigation of specific cell subtypes in the IVD requires histological studies focused on expression analysis of markers identified herein in IVD tissue sections to define their relative locations and to understand their ontogenic relationships. The availability of a functionally defined set of NP and AF cells enables systematic analysis of altered activity within gene networks, for instance, as a function of age, and is expected to significantly contribute to understanding gene-microenvironment interaction in the IVD. It is very likely that microenvironmental alterations (e.g., inflammation, oxygen tension, osmolarity) and accompanying physiological changes related to ageing or degeneration override physiological responses required to maintain cell and tissue homeostasis. Such knowledge will also be pivotal for defining appropriate conditions for isolation, culturing, and maintaining cells from the NP and AF and for application of IVD-directed differentiation of stem cells in regenerative procedures.
Supplemental Material
Supplemental material, Figure_S1 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S2 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S3 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S4 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Supp._Files for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Footnotes
Authors’ Note: This research forms part of the Project P2.01 IDiDAS of the research program of the BioMedical Materials institute, co-funded by the Dutch Ministry of Economic Affairs, Agriculture and Innovation. None of the funding sources had any role in study design, data collection, analysis and interpretation, writing of the manuscript or the decision to submit the manuscript for publication.
Author Contributions: GvdA, LMTE, LWvR, JAH, TJMW, and JWV contributed to the conception and design of the study; GvdA, LMTE, and SMR contributed to acquisition, analysis, and interpretation of data; GvdA, LMTE, SMR, JAH, TJMW, and JWV drafted and gave final approval of the submitted version.
Acknowledgments and Funding: We thank Dr. Paul Willems for providing adult human IVD tissue for histology. Approval for all experimental sections of the current study and informed consent for publication of patient details and accompanying images in this manuscript was obtained as an integral part of the MUMC-MERC Approval 08-4-021; the approval is held by the authors and is available for review. Funding: Project P2.01 IDiDAS (LWvR, TJMW); LLP14 Grant Dutch Arthritis Foundation (JWV, LWvR).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: Ethical approval for this study was obtained from the MUMC-Medical Ethical Review Committee (Approval ID 08-4-021).
Informed Consent: Written informed consent was obtained from legally authorized representatives before the study.
ORCID iD: Guus G. H. van den Akker  https://orcid.org/0000-0002-7071-1487
https://orcid.org/0000-0002-7071-1487
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Martin BI, Deyo RA, Mirza SK, Turner JA, Comstock BA, Hollingworth W, et al. Expenditures and health status among adults with back and neck problems. JAMA. 2008;299:656-64. [DOI] [PubMed] [Google Scholar]
- 2. Maniadakis N, Gray A. The economic burden of back pain in the UK. Pain. 2000;84:95-103. [DOI] [PubMed] [Google Scholar]
- 3. Luoma K, Riihimäki H, Luukkonen R, Raininko R, Viikari-Juntura E, Lamminen A. Low back pain in relation to lumbar disc degeneration. Spine (Phila Pa 1976). 2000;25:487-92. [DOI] [PubMed] [Google Scholar]
- 4. Miller JA, Schmatz C, Schultz AB. Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine (Phila Pa 1976). 1988;13:173-8. [PubMed] [Google Scholar]
- 5. Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams MA, Roughley PJ. What is intervertebral disc degeneration, and what causes it? Spine (Phila Pa 1976). 2006;31:2151-61. [DOI] [PubMed] [Google Scholar]
- 7. Adams MA, Dolan P, McNally DS. The internal mechanical functioning of intervertebral discs and articular cartilage, and its relevance to matrix biology. Matrix Biol. 2009;28:384-89. [DOI] [PubMed] [Google Scholar]
- 8. Mwale F, Roughley P, Antoniou J. Distinction between the extracellular matrix of the nucleus pulposus and hyaline cartilage: a requisite for tissue engineering of intervertebral disc. Eur Cell Mater. 2004;8:58-63. [DOI] [PubMed] [Google Scholar]
- 9. Le Maitre CL, Freemont AJ, Hoyland JA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2007;9:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang F, Cai F, Shi R, Wang XH, Wu XT. Aging and age related stresses: a senescence mechanism of intervertebral disc degeneration. Osteoarthritis Cartilage. 2016;24:398-408. [DOI] [PubMed] [Google Scholar]
- 11. Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc-content. Nat Rev Rheumatol. 2014;10:44-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phillips KL, Cullen K, Chiverton N, Michael ALR, Cole AA, Breakwell LM, et al. Potential roles of cytokines and chemokines in human intervertebral disc degeneration: interleukin-1 is a master regulator of catabolic processes. Osteoarthritis Cartilage. 2015;23:1165-77. [DOI] [PubMed] [Google Scholar]
- 13. Liu HF, Zhang H, Qiao GX, Ning B, Hu YL, Wang DC, et al. A novel rabbit disc degeneration model induced by fibronectin fragment. Joint Bone Spine. 2013;80:301-6. [DOI] [PubMed] [Google Scholar]
- 14. Sivan S, Hayes A, Wachtel E, Caterson B, Merkher Y, Maroudas A, et al. Biochemical composition and turnover of the extracellular matrix of the normal and degenerate intervertebral disc. Eur Spine J. 2014;23(Suppl 3):S344-S353. [DOI] [PubMed] [Google Scholar]
- 15. Sivan SS, Wachtel E, Roughley P. Structure, function, aging and turnover of aggrecan in the intervertebral disc. Biochim Biophys Acta. 2014;1840:3181-9. [DOI] [PubMed] [Google Scholar]
- 16. Huang YC, Urban JP, Luk KD. Intervertebral disc regeneration: do nutrients lead the way? Nat Rev Rheumatol. 2014;10:561-6. [DOI] [PubMed] [Google Scholar]
- 17. Wang D, Nasto LA, Roughley P, Leme AS, Houghton AM, Usas A, et al. Spine degeneration in a murine model of chronic human tobacco smokers. Osteoarthritis Cartilage. 2012;20:896-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van den Akker GG, Surtel DA, Cremers A, Rodrigues-Pinto R, Richardson SM, Hoyland JA, et al. Novel immortal human cell lines reveal subpopulations in the nucleus pulposus. Arthritis Res Ther. 2014;16:R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van den Akker GG, Surtel DA, Cremers A, Hoes MF, Caron MM, Richardson SM, et al. EGR1 controls divergent cellular responses of distinctive nucleus pulposus cell types. BMC Musculoskelet Disord. 2016;17:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van den Akker GG, Surtel DA, Cremers A, Richardson SM, Hoyland JA, van Rhijn LW, et al. Novel immortal cell lines support cellular heterogeneity in the human annulus fibrosus. PLoS One. 2016;11:e0144497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trout JJ, Buckwalter JA, Moore KC. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat Rec. 1982;204:307-14. [DOI] [PubMed] [Google Scholar]
- 22. Trout JJ, Buckwalter JA, Moore KC, Landas SK. Ultrastructure of the human intervertebral disc. I. Changes in notochordal cells with age. Tissue Cell. 1982;14:359-69. [DOI] [PubMed] [Google Scholar]
- 23. Lee CR, Sakai D, Nakai T, Toyama K, Mochida J, Alini M, et al. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J. 2007;16:2174-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakai D, Nakai T, Mochida J, Alini M, Grad S. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine (Phila Pa 1976). 2009;34:1448-56. [DOI] [PubMed] [Google Scholar]
- 25. Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010;62:3695-705. [DOI] [PubMed] [Google Scholar]
- 27. Power KA, Grad S, Rutges JP, Creemers LB, van Rijen MH, O’Gaora P, et al. Identification of cell surface-specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum. 2011;63:3876-86. [DOI] [PubMed] [Google Scholar]
- 28. Fujita N, Miyamoto T, Imai J, Hosogane N, Suzuki T, Yagi M, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890-96. [DOI] [PubMed] [Google Scholar]
- 29. Gilson A, Dreger M, Urban JP. Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther. 2010;12:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rutges J, Creemers LB, Dhert W, Milz S, Sakai D, Mochida J, et al. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: potential associations with aging and degeneration. Osteoarthritis Cartilage. 2010;18:416-23. [DOI] [PubMed] [Google Scholar]
- 31. Risbud MV, Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the spine research interest group at the 2014 annual ORS meeting. J Orthop Res. 2015;33:283-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodrigues-Pinto R, Berry A, Piper-Hanley K, Hanley N, Richardson SM, Hoyland JA. Spatiotemporal analysis of putative notochordal cell markers reveals CD24 and keratins 8, 18, and 19 as notochord-specific markers during early human intervertebral disc development. J Orthop Res. 2016;34:1327-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tang X, Jing L, Richardson WJ, Isaacs RE, Fitch RD, Brown CR, et al. Identifying molecular phenotype of nucleus pulposus cells in human intervertebral disc with aging and degeneration. J Orthop Res. 2016;34:1316-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorpe AA, Binch AL, Creemers LB, Sammon C, Le Maitre CL. Nucleus pulposus phenotypic markers to determine stem cell differentiation: fact or fiction? Oncotarget. 2016;7:2189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wáng YX, Wáng JQ, Káplár Z. Increased low back pain prevalence in females than in males after menopause age: evidences based on synthetic literature review. Quant Imaging Med Surg. 2016;6:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Uva P, Lahm A, Sbardellati A, Grigoriadis A, Tutt A, de Rinaldis E. Comparative membranome expression analysis in primary tumors and derived cell lines. PLoS One. 2010;5:e11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rust S, Guillard S, Sachsenmeier K, Hay C, Davidson M, Karlsson A, et al. Combining phenotypic and proteomic approaches to identify membrane targets in a “triple negative” breast cancer cell type. Mol Cancer. 2013;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ghosh D, Beavis RC, Wilkins JA. The identification and characterization of membranome components. J Proteome Res. 2008;7:1572-83. [DOI] [PubMed] [Google Scholar]
- 39. Alini M, Eisenstein SM, Ito K, Little C, Kettler AA, Masuda K, et al. Are animal models useful for studying human disc disorders/degeneration? Eur Spine J. 2008;17:2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rehman AA, Ahsan H, Khan FH. Alpha-2-macroglobulin: a physiological guardian. J Cell Physiol. 2013;228:1665-75. [DOI] [PubMed] [Google Scholar]
- 41. Binch AL, Cole AA, Breakwell LM, Michael AL, Chiverton N, Cross AK, et al. Expression and regulation of neurotrophic and angiogenic factors during human intervertebral disc degeneration. Arthritis Res Ther. 2014;16:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barcelona PF, Saragovi HU. A pro-nerve growth factor (proNGF) and NGF binding protein, α2-macroglobulin, differentially regulates p75 and TrkA receptors and is relevant to neurodegeneration ex vivo and in vivo. Mol Cell Biol. 2015;35:3396-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Osada M, Inoue O, Ding G, Shirai T, Ichise H, Hirayama K, et al. Platelet activation receptor CLEC-2 regulates blood/lymphatic vessel separation by inhibiting proliferation, migration, and tube formation of lymphatic endothelial cells. J Biol Chem. 2012;287:22241-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153-9. [DOI] [PubMed] [Google Scholar]
- 45. Tolofari SK, Richardson SM, Freemont AJ, Hoyland JA. Expression of semaphorin 3A and its receptors in the human intervertebral disc: potential role in regulating neural ingrowth in the degenerate intervertebral disc. Arthritis Res Ther. 2010;12:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bu G, Hou S, Ren D, Wu Y, Shang W, Huang W. Increased expression of netrin-1 and its deleted in colorectal cancer receptor in human diseased lumbar intervertebral disc compared with autopsy control. Spine (Phila Pa 1976). 2012;37:2074-81. [DOI] [PubMed] [Google Scholar]
- 47. Collette NM, Yee CS, Hum NR, Murugesh DK, Christiansen BA, Xie L, et al. Sostdc1 deficiency accelerates fracture healing by promoting the expansion of periosteal mesenchymal stem cells. Bone. 2016;88:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chelberg MK, Banks GM, Geiger DF, Oegema TR., Jr. Identification of heterogeneous cell populations in normal human intervertebral disc. J Anat. 1995;186:43-53. [PMC free article] [PubMed] [Google Scholar]
- 49. Sharp CA, Roberts S, Evans H, Brown SJ. Disc cell clusters in pathological human intervertebral discs are associated with increased stress protein immunostaining. Eur Spine J. 2009;18:1587-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Henriksson H, Thornemo M, Karlsson C, Hagg O, Junevik K, Lindahl A, et al. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine (Phila Pa 1976). 2009;34:2278-87. [DOI] [PubMed] [Google Scholar]
- 51. Feng G, Yang X, Shang H, Marks IW, Shen FH, Katz A. Multipotential differentiation of human anulus fibrosus cells: an in vitro study. J Bone Joint Surg Am. 2010;92:675-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu C, Guo Q, Li J, Wang S, Wang Y, Li B, et al. Identification of rabbit annulus fibrosus-derived stem cells. PLoS One. 2014;9:e108239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu LT, Huang B, Li CQ, Zhuang Y, Wang J, Zhou Y. Characteristics of stem cells derived from the degenerated human intervertebral disc cartilage endplate. PLoS One. 2011;6:e26285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Erwin WM, Islam D, Eftekarpour E, Inman RD, Karim MZ, Fehlings MG. Intervertebral disc-derived stem cells: implications for regenerative medicine and neural repair. Spine (Phila Pa 1976). 2013;38:211-6. [DOI] [PubMed] [Google Scholar]
- 55. Risbud MV, Guttapalli A, Tsai TT, Lee JY, Danielson KG, Vaccaro AR, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976). 2007;32:2537-44. [DOI] [PubMed] [Google Scholar]
- 56. Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tekari A, Chan SC, Sakai D, Grad S, Gantenbein B. Angiopoietin-1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res Ther. 2016;7:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cheng NC, Wang S, Young TH. The influence of spheroid formation of human adipose-derived stem cells on chitosan films on stemness and differentiation capabilities. Biomaterials. 2012;33:1748-58. [DOI] [PubMed] [Google Scholar]
- 59. Tesei A, Zoli W, Arienti C, Storci G, Granato AM, Pasquinelli G, et al. Isolation of stem/progenitor cells from normal lung tissue of adult humans. Cell Prolif. 2009;42:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Collins-Racie LA, Yang Z, Arai M, Li N, Majumdar MK, Nagpal S, et al. Global analysis of nuclear receptor expression and dysregulation in human osteoarthritic articular cartilage: reduced LXR signaling contributes to catabolic metabolism typical of osteoarthritis. Osteoarthritis Cartilage. 2009;17:832-42. [DOI] [PubMed] [Google Scholar]
- 61. Ythier D, Larrieu D, Binet R, Binda O, Brambilla C, Gazzeri S, et al. Sumoylation of ING2 regulates the transcription mediated by Sin3A. Oncogene. 2010;29:5946-56. [DOI] [PubMed] [Google Scholar]
- 62. Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PW. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297:14-25. [DOI] [PubMed] [Google Scholar]
- 63. Flintoff KA, Arudchelvan Y, Gong SG. FLRT2 interacts with fibronectin in the ATDC5 chondroprogenitor cells. J Cell Physiol. 2014;229:1538-47. [DOI] [PubMed] [Google Scholar]
- 64. Gruber HE, Ingram J, Hoelscher GL, Norton HJ, Hanley EN., Jr. Cell polarity in the anulus of the human intervertebral disc: morphologic, immunocytochemical, and molecular evidence. Spine (Phila Pa 1976). 2007;32:1287-94. [DOI] [PubMed] [Google Scholar]
- 65. Bronstein JM, Kozak CA, Chen XN, Wu S, Danciger M, Korenberg JR, et al. Chromosomal localization of murine and human oligodendrocyte-specific protein genes. Genomics. 1996;34:255-57. [DOI] [PubMed] [Google Scholar]
- 66. Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873-80. [DOI] [PubMed] [Google Scholar]
- 67. Strauss U, Bräuer AU. Current views on regulation and function of plasticity-related genes (PRGs/LPPRs) in the brain. Biochim Biophys Acta. 2013;1831:133-8. [DOI] [PubMed] [Google Scholar]
- 68. Somogyi CS, Matta C, Foldvari Z, Juhász T, Katona É, Takács Á, et al. Polymodal transient receptor potential vanilloid (TRPV) ion channels in chondrogenic cells. Int J Mol Sci. 2015;16:18412-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bonafe L, Cormier-Daire V, Hall C, Lachman R, Mortier G, Mundlos S, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A. 2015;167A:2869-92. [DOI] [PubMed] [Google Scholar]
- 70. Xu Y, Yang Z, Yuan H, Li Z, Li Y, Liu Q, et al. PCDH10 inhibits cell proliferation of multiple myeloma via the negative regulation of the Wnt/β-catenin/BCL-9 signaling pathway. Oncol Rep. 2015;34:747-54. [DOI] [PubMed] [Google Scholar]
- 71. Craft AM, Ahmed N, Rockel JS, Baht GS, Alman BA, Kandel RA, et al. Specification of chondrocytes and cartilage tissues from embryonic stem cells. Development. 2013;140:2597-610. [DOI] [PubMed] [Google Scholar]
- 72. Yamamoto M, Toya Y, Jensen RA, Ishikawa Y. Caveolin is an inhibitor of platelet-derived growth factor receptor signaling. Exp Cell Res. 1999;247:380-8. [DOI] [PubMed] [Google Scholar]
- 73. Joosten PH, Hol FA, van Beersum SE, Peters H, Hamel BC, Afink GB, et al. Altered regulation of platelet-derived growth factor receptor-alpha gene-transcription in vitro by spina bifida-associated mutant Pax1 proteins. Proc Natl Acad Sci U S A. 1998;95:14459-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Battegay EJ, Raines EW, Seifert RA, Bowen-Pope DF, Ross R. TGF-beta induces bimodal proliferation of connective tissue cells via complex control of an autocrine PDGF loop. Cell. 1990;63:515-24. [DOI] [PubMed] [Google Scholar]
- 75. Paglia DN, Singh H, Karukonda T, Drissi H, Moss IL. PDGF-BB delays degeneration of the intervertebral discs in a rabbit preclinical model. Spine (Phila Pa 1976). 2016;41:E449-E458. [DOI] [PubMed] [Google Scholar]
- 76. Smith RJ, Justen JM, Sam LM, Rohloff NA, Ruppel PL, Brunden MN, et al. Platelet-derived growth factor potentiates cellular responses of articular chondrocytes to interleukin-1. Arthritis Rheum. 1991;34:697-706. [DOI] [PubMed] [Google Scholar]
- 77. Ullah M, Sittinger M, Ringe J. Extracellular matrix of adipogenically differentiated mesenchymal stem cells reveals a network of collagen filaments, mostly interwoven by hexagonal structural units. Matrix Biol. 2013;32:452-65. [DOI] [PubMed] [Google Scholar]
- 78. Jorge EC, Ahmed MU, Bothe I, Coutinho LL, Dietrich S. RGMa and RGMb expression pattern during chicken development suggest unexpected roles for these repulsive guidance molecules in notochord formation, somitogenesis, and myogenesis. Dev Dyn. 2012;241:1886-900. [DOI] [PubMed] [Google Scholar]
- 79. DiPaola CP, Farmer JC, Manova K, Niswander LA. Molecular signaling in intervertebral disc development. J Orthop Res. 2005;23:1112-9. [DOI] [PubMed] [Google Scholar]
- 80. Sugiura A, Ohtori S, Yamashita M, Inoue G, Yamauchi K, Koshi T, et al. Existence of nerve growth factor receptors, tyrosine kinase a and p75 neurotrophin receptors in intervertebral discs and on dorsal root ganglion neurons innervating intervertebral discs in rats. Spine (Phila Pa 1976). 2008;33:2047-51. [DOI] [PubMed] [Google Scholar]
- 81. Stoker AW. Isoforms of a novel cell adhesion molecule-like protein tyrosine phosphatase are implicated in neural development. Mech Dev. 1994;46:201-17. [DOI] [PubMed] [Google Scholar]
- 82. Hedge TA, Mason I. Expression of Shisa2, a modulator of both Wnt and Fgf signaling, in the chick embryo. Int J Dev Biol. 2008;52:81-5. [DOI] [PubMed] [Google Scholar]
- 83. Nagano T, Takehara S, Takahashi M, Aizawa S, Yamamoto A. Shisa2 promotes the maturation of somitic precursors and transition to the segmental fate in Xenopus embryos. Development. 2006;133:4643-54. [DOI] [PubMed] [Google Scholar]
- 84. Lambert C, Dubuc JE, Montell E, Vergés J, Munaut C, Noël A, et al. Gene expression pattern of cells from inflamed and normal areas of osteoarthritis synovial membrane. Arthritis Rheumtol. 2014;66:960-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shin Y, Huh Y, Kim K, Kim S, Park K, Koh JT, et al. Low-density lipoprotein receptor-related protein 5 governs Wnt-mediated osteoarthritic cartilage destruction. Arthritis Res Ther. 2014;16:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chambrion C, Le Naour F. The tetraspanins CD9 and CD81 regulate CD9P1-induced effects on cell migration. PLoS One. 2010;5:e11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Labeur M, Wölfel B, Stalla J, Stalla GK. TMEFF2 is an endogenous inhibitor of the CRH signal transduction pathway. J Mol Endocrinol. 2015;54:51-63. [DOI] [PubMed] [Google Scholar]
- 88. Grewal T, Wason SJ, Enrich C, Rentero C. Annexins—insights from knockout mice. Biol Chem. 2016;397:1031-53. [DOI] [PubMed] [Google Scholar]
- 89. Stevens JW, Kurriger GL, Carter AS, Maynard JA. CD44 expression in the developing and growing rat intervertebral disc. Dev Dyn. 2000;219:381-90. [DOI] [PubMed] [Google Scholar]
- 90. Richardson SM, Knowles R, Tyler J, Mobasheri A, Hoyland JA. Expression of glucose transporters GLUT-1, GLUT-3, GLUT-9 and HIF-1α in normal and degenerate human intervertebral disc. Histochem Cell Biol. 2008;129:503-11. [DOI] [PubMed] [Google Scholar]
- 91. Iu J, Santerre JP, Kandel RA. Inner and outer annulus fibrosus cells exhibit differentiated phenotypes and yield changes in extracellular matrix protein composition in vitro on a polycarbonate urethane scaffold. Tissue Eng Part A. 2014;20:3261-9. [DOI] [PubMed] [Google Scholar]
- 92. Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14:700-11. [DOI] [PubMed] [Google Scholar]
- 93. Gruber H, Hoelscher GL, Ingram JA, Zinchenko N, Hanley EN., Jr. Senescent vs. non-senescent cells in the human annulus in vivo: cell harvest with laser capture microdissection and gene expression studies with microarray analysis. BMC Biotechnol. 2010;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Gruber HE, Watts JA, Hoelscher GL, Bethea SF, Ingram JA, Zinchenko NS, et al. Mitochondrial gene expression in the human annulus: in vivo data from annulus cells and selectively harvested senescent annulus cells. Spine J. 2011;11:782-91. [DOI] [PubMed] [Google Scholar]
- 95. Asai N, Ohkawara B, Ito M, Masuda A, Ishiguro N, Ohno K. LRP4 induces extracellular matrix productions and facilitates chondrocyte differentiation. Biochem Biophys Res Commun. 2014;451:302-7. [DOI] [PubMed] [Google Scholar]
- 96. Eldridge S, Nalesso G, Ismail H, Vicente-Greco K, Kabouridis P, Ramachandran M, et al. Agrin mediates chondrocyte homeostasis and requires both LRP4 and α-dystroglycan to enhance cartilage formation in vitro and in vivo. Ann Rheum Dis. 2016;75:1228-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moverare-Skrtic S, Henning P, Liu X, Nagano K, Saito H, Borjesson AE, et al. Osteoblast-derived WNT16 represses osteoclastogenesis and prevents cortical bone fragility fractures. Nat Med. 2014;20:1279-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nalesso G, Thomas BL, Sherwood JC, Yu J, Addimanda O, Eldridge SE, et al. WNT16 antagonises excessive canonical WNT activation and protects cartilage in osteoarthritis. Ann Rheum Dis. 2017;76:218-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hiyama A, Skubutyte R, Markova D, Anderson DG, Yadla S, Sakai D, et al. Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: implications in degenerative disc disease. Arthritis Rheum. 2011;63:1355-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yamada H, Yasuda T, Kotorii S, Takahashi K, Tabira T, Sunada Y. Report of a patient with CADASIL having a novel missense mutation of the Notch 3 gene-association with alopecia and lumbar herniated disc. Rinsho Shinkeigaku. 2001;41:144-6. [PubMed] [Google Scholar]
- 101. Wang H, Tian Y, Wang J, Phillips KL, Binch AL, Dunn S, et al. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 2013;288:16761-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Johnson KR, Nicodemus-Johnson J, Spindler MJ, Carnegie GK. Genome-wide gene expression analysis shows AKAP13-mediated PKD1 signaling regulates the transcriptional response to cardiac hypertrophy. PLoS One. 2015;10:e0132474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kino T, Takatori H, Manoli I, Wang Y, Tiulpakov A, Blackman MR, et al. Brx mediates the response of lymphocytes to osmotic stress through the activation of NFAT5. Sci Signal. 2009;2:ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Miyabe Y, Miyabe C, Iwai Y, Yokoyama W, Sekine C, Sugimoto K, et al. Activation of fibroblast-like synoviocytes derived from rheumatoid arthritis via lysophosphatidic acid–lysophosphatidic acid receptor 1 cascade. Arthritis Res Ther. 2014;16:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. David M, Machuca-Gayet I, Kikuta J, Ottewell P, Mima F, Leblanc R, et al. Lysophosphatidic acid receptor type 1 (LPA1) plays a functional role in osteoclast differentiation and bone resorption activity. J Biol Chem. 2014;289:6551-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Bhat R, Bissell MJ. Of plasticity and specificity: dialectics of the microenvironment and macroenvironment and the organ phenotype. Wiley Interdiscip Rev Dev Biol. 2014;3:147-63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Figure_S1 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S2 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S3 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Figure_S4 for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE
Supplemental material, Supp._Files for A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc by Guus G. H. van den Akker, Lars M. T. Eijssen, Stephen M. Richardson, Lodewijk W. van Rhijn, Judith A. Hoyland, Tim J. M. Welting and Jan Willem Voncken in CARTILAGE