Abstract
Peptide self-assembly has attracted extensive interest in the field of eco-friendly optoelectronics and bioimaging due to its inherent biocompatibility, intrinsic fluorescence, and flexible modulation. However, the practical application of such materials was hindered by the relatively low quantum yield of such assemblies. Here, inspired by the molecular structure of BFPms1, we explored the “self-assembly locking strategy” to design and manipulate the assembly of metal-stabilized cyclic(l-histidine-d-histidine) into peptide material with the high-fluorescence efficiency. We used this bioorganic material as an emissive layer in photo- and electroluminescent prototypes, demonstrating the feasibility of utilizing self-assembling peptides to fabricate a biointegrated microchip that incorporates eco-friendly and tailored optoelectronic properties. We further employed a “self-encapsulation” strategy for constructing an advanced nanocarrier with integrated in situ monitoring. The strategy of the supramolecular capture of functional components exemplifies the use of bioinspired organic chemistry to provide frontiers of smart materials, potentially allowing a better interface between sustainable optoelectronics and biomedical applications.
Keywords: supramolecular fluorescence, cyclic dipeptide, self-assembly, optoelectronic, drug release monitor
Bioorganic molecules with intrinsic fluorescence are highly desired in biomedicine and biotechnology as they allow the tracking and monitoring of fundamental biological processes without complicated and potentially toxic labeling.1−3 The search for alternatives that offer higher performance and ease of tunability, such as flexible, functional modulation and enhanced eco-friendliness, is still ongoing.4−6 Peptides, as a natural ingredient of biological systems, self-assembling through extensive and directed hydrogen bonding, and aromatic interactions, are intriguing candidates for this purpose, thus prompting extensive efforts to utilize these properties toward developing next-generation functional biomaterials.7−19 The most prominent example is diphenylalanine, a dipeptide initially identified as the smallest core recognition motif of β-amyloid, the amyloidogenic polypeptide associated with Alzheimer’s disease, which self-assembles into diverse nanostructures potentially useful in the biomedical field for biosensing.20−22 However, the majority of intrinsically fluorescent peptides have low quantum yields and photostability, which severely hinders their practical applications and specifically limits their potential as eco-friendly materials for optoelectronic devices and efficient bioimaging probes.23
Green fluorescent protein (GFP) has been extensively used as a genetically encoded fluorescent marker in biology. Specifically, a GFP mutant (BFPms1) that preferentially binds to Zn(II) has been developed. Zn(II) binding rigidifies the chromophore imidazole, reduces fluorophore mobility, and further limits the energy dissipation through thermal relaxation pathways, resulting in the enhanced fluorescence (Figure 1a).24 Inspired by this molecular structure, we aimed to design a metal-binding site on a short peptide to simulate metal coordination and design an electrostatic interaction site to generate supramolecular hosts of a β-fold barrel environment for self-locking, thereby fabricating the minimalist version of this sophisticated biological structure to provide a scalable technological solution (Figure 1b).14 Cyclic peptides derived from amino acid residues carrying complexing side chain substituents, such as imidazole, carboxylate, or thioether groups, can be used as models to mimic the coordination of metal ions in enzymes.25−30 Also, cyclic dipeptide is highly tunable due to hydrogen bonding capabilities of the skeleton and other noncovalent interactions that can be used to engineer artificial multifunctional scaffolds.31,32
Figure 1.
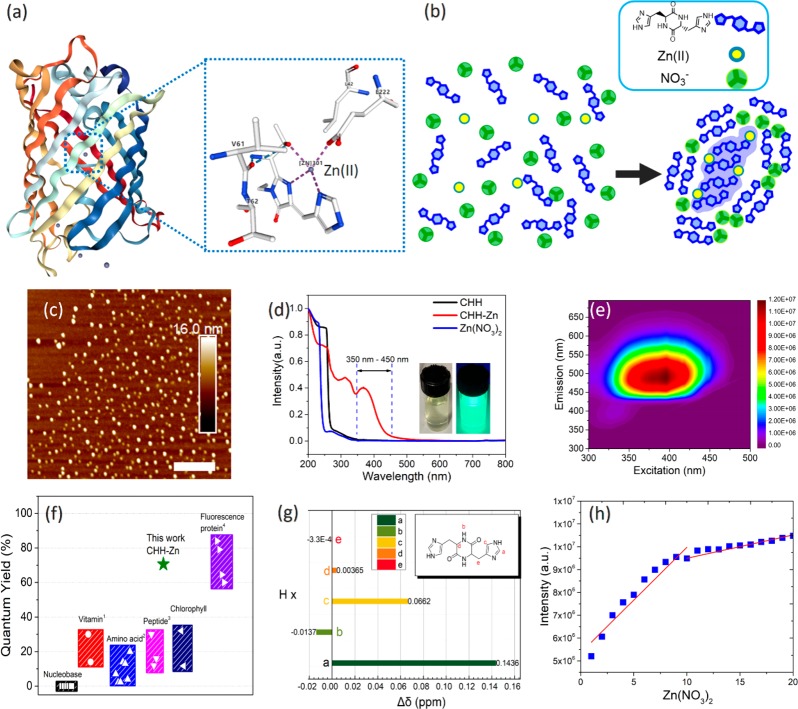
Design of self-assembled fluorescent cyclic dipeptide by the biologically inspired molecular structure of BFPms1. (a) BFPms1 overall structure and its coordination geometry with Zn(II). (b) Scheme of self-assembly mechanism of CHH, Zn(II), and NO3–. (c) AFM image showing the presence of ∼30 nm nanoparticles. Scale bar = 400 nm. (d) Normalized UV–vis spectra of CHH–Zn, CHH, and Zn(NO3)2. (Inset: CHH–Zn under daylight (left) and UV lamp (365 nm right)). (e) Excitation–emission matrix contour profiles of CHH–Zn. (f) Comparison of the quantum yield of different fluorescent biometabolites with CHH–Zn marked with a green star (vitamin, riboflavin, α-tocopherol; amino acid, Phe, Tyr, Trp, Ala, His, Lys, Ser; Peptide, Trp-Phe+Zn(II), cyclo-WW+Zn(II), Phe-Phe-Phe;20,38,39 GFP, sg11, avGFP, sapphire, H9). (g) 1H chemical shifts of CHH–Zn, compared to the peptide alone. (h) Job plot analysis of CHH with Zn(NO3)2.
Here, we explored the assembly of cyclic(l-histidine-d-histidine) (CHH) and constructed highly fluorescent peptide dots with a large quantum yield (>0.7) through “self-assembly locking strategy”. As a proof of concept, we demonstrated CHH self-assemblies to show bright fluorescence, allowing their use as an emissive layer in the photo- and electroluminescent light-emitting diodes (LEDs). Moreover, we utilized the “self-encapsulation” strategy to construct a nanocarrier to effectively deliver an anticancer drug into cancer cells with in situ monitoring. Our studies show that, analogously to diphenylalanine, CHH is a type of pioneering minimalistic self-assembling peptide, which shows that bioinspired supramolecular functional components can be applied as multifunctional nanomaterials with exceptional features for optoelectronic or biological applications.
Results and Discussion
To allow their coassembly, CHH and Zn(NO3)2 (CHH–Zn) were mixed under controlled experimental conditions, resulting in nanostructure formation. Atomic force microscopy (AFM) and transmission electron microscopy (TEM) imaging confirmed the presence of nanoparticles with an average diameter of ∼30 nm (Figures 1c and S1) which is in agreement with dynamic light scattering (DLS) data (Figure S2). Next, we explored the optical properties of the CHH–Zn nanostructures. Figure 1d shows the normalized UV–vis absorption and excitation–emission matrix contour profiles of the CHH–Zn assemblies (Figures S3 and S4). In particular, by introducing Zn ions to the cyclic dipeptide an absorption peak extending between 350 to 450 nm was observed, indicating the formation of a Zn-related coordination structure.33 Upon excitation at 390 nm, the CHH–Zn peptide nanocrystals exhibited bright fluorescence emission centered at 500 nm (inset image in Figures 1e and S5). With the excitation wavelength changes from 330 to 450 nm (Figure S6), visible photoluminescence showed a pronounced red shift from cyan to green and variation of the central peak from 490 to 520 nm along with excellent linearity of the chromaticity coordinates. Such red shift in the fluorescence emission spectra in response to a change in the excitation wavelength is termed as red edge excitation shift (REES).13,34 Notably, the maximum photoluminescence efficiency of the CHH–Zn self-assembly was about 70.6% (Figures 1f and S7), among the highest values reported so far for peptide-derived materials and even comparable to inorganic quantum dots or GFP.35−38
We further combined spectroscopic methods and control experiments to obtain specific chemical and structural information. We found that all Zn(II) present in the assembled system displayed a strong and stable fluorescence signal and similar absorption and emission spectra, whereas the sodium nitrate-related self-assembly system and CHH showed similar absorption spectra and weak fluorescence emission intensity (Figures S4 and S8). Therefore, it can be inferred that the Zn(II)-peptide coordinated structure is formed by supplying Zn(II) to the cyclic dipeptide system, which in turn determines the optical properties. NMR was further used to predict the possible coordination structure. As shown in Figure 1g and Figure S9, the imine protons of the side-chain imidazole ring downshifted Δδ = 0.143 ppm (a position) after the addition of Zn(II), implying strong coordination through imine–imidazole nitrogen.40 This coordination of the CHH imidazole ring with Zn(II) is also consistent with previous studies of carbonic anhydrase and β-amyloid peptide, where the histidine residue has a high affinity for the complexation of Zn.33,41,42 The coordination of Zn ion and imidazole ring was further verified by mass spectrometry analysis of CHH with Zn(NO3)2, showing an m/z 611.2 band corresponding to the oligomer of [2MCHH + Zn2+] (Figure S10). Moreover, a Job plot analysis determined the CHH/Zn(NO3)2 stoichiometric ratio as 2:1, which is in contrast to the 1:1 stoichiometric ratio of CHH/Zn(II) (Figure 1h and Figure S11), thus indicating the presence of another assembled structure in addition to the Zn–peptide coordination. We therefore propose a specific self-assembly mechanism mediating CHH packing with nitrate.
To validate our hypothesis and further characterize the specific self-assembly mechanism, we crystallized both CHH–Zn(II) and CHH–NaNO3 and analyzed the resulting structures via X-ray crystallography (Table S1). The CHH–Zn(II) crystallizes in orthorhombic space group Pbcn with one CHH molecule, one neutral [Zn(L)2I2] unit, and one isopropanol molecule per asymmetric unit. A perspective view of the Zn(II) center of the CHH–Zn(II) compound is illustrated in Figure 2a with a unit cell scheme. Each Zn(II) atom was coordinated with two ligands and two N-donor atoms from the imidazole groups of two different CHH molecules, occupying the apical coordination sites to generate a Zn(II) centered geometric tetrahedron. In turn, two adjacent cyclic dipeptides were connected through a β-bridgelike hydrogen bonding on the opposite sides of the backbone. The X-ray determined structure of CHH–NaNO3 revealed a packing of the cyclic-dipeptides crystal in the monoclinic space group P21/c with two CHH and four nitrates in the unit cell (Figure 2b). The components assembled to form a 1D chain with a hydrogen bond (N—H···O=C) of 2.889 Å (donor···acceptor) via a parallel β-sheet hydrogen bonding network. The adjacent chains formed the extended structure through hydrogen bonds between the imidazole ring and nitrate groups.
Figure 2.
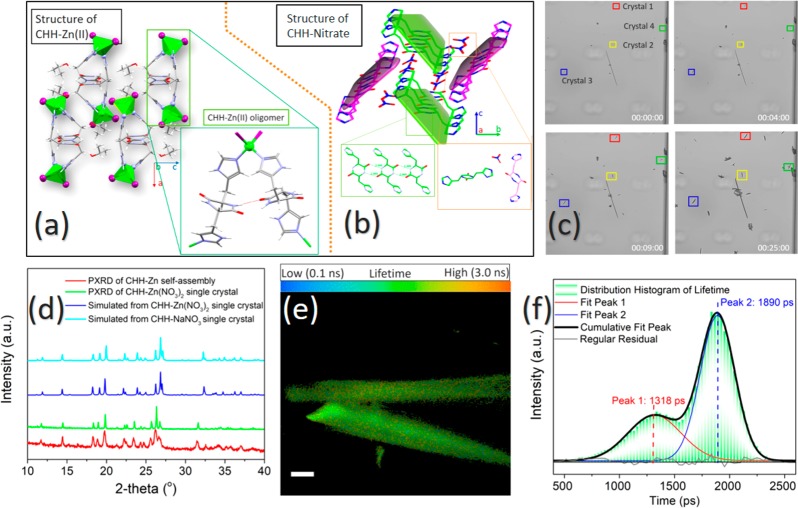
Structure analysis of the CHH–Zn assemblies. (a) Single-crystal structure of CHH–Zn(II) in Pbcn space group. Color scheme: gray, C; red, O; blue, N; green, Zn; and purple, I. (b) Single-crystal structure of CHH–NaNO3 in P21/c space group. (c) Four snapshots taken from Movie S1 demonstrating the assembly kinetics of the CHH–Zn(NO3)2 single crystal. (d) PXRD pattern of a CHH–Zn (red), the measured (green), and the simulated (blue) pattern of the single crystal structure and of CHH–Zn(NO3)2 (cyan). (e) FLIM studies of the CHH–Zn(NO3)2 single crystal. (f) Lifetime distribution histogram derived from the FLIM study, showing two peaks centered at 1318 and 1890 ps. Scale bar is 50 μm.
We further examined CHH–Zn(NO3)2 single crystals through crystallographic analysis. By employing a microfluids technique (Figure 2c, Figures S12, S13, and Movie S1), it is possible to visually observe that the Zn(NO3)2 crystals are densely packed with a growth rate of 0.01 μm s–1 along the a direction, ultimately forming a needle shape. The resulting powder X-ray diffraction (PXRD) pattern and unit cell parameters of the CHH–Zn self-assemblies highly resembled those of the formed CHH–Zn(NO3)2 crystals, indicating a similar molecular organization (Figure 2d and Figure S14). Surprisingly, the needle-shaped CHH–Zn(NO3)2 and the plaque-shaped CHH–NaNO3 crystal are isomorphous despite different crystal morphologies. Importantly, the Zn ion is not incorporated in the CHH–Zn(NO3)2 single crystal structure, suggesting that the growth of the CHH–Zn(II) oligomer may be mostly restricted by the CHH–nitrate self-assembly process. To validate our hypothesis and elucidate the effect of the CHH–Zn(II) oligomer on the CHH–Zn(NO3)2 crystal photodynamic properties, we performed confocal fluorescence lifetime microscopy (FLIM) studies of CHH–Zn(NO3)2 single crystals (Figure 2e). Figure 2f presents the lifetime distribution histogram using a multiexponential fit of the decays. Interestingly, a highly heterogeneous lifetime distribution could be observed for the CHH–Zn(NO3)2 single crystal, and the envelope of lifetime (τ) histogram could be divided into two independent fluorescence decay processes with two clearly differentiated Gaussian distributions centered at 1318 and 1890 ps. This data confirmed that the two lifetime values originated from the fluorescence decay of the two individual components, which could be assigned to the fluorescence emission within the CHH–Zn(II) oligomer and CHH–nitrate.43 In addition, CHH–Zn(NO3)2 forms yellow needle crystals, whereas CHH–NaNO3 gives rise to colorless transparent plaque crystals and the fluorescence emission intensity of CHH–Zn(NO3)2 is much higher than that of CHH–NaNO3 (Figures S15 and S16).
We computationally investigated the self-assembly of the two systems, independently comprising CHH–Zn(NO3)2 and CHH–ZnCl2, in isopropanol, using multiple explicit solvent molecular dynamics (MD) simulations in CHARMM.44 In both systems, we observed the gradual formation of dimer structures having two β-bridge-bonded CHH coordinating with one Zn(II) ion (Figure S17), similar to the X-ray crystallography data, and the gradual formation of aggregate clusters formed by the constituent elements of each system. In line with the experimental findings of the Job plot analysis (Figure 1h), mature clusters in the CHH–Zn(NO3)2 simulations showed a CHH/Zn(NO3)2 ratio of approximately 2:1, whereas mature clusters in the CHH–Zn(II) simulations displayed a CHH/ZnCl2 ratio of approximately 1:1 (Table S2). Within both sets of simulations, CHH molecules were observed to form antiparallel β-bridge conformations with the imidazole rings of two histidine molecules of opposing CHH dipeptides coordinated with the same Zn(II) ion, similar to the conformations observed in the CHH–Zn(II) crystal structure of (Figures 2a and 3a and Table S3). In the simulations comprising CHH–Zn(II) and NO3–, these antiparallel conformations were predominantly observed within the interior CHH–Zn(II) nucleus of the clusters (Figure 3d). We also observed that the radius of gyration of Zn(II) within the clusters formed in the presence of NO3– was lower compared to those formed in the absence of NO3–, showing that for clusters containing the same number of CHH molecules Zn(II) ions were more densely packed and concentrated in the clusters formed in the presence of NO3– (Figure 3b). Importantly, in the clusters formed by CHH–Zn(NO3)2 our computational analysis detected layers in which the exterior surface layer was composed of primarily CHH and NO3–, whereas the interior layer was composed primarily of CHH and Zn(II) (Figure 3c).
Figure 3.
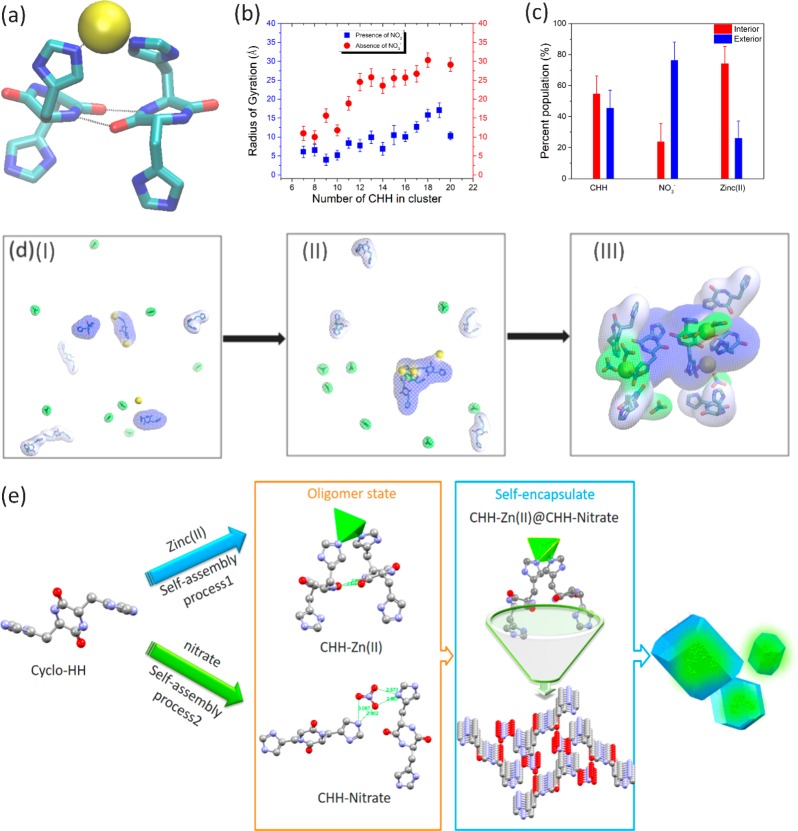
Mechanistic analysis of CHH self-assembly with Zn(NO3)2. (a) Molecular graphics image of the CHH–Zn(II) elementary structure observed in MD simulations. The CHH are shown in licorice representation. Zinc ions are shown in yellow VDW representation. Hydrogen bonds and zinc coordination are indicated with black dotted lines. (b) Radius of gyration (Å) of Zn(II) ions within the clusters observed in the simulations of CHH–Zn(II) (blue) or CHH–Zn(II) + NO3– (orange). (c) Percentage of CHH, NO3–, and Zn(II) within clusters observed in the simulations of CHH–Zn(II)–NO3–. (d) CHH–Zn(II) encased by CHH–NO3– observed in MD simulations. (I–III) Progression of CHH–Zn(II) + CHH–NO3– cluster formation observed in MD simulations. (I) Before the cluster is formed; (II) the formation of CHH–Zn(II) clusters; (III) the wrapping of CHH–Zn(II) by CHH–NO3–. The backbone atoms of the three interior CHH molecules forming antiparallel β-bridge conformations are shown in thick licorice representation. Zn(II) and NO3– are shown in licorice representation with yellow and green transparent surfaces, respectively. Interior and exterior CHH are shown with blue and light blue transparent surfaces, respectively. (e) Schematic illustration of the plausible self-assembly process of CHH–Zn assemblies.
We further aimed to provide insight into the formation of the CHH–Zn(NO3)2 clusters by performing free-energy analysis of the different pathways which may lead to their formation. On the basis of the free-energy analysis, CHH and Zn(II) coordinate first, forming the interior of the clusters, followed by individual pieces of the CHH and NO3– exterior wrapping around the preformed interior (Figures 3d and S18a). This mechanism was supported through structural analysis in which the composition of the large CHH clusters was tracked as a function of time (Figure S19a). Thus, CHH–Zn(NO3)2 self-assembles through a “self-assembly locking strategy” in which the fluorescent CHH–Zn(II) nucleus is encased by the CHH–NO3– scaffold. Figure 3d and Movie S2 show the initial formation of the internal CHH–Zn(II) nucleus, followed by the exterior CHH–NO3– wrapping around the preformed nucleus. Our combined experimental and computational analysis suggests that the CHH–Zn(II) oligomer is encapsulated into CHH–NO3– assemblies and that immobilization of the peptide oligomers by a high-stiffness scaffold would limit energy dissipation during thermal relaxation pathways for better quantum yield and fluorescence intensity.45
On the basis of our experimental and computational analyses, a plausible self-assembly mechanism of CHH and Zn(NO3)2 is depicted in Figure 3e. The self-assembly of the CHH and Zn(II) can be observed at initial oligomerization step. Following the coordination of Zn(II) with the two histidine side-chains and stabilization of the dimer, the CHH monomers begin to form hydrogen bond interactions between their backbone atoms, forming a one-dimensional chain via β-sheet bridgelike interactions and subsequently generating an extended network through the linkage of nitrates. As the CHH one-dimensional chain grows, the chelation of CHH and Zn(II) is limited. Finally, the CHH–Zn(II) oligomer clusters are encapsulated and incorporated into CHH–NO3– nanoassemblies.
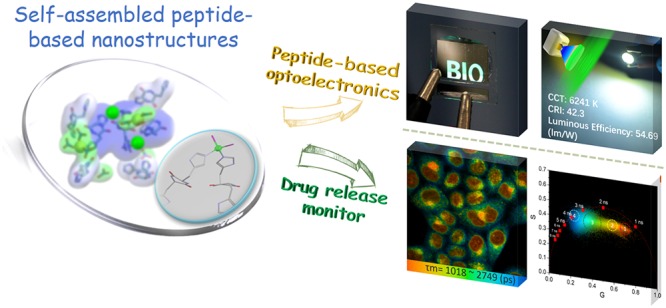
Intrigued by the optical properties of the characterized assemblies, we sought to study their capability to serve as an emissive material in photo- and electroluminescent prototypes. As shown in Figure 4a, we prepared peptide-based phosphors by embedding CHH–Zn into polyvinylpyrrolidone (PVP) at the mass ratio of 1:70. The peptide-based phosphor converted LED emitted bright green light with Commission Internationale de L’Eclairage (CIE) color coordinates of (0.31, 0.45) and achieved high luminous efficiency of 56.62 lm W–1 at 20 mA drive current (Figure S21). We further sought to study the utilization of CHH–Zn as a bio-organic light-emitting material in optoelectronics. A simple natural peptide derived bio-organic-LED (OLED) prototype was fabricated by using CHH–Zn-blended poly(N-vinyl carbazole) (PVK) as an emissive layer (Figures 4b and S22). As illustrated in Figure 4b, the operation photographs present a close-up view of the bright, uniform, and defect-free surface green electroluminescence emission from the peptide-based OLED. The typical luminance and current density curves as a function of the applied voltage for the OLED are demonstrated in Figure S23. The maximum EQE is 0.25%, corresponding to a current efficiency of 0.58 cd A–1 (the corresponding Lmax is 1385 cd m–2) (Figure S24). Because of the stable fluorescence, the bio-OLED showed no temporal degradation in the emission spectrum under the applied operating conditions, indicating significant potential for practical applications.
Figure 4.
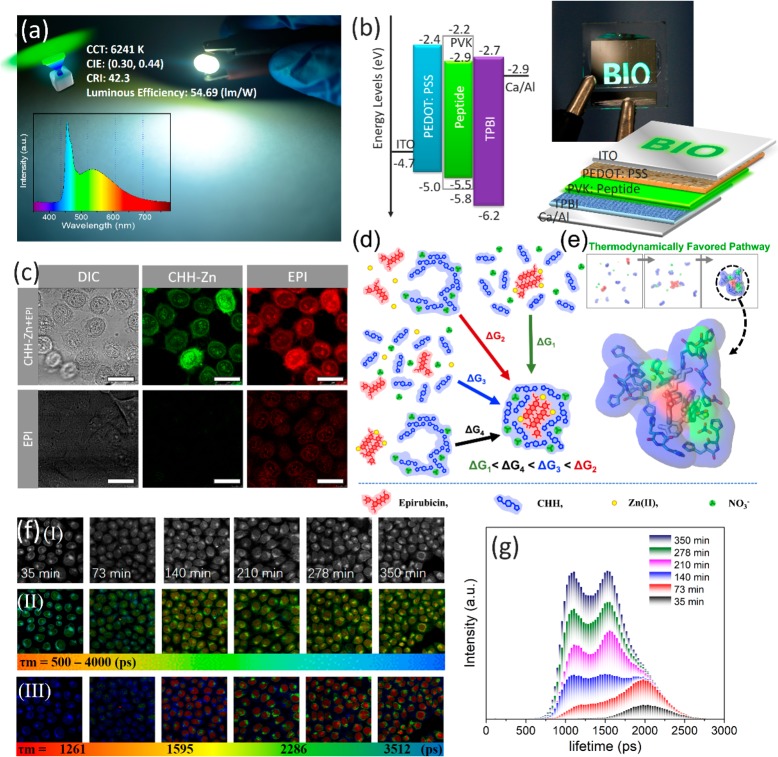
Applications of the CHH–Zn self-assemblies. (a) CHH–Zn used as a phosphor for working green LED with a luminous efficiency of 54.69 lm/W (inset: emission spectrum of working LED). (b) OLED structure, energy diagram, and operation photograph. (c) Confocal fluorescence images of HeLa cells incubated with CHH–Zn+Epirubicin (4 μg/mL) and Epirubicin (0.63 μg/mL) alone. (d) Co-assembly thermodynamic pathways of CHH–Zn+Epirubicin. (e) Epirubicin encased by CHH–Zn observed in MD simulations. (f) FLIM analysis of HeLa cells after incubation with CHH–Zn+ Epirubicin. (I) Bright-field, (II) FLIM images, and (III) phasor-separated and pseudocolored FLIM images of HeLa cells. (g) Fluorescence lifetime histogram of Epirubicin at different time points. Scale bar is 25 μm.
Because self-assembled peptide nanoparticles are composed of naturally occurring amino acids with inherent biocompatibility, peptide self-assembly with intrinsic fluorescence is suitable for bioimaging.46−48 High-resolution confocal fluorescence microscopy images of HeLa cells were collected following incubation with CHH–Zn and the DRAQ5 red DNA stain. The CHH–Zn structures were found to penetrate the cells and display bright green fluorescence under excitation of 405 nm (Figure S25). More notably, three-dimensional (3D) imaging analysis indicated that CHH–Zn could effectively transport through the nuclear pore complex of HeLa cells and accumulate within the nucleolus region (Figures S26 and S27). Also, in vitro cytotoxicity analysis shown in Figure S28 demonstrated the excellent cytocompatibility of CHH–Zn peptide nanoparticles toward HeLa cells. On the basis of the membrane permeability feature of the developed intrinsically fluorescent peptide structures, we further aimed to demonstrate their potential applications for drug delivery.49
We experimentally confirmed the coassembly of CHH–Zn and Epirubicin, an anthracycline drug used for chemotherapy, through absorbance spectra, showing a 15.67% loading capacity of Epirubicin within the CHH–Zn nanoassemblies. We computationally investigated the self-assembly properties of CHH, Zn(NO3)2 and Epirubicin in water using multiple explicit solvent runs using CHARMM.44 In all simulations, we detected the gradual formation of clusters comprising CHH, Zn(II), NO3–, and Epirubicin (Figure S19d) in which Epirubicin and Zn(II) were primarily located in the interior nucleus whereas CHH and NO3– were primarily at the exterior surface (Figures 4d, S19c, and S20), thereby depicting the self-encapsulation properties of the system. Free energy calculations (Figure S18b–d) and structural analysis (Figure S19c) show that individual or pairs of CHHs pulled Zn(II) from the solvent environment into a more peptide-like environment through an “environment-switching” mechanism.50 The Epirubicin molecules first aggregate and form the inner nucleus of the clusters which further facilitates the assembly of individual pieces of CHH and NO3– exteriorly wrapping around the preformed Epirubicin (Figure 4 panels d (ΔG1) and e, and Movie S3).
To examine the drug delivery potential of the nanoassemblies, HeLa cells incubated with CHH–Zn+Epirubicin or Epirubicin alone were examined by live-cell confocal microscopy. The fluorescence intensity of intracellular Epirubicin in cells incubated with CHH–Zn+Epirubicin was significantly higher than that of Epirubicin alone, indicating efficient Epirubicin uptake and release into the nucleolus of HeLa cells via the CHH–Zn carrier (Figure 4c). The Epirubicin release profiles (Figures S29 and S30) suggested that the release of Epirubicin from the CHH–Zn can be efficiently triggered and accelerated by an acidic stimulus, which is favored for the acidic extracellular microenvironment of tumor tissues.51,52 In order to further monitor the Epirubicin release process and eliminate autofluorescence from the biological system, we applied the two-photon FLIM technique with phasor analysis.53,54 Pixels with similar lifetimes are selected in the phasor diagram and the FLIM image is separated and painted into four subcellular compartments: cell membrane (∼3512 ps), cytoplasm (∼2286 ps), nucleus membrane (∼1595 ps), and nucleus (∼1261 ps) (Figure S31). After internalizing of Epirubicin into the cells, changes in its fluorescence lifetime can thus indicate changes in the subcellular microenvironment, reflecting drug release and transport. With elongation of incubation time (Figure 4f,g), more Epirubicin was released and, consequently, the fluorescence intensity of Epirubicin gradually increased, along with a decrease in the average lifetime. These results indicated that the CHH–Zn+Epirubicin could accumulate around and bind to the cell membrane as early as 35 min of incubation with HeLa cells and then be released in the cytoplasm due to the acidic environment and eventually accumulate in the nucleus. In addition, the release behavior of CHH–Zn+Epirubicin could be monitored by the variation of the fluorescent signal of CHH–Zn (Figure S32), showing that CHH–Zn not only promoted the transport of Epirubicin into HeLa cells but also can be acted as a real-time optical monitor for the drug release process. Thus, the fluorescence of peptide nanostructures can be used to investigate the drug release in spatiotemporal mode and metabolism kinetics of cancer drugs in a certain organ or tissue.
Conclusion
Within this study, inspired by the structure of BFPms1, we successfully constructed a fluorescent short peptide core encapsulated by the peptide scaffold building module to implement the concept of “self-assembly locking strategy”. We report the demonstration of a bright fluorescent peptide with quantum yields of up to 70% for green fluorescence, exemplifying the potential of such structures to serve as bioinspired, organic, supramolecular alternatives to complement the state-of-the-art inorganic counterparts. As a proof of concept, we demonstrate the utilization of these bright fluorescence peptide self-assemblies for eco-friendly optoelectronics and bioimaging. In particular, we show their capability to serve as an emissive layer for a bioinspired OLED prototype. Moreover, we employ the “self-encapsulation” strategy for fabricating an advanced nanocarrier for traceable intracellular drug delivery. These results allow us to envision CHH as a promising platform for further therapeutic or diagnostic applications, particularly in anticancer treatments. This efficient yet straightforward bottom-up approach for the design of highly efficient fluorescent peptide nanostructures may represent a strategy for developing peptide-based advanced nanomaterials.
Methods
Atomic Force Microscopy
Five microliters of sample solution was dropped onto a freshly cleaved mica surface and dried by N2 purge (99.99%). A topographic image was recorded under a Dimension icon AFM (Bruker) in the tapping mode at ambient temperature with a 512 × 512-pixel resolution and a scanning speed of 1.0 Hz.
UV–vis Spectra
UV–vis spectra between 200 to 800 nm were recorded on an Agilent Cary 100 UV–vis spectrophotometer with a quartz cuvette of 1 mm path length.
Fluorescence Spectroscopy and Quantum Yield (QY) Measurement
Six hundred microliters sample solution was pipetted into a 1.0 cm path-length quartz cuvette, and the spectrum was collected using a FluoroMax-4 Spectrofluorometer (Horiba Jobin Yvon, Kyoto, Japan) at ambient temperature. The excitation and emission wavelengths were set at 300–500 nm and 400–650 nm, respectively, with a slit of 2 nm. Absolute fluorescence QY measurements were performed using Quanta-Phi integrating sphere connected to Fluoromex-4.
Microfluidics Experiments
In a typical protocol, CHH–Zn(NO3)2 crystalline powder was inserted into the device. Then, a flow of fresh solutions was injected at a rate of 4 μL h–1 using Cetoni GmbH neMESYS Syringe Pumps (Korbussen, Germany) and glass HAMILTON syringes, 1,725 TLL of 250 μL. The process was examined under an Eclipse Ti-E inverted microscope (Nikon, Japan), equipped with a Zyla 4.2+ sCMOS camera (Andor, U.K.), and images were captured at different time points.
X-ray Crystallography
Crystals suitable for diffraction were coated with Paratone oil (Hampton Research), mounted on loops and flash frozen in liquid nitrogen. Single crystal X-ray diffraction data measurement was performed using a Rigaku XtaLabPro system with CuKα1 (λ = 1.5418 Å) radiation at 100(2) K. Data were collected and processed using CrysAlisPro 1.171.39.22a (Rigaku OD, 2015). The structure was solved by direct methods using SHELXT-2016/4 and refined by full-matrix least-squares against F2 with SHELXL-2013.
Photoluminescence Device Fabrication and Characterization
Commercially available InGaN chips were used at the bottom of the light-emitting diode (LED) base. For preparation of the color conversion layer, the CHH–Zn was blended into PVP at a mass ratio of 1:70, and the resulting mixtures were vacuum-dried at 60 °C for 30 min. The mixtures were applied on the InGaN chips and, following curing at 80 °C for 1 h, the LEDs peptide phosphors were obtained.
Organic LED Device Fabrication and Characterization
ITO-coated glass substrates were cleaned ultrasonically in organic solvents (acetone and isopropyl alcohol), rinsed in deionized water, and then dried in an oven at 150 °C for 10 min. The substrates were cleaned by a UV-ozone treatment to enrich the ITO surface with oxygen, thereby increasing its work function. The approximately 30 nm thick PEDOT:PSS hole injection layer was spin-coated at 3000 rpm for 30 s on the ITO, followed by annealing in an oven at 150 °C for 15 min. Subsequently, the emissive layer of CHH–Zn blended into PVK was spin-coated at 3000 rpm for 35 s over the surface of the PEDOT:PSS film from the solution of NMP, followed by baking on a hot plate at 80 °C for 15 min to form the active region of the peptide-derived bio-OLED. Finally, the substrates were transferred to a vacuum chamber, and a 30 nm thick TPBI electron transport layer was thermally deposited with base pressure of 3 × 10–4 Pa. Next, a 20 nm Ca and 100 nm thick Al cathode was deposited using a shadow mask 2 mm in width. The active area of the devices was thus 4 mm2. The thermal deposition rates for TPBI and Ca/Al were 1, 1, and 3 Å s–1, respectively. The thickness of the films was measured using a Dektak XT (Bruker) surface profilometer and a spectroscopic ellipsometer (Suntech). The luminance–current–voltage (L–I–V) characteristics were measured using a computer-controlled Keithley 236 SMU and Keithley 200 multimeter coupled with a calibrated Si photodiode. Electroluminescence spectra were measured by an Ocean Optics 2000 spectrometer, which couples a linear charge-coupled device array detector ranging from 350 to 800 nm.
Live Cells Imaging Using Confocal Microscopy
HeLa cells were grown to 70–80% confluence in glass bottom cell culture dishes. Then, the cells were cultured with media containing the CHH–Zn+EPI at a concentration of 4 μg/mL for different durations. Next, the cells were stained using a DRAQ5 dye diluted 1:1000 in PBS for 15 min at room temperature in the dark to allow staining of the nuclei. The cells were then washed twice with PBS. Imaging was performed using SP8 inverted confocal microscope (Leica Microsystems, Wetzlar, Germany). Excitation and emission ranges were the following: λex = 405 nm, λem = 420–500 nm; EPI, λex = 543 nm, λem = 550–750 nm; DRAQ5, λex = 633 nm, λem = 750–780 nm.
FLIM Analysis of Cultured Cells
HeLa cells seeded in dishes were treated with CHH–Zn+EPI at a concentration of 4 μg mL–1 for 30, 76, 125, 194, 270, and 420 min, followed by washing with PBS. The time-resolved fluorescence signal was acquired using an LSM 7 MP two-photon microscope (Carl Zeiss, Weimar, Germany) coupled to the Becker and Hickl (BH) simple-Tau-152 system. Images were acquired through a Zeiss 20×/1 NA water-immersion objective. A Zeiss dichroic mirror (T690) was used to separate the excitation and the emission light. An additional barrier filter was used to block emission light above 690 nm. Emission light was separated by a dichroic mirror (555 nm) and the two fluorescent lights were filtered by two band-pass filters (500–550 and 590–650 nm). Pseudocolored lifetime images were generated by assigning a color to the value of average fluorescence lifetime τm at each pixel. Emission light was collected via a hybrid GaAsP detector (HPM-100-40, BH, Berlin, Germany) with a Cherry bandpass filter.
Acknowledgments
This work was supported in part by the European Research Council under the European Union Horizon 2020 research and innovation program (No. 694426) (E.G.) and Huawei Technologies Co., Ltd. (E.G.). Y.C. gratefully acknowledges the Center for Nanoscience and Nanotechnology of Tel Aviv University for financial support. The authors acknowledge Texas A&M University Graduate Diversity Fellowship from the TAMU Office of Graduate and Professional Studies (A.A.O). All MD simulations and free energy calculations were conducted using the Ada supercomputing cluster and additional computational resources available to P.T. at the Texas A&M High Performance Research Computing Facility and the Chemical Engineering Department. The authors thank T. Kreiser for in vitro experiments, L. Meng and Prof. S. N. Qu for the fabrication and characterization of LEDs, and the members of the Tan, Tamamis, and Gazit laboratories for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.9b10024.
Additional experimental details, including supplementary experimental methods, TEM image, DLS result, UV–vis spectra, excitation–emission matrix contour profiles, photostability evaluation, NMR result, mass spectra, crystal morphology, XRD pattern, fluorescence microscopy images; MD simulations result: (molecular graphics image of the coordination-induced self-assembly of CHH with Zn(II), free energy of associate the clusters, time evolution percent composition of clusters); characterization of LED and OLED; confocal microscopy images, cytotoxicity result, in vitro drug release profiles, Phasor–FLIM analysis of HeLa cells cultured with CHH–Zn+EPI (PDF)
Growth of CHH–Zn(NO3)2 single crystal by microfluidic (MP4)
MD simulation of CHH–Zn(II)-NO3 (MOV)
MD simulation of CHH–Zn(II)-NO3+EPI (MOV)
Crystallographic data (CIF)
Crystallographic data (CIF)
Crystallographic data (CIF)
Author Contributions
Y.C., A.A.O., and K.T. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Gazit E. Aromatic Dipeptides Light Up. Nat. Nanotechnol. 2016, 11, 309. 10.1038/nnano.2015.321. [DOI] [PubMed] [Google Scholar]
- Hilaire M. R.; Ahmed I. A.; Lin C.-W.; Jo H.; DeGrado W. F.; Gai F. Blue Fluorescent Amino Acid for Biological Spectroscopy and Microscopy. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 6005. 10.1073/pnas.1705586114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansieri J.; Josserand V.; Lee S.-J.; Rongier A.; Imbert D.; Sallanon M. M.; Kövari E.; Dane T. G.; Vendrely C.; Chaix-Pluchery O.; Guidetti M.; Vollaire J.; Fertin A.; Usson Y.; Rannou P.; Coll J.-L.; Marquette C.; Forge V. Ultraviolet–Visible–Near-Infrared Optical Properties of Amyloid Fibrils Shed Light on Amyloidogenesis. Nat. Photonics 2019, 13, 473–479. 10.1038/s41566-019-0422-6. [DOI] [Google Scholar]
- Gu L.; Shi H.; Bian L.; Gu M.; Ling K.; Wang X.; Ma H.; Cai S.; Ning W.; Fu L.; Wang H.; Wang S.; Gao Y.; Yao W.; Huo F.; Tao Y.; An Z.; Liu X.; Huang W. Colour-Tunable Ultra-Long Organic Phosphorescence of a Single-Component Molecular Crystal. Nat. Photonics 2019, 13, 406–411. 10.1038/s41566-019-0408-4. [DOI] [Google Scholar]
- Kim D.; Yoo J. M.; Hwang H.; Lee J.; Lee S. H.; Yun S. P.; Park M. J.; Lee M.; Choi S.; Kwon S. H.; Lee S.; Kwon S.-H.; Kim S.; Park Y. J.; Kinoshita M.; Lee Y.-H.; Shin S.; Paik S. R.; Lee S. J.; Lee S.; et al. Graphene Quantum Dots Prevent α-Synucleinopathy in Parkinson’s Disease. Nat. Nanotechnol. 2018, 13, 812–818. 10.1038/s41565-018-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia-Vladu M.; Głowacki E. D.; Troshin P. A.; Schwabegger G.; Leonat L.; Susarova D. K.; Krystal O.; Ullah M.; Kanbur Y.; Bodea M. A.; Razumov V. F.; Sitter H.; Bauer S.; Sariciftci N. S. Indigo - A Natural Pigment for High Performance Ambipolar Organic Field Effect Transistors and Circuits. Adv. Mater. 2012, 24, 375–380. 10.1002/adma.201102619. [DOI] [PubMed] [Google Scholar]
- Hauser C. A. E.; Zhang S. Peptides as Biological Semiconductors. Nature 2010, 468, 516. 10.1038/468516a. [DOI] [PubMed] [Google Scholar]
- Lee H.-E.; Ahn H.-Y.; Mun J.; Lee Y. Y.; Kim M.; Cho N. H.; Chang K.; Kim W. S.; Rho J.; Nam K. T. Amino-Acid- and Peptide-Directed Synthesis of Chiral Plasmonic Gold Nanoparticles. Nature 2018, 556, 360–365. 10.1038/s41586-018-0034-1. [DOI] [PubMed] [Google Scholar]
- Fleming S.; Ulijn R. V. Design of Nanostructures Based on Aromatic Peptide Amphiphiles. Chem. Soc. Rev. 2014, 43, 8150–8177. 10.1039/C4CS00247D. [DOI] [PubMed] [Google Scholar]
- Stupp S. I.; Palmer L. C. Supramolecular Chemistry and Self-Assembly in Organic Materials Design. Chem. Mater. 2014, 26, 507–518. 10.1021/cm403028b. [DOI] [Google Scholar]
- Yan X.; Zhu P.; Li J. Self-Assembly and Application of Diphenylalanine-Based Nanostructures. Chem. Soc. Rev. 2010, 39, 1877–1890. 10.1039/b915765b. [DOI] [PubMed] [Google Scholar]
- Tao K.; Makam P.; Aizen R.; Gazit E. Self-Assembling Peptide Semiconductors. Science 2017, 358, eaam9756 10.1126/science.aam9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger O.; Adler-Abramovich L.; Levy-Sakin M.; Grunwald A.; Liebes-Peer Y.; Bachar M.; Buzhansky L.; Mossou E.; Forsyth V. T.; Schwartz T.; Ebenstein Y.; Frolow F.; Shimon L. J. W.; Patolsky F.; Gazit E. Light-Emitting Self-Assembled Peptide Nucleic Acids Exhibit Both Stacking Interactions and Watson–Crick Base Pairing. Nat. Nanotechnol. 2015, 10, 353. 10.1038/nnano.2015.27. [DOI] [PubMed] [Google Scholar]
- Bolisetty S.; Mezzenga R. Amyloid-Carbon Hybrid Membranes for Universal Water Purification. Nat. Nanotechnol. 2016, 11, 365–371. 10.1038/nnano.2015.310. [DOI] [PubMed] [Google Scholar]
- Lampel A.; McPhee S. A.; Park H.-A.; Scott G. G.; Humagain S.; Hekstra D. R.; Yoo B.; Frederix P. W. J. M.; Li T.-D.; Abzalimov R. R.; Greenbaum S. G.; Tuttle T.; Hu C.; Bettinger C. J.; Ulijn R. V. Polymeric Peptide Pigments with Sequence-Encoded Properties. Science 2017, 356, 1064. 10.1126/science.aal5005. [DOI] [PubMed] [Google Scholar]
- Jia Y.; Li J. Reconstitution of FoF1-ATPase-Based Biomimetic Systems. Nat. Rev. Chem. 2019, 3, 361–374. 10.1038/s41570-019-0100-8. [DOI] [Google Scholar]
- Sun B.; Tao K.; Jia Y.; Yan X.; Zou Q.; Gazit E.; Li J. Photoactive Properties of Supramolecular Assembled Short Peptides. Chem. Soc. Rev. 2019, 48, 4387–4400. 10.1039/C9CS00085B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan M.; Shao J.; Li J. Cell Membrane-Covered Nanoparticles as Biomaterials. Natl. Sci. Rev. 2019, 6, 551–561. 10.1093/nsr/nwz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Fei J.; Xu Y.; Li D.; Yuan T.; Li G.; Wang C.; Li J. A Photoinduced Reversible Phase Transition in a Dipeptide Supramolecular Assembly. Angew. Chem. 2018, 130, 1921–1925. 10.1002/ange.201711547. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Sun L.; Huang Y.; Wang Y.; Zhang M. Bioinspired Fluorescent Dipeptide Nanoparticles for Targeted Cancer Cell Imaging and Real-Time Monitoring of Drug Release. Nat. Nanotechnol. 2016, 11, 388. 10.1038/nnano.2015.312. [DOI] [PubMed] [Google Scholar]
- Reches M.; Gazit E. Casting Metal Nanowires within Discrete Self-Assembled Peptide Nanotubes. Science 2003, 300, 625. 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- Tamamis P.; Adler-Abramovich L.; Reches M.; Marshall K.; Sikorski P.; Serpell L.; Gazit E.; Archontis G. Self-Assembly of Phenylalanine Oligopeptides: Insights from Experiments and Simulations. Biophys. J. 2009, 96, 5020–5029. 10.1016/j.bpj.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y.; Meng L.; Chen S.; Leung C. W. T.; Da L.-T.; Faisal M.; Silva D.-A.; Liu J.; Lam J. W. Y.; Huang X.; Tang B. Z. Monitoring and Inhibition of Insulin Fibrillation by a Small Organic Fluorogen with Aggregation-Induced Emission Characteristics. J. Am. Chem. Soc. 2012, 134, 1680–1689. 10.1021/ja208720a. [DOI] [PubMed] [Google Scholar]
- Barondeau D. P.; Kassmann C. J.; Tainer J. A.; Getzoff E. D. Structural Chemistry of a Green Fluorescent Protein Zn Biosensor. J. Am. Chem. Soc. 2002, 124, 3522–3524. 10.1021/ja0176954. [DOI] [PubMed] [Google Scholar]
- Ma X.; Jia J.; Cao R.; Wang X.; Fei H. Histidine-Iridium(III) Coordination-Based Peptide Luminogenic Cyclization and Cyclo-RGD Peptides for Cancer-Cell Targeting. J. Am. Chem. Soc. 2014, 136, 17734–17737. 10.1021/ja511656q. [DOI] [PubMed] [Google Scholar]
- Clark T. D.; Buehler L. K.; Ghadiri M. R. Self-Assembling Cyclic β3-Peptide Nanotubes as Artificial Transmembrane Ion Channels. J. Am. Chem. Soc. 1998, 120, 651–656. 10.1021/ja972786f. [DOI] [Google Scholar]
- Bellezza I.; Peirce M. J.; Minelli A. Cyclic Dipeptides: From Bugs to Brain. Trends Mol. Med. 2014, 20, 551–558. 10.1016/j.molmed.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Anderson S. L.; Stylianou K. C. Biologically Derived Metal Organic Frameworks. Coord. Chem. Rev. 2017, 349, 102–128. 10.1016/j.ccr.2017.07.012. [DOI] [Google Scholar]
- Zou R.; Wang Q.; Wu J.; Wu J.; Schmuck C.; Tian H. Peptide Self-Assembly Triggered by Metal Ions. Chem. Soc. Rev. 2015, 44, 5200–5219. 10.1039/C5CS00234F. [DOI] [PubMed] [Google Scholar]
- Mannini B.; Habchi J.; Chia S.; Ruggeri F. S.; Perni M.; Knowles T. P. J.; Dobson C. M.; Vendruscolo M. Stabilization and Characterization of Cytotoxic Aβ40 Oligomers Isolated from an Aggregation Reaction in the Presence of Zinc Ions. ACS Chem. Neurosci. 2018, 9, 2959–2971. 10.1021/acschemneuro.8b00141. [DOI] [PubMed] [Google Scholar]
- Montenegro J.; Ghadiri M. R.; Granja J. R. Ion Channel Models Based on Self-Assembling Cyclic Peptide Nanotubes. Acc. Chem. Res. 2013, 46, 2955–2965. 10.1021/ar400061d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantion A.; Massüger L.; Rabu P.; Palivan C.; McCusker L. B.; Taubert A. Metal-Peptide Frameworks (MPFs): ″Bioinspired″ Metal Organic Frameworks. J. Am. Chem. Soc. 2008, 130, 2517–2526. 10.1021/ja0762588. [DOI] [PubMed] [Google Scholar]
- Lee M.; Wang T.; Makhlynets O. V.; Wu Y.; Polizzi N. F.; Wu H.; Gosavi P. M.; Stöhr J.; Korendovych I. V.; DeGrado W. F.; Hong M. Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 6191. 10.1073/pnas.1706179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko A. P. The Red-Edge Effects: 30 Years of Exploration. Luminescence 2002, 17, 19–42. 10.1002/bio.671. [DOI] [PubMed] [Google Scholar]
- Amdursky N.; Molotskii M.; Aronov D.; Adler-Abramovich L.; Gazit E.; Rosenman G. Blue Luminescence Based on Quantum Confinement at Peptide Nanotubes. Nano Lett. 2009, 9, 3111–3115. 10.1021/nl9008265. [DOI] [PubMed] [Google Scholar]
- Yuan F.; Wang Z.; Li X.; Li Y.; Tan Z. a.; Fan L.; Yang S. Bright Multicolor Bandgap Fluorescent Carbon Quantum Dots for Electroluminescent Light-Emitting Diodes. Adv. Mater. 2017, 29, 1604436. 10.1002/adma.201604436. [DOI] [PubMed] [Google Scholar]
- Taniguchi M.; Lindsey J. S. Database of Absorption and Fluorescence Spectra of > 300 Common Compounds for Use in PhotochemCAD. Photochem. Photobiol. 2018, 94, 290–327. 10.1111/php.12860. [DOI] [PubMed] [Google Scholar]
- Lapshina N.; Shishkin I. I.; Nandi R.; Noskov R. E.; Barhom H.; Joseph S.; Apter B.; Ellenbogen T.; Natan A.; Ginzburg P.; Amdursky N.; Rosenman G. Bioinspired Amyloid Nanodots with Visible Fluorescence. Adv. Opt. Mater. 2019, 7, 1801400. 10.1002/adom.201801400. [DOI] [Google Scholar]
- Tao K.; Fan Z.; Sun L.; Makam P.; Tian Z.; Ruegsegger M.; Shaham-Niv S.; Hansford D.; Aizen R.; Pan Z.; Galster S.; Ma J.; Yuan F.; Si M.; Qu S.; Zhang M.; Gazit E.; Li J. Quantum Confined Peptide Assemblies with Tunable Visible to Near-Infrared Spectral Range. Nat. Commun. 2018, 9, 3217. 10.1038/s41467-018-05568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoroddu M. A.; Medici S.; Peana M.; Anedda R. NMR Studies of Zinc Binding in a Multi-Histidinic Peptide Fragment. Dalton Trans 2010, 39, 1282–1294. 10.1039/B914296G. [DOI] [PubMed] [Google Scholar]
- Dong J.; Shokes J. E.; Scott R. A.; Lynn D. G. Modulating Amyloid Self-Assembly and Fibril Morphology with Zn(II). J. Am. Chem. Soc. 2006, 128, 3540–3542. 10.1021/ja055973j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair N. G.; Perry G.; Smith M. A.; Reddy V. P. NMR Studies of Zinc, Copper, and Iron Binding to Histidine, the Principal Metal Ion Complexing Site of Amyloid-β Peptide. J. Alzheimer's Dis. 2010, 20, 57–66. 10.3233/JAD-2010-1346. [DOI] [PubMed] [Google Scholar]
- Hosny N. A.; Mohamedi G.; Rademeyer P.; Owen J.; Wu Y.; Tang M.-X.; Eckersley R. J.; Stride E.; Kuimova M. K. Mapping Microbubble Viscosity Using Fluorescence Lifetime Imaging of Molecular Rotors. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 9225. 10.1073/pnas.1301479110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Brooks C. L. 3rd; Mackerell A. D. Jr.; Nilsson L.; Petrella R. J.; Roux B.; Won Y.; Archontis G.; Bartels C.; Boresch S.; Caflisch A.; Caves L.; Cui Q.; Dinner A. R.; Feig M.; Fischer S.; Gao J.; Hodoscek M.; Im W.; Kuczera K.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. E.; Dolgopolova E. A.; Pellechia P. J.; Palukoshka A.; Wilson T. J.; Tan R.; Maier J. M.; Greytak A. B.; Smith M. D.; Krause J. A.; Shustova N. B. Mimic of the Green Fluorescent Protein β-Barrel: Photophysics and Dynamics of Confined Chromophores Defined by a Rigid Porous Scaffold. J. Am. Chem. Soc. 2015, 137, 2223–2226. 10.1021/ja5131269. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zou Q.; Yuan C.; Li S.; Xing R.; Yan X. Amino Acid Coordination Driven Self-Assembly for Enhancing Both the Biological Stability and Tumor Accumulation of Curcumin. Angew. Chem., Int. Ed. 2018, 57, 17084–17088. 10.1002/anie.201810087. [DOI] [PubMed] [Google Scholar]
- Sun S.; Zhang L.; Jiang K.; Wu A.; Lin H. Toward High-Efficient Red Emissive Carbon Dots: Facile Preparation, Unique Properties, and Applications as Multifunctional Theranostic Agents. Chem. Mater. 2016, 28, 8659–8668. 10.1021/acs.chemmater.6b03695. [DOI] [Google Scholar]
- He H.; Wang J.; Wang H.; Zhou N.; Yang D.; Green D. R.; Xu B. Enzymatic Cleavage of Branched Peptides for Targeting Mitochondria. J. Am. Chem. Soc. 2018, 140, 1215–1218. 10.1021/jacs.7b11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard A. J.; Bierbach U. The Cell’s Nucleolus: An Emerging Target for Chemotherapeutic Intervention. ChemMedChem 2013, 8, 1441–1449. 10.1002/cmdc.201300262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K.; Chen Y.; Orr A. A.; Tian Z.; Makam P.; Gilead S.; Si M.; Rencus-Lazar S.; Qu S.; Zhang M.; Tamamis P.; Gazit E. Enhanced Fluorescence for Bioassembly by Environment-Switching Doping of Metal Ions. Adv. Funct. Mater. 2020, 1909614. 10.1002/adfm.201909614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Huang Y.; Kumar A.; Tan A.; Jin S.; Mozhi A.; Liang X.-J. pH-Sensitive Nano-Systems for Drug Delivery in Cancer Therapy. Biotechnol. Adv. 2014, 32, 693–710. 10.1016/j.biotechadv.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Clegg J. R.; Irani A. S.; Ander E. W.; Ludolph C. M.; Venkataraman A. K.; Zhong J. X.; Peppas N. A. Synthetic Networks with Tunable Responsiveness, Biodegradation, and Molecular Recognition for Precision Medicine Applications. Sci. Adv. 2019, 5, eaax7946 10.1126/sciadv.aax7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basuki J. S.; Duong H. T. T.; Macmillan A.; Erlich R. B.; Esser L.; Akerfeldt M. C.; Whan R. M.; Kavallaris M.; Boyer C.; Davis T. P. Using Fluorescence Lifetime Imaging Microscopy to Monitor Theranostic Nanoparticle Uptake and Intracellular Doxorubicin Release. ACS Nano 2013, 7, 10175–10189. 10.1021/nn404407g. [DOI] [PubMed] [Google Scholar]
- Zhou T.; Luo T.; Song J.; Qu J. Phasor–Fluorescence Lifetime Imaging Microscopy Analysis to Monitor Intercellular Drug Release from a pH-Sensitive Polymeric Nanocarrier. Anal. Chem. 2018, 90, 2170–2177. 10.1021/acs.analchem.7b04511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.