Key Points
Zoonotic viruses pose a serious threat to human and animal health. Studying the immune response to zoonotic pathogens in the natural reservoir hosts, rather than traditional animal models, offers important insights into control strategies.
Comparative studies in natural host systems have provided key information and improved our understanding of co-evolution of hosts and pathogens. This could lead to the discovery of novel immune mechanisms that control viral replication.
Understanding the differences between the immune systems of domesticated and wild animal hosts and comparing them to the human immune system is crucial for unravelling the complex disease mechanisms involved in zoonotic infections and for developing new strategies for disrupting their transmission to humans.
The use of non-traditional animal models for research poses many challenges. These include the need for specialist high-biosecurity containment facilities, a lack of species-specific reagents for immunology studies, and complex husbandry, ethics and welfare issues.
Whole-genome sequencing and comparative analysis of host species have provided key insights into how different immune responses are made to the same pathogen.
The identification of key differences in immune pathways between susceptible and non-susceptible hosts might offer clues for developing disease intervention strategies, including new antiviral vaccines and therapies, and disease-resistant animals.
Supplementary information
The online version of this article (doi:10.1038/nri3551) contains supplementary material, which is available to authorized users.
Subject terms: Infectious diseases, Viral infection, Evolutionary genetics
Immunology is traditionally viewed as a science of 'mice and men'. However, key insights can come from the study of immune responses in livestock or wild animals. The fact that the most deadly pathogens of humans are often zoonotic in nature lends further weight to the importance of this research. The authors discuss the benefits of, and challenges posed by, these studies.
Supplementary information
The online version of this article (doi:10.1038/nri3551) contains supplementary material, which is available to authorized users.
Abstract
Zoonotic viruses that emerge from wildlife and domesticated animals pose a serious threat to human and animal health. In many instances, mouse models have improved our understanding of the human immune response to infection; however, when dealing with emerging zoonotic diseases, they may be of limited use. This is particularly the case when the model fails to reproduce the disease status that is seen in the natural reservoir, transmission species or human host. In this Review, we discuss how researchers are placing more emphasis on the study of the immune response to zoonotic infections in the natural reservoir hosts and spillover species. Such studies will not only lead to a greater understanding of how these infections induce variable disease and immune responses in distinct species but also offer important insights into the evolution of mammalian immune systems.
Supplementary information
The online version of this article (doi:10.1038/nri3551) contains supplementary material, which is available to authorized users.
Main
Now, more than ever, there is a greater risk of emerging infectious diseases (EIDs) being transmitted to humans from wild and domesticated animals. Several factors have contributed to this, including the recent growth and geographic expansion of human populations, the intensification of agriculture, and the disruption of habitats owing to climate change and deforestation1,2. Moreover, increased global travel means that there is a greater likelihood that EIDs will rapidly spread. Over the past three decades the incidence of EIDs has risen in humans, with around 70% being zoonotic in nature and most being caused by viruses and drug-resistant pathogens1,2 (Box 1; Fig. 1). Containment of these EID outbreaks has often been difficult owing to their unpredictability and the absence of effective control measures, such as vaccines and antiviral therapeutics. In addition, there is a lack of essential knowledge of the immune responses that are induced by zoonotic viruses, particularly of the immune responses that provide protection.
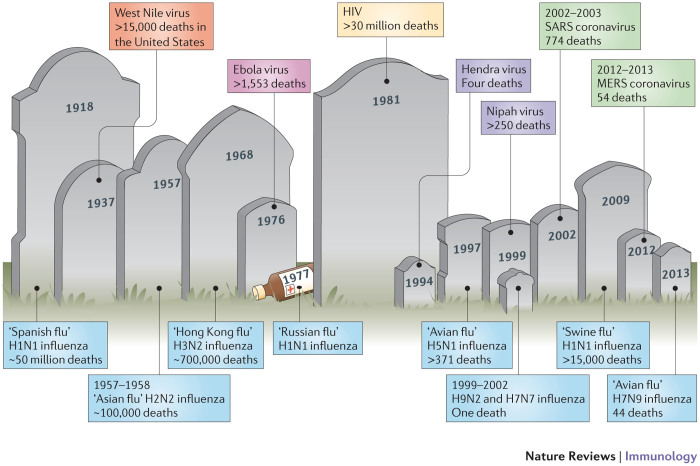
Figure 1. Emergence of zoonoses.
Over the past century, humanity has witnessed the emergence of numerous zoonotic infections that have resulted in varying numbers of human fatalities. Influenza viruses that originate from birds account for an important proportion of these deaths, and recently many new zoonotic viruses that originate in bats, such as Hendra virus, Nipah virus and severe acute respiratory syndrome (SARS) coronavirus, have caused outbreaks with high mortality rates. Hyperlinks to World Health Organization disease report updates are provided in Box 1. MERS, Middle East respiratory syndrome coronavirus.
Mouse models have been fundamental to our understanding of immune responses to infection and disease outcomes. Indeed, mice have become the traditional 'workhorse' for studying immunology because of their ease of handling, fast generation time and the ready availability of mouse-specific reagents3. However, for a better understanding of EIDs, the laboratory mouse may not be the most appropriate model. There are often many differences in the symptoms of disease between the natural, transmission and human hosts. Zoonotic infections often appear as asymptomatic and non-lethal in the natural reservoir host, but induce severe and potentially lethal disease in humans or other spillover hosts. Nevertheless, there are numerous factors that are likely to contribute to these differences, including anatomical, physiological, metabolic and behavioural traits, as well as how the immune systems of these hosts interact with the same disease agent.
In this Review, we discuss the importance of undertaking immunological research in natural reservoir and spillover host species in order to gain a better understanding of EIDs, with a focus on viral diseases. Increasingly, non-laboratory animals, such as bats, chickens and ferrets, are being used for the study of host–pathogen relationships and immune responses to EIDs. We discuss the benefits and the limitations of studying immunology in non-traditional animal species and also describe how a new approach, based on the One-Health initiative (Box 2), might provide new insights into strategies for controlling EIDs and might assist in preparing us for future pandemics.
Box 1: The impact of emerging infectious diseases.
The World Health Organization (WHO) has warned that the source of the next human pandemic is likely to be zoonotic and that wildlife is a prime culprit (see Zoonoses on the WHO website; Fig. 2).
Although the current list of known emerging infectious diseases (EIDs) is a major concern, it is the unknown EIDs with the potential for efficient human–human transmission that might pose the greatest threat. Over the past decade there have been several epidemics, raising the concern that they are precursors to a pandemic.
The severe acute respiratory syndrome (SARS) epidemic in 2003–2004 claimed more than 800 lives and cost the global economy over US$80 billion. It was shown to have involved virus transmission from bats to civet cats to humans (see severe acute respiratory syndrome (SARS) on the WHO website).
In 2012, a novel coronavirus emerged in the Middle East (Middle East respiratory syndrome coronavirus (MERS-CoV))64 with a 45% mortality rate for the 138 confirmed cases. This raised concerns that this virus might cause a SARS-like pandemic (see MERS-CoV summary updates on the WHO website).
Highly pathogenic H5N1 avian influenza virus has decimated poultry production in Asia and has claimed more than 350 human lives since 2003, with regular disease outbreaks continuing to occur (see Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO on the WHO website).
A new strain of influenza virus (H7N9), which has never previously been seen in humans, appeared in April 2013. Although this current strain of the avian influenza virus is not in a form that is able to transmit from human to human, there is still the possibility that it could mutate and trigger a serious pandemic65 (see Avian influenza A(H7N9) virus on the WHO website).
Over the past two decades, Hendra virus in Australia, Nipah virus in Malaysia and Bangladesh, and haemorrhagic fever viruses (Ebola and Marburg) have emerged from bats through intermediate hosts such as horses and pigs to infect and kill humans.
For an excellent overview of EIDs, see the chapter entitled Infectious Disease Emergence: Past, Present, and Future in the book Microbial Evolution and Co-Adaptation66.
Box 2: One-Health.
Numerous emerging disease concerns are closely connected to the ever-increasing interactions between humans and wildlife. Several factors are associated with the emergence of a disease from wildlife and its spread to and among humans67:
• The escalated need for food production to meet present and future demand has led to the intrusion of agriculture into previously untouched areas of the native environment68.
• The impact of climate change has resulted in disturbances in ecosystems and a re-distribution of disease reservoirs and vectors.
• Increased globalization and travel has increased the chance, extent and spread at which disease transmission occurs.
With this in mind, there has been a growing initiative to more closely address this animal–human–ecosystem interface. The term 'One-Health' describes a collaborative effort from multiple disciplines to support a holistic approach to the development of health strategies for humans, animals and the environment. One-Health unifies clinical and veterinary health and directly links this with environmental health research. The development of a framework directed towards strengthening alliances between these sectors has facilitated the development and application of effective and sustainable community health strategies. There is a growing view that a One-Health approach will be crucially important for our preparedness for the next zoonotic pandemic.
Evolutionary lessons from wild animals
Non-traditional animal models have had a pivotal role in shaping our current understanding of evolutionary immunology. The basic features of the innate immune system are believed to have originated in animals approximately 600 million years ago4. Studying non-traditional model organisms has led to the identification of molecules and pathways that are highly conserved through evolution, including Toll-like receptors (TLRs) and pathways such as RNA interference (RNAi)5,6. Similarly, studies of the immune systems of lower vertebrates have provided clues to the evolutionary origins of immune components. For example, B cells in teleosts and amphibians have phagocytic activity, supporting the idea that B cells evolved from ancestral phagocytic cells7.
Antigen receptors. The first features of an adaptive immune system are thought to have appeared in a shark-like ancestor approximately 500 million years ago8. Since then, natural selection processes have driven the evolution of immune systems that show new levels of complexity but that also have variations on a common theme. For example, unconventional T cell receptors (TCRs) have been discovered in several species, including certain marsupial species, sharks and frogs. TCRμ receptors in marsupials and new antigen receptor (NAR)-TCRs in sharks are unique TCR types that contain two variable domains, one of which has an antibody-like structure. Similarly, the African clawed frog (Xenopus laevis) uses a novel TCR type that contains an antibody-like variable domain9,10,11. These TCRs seem to have evolved convergently, suggesting that they might have as yet unrecognized roles in the immune responses of each of these species and might also exist in other species.
These types of discoveries from non-conventional animal models blur the boundaries between TCRs and B cell receptors and challenge traditional views of antigen recognition. Such findings are not only interesting from an evolutionary perspective but also have the potential to be translated into novel therapeutics or applied to laboratory or industrial settings. For example, the variable lymphocyte receptors found in lampreys have been used in a biotechnology setting as tools for the selective recognition of glycans that are poorly discriminated by immunoglobulins12.
Co-evolution of hosts and pathogens. One of the mechanisms responsible for shaping the immune system is the co-evolution of hosts with pathogens. In many cases, zoonotic infections that are pathogenic in humans can coexist with their natural host reservoirs in the absence of disease. This demonstrates the importance of this co-evolutionary relationship. Bats are one example of a group of mammals that has a long co-evolutionary history with the viruses that they harbour. Although infection with viruses such as severe acute respiratory syndrome (SARS) coronavirus, Hendra virus and Ebola virus seems to result in little pathology in bats, the infection of other species often causes severe disease and has fatal consequences. Bats might have become able to coexist with viruses through the evolution of unique immune mechanisms associated with the control of viral replication. It has also been suggested that the severe pathology and disease that often occurs as a result of the spillover of viruses into other vertebrate hosts may result from the disturbance of this finely tuned interaction of viral proteins with their targets in host cells13. Understanding how bats and other natural reservoirs, such as birds, coexist with human pathogens could potentially lead to the discovery of immune mechanisms that control viral replication. These mechanisms could then eventually be used to develop disease control strategies in other species.
Animal models for zoonotic pathogens
Traditional animal models. Immunologists have obtained a detailed knowledge of immune responses to pathogens through the use of mouse models, which have been used to evaluate new vaccines and therapeutics. Although there are many similarities between the immune systems of mice and humans, there are also some key differences. For example, recent genomic studies on several inflammatory diseases have shown a lack of correlation in responses between mice and humans14. Furthermore, there are many examples of successful preclinical trials of immunomodulatory drugs or vaccines in mice that have failed to translate into successful clinical trials in humans15,16,17. One issue is that some of the phenotypic markers used to discriminate immune cell populations in mice are absent in humans, and some subpopulations of immune cells differ between humans and mice (reviewed in Ref. 18). In addition, mouse and non-human primate models often incompletely reproduce human disease phenotypes, and these species are often resistant to infection with human-specific pathogens18. Researchers have circumvented this problem by the use of genetically modified or immunocompromised mice to allow the establishment of infection. Unfortunately, the results from such complex artificial systems have been difficult to interpret and translate to clinical medicine. Therefore, it has become a priority for translational medical research to focus on the development of more appropriate animal models.
Non-traditional laboratory animal models. There are many examples in the literature in which non-traditional animal models have been highly informative for our understanding of immunology19. For several decades the chicken has been used to study immunology, sexual development, and developmental biology of the limbs, nervous system and brain20. Indeed, with the exception of mice and humans, arguably the most thoroughly characterized immune system is that of the chicken. The easy accessibility to the chicken embryo has aided our understanding of immune system development and was fundamental in delineating the T cell-dependent and B cell-dependent arms of the adaptive response. Studies of the thymus have provided information about T cell maturation (reviewed in Ref. 21), and the analysis of the bursa of Fabricius has informed our understanding of B cell development and function22. Furthermore, the first interferon (IFN) was discovered in the chicken23. Other animals have also provided a wealth of knowledge concerning aspects of immunity and human disease. Natural bovine tuberculosis infection in cattle has been used as a model for human tuberculosis, and cattle are the reservoir for Mycobacterium bovis, which can be transmitted to humans24. Similarly, ferrets are widely accepted as an excellent model for influenza infection: they are naturally susceptible to infection with human influenza viruses, and the disease pathology that they develop resembles that of humans infected with influenza25. Another example is the woodchuck: these animals are a good model for studying hepatitis B virus (HBV) because they can be infected by the woodchuck hepatitis virus (WHV), which is closely related to HBV. Natural infection of woodchucks by WHV produces chronic liver disease and primary hepatocellular carcinoma, which are similar to the diseases induced by HBV in humans (reviewed in Ref. 26). Finally, studies of Tasmanian devil facial tumour disease and canine transmissible venereal tumour have helped us to understand the role of the immune system in shaping tumour evolution and have provided insights into the crucial roles of MHC genes (reviewed in Ref. 27).
Natural reservoir hosts and spillover events. Why is it important to use both the natural animal reservoir and the spillover host to study the immune responses to zoonotic pathogens? Understanding the differences between the immune systems of domesticated and wild animal hosts and comparing their immune systems to that of humans is crucial for unravelling the complex disease mechanisms that are involved in zoonotic infections (Fig. 2). Furthermore, by studying the pathogen in its natural host we might be able to devise efficient control measures in that host, thereby disrupting the transmission of the pathogen to humans. This has important implications for predicting, preventing and controlling spillover events and for the development of novel therapeutics and diagnostics. Table 1 lists selected zoonotic viruses and their reservoir hosts, susceptible hosts and transmission hosts. Supplementary information S1 (table) lists numerous other viral and non-viral zoonotic diseases.
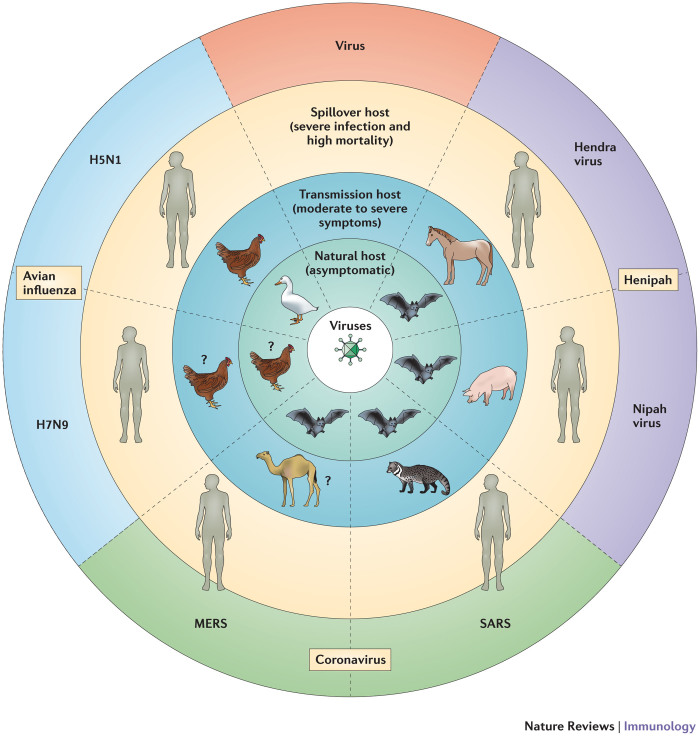
Figure 2. The severity of emerging infectious diseases is influenced by the host–pathogen interaction.
Many zoonotic agents cause little or no signs of disease in their natural hosts, such as wild birds and bats, but transmission hosts might present with disease symptoms ranging from moderate (for example, pigs infected with avian influenza virus) to severe (for example, horses infected with Hendra virus). The terminal or spillover host can present with severe symptoms and high mortality rates (for example, in the case of humans infected with H5N1 influenza and Hendra virus). For some of the most recently identified emerging infectious diseases, such as H7N9 influenza and Middle East respiratory syndrome (MERS) coronavirus, natural and transmission hosts have not been conclusively identified (indicated by a question mark). SARS, severe acute respiratory syndrome.
Table 1. Natural reservoir hosts and susceptible hosts involved in transmission of a selection of emerging zoonotic viral diseases*.
| Disease (virus) | Known reservoir hosts | Other susceptible hosts | Transmission host to humans | Refs |
|---|---|---|---|---|
| Avian influenza (H5N1, H7N9, H7N7, H9N2, H3N2 and others) | Waterfowl and wild birds | Bats, cats, dogs, ferrets, pigs, poultry (chickens, ducks and turkeys) and marine mammals | Chickens | Centers for Disease Control and Prevention‡, Zoonoses§, Animal Disease Information Summaries|| and Refs 69,70,71 |
| 'Swine flu' strains (H1N1 and H3N2) | Pigs | Ferrets, foxes, cats, dogs, poultry (chickens, ducks and turkeys) and marine mammals | Pigs | Centers for Disease Control and Prevention‡, Zoonoses§, Animal Disease Information Summaries|| and Refs 69,70,71 |
| SARS (SARS coronavirus) | Bats | Civet cats | Civet cats | Centers for Disease Control and Prevention‡ and Ref. 72 |
| Dengue fever (dengue virus) | Primates | Unknown | Mosquitoes | Centers for Disease Control and Prevention‡ and Ref. 73 |
| Hendra (Hendra virus) | Bats | Horses and ferrets | Horses | Centers for Disease Control and Prevention‡ and Ref. 74 |
| Rabies (rabies virus and other lyssaviruses) | Bats | Cats, cattle, coyotes, dogs, foxes, horses, mongooses, primates, raccoons, sheep, skunks and wolves | Bats and dogs | Centers for Disease Control and Prevention‡, Animal Disease Information Summaries|| and Refs 75,76 |
| Ebola viral haemorrhagic fever (Ebola virus) | Bats | Primates | Primates and bats | Centers for Disease Control and Prevention‡ and Zoonoses§ |
| Crimean–Congo haemorrhagic fever (Crimean–Congo haemorrhagic fever virus) | Rodents, hares, hedgehogs and ostriches | Cattle, goats, horses, pigs and sheep | Ticks | Centers for Disease Control and Prevention‡ and Zoonoses§ |
| Ross River fever (Ross River virus) | Kangaroos and wallabies | Bats, birds, cats, dogs, horses and possums | Mosquitoes | Refs 73,77,78 |
| Japanese encephalitis (Japanese encephalitis virus) | Pigs and wild birds | Horses | Mosquitoes | Centers for Disease Control and Prevention ‡ |
| West Nile virus encephalitis (West Nile virus) | Domestic and wild birds | Bats, camels, horses, marine mammals, reptiles and >30 vertebrate species | Mosquitoes and birds | Centers for Disease Control and Prevention‡ and Animal Disease Information Summaries|| |
SARS, severe acute respiratory syndrome.
*Supplementary information S1 (table) lists numerous other zoonotic diseases, including bacterial, prion, parasitic and other viral diseases.
‡See the Centers for Disease Control and Prevention website.
§See Zoonoses on the World Health Organization website.
||See Animal Disease Information Summaries on the World Organisation for Animal Health website.
One example is the case of highly pathogenic avian influenza (HPAI). Waterfowl are natural hosts for avian influenza viruses, and after infection with HPAI they develop what seems to be a limited inflammatory response of the respiratory system, usually with little or no mortality. By contrast, in chickens and in humans, infection with HPAI viruses can induce a rapid and strong inflammatory response and high levels of cytokine production, often referred to as a cytokine storm28, and the infection can become systemic and induce severe disease symptoms29,30,31. Chickens are acutely susceptible to infection with H5N1 strains of HPAI, which typically cause death within 18–36 hours. Studying waterfowl, such as ducks, and comparing their immune responses to influenza virus with those of chickens might provide invaluable insights into the 'aberrant' immune reactions that occur in influenza virus spillover hosts, such as chickens, pigs and humans (Fig. 3). Moreover, understanding the mechanisms by which some recent isolates of H5N1 viruses cause systemic disease and death in ducks32 will help to identify the elements of the immune response that are involved in disease lethality.
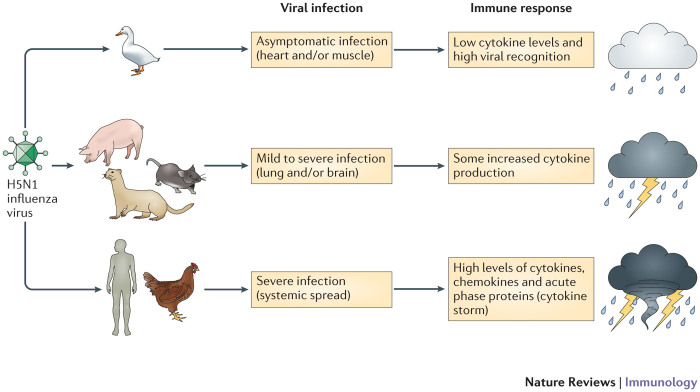
Figure 3. The host immune response to an infection influences the disease outcome.
Infection with H5N1 influenza virus can cause very different disease outcomes in different reservoir and spillover host species. Waterfowl, such as wild ducks, are the natural host for this virus and develop a limited inflammatory response that is associated with low levels of cytokine expression. Intermediate hosts, including mice, pigs and ferrets, are often used to study this infection and display mild to severe disease symptoms (depending on the H5N1 virus strain used) that are associated with increased levels of pro-inflammatory cytokines. By contrast, spillover hosts such as chickens and humans display a rapid and strong inflammatory response, often referred to as hypercytokinaemia (or cytokine storm) and the infection becomes systemic, causing severe disease symptoms and high mortality rates.
Another interesting example is Hendra virus, which does not cause disease in fruit bats, the natural reservoir for this virus, but induces severe disease in horses and humans (reviewed in Ref. 33). The study of disease pathogenesis and immune responses to Hendra virus in horses has led to the development of a horse vaccine that will help to reduce the risk of Hendra virus transmission from horses to humans33. Nevertheless, studying immunity to zoonotic viruses in wildlife is complex. For example, the fact that infectious agents, such as henipaviruses and lyssaviruses, which cause lethal diseases in humans, can coexist peacefully with bats raises many questions. It is still unclear what role the bat immune system has in keeping these pathogens under control and in enabling host survival while allowing enough viral replication to facilitate the transmission of the virus to spillover hosts. Other remaining questions are how host–pathogen interactions are influenced by the genetics of the population, environmental factors, food supply, co-infections, interactions with other species and changes in demographics, as well as by particular physiological, anatomical and metabolic features of the host.
With the constant emergence of new zoonotic pathogens, it is becoming increasingly important to establish a dialogue between scientists working in laboratory immunology (often where mouse models are used) and wildlife and livestock immunology (which avoids the need for a 'model') so that findings from mice can be confirmed in the context of natural populations. Also, immunology studies in wildlife and livestock allow us to identify the aspects of the immune response that are important for a protective phenotype in a natural setting, as well as to uncover mechanisms that increase or reduce virus replication. Such knowledge can facilitate the development of novel therapeutics for treating zoonotic infections in humans, as well as animals.
How are zoonotic research studies conducted?
Animal experimentation with zoonotic agents involves exposure to infectious microorganisms that are hazardous to human health. Microorganisms are classified into risk categories according to the hazard they pose to individuals and to communities, including livestock and the environment, and whether effective treatment and preventive measures are available. The World Health Organization (WHO) has recommended a four-tiered agent risk group classification system for laboratory work; each tier is linked to specified infrastructure and regulatory requirements, practices and procedures with the aim of ensuring the safety of research staff working with these agents, as well as the wider community (see the WHO Laboratory Biosafety Manual, Third Edition). Risk groups 3 and 4 encompass the most serious human and animal pathogens that cause severe disease for which prophylactic or therapeutic measures may not be available. More details on biocontainment laboratories are described in Box 3.
Animals such as bats and migratory water birds that serve as reservoir hosts for zoonotic pathogens are increasingly being studied in the field. These studies provide valuable information about disease distribution and transmission, in addition to informing preparedness and surveillance strategies for future pandemics. However, when the ecological relationship between host and disease agent is one of co-speciation, experimental studies in natural hosts will also be crucial for understanding virus replication, pathogenesis, transmission and persistence in animal populations. Similarly, on the occasion of spillover events and when the origin of the host–pathogen relationship is through dispersal, controlled infection studies in the relevant target species will at some point be required for the evaluation of treatments and preventives, as well as to inform methods for control of transmission risk. It is also important to determine when human pathogens are transmitted to wildlife, because the generation of new reservoirs might promote selective pressures that might increase the risk of zoonotic disease. In this regard, global initiatives such as the US Geological Survey National Wildlife Health Center and the Canadian Cooperative Wildlife Health Centre play important parts in the monitoring of wildlife diseases (Box 3).
Despite the benefits of using non-traditional animal models, working with these systems presents a number of challenges. These can include limited access to suitable subjects of the desired species, particularly in the case of fauna that might need to be wild-caught and might be members of a species of high conservation value; lack of knowledge or experience in handling and husbandry methods suitable for high levels of biocontainment that will also meet contemporary ethics and welfare requirements; a paucity of reagents such as species-specific antibodies required to carry out traditional immunological experiments; and limited or no genome sequence information for the purpose of developing molecular biology tools. For species that are not traditionally used in the laboratory, new policies and procedures for housing, husbandry, restraint and sampling might need to be developed on a case-by-case basis. In such instances, staff within the veterinary departments of zoological parks, and zoologists and veterinarians who specialize in exotic or unusual pets, are valuable resources.
Animal infection studies at a high level of biocontainment, especially those involving species other than small laboratory mammals, are particularly complex and require highly regulated facilities that have been modified to suit the needs of the animal species, highly trained and competent staff, and substantial infrastructure and resource support (Box 3). However, the value inherent to such an approach is that the data emerging from these studies should more readily translate to policies, procedures and products that will reduce the transmission of EIDs to humans.
Box 3: High-security biological containment research facilities.
Although most researchers would be familiar with Biosafety Level 1 (BSL1) and BSL2 laboratories, BSL3 and BSL4 facilities must conform to additional infrastructure requirements, policies and procedures to ensure a safe work environment. Biosafety guidelines are maintained in the United States by the Centers for Disease Control and Prevention (CDC) in partnership with the US National Institutes of Health (see Biosafety in Microbiological and Biomedical Laboratories on the CDC website), in Europe by a legislative act of the European Union and in Australia by the Department of Agriculture, Fisheries and Forestry (DAFF) and the Office of the Gene Technology Regulator (OGTR). Although regulations surrounding BSL3 and BSL4 facilities might differ from country to country, certain principles of biocontainment are consistently observed. For example, all work involving potentially infectious material must be carried out within a biological safety cabinet or another means of primary containment. In the case of BSL4 facilities, primary containment is provided by the wearing of positive pressure protective suits with independent breathing air supply (in a BSL4 suit laboratory or animal facility) or class III biological safety cabinet (in a BSL4 cabinet laboratory). Examples of BSL4 laboratories around the world include the Canadian National Microbiology Laboratory (Winnipeg); the Pirbright Institute (Pirbright, UK); the Uniformed Services University of the Health Sciences (Bethesda, USA); and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Australian Animal Health Laboratory (AAHL; Geelong, Australia). The CSIRO AAHL facility is unique in that its BSL4 animal facility is sufficiently large to allow work with Risk Group 4 agents to be carried out in diverse species, including bats, poultry, pigs, dogs, ferrets and horses, as well as small laboratory mammals. Global initiatives that monitor wildlife diseases, such as the US Geological Survey National Wildlife Health Center and the Canadian Cooperative Wildlife Health Centre have important roles in identifying the distribution of disease prevalence and reservoirs, and how these are influenced by climatic and environmental changes.
Comparative immunology studies
A major goal of studying immunology in natural hosts is to explain how infection with the same pathogen can have such vastly different outcomes in different species. Therefore, investigation of immunity in natural hosts is an area that might yield important discoveries and may illuminate the nature of successful immune responses to agents that are typically associated with adverse disease outcomes in humans and other species. There is an extensive range of technologies that have been used to understand the pathogenesis and immune responses in reservoir and transmission species. These include molecular genetics tools, such as DNA and RNA sequencing, transcriptomics, RNAi, microRNA and genome-editing meganucleases. In addition, post-translational analysis tools such as proteomics, kinomics and other protein modifications provide additional information.
Comparative genomics is a powerful approach for identifying genetic determinants underlying phenotypical differences between species. The recent advances in high-throughput sequencing techniques have facilitated whole-genome sequencing of a large number of species, including some that are reservoir hosts of important zoonotic viruses, and others that are susceptible to disease caused by those viruses. Comparative analysis of these genomes can identify gene candidates for disease-susceptibility or disease-resistance phenotypes. There are two basic approaches for identifying these candidates, both of which start by selecting two groups of species on the basis of the segregation of a particular phenotype — in this case, disease susceptibility or resistance during infection with zoonotic viruses. The first approach is to carry out high-throughput comparison of all genes across the two groups to identify features that cluster separately between the groups. The results of this type of analysis can then be subjected to categorical enrichment analysis (for example, gene ontology or pathway analysis34) to highlight gene candidates that seem to be linked to the disease phenotype that is under investigation. The second approach involves generating lists of candidate genes on the basis of a literature review. In the case of zoonotic virus infection this might begin with genes of the innate immune system, such as those related to interferon (IFN) production and signalling.
Quantitative transcriptomics by sequencing — for example, RNA sequencing (RNA-seq) and small RNA-seq — have enormous potential for identifying crucial species-specific host immune responses to virus infection. These analyses can provide unprecedented detail and can be easily carried out on species for which no species-specific reagents are available, as is the case for the natural hosts of many zoonotic viruses. Another application with strong potential in this field is genome-wide screening using RNAi (reviewed in Ref. 35). As a result of the continuous development of genome sequencing and RNAi technologies, it will soon be possible to compare host genes required for virus replication across virus-susceptible and virus-resistant species, such as chickens and ducks in the case of HPAI.
A major goal of sequencing bat genomes was to understand the genetic basis of virus–host interactions in a natural reservoir host36. More recently, the analysis of the duck genome and virus-infected duck transcriptome has continued this trend. It has been observed that avians generally encode fewer cytokines than mammals and seem to lack α-defensins and θ-defensins. However, the duck genome features lineage-specific duplications of β-defensin and butyrophilin-like genes, suggesting a possible connection with the fundamental differences in disease outcome observed between chickens and ducks infected with HPAI37. An area of potential interest is revisiting the genome sequences of animals, such as the cat, dog, goat, armadillo and camel, with the hindsight of knowing that they are natural hosts for diseases of relevance to humans (Table 1; see Supplementary information S1 (table)). Several findings from the original genome descriptions for these species are relevant to host–pathogen interactions, such as the camel trait of producing antibodies that lack light chains38 and the presence of endogenous retroviruses in the cat genome39. We provide below specific examples of some of the key insights that have been made from comparative immunology studies.
Key insights from comparative immunology
Bat immunology. Interest in the bat immune system has arisen from the findings that they are asymptomatic natural hosts for many zoonotic viruses40. Recently, whole-genome sequencing and comparative analysis of two bat species led to the identification of a large number of unique bat characteristics related to immunity and the antiviral response36. These genes represent a set of testable hypotheses that may shed light on shortcomings in our own immune responses to viruses that cause severe disease or death.
Although bats seem, in general, to have a similar immune system to other mammals, genomic and transcriptomic analyses have demonstrated that bats lack key natural killer cell receptors and several intracellular pathogen-recognition receptors, including absent in melanoma 2 (AIM2), which is a cytoplasmic receptor for double-stranded DNA36,41. As perhaps a counter-balance, several genes involved in pathogen recognition and immune responses seem to be under positive selection in bats, suggesting that these genes may increase fitness36. Such genes include those encoding DNA-damage sensors, such as DNA-dependent protein kinase catalytic subunit (DNA-PKcs; also known as PRKDC) and X-ray repair cross-complementing protein 5 (XRCC5), which have recently been identified as part of a cytoplasmic microbial nucleic acid-sensing complex42. The nuclear factor-κB (NF-κB) family member REL is also under positive selection in bats and has multiple roles in the immune system (including the regulation of type I and type III IFN production (Fig. 4)). Other immune genes under positive selection in bats include the genes encoding TLR7, NOD, LRR and pyrin-domain-containing 3 (NLRP3) and mitogen-activated protein kinase kinase kinase 7 (MAP3K7)36. NLRP3 has a role in the formation of inflammasomes and the induction of inflammation in response to infection. In vitro studies have identified several other key differences in the immune system of bats, although it is not yet clear which of these are related to disease outcome (reviewed in Ref. 43). The challenge will now be to determine the importance of these differences in relation to the observed immunological differences that underpin differential disease susceptibility.
Figure 4. Positive selection of bat genes.
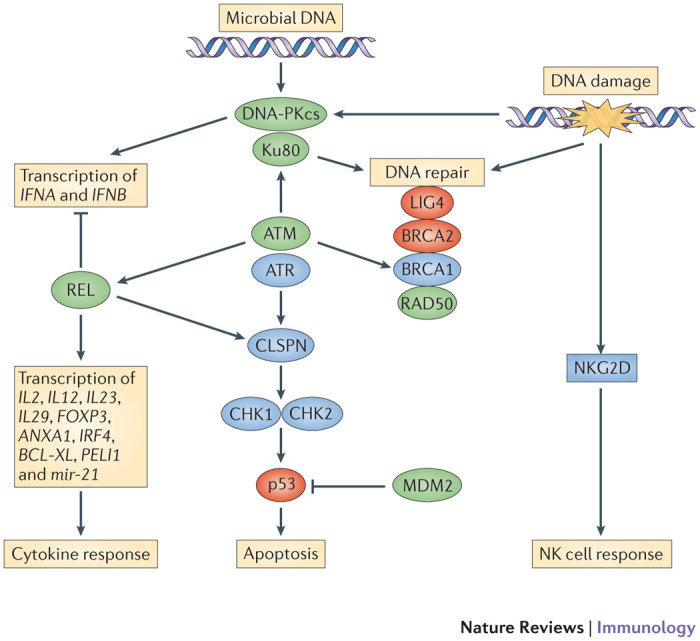
The figure illustrates key components of the DNA damage response and DNA repair pathways. Whole-genome analysis of two bat species (Pteropus alecto and Myotis davidii) showed that a high number of genes encoding components of these pathways are positively selected in P. alecto and M. davidii. Many of these genes are positively selected in both species (these encode proteins that are highlighted in green), whereas others have been positively selected in only one of the species (these encode proteins that are highlighted in red). Bat-specific differences in these genes include unique changes to the nuclear localization signals in p53 and the E3 ubiquitin-protein ligase MDM2. The impact of these changes on host–pathogen interactions is currently under investigation. The transcription factor REL (which is a member of the NF-κB family) regulates many effector proteins of the innate immune system, including type I interferons (IFNs), and it also participates in the DNA damage response. The gene encoding REL is under positive selection in bats and contains unique amino acid changes that might affect the interaction of REL with inhibitor of NF-κB (IκB). DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Ku80 are crucial to the DNA damage response but also form a microbial DNA-sensing complex connected to the type I IFN system. The relationship between innate immunity and the DNA damage response is well established, and unique changes in this pathway in bats might influence the outcomes of viral infection. It was proposed that these differences might reflect adaptations to the increased oxidative metabolism that accompanied the evolution of flight in bats. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related protein; CHK, checkpoint kinase; CLSPN, claspin; FOXP3, forkhead box P3; IL, interleukin; LIG4, DNA ligase 4; MRE11, meiotic recombination 11 homologue; NBS1, Nijmegen breakage syndrome protein 1 (also known as nibrin).
Avian influenza. Infection of both chickens30 and humans29 with H5N1 HPAI can lead to a severe inflammatory response that is associated with high levels of cytokine production (reviewed in Ref. 28). This observed aberrant immune response has been suggested to be associated with H5N1 mortalities. An intriguing observation is that with H5N1 HPAI infection there is an observed increase in mortality for patients in the middle-age bracket, unlike seasonal influenza deaths, which are mostly confined to infants and the elderly. It has been suggested that infection with H5N1 may result in a more severe disease if the patient's immune system is robust29. With this in mind, a tantalizing explanation for the differences in disease outcomes between natural hosts and spillover hosts might be that natural hosts experience less immunopathology during infection or, more specifically, that they maintain a better balance between promoting virus clearance and inducing dangerous levels of inflammation. Therefore, identifying the factors that trigger immunopathological responses, such as an overly aggressive immune response, during zoonotic infections might enable targeted therapeutic intervention and better outcomes in cases of human disease. Comparative studies between natural hosts (that are resistant to disease) and spillover hosts (that are susceptible to disease) could be very informative in uncovering such potential differences.
HIV. One illuminating example using a more established model comes from AIDS research using two different primate species with very different infection outcomes. The sooty mangabey is a natural host of simian immunodeficiency virus (SIV) but does not develop AIDS, whereas the rhesus macaque, a common laboratory model primate, develops AIDS and is used as a model for human HIV. Research has shown that a major factor related to disease outcome is the ability to resolve the initial inflammatory response44. In the non-natural host (the macaque), SIV infection leads to chronic immune activation and progressive disease. By contrast, for the natural host of SIV (the mangabey), the innate immune response resolves within 4–8 weeks despite high levels of virus replication. This example illustrates one instance whereby immunopathology might be considered a major determinant of disease outcome. Differences in immunopathology might be an important factor in explaining disease outcomes from many zoonotic virus infections and suggest that a maximum immune response is not necessarily an optimal response45.
Hantavirus. Wild rodents are natural reservoirs for hantaviruses and display no clinical signs of disease during acute or persistent infection, whereas, in humans, infection causes haemorrhagic fever or cardiopulmonary syndrome. Although laboratory mice can be used to study hantavirus infection, these models do not accurately represent viral persistence observed in wild rodent reservoir populations. Therefore, studies of the natural host using wild mouse models, including deer mice (Peromyscus spp.) and Norway rats (Rattus norvegicus), have been initiated. These studies have demonstrated differences in the immune response of these natural host species to hantavirus infection compared with humans, including evidence for reduced pro-inflammatory responses and evidence for a role of regulatory T cells in limiting immunopathology and in promoting persistent infection46,47.
Clinical implications of wild-animal studies
There are a number of key practical outcomes that research in wild and livestock animal species could achieve. For example, if we better understand influenza virus infection in pigs and birds, will we be better able to predict where the next pandemic might emerge or will we be able to develop vaccines or antivirals for these animals in order to prevent the crossover of the infection to humans? If we understand how the bat and duck immune systems respond to viruses, will this help us to develop new therapeutics and vaccines for preventing fatal infections caused by these viruses in humans? Can we engineer livestock that will be less susceptible to infection by EIDs, such as influenza and Nipah viruses, thereby blocking the transmission cycle?
The ability of the segmented influenza genome to continually re-assort within different animal host species has a crucial effect on the epidemiology of influenza outbreaks. For example, the combination of viral gene segments from four virus strains circulating in three different species (human, swine and poultry) led to the emergence of swine-origin influenza virus (S-OIV) in the human population48. The relevance of a lack of pre-existing immunity became apparent as mortality and morbidity began to rise sharply in the 20- to 40-year-old age group49, whereas the older age group was associated with a lower infection risk50 that was associated with a higher prevalence of pre-existing antibodies that cross-reacted with the 2009 H1N1 virus. The impact of a novel influenza virus arising from wildlife species was felt again in February 2013, when an avian-origin influenza virus emerged in Zhejiang, China, causing more than 130 reported human infections and 36 deaths. This virus seems to have emerged from the mixing of influenza viruses from several avian sources, including ducks and wild birds51. It was the first report of a H7N9 influenza virus infecting humans and was met with little pre-existing immunity. Furthermore, vaccines against H7N9 influenza virus are predicted to be poorly immunogenic because there are few immunogenic T cell epitopes present on the H7 molecule compared with other haemagglutinin subtypes52. Despite this particular virus being highly pathogenic in humans, natural infections with H7N9 viruses in chickens, ducks and other birds are asymptomatic and elicit an immune response that can be detected serologically53. This is in stark contrast to the H5N1 and H7N7 viruses, where disease in humans was associated with a highly pathogenic phenotype in poultry54. The immunological component to this disease in birds suggests that further study of these emerging viruses in poultry and other bird species is required to understand factors influencing disease susceptibility and transmission.
The identification of key differences in immune pathways between susceptible and non-susceptible hosts might offer clues for the development of disease intervention strategies. As mentioned above, for HPAI infection, ducks and chickens represent natural and spillover hosts, respectively. Chickens have lost expression of the innate immune sensor retinoic acid-inducible protein I (RIG-I), which may be an important clue in explaining why they suffer close to 100% mortality from HPAI. Conversely, ducks (which have intact RIG-I expression) develop only mild symptoms in response to HPAI infection and usually survive. It has been shown that the transfection of duck RIG-I into chicken cells induced IFNB promoter activity and limited the replication of low and highly pathogenic avian influenza strains, suggesting a key role of RIG-I in the ability of ducks to be resistant to influenza-mediated disease55. Although it is not understood how the absence of RIG-I in chickens affects their response to other viral infections, it would nevertheless be of interest to generate transgenic chickens that express duck RIG-I and investigate whether these animals are less susceptible to disease following HPAI infection or, indeed, following infection with other viruses.
Various tools have now been developed to allow targeted introduction of coding or non-coding sequences into the genome of animals at precise sites with high efficiency. Targeted nucleases, such as transcription activator-like effector nucleases (TALENs)56 and nucleases of the clustered regularly interspaced short palindromic repeats (CRISPR) system57, have increased targeted gene mutation efficiency to near 100%. This technology has been used in model organisms including zebrafish58,59,60, mice61, rats62 and pigs63. Using these technologies in other species will allow breakthroughs in functional genomic studies in natural reservoir and spillover species, such as ducks, bats and chickens, or in other model species, such as ferrets. This will provide the opportunity to test hypotheses of immune function and regulation in these species. The development of poultry that are less susceptible to H5N1 influenza virus is likely to help to reduce the environmental load of highly pathogenic virus and act to break the chain of infection from poultry to humans. It is hoped that the findings from immunological studies in reservoir hosts will translate to the development of therapeutics for humans and other animals. This would result in a substantial human health benefit by addressing the serious issues in those countries where these diseases are endemic.
Concluding remarks
What can we learn from nature's experiments? Studying the immune responses to zoonotic pathogens in the natural reservoir host and comparing them to the responses in spillover hosts will help to identify key processes in disease susceptibility and transmission. With the current emphasis on a One-Health approach, researchers are turning to an analysis of the immune response of natural host species for a greater understanding of the immunological basis of emerging zoonotic disease. This approach, together with the adoption of new genomic information and genome-editing technologies, will allow the identification of strategies to prevent and minimize the impact of EIDs and enhance our preparedness for pandemics.
Supplementary information
Natural reservoir hosts and susceptible hosts involved in transmission of bacterial, prion, parasitic and other viral zoonotic diseases (PDF 167 kb)
Glossary
- Variable lymphocyte receptors
Alternative forms of somatically diversified antigen receptors expressed by lymphocytes of jawless vertebrates. The combinatorial diversity of these receptors is based on variable numbers of leucine-rich repeat elements assembled by a gene-conversion process.
- Bursa of Fabricius
A primary lymphoid organ that is only found in birds. B cells mature in this organ, and indeed were first observed and named after this site: the 'B' in B cell refers to 'bursa-derived'.
Biographies
Andrew G. D. Bean obtained his Ph.D. in immunology from Monash University in Melbourne, Australia. Located at the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Australian Animal Health Laboratory in Geelong, Australia, he is using a 'One-Health' approach to study host–pathogen interactions and the development of antiviral therapeutic strategies.
Michelle L. Baker obtained her Ph.D. from the University of Queensland, Brisbane, Australia, before carrying out postdoctoral training in comparative immunogenetics at the University of New Mexico in Albuquerque, USA. She currently leads a group examining the nature of the immune response of bats to viral infection.
Cameron R. Stewart received his Ph.D. from the University of Melbourne, Australia. He is studying the immunological response to highly pathogenic viruses such as avian influenza and Hendra virus. His work includes basic research on therapeutic interventions, novel biomarkers of infection and mechanisms of disease pathogenesis.
Christopher Cowled holds a Ph.D. from the University of Queensland, Brisbane, Australia. He has worked at research institutions in Australia and overseas, and his professional interests encompass the boundaries between immunology, virology and bioinformatics, with a specific focus on bats as natural virus reservoirs.
Celine Deffrasnes received her Ph.D. in microbiology and immunology from Laval University, Quebec City, Quebec, Canada. Her main research focus is studying the host–pathogen interactions of the Hendra, dengue and avian influenza viruses. She is also investigating the role of microRNA in viral infections.
Lin-Fa Wang obtained his Ph.D. at the University of California in Davis, USA. He is currently an OCE Science Leader at the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Australian Animal Health Laboratory and the Director of the Program in Emerging Infectious Diseases at Duke-National University of Singapore Graduate Medical School in Singapore. His research is focused on emerging zoonotic viruses and host–pathogen interactions.
John W. Lowenthal obtained his Ph.D. in immunology from the University of Melbourne, Australia. He is the Theme Leader for One-Health in the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Biosecurity Flagship. Prior to joining CSIRO in 1990, he had postdoctoral positions at the Ludwig Institute for Cancer Research in Lausanne, Switzerland, and at Duke University in Durham, North Carolina, USA. His research spans veterinary immunology, disease-resistant animals, innate immune responses to viruses and developing antivirals for zoonotics. John W. Lowenthal's homepage.
Related links
FURTHER INFORMATION
Animal Disease Information Summaries
Australian Animal Health Laboratory
Biosafety in Microbiological and Biomedical Laboratories
Canadian Cooperative Wildlife Health Centre
Centers for Disease Control and Prevention
Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO
Infectious Disease Emergence: Past, Present, and Future
National Microbiology Laboratory
Severe acute respiratory syndrome (SARS)
Uniformed Services University of the Health Sciences
US Geological Survey National Wildlife Health Center
PowerPoint slides
Competing interests
The authors declare no competing financial interests.
References
- 1.Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Phil. Trans. R. Soc. Lond. B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legrand N, Weijer K, Spits H. Experimental models to study development and function of the human immune system in vivo. J. Immunol. 2006;176:2053–2058. doi: 10.4049/jimmunol.176.4.2053. [DOI] [PubMed] [Google Scholar]
- 4.Cooper MD, Herrin BR. How did our complex immune system evolve? Nature Rev. Immunol. 2010;10:2–3. doi: 10.1038/nri2686. [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Li J, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nature Immunol. 2006;7:1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- 8.Zhu C, et al. Origin of immunoglobulin isotype switching. Curr. Biol. 2012;22:872–880. doi: 10.1016/j.cub.2012.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc. Natl Acad. Sci. USA. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parra ZE, et al. A unique T cell receptor discovered in marsupials. Proc. Natl Acad. Sci. USA. 2007;104:9776–9781. doi: 10.1073/pnas.0609106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD. The dynamic TCRδ: TCRδ chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur. J. Immunol. 2010;40:2319–2329. doi: 10.1002/eji.201040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong X, et al. Sugar-binding proteins from fish: selection of high affinity “lambodies” that recognize biomedically relevant glycans. ACS Chem. Biol. 2013;8:152–160. doi: 10.1021/cb300399s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LF, Walker PJ, Poon LL. Mass extinctions, biodiversity and mitochondrial function: are bats 'special' as reservoirs for emerging viruses? Curr. Opin. Virol. 2011;1:649–657. doi: 10.1016/j.coviro.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seok J, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suntharalingam G, et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006;355:1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- 16.Benatar M. Lost in translation: treatment trials in the SOD1 mouse and in human ALS. Neurobiol. Dis. 2007;26:1–13. doi: 10.1016/j.nbd.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Tameris MD, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013;381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J. Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 19.Hein WR, Griebel PJ. A road less travelled: large animal models in immunological research. Nature Rev. Immunol. 2003;3:79–84. doi: 10.1038/nri977. [DOI] [PubMed] [Google Scholar]
- 20.Le Douarin NM, Dieterlen-Lievre F. How studies on the avian embryo have opened new avenues in the understanding of development: a view about the neural and hematopoietic systems. Dev. Growth Differ. 2013;55:1–14. doi: 10.1111/dgd.12015. [DOI] [PubMed] [Google Scholar]
- 21.Chen CL, Bucy RP, Cooper MD. T cell differentiation in birds. Semin. Immunol. 1990;2:79–86. [PubMed] [Google Scholar]
- 22.Glick B, Chang TS, Jaap G. The bursa of Fabricius and antibody production. Poult. Sci. 1956;35:224–225. doi: 10.3382/ps.0350224. [DOI] [Google Scholar]
- 23.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 24.Waters WR, et al. Tuberculosis immunity: opportunities from studies with cattle. Clin. Dev. Immunol. 2011;2011:768542. doi: 10.1155/2011/768542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belser JA, Katz JM, Tumpey TM. The ferret as a model organism to study influenza A virus infection. Dis. Model. Mech. 2011;4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menne S, Cote PJ. The woodchuck as an animal model for pathogenesis and therapy of chronic hepatitis B virus infection. World J. Gastroenterol. 2007;13:104–124. doi: 10.3748/wjg.v13.i1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belov K. Contagious cancer: lessons from the devil and the dog. Bioessays. 2012;34:285–292. doi: 10.1002/bies.201100161. [DOI] [PubMed] [Google Scholar]
- 28.Clark IA. The advent of the cytokine storm. Immunol. Cell Biol. 2007;85:271–273. doi: 10.1038/sj.icb.7100062. [DOI] [PubMed] [Google Scholar]
- 29.Lee N, et al. Hypercytokinemia and hyperactivation of phospho-p38 mitogen-activated protein kinase in severe human influenza A virus infection. Clin. Infect. Dis. 2007;45:723–731. doi: 10.1086/520981. [DOI] [PubMed] [Google Scholar]
- 30.Karpala AJ, et al. Highly pathogenic (H5N1) avian influenza induces an inflammatory T helper type 1 cytokine response in the chicken. J. Interferon Cytokine Res. 2011;31:393–400. doi: 10.1089/jir.2010.0069. [DOI] [PubMed] [Google Scholar]
- 31.Tisoncik JR, et al. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pantin-Jackwood MJ, Swayne DE. Pathobiology of Asian highly pathogenic avian influenza H5N1 virus infections in ducks. Avian Dis. 2007;51:250–259. doi: 10.1637/7710-090606R.1. [DOI] [PubMed] [Google Scholar]
- 33.Mahalingam S, et al. Hendra virus: an emerging paramyxovirus in Australia. Lancet Infect. Dis. 2012;12:799–807. doi: 10.1016/S1473-3099(12)70158-5. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meliopoulos VA, et al. Host gene targets for novel influenza therapies elucidated by high-throughput RNA interference screens. FASEB J. 2012;26:1372–1386. doi: 10.1096/fj.11-193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nature Genet. 2013;45:776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Bactrian Camels Genome Sequencing and Analysis Consortium. Genome sequences of wild and domestic bactrian camels. Nature Commun.3, 1202 (2012). [DOI] [PMC free article] [PubMed]
- 39.Pontius JU, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luis AD, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. Biol. Sci. 2013;280:20122753. doi: 10.1098/rspb.2012.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papenfuss AT, et al. The immune gene repertoire of an important viral reservoir, the Australian black flying fox. BMC Genomics. 2012;13:261. doi: 10.1186/1471-2164-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker ML, Schountz T, Wang LF. Antiviral immune responses of bats: a review. Zoonoses Publ. Health. 2013;60:104–116. doi: 10.1111/j.1863-2378.2012.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chahroudi A, Bosinger SE, Vanderford TH, Paiardini M, Silvestri G. Natural SIV hosts: showing AIDS the door. Science. 2012;335:1188–1193. doi: 10.1126/science.1217550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viney ME, Riley EM, Buchanan KL. Optimal immune responses: immunocompetence revisited. Trends Ecol. Evol. 2005;20:665–669. doi: 10.1016/j.tree.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Schountz T, et al. Regulatory T cell-like responses in deer mice persistently infected with Sin Nombre virus. Proc. Natl Acad. Sci. USA. 2007;104:15496–15501. doi: 10.1073/pnas.0707454104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Easterbrook JD, Klein SL. Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. PLoS Pathog. 2008;4:e1000172. doi: 10.1371/journal.ppat.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh Y, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chowell G, et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 2009;361:674–679. doi: 10.1056/NEJMoa0904023. [DOI] [PubMed] [Google Scholar]
- 50.Fisman DN, et al. Older age and a reduced likelihood of 2009 H1N1 virus infection. N. Engl. J. Med. 2009;361:2000–2001. doi: 10.1056/NEJMc0907256. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381:1916–1925. doi: 10.1016/S0140-6736(13)60903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Groot AS, et al. Low immunogenicity predicted for emerging avian-origin H7N9: Implication for influenza vaccine design. Hum. Vaccin. Immunother. 2013;9:950–956. doi: 10.4161/hv.24939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. Overview of the emergence and characteristics of the avian influenza A(H7N9) virus. World Health Organization[online], (2013).
- 54.Perdue ML, Swayne DE. Public health risk from avian influenza viruses. Avian Dis. 2005;49:317–327. doi: 10.1637/7390-060305R.1. [DOI] [PubMed] [Google Scholar]
- 55.Barber MR, Aldridge JR, Jr, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl Acad. Sci. USA. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cade L, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlem TJ, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotech. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carbery ID, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mashimo T, et al. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl Acad. Sci. USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 65.Yu H, et al. Human infection with avian influenza A H7N9 virus: an assessment of clinical severity. Lancet. 2013;382:138–145. doi: 10.1016/S0140-6736(13)61207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Relman DA, Hamburg MA, Choffnes ER, Mack A . : 2009. pp. 193–272. [Google Scholar]
- 67.Patz JA, et al. Unhealthy landscapes: policy recommendations on land use change and infectious disease emergence. Environ. Health Perspect. 2004;112:1092–1098. doi: 10.1289/ehp.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pulliam JR, et al. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J. R. Soc. Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swenson SL, et al. Natural cases of 2009 pandemic H1N1 Influenza A virus in pet ferrets. J. Vet. Diagn. Invest. 2010;22:784–788. doi: 10.1177/104063871002200525. [DOI] [PubMed] [Google Scholar]
- 70.Reperant LA, Kuiken T, Osterhaus AD. Influenza viruses: from birds to humans. Hum. Vaccin Immunother. 2012;8:7–16. doi: 10.4161/hv.8.1.18672. [DOI] [PubMed] [Google Scholar]
- 71.Tong S, et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carver S, Bestall A, Jardine A, Ostfeld RS. Influence of hosts on the ecology of arboviral transmission: potential mechanisms influencing dengue, Murray Valley encephalitis, and Ross River virus in Australia. Vector Borne. Zoonot. Dis. 2009;9:51–64. doi: 10.1089/vbz.2008.0040. [DOI] [PubMed] [Google Scholar]
- 74.Clayton BA, Wang LF, Marsh GA. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Publ. Health. 2013;60:69–83. doi: 10.1111/j.1863-2378.2012.01501.x. [DOI] [PubMed] [Google Scholar]
- 75.Rupprecht CE, Turmelle A, Kuzmin IV. A perspective on lyssavirus emergence and perpetuation. Curr. Opin. Virol. 2011;1:662–670. doi: 10.1016/j.coviro.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 76.Hatz CF, Kuenzli E, Funk M. Rabies: relevance, prevention, and management in travel medicine. Infect. Dis. Clin. North Am. 2012;26:739–753. doi: 10.1016/j.idc.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 77.Kay BH, Boyd AM, Ryan PA, Hall RA. Mosquito feeding patterns and natural infection of vertebrates with Ross River and Barmah Forest viruses in Brisbane, Australia. Am. J. Trop. Med. Hyg. 2007;76:417–423. doi: 10.4269/ajtmh.2007.76.417. [DOI] [PubMed] [Google Scholar]
- 78.Tong S, et al. Climate variability, social and environmental factors, and Ross River virus transmission: research development and future research needs. Environ. Health Perspect. 2008;116:1591–1597. doi: 10.1289/ehp.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Natural reservoir hosts and susceptible hosts involved in transmission of bacterial, prion, parasitic and other viral zoonotic diseases (PDF 167 kb)