Abstract
Langerhans cells are key sentinel cells of the skin and mucosal lining. They sense microorganisms through their repertoire of pattern-recognition receptors to mount and direct appropriate immune responses. We recently demonstrated that human Langerhans cells interact with the Gram-positive pathogen Staphylococcus aureus through the Langerhans cell-specific receptor langerin (CD207). It was previously hypothesized that two linked single nucleotide polymorphisms (SNPs; N288D and K313I) in the carbohydrate recognition domain of langerin would affect interaction with microorganisms. We show that recognition of S. aureus by recombinant langerin molecules is abrogated in the co-inheriting SNP variant, which is mainly explained by the N288D SNP and further enhanced by K313I. Moreover, introduction of SNP N288D in ectopically-expressed langerin affected cellular distribution of the receptor such that langerin displayed enhanced plasma membrane expression. Despite this increased binding of S. aureus by the langerin double SNP variant, uptake of bacteria by this langerin variant was compromised. Our findings indicate that in a proportion of the human population, the recognition and uptake of S. aureus by Langerhans cells may be affected, which could have important consequences for proper immune activation and S. aureus-associated disease.
Keywords: Langerin, Single nucleotide polymorphism, Langerhans cell, Staphylococcus aureus
Introduction
Langerin (CD207) is a C-type lectin receptor that, in humans, is exclusively expressed on Langerhans cells (LCs). Langerin is a recycling receptor that binds its ligands, including sulfated glycans, β-glucans, N-acetylglucosamine (GlcNAc), and complex bacterial glycans, in a Ca2+-dependent manner [1, 2, 3]. Its Ca2+ affinity stems from a fine-tuned allosteric network of communicating amino acids [4]. Engagement of langerin triggers ligand endocytosis, which is mediated by actin remodeling through the cytoplasmic proline-rich motifs of langerin [5, 6]. The internalized cargo is released from the receptor in the endosomal compartments as a consequence of lower pH and endosomal Ca2+ concentration [4, 5, 7]. This mechanism allows langerin to recycle between the plasma membrane and endosomal compartments and prevents langerin degradation.
In the human population, several single nucleotide polymorphisms (SNPs) in langerin have been described [1, 8, 9]. Interestingly, some of these SNPs are situated in the carbohydrate recognition domain of langerin and affect receptor-ligand interaction. Two particular SNPs, N288D (rs13383830) and K313I (rs57302492), are in strong linkage disequilibrium and co-inherit throughout the human population. The overall minor allele frequency of these SNPs is 13.0% (online suppl. Fig. S1; see www.karger.com/doi/10.1159/000500547) [10]. Based on the Hardy-Weinberg equilibrium, this corresponds to an estimated 1.7% of homozygotes and 22.6% heterozygous carriers. The minor allele frequency is highest in the African population (21.8%, 4.8% homozygotes), while it is lowest in populations of native American (5.3%, 0.3% homozygotes) and European (6.2%, 0.4% homozygotes) ancestry (online suppl. Fig. S1).
Based on glycan array analysis, the non-naturally occurring single K313I variant abolishes interaction of langerin with terminal sulfated glycan residues, but enhances interaction with terminal GlcNAc structures [1, 9]. SNP N288D decreases affinity for all ligands as a result of destabilized Ca2+ binding [8, 9]. Together, the SNPs skew ligand specificity from sulfated glycans to GlcNAc, with an overall decreased binding to glycan structures [9]. These findings sparked the hypothesis that these common langerin SNPs could influence recognition of GlcNAc-containing structures on microorganisms, which can be present on wall teichoic acid of Gram-positive bacteria including Staphylococcus aureus [11]. The possible influence on microbial recognition was also proposed to drive selection for K313I as compensatory mutation for N288D, explaining the strict co-inheritance of the two SNPs [9].
We recently identified langerin as a receptor for β-GlcNAc modifications on S. aureus wall teichoic acid, which strongly influenced LC activation and skin inflammation in response to S. aureus [12]. Therefore, we aimed to identify the effects of the naturally occurring combination of SNPs N288D and K313I in human langerin on S. aureus binding and internalization. Overall, our data demonstrate that uptake of S. aureus by cells expressing the langerin double SNP variant was compromised. This indicates that in a proportion of the human population, the recognition and uptake of S. aureus by LCs may be affected, which can have important consequences for proper immune activation and S. aureus-associated disease.
Material and Methods
Bacterial Strains and Culture Conditions
S. aureus strains [12] (Table 1) were grown overnight at 37°C with agitation in 5 mL Todd-Hewitt broth (THB; Oxoid). For S. aureus Newman ΔspaΔsbi pCM29, THB was supplemented with 10 µg/mL chloramphenicol (Sigma Aldrich). Overnight S. aureuscultures were subcultured the next day in fresh THB and grown to an optical density at 600 nm (OD600nm) of 0.6–0.7, which corresponds to mid-exponential growth phase. S. pyogenes MGAS315 [12] (Table 1) was grown overnight at 37°C without agitation in 5 mL THB (Becton Dickinson) supplemented with 1% yeast extract (Oxoid).
Table 1.
| Strain | Source |
| S. aureus USA300 (NRS384, CC8) | NARSA strain collection |
| S. aureus USA300 Δ tarM (WTA α-GlcNAc deficient) | [32] |
| S. aureus Newman (CC8) | ATCC, cat. No. 13420 |
| S. aureus Newman Δ spaΔ sbi pCM29 (sGFP expressing) | [12] |
| S. aureus 82086 (CC398) | [32] |
| S. aureus PS66 (CC30) | Udo Bläsi, Vienna |
| S. aureus MW2 (CC1) | [33] |
| S. aureus Wood46 (CC97) | ATCC, cat. No. 10832 |
| S. aureus Mu50 (CC5) | [33] |
| S. aureus P68 (CC25) | Udo Bläsi, Vienna |
| S. aureus NRS184 (CC22) | NARSA strain collection |
| S. aureus JH1 (CC5) | [34] |
| S. pyogenes MGAS315 | [35] |
Production of Recombinant Langerin Extracellular Domains
The extracellular domains of truncated human langerin (residues 148–328) and SNP variants thereof were recombinantly expressed from codon-optimized constructs containing a C-terminal TEV cleavage site followed by a Strep-tag II cloned into pUC19 expression vectors, expressed in E. coli BL21 (DE3; Thermo Fisher), purified and labeled as described previously [3]. SNP variants N288D and K313I and the double SNP variant were created from the langerin wildtype pUC19 expression vector by site-directed mutagenesis using QuikChange Lightning (Agilent) and primer pairs pUC19 288D and pUC19 313I [12] (Table 2), according to the manufacturer's instructions.
Table 2.
Primers used in this study
| Primer name | Sequence (5 ‘–3 ‘) |
| hLangerin Fw | GAGCTAGCAGTATTAATTAACCACCATGACTGTGGAGAAGGAG |
| hLangerin-FLAG Rv | ACGTTTCTTTTCATTTGTAAGCGACCCTATGTCCCATCAGAACCGGACTACAAAGACGATGACGACAAGTGAGCATGCATCCTAACCGGTAC |
| N288D Fw | CCAGGTGAGCCCAACGATGCTGGGAACAATGAACACTG |
| N288D Rv | CATTGTTCCCAGCATcGTTGGGCTCACCTGGAATCCAG |
| K313I Rv | GTACCGGTTAGGATGCATGCTCACGGTTCTGATGGGACATAGGGTCGCTTACAAATGAAAAGAAACGTTATGTCACATGGGGCATCATTCCAG |
| pUC19 288D Fw | ATTCCGGGCGAACCGAACAATGCCGGTAACAATGAAC |
| pUC19 288D Rv | GTTCATTGTTACCGGCATTGTTCGGTTCGCCCGGAAT |
| pUC19 313I Fw | GCATGGAATGATGCTCCGTGCGACATCACGTTTCTGTTCA |
| pUC19 313I Rv | AAATGAACAGAAACGTGATGTCGCACGGAGCATCATTCCATGCCTG |
Langerin Binding Assay
Wildtype S. aureus strains, S. aureus USA300 ΔtarM and S. pyogenes MGAS315 were harvested by centrifugation (4,000 rpm, 8 min) and resuspended at OD600nm = 0.4 in TSM buffer (2.4 g/L Tris [Roche], 8.77 g/L NaCl [Sigma Aldrich], 294 mg/L CaCl2·2H2O [Merck], 294 mg/L MgCl2·6H2O [Merck], pH = 7.4) with 0.1% bovine serum albumin (BSA; Merck). Bacteria were incubated with 0–50 µg/mL recombinant langerin-FITC (wildtype or SNP variants) for 30 min at 37°C with agitation, washed once with TSM 1% BSA, fixed in 1% formaldehyde (Brunschwig Chemie), and analyzed by flow cytometry.
Cell Culture and Lentiviral Transduction
THP1 (TIB-202, ATCC) and SupT1 (CRL-1942, ATCC) cells were cultured in RPMI (Lonza) supplemented with 5% fetal bovine serum (Biowest), 1% GlutaMAX (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco) at 37°C with 5% CO2. CHO cells (CCL-61, ATCC) were cultured in DMEM (Lonza), using the same supplements and conditions.
A TrueORF sequence-validated cDNA clone of human CD207 (OriGene Technologies) was amplified by PCR using Phusion polymerase (Thermo Fisher) and dedicated primers with or without SNPs N288D and K313I and containing a C-terminal FLAG tag (IDT) [12] (Table 2). The PCR amplicons were cloned in a BIC-PGK-Zeo-T2a-mAmetrine;EF1A [13] construct by Gibson assembly (New England Bioscience) according to the manufacturer's instructions. The vectors encoding langerin wildtype, SNP variants, or empty vector (EV) controls were introduced into THP1, SupT1, and CHO cells by lentiviral transduction, as described previously [13]. Cellular expression of langerin was verified by antibody staining with anti-CD207 (DCGM4, Beckman Coulter) and anti-FLAG (F9291, Sigma) and measured by flow cytometry. For staining of total langerin expression levels, cells were first fixed in 2% formaldehyde and permeabilized using PBS + 0.1% saponin (Sigma) + 0.5% BSA.
Binding of S. aureus to Langerin Expression Cells
Cells expressing langerin wildtype, N288D, K313I, or double SNP variant, or EV controls were harvested: suspension THP1 and SupT1 cells were washed in PBS and resuspended in TSM + 0.1% BSA; adherent CHO cells were detached by incubation in PBS + 5 mM EDTA for 15 min, washed in PBS, and resuspended in TSM + 0.1% BSA. Cells were incubated withGFP-expressing S. aureus Newman Δspa Δsbi at bacteria-to-cell ratios ranging from 0 to 6 in TSM + 0.1% BSA for 30 min at 4°C. Cells were washed in TSM + 0.1% BSA once, fixed in 1% formaldehyde, and measured by flow cytometry.
Internalization of S. aureus by THP1-Langerin Cells
S. aureus USA300 ΔtarM was grown as described. To label the bacteria with pHrodo iFL Red STP Ester (Thermo Fisher), 5 × 108 bacteria were resuspended in 750 µL and incubated for 1 h at room temperature with 100 µM pHrodo in 100 mM sodium bicarbonate, pH 8.4, as described previously [14, 15]. Bacteria were washed repeatedly and resuspended at 1 × 108 CFU/mL in RPMI 0.1% BSA. THP1-EV, THP1-langerin wildtype, and THP1-langerin double SNP variant were incubated with pHrodo-labeled S. aureus at a bacteria-to-cell ratio of 12.5 at 37°C with 5% CO2. Incubation of bacteria in citrate buffer (pH 4) was used to verify pHrodo labeling. Fluorescence was measured every 15 min for 1 h by Clariostar (BMG Labtech) using the Alexa Fluor 555 filter set. Afterwards, the cells and bacteria were transferred to Lab-Tek II borosilicate coverglass chambers (Nunc), counterstained with 10 µg/mL sWGA-FITC (Vector Labs), and imaged by confocal laser scanning microscopy (SP5, Leica).
To measure internalization by bacterial depletion from the cellular membrane, S. aureus USA300 ΔtarM was grown as described and labeled with 0.5 mg/mL FITC (Sigma Aldrich) in PBS for 30 min at 4°C, washed repeatedly, and resuspended at 1 × 108 CFU/mL in RPMI 0.1% BSA. Suspension THP1 cells expressing langerin wildtype, SNP variants, or EV controls were incubated with FITC-labeled S. aureus at a bacteria-to-cell ratio of 12.5 at 4°C. Afterwards, cells were washed three times in RPMI + 0.1% BSA to wash away unbound bacteria and the samples were split. Half of the sample was kept at 4°C, while the other half was incubated at 37°C with 5% CO2 to allow phagocytosis of adhered bacteria. The samples at 37°C were transferred to ice after 30 min and cooled to stop the reaction. Extracellular membrane-bound bacteria in all samples were stained using 1 µg/mL biotin-conjugated chicken IgY anti-protein A (CPA-65B, Immunology Consultants Laboratory) and streptavidin-Alexa Fluor 647 (Jackson Immunoresearch) and measured by flow cytometry. We assessed the internalized fraction of bacteria by measuring the loss of Alexa Fluor 647 signal after internalization of bacteria at 37°C in the FITC-positive fraction of cells, as described by Rodriguez et al. [16]:
Finternalized = (AF647FITC+, 4°C − AF647FITC+, 37°C)/AF647FITC+, 4°C
Flow Cytometry
Flow cytometry was performed on FACSVerse (BD Biosciences). Per sample, 10,000 events within the set gate were collected. Data were analyzed using FlowJo 10 (FlowJo, LLC).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7.04 (GraphPad Software). Langerin concentration curves were tested by two-way ANOVA with Tukey's multiple comparisons test, bacterial binding to cell lines by one-way ANOVA with Dunnett's multiple comparisons test, pHrodo signal by repeated measures two-way ANOVA with Dunnett's multiple comparisons test, and bacterial internalization by ANOVA with Tukey's multiple comparisons test. Correlations between bacterial binding and langerin membrane expression were analyzed by linear regression. Only statistically significant differences are indicated(p ≤ 0.05).
Data Availability
The data that support these findings are available from the corresponding author upon request.
Results
Introduction of N288D in Recombinant Human Langerin Abrogates Interaction with S. aureus
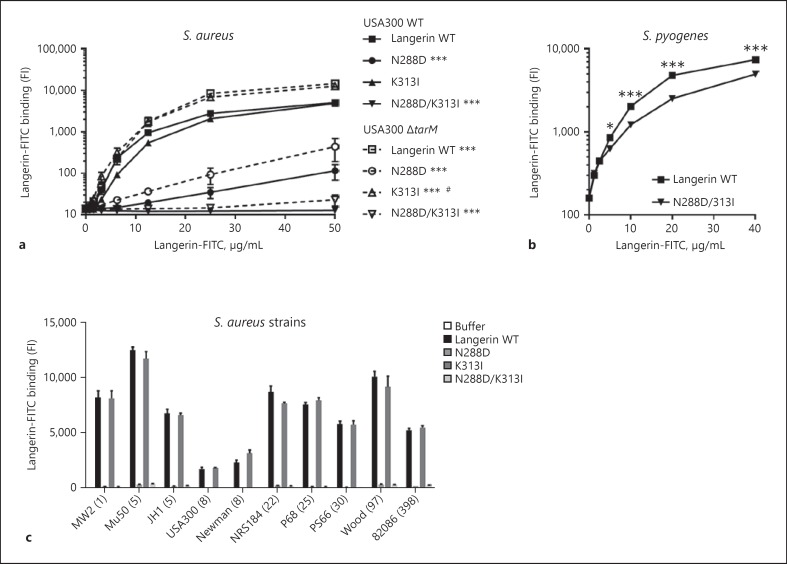
Feinberg et al. [9] showed that the ligand specificity of langerin K313I and the double SNP variant was altered compared to wildtype langerin, and hypothesized that this might affect interaction with Gram-positive pathogens. Since we recently elucidated the molecular interaction between langerin and S. aureus [12], we investigated whether this interaction would be affected by these common SNPs. We expressed and purified recombinant fluorescently labeled constructs of the extracellular domain of human langerin, containing SNPs N288D and K313I individually as well as together. Binding of recombinant langerin K313I to S. aureus USA300 wildtype was identical to the binding observed for recombinant wildtype langerin (Fig. 1a). In contrast, introduction of SNP N288D and especially the double SNP variant severely attenuated interaction with S. aureus (Fig. 1a). These results are generally consistent with the predictions by Feinberg et al. [9], except for the full loss of interaction of the double SNP variant in our experiments. Complete loss of binding could also result from problems with, e.g., protein misfolding or degradation. However, all langerin constructs had been purified in an identical manner using affinity purification on a mannan column, indicating that they are functional. In addition, the langerin double SNP variant was still able to interact with another Gram-positive pathogen, Streptococcus pyogenes (Fig. 1b). Therefore, the loss of S. aureus interaction with the recombinant langerin double SNP variant likely represents a specific S. aureus ligand binding defect.
Fig. 1.
Introduction of N288D in recombinant human langerin abrogates interaction with S. aureus. a, b Binding of FITC-labeled recombinant langerin wildtype (WT) and SNP variantsto S. aureus USA300 WT or ΔtarM (a) and S. pyogenes (b). c Binding of FITC-labeled recombinant langerin WT and SNP variants(12.5 µg/mL) to a panel of S. aureus strains from eight lineages (clonal complexes indicated in parentheses). Binding was assessed as the geometric mean fluorescence intensity. Data represent means ± standard error of the mean (SEM) of three independent experiments. Statistical differences in a are relative to the binding levels of langerin WT to the same strain, or between binding levels of langerin WT to USA300 WT or ΔtarM, at all concentrations from 12.5 to 50 µg/mL. *p< 0.05; ***p< 0.001. #25 and 50 µg/mL only.
We have previously demonstrated enhanced binding of langerin to tarM-deficient S. aureus [12]. Therefore, we examined whether this enhancement was also present in the interaction between the langerin SNP variants and S. aureus USA300 ΔtarM (Fig. 1a). Indeed, binding of the single langerin SNP variants N288D and K313I to S. aureus USA300 ΔtarM were both enhanced, whereas the double SNP variant was still unable to interact.
To test whether the observed binding pattern of langerin SNP variants relative to wildtype langerin was conserved, we tested binding of all four langerin constructs to a selection of 10 S. aureus strains from eight different clonal complexes (Fig. 1c). The binding levels of wildtype langerin were similar to previous observations [12]. However, binding of wildtype langerin and the K313I SNP variant were identical in all 10 strains. In addition, in all cases, the langerin N288D and double SNP variants displayed loss of interaction with S. aureus.
Langerin Surface Expression Is Affected by SNP N288D and Correlates with S. aureus Binding Capacity
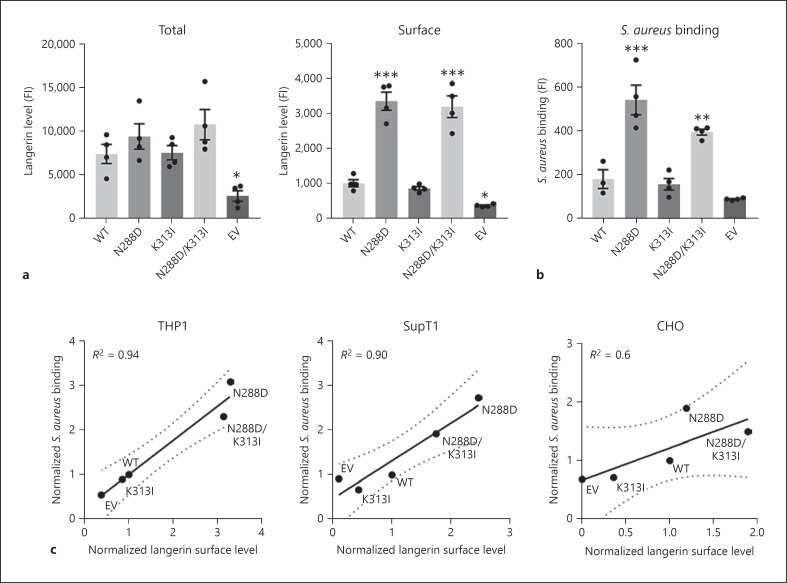
We next introduced FLAG-tagged wildtype langerin or SNP variants in THP1 cells to assess how N288D and K313I affected interaction with S. aureus in a cellular model. Total expression levels of langerin were similar in the different langerin variants as assessed by staining saponin-permeabilized cells with an anti-FLAG antibody (Fig. 2a, left). However, staining of langerin on the plasma membrane, using non-permeabilized cells, revealed striking differences in langerin surface expression between langerin variants (Fig. 2a, right). Introduction of N288D, either individually or in conjunction with K313I, caused a significant increase in langerin expression on the plasma membrane. As langerin is well-described to recycle continuously between intracellular and membrane compartments [5, 6, 17, 18], increased membrane availability of the N288D and double SNP variants of langerin is likely caused by altered recycling behavior of langerin upon SNP introduction. The altered membrane expression affected the interaction of langerin with S. aureus that we have described previously [12]; cell lines expressing langerin N288D or the double SNP variant, but not K313I on its own, showed higher binding of S. aureus compared to cells expressing wildtype langerin (Fig. 2b). Linear regression analysis of langerin surface expression and the capacity to bind S. aureus revealed a high positive correlation (R2 = 0.94), indicating that the bacterial binding capacity is highly dependent on the level of langerin expressed on the membrane (Fig. 2c).
Fig. 2.
Langerin surface expression is affected by SNP N288D and correlates with S. aureus binding capacity. a Expression levels of langerin in permeabilized (left) or non-permeabilized (right) THP1 cells expressing langerin wildtype (WT) and SNP variants or empty vector (EV) control cells. b Binding of GFP-expressing S. aureus Newman ΔspaΔsbi bacteria to the same cells at a bacteria-to-cell ratio of 3. Langerin expression was measured using an anti-FLAG antibody and assessed as the geometric mean fluorescence intensity. Means ± SEM of four independent experiments are shown. c Correlation between langerin surface expression and binding of GFP-expressing S. aureus Newman ΔspaΔsbi by THP1, SupT1, and CHO cells expressing langerin WT and SNP variants or EV controls at bacteria-to-cell ratios of 3 (THP1 and SupT1) or 6 (CHO). Data are shown as the mean values of two (SupT1 or CHO) or three (THP1) independent experiments, normalized to the values of the respective cells expressing WT langerin, and were analyzed by linear regression. *p < 0.05; **p < 0.01; ***p < 0.001.
To exclude that this effect was caused by unique behavior of langerin in THP1 cells, we repeated this experiment in two additional cell lines, suspension SupT1 cells and adherent CHO cells. In both cases, similar results were obtained, with high and moderate correlations between langerin membrane expression and S. aureus binding capacity, respectively (R2 = 0.90 in SupT1; R2 = 0.60 in CHO). We therefore conclude that S. aureus binding is highly dependent on the amount of langerin expressed on the membrane, irrespective of the introduced SNPs. Surprisingly, the SNPs did not directly influence the bacterial recognition but only did so indirectly, by altering langerin surface expression.
The Naturally Occurring Double SNP Variant in Langerin Is Defective in S. aureus Internalization
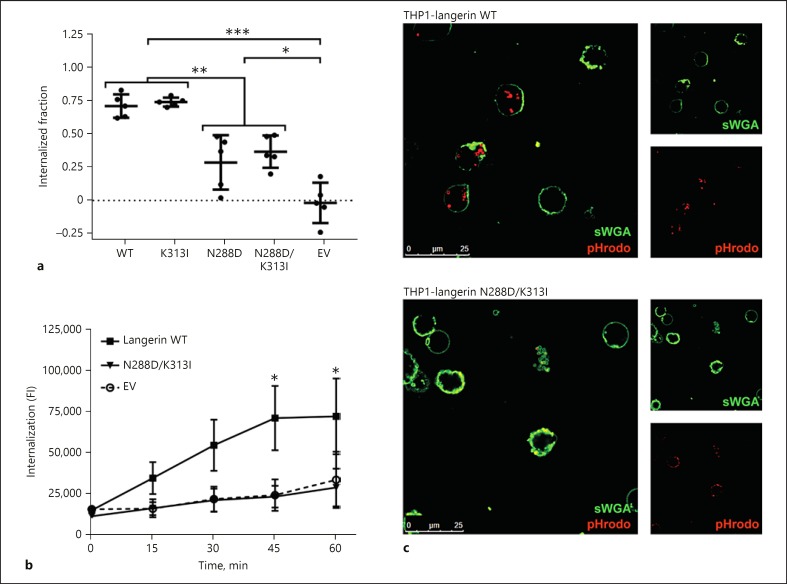
Langerin is a recycling receptor that internalizes ligands and transports them to intracellular compartments for further processing [5, 6]. As cells expressing the naturally occurring langerin double SNP variant bound more S. aureus as a consequence of altered membrane expression, we determined how this affected ligand internalization. FITC-labeled S. aureus were incubated with THP1-langerin cells at 4°C to allow bacterial binding. We used the stable USA300 ΔtarM mutant for this, as this strain behaved similarly to the wildtype strain for interaction with langerin SNP variants, but displays uniform and invariable WTA glycosylation with β-1,4-GlcNAc. Next, the samples were split and either kept at 4°C or transferred to 37°C to allow cellular uptake. Afterwards, all samples were stained using an anti-protein A antibody, which allows discrimination between adhered (stained) or internalized (non-stained) S. aureus in the split samples of 4 and 37°C, respectively [16]. Using this method, we observed that THP1 cells expressing wildtype or K313I langerin internalized approximately 75% of adhered bacteria (Fig. 3a). In contrast, the N288D and double SNP variants internalized only half of that amount (Fig. 3a).
Fig. 3.
The naturally occurring double SNP variant in langerin is defective in S. aureus internalization. a Fraction of S. aureus USA300 ΔtarM that is internalized by THP1-langerin wildtype (WT), N288D, K313I, N288D/K313I, or empty vector (EV) controls. Means ± SEM of five independent experiments are shown. b, c Internalization of pHrodo-labeled S. aureus USA300 ΔtarM by THP1-langerin WT, N288D/K313I, or EV control at a bacteria-to-cell ratio of 12.5. Internalization was assessed by mean fluorescence intensity using flow cytometry (b) and confocal microscopy (c). Data are shown as means ± SEM of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
Wildtype langerin is trafficked to early and late endosomal compartments, which are increasingly acidified [4, 5, 7]. To test whether the relatively low numbers of S. aureus internalized by cells expressing the langerin double SNP variant were still trafficked to these acidified compartments, we labeled S. aureus with the pH-sensitive dye pHrodo, which fluoresces at low pH. With this experimental setup, we observed a clear time-dependent increase in fluorescence after incubation of pHrodo-labeled S. aureus with THP1 cells expressing wildtype langerin but not EV control cells (Fig. 3b). This was complemented by confocal microscopy, confirming that pHrodo-fluorescent S. aureus was only observed in THP1 cells expressing wildtype langerin (Fig. 3c). Therefore, wildtype langerin expressed on THP1 cells is fully functional and internalized S. aureus end up in acidified endosomal compartments. In contrast, there was no increase in pHrodo signal in THP1 cells expressing the langerin double SNP variant, similar to EV controls (Fig. 3b, c). Overall, these data indicate that the naturally-occurring double SNP langerin variant displays increased surface expression, which enables increased binding of S. aureus. However, subsequent uptake and intracellular routing are highly inefficient.
Discussion
Previous studies hypothesized that the interaction between the human pattern-recognition receptor langerin and microorganisms could be affected by two co-inheriting polymorphisms in the carbohydrate recognition domain of langerin [1, 8, 9]. We demonstrate here that these SNPs indeed result in compromised uptake of S. aureus by langerin-expressing cells.
Using recombinant soluble constructs of langerin with and without these variants and S. aureus as a relevant GlcNAc-expressing human pathogen, we did not observe enhanced S. aureus binding by langerin K313I. This was surprising, since results from glycan array analysis indicated that SNP K313I enhanced detection of terminal GlcNAc residues [9]. In contrast, the langerin N288D SNP variant showed significant loss of S. aureus binding, which corresponds to observations by Feinberg et al. [9] that this SNP has reduced affinity for all langerin ligands. Surprisingly, the additional introduction of SNP K313I completely abrogated interaction with S. aureus, which counters the predictions based on glycan array analysis that K313I would partially compensate for the N288D affinity reduction [9]. There are two differences between our study and the study by Feinberg et al. [9] that are relevant to mention here. Firstly, the arrays used by Feinberg et al. [9] only contained mammalian glycan structures and could therefore only indicate general binding specificities for families of glycans, such as terminal sulfated or terminal GlcNAcylated structures. Binding to bacterial structures was not assessed [9]. Secondly, Feinberg et al. [9] used the monomeric carbohydrate recognition domain of langerin for this analysis, while we used trimeric constructs that resembles the receptor configuration as expressed by cells. Monomeric and trimeric langerin constructs are known to have different ligand binding properties, which could therefore have contributed to the discrepancies between our results [9, 19].
As a complementary approach to recombinant proteins, we introduced the langerin SNP variants in THP1 cells to test langerin-dependent S. aureus binding in a more biologically relevant model system. Expression of langerin N288D, with or without SNP K313I, on THP1 cells caused a notable increase of langerin expression on the plasma membrane. This is likely a specific effect caused by the alteration of the carbohydrate recognition domain of langerin and not an artifact caused by THP1 cells or the transduction method, as we reproduced this observation in two additional cell lines. A similar observation was reported previously for the single langerin N288D variant expressed on fibroblasts, which also displayed increased surface expression [8]. The enhanced membrane localization is potentially caused by diminished endosomal recycling of the langerin N288D variant. Due to increased surface expression of this receptor variant compared to wildtype langerin, more S. aureus bacteria adhered to the cells. In contrast, S. aureus internalization was significantly compromised in cells expressing the langerin N288D variant.
Our three main observations initially appear conflicting: on the one hand, we observed loss of binding of the recombinant langerin N288D and double SNP variant to S. aureus as well as compromised uptake by cells expressing these variants, while on the other hand, these same cells displayed higher bacterial binding. The discrepancy between the loss of binding of the recombinant N288D langerin variants (Fig. 1a) and increased bacterial binding by cells expressing these variants (Fig. 2b, c) can be explained by increased avidity in the cell-based model. When expressed on the cell, multiple receptors can cluster and engage the same bacterial particle, while such clustering is not possible for the soluble recombinant langerin constructs. The inability of cells expressing langerin containing SNP N288D to internalize S. aureus, while still being able to efficiently bind bacteria, is possibly the result of different affinity/avidity requirements for binding and phagocytosis. In the generally accepted “zipper” model of phagocytosis and endocytosis, successive zipper-like receptor-ligand interactions drive phagocytosis and endocytosis by increasingly enclosing the engulfed object [20, 21]. This process relies on sufficient affinity between the receptor and its ligand [22, 23]. Possibly, the affinity of N288D-containing langerin variants for S. aureus are still sufficient for binding of the bacteria but insufficient to trigger proper engulfment, as a result of the destabilized Ca2+ coordination that this variant displays [4]. So far, we have shown decreased internalization of S. aureus by the langerin N288D and double SNP variants, but it will be important to extrapolate this to other ligands, either expressed by other microorganisms or expressed in purified form.
A limitation of the use of cell lines overexpressing the langerin SNP variants is that the processes of engulfment and antigen processing might be different from how these processes occur in primary LCs. However, previous studies have demonstrated that the introduction of langerin in other cellular models results in the formation of Birbeck granules [5, 17, 24], the hallmark organelles of LCs. Although this suggests that the langerin-dependent processes of engulfment and antigen processing could function similarly in overexpression models, the effect of the investigated langerin SNPs in isolated LCs or in vivo is an important next step.
In this study, we have exclusively investigated homozygous langerin SNP variants. Although 0.3–4.8% of the world's population is homozygous for these variants, the majority of people will be heterozygous. Importantly, langerin is expressed as a trimeric receptor. Assuming equal expression of both alleles, heterozygotes will express a range of trimeric langerin receptors, from wildtype homotrimers (1/8 of all trimers) to heterotrimers with one or two wildtype monomers (3/8 each) to SNP variant homotrimers (1/8) [9]. Prediction of the phenotype of these heterozygous alleles is difficult. However, based on the loss of function of the langerin double SNP variant, a dose-dependent effect is one of the possibilities. In this case, langerin trimers would become less functional with incorporation of more SNP variant monomers, yielding trimers with a range of functionalities. Another possibility is a dominant-negative effect of the SNP variant over the wildtype form. Indeed, this situation occurred in an individual that was heterozygous for a particular de novo mutation in langerin (W264R). This healthy individual had fully disrupted langerin function and lacked Birbeck granule formation, leading to the conclusion that the W264R mutation had a dominant negative effect [25, 26]. Yet, experimental data on heterozygously expressed langerin SNP variants is currently lacking and will be necessary to reveal the effect that these SNPs have in heterozygous individuals.
The effects of the langerin double SNP variant on human health and disease is currently unclear. In case of S. aureus, we have previously shown that human LCs play a central role in the orchestration of the immune response against S. aureus in a langerin-dependent manner [12]. Reduced internalization by langerin and consequently diminished antigen presentation or skewed immune activation by LCs could therefore have a major impact on the health of skin and mucosa. This is further supported by the increased minor allele frequency of the double SNP variant in the African population (21.8%) compared to the European population (6.2%) and the observation that people of African descent have a lower risk for S. aureus colonization (26.8 vs. 32.9%) [27] but higher incidence of invasive S. aureus disease than European-descended populations (66.5 vs. 27.7 per 100,000 individuals) (online suppl. Fig. S1) [28, 29]. This suggests a link between the genetic background of the host and the immune response against S. aureus, which may be partially mediated by variation in langerin. Possibly, LCs expressing variant langerin are attenuated in their skin barrier gatekeeper function, thereby allowing S. aureus to penetrate to deeper tissues and cause invasive disease.
Finally, we have identified S. pyogenes as a new interaction partner of langerin, which was not affected by double SNP variant. Similar to S. aureus, S. pyogenes is a human commensal and a frequent cause of skin infections [30]. Interestingly, the interaction between LCs and S. pyogenes has been reported previously, although the molecular interaction was not clarified [31]. Possibly, langerin is one of the LC receptors involved in this interaction and warrants further investigation.
In conclusion, we have shown that, contrary to predictions based on glycan array data, the co-inheriting langerin SNPs N288D and K313I cause loss of interaction between langerin and the human pathogen S. aureus. In addition, the SNPs affected the cellular localization of langerin and compromised uptake of S. aureus. This demonstrates that binding profiles from glycan array data cannot always be directly translated to a biological situation. This complicates predictions of the phenotypic effects of genetic variation in recycling pattern-recognition receptors and is therefore an important factor to consider in similar studies.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by Vidi grant 91713303 from The Netherlands Organization for Scientific Research (NWO) to N.M.v.S. and DFG grant RA1944/2-1 to C.R. Funding sources have had no role in the preparation of the data or the manuscript.
Author Contributions
R.v.D. and N.M.v.S. planned the experiments. R.v.D. and F.F.F. performed the experiments. F.F.F. and C.R. supplied the langerin-FITC constructs. R.v.D. and N.M.v.S. wrote the manuscript. C.R. and J.A.G.v.S. provided critical feedback.
Supplementary Material
Supplementary data
References
- 1.Tateno H, Ohnishi K, Yabe R, Hayatsu N, Sato T, Takeya M, et al. Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J Biol Chem. 2010 Feb;285((9)):6390–400. doi: 10.1074/jbc.M109.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg H, Taylor ME, Razi N, McBride R, Knirel YA, Graham SA, et al. Structural basis for langerin recognition of diverse pathogen and mammalian glycans through a single binding site. J Mol Biol. 2011 Jan;405((4)):1027–39. doi: 10.1016/j.jmb.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanske J, Schulze J, Aretz J, McBride R, Loll B, Schmidt H, et al. Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent. J Biol Chem. 2017 Jan;292((3)):862–71. doi: 10.1074/jbc.M116.751750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanske J, Aleksić S, Ballaschk M, Jurk M, Shanina E, Beerbaum M, et al. Intradomain Allosteric Network Modulates Calcium Affinity of the C-Type Lectin Receptor Langerin. J Am Chem Soc. 2016 Sep;138((37)):12176–86. doi: 10.1021/jacs.6b05458. [DOI] [PubMed] [Google Scholar]
- 5.Mc Dermott R, Ziylan U, Spehner D, Bausinger H, Lipsker D, Mommaas M, et al. Birbeck granules are subdomains of endosomal recycling compartment in human epidermal Langerhans cells, which form where Langerin accumulates. Mol Biol Cell. 2002 Jan;13((1)):317–35. doi: 10.1091/mbc.01-06-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valladeau J, Ravel O, Dezutter-Dambuyant C, Moore K, Kleijmeer M, Liu Y, et al. Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity. 2000 Jan;12((1)):71–81. doi: 10.1016/s1074-7613(00)80160-0. [DOI] [PubMed] [Google Scholar]
- 7.Stambach NS, Taylor ME. Characterization of carbohydrate recognition by langerin, a C-type lectin of Langerhans cells. Glycobiology. 2003 May;13((5)):401–10. doi: 10.1093/glycob/cwg045. [DOI] [PubMed] [Google Scholar]
- 8.Ward EM, Stambach NS, Drickamer K, Taylor ME. Polymorphisms in human langerin affect stability and sugar binding activity. J Biol Chem. 2006 Jun;281((22)):15450–6. doi: 10.1074/jbc.M511502200. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg H, Rowntree TJ, Tan SL, Drickamer K, Weis WI, Taylor ME. Common polymorphisms in human langerin change specificity for glycan ligands. J Biol Chem. 2013 Dec;288((52)):36762–71. doi: 10.1074/jbc.M113.528000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015 Oct;526((7571)):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winstel V, Xia G, Peschel A. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol. 2014 May;304((3-4)):215–21. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 12.van Dalen R, De La Cruz Diaz JS, Rumpret M, Fuchsberger FF, van Teijlingen NH, Hanske J, et al. Langerhans cells sense Staphylococcus aureus wall teichoic acid through langerin to induce inflammatory responses. mBio. 2019 doi: 10.1128/mBio.00330-19. Provisionally. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Weijer ML, van Muijlwijk GH, Visser LJ, Costa AI, Wiertz EJ, Lebbink RJ. The E3 Ubiquitin Ligase TMEM129 Is a Tri-Spanning Transmembrane Protein. Viruses. 2016 Nov;8((11)):E309. doi: 10.3390/v8110309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avraham R, Haseley N, Fan A, Bloom-Ackermann Z, Livny J, Hung DT. A highly multiplexed and sensitive RNA-seq protocol for simultaneous analysis of host and pathogen transcriptomes. Nat Protoc. 2016 Aug;11((8)):1477–91. doi: 10.1038/nprot.2016.090. [DOI] [PubMed] [Google Scholar]
- 15.Lenzo JC, O'Brien-Simpson NM, Cecil J, Holden JA, Reynolds EC. Determination of Active Phagocytosis of Unopsonized Porphyromonas gingivalis by Macrophages and Neutrophils Using the pH-Sensitive Fluorescent Dye pHrodo. Infect Immun. 2016 May;84((6)):1753–60. doi: 10.1128/IAI.01482-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez ME, Van der Pol WL, Van de Winkel JG. Flow cytometry-based phagocytosis assay for sensitive detection of opsonic activity of pneumococcal capsular polysaccharide antibodies in human sera. J Immunol Methods. 2001 Jun;252((1-2)):33–44. doi: 10.1016/s0022-1759(01)00329-5. [DOI] [PubMed] [Google Scholar]
- 17.McDermott R, Bausinger H, Fricker D, Spehner D, Proamer F, Lipsker D, et al. Reproduction of Langerin/CD207 traffic and Birbeck granule formation in a human cell line model. J Invest Dermatol. 2004 Jul;123((1)):72–7. doi: 10.1111/j.0022-202X.2004.22728.x. [DOI] [PubMed] [Google Scholar]
- 18.Chabrol E, Thépaut M, Dezutter-Dambuyant C, Vivès C, Marcoux J, Kahn R, et al. Alteration of the langerin oligomerization state affects Birbeck granule formation. Biophys J. 2015 Feb;108((3)):666–77. doi: 10.1016/j.bpj.2014.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muñoz-García JC, Chabrol E, Vivès RR, Thomas A, de Paz JL, Rojo J, et al. Langerin-heparin interaction: two binding sites for small and large ligands as revealed by a combination of NMR spectroscopy and cross-linking mapping experiments. J Am Chem Soc. 2015 Apr;137((12)):4100–10. doi: 10.1021/ja511529x. [DOI] [PubMed] [Google Scholar]
- 20.Griffin FM, Jr, Griffin JA, Leider JE, Silverstein SC. Studies on the mechanism of phagocytosis. I. Requirements for circumferential attachment of particle-bound ligands to specific receptors on the macrophage plasma membrane. J Exp Med. 1975 Nov;142((5)):1263–82. doi: 10.1084/jem.142.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin FM, Jr, Griffin JA, Silverstein SC. Studies on the mechanism of phagocytosis. II. The interaction of macrophages with anti-immunoglobulin IgG-coated bone marrow-derived lymphocytes. J Exp Med. 1976 Sep;144((3)):788–809. doi: 10.1084/jem.144.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci USA. 2005 Jul;102((27)):9469–74. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herant M, Heinrich V, Dembo M. Mechanics of neutrophil phagocytosis: experiments and quantitative models. J Cell Sci. 2006 May;119((Pt 9)):1903–13. doi: 10.1242/jcs.02876. [DOI] [PubMed] [Google Scholar]
- 24.Thépaut M, Valladeau J, Nurisso A, Kahn R, Arnou B, Vivès C, et al. Structural studies of langerin and Birbeck granule: a macromolecular organization model. Biochemistry. 2009 Mar;48((12)):2684–98. doi: 10.1021/bi802151w. [DOI] [PubMed] [Google Scholar]
- 25.Mommaas M, Mulder A, Vermeer BJ, Koning F. Functional human epidermal Langerhans cells that lack Birbeck granules. J Invest Dermatol. 1994 Dec;103((6)):807–10. doi: 10.1111/1523-1747.ep12413456. [DOI] [PubMed] [Google Scholar]
- 26.Verdijk P, Dijkman R, Plasmeijer EI, Mulder AA, Zoutman WH, Mieke Mommaas A, et al. A lack of Birbeck granules in Langerhans cells is associated with a naturally occurring point mutation in the human Langerin gene. J Invest Dermatol. 2005 Apr;124((4)):714–7. doi: 10.1111/j.0022-202X.2005.23645.x. [DOI] [PubMed] [Google Scholar]
- 27.Graham PL, 3rd, Lin SX, Larson EL. A U.S. A U.S. population-based survey of Staphylococcus aureus colonization. Ann Intern Med. 2006 Mar;144((5)):318–25. doi: 10.7326/0003-4819-144-5-200603070-00006. [DOI] [PubMed] [Google Scholar]
- 28.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007 Oct;298((15)):1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 29.Kallen AJ, Mu Y, Bulens S, Reingold A, Petit S, Gershman K, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators of the Emerging Infections Program Health care-associated invasive MRSA infections, 2005-2008. JAMA. 2010 Aug;304((6)):641–8. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 30.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, et al. Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev. 2014 Apr;27((2)):264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed WP, Metzler C, Albright E. Streptococcal adherence to Langerhans cells: a possible step in the pathogenesis of streptococcal pharyngitis. Clin Immunol Immunopathol. 1994 Jan;70((1)):28–31. doi: 10.1006/clin.1994.1006. [DOI] [PubMed] [Google Scholar]
- 32.Winstel V, Kühner P, Salomon F, Larsen J, Skov R, Hoffmann W, et al. Wall Teichoic Acid Glycosylation Governs Staphylococcus aureus Nasal Colonization. MBio. 2015 Jun;6((4)):e00632. doi: 10.1128/mBio.00632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CDC From the Centers for Disease Control and Prevention From the Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—minnesota and North Dakota, 1997-1999. JAMA. 1999 Sep;282((12)):1123–5. [PubMed] [Google Scholar]
- 34.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci USA. 2007 May;104((22)):9451–6. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002 Jul;99((15)):10078–83. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Data Availability Statement
The data that support these findings are available from the corresponding author upon request.