Abstract
Tox21 and ToxCast are high-throughput in vitro screening programs coordinated by the U.S. National Toxicology Program and the U.S. Environmental Protection Agency, respectively, with the goal of forecasting biological effects in vivo based on bioactivity profiling. The present study investigated whether mechanistic insights in the biological targets of food-relevant chemicals can be obtained from ToxCast results when the chemicals are grouped according to structural similarity. Starting from the 556 direct additives that have been identified in the ToxCast database by Karmaus et al. [Karmaus, A. L., Trautman, T. D., Krishan, M., Filer, D. L., and Fix, L. A. (2017). Curation of food-relevant chemicals in ToxCast. Food Chem. Toxicol. 103, 174–182.], the results showed that, despite the limited number of assays in which the chemical groups have been tested, sufficient results are available within so-called “DNA binding” and “nuclear receptor” target families to profile the biological activities of the defined chemical groups for these targets. The most obvious activity identified was the estrogen receptor-mediated actions of the chemical group containing parabens and structurally related gallates, as well the chemical group containing genistein and daidzein (the latter 2 being particularly active toward estrogen receptor β as a potential health benefit). These group effects, as well as the biological activities of other chemical groups, were evaluated in a series of case studies. Overall, the results of the present study suggest that high-throughput screening data could add to the evidence considered for regulatory risk assessment of food chemicals and to the evaluation of desirable effects of nutrients and phytonutrients. The data will be particularly useful for providing mechanistic information and to fill data gaps with read-across.
Keywords: ToxCast, read-across, high-throughput in vitro screening, food chemicals, risk-benefit
Automatic high-throughput screening (HTS) of chemicals across a wide range of biological targets is an emerging practice in many chemical sectors. High-throughput screening plays a crucial role in the prioritization of chemicals based on toxicological mode of action as well as finding lead actives based on intended biological activity (Brunner et al., 2019; Hartman et al., 2018; Mayr and Fuerst, 2008; Olker et al., 2019). Within next-generation (non-animal) risk assessment strategies, HTS will be one of the key technologies to characterize the ability of chemicals to perturb biological pathways associated with an adverse outcome pathway (Villeneuve et al., 2019). Much effort has been devoted to the application of HTS to various sectors and regulatory environments and strategies to achieve a broader acceptance of HTS and computational approaches in regulatory decision making have recently been laid out by Thomas et al. (2019). However, little has been done to relate these approaches to the assessment of foods and food ingredients, which are often assumed to be harmless, although a variety of toxicological or beneficial biological effects can be elicited. The aim of the present study was to explore the potential of ToxCast HTS data to be integrated into regulatory safety assessment of food chemicals.
The Tox21 and ToxCast programs are high-throughput in vitro screening programs coordinated by the U.S. National Toxicology Program and the U.S. Environmental Protection Agency, respectively, with the goal to forecast biological effects in vivo, especially toxicity, based on bioactivity profiling (Kavlock et al., 2012). Tox21/ToxCast results (together referred to as ToxCast in this work) have been evaluated by several groups in various publications using clustering algorithms and self-organizing maps (Karmaus et al., 2016; Kleinstreuer et al., 2014), hierarchical clustering techniques (Sipes et al., 2013), or through links with chemical fingerprinting (Richard et al., 2016). Specific to food-relevant chemicals, Karmaus et al. (2016, 2017) identified and evaluated the activity patterns of 1211 food-use compounds within ToxCast, comprising 556 direct food additives, 371 food contact substances, and 543 pesticides.
A challenge with applying such nondirected, quantitative approaches on food-relevant chemicals is that an observed uneven coverage of chemical-endpoint combinations within this class of compounds leads to a significant bias in the results (ie, chemicals perceived to have a high biological activity are in reality those that have a broader test coverage). A second challenge of hierarchical clustering and self-organizing heatmaps is that they do not provide any direct mechanistic insights in the biological targets of a chemical relative to an adverse outcome pathway. The acquisition of such qualitative mechanistic insights is just as crucial to the consideration of ToxCast data in risk assessments of food chemicals and can be an important resource to evaluate nutrients and phytonutrients and their corresponding desirable effects.
The present study investigated whether insights in the biological targets of food-relevant chemicals can be obtained from the results of the ToxCast assays when the chemicals are grouped according to structural similarity (eg, homologous series), exploring those targets that are induced by multiple chemicals in the group. The current study focused only on the 556 direct food additives that have been identified by Karmaus et al. (2017) (chemicals that are, eg, added to foods to preserve, color and stabilize food as well as flavorings), and not on the 371 food contact substances and 543 pesticides that were identified by Karmaus et al. (2017). Both food contact substances and pesticides may lead to indirect exposures via food ingredients, but these compounds are not intended to be added to foods. The 556 direct food additives were supplemented with 7 chemicals from the original noncurated list of food-use chemicals published by Karmaus et al. (2016) to also include natural food constituents (safrole, quercetin, resveratrol, genistein, daidzein, and coumarin) as well as heptylparaben, a nonapproved food contact material that is structurally related to the approved methyl and ethyl parabens (EFSA, 2004), to give 563 reference compounds. The compounds within the dataset are clustered based on their chemical structural characteristics (eg, alcohol, aldehydes and carboxylic acids, and ketones) as well as their functional uses in food (eg, flavoring agents, nutrients, additives, and regulatory restricted). Whereas the clustering into structurally similar chemicals was used to explore the relationship between chemical homology and biological activity, the clustering into functional use categories related biological activities to current food uses. A method was set up that allows to scroll through the activities of the groups of structurally related chemicals toward different targets. Relevant biological targets of a chemical group are considered those toward which a high percentage of chemicals within a group are active. Overall, the results of the present study offer insights into the possible integration of HTS data in the safety and risk assessments of food chemical.
MATERIALS AND METHODS
Grouping of Chemicals
Grouping of the chemicals according to functional use classes
Chemical names and CAS numbers were obtained from the appendices of Karmaus et al. (2016, 2017). To obtain a link between the 563 selected direct food additives and their use in foods, particularly within the EU, the compounds were subdivided into different use categories. To this end, the CAS numbers were first matched with the European Union list of flavorings (Annex I of Regulation 1334/2008) using the R script provided https://git.wur.nl/Punt001/ilsi_toxcast.git (last accessed January 30, 2020). There were 449 compounds that matched and were categorized as EU flavorings. The majority of the remaining 114 compounds were manually categorized into “novel foods,” “nutrients,” “polyphenols,” “E-numbers” (subdivided into “sweeteners,” “antioxidants,” “preservatives,” “colors,” and “remaining E-numbers”), and flavoring oils (which were merged with the EU flavorings use class), based on an online search using particularly the EU food additives database (EU) (DG SANTE, 2011), EFSA’s OpenFoodTox database (Dorne et al., 2017; EFSA, 2017), and PubMed. The final 17 compounds that could not be linked to any known food use in the EU were categorized as “other.”
Grouping of the chemicals according to chemical structure
From the list of 563 compounds, 552 were found to correspond to discrete chemical entities with defined molecular structure. The simplified molecular-input line-entry specification (SMILES) strings of these compounds were extracted from the ToxCast Data Spreadsheets (U.S. EPA, 2018a). The remaining 11 entities correspond to mixtures (eg, peppermint oil, clover leaf oil, and polysorbate 80) and were either grouped together as structurally undefined or, in case of the flavoring oils, were assigned to the chemical group of the major constituent of oil. To this end, cornmint oil and peppermint oil are grouped in the same chemical group as menthol, whereas clove leaf oil is grouped with eugenol, anise oil with anethole, nutmeg oil with alpha-pinene, petitgrain oil with limonene, and cananga oil with beta-caryophyllene (Han et al., 2017; Jelen, 2012). Using ChemoTyper software (Molecular Networks, Erlangen, Germany) and the SMILES strings of the chemicals, the compounds were classified through application of chemical knowledge, focusing on shared structural features and, where applicable, with their known physiological roles (Mellor et al., 2019; Yang et al., 2015). During the course of this undertaking, a three-tier system of grouping was adopted, in which larger primary clusters (eg, alcohols) were further subdivided as appropriate into secondary (eg, alcohol and alkyl) and tertiary (eg, alcohol, alkyl, primary, and straight chain) groups. The final groupings can be found in Supplementary Information 1, along with additional information on the log P, log D, and the Henry’s law constants of the chemical (estimated with ACD/Labs software). In addition, an estimate of the mean similarity of the chemicals within each group was made using the ChemmineR (Cao et al., 2008) and fmcsR (Wang et al., 2013) packages in R, to calculate the maximum common substructures between the chemicals within a group and the Tanimoto coefficients based on these maximum common substructures. The average of the calculated Tanimoto coefficients (excluding the Tanimoto coefficient of the chemicals with themselves) is taken as marker for group similarity. The R codes for these calculations have been made available at https://git.wur.nl/Punt001/ilsi_toxcast.git (last accessed January 30, 2020).
The grouping according to the functional use classes (the Grouping of the Chemicals According to Functional Use Classes section) and the chemical groups were combined in a so-called circle pack plot using the igraph and ggraph libraries in R (Csardi and Nepusz, 2006; Pedersen, 2017). The R code for the circle pack graphic of the chemical groups has been made available at https://git.wur.nl/Punt001/ilsi_toxcast.git (last accessed January 30, 2020).
ToxCast Data
The ToxCast activity data of the chemicals were derived from the data spreadsheet “ac50_Matrix_180918.csv” (U.S. EPA, 2018a) containing results from 1410 different assays. These crude ToxCast data for the 563 individual chemicals provided several positive hit-calls, ie, assays for which the concentration producing 50% of maximum activity (AC50), with a value < 1 000 000 (the value used to indicate negative results), could be derived. Starting from this dataset, all assays were excluded that did not directly relate to a specific biological activity. These included all assays for which the “assay_function_type” was “background control,” the “assay_design_type” was either “background reporter” or “viability reporter,” the “intended_target_family” was “background measurement,” and the “biological_process_target” was “cell death,” “cell proliferation,” or “cytotoxicity.” These results are already taken into account during data analysis steps, eg, through the production of Z-scores, and do not represent a specific activity of toxicological interest. In addition, only assays relevant to humans were extracted by setting “species” to “human.” The remaining assay endpoints were annotated according to the targets (eg, ESR1 and ESR2, being the estrogen receptor (ER) alpha and beta, respectively), target family (eg, nuclear assays, DNA binding, or cytokines), and target subfamily (eg, nuclear assays-steroidal, nuclear assays-nonsteroidal, nuclear assays-orphan) parameters as provided in the “Assay_Summary_180918.csv” file (U.S. EPA, 2018a). By filtering out the assays that did not relate to a specific biological activity, 559 of the 1410 ToxCast assays were excluded, leaving 851 assays in the dataset.
Z-scores are utilized within the ToxCast dataset to filter out the AC50 results that were potentially affected by nonspecific effects such as cytotoxicity. Z-scores represent the number of standard deviations (on a standardized scale) that separate the potency for the specified assay from the median potency of a range of cytotoxicity assays (Houck et al., 2017; Judson et al., 2015). Assay results with a large Z-score are more likely to reflect a target specific effect that is not caused by cell stress or cytotoxicity-related processes (Kleinstreuer et al., 2014). For the present study, the available Z-scores from the “zscore_Matrix_180918.csv” (U.S. EPA, 2018a) file were used. These Z-scores were derived for chemicals with 2 or more positive responses in cytotoxicity assays. AC50 results with Z-scores < 3 were removed from the dataset as potential activity data that were affected by nonspecific effects like cytotoxicity (Judson et al., 2015). For 261 compounds out of the 563 food-relevant chemicals, this filtering based on Z-score < 3 resulted in a more than 75% reduction in positive hit-calls. For example, retinol expressed activity in 101 out of the 851 evaluated ToxCast assays, but 84 (83%) of these assay results had Z-scores < 3. The mean AC50 for retinol in the 84 assays with Z-scores < 3 was 57 ± 41 µM, whereas the mean AC50 was 6.5 ± 4.3 µM for the 17 assays with Z-score > 3. A similar result can be seen for quercetin that was active in 91 assays of which 84 assay results had Z-scores < 3. The mean AC50 value in the assays with Z-scores < 3 was 31 ± 33 µM, whereas this was 2.8 ± 2.0 µM for the assays that had Z-scores > 3. These results suggest that the specificity increases after filtering for Z-scores. On average, for all chemicals, the mean AC50 values were 10-fold lower for the results with Z-scores > 3 compared with the result with Z-scores < 3.
Warning signs (“flags”) are used in ToxCast data files to provide an indication of any unwanted influence of the method of data collection or automatic data processing on the obtained AC50 values. Possible flags include (1) “only highest concentration above baseline, active,” (2) “only one concentration above baseline, active,” (3) “multiple points above baseline, inactive,” (4) “noisy data,” (5) “borderline active,” (6) “borderline inactive,” (7) “gain AC50 < lowest concentration & loss AC50 < mean concentration,” (8) “hit-call potentially confounded by overfitting,” and (9) “biochemical assay with < 50% efficacy.” Flagged results were not filtered out from the ToxCast dataset in the present study but were considered in the different case studies to interpret the relevance of certain assays. The available flags were derived from the “AllResults_flags_180918.csv” file (U.S. EPA, 2018a). For more information please, see the U.S. EPA documentation on the data analysis steps (U.S. EPA, 2018b).
In Supplementary Information 2, the background information on the different ToxCast assays is provided, including, per assay, the number of food-relevant chemicals that were tested, the fraction of the tested food-relevant chemicals that tested positive (AC50 value < 1,000,000), and the fraction of the tested food-relevant chemicals that contained flags (specified for each of the different flags). In addition, the targets, target families, and target subfamilies to which the assays belong are provided in Supplementary Information 2.
Defining the Biological Activities of the Chemical Groups Toward Different ToxCast Targets
Within the Assay_Summary_180918.csv file, the intended biological target of each assay is defined under “technological_target_official_symbol.” For each of the tertiary chemical groups as identified with ChemoTyper (see section Grouping of Chemicals and Table 1 of the Results section), the biological activities toward the different biological targets were defined by calculating the percentage of chemicals (per tertiary chemical group) that were active in that assay of that target. To this end, the number of chemicals per tertiary chemical group that were tested in the assays of a specific target and the number of chemicals for which AC50 values (ie, the chemical tested positively) were defined, based on which the percentage activity could be calculated. For this evaluation, only those assays in which at least 3 chemicals of a group had been tested were considered. As a result, all chemical groups with < 3 chemicals were removed for further analysis. For larger groups, this means that only chemical group-assay endpoint combinations with an n > 3 were included in the dataset.
Table 1.
Defined Chemical Groups
| Primary Groupsa | Secondary Groupsa | Tertiary Groupsa (Including the Mean Tanimoto Coefficient)b |
|---|---|---|
| Alcohol | Alkenyl; Alkyl; Hydroxybenzene; Phenylalkanol | 1. Alkenyl, primary (0.44); 2. Alkenyl, secondary (0.42); 3. Alkenyl, tertiary (0.7); 4. Alkyl, diol (0.42); 5. Alkyl, primary, branched chain (0.58); 6. Alkyl, primary, straight chain (0.63); 7. Alkyl, secondary, cyclic (0.68); 8. Sugar alcohol (0.59); 9. Alkoxy phenol ether, substituted (0.57); 10. Hydroxy benzyl ketones (0.71); 11. Phenol, aliphatic substituted (0.67); 12. Phenolic aldehydes (0.73); 13. Salicyclic acid and derivatives (0.68); 14. Phenalkyl/alkenyl (0.60) |
| Aldehyde | Alkenyl; Alkyl; Aromatic | 15. Alkenyl, acyclic (0.51); 16. Alkyl, branched chain (0.50); 17. Alkyl, straight chain (0.76); 18. Benzaldehyde derivatives (0.64); 19. Phenylalkenyl (0.67); 20. Phenylalkyl (0.66) |
| Carboxylic acid | Alkenyl; Alkyl; Amino acids and derivatives; Aryl; Hydroxy acid; Keto acid; Polycarboxylic acid | 21. Alkenyl, branched chain (0.48); 22. Alkenyl, straight chain (0.37); 23. Alkyl, branched chain (0.72); 24. Alkyl, straight chain (0.59); 25. Amino acids and derivatives (0.26); 26. Benzoic acid (0.90); 27. Phenylaliphatic carboxylic acid (0.41); 28. Lactic acids; 29. Keto acid (0.53); 30. Polycarboxylic acid, aliphatic (0.52) |
| Dyes | Azo; Triarylmethane | 31. Azo (0.38); 32. Triarylmethane (0.76) |
| Ester | Aliphatic alcohol diester/triester; Alkenyl alcohol; Alkyl alcohol; Aromatic acid ester; Aromatic alcohol; Lactone | 33. Aliphatic alcohol diester/triester (0.37); 34. (3Z)-Hex-3-en-1-yl alcohol; 35. Allyl alcohol; 36. Citronellol; 37. Geraniol; 38. Linalool; 39. Branched-chain alcohol, aliphatic (0.54); 40. Branched-chain alcohol, aryl (0.69); 41. Butanol (0.51); 42. Ethanol, aliphatic (0.58); 43. Ethanol, aryl (0.55); 44. Hexanol (0.76); 45. Isobutanol (0.79); 46. Methanol, aliphatic (0.43); 47. Methanol, aryl (0.79); 48. Pentanol (0.79); 49. Propanol (0.67); 50. Straight chain (7+) alcohol, aliphatic (0.80); 51. Straight chain (7+) alcohol, aryl (0.80); 52. 2-Aminobenzoate (0.62); 53. Benzoate (0.69); 54. Cinnamate (0.57); 55. Paraben-gallate (0.72); 56. Phenylacetate (0.73); 57. Salicylate (0.67); 58. 3-Phenylpropen-2-enyl alcohol (0.76); 59. Anisyl (0.90); 60. Benzyl alcohol, aliphatic (0.85); 61. Benzyl alcohol, aryl (0.83); 62. Phenylethyl alcohol, aliphatic (0.77); 63. Phenylethyl alcohol, aryl (0.77); 64. Ascorbic acid and derivatives (0.41); 65. Lactone, five membered (0.66); 66. Lactone, six membered (0.64) |
| Ether | Alkenyl; Alkyl; Aromatic | 67. Alkenyl, acyclic (0.59); 68. Alkyl, cyclic (0.39); 69. Aryl methoxy (0.67); 70. Aryl methoxy, aliphatic substituted(0.65) |
| Heterocycles and polycycles | Hydrocarbon polycycles; Nitrogen heterocycles; Oxygen heterocycles; Sulfur-nitrogen heterocycles | 71. Bicycloheptanes and derivatives (0.67); 72. Biphenyl derivatives (0.90); 73. Naphthalene derivatives (0.50); 74. Pyrazine derivatives (0.57); 75. Pyridine derivatives (0.48); 76. Quinoline derivatives (0.56); 77. Benzodiazole (0.48); 78. Furan derivatives (0.49); 79. Pyranone (0.45); 80. Thiazole and thiazoline (0.28) |
| Hydrocarbon | Terpene | 81. Terpene (0.57) |
| Inorganic | Inorganic | 82. Inorganic (0.08) |
| Ketone | Alkenyl; Alkyl; Aryl; Jasmone derivatives | 83. Alkenyl, acyclic (0.55); 84. Cyclohexenyl (0.58); 85. Ionone/irone (0.62); 86. Alkyl, acyclic (0.61); 87. Alkyl, cyclic (0.61); 88. Benzyl (0.71); 89. Jasmone derivatives (0.42) |
| Metallic salts (organic) | Metallic salts (organic) | 90. Metallic salts (organic) (0.21) |
| Organosulfur | Alkyl thioether; Disulfide; Thiol | 91. Aliphatic thioether (0.44); 92. Disulfide (0.37); 93. Thiol (0.33) |
| Structure undefined | Structure undefined | 94. Structure undefined (NA) |
| Sugars and derivatives | Sugars and derivatives | 95. Sugars and derivatives (0.49) |
| Terpene and terpenoid derivatives | Carvone derivatives; Citronellol derivatives; Farnesene derivatives; Geraniol derivatives; Linalool derivatives; Retinol derivatives | 96. Carvone derivatives (0.67); 97. Citronellol derivatives (0.70); 98. Farnesene derivatives (0.62); 99. Geraniol derivatives (0.75); 100. Linalool derivatives (0.75); 101. Retinol derivatives (0.69) |
| Vitamins and derivatives | Vitamins and derivatives | 102. Vitamins and derivatives (0.25) |
Only those chemicals chemical groups that contain at least 3 chemicals are displayed. The full list of chemicals and their grouping is provided in Supplementary Information 1.
Mean Tanimoto coefficient, calculated based on the Maximum Common Substructures of the chemicals within a group (see Materials and Methods).
For the different target families within ToxCast (eg, DNA binding and nuclear receptor targets) and the target subfamilies of the nuclear receptor target family (being steroidal, nonsteroidal, and orphan), the percent activities of each tertiary chemical grouping per individual biological target were plotted as a heatmap using the ggplot package in R. In addition, the percentage of positive hits per target (sub)family was calculated and plotted along with the circle pack of the chemical groups. The R codes for the calculation of the biological activities per chemical group and the resulting heatmaps, and circle pack have been made available through https://git.wur.nl/Punt001/ilsi_toxcast.git (last accessed January 30, 2020). In addition, the R workflow has been made available as a web application using R Shiny (Rstudio, https://cran.r-project.org/web/packages/shiny/index.html, last accessed January 30, 2020). This web application is available through https://ilsi.eu/exploitation-of-toxcast-data-on-food-chemicals-for-safety-risk-assessment/ (last accessed January 30, 2020).
RESULTS
Grouping of the ToxCast Chemicals Based on Functional Use and Chemical Structure
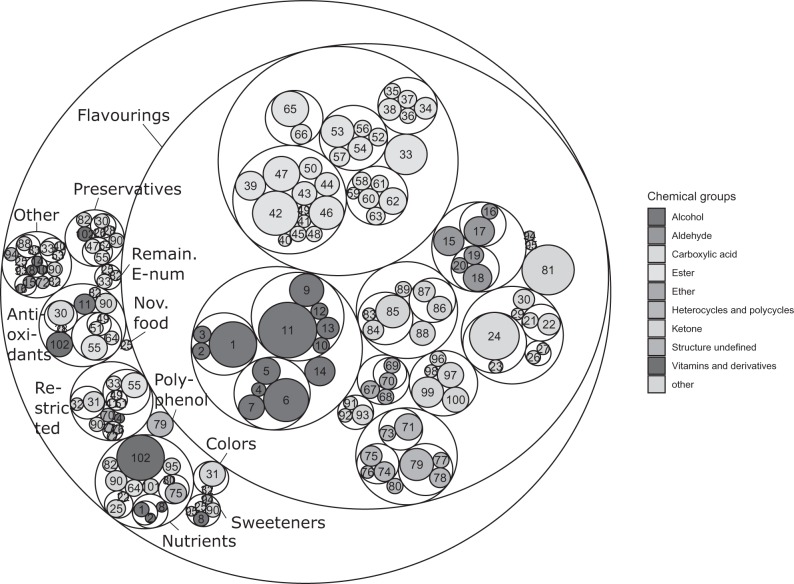
The 563 food-relevant chemicals were clustered according to their chemical structure as well as their functional use classes. The obtained groups are displayed in Figure 1 as a so-called circle pack, which displays the hierarchical architecture of the defined functional and chemical groups. Note that a given chemical group may be split across more than 1 functional group and vice versa. The first layer within Figure 1 displays the functional use classes of which the largest group consists of food flavorings (obtained after matching the CAS numbers with the EU food flavorings regulation). A total of 455 chemicals fell into the flavoring’s category. Other relevant functional use classes included the group of European E-numbers (43 chemicals, food additives that perform a certain technological function in food, subdivided into sweeteners, antioxidants, preservatives, colors, and remaining E-numbers), nutrients (31 chemicals), and regulatory restricted chemicals (19 chemicals). Chemicals that fell into multiple categories are counted multiple times, once for each category. For example, ascorbic acid is included as both an E-number (as a preservative) and nutrient (being also a vitamin). Chemicals for which no clear food use could be defined are grouped as “Other.”
Figure 1.
Circle pack plot of the defined functional groups and chemical groups. The larger a circle, the more chemicals fall into the group and closely related chemicals are packed more closely together. Tertiary groups (closest related chemicals) are labeled. The legend provides information on the primary chemical groups to which they belong. Details about the composition of the groups can be found in Table 1.
The clusters that were obtained based on the chemical structure are shown alongside the functional use classes in Figure 1. The legend provides information on the primary chemical clusters to which the chemical groups belong. Details on how the large primary clusters (eg, alcohols) are further subdivided as appropriate into secondary (eg, alcohol and alkyl) and tertiary (eg, alcohol, alkyl, primary, and straight chain) groups can be found in Table 1. Chemicals that fell into multiple chemical groups are counted multiple times, once for each group. For example, ascorbic acid falls into the “Ester, Lactone, Ascorbic acid and derivatives” group as well as the “Vitamins and derivatives” chemical group. Overall, 102 tertiary groups were defined for which the biological activity was explored. These tertiary groups consist of at least 3 closely related chemicals, with the largest chemical group consisting of 17 (group 12, phenolic aldehydes) chemicals. The majority of the defined chemical groups has a mean Tanimoto coefficient that is higher than 0.6. Some of the chemical groups are more structurally diverse and are atypical of the wider set, holding as they do compounds which exhibit unique characteristics. For example, the “amino acids and derivatives” grouping consists of a series of complex, often natural products, whereas “metallic salts organic” is founded solely upon the possession of an inorganic counter ion. Azo dyes furthermore represent a collection of compounds which may exhibit variation in wider structure despite unification by a distinctive functional group.
Global Evaluation of the Biological Activity of the Tertiary Homologous Chemical Groups
ToxCast Biological Activities Plotted as a Heatmap
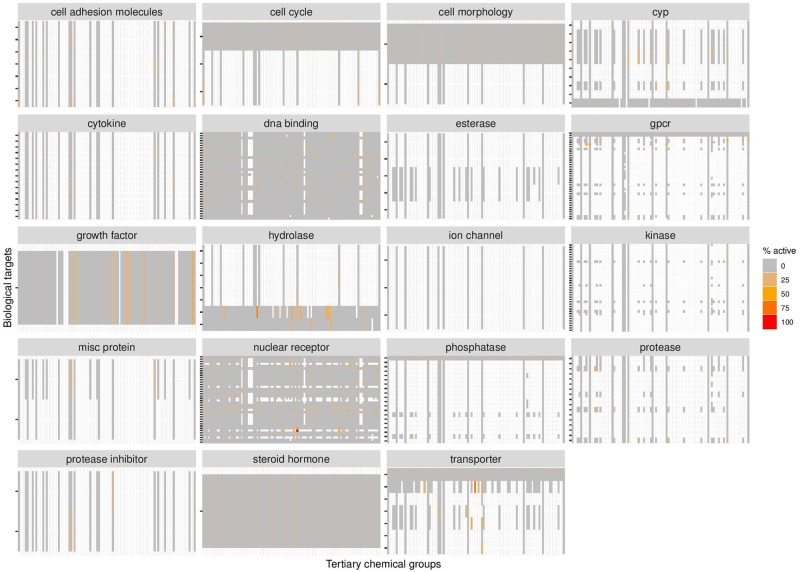
Figure 2 displays different heatmaps demonstrating the activity of the 102 defined tertiary structural groupings toward different biological targets of the different target families and Figure 3 of the “DNA binding” and “nuclear receptor target families” in detail. White spots in the heatmaps represent chemical group-biological target combinations for which insufficient data are available (n < 3 in all assays that cover that biological target). Gray means that all the chemicals within the group were inactive in the assays for that target, whereas the colors ranging from orange to red represent an increasing percentage of chemicals within the chemical group that responded in the assays of that target.
Figure 2.
Heatmaps showing coverage of biological activity for the 102 tertiary chemical groups within the different ToxCast target families. The targets are displayed on the y-axes with ticks, 1 per target. The gradient corresponds to an increasing percentage of chemicals within the chemical group that was active in the different assays of that target. White spots mean that < 3 chemicals were tested in all assays of that target. Gray spots mean that none of the chemicals in the chemical group was active in the assays of that target.
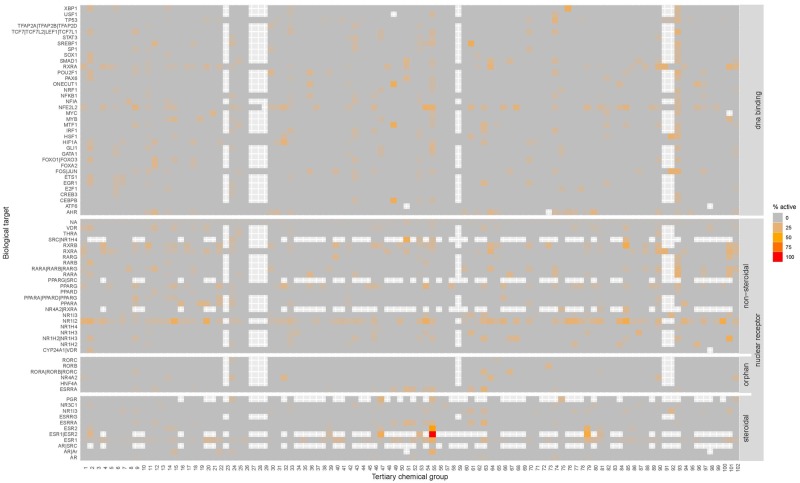
Figure 3.
Heatmaps of the biological activity of the 102 tertiary chemical groups within “DNA binding” and “nuclear receptor” target families. Each target (displayed on the y-axes with labels) is covered by 1–11 assays. The gradient corresponds to an increasing percentage of chemicals within the chemical group that showed activity in the different assays of that target. White spots mean that < 3 chemicals were tested in all assays of that target. Gray spots mean that none of the chemicals in the chemical group was active in the assays of that target. The results for all target families can be interactively viewed through www.https://ilsi.eu/exploitation-of-toxcast-data-on-food-chemicals-for-safety-risk-assessment/ (last accessed January 30, 2020).
It is clear from the number of white areas in the heatmaps of Figure 2 that there are significant data gaps in ToxCast for the food-relevant chemicals, indicating that the ToxCast dataset is not yet comprehensive for some of these types of food chemicals. Therefore, it is important to note that a lack of observed activity in the summarized findings should not be construed as indicative of inactive food-relevant chemicals, but that this is often the consequence of insufficient data. This observation may not be unique to food-relevant chemicals; other test substances within the wider ToxCast dataset beyond the scope of this inquiry may be as yet insufficiently tested for any broad conclusions to be made regarding their biological activities. Among the different target families, most of the food-relevant chemicals were tested in assays that are linked to the “DNA binding” and “nuclear receptor” target families (ie, most gray/color). The food-relevant chemicals have also been tested in assays that are linked to “cell cycle,” “growth factor,” “hydrolase,” and “steroidal hormone.” However, these latter target families consist of only 1–3 targets each (few y-axis tick marks), whereas the “DNA binding” and “nuclear receptor” target families consist of 36 and 40 targets, respectively. Further evaluations therefore focus on the activities within these latter 2 target families.
The activities within the “DNA binding” and “nuclear receptor” target families are further highlighted in Figure 3, in which the “nuclear receptor” target family is also subdivided into its 3 distinctive subfamilies (“steroidal,” “nonsteroidal,” and “orphan”). Figure 3 reveals that most tertiary chemical groups are not active toward most of biological targets (ie, 0% of the chemicals in the tertiary group showed activity in the assays of the target) within the “DNA binding” and “nuclear receptor” target families. One notable exception is chemical group 55 (containing parabens and gallates) that stands out in Figure 3 for its high activity toward ESR1|ESR2 (corresponding to ERα and ERβ), ESR1 (ERα), and ESR2 (ERβ). Other relevant chemical groups in Figure 3 are group 93 (containing thiols), which has a relatively high group activity at a variety of DNA binding targets, group 79 (containing genistein and daidzein), which has a high activity toward ESR1, ESR/ESR2, and ESR2, and group 101 (retinoids), which has a relatively high group activity toward RXRA, RAXRB, NR1I2, and NRF2.
Within the “DNA binding” and “nuclear receptor” target families, there are a few targets for which almost all the chemical groups appear to be active. Examples are the NFE2L2, RXRA, RXRB, NR1I2, and ESR1 targets (horizontal orange stripes in Figure 3). Particularly, the frequent responses toward NFE2L2, RXRA, RXRB, and NR1I2 are likely because these are relatively general endpoints that are involved in increasing metabolic capacity or oxidative stress response (Louisse et al., 2018; Mazaira et al., 2019). However, to some extent, this frequent activity also appears to be due to a proportionately high number of chemicals that are active in certain individual assays that fall under these targets (Ryan, 2017). For example, among the different assays that measure effects on ESR1, 10% of the food-relevant chemicals were active in the ATG_ERE_CIS_up assay and 7% in the TOX21_ERa_LUC_BG1_Agonist assay, whereas only 0.4%–3% of the chemicals were active in other assays that measure ESR1 (see Supplementary Information 2), suggesting that the high positive rate in some assays might be an artifact. In addition, 12% of the food-relevant chemicals were active in the ATG_NRF2_ARE_CIS_up assay (NFE2L2 target), 25% in the ATG_PXRE_CIS_up assay (NR1I2 target), TOX21_RXR_BLA_Agonist_ratio assay (RXRA target), and 7% in the ATG_RXRb_TRANS_up (RXRB target), with much lower rates in other assays for these targets. This suggests that care should be taken in the interpretation of the ToxCast results when activity toward a biological target is due to activity in one of these specific assays that generate a high number of positive results. Supplementary Information 2 provides a list of the ToxCast assays, the percentage of the food-relevant chemicals that were active in each assay, and the percentage of the results that contained flags. Based on these data, the specificity of the different assays can be assessed, which is highly relevant for the interpretation of the test results for the individual chemical groups.
Biological Activities of the Chemical Groups in the Context of Their Functional Uses
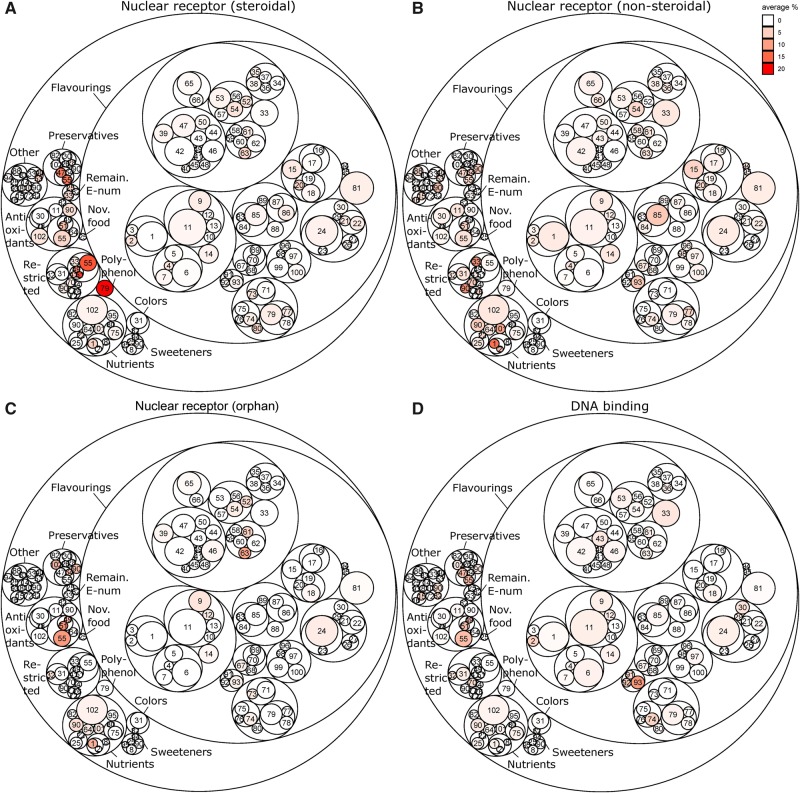
Figure 4 combines the ToxCast activity data with the circle pack of Figure 1, providing an indication of the biological activities of the groups of food-relevant chemicals in the context of their functional uses. To this end, for each chemical group, the percent activities in the assays that belong to a specific target family were calculated. For example, the distinct activities of group 55 (parabens-gallates), group 79 (containing genistein and daidzein), group 101 (retinoids), and group 93 (thiols) as were observed in Figure 3, result in an overall high activity of these groups in the “steroidal nuclear receptor” (groups 55 and 79), “nonsteroidal” (group 101), and “DNA binding” target (sub)families (group 93) in Figure 4. Figure 4 also reveals that many chemical groups are slightly active within the steroidal and nonsteroidal nuclear receptor target families. These activities generally relate to activities in the assays with a disproportionately high number of positive hits and/or assays that capture general response mechanism to chemical exposure, as discussed above.
Figure 4.
Biological activity of the tertiary chemical groups within the “nuclear receptor” (A–C) and “DNA binding” (D) target families. For each tertiary chemical group, the percent of chemicals that were active in the assays for different target families were calculated and displayed in the colors indicated. The results for all target families can be interactively viewed through www.https://ilsi.eu/exploitation-of-toxcast-data-on-food-chemicals-for-safety-risk-assessment/ (last accessed January 30, 2020).
Some of the tertiary structural groups in Figure 4 consist of chemicals that fall into different functional use classes. In those cases, the activity that is displayed in Figure 4 corresponds to percent activity of the chemicals that fall into the same use class and not the activity of the whole group. For example, Figure 4 reveals that the group of parabens (group 55) contains both restricted compounds and compounds that are used as antioxidants and preservatives. Particularly, the restricted parabens and parabens used as preservative appear to have activity in the steroidal nuclear receptor target family of assays. In comparison, group 79 consists of polyphenols (genistein, daidzein, and quercetin) and of different flavorings (eg, coumarin). Figure 4 reveals that the high steroidal nuclear receptor activity of group 79 comes only from the polyphenols of group 79 and not from the flavorings.
The results from the heat map (Figure 3) and circle pack (Figure 4) reveal that some of the key biological targets of the food-relevant chemicals can be defined by focusing on ToxCast activities of predefined homologous chemical groups. The circle pack plot (Figure 4) provides insight into the overall biological activities of the tertiary chemical groups within the “DNA binding” and “(steroidal, nonsteroidal, and orphan) nuclear receptor” target (sub)families and places the results in the context of the functional uses. The heatmap of Figure 3 provides insights into the specific targets within these target (sub)families that are affected. Based on these results, several case studies were defined to explore how the ToxCast data can be used in food safety risk evaluations and for the evaluation of desirable effects of nutrients and phytonutrients. The case studies are used to check whether the mechanistic information that is obtained from ToxCast matches with what is expected from the chemical group. To this purpose, case studies were selected around chemical groups that express a high biological activity toward a specific target (parabens), chemicals that are restricted for food use due to a specific activity (some parabens and genotoxic and carcinogenic compounds like estragole, methyleugenol, and safrole), and chemical groups are related to specific health benefits (eg, flavonoids and fatty acid). For the selected case studies, sufficient literature data are available on the mechanisms of action of the compounds. The comparison of the observed target(s) with expected target(s) is considered a crucial step to find potential caveats in the HTS data that need to be considered for future use of the data on chemicals for which little animal experimental or in vitro reference data are available.
Case Study on Regulatory Restricted Chemicals
The group of regulatory restricted chemicals provides an interesting group of food-relevant chemicals for the evaluation of ToxCast activities. Several compounds that are restricted for food use in regulations can be found within the ToxCast dataset of food-relevant compounds. Most of these compounds have an E-number (EU codes for substances that are permitted as food additive) yet have been discontinued for food use in the EU. The exact reasons for the discontinuation are not always clear but do not necessarily relate to the demonstrable toxicity of the chemical. For example, ethoxyquin (E324) was suspended from its authorization as a feed additive in EU (2017) because of a lack of data on some aspects of its safety, but it is currently being re-evaluated by EFSA again for this use (EFSA, 2019). However, for 2 chemical groups within the regulatory restricted group, demonstrable toxicological findings have played an important role in their restriction for food use. These are group 55 (containing parabens and gallates, which have estrogenic activities) and groups 70 and 77 (containing estragole, methyleugenol, and safrole, which are genotoxic and carcinogenic) (Phillips et al., 1984). It is of interest to observe whether for these groups a perturbation of the underlying biological target responsible for the restrictions can be detected with the goal of determining the potential contribution of ToxCast data in such evaluations of food safety risk.
Restricted and nonrestricted parabens and gallates
The ToxCast evaluation of the biological activities of structural groups detected a relative high activity of the paraben-gallate (group 55) toward ERα and ERβ activation (ESR1, ESR2, and ESR1|ESR2 in Figure 3). This structural group consists of the approved E-numbers methylparaben (preservative), dodecyl gallate, octyl gallate, and propyl gallate (used as antioxidants); 2 parabens that are not used in foods within the EU; butyl- and heptyl-paraben; and the restricted propylparaben (EFSA, 2004). Figure 4 reveals that the biological activity within the paraben-gallate mainly comes from the restricted and nonrestricted parabens (preservatives) rather than the gallates that are used as antioxidants. Particularly, the percentage of positive assays toward the steroidal nuclear receptors (Figure 4A) was higher for parabens than gallates. Positives were largely for estrogenic assays. The differences within the group of parabens-gallates are also reflected in the relative estrogenic potencies of these 2 groups; the mean AC50 of the positive ERα assay results for the restricted/nonapproved parabens (with Z-scores > 3) is 10.9 ± 4.7 µM for propylparaben, 5.2 ± 1.4 µM for butylparaben, and 3.7 ± 0.6 for heptylparaben, whereas the nonrestricted methylparaben has a lower potency with a mean AC50 of 53 ± 18 µM. Dodecyl and propyl gallate were not active in the ER receptor assays in the final dataset, and octyl gallate is active in only 1 out of the 8 ER receptor assays. It should, however, be noted that many of the positive ER hit-calls of the gallates appeared to have been filtered out as a result of their low Z-scores. This suggests a potential influence of, eg, cytotoxicity, on the ER results of the different gallates.
Altogether, the results obtained for group 55 reveal an interesting potency difference between the parabens and gallates within this group. These results provide relevant information that can be considered within the regulatory risk assessments of these compounds, particularly to perform a read-across. The estrogenic hazard potential of the parabens has long been included in their risk assessment (EFSA, 2004; EMA, 2015; SCCS, 2013). In case of gallates, the potential these compounds to interfere with the human ER in vitro has recently be mentioned by EFSA in re-evaluations of dodecyl, octyl, and propyl gallate but has not been included in their final risk evaluation as confirmatory in vivo data are lacking (EFSA, 2014, 2015a, b). Based on the results of the present study, the association between gallates and the ER was considered low, but follow-up in vitro studies may be needed to better understand the origin of the low Z-scores for the gallates in the ER-related assays that had to be dismissed on the basis of these low Z-scores.
Regulatory restricted genotoxic carcinogens
Evaluation of the ToxCast activities of chemical groups 70 and 77 that contain the known genotoxic and carcinogenic compounds estragole (group 70), methyleugenol (group 70), and safrole (group 77) (SCF, 2001a, b) shows that the hazard of these type of compounds cannot be adequately identified in ToxCast. In Figure 4, groups 70 and 77 can be found to have a slight activity within the nonsteroidal nuclear receptor target family. However, based on Figure 4, it can be concluded that this activity relates to activation of RXRB and NR1I2 (also called PXR), targets that induce xenobiotic metabolism enzyme synthesis and for which many noncarcinogenic chemical groups are active. Hence, this slight activity toward RXRB and NR1I2 is not considered diagnostic for the genotoxic hazard of these compounds nor for any other potential specific mechanisms of actions. Given the genotoxic mechanisms of estragole, methyleugenol, and safrole, activity in assays that include p53 tumor suppressor gene activity might be expected (Paini et al., 2011). Such activity forms part of the “DNA binding” target family. However, no such activity was found, nor was there any other indication of genotoxicity for these chemicals within the ToxCast dataset.
Because estragole, methyleugenol, and safrole require bioactivation for their genotoxic and carcinogenic effects (Punt et al., 2007), this lack of detectable activity could be due to the lack of metabolic capacity within the ToxCast assays (DeGroot et al., 2018). Moreover, genotoxicity can be difficult to detect without very tight concentration spacing, as the high-throughput assays often quantify the upregulation of DNA-repair pathways (Iyer et al., 2019). Although these assays quantify the cellular mechanisms evolved to fix low-level DNA damage, when the damage-levels are great, cells instead die without attempting repair, resulting in false-negative tests. Therefore, it is also possible that the lack of detectable genotoxic activity could be due to the large concentration spacing used for ToxCast testing and the subsequent misclassification of genotoxicity as cytotoxicity. Taken together, these findings suggest that currently genotoxicants and/or carcinogens cannot always be adequately detected within the ToxCast activity data, which is supported by other literature findings (Becker et al., 2017).
Use of ToxCast Data in Assessment of Beneficial Effects
The flavonoids, genistein, daidzein, and quercetin; as well as unsaturated fatty acids like linolenic, linoleic, and oleic acid; are all examples of compounds with health beneficial effects that can be found within the set of food-relevant chemicals used in the present study. For these substances, the relationship between beneficial and adverse biological effects is of particular interest. Therefore, the ToxCast data from these 2 substance-groups were examined to characterize the biological targets of each group of health beneficial chemicals and evaluate how the ToxCast information might be used to inform a risk-benefit assessment of the compounds.
Flavonoids
Flavonoids have been extensively studied for their biological effects against cancer, cardiovascular diseases, obesity and diabetes, as well as neurodegenerative disorders (Williamson et al., 2018). Within the set of food-relevant chemicals of the present study, 3 flavonoids are included: quercetin and the isoflavones daidzein and genistein. All are part of the “heterocycles and polycycles-oxygen heterocycles-pyranone” chemical group (group 79). Other chemicals that are part of this group are maltol, 2-ethyl-3-hydroxy-4-pyrone, maltol isobutyrate, coumarin, and 6-methyl coumarin. The molecular targets that have been suggested to play a predominant role in the health beneficial effects of flavonoids are displayed in Table 2.
Table 2.
Expected Important Biological Targets of Different Flavonoids and the Percentage Active in the Chemical Group Containing Flavonoids (Group 79) Toward These Targets
| Biological Targeta | ToxCast Targets (% Active in Group 79) | Function | ToxCast Target Family | ToxCast Target Subfamily |
|---|---|---|---|---|
| NRF2 | NFE2L2 (15) | Antioxidant | DNA binding | Basic leucine zipper |
| NF-κβ | NFKB1 (0) | Free-radical scavenging | DNA binding | NF-kappa B |
| VEGF | KDR (0), FLT1 (0), FLT4 (0) | Regulation of vascular cell development | Kinase | Receptor tyrosine kinase |
| PPAR | PPARA (0), PPARD (0), PPARG (0), PPARA|PPARD|PPARG (0), PPARG|SRC (0) | Lipid metabolism and glucose homeostasis | Nuclear receptor | Nonsteroidal |
| VCAM-1 | VCMA1 (0) | Vascular cell adhesion | Cell adhesion molecules | Immunoglobulin CAM |
| ER | ESR1 (14), ESR2 (29), ESR1|ESR2 (40) | Estrogen-dependent proliferation and differentiation | Nuclear receptor | Steroidal |
In contrast to what was expected, no activity of group 79 was found for most of the targets described in Table 2, except for activity in ER-related assays and NRF2. Similarly, when examining the data for each individual flavonoid in the group it is clear that this group is made up of diverse substances; each has relatively few identifiable activities with little overlap with those of other members of the structural group.
Interestingly, the apparent biological activities of flavonoids are significantly affected by the filtering out of results with Z-scores < 3. Without this filtering, quercetin, genistein, and daidzein are active in 19.4%, 31.3%, and 23.6% of the assays, respectively, that are part of the target (sub)families of Figure 2, whereas they are active in only 1%, 11%, and 10% of these assays after filtering. Judson et al. (2016) also identified quercetin as a highly active chemical within ToxCast, with low specificity (low Z-scores). Many of the effects of the chemicals in group 79 are thus filtered out as being nonspecific. Though results with a Z-score < 3 may reflect an indirect influence of cytotoxicity or other nonspecific mechanisms of action, the low Z-scores may also point to a nonspecific interference with the assays. For example, flavonoids are capable of stabilization of luciferase, frequently used in reporter gene assays (Prinsloo et al., 2017). The high number of assay results with low Z-scores indicates that challenges exist in using HTS to explore the biological activities of certain compound such as flavonoids.
Although the ToxCast activities of flavonoids seem uncertain due to possible nonspecific effects, group 79 does show a distinct activity within the “nuclear receptor-steroidal” target (sub)family toward the ERα and ERβ. This result is predominantly due to activity of the isoflavones genistein and daidzein (see Figure 4). The interaction of genistein and daidzein with the ER receptor has been linked to both beneficial health effects (eg, lowering menopausal symptoms, lowering cancer risks and risk for cardiovascular diseases) and adverse effects (endocrine disruption, increased hormone cancer risk) (Rietjens et al., 2017). A key hypothesis behind the benefits and risks of isoflavonoids is the differences between the activation of ERα and ERβ. ERα activation enhances cell proliferation, whereas ERβ counteracts the ERα-mediated stimulation of cell proliferation (Rietjens et al., 2017). Though many estrogenic compounds within ToxCast interact with both ERα and ERβ (including the parabens as described above), the AC50 values for genistein and daidzein were 20- and 11-fold lower for ERβ compared with ERα (based on the OT_ER_ERaERa_0480/1440 and OT_ER_ERbERb_0480/1440 assays), respectively, suggesting a predominantly ERβ-mediated effect at low concentrations (this is not the case for the parabens which have comparable AC50 values toward ERα and ERβ, eg). This selective ER modulation suggests that the risk-benefit profile of genistein and daidzein is probably dose dependent but must be extrapolated to in vivo dose-response or potency information to identify whether the effective concentrations in vitro are capable of being attained in vivo. For genistein, this has, eg, been done by Boonpawa et al. (2017), revealing that both Asian dietary intake levels and the use of genistein-containing supplements are sufficient for ERβ activation, but not for ERα modulation. Thus, the in vitro potency information over a range of ToxCast targets can be used to prioritize measurement and evaluation of in vivo biological effects within the context of risk-benefit assessments.
Fatty acids
Fatty acids, particularly unsaturated fatty acids, play a key role in reducing cardiovascular risks and anti-inflammatory effects (Williams, 2000). The set of food-relevant chemicals of the present study contains a series of both unsaturated (group 22) and saturated (group 24) fatty acids. Group 22 consists of 2-butenoic-, sorbic-, 10-undecenoic-, oleic-, linolenic-, and linoleic-acid. Group 24 consists of acetic acid, butanoic-, pentanoic-, hexanoic-, heptanoic-, octanoic-, decanoic-, dodecanoic-, tetradecanoic-, hexadecanoic (palmitic)-, and octadecanoic (stearic)-acid.
An important mode of action of fatty acids is the regulation of lipid metabolism (Varga et al., 2011). For example, Popeijus et al. (2014) have shown that fatty acid chain length and saturation influence PPARα transcriptional activation and repression in HepG2 cells, and specifically the saturated fatty acids palmitic acid (C16:0) and stearic acid (C18:0) both repress PPARα activation, whereas their unsaturated metabolites palmitoleic acid (C16:1(n‐7)) and oleic acid (C18:1(n‐9)) activate PPAR transcription. Other potentially relevant targets of fatty acids within lipid homeostasis are SREBPs, LXR, and HNF4 (Müller and Kersten, 2003). Table 3 provides an overview of the ToxCast activity of the chemical groups containing fatty acids toward these different targets.
Table 3.
Expected Important Biological Targets of Different Fatty Acids Within Lipid Homeostasis and the Percentage of Actives in the Chemical Group Containing Unsaturated Fatty Acids (Groups 22) and Saturated Fatty Acids (Group 24) Toward These Targets
| Biological Targeta | ToxCast Target (% Active in Groups 22 and 24) | Function | ToxCast Target Family | ToxCast Target Subfamily |
|---|---|---|---|---|
| PPARs | PPARA (11)(10), PPARD (0)(0), PPARG (8)(4), PPARA|PPARD|PPARG (13)(5), PPARG|SRC (13)(0) | Lipid metabolism and homeostasis, glucose utilization | Nuclear receptor | Nonsteroidal |
| SREBPs | SREBF1 (0)(6) | Lipid metabolism and homeostasis | DNA binding | Basic helix-loop-helix leucine zipper |
| LXR | NR1H2 (0)(0), NR1H3 (0)(0), NR1H2|NR1H3 (0)(0), SRC|NR1H4)(10) | Lipid metabolism and homeostasis | Nuclear receptor | Nonsteroidal |
| HNF4 | HNF4A (0)(0) | Lipid metabolism and homeostasis | Nuclear receptor | Orphan |
The ToxCast activity of both groups 22 and 24 toward the expected targets of Table 3 appears to be very limited. For example, within group 22, only 10-undecenoic acid is active in 2 PPARα-related assays, and within group, 24 the PPARα activity mainly comes from decanoic acid. The low responses seem to be partly due to the filtering based on Z-scores < 3. Without filtering, all the long chain fatty acids of the unsaturated fatty acid group (oleic acid, 10-undecenoic acid, linoleic acid, and linolenic acid) are active in the ATG_PPARa_TRANS_up assay, which is in line with what is expected (Popeijus et al., 2014). This raises questions as to whether AC50s with low Z-scores should indeed be considered to be the result of nonspecific activities and as to what causes these low Z-scores. The low observed biological activity of the saturated fatty acids group toward PPARα (either up- or down-regulation) was not affected by the Z-score filtering.
The activity toward the other potential molecular targets of saturated and unsaturated fatty acids is also limited but does not seem to be caused by the filtering based on Z-scores. In the case of SREBPs, the limited activity might be the result of the fact that unsaturated fatty acids are downregulators (Hannah et al., 2001), whereas ToxCast only contains the ATG_SREBP_CIS_up assay. The LXR receptor, which is involved in the regulation of cholesterol and fatty acid homeostasis, was not active as a ToxCast assay target for groups 22 and 25, which may be a reflection of it being responsive to HNF4A intracellular cholesterol alterations (Lund et al., 2006). Overall, the results indicate that the ToxCast dataset is at present not yet adequate to obtain insights into the biological activities of fatty acids or, eg, for the extrapolation of the potential effects over different chain lengths of fatty acids.
Remaining Relevant Groups
Table 4 provides a list of remaining relevant chemical groups that display a relatively high activity as displayed in Figures 3 and 4, but which were not assessed further as case studies. The observed activities of these groups generally relate to endpoints such as increased metabolic capacity or oxidative stress response. However, the targets that are affected by the thiol group (group 93) within the “DNA binding” target family may point at a specific activity of this chemical group that is potentially relevant for the safety evaluation of this chemical group.
Table 4.
List of Chemical Groups That Displayed a Relatively High Activity in Figures 3 and 4 and the Targets That Are Affected
| Chemical Group | Target Family and Key Targetsa |
|---|---|
| Ester-aliphatic alcohol diester/triester (group 33, n = 11 of which 1 chemical is part of the regulatory restricted group) | Nonsteroidal nuclear receptor: NR1I2 (33), PPARG (22), NR1H4 (20) |
| Metallic salts organic (group 90, n = 7 of which 1 chemical is part of the regulatory restricted group) | Nonsteroidal nuclear receptor: NR1I2 (36), RXRA (33), PPARG (30) |
| Retinol derivatives (101, n = 3) | Nonsteroidal nuclear receptor: NR1I2 (45), RXRA (40), RXRB (30), VDR (25), NR1H2|NR1H3 (20) |
| Organosulfur. thiol (group 93, n = 6) | DNA binding: TCF7|TCF7L2|LEF1|TCF7L1 (60), IRF1 (40), FOS|JUN, (33), SMAD1 (33), USF1 (33), NFKB1 (33), NFE2L2 (33), POU2F1 (33), TP53 (33), AHR (28), SREBF1 (20), HSF1 (20), XBP1ng (20) |
Only the targets with more than 20% activity are displayed.
DISCUSSION
This investigation aimed to (1) explore how HTS can be leveraged to obtain chemical-specific insights into the biological targets that may be affected by different food-relevant chemicals and (2) assess the utility of the data for the safety assessment of food chemicals as well as the evaluation of health beneficial effects of chemicals in a few case studies. A method was set up to group the chemicals according to functional use and structural similarity. For each of the tertiary chemical groups of homologous chemicals, the percent of chemicals that were active in the assays for different targets and target families were calculated. The targets that were elicited, the directionality thereof, (ie, activation vs downregulation), and the inactivity toward certain targets can provide key information in characterizing biological patterns.
A general challenge in the use of HTS data in chemical safety evaluations is the uncertainty around the individual assay results. The diverse assay space and challenges with automatic processing contribute to this uncertainty (Cox et al., 2014; Ryan, 2017; Watt and Judson, 2018). The approach of the present study, in which the focus is not on the individual chemical results but on the activity of homologs within chemical groups may contribute to reducing the uncertainty and improving the specificity when defining biological targets of chemicals based on HTS data. Taking the example of genistein, this compound is active in 114 assays within the crude ToxCast dataset, with 49% of these results containing flags. Removing all data with Z-scores < 3 as a cutoff value for nonspecific effects, the number of positive hit-calls is reduced to 16, of which 30% contain flags. Combining the results with the other flavonoids within the list of food-relevant chemicals, including daidzein, points toward the expected ER activation as the most predominant biological effect. Both genistein and daidzein tested positive in 11 different ESR1 and/or ESR2 assays with 1 flagged result for genistein.
Though these results indicate that the specificity increases by filtering the ToxCast data for Z-scores and focusing on targets that are induced by homologous chemical groups, such filtering may also result in a potential loss of information. In the different case studies, filtering of Z-scores < 3 sometimes appeared to eliminate valuable information. It is therefore also important to go back to the crude data to evaluate the filtering process prior to the use of ToxCast results for risk evaluations.
The (tertiary) chemical groups for which the ToxCast activities were assessed were obtained with ChemoTyper software (Mellor et al., 2019; Yang et al., 2015). Other methods for chemical grouping exist as well, including, eg, AMBIT (http://cefic-lri.org/toolbox/ambit/, last accessed January 30, 2020), the OECD QSAR toolbox (Dimitrov et al., 2016), and ToxMatch (https://ec.europa.eu/jrc/en/scientific-tool/toxmatch, last accessed January 30, 2020). Key to the chemical grouping is that groups should become neither too large nor too small. The highest number of chemicals within one group that was obtained using ChemoTyper contained 17 analogs. Most of the groups consist of 3–5 chemicals. Sixty-six of the originally 168 defined groups could not be used in the present study as these contained only 1 or 2 chemicals.
The fact that the number of chemicals within a group varies may pose some bias in the evaluation of the biological activity group targets. If 2 compounds in a group of 3 chemicals are active toward a specific target, this corresponds to 66% activity, whereas an activity of 2 compounds in a group of 6 would correspond to 33% activity. A similar bias in the results occurs due to the varying number of assays per target in the ToxCast dataset. Many of the biological targets as displayed in Figure 4 are covered by only 1 or 2 assays, whereas for the ESR1 target, there are 16 assays of which 11 measure ER agonism. A high percentage activity of a chemical group in the case of ESR1 will therefore occur only when the chemicals of that group are active across a wide range of assays for ESR1. Even though these results indicate that for larger chemical groups and for targets that are covered by multiple assays, it will be more difficult to pick up group activities, the results are expected to become more specific. For example, the activity of the compounds of the paraben-gallate (group 55) and combined activity of genistein and daidzein within the pyrole group (group 79) in multiple ESR1, ESR2, and ESR1|2-related assays give confidence that ER activation is an important target for these groups of chemicals. This effect of enrichment was less apparent within fatty acid and genotoxic carcinogen-case studies, due to the overall limited activity of the individual chemicals in the expected assays.
Whereas the ToxCast data may add to the evidence considered in food chemical safety evaluations, the results should serve primarily as a screening tool to set priorities for further evaluations relative to hypothesized biological targets. In addition, some care should be taken to avoid overinterpretation of the data. Not all available chemicals within ToxCast have been tested in all available assays and not all toxicity endpoints are covered by the available assays. In addition, better understanding of relevance of data with low Z-scores may minimize loss of potentially relevant information. For individual cases where low Z-scores are found, follow-up analyses may be needed to identify what causes these low Z-scores. These facts are critical aspects of the dataset which we found crucial for proper data interpretation during the present study.
A more general aspect that needs to be kept in mind is the fact that metabolic activation of chemicals is not accounted for in the ToxCast assays. This probably contributed to the observed poor prediction of genotoxicity in the case studies. Research that focuses on enhancing the metabolic capacity in HTS assays is therefore important (DeGroot et al., 2018).
Finally, it should be noted that in vitro activity data do not directly reflect in vivo biological potencies (in vivo effects will, eg, also depend on the availability of a chemical in the body). Extrapolation of the concentration-response curves to in vivo potency information is an important next step. There are an increasing number of publications that focus on establishing such an extrapolation (Becker et al., 2014; Boonpawa et al., 2017; Dent et al., 2019; Fabian et al., 2019; Punt et al., 2019; Wetmore et al., 2015). One approach is to use kinetic modeling or human biomonitoring data to compare the AC50 values with internal plasma concentrations reached during daily exposures in so-called exposure: activity ratios (EARs). EARs of different compounds can subsequently be compared in a so-called “dietary comparator ratio” approach to prioritize exposure-activity data relative a known reference compound (Becker et al., 2014; Dent et al., 2019).
To increase regulatory use of HTS, it will be important to tackle the different challenges related to HTS and quantitative in vitro to in vivo extrapolations. Recently, Thomas et al. (2019) published a blueprint to systematically address these key challenges, which can be expected to move the field forward.
Overall, the results of the present study suggest that HTS data could add to the evidence considered for regulatory risk assessment of food chemicals and to the evaluation of desirable effects of nutrients and phytonutrients. The data will be particularly useful for providing mechanistic information and to fill data gaps with read-across. Whereas the current study mainly focused on setting up a method to find key biological targets of chemical groups and the qualitative interpretation thereof, the key next step for use in risk evaluations or follow-up research is to also focus on the quantitative aspects of the results. This includes, eg, the evaluation of the (differences in) potencies of the chemicals toward targets of interest and placing the potencies in the context of human in vivo–relevant exposure.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
ILSI Europe New Approaches to Chemical Risk Assessment for Food and Food Ingredients Task Force.
Supplementary Material
ACKNOWLEDGMENTS
This work was conducted by an expert group of the European branch of the International Life Sciences Institute (ILSI Europe). We appreciate the contribution of Dr Agnes Karmaus who gave us insights in the preexisting work done on food-relevant chemicals in the ToxCast dataset. We also appreciate the contribution of Sylvain Etter (Firmenich) and Heinz Traussnig (Mayr-Melnhof Karton) to the discussions on the work of the expert group. Industry members of this task force are listed on the ILSI Europe website at www.ilsi.eu. For further information about ILSI Europe, please email info@ilsieurope.be or call + 32 2771 00 14.
Disclaimer: The opinions expressed herein and the conclusions of this publication are those of the authors and do not necessarily represent the views of ILSI Europe nor those of its member companies.
REFERENCES
- Becker R. A., Dreier D. A., Manibusan M. K., Cox L. A., Simon T. W., Bus J. S. (2017). How well can carcinogenicity be predicted by high throughput “characteristics of carcinogens” mechanistic data? Regul. Toxicol. Pharmacol. 90, 185–196. [DOI] [PubMed] [Google Scholar]
- Becker R. A., Hays S. M., Kirman C. R., Aylward L. L., Wise K. (2014). Interpreting estrogen screening assays in the context of potency and human exposure relative to natural exposures to phytoestrogens. Birth Defects Res. Part B Dev. Reprod. Toxicol. 101, 114–124. [DOI] [PubMed] [Google Scholar]
- Beekmann K., Actis-Goretta L., van Bladeren P. J., Dionisi F., Destaillats F., Rietjens I. M. C. M. (2012). A state-of-the-art overview of the effect of metabolic conjugation on the biological activity of flavonoids. Food Funct.3, 1008–1018. [DOI] [PubMed] [Google Scholar]
- Blackburn K. L., Ellison C. A., Stuard S. B., Wu S. (2019). Dosimetry considerations for in vivo and in vitro test data and a novel surrogate iTTC approach for read-across based on metabolites. Comput. Toxicol. 10, 145–157. [Google Scholar]
- Boonpawa R., Spenkelink A., Punt A., Rietjens I. M. C. M. (2017). In vitro-in silico-based analysis of the dose-dependent in vivo oestrogenicity of the soy phytoestrogen genistein in humans. Br. J. Pharmacol. 174, 2739–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. M., Dingemans M. M. L., Baken K. A., van Wezel A. P. (2019). Prioritizing anthropogenic chemicals in drinking water and sources through combined use of mass spectrometry and ToxCast toxicity data. J. Hazard. Mater. 364, 332–338. [DOI] [PubMed] [Google Scholar]
- Cao Y., Charisi A., Cheng L.-C., Jiang T., Girke T. (2008). ChemmineR: A compound mining framework for R. Bioinformatics 24, 1733–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox L. A., Popken D., Marty M. S., Rowlands J. C., Patlewicz G., Goyak K. O., Becker R. A. (2014). Developing scientific confidence in HTS-derived prediction models: Lessons learned from an endocrine case study. Regul. Toxicol. Pharmacol. 69, 443–450. [DOI] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. (2006). The igraph software package for complex network research. InterJ. Complex Syst. 1695, 1–9. [Google Scholar]
- DeGroot D. E., Swank A., Thomas R. S., Strynar M., Lee M.-Y., Carmichael P. L., Simmons S. O. (2018). mRNA transfection retrofits cell-based assays with xenobiotic metabolism. J. Pharmacol. Toxicol. Methods 92, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent M. P., Li H., Carmichael P. L., Martin F. L. (2019). Employing dietary comparators to perform risk assessments for anti-androgens without using animal data. Toxicol. Sci. 167, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DG SANTE. (2011) EU Database on Food Additives. Available at: https://webgate.ec.europa.eu/foods_system/main/? sector=FAD&auth=SANCAS. Accessed January 30, 2020.
- Dimitrov S. D., Diderich R., Sobanski T., Pavlov T. S., Chankov G. V., Chapkanov A. S., Karakolev Y. H., Temelkov S. G., Vasilev R. A., Gerova K. D., et al. (2016). QSAR Toolbox—Workflow and major functionalities. SAR QSAR Environ. Res. 27, 203–219. [DOI] [PubMed] [Google Scholar]
- Dorne J. L., Richardson J., Kass G., Georgiadis N., Monguidi M., Pasinato L., Cappe S., Verhagen H., Robinson T. (2017). Editorial: OpenFoodTox: EFSA’s open source toxicological database on chemical hazards in food and feed. EFSA J. 15, e15011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA. (2004). Opinion of the Scientific Panel on food additives, flavourings, processing aids and materials in contact with food (AFC) related to para hydroxybenzoates (E 214-219). EFSA J. 2, 83. [Google Scholar]
- EFSA. (2014). Scientific Opinion on the re-evaluation of propyl gallate (E 310) as a food additive. EFSA J 12, 46. [Google Scholar]
- EFSA. (2015a). Scientific Opinion on on the re-evaluation of dodecyl gallate (E 312) as a food additive. EFSA J. 13, 39. [Google Scholar]
- EFSA. (2015b). Scientific Opinion on the re-evaluation of octyl gallate (E 311) as a food additive. EFSA J 13, 39. [Google Scholar]
- EFSA. (2017) OpenFoodTox Database Available at: https://www.efsa.europa.eu/en/microstrategy/openfoodtox. Accessed January 30, 2020.
- EFSA. (2019). Scientific Panel on additives and products or substances used in animal feed. Minutes of the 106th Meeting of the Working Group on Technological Additives Available at: https://www.efsa.europa.eu/sites/default/files/wgs/animal-feed/wg-technological-additives-2018-2021.
- EMA. (2015). Reflection Paper on the Use of Methyl- and Propylparaben as Excipients in Human Medicinal Products for Oral Use Available at: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-use-methyl-propylparaben-excipients-human-medicinal-products-oral-use_en.pdf. Accessed January 30, 2020.
- EU. (2017). Commission Implementing Regulation (EU) 2017/962 of 7 June 2017 suspending the authorisation of ethoxyquin as a feed additive for all animal species and categories. Off. J. Eur. Comm. L 145, 13–17. [Google Scholar]
- Fabian E., Gomes C., Birk B., Williford T., Hernandez T. R., Haase C., Zbranek R., van Ravenzwaay B., Landsiedel R. (2019). In vitro-to-in vivo extrapolation (IVIVE) by PBTK modeling for animal-free risk assessment approaches of potential endocrine-disrupting compounds. Arch. Toxicol. 93, 401–416. [DOI] [PubMed] [Google Scholar]
- Han X., Beaumont C., Stevens N. (2017). Chemical composition analysis and in vitro biological activities of ten essential oils in human skin cells. Biochim. Open 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. (2001). Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276, 4365–4372. [DOI] [PubMed] [Google Scholar]
- Hartman J. K., Beames T., Parks B., Doheny D., Song G., Efremenko A., Yoon M., Foley B., Deisenroth C., McMullen P. D., et al. (2018). An in vitro approach for prioritization and evaluation of chemical effects on glucocorticoid receptor mediated adipogenesis. Toxicol. Appl. Pharmacol. 355, 112–126. [DOI] [PubMed] [Google Scholar]
- Houck K. A., Judson R. S., Knudsen T. B., Martin M. T., Richard A. M., Crofton K. M., Simeonov A., Paules R. S., Bucher J. R., Thomas R. S. (2017). Comment on “On the utility of ToxCastTM and ToxPi as methods for identifying new obesogens.” Environ. Health Perspect. 7425, A8–A11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S., Pham N., Marty M., Sandy M., Solomon G., Zeise L. (2019). An integrated approach using publicly available resources for identifying and characterizing chemicals of potential toxicity concern: Proof-of-concept with chemicals that affect cancer pathways. Toxicol. Sci. 169, 14–24. [DOI] [PubMed] [Google Scholar]
- Jelen H. (2012). Food Flavors: Chemical, Sensory and Technological Properties. CRC Press/Taylor & Francis, Boca Raton, FL. [Google Scholar]
- Judson R., Houck K., Martin M., Richard A. M., Knudsen T. B., Shah I., Little S., Wambaugh J., Setzer R. W., Kothiya P., et al. (2016). Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci. 153, 409–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Magpantay F. M., Chickarmane V., Haskell C., Tania N., Taylor J., Xia M., Huang R., Rotroff D. M., Filer D. L., et al. (2015). Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci. 148, 137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmaus A. L., Filer D. L., Martin M. T., Houck K. A. (2016). Evaluation of food-relevant chemicals in the ToxCast high-throughput screening program. Food Chem. Toxicol. 92, 188–196. [DOI] [PubMed] [Google Scholar]
- Karmaus A. L., Trautman T. D., Krishan M., Filer D. L., Fix L. A. (2017). Curation of food-relevant chemicals in ToxCast. Food Chem. Toxicol. 103, 174–182. [DOI] [PubMed] [Google Scholar]
- Kavlock R., Chandler K., Houck K., Hunter S., Judson R., Kleinstreuer N., Knudsen T., Martin M., Padilla S., Reif D., et al. (2012). Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N. C., Yang J., Berg E. L., Knudsen T. B., Richard A. M., Martin M. T., Reif D. M., Judson R. S., Polokoff M., Dix D. J., et al. (2014). Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol. 32, 583–591. [DOI] [PubMed] [Google Scholar]
- Lizarraga L. E., Dean J. L., Kaiser J. P., Wesselkamper S. C., Lambert J. C., Zhao Q. J. (2019). A case study on the application of an expert-driven read-across approach in support of quantitative risk assessment of p,p′-dichlorodiphenyldichloroethane. Regul. Toxicol. Pharmacol. 103, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louisse J., Dingemans M. M. L., Baken K. A., van Wezel A. P., Schriks M. (2018). Exploration of ToxCast/Tox21 bioassays as candidate bioanalytical tools for measuring groups of chemicals in water. Chemosphere 209, 373–380. [DOI] [PubMed] [Google Scholar]
- Lund E. G., Peterson L. B., Adams A. D., Lam M.-H. N., Burton C. A., Chin J., Guo Q., Huang S., Latham M., Lopez J. C., et al. (2006). Different roles of liver X receptor α and β in lipid metabolism: Effects of an α-selective and a dual agonist in mice deficient in each subtype. Biochem. Pharmacol. 71, 453–463. [DOI] [PubMed] [Google Scholar]
- Mayr L. M., Fuerst P. (2008). The future of high-throughput screening. J. Biomol. Screen. 13, 443–448. [DOI] [PubMed] [Google Scholar]
- Mazaira G. I., Zgajnar N. R., Lotufo C. M., Daneri-Becerra C., Sivils J. C., Soto O. B., Cox M. B., Galigniana M. D. (2019) Nuclear Receptors: A Historical Perspective, pp. 1–5. Humana, New York, NY. [DOI] [PubMed] [Google Scholar]
- Mellor C. L., Marchese Robinson R. L., Benigni R., Ebbrell D., Enoch S. J., Firman J. W., Madden J. C., Pawar G., Yang C., Cronin M. T. D. (2019). Molecular fingerprint-derived similarity measures for toxicological read-across: Recommendations for optimal use. Regul. Toxicol. Pharmacol. 101, 121–134. [DOI] [PubMed] [Google Scholar]
- Müller M., Kersten S. (2003). Nutrigenomics: Goals and strategies. Nat. Rev. Genet. 4, 315–322. [DOI] [PubMed] [Google Scholar]
- Olker J. H., Korte J. J., Denny J. S., Hartig P. C., Cardon M. C., Knutsen C. N., Kent P. M., Christensen J. P., Degitz S. J., Hornung M. W. (2019). Screening the ToxCast phase 1, phase 2, and e1k chemical libraries for inhibitors of iodothyronine deiodinases. Toxicol. Sci. 168, 430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paini A., Guignard G., Bezencon C., Latado H., Schilter B., van Bladeren P. J., Rietjens I. M. C. M., Marin-Kuan M. (2011). DNA damage response induced by 1′-hydroxyestragole in rat primary hepatocytes. In Generation of In Vitro Data to Model Dose Dependent In Vivo DNA Binding of Genotoxic Carcinogens and Its Consequences: The Case of Estragole. Available at; http://edepot.wur.nl/200999. Accessed January 30, 2020.
- Pedersen T. L. (2017). ggraph: An Implementation of Grammar of Graphics for Graphs and Networks Available at: https://cran.r-project.org/package=ggraph. Accessed January 30, 2020.
- Phillips D. H., Reddy M. V., Randerath K. (1984). 32P-post-labelling analysis of DNA adducts formed in the livers of animals treated with safrole, estragole and other naturally-occurring alkenylbenzenes. II. Newborn male B6C3F1 mice. Carcinogenesis 5, 1623–1628. [DOI] [PubMed] [Google Scholar]
- Popeijus H. E., van Otterdijk S. D., van der Krieken S. E., Konings M., Serbonij K., Plat J., Mensink R. P. (2014). Fatty acid chain length and saturation influences PPARα transcriptional activation and repression in HepG2 cells. Mol. Nutr. Food Res. 58, 2342–2349. [DOI] [PubMed] [Google Scholar]
- Prinsloo G., Papadi G., Hiben M. G., de Haan L., Louisse J., Beekmann K., Vervoort J., Rietjens I. M. C. M. (2017). In vitro bioassays to evaluate beneficial and adverse health effects of botanicals: Promises and pitfalls. Drug Discov. Today 22, 1187–1200. [DOI] [PubMed] [Google Scholar]
- Punt A., Delatour T., Scholz G., Schilter B., van Bladeren P. J., Rietjens I. M. C. M. (2007). Tandem Mass Spectrometry Analysis of N2-( trans-Isoestragol-3‘-yl)-2‘-deoxyguanosine as a Strategy to Study Species Differences in Sulfotransferase Conversion of the Proximate Carcinogen 1‘-Hydroxyestragole. Chem. Res. Toxicol.20, 991–998. [DOI] [PubMed] [Google Scholar]
- Punt A., Aartse A., Bovee T. F. H., Gerssen A., van Leeuwen S. P. J., Hoogenboom R. L. A. P., Peijnenburg A. A. C. M. (2019). Quantitative in vitro-to-in vivo extrapolation (QIVIVE) of estrogenic and anti-androgenic potencies of BPA and BADGE analogues. Arch. Toxicol. 93, 1941–1953. [DOI] [PubMed] [Google Scholar]
- Richard A. M., Judson R. S., Houck K. A., Grulke C. M., Volarath P., Thillainadarajah I., Yang C., Rathman J., Martin M. T., Wambaugh J. F., et al. (2016). ToxCast chemical landscape: Paving the road to 21st century toxicology. Chem. Res. Toxicol. 29, 1225–1251. [DOI] [PubMed] [Google Scholar]
- Rietjens I. M. C. M., Louisse J., Beekmann K. (2017). The potential health effects of dietary phytoestrogens. Br. J. Pharmacol. 174, 1263–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan N. (2017). A User’s Guide for Accessing and Interpreting ToxCast Data Available at: https://lri.americanchemistry.com/Users-Guide-for-Accessing-and-Interpreting-ToxCast-Data.pdf. Accessed January 20, 2020.
- SCCS. (2013). Scientific Committee on Consumer Safety (CSSC) Opinion on Parabens. Updated Request for a Scientific Opinion on Propyl- and Butylparaben Available at: https://ec.europa.eu/health/scientific_committees/consumer_safety/docs/sccs_o_132.pdf. Accessed September 12, 2019.
- SCF. (2001a). Opinion of the Scientific Committee on Food on Estragole (1-Allyl-4-methoxybenzene). SCF/CS/FLAV/FLAVOUR/6 ADD2 FINAL 26 September 2001. Available at: https://ec.europa.eu/food/sites/food/files/safety/docs/sci-com_scf_out104_en.pdf. Accessed January 30, 2020. [Google Scholar]
- SCF. (2001b). Opinion of the Scientific Committee on Food on Methyleugenol (4-Allyl-1,2-dimethoxybenzene) (adopted on 26 September 2001). Available at: https://ec.europa.eu/food/sites/food/files/safety/docs/sci-com_scf_out102_en.pdf. Accessed January 30, 2020. [Google Scholar]
- Sipes N. S., Martin M. T., Kothiya P., Reif D. M., Judson R. S., Richard A. M., Houck K. A., Dix D. J., Kavlock R. J., Knudsen T. B. (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol. 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Bahadori T., Buckley T. J., Cowden J., Deisenroth C., Dionisio K. L., Frithsen J. B., Grulke C. M., Gwinn M. R., Harrill J. A., et al. (2019). The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 169, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA. (2018a). ToxCast and Tox21 Summary Files. figshare. Dataset. Available at: 10.23645/epacomptox.6062479.v2. Accessed January 20, 2020. [DOI]
- U.S. EPA. (2018b). ToxCast Owner’s Manual—Guidance for Exploring Data Available at: https://www.epa.gov/sites/production/files/2018-04/documents/toxcastownermanual4252018.pdf. Accessed January 20, 2020.
- Varga T., Czimmerer Z., Nagy L. (2011). PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 1812, 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D. L., Coady K., Escher B. I., Mihaich E., Murphy C. A., Schlekat T., Garcia-Reyero N. (2019). High-throughput screening and environmental risk assessment: State of the science and emerging applications. Environ. Toxicol. Chem. 38, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Backman T. W. H., Horan K., Girke T. (2013). fmcsR: Mismatch tolerant maximum common substructure searching in R. Bioinformatics 29, 2792–2794. [DOI] [PubMed] [Google Scholar]
- Watt E. D., Judson R. S. (2018). Uncertainty quantification in ToxCast high throughput screening. PLoS One 13, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Allen B., Ferguson S. S., Sochaski M. A., Setzer R. W., Houck K. A., Strope C. L., Cantwell K., Judson R. S., et al. (2015). Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical toxicity testing. Toxicol. Sci. 148, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. M. (2000). Dietary fatty acids and human health. Ann. Zootech. 49, 165–180. [Google Scholar]
- Williamson G., Kay C. D., Crozier A. (2018). The bioavailability, transport, and bioactivity of dietary flavonoids: A review from a historical perspective. Compr. Rev. Food Sci. Food Saf. 17, 1054–1112. [DOI] [PubMed] [Google Scholar]
- Yang C., Tarkhov A., Marusczyk J., Bienfait B., Gasteiger J., Kleinoeder T., Magdziarz T., Sacher O., Schwab C. H., Schwoebel J., et al. (2015). New publicly available chemical query language, CSRML, to support chemotype representations for application to data mining and modeling. J. Chem. Inf. Model. 55, 510–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.