Abstract
Implementing screening assays that identify functional and structural cardiotoxicity earlier in the drug development pipeline has the potential to improve safety and decrease the cost and time required to bring new drugs to market. In this study, a metabolic biomarker-based assay was developed that predicts the cardiotoxicity potential of a drug based on changes in the metabolism and viability of human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM). Assay development and testing was conducted in 2 phases: (1) biomarker identification and (2) targeted assay development. In the first phase, metabolomic data from hiPSC-CM spent media following exposure to 66 drugs were used to identify biomarkers that identified both functional and structural cardiotoxicants. Four metabolites that represent different metabolic pathways (arachidonic acid, lactic acid, 2′-deoxycytidine, and thymidine) were identified as indicators of cardiotoxicity. In phase 2, a targeted, exposure-based biomarker assay was developed that measured these metabolites and hiPSC-CM viability across an 8-point concentration curve. Metabolite-specific predictive thresholds for identifying the cardiotoxicity potential of a drug were established and optimized for balanced accuracy or sensitivity. When predictive thresholds were optimized for balanced accuracy, the assay predicted the cardiotoxicity potential of 81 drugs with 86% balanced accuracy, 83% sensitivity, and 90% specificity. Alternatively, optimizing the thresholds for sensitivity yields a balanced accuracy of 85%, 90% sensitivity, and 79% specificity. This new hiPSC-CM-based assay provides a paradigm that can identify structural and functional cardiotoxic drugs that could be used in conjunction with other endpoints to provide a more comprehensive evaluation of a drug’s cardiotoxicity potential.
Keywords: cardiotoxicity, drug discovery and development, in vitro, hiPSC-CM, metabolites
Predicting the potential for cardiotoxicity of drug candidates can be challenging due to the broad range of mechanisms that can adversely affect the heart. As a result, cardiotoxicity remains a major cause of compound attrition throughout the drug discovery and development process as well as withdrawal of Food and Drug Administration approved drugs from the market (Hornberg et al., 2014; Laverty et al., 2011; Onakpoya et al., 2016; Valentin and Redfern, 2017; Weaver and Valentin, 2019). Drug-induced cardiotoxicity can occur after acute or chronic treatment and is typically categorized as functional (eg, change in mechanical function of the myocardium) or structural (eg, morphological damage or loss of cellular/subcellular components) (Cross et al., 2015; Laverty et al., 2011). Structural cardiotoxicity is a well-known adverse effect of numerous drugs, including anticancer (anthracyclines, tyrosine kinase inhibitors, and antimetabolites), antiretroviral (azidothymidine), and antidiabetic (rosiglitazone) drugs, among others (Hantson, 2019; Klimas, 2012; Mellor et al., 2011; Pai and Nahata, 2000; Shah et al., 2013; Zuppinger et al., 2007), yet it is often underassessed or not evaluated in early drug discovery (Weaver and Valentin, 2019).
The current in vitro preclinical evaluations for assessing cardiotoxicity largely focus on the impact of a drug on individual ion channels in an acute testing paradigm. In addition to these in vitro methods, animal models are often used to assess cardiotoxicity but are limited by their throughput, cost, and species-specific differences in cardiac structure, electrophysiology, signaling, and gene expression (Edwards and Louch, 2017; Force and Kolaja, 2011; Khan et al., 2013). Due to these limitations, regulatory agencies are currently reviewing alternative or complementary methods for evaluating proarrhythmia risk prior to Investigational New Drug applications (Chi, 2013; Sager et al., 2014). For example, the Comprehensive in vitro Proarrhythmia Assay initiative aims to improve current regulatory guidance by introducing alternative predictive technologies, including human stem cell-derived cardiomyocytes, into preclinical safety assessment. Successful advances have been made in preclinical functional cardiotoxicity screening; however, methods to evaluate cardiomyopathy and other more elusive, but high-risk, liabilities earlier during compound development have yet to be developed (Yang and Papoian, 2018).
Human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) are a biologically relevant in vitro model for assessing drug-induced cardiotoxicity that functionally express most of the human cardiac ion channels and sarcomeric proteins (Huo et al., 2017). The goal of this study was to develop a novel cardiotoxicity assay that identifies functional and structural drug-induced cardiotoxicity based on changes in hiPSC-CM metabolism. Our metabolomics platform profiles changes in metabolism that can be measured in the spent cell culture medium from hiPSC-CM following compound exposure. This “metabolic footprint” is a functional measurement of cellular metabolism referred to as the secretome which is composed of media components, metabolites passively and actively transported across the plasma membrane, and those produced through extracellular metabolism of enzymes. Secretome changes elicited by test compounds are indicative of alterations that occur both inside the cell and alteration of the extracellular matrix (Kell et al., 2005).
Previous in vivo and in vitro metabolomics studies have been conducted following treatment with many known structural cardiotoxicants and demonstrated that metabolic perturbations are an important mechanism underlying drug-induced cardiotoxicity; however, no metabolomics-based studies have been published evaluating the effects of functional cardiotoxicants (Chaudhari et al., 2017; Jensen et al., 2017a,b; Li et al., 2015; Niu et al., 2016; Schnackenberg et al., 2016; Tan et al., 2011; Wang et al., 2009; Yin et al., 2016). For example, metabolomic profiling of rat urine following treatment with doxorubicin was able to detect the effects of doxorubicin-induced cardiotoxicity earlier than clinical chemistry (eg, lactate dehydrogenase and creatine kinase) and histopathological assessment (Wang et al., 2009). This result was consistent with the results from Li et al., which found that metabolite biomarkers in rat plasma after exposure to doxorubicin, 5-fluorouracil, or isoproterenol were able to predict cardiotoxicity earlier than biochemical analysis and histopathological assessment (Li et al., 2015). Statistically significant changes in long-chain omega-3 fatty acids and taurine/hypotaurine metabolism were identified in mouse hearts following treatment with sunitinib and sorafenib, respectively (Jensen et al., 2017a,b). Taken together, these studies demonstrate that cardiotoxicity is associated with changes in metabolism and those changes can be detected through metabolomic analysis.
In this study, we developed a biomarker-based assay for evaluating the cardiotoxicity potential of drug candidates based on changes in the metabolism and viability of hiPSC-CM. Ultra-performance liquid chromatography-high-resolution mass spectrometry (UPLC-HRMS) was used to profile the metabolic response following exposure to a diverse set of drugs with well-characterized cardiac liabilities to identify a reproducible set of biomarkers capable of predicting cardiotoxicity potential. A rapid, targeted assay that measured changes in metabolism and cellular viability across an 8-point concentration-response curve was used to determine the exposure level at which a drug perturbs metabolism in a manner associated with cardiotoxicity potential. The predictive model for the new assay was defined and assessed with 81 drugs from several different therapeutic classes known to cause a broad range of cardiotoxic effects. The assay described here is complementary to the electrophysiological tests many companies are already conducting with hiPSC-CM and could be conducted efficiently at the same time as these tests for a more comprehensive evaluation of a drug’s cardiotoxicity potential.
MATERIALS AND METHODS
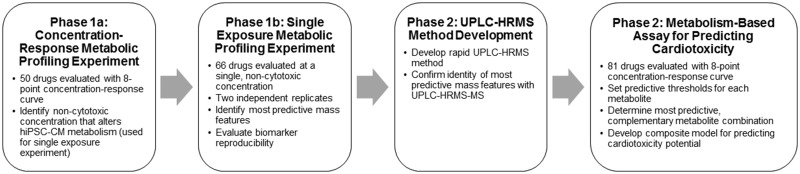
Development and evaluation of a targeted biomarker-based cardiotoxicity assay was conducted in 2 phases (Figure 1):
Figure 1.
Diagram outlining the development of the targeted biomarker assay for predicting cardiotoxicity potential.
- In the first phase, 2 metabolic profiling experiments were conducted using untargeted metabolomics methods to identify predictive metabolites that could discriminate cardiotoxicants from noncardiotoxicants independent of changes in cell viability and confirm their reproducibility.
- In the first experiment, a concentration-response study was conducted to identify secreted metabolites that changed in response to treatment and select a single, noncytotoxic exposure for each drug.
- The second experiment tested the predictivity and reproducibility of metabolite response using 2 independent replications of the drugs treated at a single exposure level.
In the second phase, the discriminatory metabolites identified in phase 1 were used to create an exposure-based, targeted assay for identifying a drug’s cardiotoxicity potential. In order to develop a rapid assay designed for screening, the sample preparation and UPLC-HRMS methods were optimized for the predictive metabolites utilized in the new assay, allowing for decreased complexity and increased throughput. The predictivity was evaluated with 81 drugs using 4-fold cross-validation. All drugs were tested using an 8-point concentration-response curve and truth was scored against the response at 10× the maximum total plasma concentration (Cmax).
Drug selection and classification
A total of 81 drugs were used for developing and assessing the targeted biomarker assay (52 cardiotoxic and 29 noncardiotoxic; Table 1). A subset of the drugs was used for the phase 1 studies to identify the predictive biomarkers and confirm their reproducibility (66 drugs; Table 1). All 81 drugs were used during phase 2 studies for prediction model training and testing.
Table 1.
Description of Drugs Tested, Reported Cardiotoxic Effects, Therapeutically Relevant Total Cmax, and Exposure Selected for Single Concentration Study in Phase 1
| Drug | Cardiotoxicity Classification | Type of Cardiotoxicity | Drug Class or Use | Cardiovascular Toxic Effects | Total Cmax (µM) | Selected Single Exposure Level (µM) | Cardiotoxicity Reference |
|---|---|---|---|---|---|---|---|
| Astemizolea | Cardiotoxic | Functional | Antihistamine | Prolonged QT, TdP | 0.008 | 0.03 | Paakkari (2002), Qureshi et al. (2011), and Colatsky et al. (2016) |
| Bepridila | Cardiotoxic | Functional | Antianginal | Prolonged QT, TdP | 3.3 | 3.3 | Clements et al. (2015) and Colatsky et al. (2016) |
| Chlorpromazine | Cardiotoxic | Functional | Antipsychotic | Prolonged QT, TdP, orthostatic hypotension, reflex tachycardia | 1.1 | N/A | Colatsky et al. (2016) and West-Ward Pharmaceuticals Corp (2018) |
| Cisapridea | Cardiotoxic | Functional | Prokinetic | Prolonged QT, TdP | 0.18 | 1.8 | Paakkari (2002), Qureshi et al. (2011), and Colatsky et al. (2016) |
| Dofetilidea | Cardiotoxic | Functional | Class III antiarrhythmic | Prolonged QT, TdP | 0.008 | 0.03 | Colatsky et al. (2016) and Aurobindo Pharma Limited (2019) |
| Encainidea | Cardiotoxic | Functional | Class Ic antiarrhythmic | Ventricular arrhythmias | 0.71 | 7.1 | Fung et al. (2001) and Qureshi et al. (2011) |
| Levomethadyl acetatea | Cardiotoxic | Functional | Analgesic | Prolonged QT, TdP | 0.6 | 6 | Roxane Laboratories Inc (2002) and Qureshi et al. (2011) |
| Nifedipinea | Cardiotoxic | Functional | Antihypertensive | Hypertension, angina | 0.58 | 0.58 | Greenstone LLC (2018) |
| Ondansetron | Cardiotoxic | Functional | Antiemetic | Prolonged QT, TdP, ventricular tachycardia | 0.7 | N/A | Colatsky et al. (2016) and The Medicines Company (2019) |
| Quinidine | Cardiotoxic | Functional | Class Ia antiarrhythmic | Prolonged QT, TdP | 21.6 | N/A | Colatsky et al. (2016) and Eon Labs Inc (2018) |
| Sertindolea | Cardiotoxic | Functional | Antipsychotic | Prolonged QT, TdP | 0.32 | 0.3 | Fung et al. (2001) and Lindström et al. (2005) |
| Sotalola | Cardiotoxic | Functional | Class III antiarrhythmic | Prolonged QT, TdP | 15 | 15 | Colatsky et al. (2016) and Bayshore Pharmaceuticals LLC (2019) |
| Terfenadinea | Cardiotoxic | Functional | Antihistamine | Prolonged QT, TdP | 0.3 | 3 | Fung et al. (2001), Paakkari (2002), Soldovieri et al. (2008), Qureshi et al. (2011), and Colatsky et al. (2016) |
| Thioridazinea | Cardiotoxic | Functional | Antipsychotic | Prolonged QT, TdP | 2.7 | 1 | Mylan Pharmaceuticals Inc (2018) |
| Verapamila | Cardiotoxic | Functional | Class IV antiarrhythmic, antihypertensive | CHF, pulmonary edema, hypotension, ventricular fibrillation | 0.815 | 0.815 | Ranbaxy Laboratories Inc (2008) |
| Azidothymidine | Cardiotoxic | Structural | Antiviral | Cardiomyopathy, syncope | 8.6 | N/A | Figueredo (2011) and Acetris Health LLC (2019) |
| Busulfana | Cardiotoxic | Structural | Antineoplastic | CHF, edema, tachycardia, cardiac tamponade, hypotension, hypertension, vasodilation | 49.6 | 50 | BC Cancer Agency (2018) |
| Daunorubicina | Cardiotoxic | Structural | Antineoplastic | Cardiomyopathy, MI, CHF, ventricular arrhythmia, pericarditis/myocarditis | 89 | 0.1 | Figueredo (2011), BC Cancer Agency (2014a), and Hikma Pharmaceuticals USA Inc (2018) |
| Dexfenfluraminea | Cardiotoxic | Structural | Anorectic | Cardiac valvular disease, cardiac fibrosis | 0.4 | 4 | Fung et al. (2001) and Bayzigitov et al. (2016) |
| Dithiazanine Iodidea | Cardiotoxic | Structural | Antihelminthic | Cardiovascular and metabolic reaction | 0.1 | 0.1 | Bayzigitov et al. (2016) |
| Doxorubicina | Cardiotoxic | Structural | Antineoplastic | CHF, decreased LVEF, sinus tachycardia, myocarditis, cardiomyopathy | 15.3 | 0.1 | Figueredo (2011), BC Cancer Agency (2016a), and Amneal Pharmaceuticals LLC (2019) |
| Idarubicina | Cardiotoxic | Structural | Antineoplastic | Cardiomyopathy, MI, CHF, ventricular arrhythmia, decreased LVEF | 0.12 | 0.1 | Figueredo (2011), BC Cancer Agency (2019a), and Hikma Pharmaceuticals USA Inc (2019) |
| Imatiniba | Cardiotoxic | Structural | Antineoplastic | CHF, decreased LVEF | 3.9 | 3 | Kerkelä et al. (2006), Xu et al. (2009), and Pun and Neilan (2016) |
| Nandrolone decanoate | Cardiotoxic | Structural | Anabolic steroid | LV hypertrophy, hypertension, impaired diastolic filling | 0.01 | N/A | Sullivan et al. (1998), Figueredo (2011), and Vasilaki et al. (2016) |
| Pergolidea | Cardiotoxic | Structural | Antidyskinetic | Cardiac valvulopathy, postural hypotension, syncope, hypertension, palpitations, vasodilatations, CHF | 0.003 | 0.03 | Amarin Pharmaceuticals Inc (2003), World Health Organization (2004), and Qureshi et al. (2011) |
| Rofecoxiba | Cardiotoxic | Structural | Nonsteroidal anti-inflammatory drug (NSAID) | MI, serious cardiovascular thrombotic events, sudden death | 1.7 | 17 | Merck & Co Inc (2002) and Qureshi et al. (2011) |
| Rosiglitazonea | Cardiotoxic | Structural | Hypoglycemic | CHF, MI | 1.7 | 1.7 | Physicians Total Care Inc (2010) |
| Tegaseroda | Cardiotoxic | Structural | Prokinetic | HF, cardiac ischemia | 0.08 | 0.8 | Qureshi et al. (2011) and Alfasigma USA Inc (2019) |
| Telmisartan | Cardiotoxic | Structural | Antihypertensive | MI (high doses) | 1.17 | N/A | H.K. Kim et al. (2012) and Jubilant Cadista Pharmaceuticals Inc (2019) |
| Trastuzumab | Cardiotoxic | Structural | Antineoplastic | Cardiomyopathy, LVD, CHF | 377 µg/ml | N/A | BC Cancer Agency (2014b) and Genentech Inc (2019) |
| Valdecoxiba | Cardiotoxic | Structural | NSAID | Cardiomyopathy, CHF, hypertension, angina, arrhythmia | 0.51 | 5.1 | G.D. Searle LLC (2001), Moodley (2008), and Qureshi et al. (2011) |
| Amiodaronea | Cardiotoxic | General | Class III antiarrhythmic | Arrhythmia, heart block, sinus bradycardia, CHF, ventricular fibrillation | 3.9 | 3 | Cameron Pharmaceuticals LLC (2019) |
| Amitriptylinea | Cardiotoxic | General | Antidepressant | Arrhythmia, prolonged QT, myopathy, MI, TdP | 3.6 | 1 | Accord Healthcare Inc (2019) |
| Amphotericin Ba | Cardiotoxic | General | Antifungal | Arrhythmia, atrial fibrillation, bradycardia, cardiac arrest, cardiomegaly | 89.8 | 3 | Intermune Inc (2006) |
| Amsacrine | Cardiotoxic | General | Antineoplastic | Atrial and ventricular tachyarrhythmia, CHF, hypotension, cardiopulmonary arrest | 15 | N/A | BC Cancer Agency (2013a) |
| Anagrelidea | Cardiotoxic | General | Antiplatelet | CHF, cardiomyopathy, atrial fibrillation, heart block, prolonged QT, TdP, arrhythmia | 0.06 | 0.6 | Figueredo (2011), BC Cancer Agency (2015), and Torrent Pharmaceuticals Limited (2019) |
| Arsenic trioxidea | Cardiotoxic | General | Antineoplastic | Prolonged QT, TdP, cardiomyopathy, tachycardia | 12.1 | 10 | BC Cancer Agency (2014c) and Ingenus Pharmaceuticals LLC (2019) |
| Bortezomiba | Cardiotoxic | General | Antineoplastic | CHF, decreased LVEF, isolated cases of prolonged QT, hypotension | 0.3 | 0.1 | BC Cancer Agency (2016b) |
| Chloroquinea | Cardiotoxic | General | Antimalarial | Prolonged QT and QRS, AV blocks, cardiomyopathy, cardiac hypertrophy | 0.96 | 3 | Figueredo (2011) and Ranbaxy Pharmaceuticals Inc (2014) |
| Clozapinea | Cardiotoxic | General | Antipsychotic | MI, myocarditis, arrhythmia, prolonged QT, TdP, cardiomyopathy (rare) | 0.95 | 9.5 | Stöllberger et al. (2005), Layland (et al.) 2009, Figueredo (2011), and Colatsky et al. (2016) |
| Crizotinib | Cardiotoxic | General | Antineoplastic | Prolonged QT, bradycardia | 0.73 | N/A | Pun and Neilan (2016) and Pfizer Laboratories (2019a) |
| Dasatiniba | Cardiotoxic | General | Antineoplastic | Prolonged QT, CHF, LVD, MI, cardiomyopathy, arrhythmia, cardiomegaly | 0.72 | 0.72 | Talpaz et al. (2006) and Xu et al. (2009), Pun and Neilan (2016), E.R. Squibb & Sons L.L.C. (2019) |
| Fluorouracila | Cardiotoxic | General | Antineoplastic | MI, CHF, coronary vasospasm, prolonged QT, (supra)ventricular tachycardia, LVD, cardiac fibrillation | 4.6 | 46 | Schimmel et al. (2004), Accord Healthcare Inc (2017), and BC Cancer Agency (2019b) |
| Isoproterenola | Cardiotoxic | General | Bronchodilator | Tachycardia, palpitations, ventricular arrhythmias, myocarditis | 0.01 | 0.1 | Zhang et al. (2008) and Cipla USA Inc (2019) |
| Lapatinib1 | Cardiotoxic | General | Antineoplastic | Prolonged QT, TdP, HF, decreased LVEF | 4.18 | 4.18 | Perez et al. (2008); Pun and Neilan (2016), and Novartis Pharmaceuticals Corporation (2019a) |
| Mitoxantronea | Cardiotoxic | General | Antineoplastic | CHF, cardiomyopathy, decreased LVEF, tachycardia, arrhythmia | 3.3 | 1 | Figueredo (2011), BC Cancer Agency (2013b), and Teva Parenteral Medicines Inc (2019) |
| Nilotiniba | Cardiotoxic | General | Antineoplastic | Prolonged QT, vascular abnormalities, decreased LVEF | 3 | 3 | Xu et al. (2009), T.D. Kim et al. (2012), Pun and Neilan (2016), and Novartis Pharmaceuticals Corporation (2019b) |
| Nortriptylinea | Cardiotoxic | General | Antidepressant | MI, arrhythmias, prolonged QT, TdP | 3.6 | 1 | BNM Group (2013) and Mayne Pharma Inc (2019) |
| Paclitaxela | Cardiotoxic | General | Antineoplastic | CHF, LVD, sinus bradycardia, atrial and ventricular arrhythmias, MI, supraventricular tachycardia, AV, left bundle branch block | 21.9 | 10 | BC Cancer Agency (2016c) and Mylan Institutional LLC (2018) |
| Sorafeniba | Cardiotoxic | General | Antineoplastic | Prolonged QT (rare), CHF, cardiac ischemia, MI, hypertension | 16.6 | 3 | Schmidinger et al. (2008), Pun and Neilan (2016), and Bayer HealthCare Pharmaceuticals Inc (2019) |
| Sunitiniba | Cardiotoxic | General | Antineoplastic | Prolonged QT, TdP, decreased LVEF, HF, hypertension, cardiomyopathy | 0.25 | 1 | Schmidinger et al. (2008), Pun and Neilan (2016), and Pfizer Laboratories (2019b) |
| Vandetaniba | Cardiotoxic | General | Antineoplastic | Cardiomyopathy, cardiac hypertrophy, myocyte degeneration, prolonged QT, TdP | 3.3 | 1 | Natale et al. (2011), Zang et al. (2012), Colatsky et al. (2016), Pun and Neilan (2016), and AstraZeneca Pharmaceuticals LP (2018) |
| Acetylsalicylic acida | Noncardiotoxic | N/A | NSAID | N/A | 10 | 10 | N/A |
| Acyclovira | Noncardiotoxic | N/A | Antiviral | N/A | 6.7 | 6.7 | N/A |
| Adipic acid | Noncardiotoxic | N/A | Food additive | N/A | 0.09 | N/A | N/A |
| Amoxicillina | Noncardiotoxic | N/A | Antibiotic | N/A | 17 | 17 | N/A |
| Ascorbic acida | Noncardiotoxic | N/A | Vitamin | N/A | 36 | 36 | N/A |
| Aspartamea | Noncardiotoxic | N/A | Food additive/sweetener | N/A | 1 | 1 | N/A |
| Axitinib | Noncardiotoxic | N/A | Antineoplastic | CHF (rare), MI (rare) | 0.07 | N/A | BC Cancer Agency (2014d), Gunnarsson et al. (2015), and Pfizer Laboratories (2019c) |
| Benzoic Acida | Noncardiotoxic | N/A | Food additive/preservative | N/A | 36 | 36 | N/A |
| Biotina | Noncardiotoxic | N/A | Vitamin | N/A | 0.01 | 0.03 | N/A |
| Cetirizinea | Noncardiotoxic | N/A | Antihistamine | N/A | 0.8 | 0.8 | N/A |
| Cimetidine | Noncardiotoxic | N/A | Antihistamine | N/A | 7 | N/A | N/A |
| Citric acida | Noncardiotoxic | N/A | Food additive/antioxidant | N/A | 128 | 100 | N/A |
| Erlotiniba | Noncardiotoxic | N/A | Antineoplastic | MI (rare) | 3.8 | 3 | Natale et al. (2011), Pun and Neilan (2016), and Armas Pharmaceuticals Inc (2019) |
| Gemfibrozil | Noncardiotoxic | N/A | Fibrate or hypolipidemic | N/A | 100 | N/A | N/A |
| Hexylresorcinola | Noncardiotoxic | N/A | Antihelminthic | N/A | 1 | 1 | N/A |
| Leucinea | Noncardiotoxic | N/A | Amino acid | N/A | 126 | 100 | N/A |
| Loratadinea | Noncardiotoxic | N/A | Antihistamine | N/A | 0.02 | 0.02 | N/A |
| Maltola | Noncardiotoxic | N/A | Food additive/flavor agent | N/A | 30 | 30 | N/A |
| Methylparabena | Noncardiotoxic | N/A | Food additive/preservative | N/A | 0.23 | 0.23 | N/A |
| Natamycina | Noncardiotoxic | N/A | Antifungal | N/A | 0.8 | 0.8 | N/A |
| Phenylphenola | Noncardiotoxic | N/A | Antifungal | N/A | 0.09 | 0.09 | N/A |
| Praziquantel | Noncardiotoxic | N/A | Antihelminthic | N/A | 0.64 | N/A | N/A |
| Ranitidinea | Noncardiotoxic | N/A | Antihistamine | N/A | 1.7 | 1.7 | N/A |
| Sildenafila | Noncardiotoxic | N/A | Vasodilator | N/A | 0.95 | 1 | N/A |
| Sucrosea | Noncardiotoxic | N/A | Food additive/sweetener | N/A | 1.8 | 1.8 | N/A |
| Tartaric acida | Noncardiotoxic | N/A | Food additive/antioxidant | N/A | 1.2 | 1.2 | N/A |
| Thiabendazolea | Noncardiotoxic | N/A | Antihelminthic | N/A | 30.8 | 30.8 | N/A |
| Tolbutamide | Noncardiotoxic | N/A | Hypoglycemic | N/A | 217 | N/A | N/A |
| Xylitola | Noncardiotoxic | N/A | Food additive/sweetener | N/A | 0.46 | 0.5 | N/A |
Abbreviations: AV, atrioventricular; CHF, chronic heart failure; HF, heart failure; LVD, left ventricle dysfunction; LVEF, left ventricle ejection fraction; MI, myocardial infarction; TdP, Torsades de Pointes.
Tested in phase 1 studies.
Each drug was classified as noncardiotoxic or cardiotoxic based on its published cardiovascular effects (Table 1). Cardiotoxic drugs were further stratified into 3 classes: functional, structural, and general (drugs that cause both structural and functional effects). The compound set contained both cardiovascular and noncardiovascular drugs known to cause cardiotoxicity in humans, including Na+, K+, and Ca2+ channel blockers, antineoplastic, antiviral, cyclooxygenase-2 inhibitors, receptor agonists and antagonists (adreno, androgen, angiotensin II, dopamine, histamine, muscarinic, peroxisome proliferator activated, and serotonin), and tyrosine kinase inhibitors. The therapeutically relevant total Cmax for each drug was identified from the literature and/or package inserts (Table 1).
Human induced pluripotent stem cell-derived cardiomyocyte culture and treatment
Cryopreserved hiPSC-CM (iCell Cardiomyocytes and iCell Cardiomyocytes2) were obtained from FUJIFILM Cellular Dynamics, Inc (FCDI, Madison, Wisconsin). The phase 1 concentration-response experiments were conducted using iCell Cardiomyocytes as these experiments were performed prior to the release of iCell Cardiomyocytes2. It was decided to use the Cardiomyocytes2 for all other experiments because they have the same metabolic profile (secretome) as the “original” iCell Cardiomyocytes (data not shown). hiPSC-CM were plated with iCell Cardiomyocytes Plating Medium (FCDI) and maintained in iCell Cardiomyocytes Maintenance Medium (FCDI) according to the manufacturer’s recommended protocols. The inner 60 wells of gelatin-coated 96-well plates were seeded with hiPSC-CM at a density of 50 000 plated cells per well. The outer wells of the plates contained an equal volume of media to minimize differences in humidity across the plate. Cell cultures were maintained in a humidified incubator at 37°C under 7% or 5% CO2 (iCell Cardiomyocytes and iCell Cardiomyocytes2, respectively) for 7 or 4 days prior to experimentation (iCell Cardiomyocytes and iCell Cardiomyocytes2, respectively). Maintenance medium was replaced every 48 h prior to beginning treatment.
Drug preparation and treatment
Drugs were purchased from Alfa Aesar (Tewksbury, Massachusetts), Selleck Chemicals (Houston, Texas), MilliporeSigma (St Louis, Missouri), TCI America (Portland, Oregon), or Toronto Research Chemicals (North York, Ontario, Canada), except for crizotinib, telmisartan, and trastuzumab, which were kindly provided by Daiichi Sankyo Co, Ltd (Tokyo, Japan) (Supplementary Table 1). An 8-point concentration-response curve, with half-log dilutions, was used for the concentration-response studies in both phases. The concentration range for each drug was selected to bracket the reported Cmax (Supplementary Table 1). A stock solution of each drug was prepared in 100% dimethyl sulfoxide (DMSO, MilliporeSigma) at a concentration 1000-fold the highest exposure level. Stock solutions were diluted 1:1000 in maintenance medium. For the concentration-response experiments, subsequent half-log serial dilutions were performed in a 2-ml deep-well plate in maintenance medium containing 0.1% DMSO. The final concentration of DMSO was 0.1% in all treatments. Functional cardiotoxicity often occurs after acute exposure, but structural cardiotoxicity can take days to months to develop clinically, so hiPSC-CM were exposed to drug for 72 h. Media and drug were replaced every 24 h and the spent media from the last 24-h treatment period were collected for UPLC-HRMS analysis. In addition to the test drug treatments, each 96-well plate included 0.1% DMSO solvent control cells and media controls to assess the impact of test drugs on the sample matrix.
Cell viability
Cell viability was assessed after UPLC-HRMS sample collection using the CellTiter-Fluor Cell Viability Assay (Promega, Madison, Wisconsin). To determine the relative fold changes for cell viability, the relative fluorescent unit (RFU) value for each sample was first background corrected by subtracting the RFU value of the treatment-specific media blank from the cell sample RFU. Next, the values were normalized to the solvent control (0.1% DMSO treated cells) by dividing the background-corrected RFU value of each sample by the average background-corrected RFU value of the solvent controls. A quality control step was included with criteria that the coefficient of variation of the measured viability RFU of the DMSO control cells could not exceed 10% for a plate to undergo UPLC-HRMS analysis.
Sample preparation
A 60% methanol/40% acetonitrile solution cooled to −20°C was added to each sample to precipitate out the proteins. For phase 1, the protein precipitation solution contained 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid (Cayman Chemical, Ann Arbor, Michigan) as an internal standard (ISTD). Samples were centrifuged at 500 × g at 4°C for 30 min. The supernatant was transferred to a UPLC-HRMS injection plate and concentrated overnight in a Savant High Capacity Speedvac Plus Concentrator. The concentrated sample was resolubilized in a 1:1 solution of 0.1% formic acid in acetonitrile: 0.1% formic acid in water mixture containing 3-isobutyl-1-methylxanthine (MilliporeSigma) and l-citrulline-4,4,5,5-d4 (Cambridge Isotope Laboratories, Inc, Tewksbury, Massachusetts) as additional ISTDs.
The precipitation solution used for phase 2 experiments contained 3 ISTDs, l-citrulline-4,4,5,5-d4, thymidine-α,α,α,6-d4 (C/D/N Isotopes Inc, Pointe-Claire, Quebec, Canada), and sodium l-lactate-3,3,3-d3 (Cambridge Isotope Laboratories, Inc). Samples were centrifuged at 2000 × g at 4°C for 10 min and the supernatant was transferred to a UPLC-HRMS injection plate for analysis.
Ultra-performance liquid chromatography-high-resolution mass spectrometry
For phase 1 biomarker discovery experiments, UPLC-HRMS data were acquired on 2 Agilent LC-HRMS systems (Agilent Technologies, Santa Clara, California), which consisted of an 1290 Infinity LC system interfaced with either an G6520A QTOF HRMS with the MassHunter Acquisition software (version B05.01, Agilent Technologies) or an G6530A QTOF HRMS with the MassHunter Acquisition software (version B 04.00, Agilent Technologies) using high-resolution exact mass conditions. Chromatographic separation of metabolites was performed with an ACQUITY BEH amide column (Waters, Milford, Massachusetts) maintained at 40°C with dimensions 2.1 × 150 mm and 1.7 μm particle size. Sample extracts were held at 4°C and 2 μl was injected for analysis. The data acquisition time was 29 min at a flow rate of 0.5 ml/min, using a 23-min solvent gradient with 5 mM ammonium acetate in water (pH 5.7, solvent A) and 5 mM ammonium acetate in 95% acetonitrile (pH 5.7, solvent B).
Targeted UPLC-HRMS data for phase 2 experiments were acquired on an Agilent 1290 Infinity LC system interfaced with an Agilent G6530A QTOF HRMS using the Agilent MassHunter Acquisition software. The same ACQUITY BEH amide column, maintained at 45°C, was used for metabolite separation. Sample (2 μl) was injected and data were collected over 7 min at a flow rate of 0.75 ml/min using a 6.1-min solvent gradient with 5 mM ammonium acetate in water or 95% acetonitrile (pH 5.7). For both study phases, electrospray ionization was employed using a dual spray electrospray ionization source operated in negative ionization mode only. A reference mass solution used for mass assignment correction is steadily pumped into the source via an isocratic pump plumbed through a 1:100 (source:waste) splitter with a flow rate of 0.40 ml/min. The mass range of the instrument was set to 60–1600 Da.
Metabolite chemical structure confirmation by UPLC-HRMS-MS
Analytical-grade chemical standards were purchased for N-acetylaspartate, alanine, arachidonic acid, 2′-deoxycytidine, lactic acid, and thymidine from MilliporeSigma. The chemical standards were spiked into the maintenance medium, and samples were prepared using the phase 1 sample preparation method. The chemical structures were confirmed using tandem mass spectrometry (UPLC-HRMS-MS) methods with the same chromatographic conditions employed in the analysis of the original samples. UPLC-HRMS-MS analyses were performed on samples spiked with chemical standards as well as spent media samples from the hiPSC-CM experiments using an Agilent G6530A QTOF HRMS. Spectra data were collected using collision energies of 10, 20, and 40 V. A metabolite was considered confirmed if the retention time, measured exact mass, and the product ion spectra of the spent media sample matched the chemical standard.
Untargeted metabolomic analysis
UPLC-HRMS data were processed as previously described (Kleinstreuer et al., 2011) using R 3.4.1. (R Core Team, 2017). Partial correlation analysis was used to identify feature cliques within a 5-s retention window in order to identify the most abundant feature and remove features that are likely adducts, fragment, or losses within a clique (Csardi and Nepusz, 2006; Watson-Haigh et al., 2010).
Secretome identification
The secretome was identified by comparing the log base 2 transformed abundance values for mass features present in the solvent control cell samples to the media control samples (lacking cells). A “mass feature” (also referred to as a “feature”) is a moiety detected by the mass spectrometer and is defined by the detected mass-to-charge ratio (m/z) and chromatographic retention time. The secretome features were tested under the null hypothesis that no difference existed between spent media samples and media controls using a Welch t test. Features with a significantly increased abundance relative to the media controls were considered secreted and those significantly decreased were considered consumed media components. The results were moderated using a fold change threshold of ±30% to select features that were predominantly present due to cardiomyocyte metabolism. The extracted ion chromatograms (EICs) of the secretome features were manually evaluated to select features that had reproducible peak shape, exhibited an observable difference in abundance between cell samples and media controls, and lacked closely eluting isobaric peaks. The secretome features were then utilized for all subsequent analyses using the automated UPLC-HRMS analysis pipeline.
Automated UPLC-HRMS analysis pipeline
An automated UPLC-HRMS data analysis pipeline was established to extract the ions associated with the secretome and spiked-in ISTDs for a UPLC-HRMS sample batch. Each 96-well plate of cell culture samples analyzed by UPLC-HRMS was processed using this pipeline. Initial UPLC-HRMS sample qualification was performed by evaluating the scan range, m/z range, intensity range, area under the total ion chromatogram, file size, and the intensity, retention time, and peak shape of the spiked-in ISTDs. Any sample failing to meet quality requirements were removed. Features were extracted with a 20-ppm m/z window and an optimized retention time window so that only a single peak was within the EIC. The EICs were smoothed and the intensity at the peak apex was used as the primary measure of abundance. The abundance values were then log base 2 transformed and run order normalized using the solvent control samples, which were spread equally throughout the UPLC-HRMS batch (Dunn et al., 2011). A Grubbs’ test was used to identify outlier samples within each treatment and exposure level (Grubbs, 1969). The abundance values were normalized to the median solvent control value for each respective mass feature. A test for ion suppression by a drug treatment with coeluting features was performed. If suppression was detected, the feature’s solvent control-normalized values were imputed with a value of 1. Finally, a batch qualification was performed based on of the coefficient of variation of the spiked-in ISTDs and number of samples removed. If the sample batch did not meet the minimum performance requirements, its UPLC-HRMS analysis was repeated. The solvent control-normalized values of each feature were then used in concentration-response modeling and to create predictive models.
Automated concentration-response analysis
Concentration-response curves were fit to the solvent control-normalized data using the R package “drc” version 3.0-1 (Ritz and Streibig, 2005). Four-parameter log-logistic and 2-phase concentration-response models were attempted to be fit to each feature and treatment. The best fitting model was selected using Akaike’s information criterion. The interpolated values from the best fitting curve of a feature were utilized to determine the response at given drug exposure in phase 1.
Manual concentration-response analysis
During phase 2, concentration-response analysis was conducted with GraphPad Prism (version 8.1, GraphPad Software, San Diego, California). Each data set was fit with a 4-parameter log-logistic, asymmetric, or multiphasic nonlinear model, and the best fitting model was selected using Akaike’s information criterion. For each drug, the selected concentration-response model was used to interpolate the response at the Cmax and 10× Cmax for each metabolite. The interpolated response values were used for cross-validation analysis and threshold settings.
Cross-validation
Stratified 4-fold cross-validation repeated 5 times was implemented using the R package “rsample” version 0.0.4 (Kuhn and Wickham, 2019). Drugs were stratified by toxicity grouping using the classes functional, structural, general, and noncardiotoxic. Classification performance metrics were based on the average of the hold-out set predictions.
Composite feature modeling
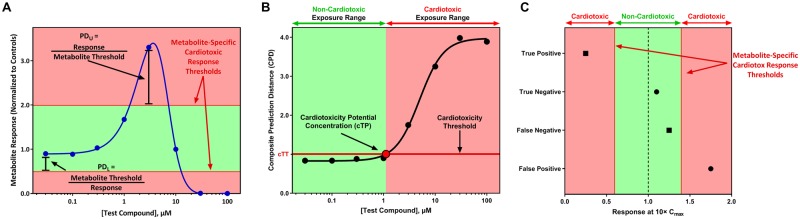
A predictive model was generated for each feature (fPM) by setting an upper and lower discriminatory threshold that maximized the balanced accuracy. Upper thresholds were generated using the value for each drug with a solvent control value of ≥ 1 and the lower thresholds used the value for each drug with a solvent control-normalized value of < 1. A drug was predicted as cardiotoxic if the response was greater than the upper threshold or less than the lower threshold. The best performing combinations of thresholds were selected based on the combination that maximized balanced accuracy or sensitvity, ties were broken using the model with the greatest sensitivity or balanced accuracy, respectively. The discriminatory thresholds were used to generate the prediction distance (PD), which is a scaled metric of response based on the magnitude at which a feature’s response exceeded their cardiotoxic response thresholds (Figure 2A). The prediction distance for upper thresholds (PDU) is median response/threshold and the prediction distance for the lower threshold (PDL) is the threshold/median response (Figure 2A). A drug with a PD > 1 is predicted to be cardiotoxic.
Figure 2.
Graphical representation of the prediction model. A, The prediction distance (PD) for each metabolite is calculated at each concentration. These results are used to determine the composite prediction distance (CPD), which is the value used in the composite model. The area above and below the “Metabolite-Specific Cardiotoxic Response Thresholds” are associated with cardiotoxicity and the area between the “Metabolite-Specific Cardiotoxic Response Thresholds” are associated with noncardiotoxicity. B, The concentration-response curve for the composite model is illustrated with the black line. The concentration predicted by the point where the concentration-response curve of the composite model crosses the cardiotoxicity threshold (horizontal line) indicates the exposure level where a compound has the potential to cause cardiotoxicity (cardiotoxicity potential concentration, black bordered circle). For (A) and (B), the x-axis is the drug concentration. The y-axis is the solvent control-normalized (fold change) values for the metabolite response (A) or the composite prediction distance (B). C, Scoring algorithm employed for known cardiotoxicants (■) and noncardiotoxicants (●) utilizing the response at 10× Cmax (x-axis) to determine the performance of the composite model. The color image is available in the online version of this article.
Composite feature models were created by combining 2–5 fPMs into a single predictive model. The composite models used a composite prediction distance (CPD), which is the maximum PD value of the combined fPMs for a given treatment. The discriminatory cutoff of the CPD was set using receiver operating characteristic (ROC) curve analysis to identify the cutoff that maximizes the balanced accuracy or sensitivity. A CPD that was greater than the discriminatory cutoff (cardiotoxicity threshold) is predicted to be cardiotoxic. Figure 2B illustrates how the assay can be applied to drugs with unknown cardiotoxicity potential. In this situation, the CPD values are fit with a nonlinear concentration-response curve, which is used to identify the exposure level where a drug perturbs metabolism in a manner indicative of cardiotoxicity and does not require any pharmacokinetic information (eg, Cmax). A drug is predicted to be cardiotoxic at the exposure level where the CPD concentration-response curve exceeds the cardiotoxicity threshold (cardiotoxicity potential concentration, Figure 2B). Exposure levels greater than the cardiotoxicity potential concentration are predicted to have cardiotoxicity potential.
In vitro responses observed between 10× and 50× the in vivo efficacious exposure are considered to be relevant for prediction of in vivo toxicity (Talbert et al., 2015). A threshold of 10× Cmax was selected as truth for cardiotoxic and noncardiotoxic drugs based on the thresholds used in previous in vitro cardiotoxicity publications (Clements and Thomas, 2014; Doherty et al., 2015; Guo et al., 2011; Sirenko et al., 2013). This range also allows for differences in the dose administered and population differences in pharmacokinetics. The scoring algorithm is illustrated in Figure 2C. A true positive (TP) was defined as cardiotoxic drug that exhibited a metabolic response that is predicted as cardiotoxic at ≤ 10× Cmax. A true negative (TN) was defined as noncardiotoxic drug that did not exhibit a metabolic response predicted to be cardiotoxic at ≤ 10× Cmax. A false negative (FN) was defined as a cardiotoxic drug that did not change metabolism or changed metabolism indicative of cardiotoxicity at exposure levels > 10× Cmax. A false positive (FP) was defined as a noncardiotoxic drug that exhibited a metabolic response that is predicted as cardiotoxic at ≤ 10× Cmax. Sensitivity was calculated as the percentage of cardiotoxic drugs correctly predicted as cardiotoxic (TP/[TP + FN]). Specificity was calculated as the percentage of noncardiotoxic drugs correctly predicted as noncardiotoxic (TN/[TN + FP]). Precision or positive predictive value (PPV) was the proportion of drugs predicted as cardiotoxic that were TPs (TP/[TP + FP]). Negative predictive value (NPV) was the proportion of drugs predicted as noncardiotoxic that were TNs (TN/[TN + FN]). Balanced accuracy was calculated by determining the average of the sensitivity and specificity. Area under the ROC curve (AUC) values were calculated using the R package “pROC” (Robin et al., 2011).
RESULTS
Phase 1: Identification of Metabolites Predictive of Cardiotoxicity
The first phase of this study was conducted to identify metabolites changed in response to treatment and a predictive metabolic signature indicative of cardiotoxicity. Characterization of the predictive metabolites led to the development of the new targeted biomarker assay for cardiotoxicity described in the second phase of this study.
Concentration-response experiment
Untargeted metabolomic data were acquired from a concentration-response experiment using spent medium of hiPSC-CM treated with 50 of the drugs (35 cardiotoxic and 15 noncardiotoxic) for 72 h to determine if a drug elicited a metabolic response at noncytotoxic exposures. To accomplish this, the maximum acceptable viability exposure (MAVE) was determined for each drug (Supplementary Table 1), which was defined as the highest exposure tested where the loss of cell viability was ≤ 10%. The MAVE was used to evaluate the metabolic response for 16 drugs where a decrease in cell viability (≥ 10%) was observed at exposure levels lower than the Cmax. The feature response was fit with a nonlinear concentration-response model to determine if the drug altered hiPSC-CM metabolism (Supplementary Figure 1A). Next, EICs of the 4 mass features with the greatest fold change from the solvent control treatment were evaluated for each drug at the exposure level closest to the Cmax or MAVE to confirm the response and feature quality (Supplementary Figure 1B). A metabolic response was observed for 31 cardiotoxic and 5 noncardiotoxic drugs at a noncytotoxic exposure within 10× Cmax. Only 2 drugs did not cause a metabolic response at any of the tested exposure levels (isoproterenol and pergolide). Using the concentration-response analysis of metabolomic and cell viability data, a single exposure was selected based on the lowest concentration that met the following criteria: (1) total Cmax concentration with evidence of changes in metabolism, (2) < 10% decrease in cell viability (MAVE), or (3) up to 10× Cmax when no evidence of metabolic changes were present at the Cmax. The final exposure levels selected for the single exposure experiments are listed in Table 1 and the descriptions of the selected exposures (ie, Cmax Total, MAVE, and 10× Cmax Total) are provided in Supplementary Table 1 and illustrated in Supplementary Figure 1A. Untargeted metabolomics data were not collected for 16 of the drugs tested in the single exposure experiments. The exposure for these drugs was selected based on the Cmax and/or MAVE.
Single exposure experiment
In order to identify predictive biomarkers of cardiotoxicity and evaluate their reproducibility, metabolic profiling was performed on 66 drugs (43 cardiotoxic and 23 noncardiotoxic) at the exposure level selected from the concentration-response experiment (Supplementary Table 1). We identified 156 features in the hiPSC-CM secretome (small molecules that are secreted and/or consumed by cells) that were present in both replicates. To reduce the feature set to the most predictive features, groups of highly correlated features were reduced to a single representative feature (Pearson correlation coefficient of ≥ 0.85, 111 features). Next, the feature set was reduced based on the predictive capacity of each feature. Using the median response of the 2 blocks for the 111 features, we evaluated the capacity of each feature to separate cardiotoxicants and noncardiotoxicants. The optimal discriminatory threshold of fold change (based on balanced accuracy) was determined for each feature based on the fold change of cells treated with a drug versus the solvent controls. Thirteen features had an AUC > 0.7, PPV > 0.8, and > 20% accuracy in each cardiotoxicity class (functional, structural, general, and non). These features were used to create composite models containing 2–5 features.
Four-fold cross-validation was used to train (based on balanced accuracy) and assess the discriminatory performance of each feature combination. Of the 2366 feature combinations, 117 had a balanced accuracy ≥ 80% (range, 0.8–0.877) in the hold-out set (sensitivity range, 0.724–0.991 and specificity range, 0.61–0.915), demonstrating the applicability of using a biomarker-based model for predicting cardiotoxicity. Balanced accuracy was maximized with only 2 features; however, sensitivity increases with the number of features included in the model.
Phase 2: Development and Evaluation of a Targeted Biomarker Assay to Predict Cardiotoxicity
Metabolite confirmation and targeted UPLC-HRMS method development
In the second phase of this study, we developed a targeted biomarker-based assay for predicting cardiotoxicity potential using the biomarkers identified in phase 1. The identities for 6 of the predictive mass features from phase 1 were confirmed by UPLC-HRMS-MS analysis (Table 2). A rapid UPLC-HRMS analysis method was developed and optimized for fast turnaround analysis of relative changes in the abundance of arachidonic acid, 2′-deoxycytidine, lactic acid, N-acetylaspartic acid, and thymidine in hiPSC-CM spent media samples. Measurement of alanine did not meet minimum performance criteria in the revised UPLC-HRMS method and it was removed from the predictive model. Removal of alanine did not alter model performance as the predictive capacity of alanine was similar to lactate and drugs predicted to be cardiotoxic by alanine were identified by the combination of the other biomarkers. The new UPLC-HRMS method allowed for a 4-fold increase in throughput (29 min vs 7 min run time) and was used to evaluate 81 drugs (52 cardiotoxic and 29 noncardiotoxic) with known cardiotoxicity potential. All drugs were tested with an 8-point concentration-response curve, and the data were used to define the final assay prediction model.
Table 2.
List of Predictive Metabolites Confirmed With UPLC-HRMS-MS
| Metabolite Name | Adduct | m/z | Retention Time (s) |
|---|---|---|---|
| Arachidonic acid | [M − H]− | 303.2332 | 55 |
| Thymidine | [M + Cl]− | 277.0618 | 132 |
| 2′-Deoxycytidine | [M + Cl]− | 262.0615 | 296 |
| Lactic acid | [M − H]− | 89.0244 | 373 |
| Alanine | [M − H]− | 88.0404 | 572 |
| N-acetylaspartic acid | [M − H]− | 174.0410 | 695 |
Metabolite response to cardiotoxic drugs
A wide range of response types were observed for lactic acid, arachidonic acid, thymidine, and 2′-deoxycytidine, which is reflective of the broad range of drugs and mechanisms of toxicities included in this study (Supplementary Figure 2). Lactic acid and arachidonic acid are both present in iCell Cardiomyocytes Maintenance Medium; however, the levels of lactic acid and arachidonic acid did not differ between the solvent controls and media blanks (no hiPSC-CM present). This indicates that the changes observed in response to drug treatment are from an increase in metabolite secretion (response values > 1) or utilization (response values < 1). In contrast, thymidine and 2′-deoxycytidine are secreted by hiPSC-CM and not components of hiPSC-CM media. For these 2 metabolites, increased and decreased levels are due to stimulatory and inhibitory effects on metabolite secretion. All the cardiotoxic drugs tested induced changes in at least 2 metabolites; however, the concentration where a drug caused an effect differed between metabolites. For example, bepridil increased lactic acid secretion at lower concentrations than the other metabolites, whereas doxorubicin decreased 2′-deoxycytidine secretion at lower concentrations than the other metabolites. Importantly, these 4 predictive metabolites respond to cardiotoxicant exposure independent of changes in cell viability.
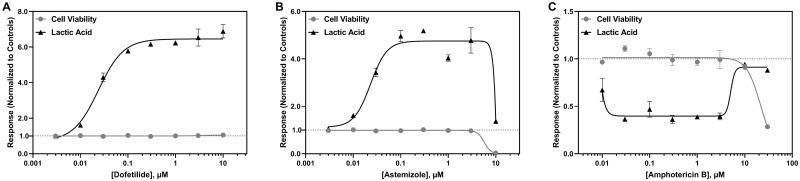
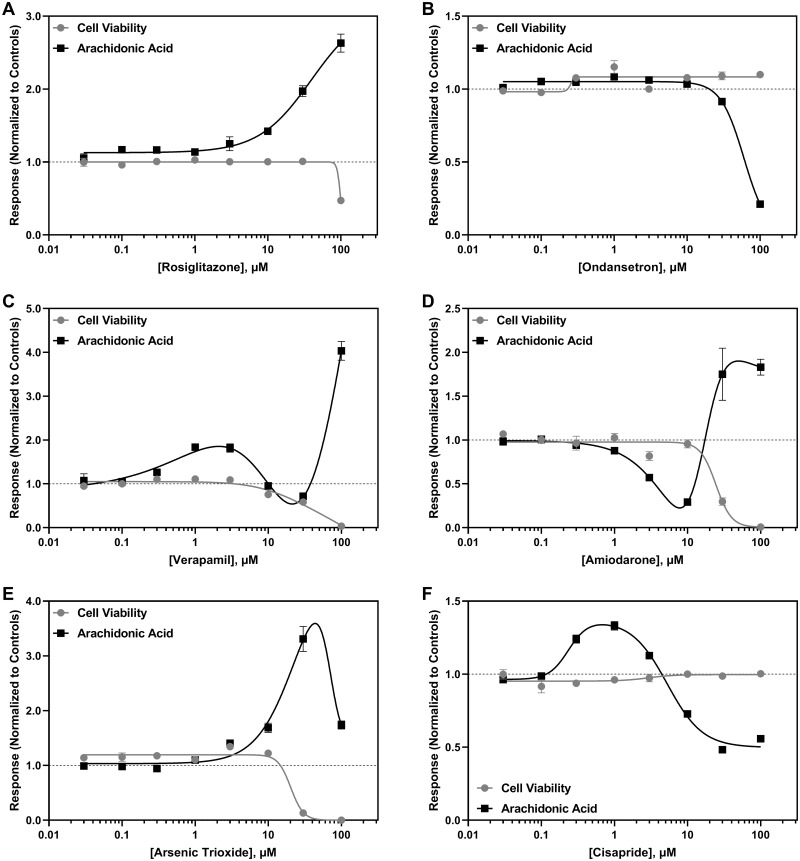
An increase in lactic acid secretion was the most common effect observed among the drugs that altered lactic acid metabolism (Figure 3A), which was followed by a decrease if a drug decreased cell viability (Figure 3B). A few cardiotoxic drugs decreased lactic acid levels relative to the solvent controls (Figure 3C). Arachidonic acid levels were increased (Figure 4A) or decreased (Figure 4B) following drug exposure independent of any change in hiPSC-CM viability. Because arachidonic acid is also a media component, a multiphasic response was also observed for cytotoxic drugs (Figs. 4C–E); however, for some drugs, a multiphasic response was observed independent of decreased cell viability (Figure 4F).
Figure 3.
Representative concentration-dependent effects on lactic acid metabolism in hiPSC-CM following cardiotoxicant exposure. Lactic acid (▲) and cell viability (●) concentration-response curves are shown for (A) dofetilide, (B) astemizole, and (C) amphotericin B. The x-axis is the drug concentration (µM) and the y-axis is the solvent control-normalized (fold change) value for lactic acid or cell viability. Data represent mean ± SEM (n = 3). If not shown, error bars are smaller than the size of the symbol.
Figure 4.
Representative concentration-dependent effects on arachidonic acid metabolism in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM) following cardiotoxicant exposure. Arachidonic acid (■) and cell viability (●) concentration-response curves are shown for (A) rosiglitazone, (B) ondansetron, (C) verapamil, (D) amiodarone, (E) arsenic trioxide, and (F) cisapride. The x-axis is the drug concentration (µM) and the y-axis is the solvent control-normalized (fold change) value for arachidonic acid or cell viability. Data represent mean ± SEM (n = 3). If not shown, error bars are smaller than the size of the symbol.
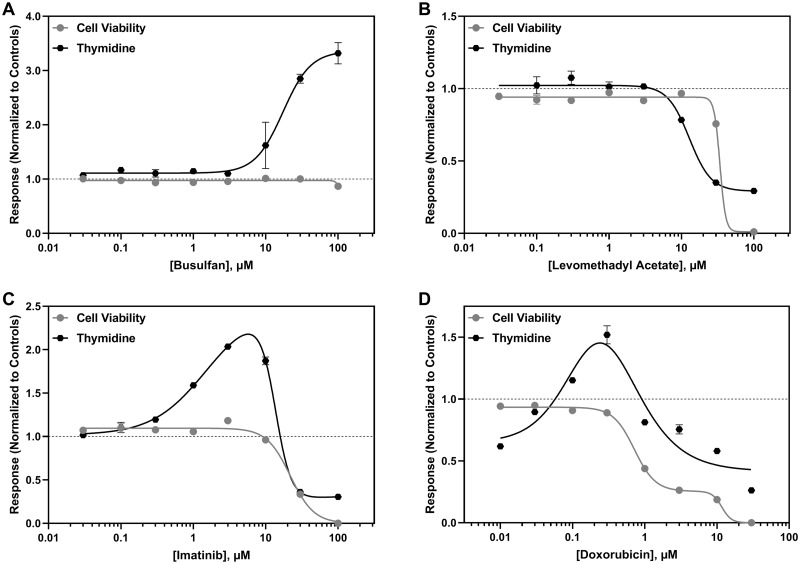
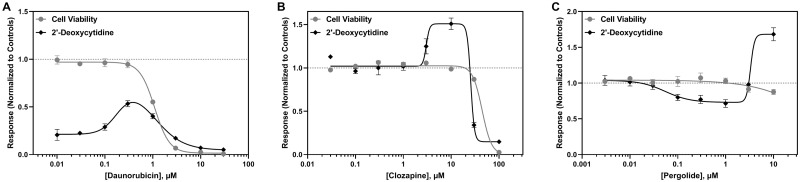
The most common response observed among drugs impacting thymidine metabolism was an increase in thymidine secretion (Figure 5A); however, some drugs decreased thymidine secretion, which was highly correlated with cell viability (Figure 5B). A biphasic response was also observed for thymidine, consisting of an increase in thymidine secretion followed by an abrupt decrease that was consistent with decreased cell viability (Figure 4C). Interestingly, the anthracyclines tested in this study first decreased thymidine secretion, followed by an increase in thymidine levels prior to another decrease at cytotoxic levels (Figure 5D). For 2′-deoxycytidine, the most common response observed was a decrease in its secretion, which occurred independent of any changes in cell viability for over half of the anticancer drugs evaluated in this study (Figure 6A). A decrease in 2′-deoxycytidine that tracked with cell viability was also observed for cytotoxic chemicals. Clozapine and thioridazine elicited a biphasic response, with a slight increase in 2′-deoxycytidine secretion followed by an abrupt decrease at cytotoxic concentrations (Figure 6B). Pergolide and azidothymidine both caused a decrease in 2′-deoxycytidine secretion that was followed by an increase (Figure 6C).
Figure 5.
Representative concentration-dependent effects on thymidine metabolism in hiPSC-CM following cardiotoxicant exposure. Thymidine ( ) and cell viability (●) are shown for (A) busulfan, (B) levomethadyl acetate, (C) imatinib, and (D) doxorubicin. The x-axis is the drug concentration (µM) and the y-axis is the solvent control-normalized (fold change) value for thymidine or cell viability. Data represent mean ± SEM (n = 3). If not shown, error bars are smaller than the size of the symbol.
) and cell viability (●) are shown for (A) busulfan, (B) levomethadyl acetate, (C) imatinib, and (D) doxorubicin. The x-axis is the drug concentration (µM) and the y-axis is the solvent control-normalized (fold change) value for thymidine or cell viability. Data represent mean ± SEM (n = 3). If not shown, error bars are smaller than the size of the symbol.
Figure 6.
Representative concentration-dependent effects on 2′-deoxycytidine metabolism in hiPSC-CM following cardiotoxicant exposure. 2′-Deoxycytidine (◆) and cell viability (●) are shown for (A) daunorubicin, (B) clozapine, and (C) pergolide. The x-axis is the drug concentration (µM) and the y-axis is the solvent control-normalized (fold change) value for 2′-deoxycytidine or cell viability. Data represent mean ± SEM (n = 3). If not shown, error bars are smaller than the size of the symbol.
Biomarker-based model of cardiotoxicity
The individual metabolites measured by the rapid UPLC-HRMS method and combinations of these metabolites with viability were evaluated for their capacity to discriminate cardiotoxic from noncardiotoxic drugs. The effect of each drug at the Cmax and 10× Cmax was determined for each metabolite by interpolating the response from the nonlinear concentration-response curve. The solvent control-normalized response was used for 2′-deoxycytidine, thymidine, and N-acetylaspartate. The metabolite response for lactic acid and arachidonic acid was normalized to cell viability because the response curve for drugs that cause cell death was multiphasic and began and ended at 1.0 (lactic acid, Supplementary Figure 3A) or above 1.0 (arachidonic acid, Supplementary Figure 3B).
The interpolated response at 10× Cmax for each drug was used to set predictive thresholds for each metabolite and score classification performance of the prediction models. A drug was scored as cardiotoxic if the interpolated value at 10× Cmax was below or above the metabolite-specific cardiotoxic response thresholds (Figure 2C). A drug was scored as noncardiotoxic if the value was between the upper and lower thresholds (Figure 2C). Four-fold cross-validation was used to assess the performance of metabolite combinations to obtain an unbiased estimate of future model performance. Composite models were created containing 2–5 metabolites (26 models). Cell viability was not included as a feature in the model because it is used as a normalizer for lactic acid and arachidonic acid. The predictive thresholds for each metabolite and the composite model were trained to either maximize balanced accuracy (BAC-trained) or sensitivity (SEN-trained). The balanced accuracy in the hold-out set for the BAC-trained composite models ranged from 0.733 to 0.796 (sensitivity, 0.595–0.832; specificity, 0.694–0.909; PPV, 0.837–0.940; NPV, 0.556–0.706; AUC, 0.661–0.816; Supplementary Table 2). For the SEN-trained composite models, the balanced accuracy of the hold-out set ranged from 0.690 to 0.788 (sensitivity, 0.653–0.889; specificity, 0.564–0.791; PPV, 0.790–0.875; NPV, 0.543–0.757; AUC, 0.694–0.834; Supplementary Table 3). The 4-metabolite model containing arachidonic acid, 2′-deoxycytidine, lactic acid, and thymidine maximized sensitivity in both the BAC-trained and SEN-trained hold-out sets was selected for further optimization. N-acetylaspartic acid was not included in the model selected for further optimization because it decreased specificity and did not increase sensitivity in the hold-out set, indicating that it did not add predictive value in combination with the other metabolites.
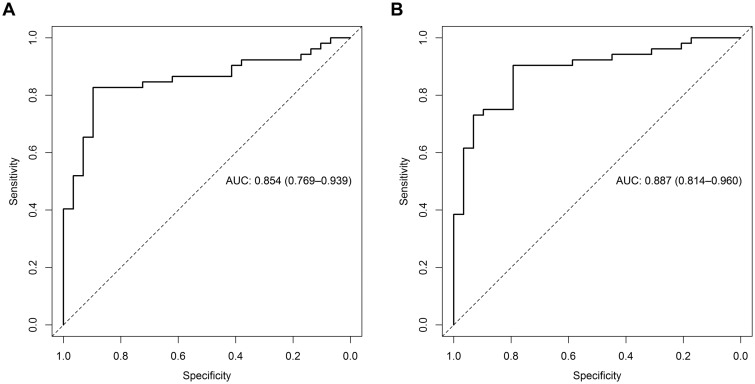
The data from all 81 drugs were used to optimize the cardiotoxicity response thresholds for each metabolite in the model selected by cross-validation (arachidonic acid, 2′-deoxycytidine, lactic acid, and thymidine). These optimized thresholds increased the predictivity of the assay above what was determined by the hold-out set from the cross-validation results. When the final model was trained to optimize balanced accuracy, it classified all 81 drugs with a balanced accuracy of 86%, sensitivity of 83%, specificity of 90%, PPV of 93%, NPV of 74%, and an AUC of 0.854 (Figure 7A and Table 3). In comparison, training the final model to maximize sensitive resulted in a balanced accuracy of 85%, sensitivity of 90%, specificity of 79%, PPV of 89%, NPV of 82%, and an AUC of 0.887 (Figure 7B and Table 4). Compared with the sensitivity of each metabolite alone, the composite models increased sensitivity by 15%–31% (Tables 3 and 4). At 10× Cmax, both models incorrectly predicted the noncardiotoxicants ascorbic acid, erlotinib, and gemfibrozil and the cardiotoxicants dexfenfluramine, isoproterenol, pergolide, telmisartan, and valdecoxib (Table 5). The BAC-trained final model also incorrectly classified the cardiotoxicants anagrelide, encainide, nandrolone decanoate, and tegaserod as noncardiotoxicants and the SEN-trained final model classified the noncardiotoxicants sildenafil, thiabendazole, and tolbutamide as cardiotoxicants. All the cardiotoxicants incorrectly classified by the composite model elicited a metabolic response indicative of cardiotoxicity (concentration-response curve crossed a predictive threshold) at concentrations > 10× Cmax.
Figure 7.
Determination of the predictivity of the (A) BAC-trained and (B) SEN-trained composite models. The highest prediction distance obtained from the 4 metabolites in the composite model (arachidonic acid, 2′-deoxycytidine, lactic acid, and thymidine) was subjected to ROC analysis. The dotted line represents the line of unity, which shows the results of random assignment of cardiotoxicity.
Table 3.
Final Prediction Model Thresholds and Performance for BAC-Trained Model
| Model | Cardiotoxicity Threshold (s) |
Balanced Accuracy | Sensitivity | Specificity | PPV | NPV | Functional | Structural | General | |
|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||||
| Viability/lactic acid | 0.325 | 1.44 | 0.827 | 0.654 | 1.000 | 1.000 | 0.617 | 0.800 | 0.500 | 0.667 |
| Viability/arachidonic acid | 0.675 | 1.72 | 0.744 | 0.558 | 0.931 | 0.935 | 0.540 | 0.467 | 0.500 | 0.667 |
| Thymidine | 0.895 | 2.27 | 0.742 | 0.519 | 0.966 | 0.964 | 0.528 | 0.333 | 0.500 | 0.667 |
| 2′-Deoxycytidine | 0.865 | 1.26 | 0.810 | 0.654 | 0.966 | 0.971 | 0.609 | 0.667 | 0.500 | 0.762 |
| Composite model | 1.025 | 0.862 | 0.827 | 0.897 | 0.935 | 0.743 | 0.933 | 0.625 | 0.905 | |
Table 4.
Final Prediction Model Thresholds and Performance for SEN-Trained Model
| Model | Cardiotoxicity Threshold (s) |
Balanced Accuracy | Sensitivity | Specificity | PPV | NPV | Functional | Structural | General | |
|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||||
| Viability/lactic acid | 0.475 | 1.44 | 0.823 | 0.750 | 0.897 | 0.929 | 0.667 | 0.867 | 0.563 | 0.810 |
| Viability/arachidonic acid | 0.675 | 1.12 | 0.777 | 0.692 | 0.862 | 0.900 | 0.610 | 0.533 | 0.688 | 0.810 |
| Thymidine | 0.895 | 1.20 | 0.756 | 0.615 | 0.897 | 0.914 | 0.565 | 0.400 | 0.500 | 0.857 |
| 2′-Deoxycytidine | 0.865 | 1.19 | 0.831 | 0.731 | 0.931 | 0.950 | 0.659 | 0.867 | 0.563 | 0.762 |
| Composite model | 1.005 | 0.848 | 0.904 | 0.793 | 0.887 | 0.821 | 1.000 | 0.750 | 0.952 | |
Table 5.
Cardiotoxicity Prediction at 10× Cmax for Each Metabolite and the Composite Models Trained for Balanced Accuracy and Sensitivity
| Drug | Cardiotoxicity Classification | BAC: Via/Lac | BAC: Via/Ara | BAC: Thy | BAC: 2dC | BAC: Composite Model | SEN: Via/Lac | SEN: Via/Ara | SEN: Thy | SEN: 2dC | SEN: Composite Model |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Astemizole | Cardiotoxic | Tox | Non | Tox | Non | Tox | Tox | Non | Tox | Tox | Tox |
| Bepridil | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Chlorpromazine | Cardiotoxic | Tox | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox |
| Cisapride | Cardiotoxic | Tox | Non | Non | Non | Tox | Tox | Non | Non | Tox | Tox |
| Dofetilide | Cardiotoxic | Tox | Tox | Tox | Non | Tox | Tox | Tox | Tox | Tox | Tox |
| Encainide | Cardiotoxic | Non | Non | Non | Non | Non | Tox | Non | Non | Non | Tox |
| Levomethadyl acetate | Cardiotoxic | Tox | Tox | Non | Non | Tox | Tox | Tox | Non | Non | Tox |
| Nifedipine | Cardiotoxic | Tox | Non | Non | Tox | Tox | Tox | Non | Non | Tox | Tox |
| Ondansetron | Cardiotoxic | Non | Non | Non | Tox | Tox | Non | Non | Non | Tox | Tox |
| Quinidine | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Sertindole | Cardiotoxic | Tox | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox |
| Sotalol | Cardiotoxic | Non | Non | Non | Tox | Tox | Non | Non | Non | Tox | Tox |
| Terfenadine | Cardiotoxic | Tox | Non | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox |
| Thioridazine | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Verapamil | Cardiotoxic | Tox | Non | Non | Tox | Tox | Tox | Non | Tox | Tox | Tox |
| Azidothymidine | Cardiotoxic | Non | Non | Tox | Tox | Tox | Non | Tox | Tox | Tox | Tox |
| Busulfan | Cardiotoxic | Tox | Tox | Tox | Non | Tox | Tox | Tox | Tox | Tox | Tox |
| Daunorubicin | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Dexfenfluramine | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Dithiazanine iodide | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Doxorubicin | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Idarubicin | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Imatinib | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Nandrolone decanoate | Cardiotoxic | Non | Non | Non | Non | Non | Non | Tox | Non | Non | Tox |
| Pergolide | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Rofecoxib | Cardiotoxic | Tox | Non | Non | Tox | Tox | Tox | Non | Non | Tox | Tox |
| Rosiglitazone | Cardiotoxic | Non | Tox | Non | Non | Tox | Non | Tox | Non | Non | Tox |
| Tegaserod | Cardiotoxic | Non | Non | Non | Non | Non | Tox | Tox | Non | Non | Tox |
| Telmisartan | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Trastuzumab | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Valdecoxib | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Amiodarone | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Amitriptyline | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Amphotericin B | Cardiotoxic | Non | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Amsacrine | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Anagrelide | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Tox | Non | Tox |
| Arsenic trioxide | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Bortezomib | Cardiotoxic | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox | Tox |
| Chloroquine | Cardiotoxic | Tox | Non | Non | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Clozapine | Cardiotoxic | Non | Tox | Non | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Crizotinib | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Dasatinib | Cardiotoxic | Tox | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox |
| Fluorouracil | Cardiotoxic | Non | Non | Tox | Tox | Tox | Non | Non | Tox | Tox | Tox |
| Isoproterenol | Cardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Lapatinib | Cardiotoxic | Non | Tox | Non | Non | Tox | Tox | Tox | Tox | Non | Tox |
| Mitoxantrone | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Nilotinib | Cardiotoxic | Non | Non | Tox | Non | Tox | Non | Tox | Tox | Non | Tox |
| Nortriptyline | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Paclitaxel | Cardiotoxic | Tox | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox |
| Sorafenib | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Sunitinib | Cardiotoxic | Tox | Non | Tox | Non | Tox | Tox | Tox | Tox | Non | Tox |
| Vandetanib | Cardiotoxic | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox | Tox |
| Acetylsalicylic acid | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Acyclovir | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Adipic acid | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Amoxicillin | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Ascorbic acid | Noncardiotoxic | Non | Tox | Non | Non | Tox | Non | Tox | Non | Tox | Tox |
| Aspartame | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Axitinib | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Benzoic acid | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Biotin | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Cetirizine | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Cimetidine | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Citric acid | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Erlotinib | Noncardiotoxic | Non | Non | Tox | Tox | Tox | Tox | Non | Tox | Tox | Tox |
| Gemfibrozil | Noncardiotoxic | Non | Tox | Non | Non | Tox | Tox | Tox | Non | Non | Tox |
| Hexylresorcinol | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Leucine | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Loratadine | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Maltol | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Methylparaben | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Natamycin | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Phenylphenol | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Praziquantel | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Ranitidine | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Sildenafil | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Tox | Non | Tox |
| Sucrose | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Tartaric acid | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
| Thiabendazole | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Tox | Tox | Non | Tox |
| Tolbutamide | Noncardiotoxic | Non | Non | Non | Non | Non | Tox | Tox | Non | Non | Tox |
| Xylitol | Noncardiotoxic | Non | Non | Non | Non | Non | Non | Non | Non | Non | Non |
Abbreviations: Non, drug was predicted to be noncardiotoxic; Tox, drug was predicted to be cardiotoxic.
DISCUSSION
This assay was developed to address the need for more accurate, complimentary alternatives for identifying structural and functional cardiotoxicants. Implementing screening assays that identify both types of cardiotoxicity, such as the one developed here, earlier during the development process has the potential to improve the cost and time required to bring new drugs to market. This is the first study to develop a model of cardiotoxicity based on changes in hiPSC-CM metabolism using a diverse set of drugs including both functional and structural cardiotoxicants. This study shows that metabolic perturbation could be used to predict a drug’s potential to cause both functional and structural cardiotoxicity. The assay’s prediction model is based on the perturbation of 4 metabolites (arachidonic acid, 2′-deoxycytidine, lactic acid, and thymidine) and cell viability in hiPSC-CM. The metabolite-specific prediction thresholds used in the model can be optimized to meet the needs of the end user. For example, the thresholds can be optimized to maximize sensitivity if the assay will be applied during early drug discovery for hazard identification and elimination, when it is more important to accurately predict TPs (Valentin et al., 2009). The SEN-trained model predicted the cardiotoxicity of 81 drugs with 90% sensitivity, 79% specificity, and 85% balanced accuracy. Alternatively, the thresholds can be optimized for balanced accuracy and higher specificity if it will be used for risk assessment after a candidate drug has been identified, when it is more important to have highly specific models (Valentin et al., 2009). In this situation, the BAC-trained model could be applied, which predicted the cardiotoxicity potential of the 81 drugs tested in this study with 90% specificity, 83% sensitivity, and 86% balanced accuracy. Using either model, these results would be classified as good to excellent according to the published framework for evaluating an in vitro assay’s ability to predict in vivo outcomes (Genschow et al., 2002).
Importantly, the assay described here predicted the cardiotoxicity potential of 81 drugs at therapeutically relevant concentrations. Although a threshold of 10× Cmax was selected for setting the response thresholds and evaluating the predictivity of the assay, it should be noted that in vitro concentrations up to 30× the in vivo Cmax have been considered relevant for predicting toxicity in multiple studies (Lin and Will, 2012; Redfern et al., 2003). Of the 5 cardiotoxic drugs predicted to be noncardiotoxic at 10× Cmax, pergolide would be predicted to be cardiotoxic at 30× Cmax and an additional 2 (dexfenfluramine and valdecoxib) are predicted as cardiotoxic with a threshold of 100× Cmax; however, increasing the margin used for prediction decreases assay specificity. Although metabolic changes were detected in vivo following isoproterenol treatment (Li et al., 2015), this drug did not impact hiPSC-CM metabolism until concentrations > 100× Cmax were reached. This discrepancy could be due to differences in chromatography methods or biological samples (plasma vs cardiomyocyte spent media) used in this study compared with the in vivo study.
It is important to note that although the therapeutic Cmax was used here as a benchmark exposure level to aid in interpreting assay performance, it is not required for data interpretation. The composite model concentration-response curve can be used to predict the concentration where a drug is expected to be cardiotoxic based on the point where it crosses the cardiotoxicity threshold (Figure 2B). For compounds that do not have an established Cmax value, changes in hiPSC-CM metabolism can be used as a signal regarding the cardiotoxic potential of the compound. It is also possible to define a concentration threshold for classification using the predicted cardiotoxicity potential concentration (cTP), as has been proposed for other in vitro cardiotoxicity testing methods (Archer et al., 2018; Clements et al., 2015; Guo et al., 2013; Pointon et al., 2013). For example, when a concentration threshold of 30 µM is applied to the cTP obtained from the BAC-trained model, the assay has a balanced accuracy of 85% (94% sensitivity, 76% specificity, 88% PPV, and 88% NPV). Using this threshold, the assay predicts functional, structural, and general cardiotoxicants with 100%, 81%, and 100% accuracy, respectively.
Changes in metabolism, as measured in the spent medium of cell culture systems, yield a distinguishable “metabolic footprint,” which is a functional measure of cellular metabolism that can be used to evaluate response to treatment. The panel of 81 drugs evaluated in this study was designed to cover a broad range of mechanisms, including cardiovascular and noncardiovascular drugs that cause a wide range of effects on the cardiovascular system. The selected metabolites were perturbed by a wide range of drug classes in this study, indicating that common metabolic pathways of toxicity are shared by many cardiotoxicants. However, the cardiotoxic drugs may impact different components in the pathways, as evidenced by the multitude of changes observed (Figures 3–6). The 4 metabolites included in the final model have key roles in modulating oxidative stress as well as mitochondrial function and replication, which have been experimentally and clinically associated with cardiotoxicity (Chaudhari et al., 2017; Varga et al., 2015; Varricchi et al., 2018). Arachidonic acid, thymidine, and 2′-deoxycytidine have not previously been identified as biomarkers for other types of toxicity and appear to be specific indicators of cardiotoxicity potential (Cuykx et al., 2019; Kleinstreuer et al., 2011; Palmer et al., 2012, 2013; van Vliet et al., 2008; West et al., 2010; Yue et al., 2019). Lactic acid has been identified as a marker of cell stress in multiple cell types (Limonciel et al., 2011; Wilmes et al., 2013); however, in this assay, its specificity (90%, Table 4) indicates that the response observed in hiPSC-CM is specific to cardiotoxic drugs and it is not behaving as a general marker of cell stress.
In this study, arachidonic acid levels were both increased and decreased following cardiotoxicant treatment, indicating its metabolism may be impacted through multiple mechanisms. Activation of the arachidonic acid cascade due to a redox state unbalance has been shown to contribute to the pathogenesis of cardiovascular disease (Mitjavila and Moreno, 2012). Arachidonic acid can be metabolized by cytochrome P450 enzymes (CYPs) into cardioprotective epoxyeicosatrienoic acids or cardiotoxic hydroxyeicosatetraenoic acids. Studies have shown that CYP-mediated arachidonic acid metabolism can be modulated by known cardiotoxic drugs (Althurwi et al., 2015; Arnold et al., 2017; Kato et al., 2017; Zhang et al., 2009). Arachidonic acid levels were increased in the plasma of rats treated with doxorubicin (Robison and Giri, 1987), which is consistent with the increased arachidonic acid levels observed in this study following doxorubicin exposure (Supplementary Figure 2). In contrast, arachidonic acid was decreased in heart tissue of rats treated with sunitinib (Jensen et al., 2017b), matching the decrease in arachidonic acid observed here following sunitinib exposure, as well as with many other cardiotoxic drugs (Figure 4 and Supplementary Figure 2). Additionally, thiazolidinediones (eg, rosiglitazone) increase arachidonic acid release from the cell membrane (Tsukamoto et al., 2004). These results are consistent with the changes in arachidonic acid metabolism observed following treatment in this study and suggest that multiple mechanisms of cardiotoxicity can be identified with single biomarker.
Exposure to cardiotoxicants increased the levels of lactic acid secreted by hiPSC-CM in most instances. Previous research has shown that hiPSC-CM cultured in media containing glucose have a mid-fetal state of energy metabolism, generating ATP primarily through glycolysis (Correia et al., 2017; Rana et al., 2012; Ulmer and Eschenhagen, forthcoming); however, when the media contains galactose (as used in this study), hiPSC-CM energy metabolism shifts to oxidative phosphorylation (Correia et al., 2017; Rana et al., 2012). Elevated levels of lactic acid reflect a shift toward anaerobic glycolysis and are well known to be associated with cardiotoxicity, especially during cardiac ischemia and heart failure (Karwi et al., 2018; Kawase et al., 2015; Lazzeri et al., 2015; Schnackenberg et al., 2016). A recent study evaluating the metabolic effects of doxorubicin treatment in rats identified decreased levels of lactic acid and fumarate, and increased malate concentrations in heart tissue, indicating that both anaerobic and aerobic metabolisms were disrupted in vivo (Niu et al., 2016). In contrast, lactic acid levels were increased in the plasma of humans treated with 5-fluorouracil (Jensen et al., 2010). The measurement of lactic acid in this assay provides insight into the state of hiPSC-CM mitochondrial energy metabolism, and its increase is reflective of the cell’s decreased ability to produce ATP via oxidative phosphorylation.
2′-Deoxycytidine and thymidine were both increased and decreased by cardiotoxicants; however, an increase in thymidine levels was the most common response observed, whereas a decrease in 2′-deoxycytidine levels was more common. Many antiviral and anticancer agents known to cause cardiotoxicity are nucleoside analogs (eg, azidothymidine and fluorouracil). These drugs have been linked to mitochondrial toxicity by inhibiting key mitochondrial enzymes required for mitochondrial DNA synthesis and mitochondrial replication (Balcarek et al., 2010; Sun et al., 2014; Varga et al., 2015). Previous research has shown that azidothymidine inhibits thymidine phosphorylation in mitochondria isolated from rat heart, as well as in the isolated perfused rat heart (McKee et al., 2004; Susan-Resiga et al., 2007) and that azidothymidine inhibits mitochondrial thymidine kinase 2 (Sun et al., 2014). Inhibition of thymidine phosphorylation is consistent with the increase in thymidine concentration following azidothymidine exposure (Supplementary Figure 2). Additionally, cardiomyocyte mitochondrial DNA damage has been proposed as a mechanism of doxorubicin-mediated cardiotoxicity (Khiati et al., 2014).
The in vitro assay developed here could easily be conducted in conjunction with impedance and multielectrode array (MEA) methods because the changes in metabolism are measured in the spent media and not destructive to the hiPSC-CM. Our biomarker-based assay is complementary to impedance and MEA approaches, and a combined approach would increase the overall accuracy of prediction. For example, bepridil, chlorpromazine, clozapine, and terfenadine, which were incorrectly predicted in the Comprehensive in vitro Proarrhythmia Assay MEA validation study (Blinova et al., 2018), all impacted hiPSC-CM metabolism and were predicted as cardiotoxic in this study. The structural cardiotoxicants azidothymidine, rofecoxib, and rosiglitazone, which are correctly predicted as cardiotoxic in our assay, were predicted to be noncardiotoxic with other in vitro assays for structural cardiotoxicity using MEA and impedance platforms, as well as additional endpoints, including high content analysis, reactive oxygen species generation, troponin secretion, lipid formation, and cell viability (Clements and Thomas, 2014; Doherty et al., 2015). On the other hand, the MEA approach correctly identified isoproterenol as cardiotoxic (Doherty et al., 2015), but this drug did not impact hiPSC-CM metabolism at therapeutically relevant concentrations in this study.
Despite the assay’s significant promise for predicting cardiotoxicity, limitations still exist. It is unlikely that any single in vitro system will be able to completely predict in vivo outcomes. In vitro models do not include biology associated with the effects of absorption, distribution, metabolism, and excretion, making it difficult to extrapolate doses, tissue/cellular drug delivery, and duration of exposure. In some cases, a drug’s cardiotoxicity may result from one of its metabolites. Prior knowledge of drug metabolism provides an opportunity to assess known metabolites in addition to the parent compound. This approach would provide information as to the relative toxicities of the metabolically related compounds, which is more difficult to ascertain using in vivo systems. When the metabolites are not known, it may be possible to use an exogenous metabolic activation system to simulate metabolism. We plan to evaluate the options for adding this to the current assay. This study evaluated a drug’s direct effect on the metabolism human cardiomyocytes from a single donor and does not account for potential contributions from other cell types found in the heart or genetic diversity. The prediction model is not able to differentiate functional from structural cardiotoxicity potential, which can partially be attributed to the promiscuity of many cardiotoxic drugs, which are known to have both functional and structural cardiotoxic effects; however, the response in specific metabolites can be used to classify a drug’s likelihood for each type of cardiotoxicity. For example, most functional cardiotoxicants caused an increase in lactic acid independent of (or prior to) changes in the other biomarkers. In contrast, changes in arachidonic acid, 2′-deoxycytidine, and thymidine prior to changes in lactic acid occurred following exposure to many of the structural cardiotoxicants tested. Future studies will focus on developing a prediction model that separates functional and structural cardiotoxicants.
In conclusion, this study demonstrates the utility of a biomarker-based in vitro assay for identifying cardiovascular liability of a broad range of drugs with a variety of mechanisms. Part of the assay’s strength is that it can use a multiexposure approach to identify the concentration where a drug displays cardiotoxicity potential. This assay, either alone or as part of a combined approach, can be used during lead identification or lead optimization to prioritize drugs with the lowest potential for cardiotoxicity for further development.
DATA AVAILABILITY
Supplementary data are available at https://doi.org/10.5061/dryad.gqnk98shs. The raw peak apex data for each metabolite and spiked-in ISTD for each sample analyzed in development of the final prediction model development is available in the Dryad repository (https://doi.org/10.5061/dryad.gqnk98shs).
DECLARATION OF CONFLICTING INTERESTS
J.A.P., A.M.S., and R.E.B. are employees of, and E.L.R.D. is an equity owner in, Stemina Biomarker Discovery Inc. J.P.V. and V.G. are employees of UCB Biopharma SPRL.
FUNDING
National Institute of General Medical Sciences of the National Institutes of Health under award number R44GM100640.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Michael Colwell for hiPSC-CM culture and sample preparation; Brandon Smart, Paul West, DelRay Sugden, and Michael Ludwig for UPLC-HRMS sample analysis; Ashley Poenitzsch Strong for assisting with grant writing and compound set development; Annie Delaunois and Andre Nogueira da Costa for intellectual support and assisting with characterizing the compound set; and Fred Kirchner, Alex Eapen, and Kiyoshi Takasuna for helpful discussions and feedback during assay development.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- Accord Healthcare Inc. (2017). Fluorouracil injection, solution In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=66d451fe-2436-494c-80c5-4528c8e34369. Accessed January 6, 2020. [Google Scholar]
- Accord Healthcare Inc. (2019). Amitriptyline hydrocloride tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=1e6d2c80-fbc8-444e-bdd3-6a91fe1b95bd. Accessed December 27, 2019. [Google Scholar]
- Acetris Health LLC. (2019). Zidovudine capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=54e21b7b-939c-4afa-9492-bde2b36dd352. Accessed January 2, 2020. [Google Scholar]
- Alfasigma USA Inc. (2019). Tegaserod tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=6a44eefe-fbe8-431c-a418-2571b26aa42e. Accessed January 3, 2020. [Google Scholar]
- Althurwi H. N., Maayah Z. H., Elshenawy O. H., El-Kadi A. (2015). Early changes in cytochrome P450s and their associated arachidonic acid metabolites play a crucial role in the initiation of cardiac hypertrophy induced by isoproterenol. Drug Metab. Dispos. 43, 1254–1266. [DOI] [PubMed] [Google Scholar]
- Amarin Pharmaceuticals Inc. (2003). Pergolide Mesylate [product information], Mill Valley, CA.
- Amneal Pharmaceuticals LLC. (2019). Doxorubicin injection, powder, lyophilized In: DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=d928625e-c1ef-4f25-8d25-764c8c6aab67. Accessed January 3, 2020. [Google Scholar]
- Archer C. R., Sargeant R., Basak J., Pilling J., Barnes J. R., Pointon A. (2018). Characterization and validation of a human 3D cardiac microtissue for the assessment of changes in cardiac pathology. Sci. Rep. 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armas Pharmaceuticals Inc. (2019). Erlotinib hydrochloride tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=2c4c7353-b81c-4651-8aac-6d15e88145af. Accessed January 6, 2020. [Google Scholar]
- Arnold W. R., Baylon J. L., Tajkhorshid E., Das A. (2017). Arachidonic acid metabolism by human cardiovascular CYP2J2 is modulated by doxorubicin. Biochemistry 56, 6700–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AstraZeneca Pharmaceuticals LP. (2018). Vandetanib tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=4dc7f0af-77fb-4eec-46b9-dd1c2dcb4525. Accessed January 6, 2020. [Google Scholar]
- Aurobindo Pharma Limited. (2019). Dofetilide capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=85f2b35c-31d5-4161-b342-c781d23945a5. Accessed January 3, 2020. [Google Scholar]
- Balcarek K., Venhoff N., Deveaud C., Beauvoit B., Bonnet J., Kirschner J., Venhoff A. C., Lebrecht D., Walker U. A. (2010). Role of pyrimidine depletion in the mitochondrial cardiotoxicity of nucleoside analogue reverse transcriptase inhibitors. J. Acquir. Immune Defic. Syndr. 55, 550–557. [DOI] [PubMed] [Google Scholar]
- Bayer HealthCare Pharmaceuticals Inc. (2019). Sorafenib tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=b50667e4-5ebc-4968-a646-d605058dbef0. Accessed January 6, 2020. [Google Scholar]
- Bayshore Pharmaceuticals LLC. (2019). Sotalol hydrochloride tablet In DailyMed [Internet]. . National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=56879738-8662-4f5c-8386-761ab2b5e46f. Accessed January 3, 2020. [Google Scholar]
- Bayzigitov D. R., Medvedev S. P., Dementyeva E. V., Bayramova S. A., Pokushalov E. A., Karaskov A. M., Zakian S. M. (2016). Human induced pluripotent stem cell-derived cardiomyocytes afford new opportunities in inherited cardiovascular disease modeling. Cardiol. Res. Pract. 2016, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BC Cancer Agency. (2013a). Amsacrine monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2013b). Mitoxantrone monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2014a). Daunorubicin monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2014b). Trastuzumab monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2014c). Arsenic trioxide monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2014d). Axitinib monograph In: BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2015). Anagrelide monograph In: BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2016a). Doxorubicin monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2016b). Bortezomib monograph In BC Cancer Drug Manual® (Badry N., Ed.).BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2016c). Paclitaxel monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2018). Busulfan monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2019a). Idarubicin monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- BC Cancer Agency. (2019b). Fluorouracil monograph In BC Cancer Drug Manual® (Badry N., Ed.). BC Cancer, Vancouver, British Columbia, Canada. [Google Scholar]
- Blinova K., Dang Q., Millard D., Smith G., Pierson J., Guo L., Brock M., Lu H. R., Kraushaar U., Zeng H., et al. (2018). International multisite study of human-induced pluripotent stem cell-derived cardiomyocytes for drug proarrhythmic potential assessment. Cell Rep. 24, 3582–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BNM Group. (2013). Nortriptyline NRIM [New Zealand Data Sheet], Auckland, New Zealand.
- Cameron Pharmaceuticals LLC. (2019). Amiodarone hydrochloride tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=d779a746-0bdd-49cc-b0d8-5f9a72524b8c. Accessed December 27, 2019. [Google Scholar]
- Chaudhari U., Ellis J. K., Wagh V., Nemade H., Hescheler J., Keun H. C., Sachinidis A. (2017). Metabolite signatures of doxorubicin induced toxicity in human induced pluripotent stem cell-derived cardiomyocytes. Amino Acids 49, 1955–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi K. R. (2013). Revolution dawning in cardiotoxicity testing. Nat. Rev. Drug Discov. 12, 565–567. [DOI] [PubMed] [Google Scholar]
- Cipla USA Inc. (2019). Isoproterenol hydrochloride injection, solution In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=f724120d-da28-4065-9e05-d7dd6afbb546. Accessed January 3, 2020. [Google Scholar]
- Clements M., Millar V., Williams A. S., Kalinka S. (2015). Bridging functional and structural cardiotoxicity assays using human embryonic stem cell-derived cardiomyocytes for a more comprehensive risk assessment. Toxicol. Sci. 148, 241–260. [DOI] [PubMed] [Google Scholar]
- Clements M., Thomas N. (2014). High-throughput multi-parameter profiling of electrophysiological drug effects in human embryonic stem cell derived cardiomyocytes using multi-electrode arrays. Toxicol. Sci. 140, 445–461. [DOI] [PubMed] [Google Scholar]
- Colatsky T., Fermini B., Gintant G., Pierson J. B., Sager P., Sekino Y., Strauss D. G., Stockbridge N. (2016). The Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative—Update on progress. J. Pharmacol. Toxicol. Methods 81, 15–20. [DOI] [PubMed] [Google Scholar]
- Correia C., Koshkin A., Duarte P., Hu D., Teixeira A., Domian I., Serra M., Alves P. M. (2017). Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci. Rep. 7, 8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross M. J., Berridge B. R., Clements P. J. M., Cove-Smith L., Force T. L., Hoffmann P., Holbrook M., Lyon A. R., Mellor H. R., Norris A. A., et al. (2015). Physiological, pharmacological and toxicological considerations of drug-induced structural cardiac injury. Br. J. Pharmacol. 172, 957–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G., Nepusz T.. 2006. The igraph software package for complex network research. InterJ. Complex Syst. 1695, 1–9. [Google Scholar]
- Cuykx M., Beirnaert C., Rodrigues R. M., Laukens K., Vanhaecke T., Covaci A. (2019). Untargeted liquid chromatography-mass spectrometry metabolomics to assess drug-induced cholestatic features in HepaRG® cells. Toxicol. Appl. Pharmacol. 379, 114666. [DOI] [PubMed] [Google Scholar]
- Doherty K. R., Talbert D. R., Trusk P. B., Moran D. M., Shell S. A., Bacus S. (2015). Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol. Appl. Pharmacol. 285, 51–60. [DOI] [PubMed] [Google Scholar]
- Dunn W. B., Broadhurst D., Begley P., Zelena E., Francis-McIntyre S., Anderson N., Brown M., Knowles J. D., Halsall A., Haselden J. N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083. [DOI] [PubMed] [Google Scholar]
- E.R. Squibb & Sons L.L.C. (2019). Dasatinib tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=4764f37b-c9e6-4ede-bcc2-8a03b7c521df. Accessed January 6, 2020. [Google Scholar]
- Edwards A. G., Louch W. E. (2017). Species-dependent mechanisms of cardiac arrhythmia: A cellular focus. Clin. Med. Insights Cardiol. 11, 1179546816686061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eon Labs Inc. (2018). Quinidine sulfate tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=a10b6ded-4fe0-4059-bd77-c45acc3c876d. Accessed January 3, 2020. [Google Scholar]
- Figueredo V. M. (2011). Chemical cardiomyopathies: The negative effects of medications and nonprescribed drugs on the heart. Am. J. Med. 124, 480–488. [DOI] [PubMed] [Google Scholar]
- Force T., Kolaja K. L. (2011). Cardiotoxicity of kinase inhibitors: The prediction and translation of preclinical models to clinical outcomes. Nat. Rev. Drug Discov. 10, 111–126. [DOI] [PubMed] [Google Scholar]
- Fung M., Thornton A., Mybeck K., Wu J. H., Hornbuckle K., Muniz E. (2001). Evaluation of the characteristics of safety withdrawal of prescription drugs from worldwide pharmaceutical markets-1960 to 1999. Drug Inf. J. 35, 293–317. [Google Scholar]
- G.D. Searle LLC. (2001). Bextra [product information], Chicago, IL.
- Genentech Inc. (2019). Trastuzumab injection, powder, lyophilized, for solution In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=492dbdb2-077e-4064-bff3-372d6af0a7a2. Accessed January 3, 2020. [Google Scholar]
- Genschow E., Spielmann H., Scholz G., Seiler A., Brown N., Piersma A., Brady M., Clemann N., Huuskonen H., Paillard F., et al. (2002). The ECVAM international validation study on in vitro embryotoxicity tests: Results of the definitive phase and evaluation of prediction models. Altern. Lab. Anim. 30, 151–176. [DOI] [PubMed] [Google Scholar]
- Greenstone LLC. (2018). Nifedipine capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=9e5df4e1-be0d-4026-a7b0-3679b1effbde. Accessed January 3, 2020. [Google Scholar]
- Grubbs F. E. (1969). Procedures for detecting outlying observations in samples. Technometrics 11, 1–21. [Google Scholar]
- Gunnarsson O., Pfanzelter N. R., Cohen R. B., Keefe S. M. (2015). Evaluating the safety and efficacy of axitinib in the treatment of advanced renal cell carcinoma. Cancer Manag. Res. 7, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Abrams R. M., Babiarz J. E., Cohen J. D., Kameoka S., Sanders M. J., Chiao E., Kolaja K. L. (2011). Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol. Sci. 123, 281–289. [DOI] [PubMed] [Google Scholar]
- Guo L., Coyle L., Abrams R. M. C., Kemper R., Chiao E. T., Kolaja K. L. (2013). Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol. Sci. 136, 581–594. [DOI] [PubMed] [Google Scholar]
- Hantson P. (2019). Mechanisms of toxic cardiomyopathy. Clin. Toxicol. 57, 1–9. [DOI] [PubMed] [Google Scholar]
- Hikma Pharmaceuticals USA Inc. (2018). Daunorubicin hydrochloride injection In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=227784a8-ce68-4dd4-8ac5-a65265969677. Accessed January 3, 2020. [Google Scholar]
- Hikma Pharmaceuticals USA Inc. (2019). Idarubicin hydrochloride injection, solution In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=7c20b574-285b-49eb-a98e-6752e4bb481d. Accessed January 3, 2020. [Google Scholar]
- Hornberg J. J., Laursen M., Brenden N., Persson M., Thougaard A. V., Toft D. B., Mow T. (2014). Exploratory toxicology as an integrated part of drug discovery. Part II: Screening strategies. Drug Discov. Today 19, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Huo J., Kamalakar A., Yang X., Word B., Stockbridge N., Lyn-Cook B., Pang L. (2017). Evaluation of batch variations in induced pluripotent stem cell-derived human cardiomyocytes from 2 major suppliers. Toxicol. Sci. 156, 25–38. [DOI] [PubMed] [Google Scholar]
- Ingenus Pharmaceuticals LLC. (2019). Arsenic trioxide injection In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=c99b01b4-67ff-4793-8375-fe3a21c6c9be. Accessed December 27, 2019. [Google Scholar]
- Intermune Inc. (2006). Amphotericin B injection, lipid complex In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=9b8ea543-1de8-472f-9666-34f99ca2f183&audience=consumer. Accessed December 27, 2019. [Google Scholar]
- Jensen B. C., Parry T. L., Huang W., Beak J. Y., Ilaiwy A., Bain J. R., Newgard C. B., Muehlbauer M. J., Patterson C., Johnson G. L., et al. (2017a). Effects of the kinase inhibitor sorafenib on heart, muscle, liver and plasma metabolism in vivo using non-targeted metabolomics analysis. Br. J. Pharmacol. 174, 4797–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B. C., Parry T. L., Huang W., Ilaiwy A., Bain J. R., Muehlbauer M. J., O’Neal S. K., Patterson C., Johnson G. L., Willis M. S. (2017b). Non-targeted metabolomics analysis of the effects of tyrosine kinase inhibitors sunitinib and erlotinib on heart, muscle, liver and serum metabolism in vivo. Metabolites 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. A., Hasbak P., Mortensen J., Sørensen J. B. (2010). Fluorouracil induces myocardial ischemia with increases of plasma brain natriuretic peptide and lactic acid but without dysfunction of left ventricle. J. Clin. Oncol. 28, 5280–5286. [DOI] [PubMed] [Google Scholar]
- Jubilant Cadista Pharmaceuticals Inc. (2019). Telmisartan tablet In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=e1a907a7-fad9-a860-8639-4c6136c85734. Accessed January 3, 2020. [Google Scholar]
- Karwi Q. G., Uddin G. M., Ho K. L., Lopaschuk G. D. (2018). Loss of metabolic flexibility in the failing heart. Front. Cardiovasc. Med. 5, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Mukai Y., Rane A., Inotsume N., Toda T. (2017). The inhibitory effect of telmisartan on the metabolism of arachidonic acid by CYP2C9 and CYP2C8: An in vitro study. Biol. Pharm. Bull. Pharm. Bull. 40, 1409–1415. [DOI] [PubMed] [Google Scholar]
- Kawase T., Toyofuku M., Higashihara T., Okubo Y., Takahashi L., Kagawa Y., Yamane K., Mito S., Tamekiyo H., Otsuka M., et al. (2015). Validation of lactate level as a predictor of early mortality in acute decompensated heart failure patients who entered intensive care unit. J. Cardiol. 65, 164–170. [DOI] [PubMed] [Google Scholar]
- Kell D. B., Brown M., Davey H. M., Dunn W. B., Spasic I., Oliver S. G. (2005). Metabolic footprinting and systems biology: The medium is the message. Nat. Rev. Microbiol. 3, 557–565. [DOI] [PubMed] [Google Scholar]
- Kerkelä R., Grazette L., Yacobi R., Iliescu C., Patten R., Beahm C., Walters B., Shevtsov S., Pesant S., Clubb F. J., et al. (2006). Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat. Med. 12, 908–916. [DOI] [PubMed] [Google Scholar]
- Khan J. M., Lyon A. R., Harding S. E. (2013). The case for induced pluripotent stem cell-derived cardiomyocytes in pharmacological screening. Br. J. Pharmacol. 169, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiati S., Dalla Rosa I., Sourbier C., Ma X., Rao V. A., Neckers L. M., Zhang H., Pommier Y. (2014). Mitochondrial topoisomerase I (Top1mt) is a novel limiting factor of doxorubicin cardiotoxicity. Clin. Cancer Res. 20, 4873–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K., Youm J. B., Lee S. R., Lim S. E., Lee S. Y., Ko T. H., Long L. T., Nilius B., Won D. N., Noh J. H., et al. (2012). The angiotensin receptor blocker and PPAR-γ agonist, telmisartan, delays inactivation of voltage-gated sodium channel in rat heart: Novel mechanism of drug action. Pflugers Arch. 464, 631–643. [DOI] [PubMed] [Google Scholar]
- Kim T. D., le Coutre P., Schwarz M., Grille P., Levitin M., Fateh-Moghadam S., Giles F. J., Dorken B., Haverkamp W., Kohncke C. (2012). Clinical cardiac safety profile of nilotinib. Haematologica 97, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer N. C., Smith A. M., West P. R., Conard K. R., Fontaine B. R., Weir-Hauptman A. M., Palmer J. A., Knudsen T. B., Dix D. J., Donley E. L. R., et al. (2011). Identifying developmental toxicity pathways for a subset of ToxCast chemicals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 257, 111–121. [DOI] [PubMed] [Google Scholar]
- Klimas J. (2012). Drug-induced cardiomyopathies In Cardiomyopathies—From Basic Research to Clinical Management (Veselka J., Ed.), pp. 581–620. InTech, Rijeka, Croatia. [Google Scholar]
- Kuhn M., Wickham H.. 2019. rsample: General Resampling Infrastructure [R package rsample version 0.0.4]. Available at: https://CRAN.R-project.org/package=rsample. Accessed July 5, 2019.
- Laverty H., Benson C., Cartwright E., Cross M., Garland C., Hammond T., Holloway C., McMahon N., Milligan J., Park B., et al. (2011). How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol. 163, 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layland J. J., Liew D., Prior D. L. (2009). Clozapine-induced cardiotoxicity: A clinical update. Med. J. Aust. 190, 190–192. [DOI] [PubMed] [Google Scholar]
- Lazzeri C., Valente S., Chiostri M., Gensini G. F. (2015). Clinical significance of Lactate in acute cardiac patients. World J. Cardiol. 7, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ju L., Hou Z., Deng H., Zhang Z., Wang L., Yang Z., Yin J., Zhang Y. (2015). Screening, verification, and optimization of biomarkers for early prediction of cardiotoxicity based on metabolomics. J. Proteome Res. 14, 2437–2445. [DOI] [PubMed] [Google Scholar]
- Limonciel A., Aschauer L., Wilmes A., Prajczer S., Leonard M. O., Pfaller W., Jennings P. (2011). Lactate is an ideal non-invasive marker for evaluating temporal alterations in cell stress and toxicity in repeat dose testing regimes. Toxicol. In Vitro 25, 1855–1862. [DOI] [PubMed] [Google Scholar]
- Lin Z., Will Y. (2012). Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicol. Sci. 126, 114–127. [DOI] [PubMed] [Google Scholar]
- Lindström E., Farde L., Eberhard J., Haverkamp W. (2005). QTc interval prolongation and antipsychotic drug treatments: Focus on sertindole. Int. J. Neuropsychopharmacol. 8, 615–629. [DOI] [PubMed] [Google Scholar]
- Mayne Pharma Inc. (2019). Nortriptyline hydrochloride capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=765d726b-fd4b-4ef7-afd7-9e7e9bf8cae6. Accessed January 6, 2020. [Google Scholar]
- McKee E. E., Bentley A. T., Hatch M., Gingerich J., Susan-Resiga D. (2004). Phosphorylation of thymidine and AZT in heart mitochondria: Elucidation of a novel mechanism of AZT cardiotoxicity. Cardiovasc. Toxicol. 4, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H. R., Bell A. R., Valentin J. P., Roberts R. (2011). Cardiotoxicity associated with targeting kinase pathways in cancer. Toxicol. Sci. 120, 14–32. [DOI] [PubMed] [Google Scholar]
- Merck & Co Inc. (2002). VIOXX® [product information], Whitehouse Station, NJ.
- Mitjavila M. T., Moreno J. J. (2012). The effects of polyphenols on oxidative stress and the arachidonic acid cascade. Implications for the prevention/treatment of high prevalence diseases. Biochem. Pharmacol. 84, 1113–1122. [DOI] [PubMed] [Google Scholar]
- Moodley I. (2008). Review of the cardiovascular safety of COXIBs compared to NSAIDS. Cardiovasc. J. Afr. 19, 102–107. [PMC free article] [PubMed] [Google Scholar]
- Mylan Institutional LLC. (2018). Paclitaxel injection In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=9ffd3e34-537f-4f65-b00e-57c25bab3b01. Accessed January 3, 2020. [Google Scholar]
- Mylan Pharmaceuticals Inc. (2018). Thioridazine hydrochloride tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=56b3f4c2-52af-4947-b225-6808ae9f26f5. Accessed January 3, 2020. [Google Scholar]
- Natale R. B., Thongprasert S., Greco F. A., Thomas M., Tsai C. M., Sunpaweravong P., Ferry D., Mulatero C., Whorf R., Thompson J., et al. (2011). Phase III trial of vandetanib compared with erlotinib in patients with previously treated advanced non–small-cell lung cancer. J. Clin. Oncol. 29, 1059–1066. [DOI] [PubMed] [Google Scholar]
- Niu Q. Y., Li Z. Y., Du G. H., Qin X. M. (2016). 1H NMR based metabolomic profiling revealed doxorubicin-induced systematic alterations in a rat model. J. Pharm. Biomed. Anal. 118, 338–348. [DOI] [PubMed] [Google Scholar]
- Novartis Pharmaceuticals Corporation. (2019a). Lapatinib tablet In: DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=eee37f88-ec6a-4c30-b8aa-e2c71f93088c. Accessed January 6, 2020. [Google Scholar]
- Novartis Pharmaceuticals Corporation. (2019b). Nilotinib capsule In: DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=6093952a-5248-45cb-ad17-33716a411146. Accessed January 6, 2020. [Google Scholar]
- Onakpoya I. J., Heneghan C. J., Aronson J. K. (2016). Post-marketing withdrawal of 462 medicinal products because of adverse drug reactions: A systematic review of the world literature. BMC Med. 14, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakkari I. (2002). Cardiotoxicity of new antihistamines and cisapride. Toxicol. Lett. 127, 279–284. [DOI] [PubMed] [Google Scholar]
- Pai V. B., Nahata M. C. (2000). Cardiotoxicity of chemotherapeutic agents: Incidence, treatment and prevention. Drug Saf. 22, 263–302. [DOI] [PubMed] [Google Scholar]
- Palmer J. A., Poenitzsch A. M., Smith S. M., Conard K. R., West P. R., Cezar G. G. (2012). Metabolic biomarkers of prenatal alcohol exposure in human embryonic stem cell-derived neural lineages. Alcohol Clin. Exp. Res. 36, 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. A., Smith A. M., Egnash L. A., Conard K. R., West P. R., Burrier R. E., Donley E. L. R., Kirchner F. R. (2013). Establishment and assessment of a new human embryonic stem cell-based biomarker assay for developmental toxicity screening. Birth Defects Res. B Dev. Reprod. Toxicol. 98, 343–363. [DOI] [PubMed] [Google Scholar]
- Perez E. A., Koehler M., Byrne J., Preston A. J., Rappold E., Ewer M. S. (2008). Cardiac safety of lapatinib: Pooled analysis of 3689 patients enrolled in clinical trials. Mayo Clin. Proc. 83, 679–686. [DOI] [PubMed] [Google Scholar]
- Pfizer Laboratories. (2019a). Crizotinib capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=2a51b0de-47d6-455e-a94c-d2c737b04ff7. Accessed January 6, 2020. [Google Scholar]
- Pfizer Laboratories. (2019b). Sunitinib malate capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=43a4d7f8-48ae-4a63-9108-2fa8e3ea9d9c. Accessed January 6, 2020. [Google Scholar]
- Pfizer Laboratories. (2019c). Axitinib tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=84137882-e000-47da-bd5b-fa76ab3c76f9. Accessed January 6, 2020. [Google Scholar]
- Physicians Total Care Inc. (2010). Rosiglitazone maleate tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=ef14122b-7fff-45fa-b13b-0ea9e48bd57d. Accessed January 3, 2020. [Google Scholar]
- Pointon A., Abi-gerges N., Cross M. J., Sidaway J. E. (2013). Phenotypic profiling of structural cardiotoxins in vitro reveals dependency on multiple mechanisms of toxicity. Toxicol. Sci. 132, 317–326. [DOI] [PubMed] [Google Scholar]
- Pun S. C., Neilan T. G. (2016). Cardiovascular side effects of small molecule therapies for cancer. Eur. Heart J. 37, 2742–2745. [DOI] [PubMed] [Google Scholar]
- Qureshi Z. P., Seoane-Vazquez E., Rodriguez-Monguio R., Stevenson K. B., Szeinbach S. L. (2011). Market withdrawal of new molecular entities approved in the United States from 1980 to 2009. Pharmacoepidemiol. Drug Saf. 20, 772–777. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Rana P., Anson B., Engle S., Will Y. (2012). Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: Bioenergetics and utilization in safety screening. Toxicol. Sci. 130, 117–131. [DOI] [PubMed] [Google Scholar]
- Ranbaxy Laboratories Inc. (2008). Verapamil hydrochloride tablet, coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=6ae13cb4-0316-40d1-9216-c7d5556aaed3. Accessed January 3, 2020. [Google Scholar]
- Ranbaxy Pharmaceuticals Inc. (2014). Chloroquine phosphate tablet, film coated In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=f398f8a9-92f3-47cb-81c2-6078806a464d##. Accessed January 3, 2020. [Google Scholar]
- Redfern W. S., Carlsson L., Davis A. S., Lynch W. G., MacKenzie I., Palethorpe S., Siegl P. K. S., Strang I., Sullivan A. T., Wallis R. (2003). Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: Evidence for a provisional safety margin in drug development. Cardiovasc. Res. 58, 32–45. [DOI] [PubMed] [Google Scholar]
- Ritz C., Streibig J. C. (2005). Bioassay analysis using R. J. Stat. Softw. 12, 1–12. [Google Scholar]
- Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. (2011). pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison T. W., Giri S. N. (1987). Effects of chronic administration of doxorubicin on plasma levels of prostaglandins, thromboxane B2, and fatty acids in rats. Cancer Chemother. Pharmacol. 19, 213–220. [DOI] [PubMed] [Google Scholar]
- Roxane Laboratories Inc. (2002). Levomethadyl Acetate Hydrochloride Oral Solution [product information], Columbus, OH.
- Sager P. T., Gintant G., Turner J. R., Pettit S., Stockbridge N. (2014). Rechanneling the cardiac proarrhythmia safety paradigm: A meeting report from the Cardiac Safety Research Consortium. Am. Heart J. 167, 292–300. [DOI] [PubMed] [Google Scholar]
- Schimmel K. J. M., Richel D. J., van den Brink R. B. A., Guchelaar H. J. (2004). Cardiotoxicity of cytotoxic drugs. Cancer Treat. Rev. 30, 181–191. [DOI] [PubMed] [Google Scholar]
- Schmidinger M., Zielinski C. C., Vogl U. M., Bojic A., Bojic M., Schukro C., Ruhsam M., Hejna M., Schmidinger H. (2008). Cardiac toxicity of sunitinib and sorafenib in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 26, 5204–5212. [DOI] [PubMed] [Google Scholar]
- Schnackenberg L. K., Pence L., Vijay V., Moland C. L., George N., Cao Z., Yu L. R., Fuscoe J. C., Beger R. D., Desai V. G. (2016). Early metabolomics changes in heart and plasma during chronic doxorubicin treatment in B6C3F1 mice. J. Appl. Toxicol. 36, 1486–1495. [DOI] [PubMed] [Google Scholar]
- Shah D. R., Shah R. R., Morganroth J. (2013). Tyrosine kinase inhibitors: Their on-target toxicities as potential indicators of efficacy. Drug Saf. 36, 413–426. [DOI] [PubMed] [Google Scholar]
- Sirenko O., Crittenden C., Callamaras N., Hesley J., Chen Y. W., Funes C., Rusyn I., Anson B., Cromwell E. F. (2013). Multiparameter in vitro assessment of compound effects on cardiomyocyte physiology using iPSC cells. J. Biomol. Screen. 18, 39–53. [DOI] [PubMed] [Google Scholar]
- Soldovieri M. V., Miceli F., Taglialatela M. (2008). Cardiotoxic effects of antihistamines: From basics to clinics (…and back). Chem. Res. Toxicol. 21, 997–1004. [DOI] [PubMed] [Google Scholar]
- Stöllberger C., Huber J. O., Finsterer J. (2005). Antipsychotic drugs and QT prolongation. Int. Clin. Psychopharmacol. 20, 243–251. [DOI] [PubMed] [Google Scholar]
- Sullivan M. L., Martinez C. M., Gennis P., Gallagher E. J. (1998). The cardiac toxicity of anabolic steroids. Prog. Cardiovasc. Dis. 41, 1–15. [DOI] [PubMed] [Google Scholar]
- Sun R., Eriksson S., Wang L. (2014). Zidovudine induces downregulation of mitochondrial deoxynucleoside kinases: Implications for mitochondrial toxicity of antiviral nucleoside analogs. Antimicrob. Agents Chemother. 58, 6758–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susan-Resiga D., Bentley A. T., Lynx M. D., LaClair D. D., McKee E. E. (2007). Zidovudine inhibits thymidine phosphorylation in the isolated perfused rat heart. Antimicrob. Agents Chemother. 51, 1142–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert D. R., Doherty K. R., Trusk P. B., Moran D. M., Shell S. A., Bacus S. (2015). A multi-parameter in vitro screen in human stem cell-derived cardiomyocytes identifies ponatinib-induced structural and functional cardiac toxicity. Toxicol. Sci. 143, 147–155. [DOI] [PubMed] [Google Scholar]
- Talpaz M., Shah N. P., Kantarjian H., Donato N., Nicoll J., Paquette R., Cortes J., O’Brien S., Nicaise C., Bleickardt E., et al. (2006). Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 354, 2531–2541. [DOI] [PubMed] [Google Scholar]
- Tan G., Lou Z., Liao W., Zhu Z., Dong X., Zhang W., Li W., Chai Y. (2011). Potential biomarkers in mouse myocardium of doxorubicin-induced cardiomyopathy: A metabonomic method and its application. PLoS One 6, e27683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teva Parenteral Medicines Inc. (2019). Mitoxantrone injection, solution, concentrate In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=4d0f0f1a-31af-40fa-9c64-e90891fa6ce4. Accessed January 3, 2020. [Google Scholar]
- The Medicines Company. (2019). Ondansetron hydrochloride injection In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=dc3dc0e4-b7da-4da4-afa6-d57e4ee42339. Accessed January 3, 2020. [Google Scholar]
- Torrent Pharmaceuticals Limited. (2019). Anagrelide capsule In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=875cc32a-219c-4b6b-802a-98a47cbc9a78&audience=professional. Accessed December 27, 2019. [Google Scholar]
- Tsukamoto H., Hishinuma T., Suzuki N., Tayama R., Hiratsuka M., Yoshihisa T., Mizugaki M., Goto J. (2004). Thiazolidinediones increase arachidonic acid release and subsequent prostanoid production in a peroxisome proliferator-activated receptor gamma-independent manner. Prostaglandins Other Lipid Mediat. 73, 191–213. [DOI] [PubMed] [Google Scholar]
- Ulmer B. M., Eschenhagen T.. Human pluripotent stem cell-derived cardiomyocytes for studying energy metabolism. Biochim. Biophys. Acta Mol. Cell. Res. (forthcoming). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin J. P., Bialecki R., Ewart L., Hammond T., Leishmann D., Lindgren S., Martinez V., Pollard C., Redfern W., Wallis R. (2009). A framework to assess the translation of safety pharmacology data to humans. J. Pharmacol. Toxicol. Methods 60, 152–158. [DOI] [PubMed] [Google Scholar]
- Valentin J. -P., Redfern W. S. (2017). Prevalence, frequency and impact of safety related issues throughout the pharmaceutical life cycle. Toxicology 150, 170. [Google Scholar]
- van Vliet E., Morath S., Eskes C., Linge J., Rappsilber J., Honegger P., Hartung T., Coecke S. (2008). A novel in vitro metabolomics approach for neurotoxicity testing, proof of principle for methyl mercury chloride and caffeine. Neurotoxicology 29, 1–12. [DOI] [PubMed] [Google Scholar]
- Varga Z. V., Ferdinandy P., Liaudet L., Pacher P. (2015). Drug-induced mitochondrial dysfunction and cardiotoxicity. Am. J. Physiol. Circ. Physiol. 309, H1453–H1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varricchi G., Ameri P., Cadeddu C., Ghigo A., Madonna R., Marone G., Mercurio V., Monte I., Novo G., Parrella P., et al. (2018). Antineoplastic drug-induced cardiotoxicity: A redox perspective. Front. Physiol. 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilaki F., Tsitsimpikou C., Tsarouhas K., Germanakis I., Tzardi M., Kavvalakis M., Ozcagli E., Kouretas D., Tsatsakis A. M. (2016). Cardiotoxicity in rabbits after long-term nandrolone decanoate administration. Toxicol. Lett. 241, 143–151. [DOI] [PubMed] [Google Scholar]
- Wang J., Reijmers T., Chen L., Van Der Heijden R., Wang M., Peng S., Hankemeier T., Xu G., Van Der Greef J. (2009). Systems toxicology study of doxorubicin on rats using ultra performance liquid chromatography coupled with mass spectrometry based metabolomics. Metabolomics 5, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson-Haigh N. S., Kadarmideen H. N., Reverter A. (2010). PCIT: An R package for weighted gene co-expression networks based on partial correlation and information theory approaches. Bioinformatics 26, 411–413. [DOI] [PubMed] [Google Scholar]
- Weaver R. J., Valentin J. P. (2019). Today’s challenges to de-risk and predict drug safety in human “mind-the-gap.” Toxicol. Sci. 167, 307–321. [DOI] [PubMed] [Google Scholar]
- West P. R., Weir A. M., Smith A. M., Donley E. L. R., Cezar G. G. (2010). Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharmacol. 247, 18–27. [DOI] [PubMed] [Google Scholar]
- West-Ward Pharmaceuticals Corp. (2018). Chlorpromazine hydrochloride injection In DailyMed [Internet]. 2005. National Library of Medicine (US; ), Bethesda, MD: Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=c0f1fcfd-8863-4032-bf80-185df60a4ba0. Accessed January 3, 2020. [Google Scholar]
- Wilmes A., Limonciel A., Aschauer L., Moenks K., Bielow C., Leonard M. O., Hamon J., Carpi D., Ruzek S., Handler A., et al. (2013). Application of integrated transcriptomic, proteomic and metabolomic profiling for the delineation of mechanisms of drug induced cell stress. J. Proteomics 79, 180–194. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2004). Cardiac valvulopathy and pergolide. WHO Drug Inf. 18, 208. [Google Scholar]
- Xu Z., Cang S., Yang T., Liu D. (2009). Cardiotoxicity of tyrosine kinase inhibitors in chronic myelogenous leukemia therapy. Hematol. Rep. 1, e4. [Google Scholar]
- Yang X., Papoian T. (2018). Moving beyond the comprehensive in vitro proarrhythmia assay: Use of human-induced pluripotent stem cell-derived cardiomyocytes to assess contractile effects associated with drug-induced structural cardiotoxicity. J. Appl. Toxicol. 38, 1166–1176. [DOI] [PubMed] [Google Scholar]
- Yin J., Xie J., Guo X., Ju L., Li Y., Zhang Y. (2016). Plasma metabolic profiling analysis of cyclophosphamide-induced cardiotoxicity using metabolomics coupled with UPLC/Q-TOF-MS and ROC curve. J. Chromatogr. B 1033–1034, 428–435. [DOI] [PubMed] [Google Scholar]
- Yue S., Yu J., Kong Y., Chen H., Mao M., Ji C., Shao S., Zhu J., Gu J., Zhao M. (2019). Metabolomic modulations of HepG2 cells exposed to bisphenol analogues. Environ. Int. 129, 59–67. [DOI] [PubMed] [Google Scholar]
- Zang J., Wu S., Tang L., Xu X., Bai J., Ding C., Chang Y., Yue L., Kang E., He J. (2012). Incidence and risk of QTc interval prolongation among cancer patients treated with vandetanib: A systematic review and meta-analysis. PLoS One 7, e30353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Knapton A., Lipshultz S. E., Weaver J. L., Herman E. H. (2008). Isoproterenol-induced cardiotoxicity in Sprague-Dawley rats: Correlation of reversible and irreversible myocardial injury with release of cardiac troponin T and roles of iNOS in myocardial injury. Toxicol. Pathol. 36, 277–288. [DOI] [PubMed] [Google Scholar]
- Zhang Y., El-Sikhry H., Chaudhary K. R., Batchu S. N., Shayeganpour A., Jukar T. O., Bradbury J. A., Graves J. P., DeGraff L. M., Myers P., et al. (2009). Overexpression of CYP2J2 provides protection against doxorubicin-induced cardiotoxicity. Am. J. Physiol. Circ. Physiol. 297, H37–H46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppinger C., Timolati F., Suter T. M. (2007). Pathophysiology and diagnosis of cancer drug induced cardiomyopathy. Cardiovasc. Toxicol. 7, 61–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary data are available at https://doi.org/10.5061/dryad.gqnk98shs. The raw peak apex data for each metabolite and spiked-in ISTD for each sample analyzed in development of the final prediction model development is available in the Dryad repository (https://doi.org/10.5061/dryad.gqnk98shs).