Abstract
Background
Heat shock protein family A member 1 like (Hspa1l) is a member of the 70kD heat shock protein (Hsp70) family. HSPA1L is an ancient, evolutionarily conserved gene with a highly conserved domain structure. The gene is highly abundant and constitutively expressed in the mice testes. However, the role of Hspa1l in the testes has still not been elucidated.
Methods
Hspa1l-mutant mice were generated using the CRISPR/Cas9 system. Histological and immunofluorescence staining were used to analyze the phenotypes of testis and epididymis. Apoptotic cells were detected through TUNEL assays. Fertility and sperm motilities were also tested. Quantitative RT-PCR was used for analyzing of candidate genes expression. Heat treatment was used to induce heat stress of the testis.
Results
We successfully generated Hspa1l knockout mice. Hspa1l-/- mice exhibited normal development and fertility. Further, Hspa1l-/- mice shown no significant difference in spermatogenesis, the number of apoptotic cells in testes epididymal histology, sperm count and sperm motility from Hspa1l+/+ mice. Moreover, heat stress does not exacerbate the cell apoptosis in Hspa1l-/- testes. These results revealed that HSPA1L is not essential for physiological spermatogenesis, nor is it involved in heat-induced stress responses, which provides a basis for further studies.
Keywords: HSPA1L, Spermatogenesis, Male infertility, Gene knockout
Introduction
Infertility is a widespread issue that affects approximately 70 million people worldwide. Approximately 9% of couples are affected by infertility, with male infertility thought to play a role in 50% of infertile couples estimated by the World Health Organization (WHO) (Barratt et al., 2017). Male infertility attributed to a variety of reasons including: genetic factors such as mutations (Fainberg & Kashanian, 2019; Signore et al., 2019); diseases such as testicular cancer, cryptorchidism (Skakkebaek et al., 2016); unhealthy habits such as alcohol, tobacco and drug addiction (Sansone et al., 2018); environmental stress such as intense exposure to heat, pesticides, radiation, radioactivity and other harmful substances (Durairajanayagam, Agarwal & Ong, 2015; Kesari, Agarwal & Henkel, 2018). Spermatogenesis is a precisely regulated and complex process. Adult male testicular tissue consists of somatic and spermatogenic cells. Somatic cells in the testis mainly include Sertoli cells, Leydig cells, and testicular peritubular cells (TPCs). These somatic cells form the testicular microenvironment, and play an important role in regulating spermatogenesis. Spermatogenic cells mainly include spermatogonia (SPG), which are located in the basal compartment of seminiferous tubules. Human SPG can be divided into three different types: dark type A, light type A and B type SPG. Spermatogonial stem cells (SSCs) are self-renewing undifferentiated SPGs, and are thought to exist in type A SPGs (Fayomi & Orwig, 2018). Primary spermatocytes (SPCs) extend into the lumen of the seminiferous tubules, and are generated by SPGs through the transition process from mitosis to meiosis. SPCs can be divided into five stages: leptotene, zygotene, pachytene, diplotene and diakinesis. Primary SPCs undergo the first meiotic division to form secondary SPCs, then undergo a second meiosis to produce the round spermatids. These round spermatids further change shape, forming elongated spermatids that are then discharged into the lumen (Hermo et al., 2010; Neto et al., 2016; Wen et al., 2020). Germ cells development in the first wave of spermatogenesis in mice can be divided into the following stages: cells exhibiting stem-cell properties at postnatal day (PND) 0, spermatogonial mitosis at PND 7, pachyetene spermatocytes at PND 14, round spermatids at PND 20, and elongated spermatids at PND 28 (Chen et al., 2018b; Huang et al., 2008; Pence et al., 2019).
The HSP70 family is one of the most conserved protein families through evolution, from archaebacteria to plants and humans (Daugaard, Rohde & Jaattela, 2007; Radons, 2016). The HSP70s have a highly conserved domain structure containing a N-terminal nucleotide-binding domain (NBD) that exhibits ATPase activity, a middle flexible linker region, a substrate binding domain (SBD) that binds hydrophobic polypeptides, and an α-helical C-terminal domain (CTD) that seals the substrate and also mediates the binding of co-chaperone (Daugaard, Rohde & Jaattela, 2007; Radons, 2016; Vos et al., 2008; Wisniewska et al., 2010; Zuiderweg, Hightower & Gestwicki, 2017). The NBD and SBD always work together. The NBD binds and hydrolyzes ATP to regulate the affinity for substrates, and the binding of hydrophobic substrates to SBD accelerates the hydrolysis of ATP (Mayer, 2013; Mayer & Bukau, 2005).
Due to the ability of HSP70 to bind ATP and hydrophobic protein sequences, HSP70s play a crucial role in refolding denatured proteins, disaggregation, protein degradation, and maintaining protein homeostasis (Fernandez-Fernandez & Valpuesta, 2018). HSP70s also play a role in cell survival during stresses, including heat shock, oxidative stress, hypoxia, heavy metals, altered pH, inflammation, ischemia, fever, malignancy, etc. (Chen, Feder & Kang, 2018a; Ikwegbue et al., 2017; Radons, 2016; Singh et al., 2010). The changes in expression of HSP70s has been indicated in various diseases, including cancer and nervous system diseases (Lee et al., 2001; Perez et al., 1991; Turturici, Sconzo & Geraci, 2011).
The human HSP70 family consists of 13 protein members which differ from each other in their amino acid sequences, subcellular locations and expression level (Daugaard, Rohde & Jaattela, 2007; Radons, 2016). In the HSP70 family, only HSPA2 and HSPA1L are mainly expressed in the testis. HSPA2 has been suggested to play an essential role in spermatogenesis, and is linked to male infertility (Dix et al., 1996; Erata et al., 2008; Nixon et al., 2015; Zhu, Dix & Eddy, 1997). Hspa1l is located in a major histocompatibility class (MHC) III cluster on 6p21.33 in human, where Hspa1a and Hspa1b are also located. Hspa1a and Hspa1b are more than 99% identical, and Hspa1l is 91% identical to Hspa1a (Daugaard, Rohde & Jaattela, 2007). HSPA1A and HSPA1B differ by only two amino acids. They are both stress-inducible and are functionally interchangeable so they are jointly named as HSP70 or HSP70-1 (Daugaard, Rohde & Jaattela, 2007; Radons, 2016). The basal mRNA expression of HSPA1A or HSPA1B varies in most tissues, and can be rapidly induced by stress (Murphy, 2013). The dysfunction of HSPA1A/B increases cells’ sensitivity to stress, and also affects the stability of vital organelles such as lysosomes and mitotic centrosomes (Dix, Garges & Hong, 1998; Fang et al., 2016; Huang, Mivechi & Moskophidis, 2001; Yamashima, 2012). Unlike Hspa1a and Hspa1b, Hspa1l is constitutively expressed, and is highly abundant in the testis (Daugaard, Rohde & Jaattela, 2007; Radons, 2016). The mutations of Hspa1l is supposed to be associated with inflammatory bowel disease, an increased risk of male infertility and spontaneous preterm births (Ciftci et al., 2015; Huusko et al., 2018; Kohan, Tabiee & Sepahi, 2019; Takahashi et al., 2017). HSPA1L has been thought to play important functions in testis similar to HSPA2 (Tsunekawa et al., 1999). However, the in vivo function of HSPA1L in testis has still not been elucidated in detail yet. Thus, in this study, we used the CRISPR/Cas9 system to generate Hspa1l-knockout mice to investigate the role of HSPA1L in testicular development and spermatogenesis.
Materials & Methods
Animals
Protocols for mice care, feeding, housing and treatments were drafted and implemented under the guidance of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University. The procedures of using and treating mice were approved by the Animal Ethical and Welfare Committee (Approval No. IACUC-1601117).
All mice were maintained under SPF conditions (12 h light/dark cycle, 20−26 °C, 50–55% humidity) with free access to water and food in Laboratory Animal Center of Nanjing Medical University. All mice were treated humanely and with efforts to minimize suffering. To induce loss of consciousness and death with a minimum of pain and distress, all mice were euthanized by cervical dislocation to collected tissue samples for further analyses. There were no surviving animals at the end of study.
Generation of Hspa1l-mutant mice
We used the CRISPR/Cas9 technology to generate Hspa1l-knockout mice. The single-guide RNAs (sgRNAs) has been devised in the light of exon2 of Hspa1l. The sgRNA target sequences were 5′-CCACCAAGGATGCAGGTGTCATC-3 ′ and 5′- CGTGCACGAGTAGAAGCTGGACC-3 ′. The Cas9 plasmid and sgRNA plasmid were linearized by AgeI and Dra I respectively, and purified by MinElute PCR Purification Kit (Qiagen, Duesseldorf, Germany). MMESSAGE mMACHINE T7 Ultra Kit (Ambion, TX, USA) were used to produce Cas9 mRNA. MEGA Shortscript and Clear Kit (Ambion, TX, USA) were used to produce and purify the sgRNA. Wild-type C57BL/6 superovulated females were mated with C57BL/6 males to obtain zygotes for Cas9 mRNA and sgRNA injection.
Genotyping
Edited founders with Hspa1l frameshift mutations were mated with Hspa1l+∕+ for at least three generations to avoid off-targets. The genotype identification of their offspring was completed by PCR amplification (primers: Forward, 5′-TACTGACGAAGATGAAGGAGACT-3′; Reverse, 5′-CGCTTGTTCTGGCTGATGTC-3′) and Sanger sequencing. The results of sequencing were analyzed by SnapGene (version 3.2.1).
Heat treatment
Mice used for heat treatment were anaesthetized by 1.2% Avertin (Sigma-Aldrich, St. Louis, USA), and lower half of the body of adult (8–10 weeks) Hspa1l−∕− andHspa1l+∕+ male mice were subjected to water bath for 30 min at 42 °C after narcotization. Animals were dried and returned to their cages for another 4 h. Mice narcotized and left in their cages at normal temperature were used as controls. All mice were euthanized for further analyses after the treatment (n = 5).
Western blot
Testicular protein (n = 5 per group) was extracted by RIPA buffer (P0013C, Beyotime, Shanghai, China) containing protease inhibitor cocktail (B14002, Bimake, Houston, USA). The Bradford method was used to measure the concentration of protein in each sample. Samples were separated by SDS-PAGE of 10% acrylamide gel and electrotransferred to a PVDF membrane. The membranes were then blocked with 5% skim milk in TBS for 1 h at room temperature and followed by 2 h incubation with the primary rabbit anti-HSPA1L (1:1500 dilution, 13970-1, Proteintech, PA, USA) and anti-GAPDH (1:1500 dilution, 14C10, Cell Signaling Technology, MA, USA) at room temperature. After three washes with TBST, the membranes were incubated with secondary antibodies (1:2000 dilution, 31,460, Thermo Fisher, NY, USA) for 40 min at room temperature. ChemiDoc XRS+ System (Bio-Rad, CA, USA) were used for visualizing specific proteins bands.
Histology
The tissues from mouse testes or epididymis were fixed in modified Davidson’s fluid overnight, and stored in 70% ethanol. This was followed by a series of ethanol dehydration steps. Finally, the tissue was embedded in paraffin. Sections (5 μm) were spread onto slides then dried overnight at 65 °C. Tissue sections were stained with hematoxylin and eosin (H & E) after deparaffination by immersing in serial concentrations of ethanol. Sperm samples obtained from the cauda epididymis were spread onto slides and fixed with 4% paraformaldehyde in PBS for 30 mins, then washed and stained with H&E for histological analysis. Mammalian sperms are considered abnormal when some certain morphology appears, including head abnormal and tail abnormal. Head abnormal includes a hairpin at the neck, headless, and a hammer like, collapsed, triangular or thin elongated head. Absent, short, angular or irregular tails are assessed as tails abnormal (Gatimel et al., 2017; Kawai et al., 2006). All observations were made using bright field microscopy (ZEISS Axio Skop Plus2, Jena, Germany).
Immunofluorescence and TUNEL assay
Testis sections were rehydrated and antigens were retrieved in sodium citrate buffer by boiling for 10 min. 5% BSA was used to block the sections at room temperature for 2 h after cooled. Incubation with primary antibodies (list in Table S1) were done overnight at 4 °C. The membranes were then washes three times with PBST (0.02% Tween 20 in PBS) before incubation with secondary antibody (list in Table S1) and Hoechst 33342 (1:1000 dilution, Invitrogen, CA, USA) at 37 °C for 1 h. Sections were then washed and mounted. Terminal deoxynucleotidyl transferase mediated nick-end labeling (TUNEL) assay (Vazyme, Nanjing, China) was used to detected apoptotic cells in testis according to the manufacturer’s protocols (n = 5 per group). LSM800 confocal microscope (Carl Zeiss AG, Jena, Germany) was used for Images capture.
Fertility test
Adult Hspa1l−∕− and Hspa1l+∕+ male mice were bred with Hspa1l+∕+ C57BL/6 females, respectively, and Hspa1l−∕− female mice were also bred with Hspa1l+∕+ C57BL/6 males to carry out fertility tests. The mice were checked every morning for a vaginal plug and the date and number of pups were recorded from each litter (n = 5).
Epididymal sperm analysis
Several cuts were made throughout the cauda epididymis and suspended in 10% FBS in modified HTF medium (FUJIFILM Irvine Scientific, CA, USA) at 37 °C for 5 min to obtain mature sperm. 10 µl sperm samples were analyzed using computer assisted semen analysis (Hamilton Thorne Research, MA, USA). The motility, progressive motility, and sperm concentration of the Hspa1l−∕− mice and controls were then measured and analyzed (n = 5).
Quantitative RT-PCR
Total RNA of tissues (n = 5 per group) was extracted by TRIzol reagent (Invitrogen, CA, USA). NanoDrop 2000C (Thermo, MA, USA) was used to determine the RNA concentration. PrimeScript RT Master Mix (Takara, CA, USA) was used for reverse transcription of 500 ng total RNA according to the standard protocol. The cDNA was diluted (1:4) and detected by quantitative RT-PCR using a AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China) set to the following conditions: 95 °C denatured for 5 min, 95 °C denatured for 10 s for 40 amplification cycles and annealing and extension at 60 °C for 30 s. 18s rRNAwas the positive control used for normalizing the gene expression and the sequences of primers used in this paper are listed in Table S2.
Statistical analysis
All data/experiments were repeated at least 5 times, and were represented as the mean ± SD. Independent Student’s t-test or one-way ANOVA were used for statistic comparison. P-value <0.05 was considered statistically significant. GraphPad Prism 6.02 was used to analyze data and draw graphs.
Results
Expression analysis of Hspa1l in mice
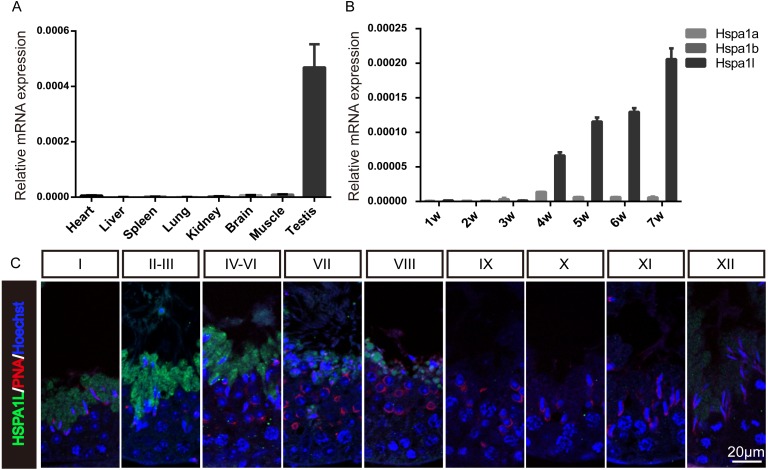
To investigate the expression of Hspa1l in mice, multi-tissue expression analysis were used, which showed that Hspa1l was predominantly expressed in testis (Fig. 1A and Fig. S1). The Hspa1l mRNA levels start to build up in the testis of 4 weeks old mice pups followed by high expression levels as they grow (Fig. 1B). It is the most abundant of the 3 genes from the MHC III cluster in 17B1 in adult testis, significantly higher than Hspa1a and Hspa1b. Immunofluorescence analysis showed that HSPA1L was expressed only in spermatids in testes, as it was weakly detectable from step 12 spermatids of stage I, and was the strongest expressed in step 14 and 15 spermatids at stages II–VI (Fig. 1C).
Figure 1. Expression of Hspa1l in mouse.
(A) Expression analysis of hspa1l in mouse tissues, including heart, liver, spleen, lung, kidney, brain, muscle and testis by RT-qPCR. (B) Expression of hspa1a, hspa1b and hspa1l in mouse testes at different developmental stages, including 1, 2, 3, 4, 5, 6 and 7 weeks by RT-qPCR. 18s served as a cDNA loading control in (A–B). (C) HSPA1L staining at specific developmental stages of the mouse male germ cells by immunofluorescence. PNA, shows spermatids.
Generation of Hspa1l−∕− mice
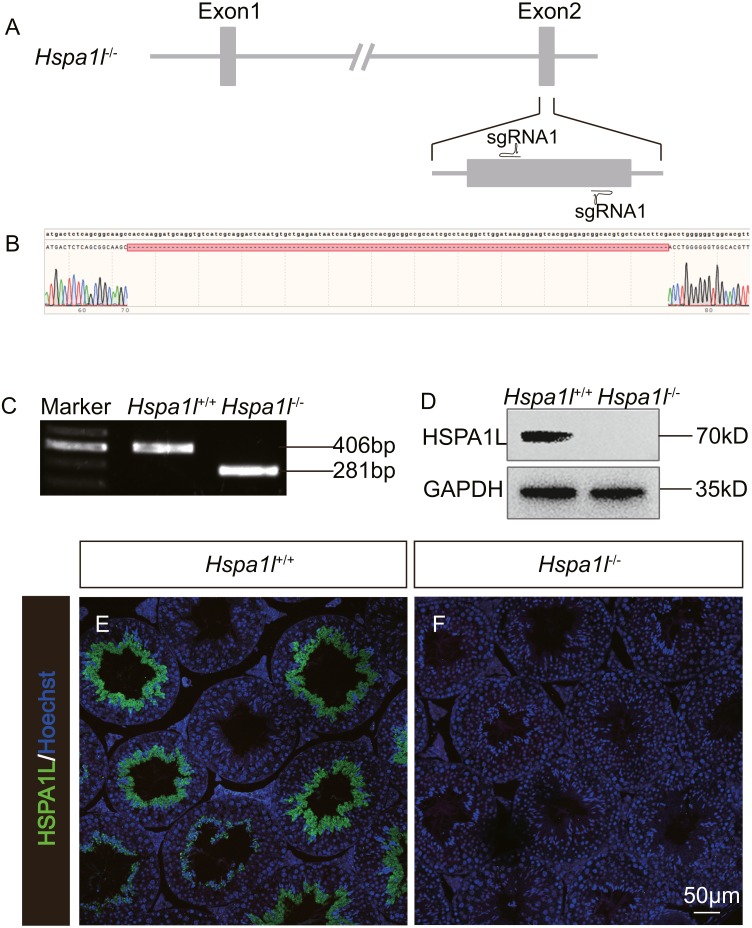
In order to investigate the function of Hspa1l in testis, we generated a Hspa1l mutant mouse model using the CRISPR/Cas9 system. We created a frameshift by deleting 125 base pairs (bp) of exon 2 of Hspa1l (Fig. 2A). The resultant deletion was confirmed by PCR and Sanger sequencing (Figs. 2B and 2C). Western blot and immunofluorescence analyses showed that HSPA1L was undetectable in Hspa1l−∕− homozygote testis (Figs. 2D–2F). All Hspa1l −∕− mice showed normal development and were viable.
Figure 2. Generation of Hspa1l−∕− mice.
(A) Schematic representation of targeting strategy using CRISPR/Cas9 system; (B) Sanger sequencing shows a 125-bp deletion were detected in Hspa1l−∕− mice; (C) Agarose gel electrophoresis analysis proved the 125-bp deletion in Hspa1l−∕− mice; (D) Western blot showing no band at expected size (70 kD) in Hspa1l−∕− testis; (E–F) HSPA1L expression from Hspa1l+∕+ and Hspa1l−∕− adult mice by immunofluorescence.
Hspa1l−∕− mice are fertile with normal spermatogenesis
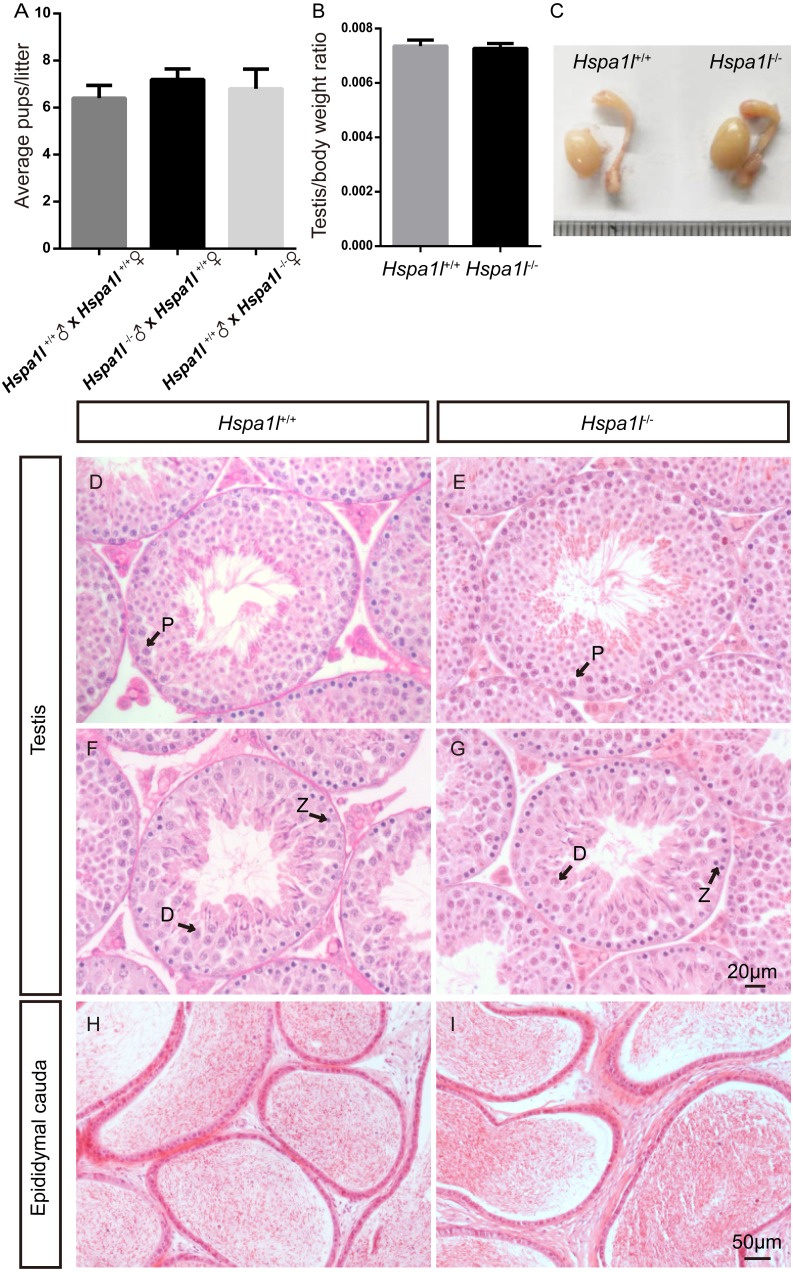
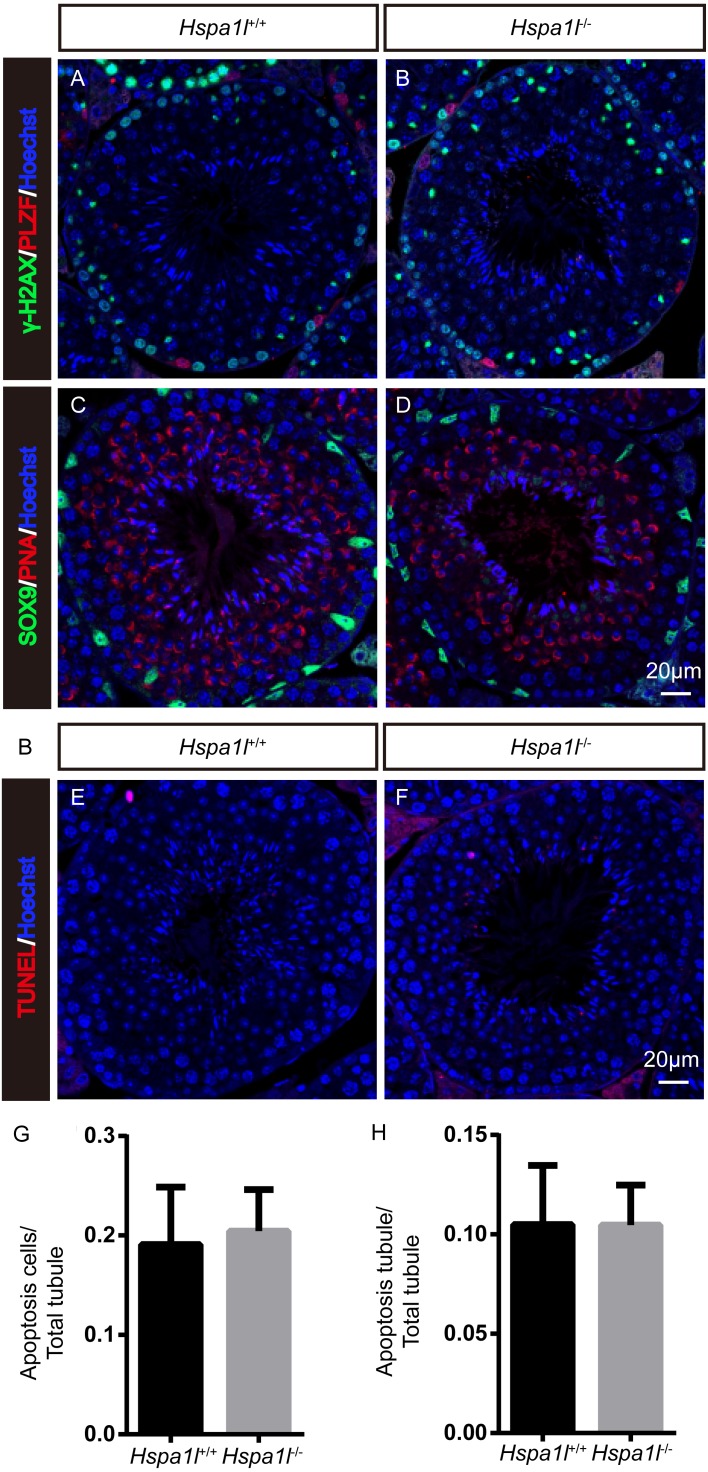
Unlike the Hspa2 mutant mice (Dix et al., 1996), Hspa1l−∕− males were fertile (n = 5, P = 0.344) (Fig. 3A). Intercrossed Hspa1l+∕− mice offspring show normal litter size and sex ratios under the expected Mendelian distribution. There was no difference in size of testes (n = 5, P = 0.262) and epididymides between Hspa1l−∕− and Hspa1l+∕+ male mice (Figs. 3B and 3C). In addition, normal spermatogenic cells of all stages were observed in the seminiferous tubules of adult Hspa1l−∕− mice by H&E staining (Figs. 3D–3I). Immunofluorescence analysis showed the presence of normal PLZF-positive spermatogonia, γ-H2AX-positive spermatocytes, and PNA-positive acrosomes in spermatids (Figs. 4A–4D). TUNEL analysis of testicular sections showed that the number of apoptotic cells per tubule (n = 5, P = 0.687) and the ratio of apoptotic tubules (n = 5, P = 0.986) were similar between Hspa1l−∕− and Hspa1l+∕+ control testes (Figs. 4E–4H).
Figure 3. Hspa1l−∕− mice showed normal spermatogenesis.
(A) Fertility tests show average pups per litter of Hspa1l+∕+ and Hspa1l−∕− mice, n = 5. (B) Average testis weight/body weight of Hspa1l+∕+ and Hspa1l−∕− male mice, n = 5; (C) Morphology of Hspa1l+∕+ and Hspa1l−∕− adult mice in testis and epididymis; (D–I) H&E-stained sections of testes and epididymal cauda from Hspa1l+∕+ and Hspa1l−∕− mice; P, pachytene; Z, zygotene; D, diplotene.
Figure 4. Immunofluorescence and apoptosis analysis of Hspa1l+∕+ and Hspa1l−∕− testes.
(A–D) The PLZF, γ-H2AX, PNA and Sox9 signals shows the locations of spermatogonia, spermatocytes, spermatids and Sertoli cells in testis sections from both Hspa1l+∕+ and Hspa1l−∕− mice, respectively. (E–F) Testicular sections of TUNEL assay in Hspa1l+∕+ and Hspa1l−∕− mice; (G) Average apoptotic cells per seminiferous tubule in Hspa1l+∕+ and Hspa1l−∕− testicular sections; (H) Average apoptotic cells per seminiferous tubules in Hspa1l+∕+ and Hspa1l−∕− testicular sections, n = 5.
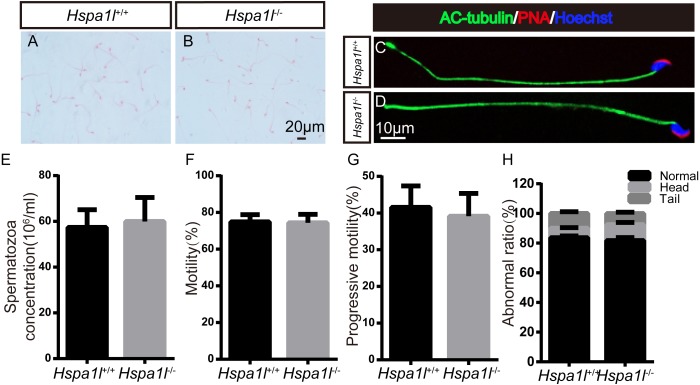
Spermatozoa are normal in Hspa1l−∕− mice
There was no significant difference between the whole epididymal sperm count (n = 5, P = 0.669) and the morphology of spermatozoa from the epididymal cauda between Hspa1l−∕− and Hspa1l+∕+ mice (Figs. 5A–5E). The sperm motility (n = 5, P = 0.842) and progressive motility (n = 5, P = 0.546) in Hspa1l−∕− mice were comparable to the Hspa1l+∕+ controls. Morphological analysis also showed the ratio of normal sperm (n = 5, P = 0.445) were unaffected in Hspa1l−∕− male mice (Figs. 5F–5H). These results show that the deletion of Hspa1l does not affect spermatogenesis and fertility in Hspa1l−∕− mice.
Figure 5. Spermatozoa appear normal in Hspa1l−∕− mice.
(A–B) H&E-stained spermatozoa from Hspa1l +∕+ and Hspa1l−∕− mice; (C–D) AC-tubulin, PNA signals from Hspa1l+∕+ and Hspa1l−∕− spermatozoa by immunofluorescence; (E) Sperm concentration of the cauda epididymal from Hspa1l+∕+ and Hspa1l−∕− mice, n = 5; (F) Average rate of motile sperm and (G) progressive sperm from Hspa1l+∕+ and Hspa1l−∕− mice, n = 5; (H) Abnormal epididymal sperm count from Hspa1l+∕+ and Hspa1l−∕− mice, n = 5.
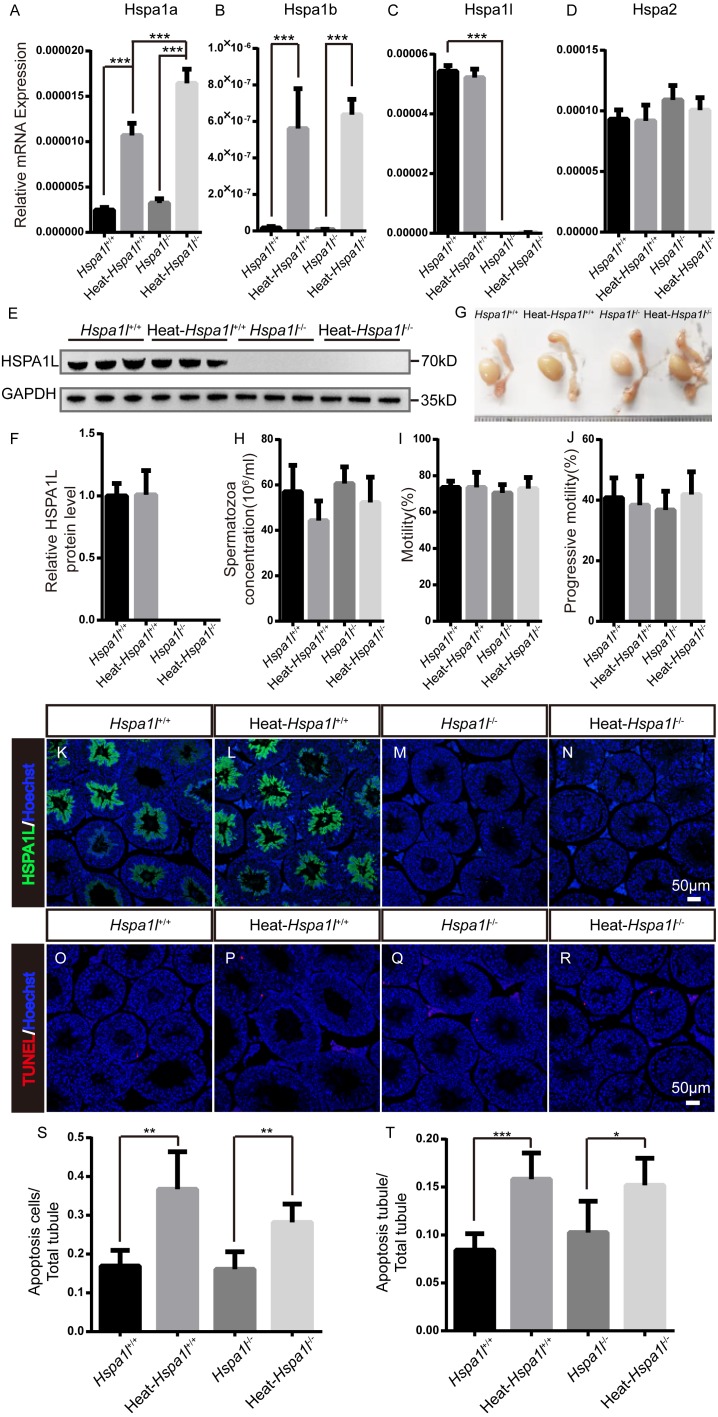
Hspa1l is not essential in responses to heat stress
HSP70s are strong anti-apoptotic proteins which can prevent apoptosis from different stresses, such as heat shock (Durairajanayagam, Agarwal & Ong, 2015), oxidative stress (Ikwegbue et al., 2017), infection (Mays et al., 2019), etc. We applied heat stress to test the anti-apoptotic ability of HSPA1L in testis. We found that Hspa1a and Hspa1b mRNA levels increased after heat treatment, while those of Hspa1l and Hspa2 were not affected (Figs. 6A–6D). We measured the expression of other Hsp70s, and found that Hspa5 is down-regulated after heat stress. However, Hspa9 and Hspa12b were downregulated only in heat treated Hspa1l−∕− testes (Fig. S2). Further analysis of protein expression levels showed that HSPA1L remained unaffected under heat stress (n = 5, P = 0.925), consistent with its RNA levels (Figs. 6E–6F and Fig. S3). Furthermore, compared to the Hspa1l+∕+ control, there was no significant differences in testes and epididymides size (Fig. 6G), sperm count (n = 5, P = 0.091), sperm motility (n = 5, P = 0.809) and progressive motility (n = 5, P = 0.708) with or without heat stress in Hspa1l−∕− (Figs. 6H–6J). TUNEL analysis of testicular sections showed no difference between the Hspa1l−∕− and control testes after heat stress for both the number of apoptotic cells (n = 5, P = 0.110) and the number of apoptotic tubules (n = 5, P = 0.312). Hspa1l−∕− testis responded to heat stress in a way comparable to controls (Figs. 6K–6T). Thus, Hspa1l doesn’t play an important role during spermatogenesis or in the anti-apoptotic response to stress, as suggested in the earlier reports.
Figure 6. Analysis of testis and sperm in Hspa1l−∕− mice after heat stress.
(A–D) Expression of Hspa1a, Hspa1b, Hspa1l and Hspa2 in Hspa1l+∕+ and Hspa1l−∕− mice testis with or without heat treatment, n = 5, *** P < 0.001; (E) HSPA1L expression in Hspa1l+∕+ and Hspa1l−∕− mice testis with or without heat treatment by western blot and (F) quantitated by densitometric analysis, n = 5; (G) Testes and epididymides from Hspa1l+∕+ and Hspa1l−∕− adult mice with or without heat treatment; (H) Cauda epididymal sperm concentration from Hspa1l+∕+ and Hspa1l−∕− mice with or without heat treatment, n = 5; (I) Average sperm motility and (J) progressive motility from Hspa1l+∕+ and Hspa1l−∕− mice with or without heat treatment, n = 5; (K–N) HSPA1L staining in Hspa1l +∕+ and Hspa1l−∕− mice testis with or without heat treatment by immunofluorescence; (O–R) TUNEL assay of Hspa1l+∕+ and Hspa1l −∕− testes with or without heat treatment; (S) Average apoptotic cells per seminiferous tubule in testicular sections, n = 5, ** P < 0.01; (T) Average apoptotic cells per seminiferous tubules in testicular sections, n = 5. * P < 0.05, *** P < 0.001.
Discussion
In this study, we examined the possible function of Hspa1l, a member of Hsp70 family of proteins, with testis-predominant expression. Hspa1l showed developmental stage-specific expression pattern during testis development. It typically starts to express from step 12 spermatids of stage I spermatogenesis. Previously, it was thought to have important functions in spermatogenesis and was therefore associated with an increased risk of idiopathic male infertility (Kohan, Tabiee & Sepahi, 2019). However, our studies demonstrate that Hspa1l−∕− Hspa1l−∕− mice have normal spermatogenesis with appearance of all types and stages of spermatogenic cells. Both male and female Hspa1l−∕− mice were fertile without any apparent developmental abnormalities.
HSPA1A/B, two members of HSP70 family from the same cluster as HSPA1L, are normally located in cytosol, and translocate to the nucleus rapidly under emergency circumstances, such as heat stress, to protect the chromatin (Lindquist & Craig, 1988). HSPA1L is highly expressed in testis, and located in cytosol as well. Testis is susceptible to hyperthermia, which could damage spermatogenesis and lead to male infertility (Durairajanayagam, Agarwal & Ong, 2015). We found that while Hspa1l is a testis-predominant gene, it is not essential to spermatogenesis. Therefore, we investigated whether it might play an important role in heat stress-induced responses. However, unlike HSPA1A/B, HSPA1L did not translocate to the nucleus after heat stress, indicating that although these three proteins are highly analogous, the function of HSPA1L may be very different from HSPA1A/B.
Cells enhance their expression of stress- inducible HSP70s to respond to environmental stresses (Chen, Feder & Kang, 2018a; Ikwegbue et al., 2017; Radons, 2016; Singh et al., 2010). HSPA1L was thought to have the ability to enhance cell survival in testes following multiple stresses, such as a heat stress. By that reasoning, the Hspa1l−∕− testis may be more sensitive to heat stress. Also, the expression of HSPA1L is focused in spermatids, so it was reasonable to speculate that heat treated Hspa1l−∕− testis might show an increase of apoptotic spermatids. However, TUNEL analysis of Hspa1l−∕− testicular sections showed that no increase in the number of apoptotic cells compared to the controls after the heat treatment. Thus, the deletion of Hspa1l does not affect the germ cells’ response to heat stress.
It has been reported that Hspa1l mutants have increased risk of male infertility (Huusko et al., 2018; Takahashi et al., 2017), but our studies show that there was no difference in the fertility of Hspa1l−∕− mutant mice compared to Hspa1l+∕+ controls. Analysis of the expression levels of other Hsp70s showed no up-regulation in Hspa1l−∕− testes. However, the expression levels of Hspa1a and Hspa1b increased in Hspa1l−∕− testes compared with Hspa1l+∕+ testis after heat treatment, which implied that Hspa1a and Hspa1b possibly compensated for Hspa1l function under heat stress. Interestingly, we found that Hspa5 was downregulated in Hspa1l−∕− testes, and Hspa9 and Hspa12b were downregulated in Hspa1l −∕− testes after heat stress compared to Hspa1l+∕+. The expression of these Hsp70 family members were affected by Hspa1l expression, the biological significance of which needs to be examined further.
As we investigated that heat stress does not affect cells survival in Hspa1l−∕− mutants, the affects from other stresses have not yet been studied. Especially, the study of inflammation in Hspa1l−∕− mutants because Hspa1l and Hspa1a/b all fall under the MHC class III family. Testis is a tissue with immune privilege thanks to the blood-testis barrier (BTB) (Fijak, Bhushan & Meinhardt, 2011). Considering the different expression pattern of Hspa1l from Hspa1a/b, it indicated that Hspa1l may has a crucial role in testes when encounter inflammation, which needs further investigation.
Conclusions
Hspa1l is highly and constitutively expressed in testis. However, Hspa1l−∕− mice are fertile, and showed no abnormality in spermatogenesis. Further, heat stress does not exacerbate the germ cells’ apoptosis in Hspa1l−∕− testes. This suggests that there is no essential function of Hspa1l in response to heat stress, possibly compensated by other members of the family, Hspa1a and Hspa1b. The in vivo functional studies of HSPA1L could provide a basis for further elucidation of functions of the highly conserved HSP70s family.
Supplemental Information
n = 5, * P < 0.05, *** P < 0.001.
Acknowledgments
We thank Jinyang Cai for continuous support with microscopy.
Funding Statement
This work was supported by the National Key R&D Program (2017YFA0103803, 2016YFA0500903, 2016YFA0503300), the National Natural Science Foundation of China (31571538, 31771651, 31890784 and 31701300), and Natural Science Foundation of Jiangsu Province (Grants No. BK20190081). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Zuomin Zhou, Email: zhouzm@njmu.edu.cn.
Mingxi Liu, Email: mingxi.liu@njmu.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Xin Wang performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Wenxiu Xie, Yejin Yao, Yunfei Zhu, Jianli Zhou and Yiqiang Cui performed the experiments, prepared figures and/or tables, and approved the final draft.
Xuejiang Guo and Yan Yuan analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Zuomin Zhou and Mingxi Liu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The mice use and treated agreement were checked and authorized under the Animal Ethical and Welfare Committee (Approval No. IACUC-1601117). Protocols for mice feeding and using are under the guiding principles of the Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University.
Data Availability
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.
References
- Barratt et al. (2017).Barratt CLR, Bjorndahl L, De Jonge CJ, Lamb DJ, Osorio Martini F, McLachlan R, Oates RD, Van der Poel S, St John B, Sigman M, Sokol R, Tournaye H. The diagnosis of male infertility: an analysis of the evidence to support the development of global WHO guidance-challenges and future research opportunities. Human Reproduction Update. 2017;23:660–680. doi: 10.1093/humupd/dmx021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Feder & Kang (2018a).Chen B, Feder ME, Kang L. Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Molecular Ecology. 2018a;27:3040–3054. doi: 10.1111/mec.14769. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2018b).Chen Y, Zheng Y, Gao Y, Lin Z, Yang S, Wang T, Wang Q, Xie N, Hua R, Liu M, Sha J, Griswold MD, Li J, Tang F, Tong MH. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Research. 2018b;28:879–896. doi: 10.1038/s41422-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci et al. (2015).Ciftci H, Celepkolo B, Dilmec F, Koksal M, Yeni E, Yagmur I, Gumus K. Genetic polymorphisms of hspa1b and hspa1l in infertile men. Journal of Pakistan Medical Association. 2015;65:701–704. [PubMed] [Google Scholar]
- Daugaard, Rohde & Jaattela (2007).Daugaard M, Rohde M, Jaattela M. The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Letters. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Dix et al. (1996).Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, Goulding EH, Eddy EM. Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3264–3268. doi: 10.1073/pnas.93.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, Garges & Hong (1998).Dix DJ, Garges JB, Hong RL. Inhibition of hsp70-1 and hsp70-3 expression disrupts preimplantation embryogenesis and heightens embryo sensitivity to arsenic. Molecular Reproduction and Development. 1998;51:373–380. doi: 10.1002/(SICI)1098-2795(199812)51:4<373::AID-MRD3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Durairajanayagam, Agarwal & Ong (2015).Durairajanayagam D, Agarwal A, Ong C. Causes, effects and molecular mechanisms of testicular heat stress. Reproductive BioMedicine Online. 2015;30:14–27. doi: 10.1016/j.rbmo.2014.09.018. [DOI] [PubMed] [Google Scholar]
- Erata et al. (2008).Erata GO, Kocak Toker N, Durlanik O, Kadioglu A, Aktan G, Aykac Toker G. The role of heat shock protein 70 (Hsp 70) in male infertility: is it a line of defense against sperm DNA fragmentation? Fertility and Sterility. 2008;90:322–327. doi: 10.1016/j.fertnstert.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Fainberg & Kashanian (2019).Fainberg J, Kashanian JA. Recent advances in understanding and managing male infertility. F1000Research. 2019;8(F1000 Faculty Rev):670. doi: 10.12688/f1000research.17076.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang et al. (2016).Fang CT, Kuo HH, Pan TS, Yu FC, Yih LH. HSP70 regulates the function of mitotic centrosomes. Cellular and Molecular Life Science. 2016;73:3949–3960. doi: 10.1007/s00018-016-2236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayomi & Orwig (2018).Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Research. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Fernandez & Valpuesta (2018).Fernandez-Fernandez MR, Valpuesta JM. Hsp70 chaperone: a master player in protein homeostasis. F1000Research. 2018;7(F1000 Faculty Rev):1497. doi: 10.12688/f1000research.15528.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijak, Bhushan & Meinhardt (2011).Fijak M, Bhushan S, Meinhardt A. Immunoprivileged sites: the testis. Methods in Molecular Biology. 2011;677:459–470. doi: 10.1007/978-1-60761-869-0_29. [DOI] [PubMed] [Google Scholar]
- Gatimel et al. (2017).Gatimel N, Moreau J, Parinaud J, Leandri RD. Sperm morphology: assessment, pathophysiology, clinical relevance, and state of the art in 2017. Andrology. 2017;5:845–862. doi: 10.1111/andr.12389. [DOI] [PubMed] [Google Scholar]
- Hermo et al. (2010).Hermo L, Pelletier RM, Cyr DG, Smith CE. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 1: background to spermatogenesis, spermatogonia, and spermatocytes. Microscopy Research and Technique. 2010;73:241–278. doi: 10.1002/jemt.20783. [DOI] [PubMed] [Google Scholar]
- Huang, Mivechi & Moskophidis (2001).Huang L, Mivechi NF, Moskophidis D. Insights into regulation and function of the major stress-induced hsp70 molecular chaperone in vivo: analysis of mice with targeted gene disruption of the hsp70.1 or hsp70.3 gene. Molecular and Cellular Biology. 2001;21:8575–8591. doi: 10.1128/mcb.21.24.8575-8591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2008).Huang XY, Guo XJ, Shen J, Wang YF, Chen L, Xie J, Wang NL, Wang FQ, Zhao C, Huo R, Lin M, Wang X, Zhou ZM, Sha JH. Construction of a proteome profile and functional analysis of the proteins involved in the initiation of mouse spermatogenesis. Journal of Proteome Research. 2008;7:3435–3446. doi: 10.1021/pr800179h. [DOI] [PubMed] [Google Scholar]
- Huusko et al. (2018).Huusko JM, Karjalainen MK, Graham BE, Zhang G, Farrow EG, Miller NA, Jacobsson B, Eidem HR, Murray JC, Bedell B, Breheny P, Brown NW, Bodker FL, Litterman NK, Jiang PP, Russell L, Hinds DA, Hu Y, Rokas A, Teramo K, Christensen K, Williams SM, Ramet M, Kingsmore SF, Ryckman KK, Hallman M, Muglia LJ. Whole exome sequencing reveals HSPA1L as a genetic risk factor for spontaneous preterm birth. PLOS Genetics. 2018;14:e1007394. doi: 10.1371/journal.pgen.1007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikwegbue et al. (2017).Ikwegbue PC, Masamba P, Oyinloye BE, Kappo AP. Roles of heat shock proteins in apoptosis, oxidative stress, human inflammatory diseases, and cancer. Pharmaceuticals. 2017;11(1):2. doi: 10.3390/ph11010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai et al. (2006).Kawai Y, Hata T, Suzuki O, Matsuda J. The relationship between sperm morphology and in vitro fertilization ability in mice. Journal of Reproduction and Development. 2006;52:561–568. doi: 10.1262/jrd.18023. [DOI] [PubMed] [Google Scholar]
- Kesari, Agarwal & Henkel (2018).Kesari KK, Agarwal A, Henkel R. Radiations and male fertility. Reproductive Biology and Endocrinology. 2018;16:118. doi: 10.1186/s12958-018-0431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohan, Tabiee & Sepahi (2019).Kohan L, Tabiee O, Sepahi N. HSPA1L and HSPA1B gene polymorphisms and haplotypes are associated with idiopathic male infertility in Iranian population. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2019;240:57–61. doi: 10.1016/j.ejogrb.2019.06.014. [DOI] [PubMed] [Google Scholar]
- Lee et al. (2001).Lee SH, Kim M, Yoon BW, Kim YJ, Ma SJ, Roh JK, Lee JS, Seo JS. Targeted hsp70.1 disruption increases infarction volume after focal cerebral ischemia in mice. Stroke. 2001;32:2905–2912. doi: 10.1161/hs1201.099604. [DOI] [PubMed] [Google Scholar]
- Lindquist & Craig (1988).Lindquist S, Craig EA. The heat-shock proteins. Annual Review of Genetics. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Mayer (2013).Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends in Biochemical Sciences. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Mayer & Bukau (2005).Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cellular and Molecular Life Science. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays et al. (2019).Mays CE, Armijo E, Morales R, Kramm C, Flores A, Tiwari A, Bian J, Telling GC, Pandita TK, Hunt CR, Soto C. Prion disease is accelerated in mice lacking stress-induced heat shock protein 70 (HSP70) Journal of Biological Chemistry. 2019;294:13619–13628. doi: 10.1074/jbc.RA118.006186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy (2013).Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013;34:1181–1188. doi: 10.1093/carcin/bgt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto et al. (2016).Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Spermatogenesis in humans and its affecting factors. Seminars in Cell & Developmental Biology. 2016;59:10–26. doi: 10.1016/j.semcdb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Nixon et al. (2015).Nixon B, Bromfield EG, Dun MD, Redgrove KA, McLaughlin EA, Aitken RJ. The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian Journal of Andrology. 2015;17:568–573. doi: 10.4103/1008-682x.151395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence et al. (2019).Pence LM, Schmitt TC, Beger RD, Del Valle PL, Nakamura N. Testicular function in cultured postnatal mouse testis fragments is similar to that of animals during the first wave of spermatogenesis. Birth Defects Research. 2019;111:270–280. doi: 10.1002/bdr2.1451. [DOI] [PubMed] [Google Scholar]
- Perez et al. (1991).Perez N, Sugar J, Charya S, Johnson G, Merril C, Bierer L, Perl D, Haroutunian V, Wallace W. Increased synthesis and accumulation of heat shock 70 proteins in Alzheimer’s disease. Brain Research. Molecular Brain Research. 1991;11:249–254. doi: 10.1016/0169-328x(91)90033-t. [DOI] [PubMed] [Google Scholar]
- Radons (2016).Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone et al. (2018).Sansone A, Di Dato C, De Angelis C, Menafra D, Pozza C, Pivonello R, Isidori A, Gianfrilli D. Smoke, alcohol and drug addiction and male fertility. Reproductive Biology and Endocrinology. 2018;16:3. doi: 10.1186/s12958-018-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signore et al. (2019).Signore F, Gulia C, Votino R, De Leo V, Zaami S, Putignani L, Gigli S, Santini E, Bertacca L, Porrello A, Piergentili R. The role of number of copies, structure, behavior and copy number variations (CNV) of the Y chromosome in male infertility. Gene. 2019;11(1):40. doi: 10.3390/genes11010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh et al. (2010).Singh R, Kolvraa S, Bross P, Christensen K, Bathum L, Gregersen N, Tan Q, Rattan SI. Anti-inflammatory heat shock protein 70 genes are positively associated with human survival. Current Pharmaceutical Design. 2010;16:796–801. doi: 10.2174/138161210790883499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skakkebaek et al. (2016).Skakkebaek NE, Rajpert-De Meyts E, Buck Louis GM, Toppari J, Andersson AM, Eisenberg ML, Jensen TK, Jorgensen N, Swan SH, Sapra KJ, Ziebe S, Priskorn L, Juul A. Male reproductive disorders and fertility trends: influences of environment and genetic susceptibility. Physiological Reviews. 2016;96:55–97. doi: 10.1152/physrev.00017.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi et al. (2017).Takahashi S, Andreoletti G, Chen R, Munehira Y, Batra A, Afzal NA, Beattie RM, Bernstein JA, Ennis S, Snyder M. De novo and rare mutations in the HSPA1L heat shock gene associated with inflammatory bowel disease. Genome Medicine. 2017;9:8. doi: 10.1186/s13073-016-0394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa et al. (1999).Tsunekawa N, Matsumoto M, Tone S, Nishida T, Fujimoto H. The Hsp70 homolog gene, Hsc70t, is expressed under translational control during mouse spermiogenesis. Molecular Reproduction and Development. 1999;52:383–391. doi: 10.1002/(SICI)1098-2795(199904)52:4<383::AID-MRD7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Turturici, Sconzo & Geraci (2011).Turturici G, Sconzo G, Geraci F. Hsp70 and its molecular role in nervous system diseases. Biochemistry Research International. 2011;2011:618127. doi: 10.1155/2011/618127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos et al. (2008).Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry. 2008;47:7001–7011. doi: 10.1021/bi800639z. [DOI] [PubMed] [Google Scholar]
- Wen et al. (2020).Wen L, Liu Q, Xu J, Liu X, Shi C, Yang Z, Zhang Y, Xu H, Liu J, Yang H, Huang H, Qiao J, Tang F, Chen ZJ. Recent advances in mammalian reproductive biology. Science China Life Sciences. 2020;63:18–58. doi: 10.1007/s11427-019-1572-7. [DOI] [PubMed] [Google Scholar]
- Wisniewska et al. (2010).Wisniewska M, Karlberg T, Lehtio L, Johansson I, Kotenyova T, Moche M, Schuler H. Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B’, and HSPA5/BiP/GRP78. PLOS ONE. 2010;5:e8625. doi: 10.1371/journal.pone.0008625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashima (2012).Yamashima T. Hsp70.1 and related lysosomal factors for necrotic neuronal death. Journal of Neurochemistry. 2012;120:477–494. doi: 10.1111/j.1471-4159.2011.07596.x. [DOI] [PubMed] [Google Scholar]
- Zhu, Dix & Eddy (1997).Zhu D, Dix DJ, Eddy EM. HSP70-2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development. 1997;124:3007–3014. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- Zuiderweg, Hightower & Gestwicki (2017).Zuiderweg ER, Hightower LE, Gestwicki JE. The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones. 2017;22:173–189. doi: 10.1007/s12192-017-0776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
n = 5, * P < 0.05, *** P < 0.001.
Data Availability Statement
The following information was supplied regarding data availability:
The raw measurements are available in the Supplementary Files.