Abstract
Small RNAs comprise three families of noncoding regulatory RNAs that control gene expression by blocking mRNA translation or leading to mRNA cleavage. Such post‐transcriptional negative regulation is relevant for both plant development and environmental adaptations. An important biotechnological application of miRNA identification is the discovery of regulators and effectors of cell wall degradation, which can improve/facilitate hydrolysis of cell wall polymers for second‐generation bioethanol production. The recent characterization of plant innate cell wall modifications occurring during root aerenchyma development triggered by ethylene led to the possibility of prospection for mechanisms of cell wall disassembly in sugarcane. By using next‐generation sequencing, 39 miRNAs were identified in root segments along the process of aerenchyma development. Among them, 31 miRNAs were unknown to the sugarcane miRBase repository but previously identified as produced by its relative Sorghum bicolor. Key putative targets related to signal transduction, carbohydrate metabolic process, and cell wall organization or biogenesis were among the most representative gene categories targeted by miRNA. They belong to the subclasses of genes associated with the four modules of cell wall modification in sugarcane roots: cell expansion, cell separation, hemicellulose, and cellulose hydrolysis. Thirteen miRNAs possibly related to ethylene perception and signaling were also identified. Our findings suggest that miRNAs may be involved in the regulation of cell wall degradation during aerenchyma formation. This work also points out to potential molecular tools for sugarcane improvement in the context of second‐generation biofuels.
Keywords: aerenchyma, bioenergy, cell wall, miRNA, sugarcane
1. INTRODUCTION
miRNAs are a class of relevant plant development regulators (Borges & Martienssen, 2015; Chen, 2004; Guleria & Yadav, 2011). Post‐transcriptional regulation is achieved through sequence‐specific base‐pairing of 20 to 22 nucleotides of a small RNA and a mRNA. Dicer‐like proteins are plant ribonucleases responsible for processing primary miRNAs leading to its mature functional conformation. Within the cytoplasm, the active guide strand (i.e., the one that effectively binds to the target mRNA) is then separated from its complementary strand (named miRNA* or star miRNA). miRNA* is degraded and mature miRNA is loaded onto the argonaute protein, giving rise to the RNA‐induced silencing complex (RISC). This protein complex leads the miRNA guide strand to target mRNAs due to base complementarity within its 3’ UTR sequence (Li & Zhang, 2016). The mRNA can be then targeted for degradation, or its translational process can be blocked (Borges & Martienssen, 2015).
The wide range of pathways regulated by miRNA within cellular metabolism has led to the assumption that plant miRNA can serve as important tools for plant improvement (Zhang, 2015). miRNA identification involved in plant tolerance to environmental stresses and carbohydrate metabolism can be especially relevant for crops, since increased biomass, higher digestibility, and stress tolerance are desirable traits, especially for biofuel industries. Some of these crops are Panicum virgatum, Sorghum bicolor, Zea mays, and Saccharum sp. For instance, P. virgatum plants overexpressing Z. mays miR156 displayed distinct biomass composition and presented higher saccharification yields due to increased starch content (Chuck et al., 2011; Fu et al., 2012). Data mining for miRNA regulation among cell wall‐related transcripts in Sorghum bicolor has led to the identification of 10 putative targets, including laccases and cellulose synthases (Rai et al., 2016). However, the underlying miRNA regulation of cell wall metabolism in sugarcane lacks experimental evidence.
Four miRNA experimental procedures have already been performed with sugarcane miRNA using leaves (Ferreira et al., 2012; Thiebaut et al., 2014; Thiebaut, Rojas, et al., 2012; Zanca et al., 2010), buds (Ortiz‐Morea et al., 2013; Zanca et al., 2010), roots (Thiebaut et al., 2014; Thiebaut, Rojas, et al., 2012), and whole plant (Thiebaut, Rojas, et al., 2012), and a sugarcane EST database (Vettore, Silva, Kemper, & Arruda, 2001) was also subjected to miRNA identification (Ortiz‐Morea et al., 2013). Surprisingly, only 19 miRNA precursors and 20 mature sugarcane sequences are currently available in the miRBase repository v.21, which represents less than 1% of monocot miRNAs (release April 21, 2017; http://www.mirbase.org/index.shtml). It inevitably underestimates sugarcane miRNA diversity, and it also represents the need for further investigation of miRNA function and identification. Sugarcane features directly related to bioenergy, such as cell wall composition and modifications, lack miRNA regulation studies.
Sugarcane is an essential crop in Brazil due to its widespread use in sugar and ethanol production. Also, further expansion of production is possible without need to use land presently occupied by preserved biomes or food production as projected to 2045 (Jaiswal et al., 2017). Sugar is obtained through culm milling followed by sucrose extraction, whereas ethanol results from sugar fermentation. Fermentation can occur using two different substrates, which defines first‐generation (1G) and second‐generation (2G) ethanol. The fermentation of the sucrose stored within sugarcane culm has supported Brazil as the second larger 1G producer in the world. Although available commercially, 2G ethanol production still needs further improvements, since it requires more effective depolymerization of structural polysaccharides of the cell wall (Buckeridge & De Souza, 2017).
Recently, the characterization of aerenchyma formation within sugarcane roots uncovered the potential use of innate pathways for cell wall deconstruction, facilitating the access to cell wall polymers and paving the way toward successful 2G ethanol production (Leite et al., 2017; Tavares, Souza, & Buckeridge, 2015). Lysigenous aerenchyma is a developmental process activated by the hormone ethylene. Its action leads to the opening of gas spaces within parenchymatic tissues due to programmed cell death and cell wall modifications (Takahashi, Yamauchi, Colmer, & Nakazono, 2014; Tavares et al., 2018; Yamauchi, Colmer, Pedersen, & Nakazono, 2018; Yamauchi, Shimamura, Nakazono, & Mochizukic, 2013). Immunolocalization showed arabinoxylan debranching, homogalacturonan hydrolysis from the middle lamella, and β‐glucan mobilization during the formation of aerenchyma in sugarcane roots (Leite et al., 2017). The negative regulation played upon homogalacturonan hydrolysis by an ethylene response factor, RAV1, represents an important trigger to cell wall attack (Rahji et al., 2011; Tavares et al., 2019). Thus, aerenchyma formation seems to rely on cell targeting induced by ethylene and auxin balance. This is followed by cell expansion and separation, programmed cell death, and hemicellulose and cellulose hydrolysis. Each step configures a conserved set of pathways—named “modules”—shared between other endogenous cell wall degradation events (Grandis, Souza, Tavares, & Buckeridge, 2014; Tavares et al., 2015). Although cell wall changes are subtle and sugarcane pectin content is rather low (De Souza, Leite, Pattathil, Hahn, & Buckeridge, 2013), the effects of partial pectin degradation within plant tissues, especially in the middle lamella, may be quite relevant for saccharification and bioenergy production (Latarullo, Tavares, Maldonado, Leite, & Buckeridge, 2016). During aerenchyma formation in sugarcane roots, pectin degradation is thought to be a result of the attack by acetyl esterases, endopolygalacturonases, β‐galactosidases, and α‐arabinofuranosidases, followed by the action of β‐glucan‐/callose‐hydrolyzing enzymes (Grandis et al., 2019; Tavares et al., 2018, 2019). Concomitantly, there are modifications in arabinoxylan (by α‐arabinofuranosidases), xyloglucan (by xyloglucan endotransglucosylase/hydrolase), xyloglucan–cellulose interactions (by expansins), and partial hydrolysis of cellulose (Grandis et al., 2019). Thus, the precise control underlying this process might be a key factor to understand modulation of cell wall changes in sugarcane. One question is to what extent epigenetics might be involved in the control of aerenchyma formation in sugarcane and other grasses.
In this study, we identified 39 expressed miRNAs within sugarcane roots undergoing aerenchyma formation. Transcripts related to carbohydrate metabolic process and cell wall organization or biogenesis were predicted to be targeted by the sequenced miRNA. Among those, miRNAs expressed during aerenchyma formation and presumably target transcripts related to several cell wall polysaccharides previously detected in sugarcane (Leite et al., 2017). A more significant fraction of these transcripts is related to pectin degradation. The diversity of miRNA targeting pectin‐related and ethylene regulation‐related transcripts corroborates previous data showing decreasing cellular adhesion and hormone signaling as fundamental mechanisms during the onset of aerenchyma formation (Grandis et al., 2019; Leite et al., 2017; Tavares et al., 2018, 2019). Our results represent an important step toward the identification of miRNAs involved in sugarcane cell wall degradation, as well as its hormonal control. It also paves the way for the development of biotechnology in the biofuel field.
2. MATERIALS AND METHODS
2.1. Plant samples and RNA extraction
All experiments employed sugarcane (Saccharum sp.) variety SP80‐3280 grown in Piracicaba, São Paulo, Brazil. Lateral buds from harvested culms were planted in vermiculite and grown for 4 months supplied with 40 g NPK (30:20:30) fertilizer. Plants were watered weekly with 100 ml of water and grown under natural environmental conditions including periodic rain and natural South Hemisphere summer climate fluctuations (from December 2013 until March 2014). Thirty plants were pooled in three biological replicates, with ten plants each. The first 4 cm of tiller roots was collected and separated into 4 one‐centimeter sections from the apex containing the meristem (S1) toward the base (S4) (Leite et al., 2017) (Figure 1).
Figure 1.
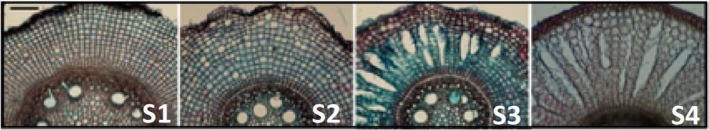
Aerenchyma formation in sugarcane roots from apex to 4 cm divided into 1 cm segments (S1–S4). Sections were stained with 1% safrablau (Bukatsch, 1972). Scale bar = 20 µm
Total RNA extraction proceeded with 100 mg of each replicate further homogenized using mirVana™ miRNA Isolation Kit (Thermo Fisher Scientific®), according to manufacturer's instructions. The same set of samples was used for qRT‐PCR validation, according to Tavares et al. (2019).
2.2. miRNA sequencing
RNAs shorter than 200 nt were first enriched by Magnetic Bead Purification Module (Thermo Fisher Scientific®) and next used on Ion Total RNA‐Seq Kit v2 (Thermo Fisher Scientific®), according to manufacturer's instructions. Samples were sequenced on Ion Proton™ Sequencer, with 500 flows for 200‐base read sequencing. Pools of three bar‐coded sample libraries were loaded onto each Ion PI sequencing chip v2.
2.3. Bioinformatic analyses
Sequence quality was first evaluated by using FastQC (Andrews, 2010) with default settings. Ribosome, transfer, small nuclear, small nucleolar, long no‐coding, and trans‐acting small interfering RNAs (rRNA, tRNA, snRNA, snoRNA, lncRNA, and tasiRNA) were removed by Blastn tool. Sequencing reads were mapped by mirDeep2 (Friedländer, Mackowiak, Li, Chen, & Rajewsky, 2012), using S. bicolor v.3.1 as reference genome (Paterson et al., 2009). Reads shorter than 17 nt were not used during mapping step. A 250‐nt window around mapped reads was used for precursor prediction and excision. miRNA mature sequences were identified based on sugarcane and S. bicolor mature and precursor sequences deposited on miRBase v.21 onto miRDeep2. miRDeep2 tools were manipulated through Galaxy platform 7.221.3 (Afgan et al., 2016).
Precursor sequence displaying A/U percentage outside 30%–70% range (Zhang, Pan, Cox, Cobb, & Anderson, 2006) and those with minimum fold energy index (MFEI) values below 0.7 (Zhang et al., 2006) or below the minimum free energy of folding randomization (computed by Randfold; Bonnet, Wuyts, Rouze, & Peer, 2004) were discarded. Precursor miRNA not present in all biological replicates from the same root segment was not used in the data analysis. miRNA abundance was expressed in terms of reads per million (RPM).
Target prediction was performed by using novel and conserved miRNA sequences and the psRNATarget (Dai & Zhao, 2011) tool, with default configurations. Sugarcane ESTs were used as target database (Vettore et al., 2001). Alternatively, Arabidopsis thaliana genome (TAIR release 2004/01/22) was also used for target prediction. The experimental design is summarized in Figure 2.
Figure 2.
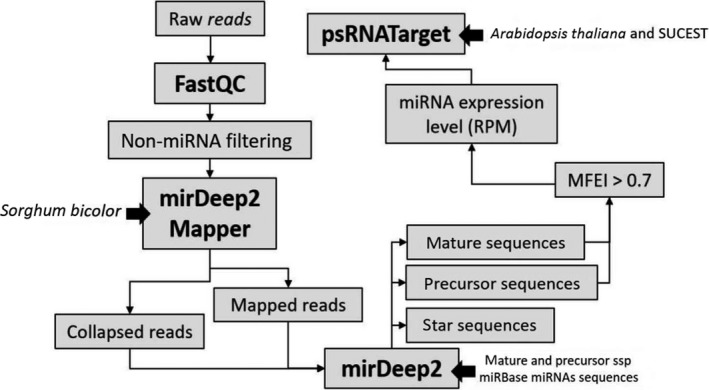
Pipeline for miRNA prediction using mirDeep2 and target identification through psRNATarget. Ion Proton raw reads were uploaded on FastQC for quality analysis, and further, rRNA, tRNA, snRNA, snoRNA, lncRNA, and tasiRNA were removed. Filtered reads were uploaded on miRDeep2 mapper module together with Sorghum bicolor as the reference genome. Collapsed and mapped reads output were used as input on miRDeep2 core together with mature and precursor miRNA sequences from S. bicolor and Saccharum sp retrieved from miRBase v. 21. Precursor, mature, and star miRNA sequences were predicted. The mature miRNA expression level was measured as reads per million if the corresponding precursor presented a minimum fold energy index (MFEI) higher than 0.7. Target prediction was performed by uploading mature miRNA sequences from previous steps and using the sugarcane EST database (SUCEST) and Arabidopsis thaliana as queries
2.4. Stem‐loop reverse transcription and qRT‐PCR for target validation
Stem‐loop primers were designed according to Varkonyi‐Gasic, Wu, Wood, Walton, and Hellens (2007). Reverse transcription was performed using SuperScript® III First‐Strand Synthesis System (Thermo Fisher Scientific®), with 2 µg of total RNA, 200 ng random hexamers, and 20 µM stem‐loop primer. Primers for target sugarcane ESTs were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/) and the following parameters: Tm 58°C to 60°C, 40%–60% GC, and amplicons ranging from 50 to 150 bp. EST primer efficiencies were calculated on the basis of standard curve dilutions, and only those with efficiency {[10(−1/slope)‐1] × 100} within the 90%–110% range were used.
qRT‐PCR was performed using SYBR Green PCR Master Mix (Thermo Fisher Scientific®) according to the manufacturer's instructions. All reactions were made in 3 technical replicates. The cycling amplification consisted of denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C, primer annealing at 60°C, and extension at 72°C for 1 min each step. Expression ratio was determined by qBase Plus 2.0 software (Biogazelle, Zwijnaarde, Belgium; Hellemans, Mortier, Paepe, Speleman, & Vandesompele, 2007). The reference primers [polyubiquitin (SCCCST2001G02.g), rRNA 60S (SCJFRZ2009G01.g), ubiquitin (SCBGLR1002D06.g, SCCCCL3080A11.g), and actin (SCRFLR1012H05.g)] were selected using the geNormPLUS tool (Biogazelle, Zwijnaarde, Belgium; Vandesompele et al., 2002). An aliquot of treated RNA was used in qRT‐PCR to rule out DNA contamination using GAPDH primer (Iskandar et al., 2004). Sugarcane EST sequences are available at http://sucest-fun.org (Vettore et al., 2001). All primer sequences used in this study are listed in Table S1.
3. RESULTS
3.1. Root miRNA sequencing and precursor prediction
The development of the lysigenous aerenchyma was observed along the root segments. The root cortex was intact within S1 and S2, and the aerenchyma area was noticeable in S3 by the opening of gas spaces, which increased in S4 (Figure 1). This was in accordance with previous results (Grandis et al., 2019; Leite et al., 2017; Tavares et al., 2018).
Aiming at characterizing sugarcane miRNA transcriptome and identifying candidates related to aerenchyma formation, 4 miRNA libraries were obtained from sequenced RNA extracts from sugarcane roots with developing aerenchyma. A total of 12 libraries (3 biological replicates from 4 root segments) produced 180435212 reads after Ion Proton sequencing, ranging from 73 (S1) to 31.5 (S4) million reads (Table S2). Non‐miRNA filtering (taking out rRNA, tRNA, snRNA, snoRNA, lncRNA, and tasiRNA) led to 119961617 total reads in all libraries.
The remaining sequences after the filtering and trimming steps showed lengths ranging from 20 to 22 nt (Table S3), which is in accordance with the expected range for miRNA (Li & Zhang, 2016). The dominance of 21‐nt length sequences was similar to the tendency observed elsewhere for sugarcane miRNA sequences from four different cultivars (Ferreira et al., 2012) but distinct from the 22‐ and 24‐nt bias shown by other reports with the same plant (Carnavale‐Bottino et al., 2013; Lin, Chen, Qin, & Lin, 2014; Thiebaut, Grativol, et al., 2012; Thiebaut et al., 2014). Furthermore, the majority of mature miRNA presented U as the first nucleotide (Table S3), which is in accordance with previously reported data on sugarcane miRNA (Ferreira et al., 2012; Thiebaut, Rojas, et al., 2012).
3.2. Reference genome read mapping and miRNA prediction
The pipeline described above allowed the identification of 39 miRNAs, from 19 different families. Five out of 39 have already been deposited as sugarcane miRNAs in miRBase (Table 1). The remaining 34 miRNAs that have been identified in S. bicolor were named on the basis of the observed homology between predicted sugarcane miRNA and those from S. bicolor available on miRBase.
Table 1.
Sugarcane miRNA families, mature sequence, and genomic locations according to Sorghum bicolor reference genome
| miRNA family | miRNA | Mature sequence | Genomic coordinateb | Genomic locationb | Sugarcane EST |
|---|---|---|---|---|---|
| miR156 | miR156aa | gcucacucucuaucugucagc | 3:3473045.0.3473132:− | Intergenic | SCQGLV1015B11.g |
| miR156b | gcucacuucucuuucugucagc | 4:5373544.0.5373631:− | Intergenic | No | |
| miR156e | gcucgcuucucuuucugucagc | 10:55009890.0.55009977:+ | Intergenic | No | |
| miR159 | miR159b | cuuggauugaagggagcucc | 3:1225075.0.1225121:− | Exon | SCAGFL3025B10.g |
| miR160 | miR160a | ugccuggcucccuguaugcca | 4:4236166.0.4236252:− | Intergenic | No |
| miR164 | miR164b | uggagaagcagggcacgugcu | 4:64881688.0.64881762:‐ | Exon | No |
| miR166 | miR166b | ucggaccaggcuucauucccc | 1:7426523.0.7426592:+ | Intron | SCQGAD1065C10.g |
| miR166d | ucggaccaggcuucauucccc | 4:63283311.0.63283396:− | Intergenic | SCQGAD1065C12.g | |
| miR167 | miR167ba | ugaagcugccagcaugaucuga | 3:64088380.0.64088466:− | Intergenic | SCSFSD1065B12.g |
| miR168 | miR168aa | ucgcuuggugcagaucgggac | 4:2246332.0.2246401:‐ | utr | SCEPRZ3087H11.g |
| miR171 | miR171a | ugauugagccgugccaauauc | 1:7845729.0.7845804:− | Intergenic | No |
| miR171c | ugagccgagccaauaucacuuc | 2:17125742.0.17125820:− | Intergenic | No | |
| miR171e | ugagccgaaccaauaucacuc | 6:54609050.0.54609135:+ | Intergenic | No | |
| miR171f | ugagccgaaccaauaucacuc | 4:62099920.0.62099999:− | Intergenic | No | |
| miR171h | uugagccgcgucaauaucucc | 1:15608733.0.15608816:− | Intergenic | No | |
| miR171i | ugauugagccgugccaauauc | 1:52558150.0.52558237:− | Intergenic | No | |
| miR171j | uugagccgcgccaauaucucu | 10:54088664.0.54088747:+ | Intergenic | SCJFAD1013C10.g | |
| miR171k | ugauugagccgugccaauauc | 6:57730667.0.57730744:− | Intergenic | No | |
| miR172 | miR172e | ugaaucuugaugaugcugcac | 2:14181333.0.14181407:− | Intergenic | No |
| miR393 | miR393b | ucagugcaaucccuuuggaau | 6:61406226.0.61406311:− | Intergenic | No |
| miR394 | miR394a | uuggcauucuguccaccucc | 2:66910981.0.66911054:+ | Intergenic | SCSBFL1108F06.g |
| miR395 | miR395b | guucucugcaagcacuucacg | 6:58761024.0.58761088:+ | Intergenic | No |
| miR395c | guucccuacaagcacuucacg | 6:58197026.0.58197095:− | Intergenic | No | |
| miR395e | guucucugcaagcacuucacg | 6:58197552.0.58197616:− | Intergenic | No | |
| miR395f | ugaaguguuugggggaacuc | 6:58196851.0.58196932:‐ | Intergenic | SCSGAD1006A12.g | |
| miR395h | guucccuucaagcacuucaca | 6:58761342.0.58761423:+ | Intergenic | No | |
| miR395j | guucccuucaagcacuucaca | 7:4658065.0.4658152:+ | Intergenic | No | |
| miR395l | guucccuucaagcacuucaca | 7:4658541.0.4658625:+ | Intergenic | No | |
| miR396 | miR396a | uuccacagcuuucuugaacug | 4:66092530.0.66092618:− | Intron | SCCCCL7C05F04.g |
| miR397 | miR397−5p | ucaccggcgcugcacucaauu | 4:4027096.0.4027184:− | Exon | No |
| miR399 | miR399b | gugcagcucuccucuggcaug | 4:9842733.0.9842814:− | Intergenic | No |
| miR399i | ugccaaaggagaguugcccug | 6:55042944.0.55043024:+ | Intergenic | No | |
| miR399j | ugccaaaggagaauugcccug | 4:9862937.0.9863027:− | Intergenic | No | |
| miR399k | ugccaaaggggauuugcccgg | 4:9868286.0.9868347:+ | Intron | No | |
| miR528 | miR528a | uggaaggggcaugcagaggag | 1:71476710.0.71476794:− | Intergenic | SCUTSD1026H02.g |
| miR2118 | miR2118−5p | ggcaugggaacauguaggaagg | 6:46386348.0.46386421:− | Intergenic | No |
| miR6222 | miR6222−3p | uagcugauccaaacaggcccu | 1:40668641.0.40668705:− | Intergenic | SCEZRT3070B02.gc |
| miR6222−5p | uagcugauccaaacaggcccu | 1:40668684.0.40668772:‐ | Intergenic | SCEZRT3070B02.gc | |
| miR6223 | miR6223−5p | cuagcauguuccuccuaagag | 7:8092842.0.8092921:+ | Exon | No |
miRNAs previously identified for sugarcane and deposited onto miRBase v.21;
Genome coordinate and location according to S. bicolor v. 3.1;
Corresponds to the same EST.
By mapping miRNAs onto S. bicolor genome, it was observed that the majority of the miRNAs are predicted to be encoded by an independent transcriptional unit (intergenic genomic location), whereas only 8 are located into gene sequences (exon, intron, or utr according to S. bicolor genomic location) (Table 1). Half of those are predicted to be encoded by mRNA coding sequences. Thirteen out of 39 miRNAs were assigned to putative correspondent sugarcane ESTs, but 2 miR6222 coincided with the same EST. Seven ESTs correspond to 8 miRNAs not yet deposited into miRBase. miR159b, comprised within EST SCAGFL3025B10.g sequence, was one of the few precursors predicted to be encoded by an exon (according to S. bicolor genomic coordinates).
3.3. miRNA expression profiles and target prediction by using A. thaliana genome and sugarcane EST as queries
The majority of the identified mature sequences were expressed in all root segments (Figure 3), which represents more than 86% of the total miRNA identified in the present study. This indicates that miRNAs are present during aerenchyma development. The expression level in terms of reads per million (Figure 4 and Table S4) shows four general expression patterns regarding the range of expression (blue boxes). However, there is an overall tendency of lower miRNA expression levels on S1, except for the most basal clade. Furthermore, miRNAs from the same family do not present similar expression levels.
Figure 3.
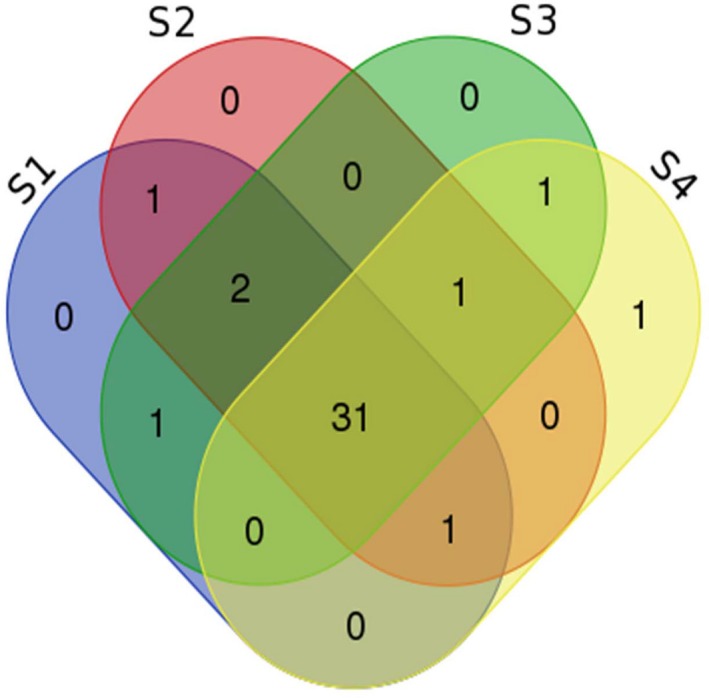
Distribution of the number of shared and exclusive expressed miRNAs during aerenchyma formation in sugarcane root segments (S1 to S4)
Figure 4.
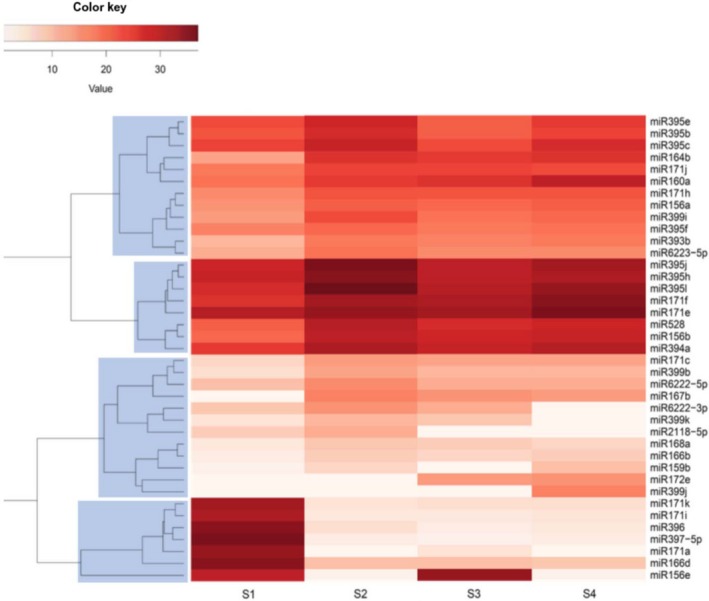
Hierarchical heatmap clustering miRNA expression level along with the sugarcane root segments during aerenchyma formation (S1 to S4). Blue boxes enclose the four major expression clusters
The set of identified miRNA was used to search for predicted targets by using as queries the SUCEST database and the A. thaliana genome. Putative targets were predicted for all the 39 miRNAs identified in the present study [747 (Table S5) and 4,728 (Table S6) targets] for SUCEST and A. thaliana genome, respectively. In most cases, the predicted targets are inhibited by cleavage instead of translation blockage. Five miRNAs and 8 putative targets were selected for qRT‐PCR validation (Tables S7 and S8), but there were no significant differences among root segments using one‐way ANOVA.
Gene ontology term enrichment for the complete set of putative targets identified in A. thaliana recovered several enriched functional categories and processes, including biosynthetic process (14.5%), cellular nitrogen compound metabolic process (13.7%), anatomical structure development (8.8%), response to stress (7.6%), and transport (5.8%) (Figure 5 and Table S9). Cell wall organization or biogenesis and carbohydrate metabolic process were the 15th and 12th most represented categories, with 2.3% and 3.5% of assigned targets respectively.
Figure 5.
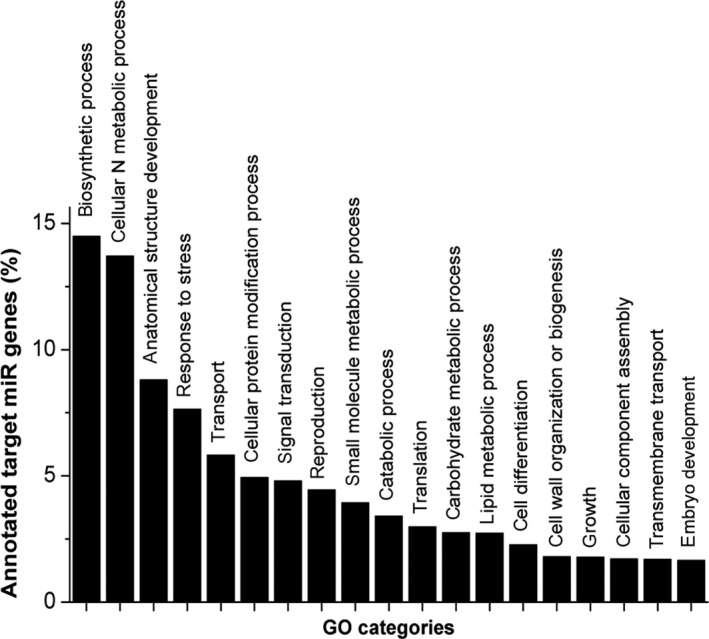
Gene ontology usage of miRNA targets. Only the 19 first categories are shown. Bars indicate the percentage of annotated GO terms for predicted targets of sugarcane miRNA using Arabidopsis thaliana as a query
All of the hydrolysis modules (cell expansion, cell separation, hemicellulose, and cellulose hydrolyzes) directly related to the cell wall (Grandis et al., 2014) are predicted to be targeted by miRNAs (Table 2). Cell separation, which comprises pectin hydrolysis, presented the highest number and diversity of predicted targets (50%), followed by hemicellulose hydrolysis (22.4%), cellulose modifications (15.5%), and cell expansion (12.1%). Remarkably, it was observed that pectins are indeed somehow subjected to structural changes during aerenchyma formation in sugarcane (Leite et al., 2017). This is a committed step for the attack of specific enzymes to the other polysaccharides, hemicelluloses, and cellulose that culminates in aerenchyma formation (Tavares et al., 2015, 2019) and production of a cell wall composite (Grandis et al., 2019; Leite et al., 2017).
Table 2.
Predicted cell wall‐related transcripts as putative targets for sugarcane miRNAs using Arabidopsis thaliana genome and SUCEST database as queries. The targets were distributed among cell wall modules according to Grandis et al. (2014)
| Module | Target description | CAZy or annotation | Target transcript (miRNA) |
|---|---|---|---|
| Cell expansion | Expansin | Expansin A17 | AT4G01630 (miR6222−3p, miR6222−5p) |
| Extensin | (Pro‐rich) extensin‐like; Pro‐rich extensin receptor kinase | AT4G13340 (miR397−5p); AT3G22800 (miR171a, miR171i, miR171k); AT4G08410 (miR395c, miR395h, miR395j, miR395l); AT5G59170 (miR393b); AT3G24550 (miR171c); AT2G18470 (miR6222−3p, miR6222−5p) | |
| Cell separation | Pectate lyase | PL1_1 | AT4G22090 (miR394a); AT5G15110 (miR168a); AT5G55720 (miR171a, miR171i); AT3G55140 (miR168a, miR2118−5p) |
| Pectin lyase‐like protein | GH28 | AT1G43100 (miR2118−5p) | |
| Pectin methylesterase | CE8 | AT3G17060 (miR171j, miR2118−5p); AT2G21610 (miR159b); AT4G00190 (miR167b); AT1G44980 (miR397−5p) | |
| Pectin methylesterase inhibitor | PMEI2 | AT3G17220 (miR399i, miR2118−5p) | |
| Pectin acetyl esterase | CE13 | AT3G05910 (miR6222−3p, miR6222−5p) | |
| Polygalacturonase | GH28, PG1 | AT4G20040 (miR159b); AT1G43100 (miR2118−5p); AT1G17150 (miR528); AT1G60390 (miR395b, miR395e); AT3G07970 (miR395h, miR395j, miR395l); AT4G01890 (miR167b); AT4G23820 (miR395f) | |
| β‐galactosidase | GH35; β‐gal‐related protein | AT4G38590 (miR395f); AT3G52840 (miR156a); AT4G36360 (miR395b, miR395e); AT5G63800 (miR2118−5p); AT5G49250 (miR166b, miR166d) | |
| Fasciclin‐like arabinogalactan protein | FLA1; FLA3; FLA8; FLA14; FLA17 | AT5G55730 (miR171a, miR171i, miR171k); AT5G44130 (miR166b, miR166d); AT3G12660 (miR156e); AT5G06390 (miR395b, miR395e); AT2G24450 (miR395f); AT2G45470 (miR396) | |
| Hemicellulose hydrolysis | Mannosidase | GH5 | AT4G28320 (miR395h, miR395j, miR395l) |
| Feruloyl esterase | Putative esterase‐like | AT5G11910 (miR6222−3p, miR6222−5p) | |
| Fucosidase | α‐L‐fucosidase 2 | SCACLR1036F01.g (miR6223−5p) | |
| β−1,3‐glucosidase | GH17; GH3 | AT1G77780 (miR156e); AT5G20950 (miR395f); AT4G18340 (miR171a, miR171i, miR171j, miR171k); AT1G32860 (miR166b, miR166d); AT3G57270 (miR171a, miR171i, miR171k); SCEZLB1006G08.g (miR395b, miR395l); SCCCSD2003H02.g (miR159b); SCEZRT2022D09.g (miR395f) | |
| Xyloglucan endotransglucosylase/ hydrolase | GH16 | AT4G30290 (miR167b) | |
| Xyloglucanase | GT77 | AT2G35610 (miR399i) | |
| Cellulose hydrolysis | Endo‐β‐glucanase | GH5; GH5_11; GH9 | AT1G19940 (miR6223−5p); AT2G44540 (miR171a, miR171i, miR171k); AT2G44550 (miR171a, miR171i, miR171k); AT2G44560 (miR171a, miR171i, miR171k); AT3G26140 (miR6223−5p); AT1G22880 (miR395f) |
| β −1,4‐glucosidase | GH1 | AT3G18080 (miR159b); AT1G26560 (miR171e, miR171f); AT5G54570 (miR156b, miR156e) |
Besides cell wall modifications, ethylene regulation is also one of the hallmarks underlying aerenchyma formation (Tavares et al., 2019; Yamauchi et al., 2013). Target prediction analysis showed that virtually all steps from ethylene production to transcriptional activity mediated by this phytohormone could be negatively regulated by a subset of the identified miRNAs (Table 3). Eleven families of ethylene‐responsive factors (ERFs) are predicted targets for 11 families of miRNAs. Although they are different subfamilies, presenting at least one AP2 DNA‐binding domain (Licausi, Ohme‐Takagi, & Perata, 2013), they are not all involved in ethylene signal transduction. Indeed, it has been observed that CYTOKININ RESPONSE FACTOR (CRF) and ABA REPRESSOR (ABR) mediate transcriptional response to cytokinin and abscisic acid, respectively (Pandey et al., 2005; Rashotte et al., 2006). Furthermore, most of the ERFs predicted to be negatively regulated by miRNAs display a repression domain within the protein sequence (Licausi et al., 2013).
Table 3.
Predicted ethylene‐related transcripts as putative targets for sugarcane miRNAs using Arabidopsis thaliana genome and SUCEST database as queries
| Category | Target description | Target | Target transcript (miRNA) |
|---|---|---|---|
| Ethylene overproduction protein | ETO | ETO1 | AT3G51770 (miR396) |
| Ethylene perception | ERS | ERS2 | AT1G04310 (miR171j) |
| Ethylene signaling | ETHYLENE INSENSITIVE | EIN3 | AT3G20770 (miR172e) |
| Ethylene‐responsive transcription factor | WAX INDUCER | WIN1 | AT1G15360 (miR6223−5p) |
| RELATIVE TO AB3/VP1 | RAV1 | AT1G51120 (miR156b) | |
| Ethylene response factors | ERF | AT1G68550 (miR394a); AT1G71130 (miR528); AT2G20880 (miR395f); AT1G22810 (miR395f); AT2G31230 (miR528); AT4G17490 (miR528); AT4G18450 (miR166b, miR166d); AT5G07580 (miR395f); AT5G21960 (miR166b, miR166d); AT5G47230 (miR528); AT2G41710 (miR171h); AT1G16060 (miR394a); SCUTLR1058E01.g (miR172e); SCVPCL6044F03.g (miR172e); | |
| Floral homeotic protein APETALA 2 | AP2 | AT4G36920 (miR172e) | |
| SCHNARCHZAPFEN | SNZ | AT2G39250 (miR172e) | |
| WRINKLED | WRI1; WRI4 | AT3G54320 (miR164b); AT1G79700 (miR164b) | |
| SCHLAFMUTZE | SMZ | AT3G54990 (miR172e) | |
| CYTOKININ RESPONSE FACTOR | CRF6; CRF2 | AT3G61630 (miR171h); AT4G23750 (miR164b) | |
| AINTEGUMENTA‐LIKE | AIL5 | AT5G57390 (miR156b) | |
| TARGET OF EAT | TOE2; TOE3 | AT5G60120 (miR172e); SCSBHR1050A07.g (miR172e) | |
| ABA REPRESSOR | ABR1 | AT5G64750 (miR393b) |
In order to have an overview of miRNA profiles along with the different root segments, a principal component analysis (PCA) was performed (Figure 6a). The PCA allowed for quite clear discrimination among samples, with PC1 (accounting for 53.2% of variation) separating S1 from the other root segments and PC2 (35.2% of variation) splitting root segments consistently with the order of aerenchyma formation, that is, from S1 to S4. The miRNA species that most contributed to the separation of S1 samples in PC1 were miR6222‐5p, miR395f, and miR2118‐5p, whereas miR396, miR166b and miR166d, miR393b, miR397‐5p, miR167b, miR164b, and some miR171 (e, h, and j) were important for segregating samples from the other segments (Figure 6b and Table S10). In PC2, miR172e and miR399j influenced the separation of S1 and S2, while miR528, miR6223‐5p, and most of the members of the miR395 family were important for segregating S3 and S4 (Figure 6b and Table S10). Although it is difficult to find clear patterns of miRNA expression among the identified miRNA members/families and segments (Figure 4), the PCA confirms that their global expression differs along the aerenchyma formation. Indeed, S1 is the meristem, S2 is the beginning of the transformation of the parenchymatic cells into aerenchyma, and S3 and S4 are tissues already displaying aerenchyma regions and therefore the action of the cell wall degrading enzymes.
Figure 6.
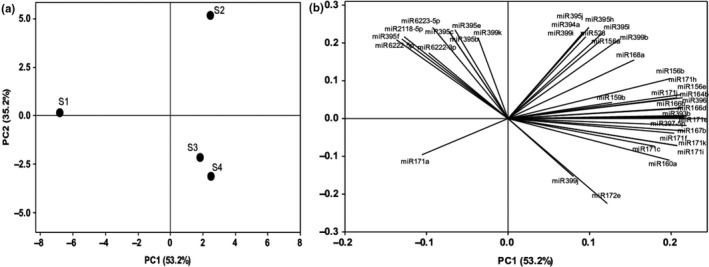
Distance biplots from 39 miRNAs found in sugarcane roots segments during aerenchyma formation. (a) The centroid separation corresponds to data average for roots segments in the plane defined by the first and second main components (PC1 and PC2). Percentage values in parentheses (x and y axes) show the proportion of the variance explained by axis. (b) The plot of the PC1 and PC2 loading vectors, describing the relationship among the miRNAs and roots segments. The mRNAs analyzed from Table 1 were expressed in descriptor vectors all PCs loading from PC1 and PC2 showed in Table S10. (n = 3)
4. DISCUSSION
Several publications reported the identification of miRNAs in biofuel crops such as Miscanthus giganteus (Jha & Shankar, 2013), Setaria italica (Yadav, Muthamilarasan, Pandey, Khan, & Prasad, 2014; Yi, Xie, Liu, Qi, & Yu, 2013), Sorghum bicolor (Calviño, Bruggmann, & Messing, 2011), Z. mays (Aravind et al., 2017; Liu et al., 2019), and Saccharum sp. (Gentile, Dias, Mattos, Ferreira, & Menossi, 2015 and references therein; Li et al., 2017; Thiebaut et al., 2017; Su et al., 2017; Yang et al., 2017). However, only a few of them addressed specific miRNAs predicted to be involved in cell wall construction or remodeling. In the present work, we identified 39 miRNAs expressed within the roots of the biofuel crop sugarcane undergoing cell wall changes due to aerenchyma formation. In this fine‐tuned process, several classes of enzymes act in a coordinated fashion on the wall polysaccharides of cortex cells to modify and/or degrade them, so that the gas spaces in the roots are formed (Grandis et al., 2019; Leite et al., 2017; Rahji et al., 2011). The identified miRNAs are comprised within 19 families, which are comparable to sugarcane miRNA previously published data [14 (Zanca et al., 2010), 18 (Ferreira et al., 2012), 25 (Carnavale‐Bottino et al., 2013), and 26 (Ortiz‐Morea et al., 2013)]. From the 39 miRNAs, 31 have not yet been identified for sugarcane according to miRBase database release v. 21, but were previously reported in the close relative species S. bicolor (Table 1).
The majority of mature sequences presented 21 nt length and U as the 5’ terminal nucleotide (Table S3). The identity of the first nucleotide within a mature sequence represents an important issue regarding DCL cleavage. Usually, 21‐nt‐long mature sequences are a product of DCL1 or DCL4 (Voinnet, 2009). One of the nuclease domains, RNase III‐A, releases preferentially miRNAs with U as the 5’ terminal nucleotide (Starega‐Roslan, Galka‐Marciniak, & Krzyzosiak, 2015). Furthermore, miRNAs loading onto the Argonaute are also directed by the identity of the 5’ nucleotide in plants (Mi et al., 2008), humans (Frank, Sonenberg, & Nagar, 2010), and flies (Okamura, Liu, & Lai, 2009). The strong dominance of 21 nt 5’ U sequences has also been observed for other unrelated species such as A. thaliana (Rajagopalan, Vaucheret, Trejo, & Bartel, 2006), Physcomytrella patens (Cho et al., 2008), Z. mays (Nobuta et al., 2008), and Populus trichocarpa (Klevebring et al., 2009).
The knowledge about ethylene signaling makes aerenchyma formation an interesting subject for miRNA transcriptomic studies. Although ethylene has been pointed out as the major hormonal trigger of aerenchyma formation, its levels decrease as the aerenchyma develops (Tavares et al., 2018). Key players of the signal cascade have been described (He, Morgan, & Drew, 1996) and one scERF1 named scRAV1 acting as negative regulator upon pectin hydrolysis at the onset of aerenchyma formation was identified (Tavares et al., 2019). miR156 and miR172 are known negative regulators of AP2/ERF transcription factors implicated in flower organogenesis (Yant et al., 2010), developmental timing (Wu et al., 2009), nodulation (Nova‐Franco et al., 2015; Wang et al., 2014), ripening (Bi, Meng, Ma, & Yi, 2015), and apical dominance (Schwab et al., 2005). Thirteen miRNAs were predicted to target transcripts related to ethylene production, perception, signal transduction, and ethylene‐mediated transcriptional activity (Table 3). Among those, miR172, which was only detected when the aerenchyma is already visible (S3 and S4), targets transcripts from the last two of the mentioned categories. As a consequence, the role played by the targeted ERFs might take place at the onset of aerenchyma formation (S1 and S2), what is expected according to previous observations of our group using the same type of experiment (Tavares et al., 2019). At this point, miR172 transcription is rather low or absent, and cell wall changes are not yet visible (Figure 4). EIN3 is a known positive component in ethylene response (Takahashi, Yamauchi, Rajhi, Nishizawa, & Nakazono, 2015; Tavares et al., 2019). It is possible that EIN3 is not repressed by miR172 at the initial stages of aerenchyma development, and when PCD starts, the expression levels of miRNA172 increase and repress EIN3 in S3 and S4. This would possibly happen after ethylene signaling to PCD.
It has been hypothesized that cell wall modifications rely on a general succession of events named modules (Grandis et al., 2014). Here, we observed that cell wall organization or biogenesis and carbohydrate metabolic process were well‐represented ontology categories among putative miRNA target genes (Figure 5 and Table S9). Those include transcripts related to cell expansion and separation, and cellulose and hemicellulose hydrolysis modules. Thirty‐one out of 39 miRNAs are predicted to target at least one cell wall‐modification‐related transcript (Table 2). miRNA6222‐3p (Tables S5 and S6) stands out for targeting transcripts coding for 5 different enzyme activities. Except for cellulose hydrolysis, all other cell wall degradation modules are predicted to be targeted by this miRNA. In addition, all degradation modules are targeted by members from miR171 and miR395 families (Table 2).
Hemicellulose hydrolysis‐related targets are around 22% of cell wall‐related transcripts targeted by miRNA during aerenchyma formation. Arabinoxylan debranching and β‐glucan hydrolysis occur during this process (Grandis et al., 2019; Leite et al., 2017). miR6223‐5p showed decreasing levels along the root segments, which suggests that the targeted feruloyl esterase mRNA would be de‐repressed as the aerenchyma develops. This esterase breaks ferulic acid bridges between arabinose residues that decorate arabinoxylan backbones. Besides being responsible for the cross‐linking among arabinoxylan chains, ferulic bridges may also anchor lignin to arabinoxylan in grasses (Buanafina, 2009). As a result, when ferulic acid is absent, arabinosyl residues can be further hydrolyzed, allowing the enzyme attack on xylan. Consistent with this, the levels of arabinose decrease from S1 to S4, whereas the lignin content remains stable along the root (Leite et al., 2017). Thus, the control of expression of feruloyl esterases by miRNAs might be important because it is likely to promote loosening of interchain connections. Such modification would allow the action of hydrolases, producing the composite that confers impermeability to gasses in the aerenchyma (Leite et al., 2017).
Cell separation encompasses half of the targeted cell wall‐related transcripts (Table 2) and mostly relies on pectin degradation (Grandis et al., 2019; Leite et al., 2017; Tavares et al., 2019). This module includes carbohydrate‐active enzymes such as glycosyl hydrolases, esterases, and lyases (Grandis et al., 2014, 2019).
The most abundant pectin in plant cell walls is homogalacturonan and its de‐acetylation and de‐methylation by pectin acetyl and methylesterases, respectively. This occurs prior to the attack on α‐1,4 linkages between galacturonic acids by endopolygalacturonases or by eliminative cleavage by pectate lyase (Kohli & Gupta, 2015; Latarullo et al., 2016; Yadav, Yadav, Yadav, & Yadav, 2009). β‐galactosidase is also thought to increase cell wall porosity through the hydrolysis of galactan side chains from heavily decorated pectin (Grandis et al., 2019; Leite et al., 2017). Transcripts coding for all 5 enzyme activities are among putative targets for miRNA expressed during aerenchyma formation (Table 2).
Some of the pectin degradation‐related targets of miRNA regulation have been reported elsewhere, in accordance with what we observed in sugarcane roots. miR156, miR167, miR171, miR394, miR395, and miR2118 are expressed in Panicum virgatum, lotus, and cotton when subjected to abiotic stress. These miRNAs were predicted to target pectinesterase, β‐galactosidase, pectin lyase, and endopolygalacturonase mRNAs (Xie et al., 2014; Xie, Wang, Sun, & Zhang, 2015). Also, overexpression of miR156 in alfalfa led to disturbed pectin content (Aung et al., 2015). These findings, together with the observation by Leite et al. (2017) that galactose consistently decreases and β‐galactosidase increases (Grandis et al., 2019) during aerenchyma formation are consistent with the hypothesis that miR156 might be a negative regulator of pectin degradation through β‐galactosidase within sugarcane roots ongoing aerenchyma formation.
The cell separation module comprises not only pectinases but also fasciclin‐like arabinogalactan proteins. They are a class of hydroxyproline‐rich proteins highly abundant within cell walls and plasma membrane (Harpaz‐Saad et al., 2011; Shi, Kim, Guo, Stevenson, & Zhu, 2003). Ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid is a putative key mediator of the role played by fasciclin‐like proteins upon cell wall regulation within roots (Xu, Rahman, Baskin, & Kieber, 2008). These proteins have also been implicated in programmed cell death (Gao & Showalter, 1999), which is a feature of aerenchyma formation in sugarcane roots (Leite et al., 2017). Fasciclin‐like arabinogalactan‐related mRNAs alone were predicted to be targeted by 12 miRNAs. It has been suggested that the role played by β‐galactosidases on pectin porosity could be mediated by the interaction with arabinogalactan‐like proteins due to the removal of galactosyl side chains (Dean et al., 2007; Lamport, Kieliszewski, & Showalter, 2006; Seifert & Roberts, 2007). The transcriptional pattern of miR171i, miR171k, and miR166d displayed a tendency to increase along the sugarcane root segments (Figure 4). This might suggest enhanced translational blockage of the putative targets β‐galactosidase and fasciclin‐like arabinogalactan mRNAs. The lower miRNA levels closer to the root apex are consistent with the expected role played by β‐galactosidase and fasciclin arabinogalactan‐like proteins prior to major cell wall changes. Altogether, miRNA transcription activity supports the idea of pectin porosity (Baron‐Epel, Gharyal, & Schindler, 1988) as a preparatory step for further cell wall modifications during aerenchyma formation in sugarcane roots. Hypothetically, an increase in porosity due to the action of β‐galactosidase on the walls of sugarcane roots, especially at S2, might be responsible for opening the access to other wall‐modifying enzymes and produce the composite that is the final product of the aerenchyma formation process (Grandis et al., 2019; Leite et al., 2017).
Endopolygalacturonase is also a key enzyme for aerenchyma development in sugarcane roots (Tavares et al., 2019). Endopolygalacturonase mRNA levels increase upon transcriptional de‐repression by scRAV1 at the same time as ethylene declines, and major changes regarding pectins become visible (Leite et al., 2017; Tavares et al., 2018). Curiously, the blockage of ethylene perception using the drug 1‐methylcyclopropene (1‐MCP) delays but is unable to prevent aerenchyma development (Tavares et al., 2018). miR6223‐5p, miR528, and miR2118 decreasing levels from S1 to S4 might indicate that translational activity upon target transcripts (pectin acetyl esterase and endopolygalacturonase) is de‐repressed (Figure 4). These results suggest that, besides the hormone and transcriptional repression played by ethylene through scRAV1, endopolygalacturonase hydrolysis could also be subjected to miRNA regulation. Furthermore, the connection between ethylene perception and action, and pectin degradation through scRAV repression might also be influenced by miRNA regulation. The present work showed that miR156 is predicted to target scRAV1 (Tavares et al., 2019) transcription factor. This result is consistent with the observation that RAV1 expression was affected under miR156 overexpression in rice (Xie et al., 2012). Moreover, alteration in this miRNA led to a reduction in lignin content, higher cellulose accumulation, increased sugar yield, and improved digestibility in P. virgatum, maize, alfalfa, and poplar (Chuck et al., 2011; Fu et al., 2012; Rubinelli, Chuck, Li, & Meilan, 2013). miR156‐overexpressing plants of Medicago sativa were shown to have contrasting expression patterns for different endopolygalacturonases (Gao, Austin, Amyot, & Hannoufa, 2016). Whether miR156 regulates pectinase genes directly or via RAV1 inhibition remains to be solved.
The global analysis of miRNA expression levels revealed a clear distinction among all root segments, and S1 was completely separated from S2, S3, and S4 in PC1 (Figure 6). Several miRNAs influenced this separation (Table S10), but, probably, they are more related to the different developmental root zones rather than with the formation of aerenchyma itself. PC2 discriminates S2 completely from the other segments, suggesting that this segment is where most of the epigenetic control via miRNA of aerenchyma formation takes place in sugarcane roots. S3 and S4 were closely grouped, with miR172e and miR399j being relevant in this separation. In these segments, the ethylene signaling, programmed cell death, and pectin degradation modules are already advanced, and the formation of air spaces is visible (Leite et al., 2017). The miRNA expression levels alone (Figure 4) make it difficult to define clear patterns able to explain the aerenchyma formation. However, the global analysis reinforces the fact that the segments differ among each other and miRNA expression possibly contributes to this process.
5. CONCLUSIONS
Our results show that miRNA epigenetic control in sugarcane is predicted to target several genes within a range of GO categories, including biosynthetic process, cellular N metabolic process, anatomical structure development, to name but a few. Our analyses, focused on the cell wall‐ and hormone‐related processes in the roots of sugarcane, suggest the existence of a layer of epigenetic control via miRNA. Considering the modularity of aerenchyma formation (ethylene action, cell expansion, cell separation, hemicellulose, and cellulose hydrolysis), the sugarcane miRNAs found in this work seem to be involved in hormone perception, cell separation, and hemicellulose hydrolysis.
Since sugarcane is an important bioenergy crop, our findings may have some impact on the production of bioenergy in the future, as the aerenchyma formation process offers opportunities to gain control of the whole‐plant cell wall hydrolysis and biomass saccharification.
CONFLICT OF INTEREST
The authors declare no conflict of interest associated with the work described in this manuscript.
AUTHOR CONTRIBUTIONS
E.Q.P.T performed most of the experiments; E.Q.P.T, G.H.R, and M.S.B designed the experiments and analyzed the data; A.R.P and J.W.G provided technical assistance with bioinformatic analysis and figure design; E.Q.P.T and M.S.B conceived the project and wrote the manuscript; A.G and M.C.M.M incorporated new data analyses; A.G, M.C.M.M, and M.S.B rewrote the manuscript; and M.S.B agrees to serve as the author responsible for contact and ensures communication.
Supporting information
ACKNOWLEDGMENTS
The authors thank the Tropical Medicine Institute facility from the University of São Paulo (USP) for sequencing services and Centro de Facilidades de Apoio à Pesquisa from USP for the use of computational and bioinformatics resources. This work was supported by fellowships (to E.Q.P.T.) from the São Paulo Research Foundation (FAPESP 2011/52065‐3 and 2015/13479‐8) and the National Institute of Science and Technology of Bioethanol (INCT‐Bioethanol) (FAPESP 2008/57908‐6 and 2014/50884‐5; National Council for Scientific and Technological Development CNPq 574002/2008‐1 and 465319/2014‐9).
Tavares Queiroz de Pinho E, Martins Camara Mattos M, Grandis A, et al. Newly identified miRNAs may contribute to aerenchyma formation in sugarcane roots. Plant Direct. 2020;4:1–14. 10.1002/pld3.204
REFERENCES
- Afgan, E. , Baker, D. , van den Beek, M. , Blankenberg, D. , Bouvier, D. , Čech, M. , … Goecks, J. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research, 44, 3–10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- Aravind, J. , Rinku, S. , Pooja, B. , Shikha, M. , Kaliyugam, S. , Mallikarjuna, M. G. , … Nepolean, T. (2017). Identification, characterization, and functional validation of drought‐responsive microRNAs in subtropical maize inbreds. Frontiers in Plant Science, 8, 941 10.3389/fpls.2017.00941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung, B. , Gruber, M. Y. , Amyot, L. , Omari, K. , Bertrand, A. , & Hannoufa, A. (2015). MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnology Journal, 13, 779–790. 10.1111/pbi.12308 [DOI] [PubMed] [Google Scholar]
- Baron‐Epel, O. , Gharyal, P. K. , & Schindler, M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta, 175, 389–395. 10.1007/BF00396345 [DOI] [PubMed] [Google Scholar]
- Bi, F. , Meng, X. , Ma, C. , & Yi, G. (2015). Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genomics, 16, 776 10.1186/s12864-015-1995-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet, E. , Wuyts, J. , Rouze, P. , & Van de Peer, Y. (2004). Evidence that microRNA precursors, unlike other non‐coding RNAs, have lower folding free energies than random sequences. Bioinformatics, 20, 2911–2917. 10.1093/bioinformatics/bth374 [DOI] [PubMed] [Google Scholar]
- Borges, F. , & Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nature Reviews Molecular Cell Biology, 16, 727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buanafina, M. (2009). Feruloylation in grasses: Current and future perspectives. Molecular Plant, 2, 861–872. 10.1093/mp/ssp067 [DOI] [PubMed] [Google Scholar]
- Buckeridge, M. S. , & De Souza, A. P. (2017). Advances of basic science for second generation bioethanol from sugarcane (p. 219). New York, NY: Springer. [Google Scholar]
- Bukatsch, F. (1972). Bemerkungen zur Doppelfarbung: Astrablau‐Safranin. Mikrokosmos, 61, 255. [Google Scholar]
- Calviño, M. , Bruggmann, R. , & Messing, J. (2011). Characterization of the small RNA component of the transcriptome from grain and sweet sorghum stems. BMC Genomics, 12, 356 10.1186/1471-2164-12-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnavale‐Bottino, M. , Rosario, S. , Grativol, C. , Thiebaut, F. , Rojas, C. A. , Farrineli, L. , … Ferreira, P. C. G. (2013). High‐throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS ONE, 8, e59423 10.1371/journal.pone.0059423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, 303, 2022–2025. 10.1126/science.1088060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S. H. , Addo‐Quaye, C. , Coruh, C. , Arif, M. A. , Ma, Z. , Frank, W. , & Axtell, M. J. (2008). Physcomitrella patens DCL3 is required for 22–24 nt siRNA accumulation, suppression of retrotransposon‐derived transcripts, and normal development. PLoS Genetics, 4, e1000314 10.1371/journal.pgen.1000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G. S. , Tobias, C. , Sun, L. , Kraemer, F. , Li, C. , Dibble, D. , … Hake, S. (2011). Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proceedings of the National Academy of Sciences, 108, 17550–17555. 10.1073/pnas.1113971108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. , & Zhao, P. X. (2011). PsRNATarget: A plant small RNA target analysis server. Nucleic Acids Research, 39, 155–159. 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, A. P. , Leite, D. C. C. , Pattathil, S. , Hahn, M. G. , & Buckeridge, M. S. (2013). Composition and structure of sugarcane cell wall polysaccharides: Implications for second‐generation bioethanol production. BioEnergy Research, 6, 564–579. 10.1007/s12155-012-9268-1 [DOI] [Google Scholar]
- Dean, G. H. , Zheng, H. , Tewari, J. , Huang, J. , Young, D. S. , Hwang, Y. T. , … Haughn, G. W. (2007). The Arabidopsis MUM2 gene encodes a beta‐galactosidase required for the production of seed coat mucilage with correct hydration properties. The Plant Cell, 19, 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, T. H. , Gentile, A. , Vilela, R. D. , Costa, G. G. , Dias, L. I. , Endres, L. , & Menossi, M. (2012). MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE, 7, 46703–46717. 10.1371/journal.pone.0046703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, F. , Sonenberg, N. , & Nagar, B. (2010). Structural basis for 5'‐nucleotide base‐specific recognition of guide RNA by human AGO2. Nature, 465, 818–822. 10.1038/nature09039 [DOI] [PubMed] [Google Scholar]
- Friedländer, M. R. , Mackowiak, S. D. , Li, N. , Chen, W. , & Rajewsky, N. (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Research, 40, 37–52. 10.1093/nar/gkr688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C. , Sunkar, R. , Zhou, C. , Shen, H. , Zhang, J. Y. , Matts, J. , … Wang, Z. Y. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnology Journal, 10, 443–452. 10.1111/j.1467-7652.2011.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M. , & Showalter, A. M. (1999). Yariv reagent treatment induces programmed cell death in Arabidopis cell cultures and implicates arabinogalactan protein involvement. The Plant Journal, 19, 321–331. [DOI] [PubMed] [Google Scholar]
- Gao, R. , Austin, R. S. , Amyot, L. , & Hannoufa, A. (2016). Comparative transcriptome investigation of global gene expression changes caused by miR156 overexpression in Medicago sativa . BMC Genomics, 17, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile, A. , Dias, L. I. , Mattos, R. S. , Ferreira, T. H. , & Menossi, M. (2015). MicroRNAs and drought responses in sugarcane. Frontiers in Plant Science, 6, 58 10.3389/fpls.2015.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandis, A. , De Souza, A. P. , Tavares, E. Q. P. , & Buckeridge, M. S. (2014). Using natural plant cell wall degradation mechanisms to improve second generation bioethanol In McCann M. C., Buckeridge M. S., & Carpita N. C. (Eds.), Plants and bioenergy (pp. 211–230). New York, NY: Springer. [Google Scholar]
- Grandis, A. , Leite, D. C. C. , Tavares, E. Q. P. , Arenque‐Musa, B. C. , Gaiarsa, J. W. , Martins, M. C. M. , … Buckeridge, M. S. (2019). Cell wall hydrolases act in concert during aerenchyma development in sugarcane roots. Annals of Botany, 124, 1067–1089. 10.1093/aob/mcz099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria, P. , & Yadav, S. K. (2011). Identification of miR414 and expression analysis of conserved miRNAs from Stevia rebaudiana . Genomics, Proteomics & Bioinformatics, 9, 211–217. 10.1016/S1672-0229(11)60024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz‐Saad, S. , McFarlane, H. E. , Xu, S. , Divi, U. K. , Forward, B. , Western, T. L. , & Kieber, J. J. (2011). Cellulose synthesis via the FEI2 RLK/SOS5 pathway and cellulose synthase 5 is required for the structure of seed coat mucilage in Arabidopsis. The Plant Journal, 68, 941–953. 10.1111/j.1365-313X.2011.04760.x [DOI] [PubMed] [Google Scholar]
- He, C. J. , Morgan, P. W. , & Drew, M. C. (1996). Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Journal of Plant Physiology, 112, 463–472. 10.1104/pp.112.2.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans, J. , Mortier, G. , De Paepe, A. , Speleman, F. , & Vandesompele, J. (2007). qBase relative quantification framework and software for management and automated analysis of real‐time quantitative PCR data. Genome Biology, 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskandar, H. M. , Simpson, R. S. , Casu, R. E. , Bonnett, G. D. , MaCLean, D. J. , & Manners, J. M. (2004). Comparison of reference genes for quantitative real‐time polymerase chain reaction analysis of gene expression in sugarcane. Plant Molecular Biology Reporter, 22, 325–337. 10.1007/BF02772676 [DOI] [Google Scholar]
- Jaiswal, D. , De Souza, A. P. , Larsen, S. , LeBauer, D. , Miguez, F. E. , Sparovek, G. , … Long, S. P. (2017). Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nature Climate Change, 7, 788–792. 10.1038/nclimate3410 [DOI] [Google Scholar]
- Jha, A. , & Shankar, R. (2013). miReader: Discovering novel miRNAs in species without sequenced genome. PLoS ONE, 8, e66857 10.1371/journal.pone.0066857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevebring, D. , Street, N. R. , Fahlgren, N. , Kasschau, K. D. , Carrington, J. C. , Lundeberg, J. , & Jansson, S. (2009). Genome‐wide profiling of Populus small RNAs. BMC Genomics, 10, 620 10.1186/1471-2164-10-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, P. , & Gupta, R. (2015). Alkaline pectinases: A review. Biocatalysis and Agricultural Biotechnology, 4, 279–285. 10.1016/j.bcab.2015.07.001 [DOI] [Google Scholar]
- Lamport, D. T. , Kieliszewski, M. J. , & Showalter, A. M. (2006). Salt stress upregulates periplasmic arabinogalactan proteins: Using salt stress to analyse AGP function. New Phytologist, 169, 479–492. 10.1111/j.1469-8137.2005.01591.x [DOI] [PubMed] [Google Scholar]
- Latarullo, M. B. , Tavares, E. Q. , Maldonado, G. P. , Leite, D. C. , & Buckeridge, M. S. (2016). Pectins, endopolygalacturonases, and bioenergy. Frontiers in Plant Science, 20, 1401 10.3389/fpls.2016.01401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, D. C. C. , Grandis, A. , Tavares, E. Q. P. , Piovezani, A. R. , Pattathil, S. , Avci, U. , … Buckeridge, M. S. (2017). Cell wall changes during the formation of aerenchyma in sugarcane roots. Annals of Botany, 120, 693–708. 10.1093/aob/mcx050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , & Zhang, B. (2016). MicroRNAs in control of plant development. Journal of Cellular Physiology, 231, 303–313. 10.1002/jcp.25125 [DOI] [PubMed] [Google Scholar]
- Li, M. , Liang, Z. , He, S. , Zeng, Y. , Jing, Y. , Fang, W. , … Tan, F. (2017). Genome‐wide identification of leaf abscission associated microRNAs in sugarcane (Saccharum officinarum L.). BMC Genomics, 18, 754 10.1186/s12864-017-4053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi, F. , Ohme‐Takagi, M. , & Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytologist, 199, 639–649. 10.1111/nph.12291 [DOI] [PubMed] [Google Scholar]
- Lin, S. , Chen, T. , Qin, X. , & Lin, W. (2014). Identification of microRNA families expressed in sugarcane leaves subjected to drought stress and the targets thereof. Pakistan Journal of Agricultural Sciences, 51, 925–934. [Google Scholar]
- Liu, X. , Zhang, X. , Sun, B. , Hao, L. , Liu, C. , Zhang, D. , … Li, Y. (2019). Genome‐wide identification and comparative analysis of drought‐related microRNAs in two maize inbred lines with contrasting drought tolerance by deep sequencing. PLoS ONE, 14, e0219176 10.1371/journal.pone.0219176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S. , Cai, T. , Hu, Y. , Chen, Y. , Hodges, E. , Ni, F. , … Qi, Y. (2008). Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5' terminal nucleotide. Cell, 133, 116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta, K. , Lu, C. , Shrivastava, R. , Pillay, M. , De Paoli, E. , Accerbi, M. , … Meyers, B. C. (2008). Distinct size distribution of endogenous siRNAs in maize: Evidence from deep sequencing in the mop1‐1 mutant. Proceedings of the National Academy of Sciences, 105, 14958–14963. 10.1073/pnas.0808066105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nova‐Franco, B. , Íñiguez, L. P. , Valdés‐López, O. , Alvarado‐Affantranger, X. , Leija, A. , Fuentes, S. I. , … Hernández, G. (2015). The micro‐RNA72c‐APETALA2‐1 node as a key regulator of the common bean‐Rhizobium etli nitrogen fixation symbiosis. Plant Physiology, 168, 273–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, K. , Liu, N. , & Lai, E. C. (2009). Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Molecular Cell, 36, 431–444. 10.1016/j.molcel.2009.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Morea, F. A. , Vicentini, R. , Silva, G. F. , Silva, E. M. , Carrer, H. , Rodrigues, A. P. , & Nogueira, F. T. (2013). Global analysis of the sugarcane microtranscriptome reveals a unique composition of small RNAs associated with axillary bud outgrowth. Journal of Experimental Botany, 64, 2307–2320. 10.1093/jxb/ert089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, G. K. , Grant, J. J. , Cheong, Y. H. , Kim, B. G. , Li, L. , & Luan, S. (2005). ABR1, an APETALA2‐domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiology, 139, 1185–1193. 10.1104/pp.105.066324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H. , Bowers, J. E. , Bruggmann, R. , Dubchak, I. , Grimwood, J. , Gundlach, H. , … Rokhsar, D. S. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature, 29, 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Rahji, I. , Yamauchi, T. , Takahashi, H. , Nishiuchi, S. , Shiono, K. , Watanabe, R. , … Nakazono, M. (2011). Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytologist, 190, 351–368. 10.1111/j.1469-8137.2010.03535.x [DOI] [PubMed] [Google Scholar]
- Rai, K. M. , Thu, S. W. , Balasubramanian, V. K. , Cobos, C. J. , Disasa, T. , & Mendu, V. (2016). Identification, characterization, and expression analysis of cell wall related genes in Sorghum bicolor (L.) Moench, a food, fodder, and biofuel crop. Frontiers in Plant Science, 7, 1287 10.3389/fpls.2016.01287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, R. , Vaucheret, H. , Trejo, J. , & Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Development, 20, 3407–3425. 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte, A. M. , Mason, M. G. , Hutchison, C. E. , Ferreira, F. J. , Schaller, G. E. , & Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two‐component pathway. Proceedings of the National Academy of Sciences, 103, 11081–11085. 10.1073/pnas.0602038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinelli, P. M. , Chuck, G. , Li, X. , & Meilan, R. (2013). Constitutive expression of the Corngrass1 microRNA in poplar affects plant architecture and stem lignin content and composition. Biomass and Bioenergy, 54, 312–321. 10.1016/j.biombioe.2012.03.001 [DOI] [Google Scholar]
- Schwab, R. , Palatnik, J. F. , Riester, M. , Schommer, C. , Schmid, M. , & Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Developmental Cell, 8, 517–527. [DOI] [PubMed] [Google Scholar]
- Seifert, G. J. , & Roberts, K. (2007). The biology of arabinogalactan proteins. Annual Review of Plant Biology, 58, 137–161. 10.1146/annurev.arplant.58.032806.103801 [DOI] [PubMed] [Google Scholar]
- Shi, H. , Kim, Y. , Guo, Y. , Stevenson, B. , & Zhu, J. K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. The Plant Cell, 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starega‐Roslan, J. , Galka‐Marciniak, P. , & Krzyzosiak, W. J. (2015). Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer. Nucleic Acids Research, 43, 10939–10951. 10.1093/nar/gkv968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Zhang, Y. , Huang, N. , Liu, F. , Su, W. , Xu, L. , … Que, Y. (2017). Small RNA sequencing reveals a role for sugarcane miRNAs and their targets in response to Sporisorium scitamineum infection. BMC Genomics, 18, 325 10.1186/s12864-017-3716-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H. , Yamauchi, T. , Colmer, T. D. , & Nakazono, M. (2014). Aerenchyma formation in plants In van Dongen J. T., & Licausi F. (Eds.), Low‐oxygen stress in plants, plant cell monographs, Vol. 21 (pp. 247–265). Wein: Springer‐Verlag. [Google Scholar]
- Takahashi, H. , Yamauchi, T. , Rajhi, I. , Nishizawa, N. K. , & Nakazono, M. (2015). Transcript profiles in cortical cells of maize primary root during ethylene‐induced lysigenous aerenchyma formation under aerobic conditions. Annals of Botany, 115, 879–894. 10.1093/aob/mcv018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares, E. Q. P. , De Souza, A. P. , & Buckeridge, M. S. (2015). How endogenous plant cell‐wall degradation mechanisms can help achieve higher efficiency in saccharification of biomass. Journal of Experimental Botany, 66, 4133–4143. 10.1093/jxb/erv171 [DOI] [PubMed] [Google Scholar]
- Tavares, E. Q. P. , De Souza, A. P. , Romim, G. H. , Grandis, A. , Plasencia, A. , Gaiarsa, J. W. , … Buckeridge, M. S. (2019). The control of endopolygalacturonase expression by sugarcane RAV transcription factor during aerenchyma formation. Journal of Experimental Botany, 70, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares, E. Q. P. , Grandis, A. , Lembke, C. G. , Souza, G. M. , Purgatto, E. , De Souza, A. P. , & Buckeridge, M. S. (2018). Roles of auxin and ethylene in aerenchyma formation in sugarcane roots. Plant Signaling & Behavior, 13, e1422464 10.1080/15592324.2017.1422464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut, F. , Grativol, C. , Carnavale‐Bottino, M. , Rojas, C. A. , Tanurdzic, M. , Farinelli, L. , … Ferreira, P. C. (2012). Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics, 13, 290–304. 10.1186/1471-2164-13-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut, F. , Grativol, C. , Tanurdzic, M. , Carnavale‐Bottino, M. , Vieira, T. , Motta, M. R. , … Ferreira, P. C. (2014). Differential sRNA regulation in leaves and roots of sugarcane under water depletion. PLoS ONE, 9, e93822 10.1371/journal.pone.0093822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut, F. , Rojas, C. A. , Almeida, K. L. , Grativol, C. , Domiciano, G. C. , Lamb, C. R. , … Ferreira, P. C. (2012). Regulation of miR319 during cold stress in sugarcane. Plant Cell and Environment, 35, 502–512. 10.1111/j.1365-3040.2011.02430.x [DOI] [PubMed] [Google Scholar]
- Thiebaut, F. , Rojas, C. A. , Grativol, C. , Calixto, E. P. D. R. , Motta, M. R. , Ballesteros, H. G. F. , … Ferreira, P. C. G. (2017). Roles of non‐coding RNA in sugarcane‐microbe interaction. Non Coding RNA, 3, 25 10.3390/ncrna3040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele, J. , De Preter, K. , Pattyn, F. , Poppe, B. , Van Roy, N. , De Paepe, A. , & Speleman, F. (2002). Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi‐Gasic, E. , Wu, R. , Wood, M. , Walton, E. F. , & Hellens, R. P. (2007). Protocol: A highly sensitive RT‐PCR method for detection and quantification of microRNAs. Plant Methods, 12, 3–12. 10.1186/1746-4811-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore, A. L. , Silva, F. R. , Kemper, E. L. , & Arruda, P. (2001). The libraries that made SUCEST. Genetics and Molecular Biology, 24, 1–7. 10.1590/S1415-47572001000100002 [DOI] [Google Scholar]
- Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell, 136, 669–687. 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, L. , Zou, Y. , Chen, L. , Cai, Z. , Zhang, S. , … Li, X. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. The Plant Cell, 26, 4782–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Park, M. Y. , Conway, S. R. , Wang, J. W. , Weigel, D. , & Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell, 138, 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, F. , Stewart, C. N. J. , Taki, F. A. , He, Q. , Liu, H. , & Zhang, B. (2014). High‐throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress. Plant Biotechnology Journal, 12, 354–366. 10.1111/pbi.12142 [DOI] [PubMed] [Google Scholar]
- Xie, F. , Wang, Q. , Sun, R. , & Zhang, B. (2015). Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. Journal of Experimental Botany, 66, 789–804. 10.1093/jxb/eru437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Shen, J. , Hou, X. , Yao, J. , Li, X. , Xiao, J. , & Xiong, L. (2012). Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiology, 158, 1382–1394. 10.1104/pp.111.190488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S. L. , Rahman, A. , Baskin, T. I. , & Kieber, J. J. (2008). Two leucine‐rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. The Plant Cell, 20, 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, C. B. , Muthamilarasan, M. , Pandey, G. , Khan, Y. , & Prasad, M. (2014). Development of novel microRNA‐based genetic markers in foxtail millet for genotyping applications in related grass species. Molecular Breeding, 34, 2219–2224. 10.1007/s11032-014-0137-9 [DOI] [Google Scholar]
- Yadav, S. , Yadav, P. K. , Yadav, D. , & Yadav, K. D. S. (2009). Pectin lyase: A review. Process Biochemistry, 44, 1–10. 10.1016/j.procbio.2008.09.012 [DOI] [Google Scholar]
- Yamauchi, T. , Colmer, T. D. , Pedersen, O. , & Nakazono, M. (2018). Regulation of root traits for internal aeration and tolerance to soil waterlogging‐flooding stress. Plant Physiology, 176, 1118–1130. 10.1104/pp.17.01157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, T. , Shimamura, S. , Nakazono, M. , & Mochizukic, T. (2013). Aerenchyma formation in crop species: A review. Field Crops Research, 152, 8–16. 10.1016/j.fcr.2012.12.008 [DOI] [Google Scholar]
- Yang, Y. , Zhang, X. , Su, Y. , Zou, J. , Wang, Z. , Xu, L. , & Que, Y. (2017). miRNA alteration is an important mechanism in sugarcane response to low‐temperature environment. BMC Genomics, 18, 833 10.1186/s12864-017-4231-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant, L. , Mathieu, J. , Dinh, T. T. , Ott, F. , Lanz, C. , Wollmann, H. , … Schmid, M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2 . The Plant Cell, 22, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, F. , Xie, S. , Liu, Y. , Qi, X. , & Yu, J. (2013). Genome‐wide characterization of microRNA in foxtail millet (Setaria italica). BMC Plant Biology, 13, 212 10.1186/1471-2229-13-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanca, A. S. , Vicentini, R. , Ortiz‐Morea, F. A. , Del Bem, L. E. , da Silva, M. J. , Vincentz, M. , & Nogueira, F. T. (2010). Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biology, 10, 260–273. 10.1186/1471-2229-10-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. (2015). MicroRNA: A new target for improving plant tolerance to abiotic stress. Journal of Experimental Botany, 66, 1749–1761. 10.1093/jxb/erv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. H. , Pan, X. P. , Cox, S. B. , Cobb, G. P. , & Anderson, T. A. (2006). Evidence that miRNAs are different from other RNAs. Cellular and Molecular Life Sciences, 63, 246–254. 10.1007/s00018-005-5467-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


