Supplemental Digital Content is available in the text.
Keywords: adipose tissue, inflammation, lncRNA, macrophage, obesity, transcriptome
Abstract
Objective:
Systemic low-grade inflammation associated with obesity and metabolic syndrome is a strong risk factor for the development of diabetes mellitus and associated cardiovascular complications. This inflammatory state is caused by release of proinflammatory cytokines by macrophages, especially in adipose tissue. Long noncoding RNAs regulate macrophage activation and inflammatory gene networks, but their role in macrophage dysfunction during diet-induced obesity has been largely unexplored.
Approach and Results:
We sequenced total RNA from peritoneal macrophages isolated from mice fed either high-fat diet or standard diet and performed de novo transcriptome assembly to identify novel differentially expressed mRNAs and long noncoding RNAs. A top candidate long noncoding RNA, macrophage inflammation-suppressing transcript (Mist), was downregulated in both peritoneal macrophages and adipose tissue macrophages from high-fat diet–fed mice. GapmeR-mediated Mist knockdown in vitro and in vivo upregulated expression of genes associated with immune response and inflammation and increased modified LDL (low-density lipoprotein) uptake in macrophages. Conversely, Mist overexpression decreased basal and LPS (lipopolysaccharide)-induced expression of inflammatory response genes and decreased modified LDL uptake. RNA-pull down coupled with mass spectrometry showed that Mist interacts with PARP1 (poly [ADP]-ribose polymerase-1). Disruption of this RNA-protein interaction increased PARP1 recruitment and chromatin PARylation at promoters of inflammatory genes, resulting in increased gene expression. Furthermore, human orthologous MIST was also downregulated by proinflammatory stimuli, and its expression in human adipose tissue macrophages inversely correlated with obesity and insulin resistance.
Conclusions:
Mist is a novel protective long noncoding RNA, and its loss during obesity contributes to metabolic dysfunction and proinflammatory phenotype of macrophages via epigenetic mechanisms.
Highlights.
Twelve novel long noncoding RNAs are dysregulated in macrophages from high-fat diet–fed obese mice.
Macrophage-expressed long noncoding RNA macrophage inflammation-suppressing transcript (Mist) is downregulated in mouse adipose tissue macrophages on diet-induced obesity.
Mist attenuates macrophage inflammatory response and uptake of modified LDL (low-density lipoprotein).
MIST is decreased in stromal vascular fraction from omentum of metabolically unhealthy human subjects.
Mist-PARP1 (poly(ADP-ribose) polymerase 1) interaction prevents PARylation and activation of proinflammatory gene promoters.
Low-grade chronic inflammation is a hallmark of obesity and an important risk factor for insulin resistance and the development of type 2 diabetes mellitus.1,2 Elevated free fatty acids and dysregulation of key adipokines in obesity and diabetes mellitus leads to altered metabolic function, activation of TF (transcription factor) NF-κB (nuclear factor-kappa B) in adipose tissue and macrophages, and increased expression of proinflammatory cytokines including TNF-α (tumor necrosis factor-α) and IL-6 (interleukin-6).3,4 This proinflammatory signaling can block insulin actions in metabolic tissues and induce systemic insulin resistance, as well as contribute to other obesity-related cardiovascular complications such as atherosclerosis.5
It has been recognized that this inflammatory state is primarily caused by macrophages undergoing phenotypic changes in response to excess lipid levels.1,2 Obesity induces a phenotypic switch in macrophages from an alternately activated M2 state to a classically activated proinflammatory M1 phenotype.6 Studies by our laboratory and others have found that monocytes cultured with high glucose or advanced glycation end products have increased NF-κB activity and express higher levels of inflammatory genes such as C-C motif chemokine ligand 2 (Ccl2), Tnf, and interleukin-1β (Il1b), which promote monocyte activation and adhesion to endothelium.7,8 Recent studies have also associated obesity with a state of metabolic endotoxemia, described as elevated circulating levels of LPS (lipopolysaccharide) derived from gut microbiota, which likely induces M1 phenotypic switch in macrophages and causes inflammation.9,10 Obesity is associated with increased macrophage infiltration into adipose tissue, liver, and skeletal muscle. In particular, macrophage accumulation and polarization in adipose tissue during obesity plays a critical role in promoting systemic proinflammatory signaling and insulin resistance in a paracrine and likely endocrine fashion.2,3,6,11,12 Notably, the metabolically activated macrophage could have a distinct proinflammatory phenotype distinct from the canonical M1/M2 paradigm.13
Next-generation sequencing studies have identified thousands of transcripts called long noncoding RNAs (lncRNAs), which control gene expression, cellular phenotypes, developmental processes, and disease.14,15 LncRNAs can act in cis and in trans to affect gene expression through diverse mechanisms, including altering the recruitment of TFs and chromatin remodelers to specific genomic loci, acting as scaffolds in ribonucleoprotein particles, and regulating mRNA and microRNA functions/stability.16 LncRNAs have also been identified as regulators of key macrophage functions including reactive oxygen species production, cholesterol homeostasis, inflammation, and phenotypic polarization.17–21 We previously used RNA-seq to generate the first profile of lncRNAs differentially regulated in bone marrow–derived macrophages (BMDMs) from diabetic db/db mice versus nondiabetic db/+ mice.22 One of these lncRNAs, E330013P06, was upregulated under diabetic conditions and was shown to promote inflammation and macrophage lipid uptake. Recently, we showed that another diabetes mellitus–induced macrophage lncRNA, Dnm3os, controls macrophage inflammatory phenotype via trans mechanisms by interacting with nucleolin and through chromatin remodeling.23 However, the potential role of lncRNAs in directing macrophage phenotype during diet-induced obesity remains unexplored.
Because lncRNA expression is highly specific to cell type and environmental cues,24 there likely remain undiscovered functional lncRNA transcripts. In this study, we identified several novel lncRNAs in macrophages that are differentially expressed in a mouse model of diet-induced obesity and pre–diabetes mellitus. One of these lncRNAs, which we named as macrophage inflammation-suppressing transcript (Mist), was downregulated in peritoneal macrophages (PMs) and adipose tissue macrophages (ATMs) from diet-induced obesity mice as well as in human stromal vascular fraction isolated from adipose tissue from obese metabolically unhealthy donors relative to controls. Gain- and loss-of-function studies showed a functional role for Mist in anti-inflammatory and antiatherosclerotic phenotype of macrophages. Mechanistically, we identified that Mist interacts with PARP1 (poly ADP-ribose polymerase-1) in the nucleus and blocks PARP1 recruitment at inflammatory gene promoters. Disruption of this RNA-protein interaction promotes PARP1 recruitment and chromatin PARylation to enhance inflammatory gene expression. These data reveal that Mist is a novel protective lncRNA, and its loss contributes to metabolic dysfunction and proinflammatory phenotype of macrophages in the context of obesity and diabetes mellitus.
Methods
The RNA-seq and microarray data have been made publicly available at the Gene Expression Omnibus with the accession numbers GSE126887 and GSE126839, respectively. All other supporting data are available within the article and in the Data Supplement.
Isolation and Culture of Primary Macrophages and Cell Lines
All animal experiments were performed with protocols approved by City of Hope Institutional Animal Care and Use Committee. Male C57BL/6J mice (8 weeks old) were fed with standard laboratory diet or high-fat diet (HFD, 60% kcal, Research Diets Inc, D12492i) for 12 or 16 weeks for PM and ATM collection, respectively. Male mice were used because female mice are relatively less susceptible to HFD-induced systemic inflammation and develop only some components of the metabolic syndrome.25 Mice were weighed weekly and glucose tolerance tests performed immediately before macrophage collection (Figure I in the Data Supplement). For glucose tolerance tests, mice were fasted for 4 to 6 hours but provided with water ad libitum; mice were then injected with sterilized D-glucose solution (0.1 g/mL stock) at 1 g/kg of body weight. Tail vein blood samples were collected at indicated time intervals post-injection and blood glucose levels determined using a glucometer. PMs were collected 4 days after intraperitoneal injection of 3% thioglycollate medium. BMDMs were obtained from femurs and tibia and cultured as described.22 ATMs were obtained from epididymal visceral adipose tissue. Briefly, dissected fat pads were minced and gently agitated for 1 hour at 37°C in digestion buffer (100 mmol/L HEPES pH 7.4; 120 mmol/L NaCl; 50 mmol/L KCl; 5 mmol/L glucose; 1 mmol/L CaCl2; and 1.5% BSA) containing 1 mg/mL collagenase D (Roche, Basel, Switzerland). Cell suspension was strained through 100 µm filter, centrifuged for 10 minutes at 4°C, and resuspended in 3 mL RBC lysis buffer. Twelve milliliters MACS buffer (PBS; 0.5% BSA; 2 mmol/L EDTA) was added, and cells were centrifuged again. Stromal vascular fraction (SVF) pellet was resuspended in 1 mL MACS buffer, and macrophages were positively selected using anti-F4/80 microbeads and MACS columns (Miltenyi Biotec, Bergisch Gladbach, Germany).
RAW 264.7 (RAW) mouse macrophage cell line (ATCC TIB-71) was used for additional experiments. Where indicated, cells were treated with LPS (100 ng/mL). Human THP-1 monocytes (ATCC TIB-202) were differentiated into macrophages with phorbol 12-myristate 13-acetate at 20 ng/mL for 48 hours before LPS treatment as indicated.
Human Subjects and Tissue Processing
All human studies were conducted according to approved Institutional Review Board protocols. De-identified human RNA was used for gene expression studies. Visceral adipose tissue samples were collected from the greater omentum at the time of endoscopic hernia repair or nissen fundoplication (nonobese subjects, body mass index <30) and gastric bypass surgery (subjects with obesity, body mass index >40) between the years 2011 and 2013. A nonhuman subject research determination was granted by the University of Maryland, Baltimore, Institutional Review Board (HP-00058301) to J.A. Deiuliis where the human quantitative polymerase chain reaction (qPCR) data was generated and RNA sample–specific biometric characteristics compiled. A nonhuman subject research determination was also granted by Case Western Reserve University Institutional Review Board (STUDY20190017). Fasting blood was drawn on the morning of surgery, and blood chemistry values are provided in Table I in the Data Supplement.
After excision, the omental adipose tissue sample was rinsed with PBS, minced, and digested in sterile solution of collagenase type II from Clostridium histolyticum (1 mg/mL) at 37°C. The adipose tissue digest was passed through a cell strainer, centrifuged at 300g for 10 minutes. The resulting SVF pellet was washed with ice-cold PBS and homogenized in TRIzol (Life Technologies, Carlsbad, CA). RNA was further purified using total RNA isolation system (Exiqon, Foster City, CA). RNA quality was determined by Bioanalyzer (Agilent, Santa Clara, CA); only samples with RIN scores >7 were used.
RNA Isolation and Quantitative Reverse Transcription PCR
Total RNA was isolated from mouse PMs using TRIzol and from BMDMs, ATMs, RAW cells and THP-1 cells using RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized with high capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Waltham, MA). Gene expression was analyzed using 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA) with KAPA SYBR FAST qPCR Master Mix (Roche, Basel, Switzerland) and gene-specific primers (Table II in the Data Supplement) in triplicate. Relative gene expression levels were determined using 2−ΔΔCt method, normalized against internal controls Actb (mouse) and GAPDH (human) unless specified otherwise.
RNA Sequencing, Ribosomal Profiling, and Downstream Analysis
Whole transcriptome analysis of RNA from mouse PMs was performed using total RNA depleted of rRNA (Ribo-Zero, Illumina). Paired-end libraries were prepared and sequenced at City of Hope Integrative Genomics Core using an Illumina HiSeq 2000 system. Reads were mapped to the mouse genome assembly mm9 using TopHat2 aligner. Cufflinks and Cuffmerge software26 was used to generate de novo transcript assemblies. Novel genes were classified using the pipeline in Figure IIA in the Data Supplement as described22; briefly, transcripts that overlapped with RefSeq genes of mouse and 7 other organisms (human, chimp, rat, rabbit, orangutan, rhesus, and marmoset) were filtered out. Filtered transcripts were further assessed for potential open reading frames by using PhyloCSF and searching PfamA/B databases. Multiexonic transcripts with a score of >100 or with encoded PfamA/B protein domains were classified as novel protein-encoding transcripts. Transcripts of >200 bp containing at least 2 exons, having a PhyloCSF score of <100 and lacking PfamA/B domains, were defined as novel lncRNAs. Gene counts were generated with HTseq27 and differentially expressed genes (DEGs) identified with DEseq2.28 Biological functions and network analysis of DEGs with FPKM (fragments per kilobase per transcript) >1 and P<0.1 (481 genes) were performed using ingenuity pathway analysis (IPA, Qiagen) and DAVID (Database for Annotation, Visualization and Integrated Discovery) gene ontology analysis. Raw sequencing data and gene counts are deposited to NCBI GEO (Gene Expression Omnibus) repository GSE126887. For ribosomal profiling data, raw data from Wang et al29 was aligned to mm9 genome assembly using HISAT2, and gene FPKM values were generated using Cufflinks.
Nuclear Fractionation
RAW cells (≈40 million) were detached from plates by accutase treatment (Innovative Cell Technologies, San Diego, CA). Cells were washed in ice-cold PBS twice and resuspended in 1 mL of Lysis Buffer B (10 mmol/L Tris-HCl, 140 mmol/L NaCl, 1.5 mmol/L MgCl2, and 0.5% NP-40, RNAse inhibitor). Nuclei were pelleted at 1000 g, and supernatant containing cytoplasmic fraction was collected. Nuclei were resuspended in Lysis Buffer B and slowly mixed while adding 100 μL detergent stock (3.3% sodium deoxycholate, 6.6% tween 20). Nuclei were incubated on ice for 5 minutes, pelleted, and washed with Lysis Buffer B. RNA from remaining nuclear and cytoplasmic fractions was extracted using Trizol followed by RNA cleanup using Qiagen RNEasy columns. cDNA was synthesized using High Capacity cDNA Reverse Transcription kit and quantified by real-time (RT)-qPCR.
RNA-Fluorescent In Situ Hybridization
Mist subcellular localization was visualized using ViewRNA ISH Cell Assay kit (Affymetrix, Santa Clara, CA) and custom-designed Mist probe. Macrophages were fixed according to manufacturer’s protocol, with 4% formaldehyde for 30 minutes at protease dilution of 1:2000. Images were acquired on Zeiss Observer II microscope and processed with ZEN Blue software and ImageJ.
GapmeR-Mediated Mist Knockdown (In Vitro and In Vivo) and Microarray Profiling
RAW cells were transfected with antisense locked nucleic acid–modified GapmeRs (Exiqon) targeting Mist or a nontargeting control using Lipofectamine RNAiMAX (Invitrogen), and RNA was collected 48 hours post-transfection. For initial GapmeR screening, cells were transfected at 100 nmol/L. Subsequent experiments showed optimal knockdown at 50 nmol/L, which was used for all other Mist knockdown experiments in RAW cells. Global gene expression was analyzed using GeneChip Mouse Gene 2.0 ST whole-transcript array. DEGs were identified as absolute fold change >1.5 and P<0.05. Gene ontology and TF enrichment of DEGs was performed using DAVID30 and ENRICHR,31 respectively. Enriched TFs were identified from chromatin immunoprecipitation (ChIP)-X experiments culled from ENCODE (Encyclopedia of DNA Elements) and ChEA databases, as described.31 Microarray data are deposited to NCBI GEO repository GSE126839. For in vivo knockdown of Mist, GapmeRs were administered to male standard diet–fed WT C57BL/6 mice (8 weeks old) intraperitoneally at 5 mg/kg as described under Results.
Modified LDL Uptake Assays
DiI-ac-LDL (acetylated low-density lipoprotein labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine) was added at 10 µg/mL to RAW macrophages plated in DMEM+0.1% BSA. After 4 hours at 37°C, fluorescent images were collected with Evos FL Cell Imaging System (ThermoFisher Scientific). Alternately, cells were washed, detached using Accutase and resuspended in staining buffer+DAPI, and LDL uptake assessed by flow cytometry using BD LSRFortessa cell analyzer. Additional quantitative analysis was performed by comparing normalized fluorescence of cell lysates following published protocol.32
5′ and 3′ Rapid Amplification of cDNA Ends PCR, Cloning, and Overexpression of Mist in Macrophages
5′ and 3′ Rapid amplification of cDNA ends experiments were performed using FirstChoice RLM-RACE Kit (ThermoFischer). After identification of transcript ends, full-length Mist was amplified from RAW cell cDNA and cloned into pcDNA3.1(+) and in reverse orientation in pcDNA3.1(−) expression vectors using In-Fusion HD Cloning (Clontech). Transient overexpression in RAW cells was performed by transfection with Lipofectamine LTX (0.3 µg DNA/well in 24-well plates) and RNA collected after 72 hours.
ELISAs in BMDMs
BMDMs were transiently transfected by electroporation using Mouse Macrophage Nucleofector Kit (Lonza). Forty-eight hours post-transfection, complete medium was replaced with 0.5% FBS containing medium. Twenty-four hours later, supernatants were collected, spun down, and subjected to ELISA cytokine analysis for TNF-α and IL-1β (R&D Systems). RNA was collected for RT-qPCR analysis.
Chromatin Immunoprecipitation
ChIP was performed as previously described.23,33 Sheared and precleared chromatin (1×106 cell equivalents) was immunoprecipitated with 3 µg of H3K4me3 (H3 lysine-4 monomethylation) antibody (Millipore 17-614) or control Rabbit IgG, overnight at 4°C. ChIP-enriched DNA was analyzed by qPCR with promoter-specific primers (Table II in the Data Supplement). ChIP DNA enrichment was first calculated by % Input and then represented as fold-over IgG.
ChIP assays for PAR (poly(ADP)-ribosylation; Trevigen anti-PAR Cat number 4335-MC-100, Lot number 39816F17) and PARP1 (Abcam Anti-PARP1, ab227244, Lot GR3226439-13) were performed with modified protocol to minimize generation of PAR artifacts.34 Sonication was increased to 16 cycles to achieve similar fragment sizes, as verified by reverse crosslinking and running sheared chromatin on agarose gel. ChIP enrichment relative to input (% Input) was calculated using the formula 100×2(Ct [adjusted input to 100%]−Ct [IP]).
RNA Pulldown Coupled With Mass Spectrometry to Identify Mist-Interacting Proteins
RNA pulldown was performed as previously described with some modifications.23,35 Briefly, precleared nuclear extract from RAW macrophages (1 mg) was incubated with biotin-labeled full-length Mist sense or antisense RNA probes (3 µg) and tRNA (30 µg) at 4°C for 2 hours. Washed Streptavidin agarose beads were added to each binding reaction and further incubated at 4°C for 2 hours. Beads were washed, RNA-protein complexes were resolved on SDS-PAGE, bands were excised, and proteins were identified by Mass Spectrometry at City of Hope’s Mass Spectrometry Core. Scaffold (Proteome Software Inc, Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptides were identified using Scaffold (Proteome Software Inc, Portland, OR) and were accepted if they could be established at minimum 99.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at minimum 99.0% probability and contained at least 2 identified peptides. For validation experiments, eluted proteins were also analyzed by Western blotting with antibodies targeting PARP1 (Abcam, ab227244, Lot GR3226439-13) and HNRNPA3 (heterogeneous nuclear ribonucleoprotein A3; Proteintech, Cat number 25142-1-AP).
Nuclear RNA Immunoprecipitation
Native RNA immunoprecipitation (RIP) was performed using published protocol.36 Briefly, nuclear lysates from RAW cells were diluted in RIP buffer and incubated with indicated antibodies at manufacturer-recommended dilutions. RNA-protein complexes were captured on magnetic-IgG beads, and bound RNA was isolated and analyzed by RT-qPCR. Antibodies used were PARP1 (1:100), HNRNPA3 (1:1000), SUPT16H (1:500, Proteintech, Cat number 20551-1-AP), SMARCA5 (1:500, Proteintech, Cat number 13066-1-AP), and Rabbit IgG control (1:1000). Cross-linked RIP was performed following published protocol.37
Statistical Analysis
Gene expression values are expressed as mean+SD. Unless noted otherwise, statistical significance was calculated using unpaired 2-tailed Student t test to compare 2 groups or ANOVA followed by Fisher least significant difference test to compare multiple groups using Graphpad PRISM 7.0 software. All expression values from RT-qPCR were log-transformed and checked for normal distribution before statistical testing. Post hoc analysis of MIST expression from human SVF was calculated using Student t test or 1-way ANOVA followed by Tukey multiple comparisons test. Before correlation testing for human SVF data, health parameters were first checked for Gaussian distribution using D’Agostino and Pearson normality test, followed by Pearson correlation. Differences in health parameters across cohorts were tested using Welch test for normally distributed parameters and Mann-Whitney test for non-normal distributed parameters. To exclude treatment intervention as a variable affecting metabolic health, individuals prescribed metformin or thiazolidinedione were excluded from analysis. P<0.05 were considered statistically significant.
Results
LncRNA Mist Is Downregulated in Macrophages From HFD-Fed Obese Mice
To examine whether key macrophage lncRNAs are dysregulated during obesity and can contribute to increased inflammation, we used thioglycollate-elicited PMs from C57BL/6 mice fed HFD for 12 weeks and control littermates on standard laboratory diet (ND). HFD led to significant increases in body weight and impaired glucose tolerance (Figure IA through IC in the Data Supplement). RNA-seq analysis of total RNA from these PMs followed by de novo transcriptome assembly revealed 510 DEGs in macrophages from HFD versus ND mice (Figure 1A), the majority of which were protein-coding (Figure 1B). As expected, HFD-induced genes showed enriched association with inflammatory response (Figure ID through IE in the Data Supplement).
Figure 1.
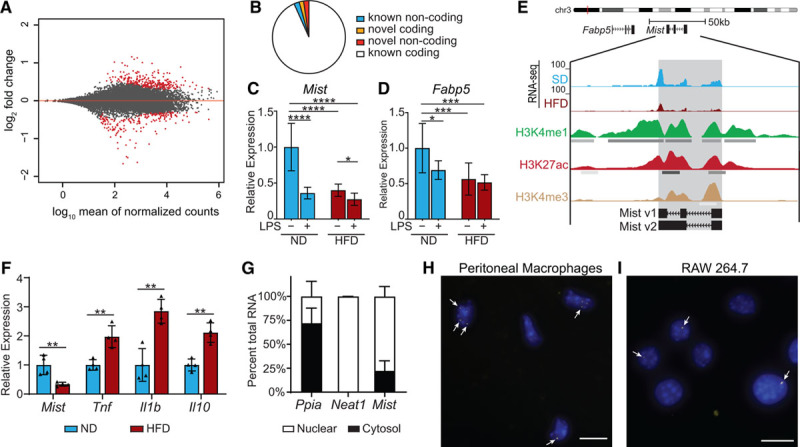
Macrophage inflammation-suppressing transcript (Mist) is suppressed in macrophages by high-fat diet (HFD) in vivo and LPS (lipopolysaccharide) treatment in vitro. A, MA plot of log2 ratio fold change (HFD/normal diet) vs normalized mean counts. Points for differentially expressed genes (false discovery rate <0.1) are in red. B, Classification of differentially expressed genes. C and D, Real-time quantitative polymerase chain reaction (RT-qPCR) analysis of Mist and Fabp5 in peritoneal macrophages from normal diet (ND) or HFD mice treated with or without LPS (100 ng/mL) for 3 h (n=6 mice/group). E, Representative tracks of our RNA-seq data at Mist genomic locus, and chromatin immunoprecipitation-seq tracks of overlapping histone modifications from bone marrow–derived macrophage (BMDMs; ENCODE [Encyclopedia of DNA Elements] Consortium). F, RT-qPCR analysis of genes expressed in adipose tissue macrophages (ATMs) from mice fed ND or HFD for 16 wk (n=4 mice/group). G, Nuclear localization of Mist relative to cytosol in macrophages. Subcellular fractionation of RAW 264.7 cells followed by RT-qPCR for indicated genes, including Neat1, a known nuclear lncRNA. H and I, RNA-fluorescent in situ hybridization in mouse peritoneal macrophages and RAW cells for Mist (yellow foci, denoted by white arrows); nuclei are counterstained with DAPI (blue). Scale bar represents 10 μm. Bar graphs represent mean±SD. H3K27ac indicates lysine-27 acetylation; and H3K4me1, H3 lysine-4 monomethylation. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 calculated using Fisher least significant difference test on log-transformed expression values.
We identified 37 novel macrophage lncRNAs from this data set using our data analysis pipeline (Figure IIA in the Data Supplement), of which 12 were differentially expressed in HFD-fed mice (Figure IIB in the Data Supplement). One of these lncRNAs, XLOC_019580, which we renamed Mist, overlaps with cDNA AK078702 on chromosome 3 originally identified in the FANTOM (Functional Annotation of the Mammalian Genome) consortium38 (Gencode ID: Gm38335) but lacks an overlapping Refseq annotation and has not been previously studied. Mist was chosen for further study because it is an intergenic lncRNA that lies ≈5.6 kb downstream of the protein-coding gene fatty acid binding protein 5 (Fabp5; Table III in the Data Supplement), an intracellular lipid chaperone with known roles in atherosclerosis, insulin sensitivity, and inflammation.39,40 Expression of Mist and Fabp5 were reduced in macrophages from HFD mice compared with control mice, which was confirmed by RT-qPCR (Figure 1C and 1D). Because metabolic syndrome is associated with increased inflammation, we examined whether Mist was similarly regulated by proinflammatory M1 stimuli. PMs treated with LPS in vitro also showed downregulation of Mist and Fabp5, and PMs from HFD mice showed further reduction in Mist expression (Figure 1C and 1D). Interestingly, Mist locus also overlaps with enhancer marks histone H3K4me1 and H3K27ac (lysine-27 acetylation; Figure 1E), suggesting a potential function related to chromatin and gene regulation.
ATMs are considered important regulators of obesity-induced inflammation and insulin resistance. To determine the expression pattern of Mist in ATMs, we isolated epididymal visceral white adipose tissue after 16 weeks HFD feeding, followed by extraction of RNA from F4/80+ cells enriched from SVF. Interestingly, expression of Mist was also reduced in ATMs from HFD-fed mice compared with ND, while Tnf, Il1b, and Il10 were upregulated (Figure 1F), as expected.13
Mist Is a Nuclear Transcript That Lacks Coding Potential
RT-qPCRs of subcellular fractions showed that Mist is localized primarily in the nuclei of macrophages (Figure 1G). This was further confirmed using RNA-fluorescent in situ hybridization with branched-DNA signal amplification probes, which showed Mist foci overlapping with nuclear DAPI staining in both primary PMs and RAW macrophage cell lines (Figure 1H and 1I). Cells were also probed for Ppia as a positive cytoplasmic RNA control, and cells with no probe were processed in parallel as negative controls (Figure IIIA through IIID in the Data Supplement). Further characterization by 5′ and 3′ rapid amplification of cDNA ends PCRs revealed 2 alternatively spliced variants of Mist, 1460 bp (v1) and 2197 bp (v2) long, containing 3 and 2 exons, respectively (Figure IIIE through IIIF in the Data Supplement, Figure 1E, and Table IV in the Data Supplement). For functional studies, we cloned Mist v1 and v2 variants into expression vectors and a vector expressing antisense Mist v1 transcript (Mist-AS; Figure IIIG through IIII in the Data Supplement). We also determined that Mist is a polyadenylated RNA (Figure IVA in the Data Supplement). To verify that Mist lacks coding potential, we performed in vitro transcription/translation assays with Mist_v1 construct. Results showed that Mist does not yield a peptide product (Figure IVB in the Data Supplement). Analysis of ribosomal profiling data from mouse PMs29 showed relatively low ribosomal enrichment of Mist (Figure IVC in the Data Supplement), further demonstrating that Mist lacks coding potential. We also searched for expression of the partially overlapping Gencode gene Gm38335 (Figure IIIE in the Data Supplement) among EMBL-EBI Baseline Expression Atlas and found that the only cell types with transcripts per million counts >1 were macrophages (Figure IVD in the Data Supplement), indicating that Mist is indeed a macrophage-specific transcript.
Mist Knockdown Upregulates Genes Involved in Inflammation and Lipid Metabolism
Metabolic syndrome dysregulates macrophage polarization and promotes proinflammatory M1 phenotype. To test our hypothesis that Mist regulates genes involved in macrophage polarization and function, we employed a loss-of-function approach using LNA-modified ASOs (antisense oligonucleotides, GapmeRs). We tested 4 different GapmeRs targeting Mist in RAW macrophages (Figure VA and VB in the Data Supplement) and used the most effective GapmeR (ASO-Mist) relative to control Gapmer (ASO-NC) in succeeding knockdown experiments. In subsequent experiments, we used optimized transfection conditions to achieve close to 80% knockdown in RAW cells (Figure VC in the Data Supplement).
To determine Mist targets in an unbiased manner, we silenced Mist expression with ASO-Mist in RAW cells and performed microarray profiling of gene expression (Figure VI in the Data Supplement) and subsequent RT-qPCR validation. Mist knockdown significantly altered expression of genes associated with macrophage functions including immune response, interferon signaling, and lipid metabolism (Figure 2A). Gene ontology analysis showed an enrichment of innate immune response, TNF production, and macrophage foam cell formation (Figure 2B) in upregulated genes and those related to lipid metabolism and cholesterol esterification among downregulated genes (Figure 2C). Upregulated gene promoters showed enrichment of TF IRF8 (Figure 2D), while the downregulated gene set was significantly enriched for targets of TF CEBPβ (Figure 2E), important drivers of M1- and M2-specific gene expression, respectively.41,42 Further RT-qPCR validation showed that Mist knockdown also suppressed expression of its neighboring gene Fabp5 (Figure 2F). Interestingly, proinflammatory cytokines (Tnf, Il6, Il1b, and Ccl2) associated with macrophage M1 phenotype were upregulated by Mist knockdown, while markers of macrophage M2 activation such as Egr2, Cd83,43 and Ppard were suppressed by Mist knockdown. Mist knockdown also enhanced expression of Il10 (Figure 2F), which is often upregulated in activated macrophages to curtail excessive inflammatory response.44 Notably, scavenger receptor Cd36 was also highly induced on Mist knockdown (Figure 2F). Overall, these data show that Mist knockdown increases genes associated with inflammation and M1 polarization but inhibits markers associated with alternative activation, suggesting an anti-inflammatory function for Mist in macrophages.
Figure 2.
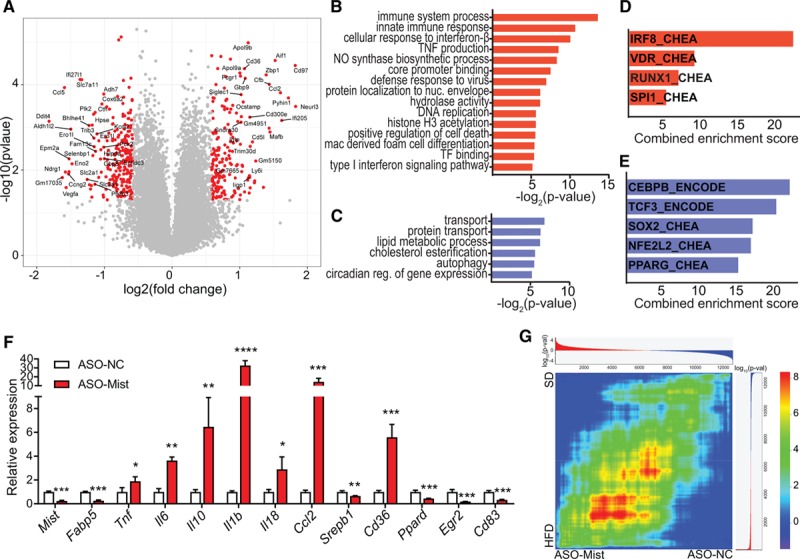
Macrophage inflammation-suppressing transcript (Mist) knockdown induces global changes in transcriptome and macrophage phenotype. A, Volcano plot of differential gene expression in RAW 264.7 macrophages transfected with Mist GapmeR (ASO-Mist) vs nontargeting control antisense oligonucleotide (ASO-NTC) GapmeR. Global gene expression was determined by microarray analysis using GeneChip Mouse Gene 2.0 ST array. Differentially expressed genes (DEGs) with absolute log2 (fold change) >1.5 and P<0.05 are in red and labeled. B and C, DAVID (Database for Annotation, Visualization and Integrated Discovery) gene ontology terms enriched in up- (red) and downregulated (blue) gene sets. D and E, Transcription factors associated with activation of DEGs up- (red) and downregulated (blue) by Mist-targeted GapmeR. Enriched transcription factors were identified from chromatin immunoprecipitation-X experiments culled from ENCODE (Encyclopedia of DNA Elements) and ChEA databases, as described.31 Combined score was computed by multiplying log of Fisher exact test P value by z score of deviation from expected rank. F, Quantitative polymerase chain reaction validation analysis of indicated genes from RAW 264.7 macrophages transfected with indicated GapmeRs (ASO); n=3. Bar graphs represent mean±SD. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 calculated using Student t test on log-transformed values. G, Rank-rank hypergeometric overlap (RRHO) heat map, comparing the degree of overlap between ranked lists of signed log10-transformed P values for changes in gene expression in high-fat diet (HFD) PMs (y axis) or ASO-Mist -transfected macrophages (x axis). Colors indicate log-transformed hypergeometric P value showing strength of overlap as positive (upper-right and lower-left quadrants) or negative enrichment (upper-left and lower-right), after Benjamini-Yekutieli multiple hypothesis correction.
To further determine whether Mist silencing in macrophages causes gene expression changes that resemble those induced by HFD, we compared DEGs on Mist knockdown with DEGs identified in our RNA-seq data from HFD-fed PMs. The 2 DEG lists shared a significant number of up- and downregulated genes, with hypergeometric P values of 2.47×10−4 and 2.15×10−5, respectively. To account for the differences in confidence threshold distribution between these data sets, we also used rank-rank hypergeometric overlap45 to compare expression profiles and identify shared gene expression patterns (Figure 2G). The rank-rank hypergeometric overlap heatmap shows both a global trend of positive correlation as well as specific gene sets similarly enriched in both HFD and Mist knockdown.
Loss of Mist Increases Modified LDL Uptake In Vitro and Increases Inflammatory Genes in Standard Diet–Fed Mice In Vivo
CD36 promotes macrophage foam cell formation via uptake of oxidized LDL and is highly expressed in macrophages in atherosclerotic lesions.46 CD36 is also upregulated in ATMs during obesity13 and in PMs from our HFD-fed mice. Our observations that Mist negatively regulates Cd36 expression and genes associated with cholesterol esterification led us to hypothesize that Mist influences modified LDL uptake. Indeed, Mist silencing significantly increased ac-LDL (acetylated low-density lipoprotein) uptake (Figure 3A and 3B).
Figure 3.
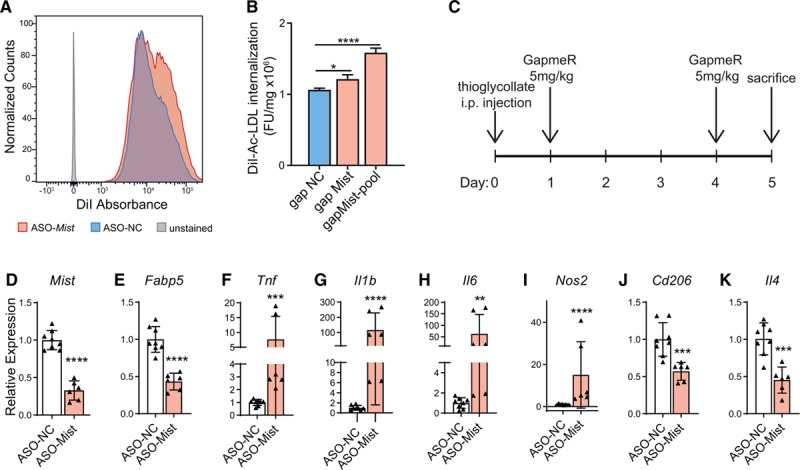
Macrophage inflammation-suppressing transcript (Mist) silencing increases uptake of modified LDL (low-density lipoprotein) and affects macrophage gene expression profile in vivo. A, Histogram showing fluorescence-activated cell sorter absorbance of DiI-labelled ac-LDL (acetylated LDL) after GapmeR ASO-mediated Mist knockdown in RAW 264.7 cells. B, Alternative spectrofluorometric assay of DiI-ac-LDL absorbance in RAW macrophages after Mist knockdown from a separate experiment. Bars represent mean+SD of normalized DiI-ac-LDL internalization, n=3. C),Experimental protocol for Mist knockdown in vivo. GapmeR was injected intraperitoneally (5 mg kg−1) in mice with control GapmeR (ASO-NC) and mice with Mist-target GapmeR (ASO-Mist) on days 1 and 4 after thioglycollate injection, and PMs were harvested on day 5. D–K, Real-time quantitative polymerase chain reaction (RT-qPCR) analysis of gene expression in peritoneal macrophages collected from mice injected with Mist-targeted (ASO-Mist) or control (ASO-NC) GapmeR. ASO-Mist Gapmer caused significant decrease in Mist expression (D), as well as neighboring gene Fabp5 (E). increased expression of M1 genes Tnf (F), Il1b (G), Il6 (H), and Nos2 (I) were observed, as well as decreases in M2 macrophage markers Cd206 (J) and Il4 (K). n=4 and n=3 mice for ASO-NC and ASO-Mist-injected mice, respectively. Macrophages were processed in duplicate per mouse, for total of n=8 and n=6 for ASO-NC and ASO-Mist, respectively. Bar graphs represent mean±SD. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 calculated using 1-way ANOVA followed by Tukey multiple comparisons test (B) or Student t test on log-transformed expression values (D–K).
To investigate whether Mist has similar anti-inflammatory functions in macrophages in vivo, we injected ASO-Mist and ASO-NC GapmeRs into normal C57BL/6J mice and examined gene expression in thioglycollate-elicited PMs 5 days later (Figure 3C). Two ASO-Mist injections were sufficient to confer significant reduction in Mist expression versus PMs from ASO-NC injected mice (Figure 3D). Similar to our observations in vitro, Mist silencing in vivo caused a reduction in Fabp5 expression (Figure 3E), significant upregulation of M1 genes Tnf, Il1b, Il6, and Nos2 (Figure 3F through 3I), and downregulation of important M2 marker Cd206 (Figure 3J) and driver of alternative activation Il4 (Figure 3K), indicating that Mist knockdown promotes M1 polarization and proinflammation in vivo.
Mist Gain-of-Function Leads to Reduction in Inflammatory Response and Increases ac-LDL Uptake
We next examined whether, conversely, Mist overexpression can have protective effects in macrophages. Transfection of RAW macrophages with plasmids expressing Mist_v1 and Mist_v2 splice variants (Figure IIIH and IIII in the Data Supplement) showed that Mist overexpression (6-fold increase) reduced basal levels of genes associated with inflammation and lipid metabolism (Figure 4A through 4D), most of which were upregulated by Mist knockdown (Figure 2F). Additionally, Mist overexpression partially blunted the LPS-mediated induction of inflammatory response in RAW cells (Figure VIIA through VIID in the Data Supplement). This effect was even more pronounced in primary BMDMs on Mist_v1 overexpression, which inhibited the expression of Cd36 and key inflammatory genes in response to LPS treatment (Figure 4E through 4J), as well as secretion of inflammatory cytokines IL-1β and TNF-α (Figure 4K and 4L). Additionally, ectopic overexpression of Mist inhibited ac-LDL uptake (Figure 4M and 4N), demonstrating a potential role for Mist in protecting against macrophage foam cell formation. These results collectively indicate that changes in Mist expression can influence genes associated with macrophage polarization and that Mist exhibits anti-inflammatory functions.
Figure 4.
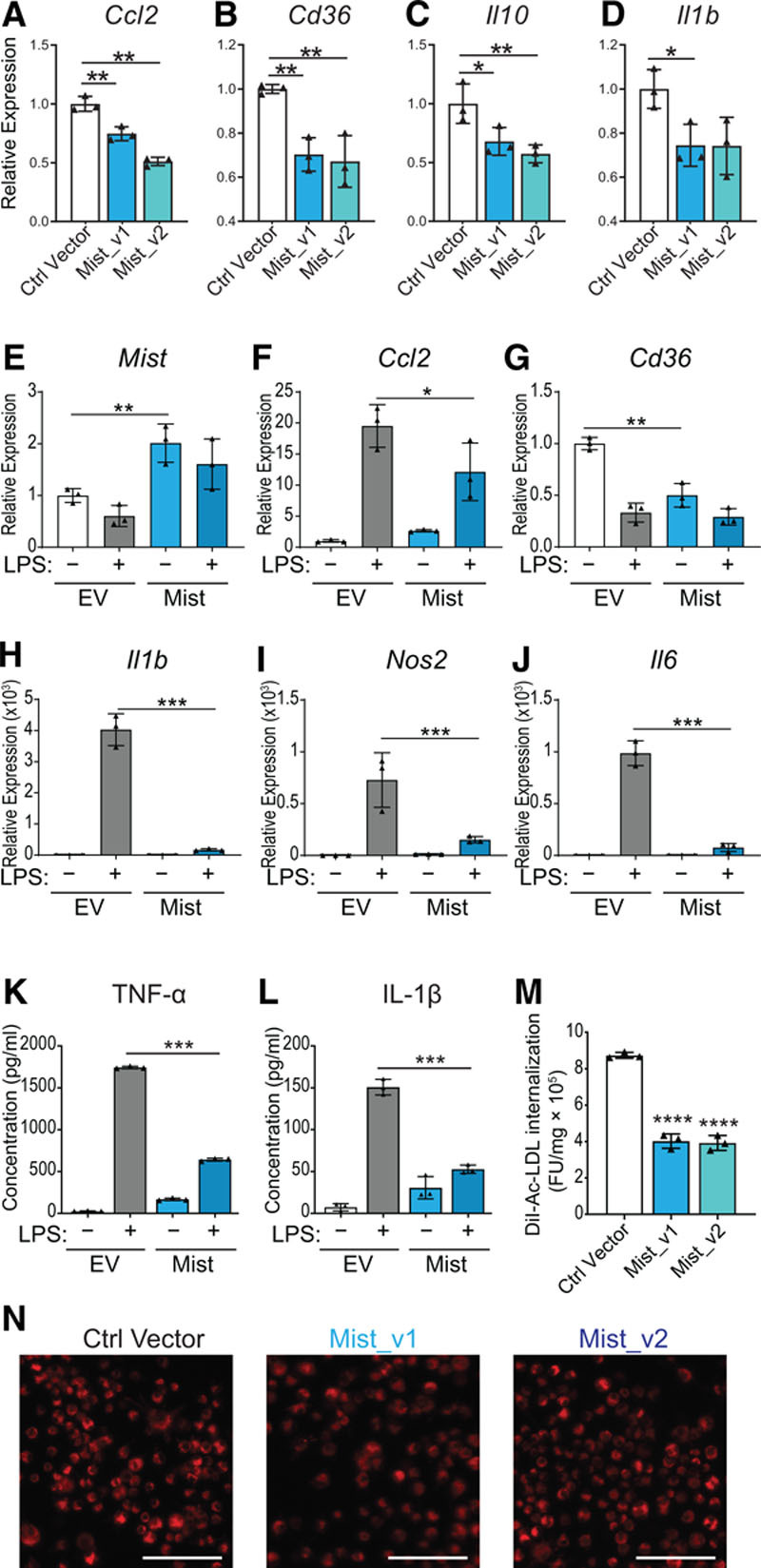
Macrophage inflammation–suppressing transcript (Mist) gain-of-function blunts inflammatory response in macrophages and decreases modified LDL (low-density lipoprotein) uptake. A–D, Quantitative polymerase chain reaction (qPCR) of indicated genes after Mist transient overexpression in RAW cells. Mist splice variants (v1 and v2) were cloned into pcDNA3.1 expression vectors (Figure IIIC through IIIE in the Data Supplement). Ctrl vector refers to Mist antisense sequence cloned into pcDNA3.1(-) vector, n=3. E–J, Mist overexpression in bone marrow–derived macrophages (BMDMs). RT-qPCR of select genes in BMDMs after transfection with pcDNA3.1_Mist_v1 construct (Mist) or empty vector control (EV). Cells were treated with or without LPS (10 ng/mL) 72 h post-transfection, and RNA collected 16 h later, n=3. K, TNF-α (tumor necrosis factor α) and (L) IL (interleukin)-1β levels, analyzed by ELISA, in supernatants from mouse BMDMs transfected with Mist overexpression vector or EV. M, Spectrofluorometric assay of RAW cells stably overexpressing Mist splice variants or Mist antisense sequence (Ctrl Vector), exposed to DiI-ac-LDL (red) for 4 h (n=3). N, Representative images of DiI-ac-LDL internalization in RAW cells. Scale bar represents 50 µm. All bar graphs represent mean values, error bars=SD. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 calculated using Fisher least significant difference test on log-transformed expression values (A–J) or Student t test (K–M).
Mist Effects Are Only Partially Mediated by Nearby Fabp5
Because lncRNAs can affect expression of nearby genes in cis, and Fabp5 was downregulated after Mist knockdown (Figure 2F and 3E), we examined whether Mist functional effects on macrophage phenotype were due to Fabp5 downregulation. Fabp5 knockdown using siRNAs in RAW cells increased expression of Il1b, Il10, and Ccl2 but had no effect on Mist and several other Mist-regulated genes (Figure VIIIA through VIIIH in the Data Supplement). These results indicate that Mist does not simply function as an epistatic regulator of Fabp5 (in cis) and can affect gene expression in trans.
Mist Interacts With PARP1 to Epigenetically Influence Inflammatory Gene Expression
LncRNAs often exert their functions via interactions with nuclear proteins and coordinately modify nucleosomes and influence chromatin structure.16 To understand how Mist might mechanistically exert its effects in macrophages, we performed RNA pulldown experiments using biotinylated Mist sense or antisense RNA (Mist-AS) probes followed by mass spectrometry to find candidate nuclear protein interactors of Mist (Figure 5A). The probes pulled down a total of 296 proteins, of which 86 had normalized spectra counts enriched >2-fold compared with Mist-AS probe (Table V in the Data Supplement). We focused on PARP1 and HNRNPA3 proteins which were highly enriched in Mist pulldown due to their reported roles in cellular function and inflammation regulation.47,48 We validated the mass spectrometry results using RNA pulldown followed by Western blot analysis with HNRNPA3 (Figure IXA in the Data Supplement) and PARP1 antibody (Figure 5B). These interactions were further assessed by native RIP followed by RT-qPCR. PARP1 showed strong interaction with Mist, while other candidate proteins failed to immunoprecipitate with Mist (Figure IXB through IXC in the Data Supplement). Additional UV crosslink RIP experiments confirmed the enrichment of Mist in PARP1 co-purified RNA (Figure 5C). Macrophage lncRNA Dnm3os was used as a control in Figure 5B and 5C, in addition to U6 and Ppia in Figure 5C.
Figure 5.
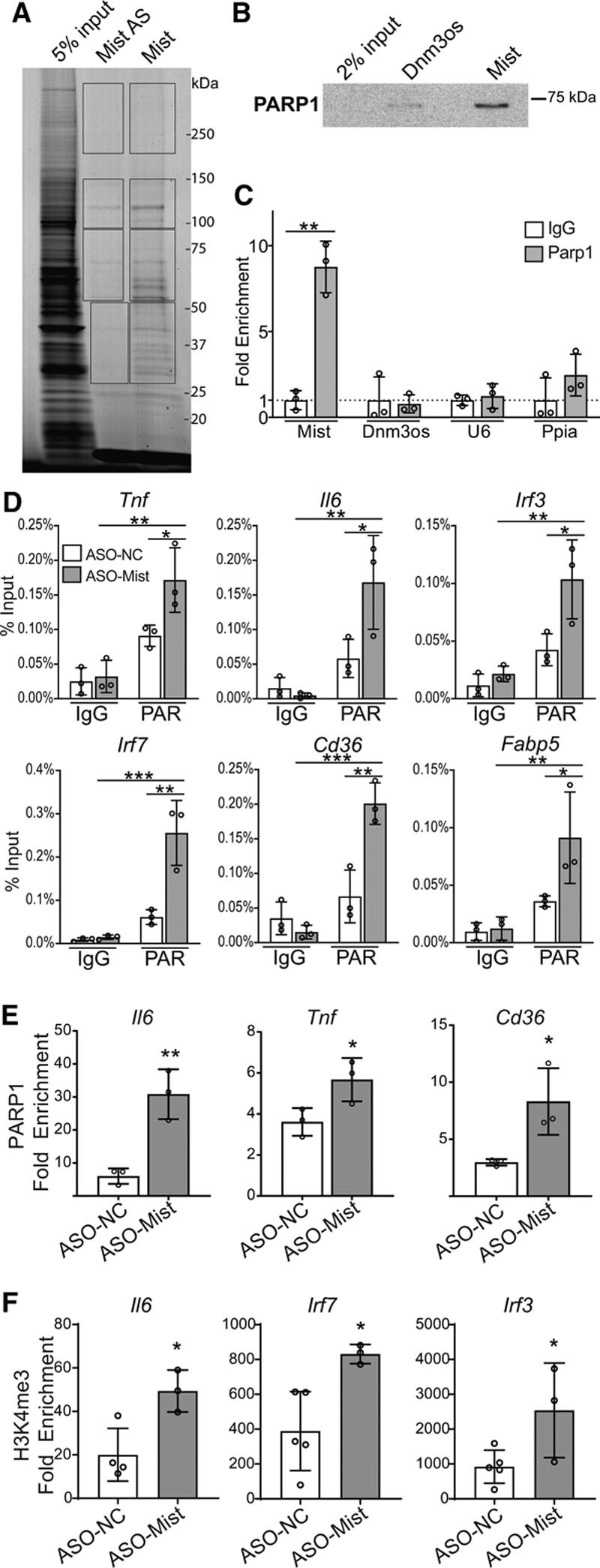
Macrophage inflammation–suppressing transcript (Mist) interacts with nuclear proteins to regulate gene expression and promoter histone poly(ADP)-ribosylation. A, RNA pulldown of Mist-interacting protein partners. Biotinylated Mist sense (Mist) and antisense control (Mist-AS) RNA transcripts were incubated with nuclear lysates from PMs, fractionated on SDS-PAGE gel and visualized with Simply Blue Safestain. Protein bands in different regions from each lane (indicated by rectangular boxes) were subjected to mass spectrometry to identify Mist-interacting proteins. B, Western blot of RNA-pulldown protein using antibodies specific for PARP1 (poly(ADP-ribose) polymerase 1). RNA-protein complexes were run on SDS-PAGE gel and probed using indicated antibodies. Dnm3os probe used as a control. C, Cross-link RNA-immunoprecipitation using PARP1 antibody or isotype control (IgG), followed by RT-qPCR (real-time quantitative polymerase chain reaction) for detection of associated RNAs. Dnm3os, U6, and Ppia used as controls. D, Chromatin immunoprecipitation (ChIP)-qPCR of poly(ADP)-ribosylation (PAR) at promoters of indicated genes (n=3). RAW 264.7 cells were treated with Mist-targeting or nontargeting ASOs. E, ChIP-qPCR for PARP1 enrichment at promoters of indicated genes. ChIP assay was performed using PAR-optimized protocol (see Methods), n=3 per group. F, ChIP-qPCR of H3K4me3 enrichment at promoters of indicated genes in RAW cells transfected with ASO-Mist (n=3) or nontargeting ASO (ASO-NC; n=5). RAW 264.7 cells were treated with Mist-targeting or nontargeting ASOs. Fold enrichment values represent enrichment relative to IgG. Bar graphs represent mean values, error bars=SD. ns P>0.05, *P<0.05, **P<0.01, and ***P<0.001 calculated using Student t test (C, E, and F) or 2-way ANOVA followed by Tukey multiple comparisons test (D).
PARP1 is a known transcriptional regulator and modifier of chromatin architecture. TNF-α-induced NF-kB transcriptional activation also involves PARP1 activity.49 Enrichment of PARP1 and histone PARylation at promoters is highly correlated with enrichment of H3K4me3, a chromatin mark associated with active transcription.50 Notably, IPA Upstream Regulator Analysis of Mist-regulated genes (Figure 2A) predicted an increase of PARP1 activity (bias-corrected z-score=2.18, P=5.57×10−11) but our microarray data showed that Mist knockdown did not affect Parp1 mRNA levels, suggesting that Mist might regulate PARP1 function. Because lncRNAs can inhibit binding of transcriptional machinery at promoters by acting as decoys,51 we hypothesized that Mist might inhibit PARP1 binding and activity at promoters of proinflammatory genes to repress their expression via epigenetic mechanisms. Indeed, ChIP assays showed that Mist knockdown significantly increased chromatin PARylation (Figure 5D), PARP1 occupancy (Figure 5E), and H3K4me3 (Figure 5F) at promoters of key Mist-responsive genes, relative to control GapmeR. Irf7, Irf3, and Fabp5 promoters showed increased PARylation on Mist knockdown (Figure 5D) but did not show increased PARP1 occupancy (Figure XA through XC in the Data Supplement), which could be due to cooperation with other chromatin marks at these promoters. Overall, these results clearly demonstrate that Mist regulates PARP1 function at chromatin in macrophages.
MIST Levels Are Inversely Correlated With Obesity and Metabolic Dysfunction in Humans
While most lncRNAs have evolutionarily conserved genomic locations, promoter sequences, and tissue-specific expression patterns, levels of sequence conservation are variable and often less significant compared with protein-coding genes.52 The predicted human MIST locus shows evolutionarily conserved regions overlapping with the aligned exons and promoter from mouse (Figure XIA in the Data Supplement). Transmap, a cross-species transcript alignment tool,53 shows 86.7% alignment of Mist-overlapping RNA Gm38335 to its syntenic region in the human genome with 67.1% conserved sequence identity. Notably, this region overlaps with 4 human ESTs, further evidence of an underlying transcript in humans (Figure XIB in the Data Supplement). Human monocyte data from ENCODE shows a similar histone signature as seen in mouse macrophages. Additionally, PhyloCSF tracks overlapping and flanking this locus show no evidence of protein-coding potential in any reading frame (Figure XIB in the Data Supplement).
We used NCBI’s Evolutionarily Conserved Regions Browser to design RT-qPCR primers for human orthologous MIST and could amplify human MIST using RNA from human THP-1 monocytes and primary human monocyte-derived macrophages. We next examined whether MIST exhibits a similar expression pattern as its mouse homolog. Indeed, LPS treatment of phorbol 12-myristate 13-acetate-differentiated THP-1 macrophages caused a significant reduction in MIST expression (Figure 6A), similar to that seen in mouse macrophages (Figure 1C). Notably, treatment with anti-inflammatory cytokine IL-4 caused an increase in MIST expression in macrophages differentiated from CD14+ monocytes from healthy donors (4.21±1.26-fold over control, n=6; P<0.01). However, we did not observe similar results in mouse macrophages.
Figure 6.
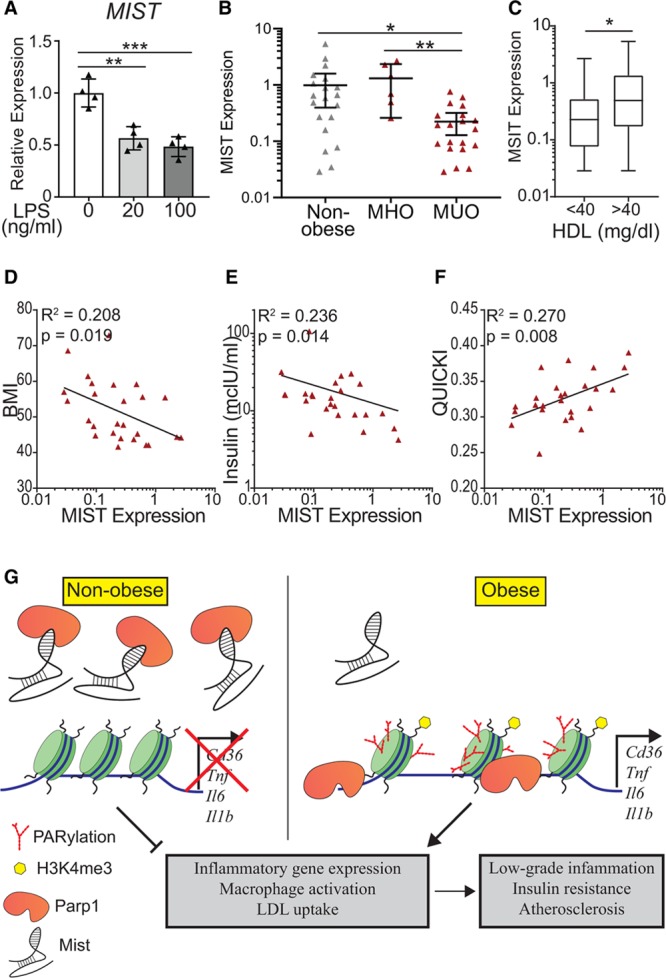
Macrophage inflammation-suppressing transcript (Mist) expression in human macrophages. A, Quantitative polymerase chain reaction (qPCR) analysis of MIST expression in THP-1 macrophages treated with LPS (lipopolysaccharide) for 3 h. n=4, bar graphs represent mean±SD. B, MIST expression in stromal vascular fraction (SVF) of adipose tissue samples collected from patients with obesity undergoing bariatric surgery and nonobese donors. SVF was isolated and RNA was extracted. qPCR was performed using primers for MIST homolog. Obese patients were stratified into metabolically healthy obese (MHO, n=6) and metabolically unhealthy obese (MUO, n=19) pools. MHO was defined by the following criteria: body mass index (BMI) >40, triglyceride <150 mg/dL, fasting glucose <101 mg/dL, fasting insulin <10 mcIU/mL, and HbA1C <5.7% (39 mmol/mol). Bar graphs represent mean±95% CI. C, MIST expression in patients binned by high-density lipoprotein (HDL) levels independent of obesity; HDL <40 mg/dL (n=21) and HDL >40 mg/dL (n=20). D–F, MIST expression within obese cohort in relationship with physiological measurements, including BMI (D), fasting insulin levels (E), and QUICKI (Quantitative Insulin-Sensitivity Check Index) measurement (F). Lines represent nonlinear semilog or log-log regression. (G) Model for Mist regulation of gene expression in macrophages and downstream effects on macrophage phenotype during obesity. Green cylinders represent nucleosomal histones, red branched marks represent PARylation, and yellow hexagons symbolize histone posttranslational modification H3K4me3. *P<0.05, **P<0.01, and ***P<0.001 calculated using 1-way ANOVA followed by Tukey multiple comparisons test (A–B) or Student t test (C) on log-transformed expression values. Gaussian distribution was verified by D’Agostino and Pearson normality test for values of log(Mist), BMI, log(Insulin), and QUICKI before all comparison and correlation tests.
We next examined whether changes in MIST expression in vivo are associated with human obesity and metabolic dysfunction. To examine relevance to human clinical obesity, insulin resistance, and metabolic syndrome, we compared human MIST RNA levels in SVF isolated from omentum of patients with obesity undergoing bariatric surgery versus nonobese donors (Table I in the Data Supplement). MIST expression tended to be lower in SVF from all subjects with obesity versus nonobese individuals (Figure XII in the Data Supplement). Furthermore, when individuals with obesity were further stratified into metabolically unhealthy obese and metabolically healthy obese groups (defined in Table I in the Data Supplement), MIST expression was significantly downregulated in metabolically unhealthy obese relative to metabolically healthy obese and nonobese individuals (Figure 6B). Additionally, patients with HDL levels <40 mg/dL showed significantly lower levels of MIST (Figure 6C). Among the obese cohort, MIST levels were also negatively correlated with individual body mass index and fasting insulin levels but positively correlated with QUICKI (Quantitative Insulin-Sensitivity Check Index) index of insulin sensitivity (Figure 6D through 6F), providing further evidence that repressed MIST expression in human ATMs correlates with increased severity of obesity and metabolic dysfunction.
Discussion
Because macrophages are important players in obesity-induced inflammation, metabolic syndrome, subsequent diabetes mellitus, and its complications, it is critical to identify the regulatory factors involved in macrophage dysfunction during obesity and insulin resistance. Here, we identify a previously unknown lncRNA, Mist, that is downregulated in multiple macrophage populations from obese insulin resistance mice and SVF from metabolically unhealthy obese humans. Mist is primarily localized in the nucleus and transcribed from a putative enhancer region based on H3K4me1 and H3K27ac enrichment but appears to affect gene expression beyond its immediate genomic neighborhood. Mist inhibits macrophage inflammatory response and foam cell formation, suggesting a protective role that is lost on metabolic dysfunction during obesity. We found that knockdown of Mist with specific GapmeR antisense oligonucleotides could enhance the expression of macrophage inflammatory genes in vitro as well as in vivo in normal mice, supporting the anti-inflammatory role of Mist under normal conditions that is lost during diet-induced obesity. This was further supported by our reciprocal data showing overexpression of Mist in vitro attenuates the inflammatory effects of LPS in cultured macrophages, including bone marrow macrophages. However, we were unable to test whether Mist overexpression has similar protective effects in vivo in HFD-fed mice because the technology for lncRNA overexpression in vivo is not yet mature.
We also identified a human ortholog of MIST that was expressed in human macrophages. Interestingly, previously published data of chromatin accessibility across various stages of hematopoietic differentiation54 shows open chromatin at the conserved MIST promoter locus in most progenitor cells, but among 6 mature differentiated populations, open chromatin is only present in monocytes (Figure XIII in the Data Supplement, red boxed area), suggesting monocyte/macrophage-type specific expression of MIST in humans. Importantly, in human omentum SVF, we found MIST expression was not only downregulated in metabolically unhealthy obese compared with metabolically healthy obese and nonobese donors but also showed a negative relationship with severity of obesity and positive correlation with markers of metabolic health and insulin sensitivity. One limitation of our study is that SVF contains a heterogeneous mix of immune and stem cells. Additionally, human MIST is unannotated in most gene databases, making it difficult to validate the specificity of MIST expression in human macrophages. However, overall, these results suggest that MIST levels could play a role in altering macrophage phenotype under obese conditions in humans. A number of human macrophage lncRNAs have recently been associated with cardiometabolic disorders,55 and we also found human orthologs of lncRNAs E330013P06 and DNM3OS in human monocytes/macrophages that were upregulated under diabetic conditions.22,23 These reports together with our current data suggest macrophage lncRNAs could be novel therapeutic targets for human metabolic disease and diabetes mellitus.
Mist affected the expression of its neighboring gene Fabp5 in a manner consistent with cis-mediated regulation56; however, Fabp5 silencing recapitulated some but not all the effects of Mist knockdown. Although our results do not preclude mechanisms involving underlying regulatory elements including enhancers at the Mist locus, our data indicate a functional role for the Mist transcript itself. Further studies are necessary to elucidate the overlap between these potential mechanistic pathways.
Interaction with histone modifiers is a well-established epigenetic mechanism for lncRNA regulation of putative target gene transcription. Our RNA-pulldown experiments identified PARP1 as a lead Mist-interacting partner. Furthermore, Mist knockdown potentiated PARP1 recruitment and PARylation of chromatin at inflammatory gene promoters. PARP1 is involved in maintenance of multiple chromatin accessibility states, including euchromatin at genomic loci of active genes.50 PARylation of histones can help maintain H3K4me3 at the transcription start site of active genes.57 We observed increased H3K4me3 at some but not all gene promoters tested, suggesting potential involvement of other histone modifications regulated by PARylation.50 Importantly, PARylation was increased at all tested Mist targets after Mist knockdown, indicating that interaction with PARP1 plays key role in gene regulation by Mist. Previous studies showed PARP1 binding to lncRNAs58; however, only recently has PARP1 interaction with an lncRNA shown to promote PARP1 recruitment and histone PARylation at target gene promoters.59 In contrast, our data shows that Mist sequesters or blocks PARP1 binding and inhibits PARylation at inflammatory gene promoters. This is reminiscent of other lncRNAs with opposing functions on chromatin modifiers, such as lncRNAs Kcnq1ot1 and ROR acting on histone methyltransferase G9a as recruiter60 or decoy,51 respectively. Although several proinflammatory macrophage lncRNAs have been reported, much less is known about anti-inflammatory lncRNAs, especially under insulin-resistant conditions. Our data show for the first time that a novel lncRNA, Mist, may restrain inflammation under normal conditions, and its downregulation during obesity can de-repress inflammatory pathways at least in part via epigenetic mechanisms and contribute to metabolic dysfunction (Figure 6G). Thus, macrophage lncRNAs such as Mist and their targets should be evaluated as potential therapeutic targets for insulin resistance/pre–diabetes mellitus, inflammatory metabolic disorders, and diabetes mellitus.
Acknowledgments
We are very grateful to Dr Dustin E. Schones and Candi Trac for epididymal adipose tissue from high-fat diet and normal diet mice and to Dr Parijat Senapati (Department of Diabetes Complications and Metabolism, Beckman Research Institute) for assistance with experimental design. Research reported in this publication included work performed in the following City of Hope Campus Cores: Integrative Genomics, DNA/RNA synthesis, and Mass Spectrometry and Proteomics (supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572), and The Light Microscopy Core. K. Stapleton conceptualized the work, designed and performed most experiments, and wrote the article. S. Das and M.A. Reddy performed experiments and edited the article. A. Leung assisted with experimental design. L. Lanting and V. Amaram performed mouse studies. Z. Chen designed the novel lncRNA identification pipeline. L. Zhang performed quantitative polymerase chain reaction for bone marrow–derived macrophage experiments. R. Palanivel and J.A. Deiuliis conducted the human stromal vascular fraction experiments. J.A. Deiuliis edited the article. R. Natarajan conceptualized the work, edited the article, and supervised the work. R. Natarajan is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Sources of Funding
This study was supported by grants from the National Institutes of Health (NIH): R01 DK065073, R01 HL106089, and R01 DK081705 (to R. Natarajan), K01 DK104993 (A. Leung), and K01 DK099475 (to J.A. Deiuliis).
Disclosures
None.
Supplementary Material
Footnotes
Nonstandard Abbreviations and Acronyms
- ac-LDL
- acetylated low-density lipoprotein
- ASO
- antisense oligonucleotide
- ATM
- adipose tissue macrophage
- BMDM
- bone marrow–derived macrophage
- ChIP
- chromatin immunoprecipitation
- DEG
- differentially expressed gene
- H3K27ac
- lysine-27 acetylation
- H3k4me3
- H3 lysine-4 monomethylation
- HFD
- high-fat diet
- HNRNPA3
- heterogeneous nuclear ribonucleoprotein A3
- IL-6
- interleukin-6
- lncRNA
- long noncoding RNA
- LPS
- lipopolysaccharide
- NF-κB
- nuclear factor-kappa B
- PARP1
- poly(ADP-ribose) polymerase 1
- PM
- peritoneal macrophage
- RIP
- RNA immunoprecipitation
- RT-qPCR
- real-time quantitative PCR
- SVF
- stromal vascular fraction
- TF
- transcription factor
- TNF-α
- tumor necrosis factor α
For Sources of Funding and Disclosures, see page 927.
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.119.313359.
References
- 1.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 4.Li SL, Reddy MA, Cai Q, Meng L, Yuan H, Lanting L, Natarajan R. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes. 2006;55:2611–2619. doi: 10.2337/db06-0164. doi: 10.2337/db06-0164. [DOI] [PubMed] [Google Scholar]
- 5.Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 6.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasu MR, Devaraj S, Jialal I. High glucose induces IL-1beta expression in human monocytes: mechanistic insights. Am J Physiol Endocrinol Metab. 2007;293:E337–E346. doi: 10.1152/ajpendo.00718.2006. doi: 10.1152/ajpendo.00718.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shanmugam N, Reddy MA, Guha M, Natarajan R. High glucose-induced expression of proinflammatory cytokine and chemokine genes in monocytic cells. Diabetes. 2003;52:1256–1264. doi: 10.2337/diabetes.52.5.1256. doi: 10.2337/diabetes.52.5.1256. [DOI] [PubMed] [Google Scholar]
- 9.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster GI, Langley KG, Berglund NA, Kammoun HL, Reibe S, Estevez E, Weir J, Mellett NA, Pernes G, Conway JRW, et al. Evidence that TLR4 is not a receptor for saturated fatty acids but mediates lipid-induced inflammation by reprogramming macrophage metabolism. Cell Metab. 2018;27:1096–1110.e5. doi: 10.1016/j.cmet.2018.03.014. doi: 10.1016/j.cmet.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Weinstock A, Brown EJ, Garabedian ML, Pena S, Sharma M, Lafaille J, Moore KJ, Fisher EA. Single-cell RNA sequencing of visceral adipose tissue leukocytes reveals that caloric restriction following obesity promotes the accumulation of a distinct macrophage population with features of phagocytic cells. Immunometabolism. 2019;1:e190008. doi: 10.20900/immunometab20190008. doi: 10.20900/immunometab20190008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 16.Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Chao T-C, Chang K-Y, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM. The long noncoding RNA THRIL regulates TNF expression through its interaction with hnRNPL. Proc Natl Acad Sci. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, Sandhu J, Casero D, Vallim TQ, Hong C, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124–128. doi: 10.1038/nature17674. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathy NW, Chen XM. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem. 2017;292:12375–12382. doi: 10.1074/jbc.R116.760884. doi: 10.1074/jbc.R116.760884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hennessy EJ, van Solingen C, Scacalossi KR, Ouimet M, Afonso MS, Prins J, Koelwyn GJ, Sharma M, Ramkhelawon B, Carpenter S, et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab. 2019;1:98–110. doi: 10.1038/s42255-018-0004-9. doi: 10.1038/s42255-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy MA, Chen Z, Park JT, Wang M, Lanting L, Zhang Q, Bhatt K, Leung A, Wu X, Putta S, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes. 2014;63:4249–4261. doi: 10.2337/db14-0298. doi: 10.2337/db14-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M, Amaram V, Ganguly R, Zhang L, Devaraj S, et al. Diabetes mellitus-induced long noncoding RNA dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler Thromb Vasc Biol. 2018;38:1806–1820. doi: 10.1161/ATVBAHA.117.310663. doi: 10.1161/ATVBAHA.117.310663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111. doi: 10.1126/science.aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersson US, Waldén TB, Carlsson PO, Jansson L, Phillipson M. Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS One. 2012;7:e46057. doi: 10.1371/journal.pone.0046057. doi: 10.1371/journal.pone.0046057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Xu J, Wang Y, Cao X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358:1051–1055. doi: 10.1126/science.aao0409. doi: 10.1126/science.aao0409. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teupser D, Thiery J, Walli AK, Seidel D. Determination of LDL- and scavenger-receptor activity in adherent and non-adherent cultured cells with a new single-step fluorometric assay. Biochim Biophys Acta. 1996;1303:193–198. doi: 10.1016/0005-2760(96)00094-x. doi: 10.1016/0005-2760(96)00094-x. [DOI] [PubMed] [Google Scholar]
- 33.Cong R, Das S, Ugrinova I, Kumar S, Mongelard F, Wong J, Bouvet P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012;40:9441–9454. doi: 10.1093/nar/gks720. doi: 10.1093/nar/gks720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beneke S, Meyer K, Holtz A, Hüttner K, Bürkle A. Chromatin composition is changed by poly(ADP-ribosyl)ation during chromatin immunoprecipitation. PLoS One. 2012;7:e32914. doi: 10.1371/journal.pone.0032914. doi: 10.1371/journal.pone.0032914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Feng Y, Hu Z, Hu X, Yuan CX, Fan Y, Zhang L. Characterization of long noncoding RNA-associated proteins by RNA-immunoprecipitation. Methods Mol Biol. 2016;1402:19–26. doi: 10.1007/978-1-4939-3378-5_3. doi: 10.1007/978-1-4939-3378-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gagliardi M, Matarazzo MR. RIP: RNA Immunoprecipitation. Methods Mol Biol. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- 38.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 39.Babaev VR, Runner RP, Fan D, Ding L, Zhang Y, Tao H, Erbay E, Görgün CZ, Fazio S, Hotamisligil GS, et al. Macrophage Mal1 deficiency suppresses atherosclerosis in low-density lipoprotein receptor-null mice by activating peroxisome proliferator-activated receptor-γ-regulated genes. Arterioscler Thromb Vasc Biol. 2011;31:1283–1290. doi: 10.1161/ATVBAHA.111.225839. doi: 10.1161/ATVBAHA.111.225839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda K, Cao H, Kono K, Gorgun CZ, Furuhashi M, Uysal KT, Cao Q, Atsumi G, Malone H, Krishnan B, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005;1:107–119. doi: 10.1016/j.cmet.2004.12.008. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 41.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015;10:e0145342. doi: 10.1371/journal.pone.0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 45.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ba X, Garg NJ. Signaling mechanism of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammatory diseases. Am J Pathol. 2011;178:946–955. doi: 10.1016/j.ajpath.2010.12.004. doi: 10.1016/j.ajpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vuong B, Hogan-Cann AD, Alano CC, Stevenson M, Chan WY, Anderson CM, Swanson RA, Kauppinen TM. NF-κB transcriptional activation by TNFα requires phospholipase C, extracellular signal-regulated kinase 2 and poly(ADP-ribose) polymerase-1. J Neuroinflammation. 2015;12:229. doi: 10.1186/s12974-015-0448-8. doi: 10.1186/s12974-015-0448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciccarone F, Zampieri M, Caiafa P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin Cell Dev Biol. 2017;63:123–134. doi: 10.1016/j.semcdb.2016.11.010. doi: 10.1016/j.semcdb.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Fan J, Xing Y, Wen X, Jia R, Ni H, He J, Ding X, Pan H, Qian G, Ge S, et al. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16:139. doi: 10.1186/s13059-015-0705-2. doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta. 2014;1840:1063–1071. doi: 10.1016/j.bbagen.2013.10.035. doi: 10.1016/j.bbagen.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 54.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, Xue C, Wang Y, Shi J, Zhang X, Li W, Nunez S, Foulkes AS, Lin J, Hinkle CC, et al. Deep RNA sequencing uncovers a repertoire of human macrophage long intergenic noncoding RNAs modulated by macrophage activation and associated with cardiometabolic diseases. J Am Heart Assoc. 2017;6:e007431. doi: 10.1161/JAHA.117.007431. doi: 10.1161/JAHA.117.007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melikishvili M, Chariker JH, Rouchka EC, Fondufe-Mittendorf YN. Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov. 2017;3:17043. doi: 10.1038/celldisc.2017.43. doi: 10.1038/celldisc.2017.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Man HSJ, Sukumar AN, Lam GC, Turgeon PJ, Yan MS, Ku KH, Dubinsky MK, Ho JJD, Wang JJ, Das S, et al. Angiogenic patterning by STEEL, an endothelial-enriched long noncoding RNA. Proc Natl Acad Sci U S A. 2018;115:2401–2406. doi: 10.1073/pnas.1715182115. doi: 10.1073/pnas.1715182115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


