Abstract
Hepatic inflammasome activation is considered a major contributor to liver fibrosis in NASH. Apoptosis signal–regulating kinase 1 (ASK1) is an apical mitogen-activated protein kinase that activates hepatic JNK and p38 to promote apoptosis, inflammation, and fibrosis. The aim of the current study was to investigate whether pharmacologic inhibition of ASK1 could attenuate hepatic fibrosis driven by inflammasome activation using gain-of-function NOD-like receptor protein 3 (Nlrp3) mutant mice. Tamoxifen-inducible Nlrp3 knock-in (Nlrp3A350V/+CreT-KI) mice and WT mice were administered either control chow diet or diet containing the selective ASK1 inhibitor GS-444217 for 6 weeks. Livers of Nlrp3-KI mice had increased inflammation, cell death, and fibrosis and increased phosphorylation of ASK1, p38, and c-Jun. GS-444217 reduced ASK1 pathway activation, liver cell death, and liver fibrosis. ASK1 inhibition resulted in a significant downregulation of genes involved in collagen production and extracellular matrix deposition, as well as in a reduced hepatic TNF-α expression. ASK1 inhibition also directly reduced LPS-induced gene expression of Collagen 1A1 (Col1a1) in hepatic stellate cells isolated from Nlrp3-KI mice. In conclusion, ASK1 inhibition reduced liver cell death and fibrosis downstream of inflammatory signaling induced by NLRP3. These data provide mechanistic insight into the antifibrotic mechanisms of ASK1 inhibition.
Keywords: Hepatology, Therapeutics
Keywords: Apoptosis pathways, Fibrosis
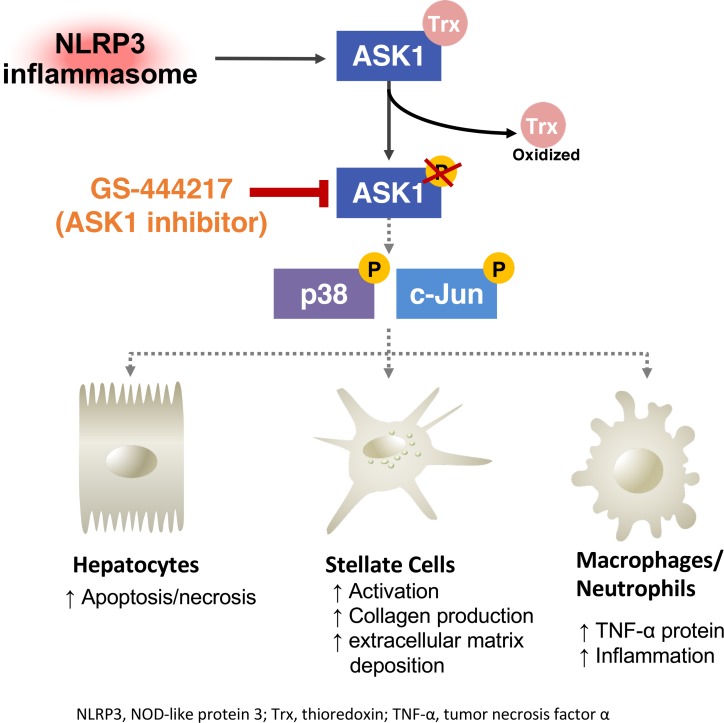
ASK1 inhibition reduced fibrosis and liver cell death downstream of inflammatory signaling in tamoxifen-inducible Nlrp3 knock-in (KI) mice.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is a multifactorial disease, characterized by a broad spectrum of symptoms ranging from simple steatosis to nonalcoholic steatohepatitis (NASH) with fibrosis, liver cirrhosis, end-stage liver disease, and hepatocellular carcinoma (HCC) (1). NAFLD has become the most common cause of chronic liver disease in the United States, and NASH is currently the leading cause of liver transplantation in women and the second leading cause for men (2–5). Because liver fibrosis is considered a strong predictor of mortality and morbidity in patients with NASH (6–8), there is an urgent medical need to identify novel therapies, which can reduce the progression of hepatic fibrosis in this growing population of patients.
Apoptosis signal–regulating kinase 1 (ASK1) is a ubiquitously expressed apical mitogen-activated kinase kinase kinase (MAP3K) that is activated by pathological stimuli that exist in human NASH liver, including oxidative and ER stress, and proinflammatory stimuli such as TNF-α and LPS (9–12). On activation, ASK1 phosphorylates MAP2K-3, -4, -6, and -7, which in turn phosphorylate and activate effector MAPKs p38 and c-Jun N-terminal kinase (JNK) to promote apoptosis, inflammatory cytokine expression, and induction of fibrogenic genes in the liver (13, 14). Emerging data have revealed that the ASK1-p38/JNK pathway is increased in livers of patients with NASH and that ASK1 plays a causal role in NASH pathogenesis in murine models (15, 16).
Our group has previously demonstrated that livers of global NOD-like receptor protein 3 (Nlrp3A350V) mutant mice develop fibrosis and severe hepatic inflammation characterized by elevated levels of TNF-α (17–19). The NLRP3 inflammasome activation plays an important role in NASH progression and is increased in livers of patients with NASH (19). Thus, Nlrp3A350V mutant mice represent a tool to study inflammasome-driven liver injury and fibrosis and to evaluate therapeutic strategies. In the present study, we evaluated the efficacy of a selective ASK1 inhibitor (GS-444217), to reduce fibrosis development in Nlrp3 mutant mice. This study aimed to investigate whether ASK1 inhibition could reduce liver cell death, inflammation, and fibrogenesis in the Nlrp3 mutant mouse liver and to identify major target cells of ASK1 inhibition and hepatic transcriptional pathways regulated by ASK1.
Results
Oral administration of GS-444217 inhibits ASK1 pathway activity in Nlrp3-KI liver.
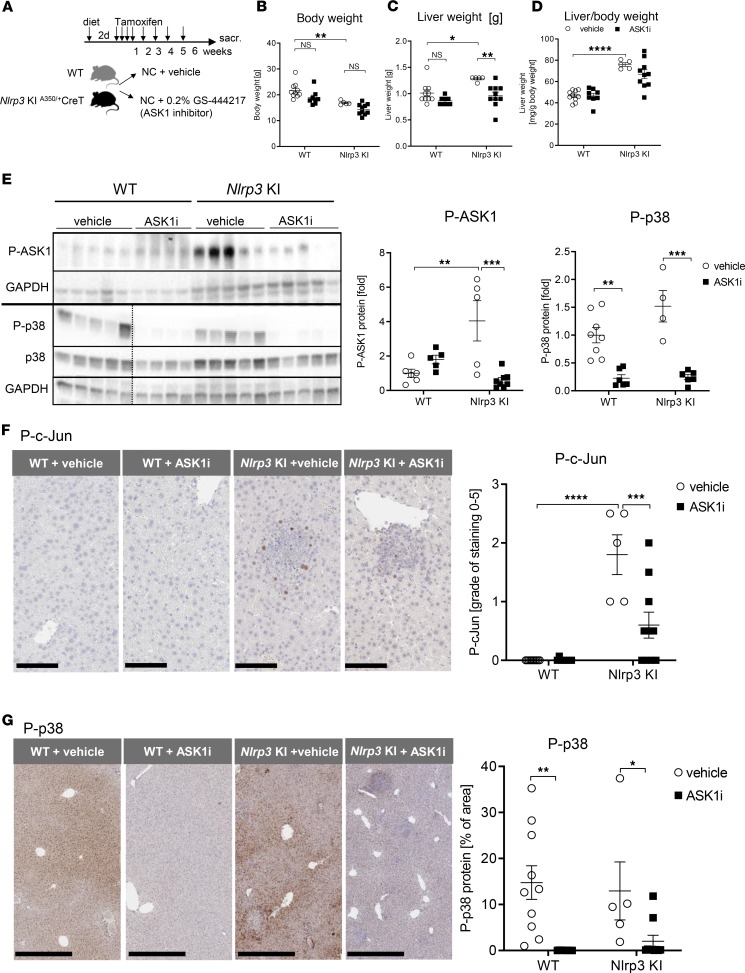
Tamoxifen-inducible Nlrp3-KI and WT mice (~8 weeks of age) were administered diet containing the ASK1 inhibitor GS-444217 (administered as a 0.2% in chow) or repelleted control chow diet, starting from 2 days before tamoxifen injection and continued for 6 weeks (study design shown in Figure 1A). Steady-state serum levels of GS-444217 in Nlrp3-KI mice after 6 weeks of continuous feeding were 7.5 ± 2.3 μM, which would be expected to effectively suppress ASK1 activity in mice (protein adjusted EC50 of GS-444217 in mice, 2.8 μM). There was no significant difference in the serum drug concentration between WT and mutant mice (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123294DS1). NLRP3 overactivation for 6 weeks resulted in significant reductions in body weight (WT 21.7 g vs. Nlrp3-KI 16.9 g, P < 0.01) (Figure 1B) and increased total liver weight (WT 1.0 ± 0.06 g vs. Nlrp3-KI 1.3 ± 0.07 g, P < 0.05) (Figure 1C) and liver/body weight ratio (WT 46.6 mg/g body weight vs. Nlrp3-KI 75.9 mg/g body weight, P < 0.001) (Figure 1D). GS-444217 significantly reduced liver weight compared with vehicle-treated mice (Figure 1, B–D). To investigate ASK1 pathway activity in Nlrp3-KI mouse liver, we analyzed the phosphorylation of ASK1 and its downstream targets p38 and c-Jun by Western blot and IHC staining. Phosphorylation of ASK1 and c-Jun were significantly increased in Nlrp3-KI liver when compared with WT (Figure 1, E–G). IHC analysis showed a significant upregulation of nuclear phosphorylated c-Jun (P–c-Jun) in Nlrp3-KI liver, which was localized to focal areas of hepatocyte injury and cell death (Figure 1F). IHC analysis of P-p38 demonstrated extensive nuclear and cytosolic staining throughout the liver in both WT and Nlrp3-KI liver that was not significantly increased in the model (Figure 1G). GS-444217 significantly reduced the phosphorylation of ASK1, p38, and c-Jun as demonstrated by Western blot (Figure 1E) and IHC staining (Figure 1, F and G). Taken together, these data demonstrate that ASK1 pathway activity is significantly elevated in the Nlrp3-KI mouse liver and is reduced by oral administration of GS-444217.
Figure 1. GS-444217 inhibits hepatic ASK1 signaling in Nlrp3-KI mice.
(A) Schematic overview of the experimental set up. Nlrp3-KI mice and WT mice were injected i.p. with tamoxifen daily for 4 days, followed by a once-weekly injection. Mice were administered control chow (NC) diet (+ vehicle) or diet containing the ASK1 inhibitor (ASK1i) GS-444217 (0.2% in chow) starting 2 days before tamoxifen injection and continuing for 6 weeks. (B–D) Body weight [g] (B), total liver weight [g] (C), and liver/body weight [mg/g body weight] (D) of WT and Nlrp3-KI mice treated with vehicle (white, WT n = 10, Nlrp3-KI n = 5) or ASK1i (black, WT n = 8, Nlrp3-KI n = 10). (E) Immunoblot analysis of liver lysates from WT and Nlrp3-KI mice treated with vehicle (white, WT n = 5, Nlrp3-KI n = 5) or ASK1i (black, WT n = 5, Nlrp3-KI n = 6) for phosphorylated ASK1 (P-ASK1) (WT + vehicle n = 6, WT + ASK1i n = 5, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 8), P-p38, and p38 (WT + vehicle n = 8, WT + ASK1i n = 8, Nlrp3-KI + vehicle n = 4, Nlrp3-KI + ASK1i n = 6). Western blot of P-ASK1 and P-p38 run on 2 different gels indicated by the black horizontal line. Therefore, 2 loading controls (GAPDH) are shown. In the lower blot, a dashed black line is used to indicate splicing of noncontiguous lanes of the same blot. Densitometric analysis was performed on background-substracted blots and was normalized on GAPDH. WT + vehicle was used as reference control and was set at 1. (F and G) Representative immunohistochemical staining of P–c-Jun (magnification, 40×; scale bar: 100 μm; WT + vehicle n = 10; WT + ASK1i n = 8; Nlrp3-KI + vehicle n = 5; Nlrp3-KI + ASK1i n = 10) (F) and P-p38 (magnification, 10×; scale bar: 500 μm; WT + vehicle n = 10; WT + ASK1i n = 8; Nlrp3-KI + vehicle n = 8; Nlrp3-KI + ASK1i n = 5) (G) on formalin-fixed paraffin-embedded liver tissue slides. Staining grade of P–c-Jun and percentage of area of P-p38 was calculated by using whole tissue slide. Treatment with ASK1i significantly reduced P–c-Jun+ and P-p38+ cells in Nlrp3 mutant mice. P–c-Jun was not detectable in WT mice (vehicle and ASK1i). Data represent mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ***P < 0.0001 (1-way ANOVA with Bonferroni post hoc test).
ASK1 inhibition moderately reduces liver inflammation score and significantly reduces TNF-α expression in Nlrp3-KI mice.
Nlrp3-KI livers developed severe hepatic inflammation with prominent inflammatory foci composed of polymorphonuclear cells, microabscesses, and liver cell death (Figure 2A). Hepatic neutrophilic infiltration was significantly elevated in Nlrp3-KI liver, as demonstrated by IHC staining for Myeloperoxidase (MPO), a specific component of neutrophil azurophilic granules (Figure 2B) (20). ASK1 inhibition in Nlrp3-KI mice caused a moderate, nonsignificant reduction of hepatic inflammation, as assessed by pathology read (P = 0.05 vs. Nlrp3-KI + vehicle) (Figure 2A), MPO+ staining (Nlrp3-KI + vehicle 0.52% of area vs. Nlrp3-KI + ASK1 inhibitor [ASK1i] 0.34% of area, P = 0.23) (Figure 2B), or Mpo mRNA levels (Nlrp3-KI + vehicle 238.8-fold vs. Nlrp3-KI + ASK1i 145.4-fold, P = 0.08) (Figure 2C). We further investigated the effect of ASK1 inhibition on hepatic inflammatory signaling by analyzing protein expression of TNF-α (Figure 2D). TNF-α is produced by activated macrophages and neutrophils (21) and plays a central role in NLRP3 inflammasome–induced liver inflammation and fibrosis (18). Nlrp3-KI mice had a 200-fold increase in hepatic expression of the TNF-α soluble homotrimer (52 kDa) and 3.8-fold increase in its precursor form (transmembrane TNF-α), which is expressed as a cell surface type II polypeptide (26 kDa) (both P < 0.0001 vs. WT) (Figure 2D). ASK1 inhibition significantly reduced expression of both the soluble homotrimer and membrane-bound TNF-α (P < 0.0001 vs. Nlrp3-KI + vehicle).
Figure 2. ASK1 inhibition reduced hepatic TNF-α protein expression and moderately improved inflammation score.
(A) H&E staining of liver sections (thickness 5 μm) of WT and Nlrp3-KI treated with vehicle and GS-444217 (magnification, 10×; scale bar: 250 μm) (WT + vehicle n = 10, WT + ASK1i n = 7, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). Arrows point to areas with inflammatory cell infiltration. Nlrp3 mutant mice treated with vehicle showed severe liver inflammation, while mice that received the ASK1i showed mild improvements in the grade of inflammation (P = 0.05 vs. Nlrp3-KI + vehicle; Mann-Whitney U test). (B) As a marker for neutrophil cell infiltration, we used Myeloperoxidase (MPO), which was significantly increased in Nlrp3 mutant mice (+ vehicle) and was reduced by GS-444217 (not significant) (magnification, 20×; scale bar: 100 μm) (WT + vehicle n = 2, WT + ASK1i n = 4, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). (C) The same trend was observed on Mpo gene expression level (P = 0.08 vs. Nlrp3-KI + vehicle) (WT + vehicle n = 5, WT + ASK1i n = 4, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). (D) Immunoblot analysis of liver lysates of WT and Nlrp3-KI mice treated with vehicle and ASK1i for detecting the active homotrimer (52 kDa) and the membrane-bound (26 kD) TNF-α (WT + vehicle n = 7, WT + ASK1i n = 7, Nlrp3-KI + vehicle n = 4, Nlrp3-KI + ASK1i n = 6). Data were normalized on GAPDH and are expressed as the mean ± SEM. WT + vehicle was set at 1. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (1-way ANOVA with Bonferroni post hoc test or otherwise stated).
ASK1 inhibition reduces hepatocellular death in Nlrp3-KI mice.
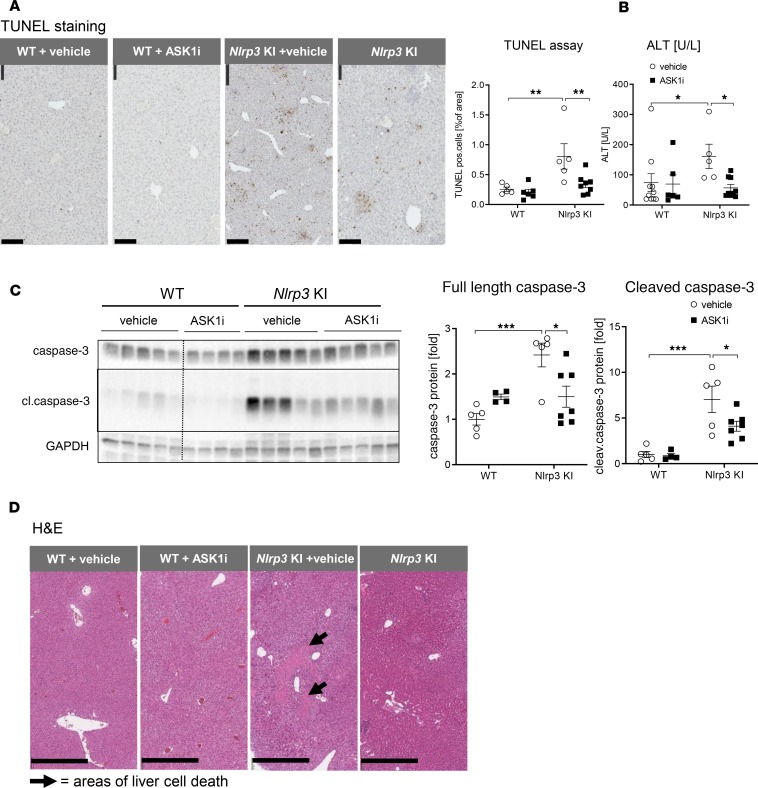
Hepatocellular cell death represents a major pathogenic event in NASH and is considered a key driver in the transition from steatosis to steatohepatitis (22–25). Nlrp3 mutant mice had a significant increase in hepatocellular death, as demonstrated by increased TUNEL+ cells (increased from 0.3% of area in WT to 0.8% of area in Nlrp3-KI, P < 0.01) (Figure 3A), increased serum levels of ALT (increased from 74.1 ± 29.8 U/L in WT to 161.3 ± 40.1 U/L in Nlrp3-KI, P < 0.05) (Figure 3B), increased cleavage of hepatic caspase-3 (increased by 7.0-fold in Nlrp3-KI compared with WT, P < 0.001) (Figure 3C), and atypical morphological H&E staining (Figure 3D). TUNEL+ staining in Nlrp3-KI livers was identified predominately in the nucleus of hepatocytes in areas of inflammatory foci (Figure 3A). ASK1 inhibition in Nlrp3-KI mice markedly reduced hepatocellular death, as demonstrated by reduced TUNEL staining (0.3% of area in Nlrp3-KI + ASK1i vs. 0.8% of area in Nlrp3-KI, P < 0.01) (Figure 3A), reduced serum ALT (56.9 U/L in Nlrp3-KI + ASK1i vs. 161.3 U/L in Nlrp3-KI, P < 0.05) (Figure 3B), and reduced caspase-3 cleavage (4.1-fold in Nlrp3-KI + ASK1i vs. 7.0-fold in Nlrp3-KI, P < 0.05) (Figure 3C), as well as H&E staining (Figure 3D).
Figure 3. ASK1 inhibition significantly reduced liver injury and cell death in Nlrp3-KI mice.
(A) TUNEL staining showed increased percentage of TUNEL+ cells in Nlrp3-KI mice, and this was ameliorated by GS-444217 (magnification, 20×; scale bar: 100 μm) (WT + vehicle n = 5, WT + ASK1i n = 6, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 8). (B)Nlrp3 mutant mice showed higher levels of ALT than WT + vehicle, indicating increased liver cell death, which was reduced by GS-444217. (WT + vehicle n = 10, WT + ASK1i n = 6, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). (C) Immunoblot analysis of liver lysates of WT and Nlrp3-KI mice treated with vehicle and ASK1i for caspase-3 and its active cleaved form (cl–caspase-3). Data were normalized on GAPDH, and WT + vehicle was set at 1 (WT + vehicle n = 5, WT + ASK1i n = 4, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 7). A dashed vertical black line is used to indicate splicing of noncontiguous lanes of the same blot. These experiments were performed at the same time and on the same gels as those shown in Figure 1. (D) H&E staining of liver sections with arrows pointing on areas with liver cell death (magnification, 10×; scale bar: 500 μm). *P < 0.05; **P < 0.01; ***P < 0.001 (1-way ANOVA with Bonferroni post hoc test unless otherwise stated).
ASK1 inhibition reduced liver fibrosis in Nlrp3-KI mice.
Nlrp3-KI mice developed significant liver fibrosis as demonstrated by increased collagen deposition measured by Picrosirius red (PSR) staining (increased from 0.6% of area in WT to 2.0% of area in Nlrp3-KI, P < 0.001) (Figure 4A) and by hepatic hydroxyproline (Hyp) levels (increased from 272.0 ± 10.0 μg Hyp/liver in WT to 510.1 ± 42.3 μg Hyp/liver in Nlrp3-KI, P < 0.0001) (Figure 4D). These changes were accompanied by increased markers of activated HSCs. α–Smooth muscle actin+ (α-SMA+) cells increased from 0.6% of area in WT to 6.6% of area in Nlrp3-KI (P < 0.001) (Figure 4B) and Desmin protein amount increased by 2.5-fold in Nlrp3-KI compared with WT (P < 0.001) (Figure 4C). Collectively, these data demonstrate that NLRP3 hyperactivation promotes significant liver fibrosis, consistent with previous observations in the model. ASK1 inhibition markedly reduced liver fibrosis, as shown by reduced percentage of area of PSR staining (reduced from 2.0% in Nlrp3-KI + vehicle vs. 1.2% in Nlrp3-KI + ASK1i, P < 0.05), reduced α-SMA+ staining (6.6% in Nlrp3-KI + vehicle vs. 1.6% in Nlrp3-KI + ASK1i, P < 0.001) and reduced hepatic Desmin protein (2.5-fold in Nlrp3-KI + vehicle vs. 0.8-fold, in Nlrp3-KI + ASK1i, P < 0.01) and total Hyp level in the liver (Hyp conc. 510.1 μg Hyp/liver in Nlrp3-KI + vehicle vs. 353.2 μg Hyp/liver in Nlrp3-KI + ASK1i, P < 0.01) (Figure 4, A–D).
Figure 4. ASK1 inhibition reduced hepatic fibrosis in Nlrp3-KI mice.
(A and B) We analyzed the antifibrotic effect of ASK1i by measuring Picrosirius Red (PSR) (A) (magnification, 20×; scale bar: 100 μm) (WT + vehicle n = 8, WT + ASK1i n = 7, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 6) and α-SMA in paraffin-embedded liver sections of WT and Nlrp3-KI mice (B) (magnification 20×, scale bar 100 μm) (WT + vehicle n = 4, WT + ASK1i n = 5, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). Nlrp3-KI mice treated with vehicle had significant hepatic fibrosis, which was ameliorated by GS-444217. (C) Immunoblot analysis of Desmin, a marker for hepatic stellate cell activation, showed reduced Desmin level in Nlrp3-KI mice treated with GS-444217. Data were normalized on GAPDH, and WT + vehicle was set at 1 (WT + vehicle n = 5, WT + ASK1i n = 4, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 7). A dashed vertical black line is used to indicate splicing of noncontiguous lanes of the same blot. These experiments were performed at the same time and on the same gels as those shown in Figure 1. (D) To analyze collagen production in the liver, we measured total hydroxyproline level [μg/ liver] in WT and Nlrp3-KI mice treated with vehicle and GS-444217. ASK1 inhibition significantly reduced liver hydroxyproline content in Nlrp3-KI. Data are expressed as the mean ± SEM (WT + vehicle n = 10, WT + ASK1i n = 8, Nlrp3-KI + vehicle n = 5, Nlrp3-KI + ASK1i n = 9). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (1-way ANOVA with Bonferroni post hoc test).
Effects of ASK1 inhibition in isolated primary liver cells from Nlrp3-KI mice.
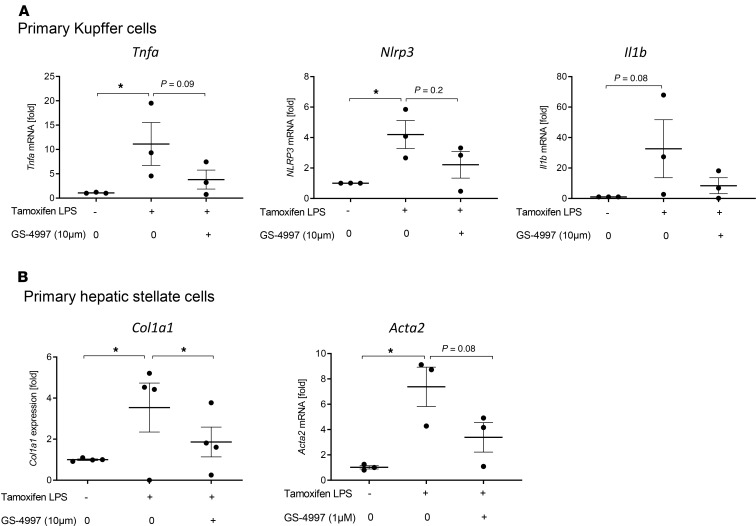
To identify downstream signaling pathways and major target cells of ASK1 inhibition, we isolated primary Kupffer cells (Figure 5A), hepatic stellate cells (Figure 5B), and hepatocytes from Nlrp3-KI mice. Because activation of the inflammasome pathway requires both upregulation of NLRP3 protein expression followed by inflammasome assembly and activation, we treated cells with LPS, to induce Nlrp3 gene expression, and with tamoxifen, to activate the Nlrp3 mutation and the inflammasome pathway. Cells were treated with tamoxifen and LPS for 24 hours with or without preincubation of the ASK1i GS-4997 (selonsertib). Activation of the Nlrp3 mutation by tamoxifen and LPS led to increased expression of proinflammatory genes (Tnfa, Nlrp3, Il1b) in Kupffer cells (Figure 5A) and profibrotic genes (Col1a1, Acta2) in hepatic stellate cells (Figure 5B). We found the most dominant effects of ASK1 inhibition on isolated hepatic stellate cells where it downregulated Col1a1 and Acta2 gene expression (Figure 5B). Treatment of KC with the ASK1i reduced expression of Tnfa and Nlrp3, though it did not reach significance (Figure 5A). We also found no significant changes in gene expression in isolated hepatocytes (data not shown). The significant effect of ASK1 inhibition to reduce collagen production in isolated hepatic stellate cells suggest that ASK1 may play a fibrogenic role through promoting hepatic collagen deposition in response to inflammasome activation
Figure 5. Effects of ASK1 inhibition in isolated primary liver cells from Nlrp3-KI mice.
(A and B) Isolated primary Kupffer cells (A) and hepatic stellate cells (B) from Nlrp3-KI mice were treated with tamoxifen and LPS for 24 hours with our without preincubation of the ASK1 inhibitor GS-4997 (1 μM, 10 μM) and gene expression of Tnfa, Nlrp3, and Il1b in Kupffer cells and Col1a1 and Acta2 in hepatic stellate cells were analyzed. Data are expressed as the mean ± SEM of 3–4 individual mice and were normalized to WT. *P < 0.05 (1-way ANOVA with Bonferroni post hoc test).
Identification of differentially expressed genes in livers of ASK1i-treated Nlrp3-KI mice via RNA sequencing.
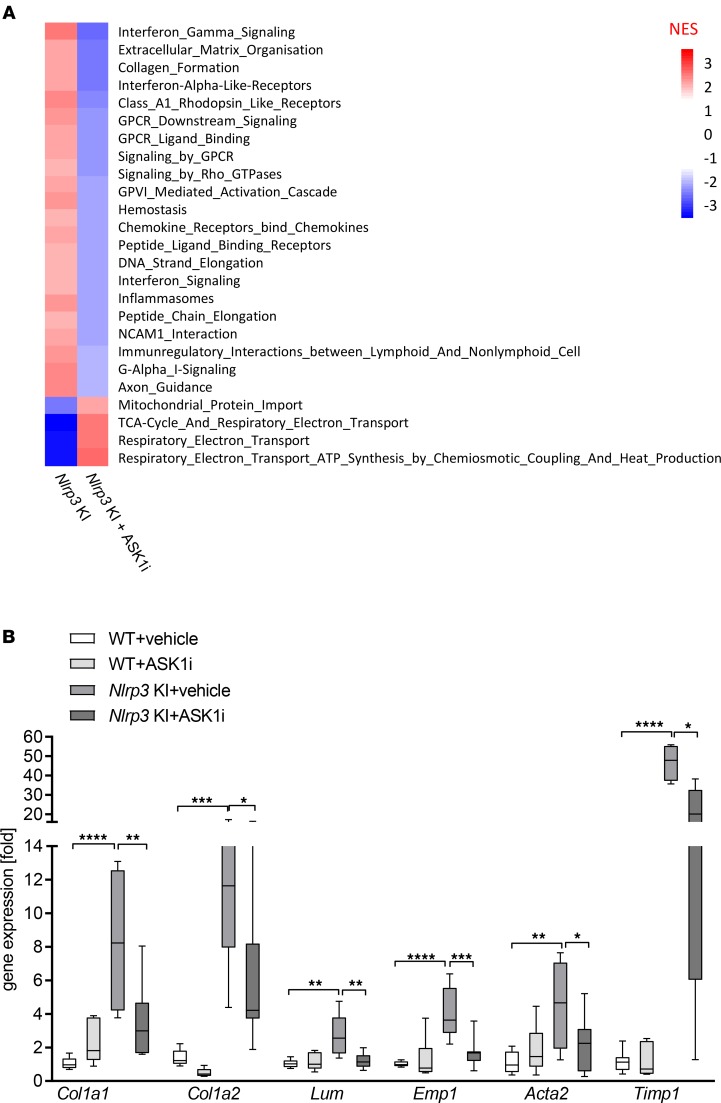
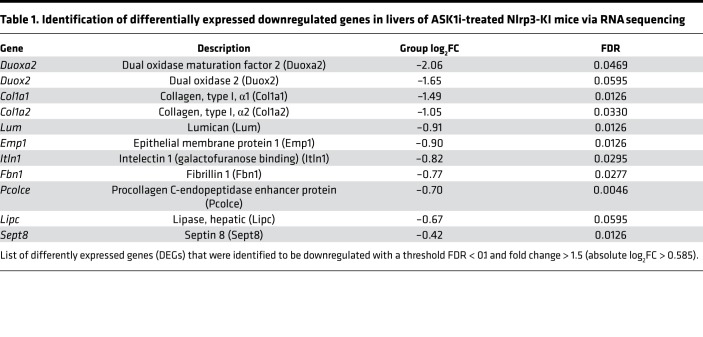
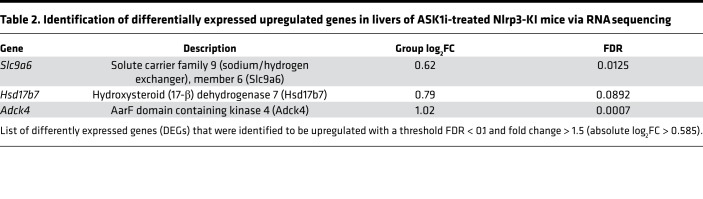
To characterize hepatic transcriptional signatures affected by ASK1 inhibition, we performed RNA sequencing (RNA-seq) followed by gene set enrichment analysis (GSEA). Comparison of Nlrp3-KI + ASK1i vs. Nlrp3-KI + vehicle revealed that ASK1 inhibition caused a significant downregulation of multiple gene sets involved in extracellular matrix (ECM) organization and collagen production (Figure 6A and Table 1). Gene signatures that were upregulated by ASK1 inhibition included those involving metabolic processes such as mitochondrial protein import and tricarboxylic acid (TCA) cycle and electron transport chain regulation (Figure 6A and Table 2). Differentially expressed genes (DEGs) were classified as those being significantly modulated by Nlrp3-KI and reversed by ASK1 inhibition. A total of 14 DEGs (11 down- and 3 upregulated) were identified in livers from Nlrp3-KI + ASK1i–treated mice compared with vehicle only. The list of down- and upregulated genes are presented in Tables 1 and 2. ASK1 inhibition significantly reduced hepatic expression of profibrogenic genes including Col1a1, Col1a2, lumican (Lum), epithelial membrane protein (Emp1), procollagen C-endopeptidase enhancer (Pcolce), and fibrillin1 (Fbn1). Real-time PCR was also used to quantify 4 DEGs (Col1a1, Col1a2, Lumican, Emp1) identified by RNA-seq (Figure 6B) and on additional, well-characterized fibrosis markers (Acta2 and Timp1). All of the hepatic fibrogenic genes measured were increased in Nlrp3-KI mice and were significantly reduced by ASK1 inhibition (Figure 6B). Gene ontology (GO) analysis for biological processes, molecular function, and cell compartment is given in Supplemental Figure 2.
Figure 6. ASK1 inhibition significantly reduced hepatic expression of fibrosis genes.
(A) Gene set enrichment analysis in livers of ASK1i-treated Nlrp3 mutant mice compared with vehicle-treated Nlrp3 mutant mice. Heatmap of normalized enrichment score (NES) are shown for significantly enriched REACTOME pathways (absolute NES > 2 and FDR < 0.05). (B) RT-PCR analysis of selected DEGs to validate RNA sequencing data. Col1a1, Col1a2, Lum, and Emp1 were positively validated and showed, together with Acta2 and Timp1, a downregulation in ASK1i-treated Nlrp3 mutant mice. Data are expressed as the mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (1-way ANOVA with Bonferroni post hoc test).
Table 1. Identification of differentially expressed downregulated genes in livers of ASK1i-treated Nlrp3-KI mice via RNA sequencing.
Table 2. Identification of differentially expressed upregulated genes in livers of ASK1i-treated Nlrp3-KI mice via RNA sequencing.
Discussion
In the present study, we investigated whether pharmacological inhibition of ASK1 could reduce the progression of hepatic fibrosis in Nlrp3 mutant mice. Our data demonstrate that GS-444217 suppressed hepatic ASK1 activity, resulting in significantly reduced liver injury, hepatocyte cell death, and fibrosis with a moderate (nonsignificant) reduction in lobular inflammation. Transcriptomic analysis revealed that ASK1 inhibition significantly reduced hepatic expression of fibrogenic genes involved in collagen synthesis and matrix deposition, while in vitro analysis demonstrated that ASK1 inhibition suppressed Col1a1 expression in LPS-treated hepatic stellate cells isolated from Nlrp3-KI mice. Taken together, these data demonstrate a causal role for ASK1 in promoting liver fibrosis and suggest that ASK1 mediates hepatic stellate cell activation downstream of NLRP3 activation.
Experimental and clinical NASH are associated with increased expression/activity of hepatic NLRP3 inflammasome components (17, 19, 26, 27). Mice deficient in Nlrp3 or its essential components (Asc−/− and Casp1−/−) are significantly protected against high-fat diet–induced NAFLD. TNF-α is a crucial mediator of liver inflammation and fibrosis in Nlrp3-KI mice (18) and also acts as an upstream activator of ASK1 (12, 28, 29); therefore, we hypothesized that ASK1 may play a causal role in the progression of liver fibrosis in Nlrp3 mutant mice. Nlrp3 mutant mice had increased hepatic ASK1 phosphorylation, indicative of increased ASK1 kinase activity. ASK1 inhibition significantly attenuated NLRP3-induced liver injury and reduced cleavage of caspase-3, in agreement with previous reports that ASK1 mediates apoptotic and necrotic cell death (12, 28, 30).
Histopathological and transcriptome analysis revealed a major therapeutic benefit of ASK1 inhibition on regulation of hepatic fibrogenesis. Both p38 and JNK can promote fibrogenic gene expression through phosphorylating nuclear transcription factors ATF2 and c-Jun (31–35), respectively. In mouse models of NAFLD and liver fibrosis, JNK promotes production of proinflammatory cytokines such as TNF-α; IL-1β; IL‑6; chemokines such as CCL2, CCL3, and CCL4; and profibrotic mediators such as TGF‑β (36, 37). In hepatic stellate cells, JNK signaling has been shown to be required for differentiation into myofibroblasts (38). Here, ASK1 inhibition significantly reduced collagen deposition and reduced accumulation of Hyp. In addition, ASK1 inhibition reduced markers of activated hepatic myofibroblasts, including α-SMA and Desmin. Transcriptomic analysis demonstrated that ASK1 inhibition was associated with downregulation of fibrogenic genes involved in collagen synthesis (including Col1a1, Col1a2, and Pcolce) and ECM deposition (including Lum, Emp1, and Fbn1). GO analysis revealed that major hepatic transcriptional signatures downregulated by ASK1 inhibition included those involved in collagen fibril assembly, matrix adhesion, and ECM organization. Furthermore, studies in isolated primary liver cells from Nlrp3-KI mice demonstrated a prominent role for ASK1 in promoting Col1a1 expression in hepatic stellate cells. Collectively, these data suggest that ASK1 plays a pathological role in promoting hepatic fibrogenesis induced by NLPR3 inflammasome activation.
In contrast to the effects on liver fibrosis in Nlrp3-KI mice, ASK1 inhibition conferred only modest (and nonsignificant) improvements in liver inflammation score, although there was a significant reduction in TNF-α gene and protein expression. The reduction of TNF-α suggests an effect on resident immune cells (Kupffer cells) without affecting infiltration of circulating leukocytes and changing lobular inflammation score. TNF-α is a characteristic feature of hepatic inflammation; however, when this was reduced to baseline with ASK1i, there was no effect on lobular inflammation. Thus, ASK1i in combination with an agent that blocks infiltrating immune cells could be an appropriate combination approach. Our data also provide insights into biomarkers of hepatic fibrogenesis and fibrosis that can be impacted by therapeutic intervention. For example, clinical studies recently demonstrated that lumican, downregulated by ASK1 inhibition in the current study, is a promising noninvasive biomarker for an early profibrotic state in patients with NAFLD (39), as its circulating level correlates with hepatic collagen fractional synthesis rate (40).
In summary, we report a therapeutic effect of the ASK1i GS-444217 to reduce liver cell death and progressive liver fibrosis in an Nlrp3-KI model of liver injury. Together, these data demonstrate a causal role for ASK1 activity in liver fibrosis progression and support ASK1 inhibition as a therapeutic target. Recently, 2 randomized, double-blind, placebo-controlled clinical trials were completed that evaluated the safety and efficacy of the ASK1i selonsertib in patients with either bridging fibrosis (F3) or compensated cirrhosis (F4) due to NASH. Unfortunately, topline data from both of these trials indicate that the primary outcome measure of a ≥ 1-stage improvement in fibrosis within 48 weeks was not achieved. Therefore, despite our preclinical findings in the current study demonstrating that ASK1 inhibition can reduce fibrosis progression in the Nlrp3-KI model of liver injury, ASK1 inhibition alone was not sufficient to reverse liver fibrosis in patients with advanced NASH.
Several potential explanations for these negative trial results are possible, and detailed interpretation awaits publication of the clinical data. However, there are various possible reasons why the antifibrotic efficacy observed in the Nlrp3-KI model did not translate into clinical efficacy. Firstly, NASH is a complex, multifactorial disease, and liver fibrosis in NASH liver is likely driven by multiple distinct mechanisms, including hepatic lipotoxicity, inflammation, and fibrogenesis. The preclinical model employed in the current study, induced through inflammasome-driven liver injury, does not fully recapitulate all of the metabolic drivers that exist in human NASH liver. Secondly, the phase III clinical trials evaluated whether ASK1 inhibition could regress fibrosis in patients with advanced NASH with bridging fibrosis and cirrhosis, while the current model evaluated whether ASK1 inhibition could reduce fibrosis progression. Therefore, another possibility is that the extent of preexisting fibrosis in the NASH patients may have been too advanced and, thus, not amenable to regression by treatment with selonsertib alone over a 48-week treatment period. Finally, it may be that a combination strategy that targets multiple distinct disease drivers may be required to regress liver fibrosis in patients with advanced NASH. In our current study, the beneficial effect of ASK1 inhibition to prevent NLRP3-driven fibrosis appears to have been mediated predominately through reduced cell death and fibrogenesis, with only moderate effects observed on lobular inflammation. Based on these data, it may be hypothesized that ASK1 inhibition used in combination with a direct antiinflammatory mechanism may confer superior efficacy to reverse established fibrosis. Emergent preclinical data has demonstrated that the antifibrotic efficacy of ASK1 inhibition in preclinical models of diet-induced NASH is significantly improved when administered in combination with other therapeutic agents with distinct mechanism of actions including Farnesoid X receptor (FXR) agonism and Acetyl CoA carboxylase (ACC) inhibition (41, 42). An ongoing study (NCT03449446) is currently evaluating the safety and efficacy of various combinations of NASH drugs, including selonsertib, in patients with advanced fibrosis due to NASH, which will address this hypothesis clinically.
In conclusion, NASH remains a highly debilitating disease that is increasing globally, and there is a significant unmet medical need to develop novel therapeutic strategies. The data herein demonstrate a causal role for ASK1 activity in promoting hepatic fibrogenesis induced by NLRP3 inflammasome activation and provide mechanistic insight into the fibrogenic pathways downstream of ASK1. However, while these findings support ASK1 as a target for liver fibrosis, additional basic and clinical research is required to translate these findings to develop a therapeutic regime that can promote regression of established liver fibrosis in NASH.
Methods
Mouse strains.
We used Nlrp3A350V/+CreT knock-in mice (Nlrp3-KI) that were generated as previously described, with an alanine 350 to valine (A350V) substitution and the presence of an intronic floxed neomycin resistance cassette, in which expression of the mutation does not occur unless the Nlrp3 mutants are first bred with mice expressing Cre recombinase (43). Nlrp3-KI mice were bred to B6.Cg-Tg (Cre/Esr1)5Amc/J mice (obtained from the Jackson Laboratory) to allow for mutant Nlrp3 expression in adult models after administration of tamoxifen (44).
Temporal induction of mutant Nlrp3 expression.
For the temporal induction of mutant Nlrp3 expression, Nlrp3-KI mice and WT mice were injected i.p. with 50 mg/kg tamoxifen-free base (MP Biomedicals) in 90 % sunflower seed oil from Helianthus annus (MilliporeSigma) and 10 % ethanol daily for 4 days, followed by a once-weekly injection as previously described (ref. 45 and Figure 1A).
ASK1i treatment.
Eight- to 12-week-old mice were administered control chow diet (vehicle) (n = 10 WT + vehicle, n = 5 Nlrp3-KI + vehicle) or diet containing the ASK1i GS-444217 (0.2% in chow) (n = 8 WT + ASK1i, n = 9 Nlrp3-KI + ASK1i) starting 2 days before tamoxifen injection and continuing for 6 weeks (Figure 1A).
Liver sample preparation.
Nlrp3 mutant (Nlrp3-KI) and WT mice were sacrificed after 6 weeks of ASK1i treatment. Blood samples (~0.2 mL) were obtained by heart puncture. Liver tissue was harvested and representative pieces were either (a) fixed in 10% formalin for 24 hours, (b) embedded in OCT on n-Butan nitrogen and then frozen at –80°C, (c) placed in 0.5 mL RNAlater Solution (Invitrogen), or (d) snap-frozen in liquid nitrogen and stored at –80°C.
Isolation of primary liver cells.
Simultaneous cell isolation of liver cell populations has been described elsewhere (46). For isolation of hepatic stellate cells, 18- to 24-week-old female or male mice were used. Briefly, mice were anesthetized by ketamine/xylazine injection and perfused in situ through the inferior vena cava with sequential Pronase E (0.4 mg/mL, MilliporeSigma) and Collagenase D (0.8 mg/mL, MilliporeSigma) solutions. Liver was removed and digested in vitro with Collagenase D (0.5 mg/mL), Pronase E (0.5 mg/mL) and DNAse I (0.02 mg/mL, MilliporeSigma). After 20 minutes, tissue was filtered through a 70-μm mesh. Cells were separated using a Nycodenz gradient centrifugation. The HSC/hepatocytes (DMEM 10% FCS 1% penicillin and streptomycin [P/S]) or Kuppfer cells (RPMI 10% FCS 1% P/S, Thermo Fisher Scientific) were seeded into plastic tissue culture flasks and incubated at 37°C with 5% CO2 overnight. The next morning, the culture medium was changed and cells were treated with GS-4997 thirty minutes before inflammasome activation with 4-OH tamoxifen (MilliporeSigma) plus LPS (1 μg/mL MilliporeSigma) for 24 hours.
Liver histology and immunostaining.
Livers were sliced in 5-μm sections and were routinely stained for H&E. H&E staining was used to score the grade of liver inflammation and necrosis regarding the NAFLD activity score (47) and the amount of necrotic lesions. Liver fibrosis was assessed with PSR staining and was quantitated by digital image analysis (ImageJ, NIH). For PSR staining, liver sections were incubated for 30 minutes at room temperature with an aqueous solution of saturated picric acid containing 0.1% Fast Green FCF and 0.1% Direct Red (MilliporeSigma). To study liver cell death, TUNEL assay was performed using manufacturer’s instructions (ApopTag Peroxidase In Situ Apoptosis Detection Kit, MilliporeSigma) and quantitated by ImageJ using the plugin color deconvolution as described in ref. 18. Percentage of area was calculated from gray-scaled pictures. IHC staining for α-SMA (1:250, Abcam, catalog 124964) and MPO (1:200, Thermo Fisher Scientific, catalog RB-373) was performed on formalin-fixed, paraffin-embedded livers according to manufacturer’s instruction and counterstained with Mayer’s Hematoxylin solution (Sigma-Aldrich). IHC analysis of P-p38 (P-p38 Thr180/Tyr182, Cell Signaling Technology, catalog 4631, Danvers, MA, USA), and P–c-Jun (P–c-Jun Ser63, Cell Signaling Technology, catalog 2361) was performed on the Discovery Ultra autostainer (Ventana Medical Systems, Roche) using HQ detection and the DAB chromogen kit (BD Biosciences). Liver slices were incubated with the primary antibody for 40 minutes at room temperature and then incubated with the secondary antibody, anti–rabbit HQ (catalog 760-4815, Roche), for 30 minutes, followed by detection with DAB chromogen for 5 minutes. The slides were then counterstained with CAT hematoxylin and mounted with EcoMount media (BioCare Medical). Whole slide scans were acquired using the AT2 Leica Slide Scanner.
Hepatic Hyp determination.
Hyp levels were determined biochemically as described before with some minor modifications (48). Briefly, snap-frozen liver pieces (70–100 mg total) were hydrolyzed in 2 mL 6N HCl (MilliporeSigma) at 110°C for 16 hours. After samples were cooled to room temperature, we centrifuged them for 2 minutes at 12,000 g and let them sit for another 10 minutes to sediment the particles. Clear hydrolysate (5 μL) was used for the assay. Standard curve was prepared with Hyp solved in 6N HCl (10–200 μg/mL). A total of 50 μL of citrate acetate buffer (pH 6.0, MilliporeSigma) and 100 μL of chloramine T was added to each well as previously described (48). After 20 minutes incubation at room temperature, 100 μL Ehrlich’s reagent (MilliporeSigma) solved in n-propanol and 70% perchloric acid (MilliporeSigma) was added and heated to 65°C for 15 minutes. After cool down to room temperature, the absorbance was measured at 558 nm. Based on relative hepatic Hyp (per mg of wet liver), total hepatic Hyp was calculated (per total liver, as obtained by multiplying liver weights with relative hepatic HYP).
Serum analysis.
Blood samples were clotted at room temperature for 40 minutes and centrifuged at 1,500 g for 10 minutes at 4°C. Supernatant was transferred to a new tube and centrifuged for 5 minutes at 10,000 g. Serum was then stored at –80°C. Serum levels of ALT [U/L] were measured in serum samples according to the manufacturer’s instruction (Infinity ALT, Thermo Fisher Scientific) using an ALT standard curve.
Immunoblot analysis.
For immunoblot analysis, 30–50 μg of whole-liver lysate was resolved by a 4–20% gradient gel, transferred to nitrocellulose membrane, blocked in 5% Blotting-grade Blocker (Bio-Rad), and incubated with appropriate primary antibodies. Membranes were incubated with peroxidase-conjugated secondary antibodies (Cell Signaling Technology). Protein bands were visualized with the enhanced chemiluminescence (Pico or Femto) reagent and digitized using a CCD camera (ChemiDoc, Bio-Rad). Expression intensity was quantified by ImageJ after background substraction. Anti–P-ASK1 (T845) (1:250, Cell Signaling Technology, catalog 3765), anti-ASK1 (D11C9) (1:1000, Cell Signaling Technology, catalog 8662), anti–P-p38 (Thr180/Tyr182) (1:1000, Cell Signaling Technology, catalog 9211), anti–p38 MAP kinase (1:1000, Cell Signaling Technology, catalog 9212), anti–cleaved caspase 3 (D175) (1:500, Cell Signaling Technology, catalog 9661), anti–caspase 3 (1:1000, Cell Signaling Technology, catalog 9662), anti-Desmin (1:1000, Thermo Fisher, catalog R0-9014-P0), and anti–TNF-α (1:500, Abcam, catalog 9579) were used. Protein load was verified with an anti-GAPDH (1:10,000; Genetex; catalog GTX627408) or anti–β-actin (1:5000, Abgent, catalog AM1829b) antibody.
Real-time PCR.
Total RNA was isolated from liver tissue (50–100 mg), homogenized in TRIzol (Thermo Fisher Scientific), and incubated for 5 minutes at room temperature to allow complete tissue dissociation. A total of 200 μL of chloroform (MilliporeSigma) was added per 1 mL of TRIzol used and vortexed for 15 seconds. After 3 minutes at room temperature, the samples were centrifuged at 12,000 g for 15 minutes at 4°C. The clear phase from the TRIzol/Chloroform Mix was transferred to a new tube with 500 μL of isopropanol (MilliporeSigma). Samples were mixed, incubated for 10 minutes, and centrifuged at 12,000 g for 10 minutes. After washing the pellet 2 times with 75% ethanol, RNA was dissolved in appropriate volume of ddH2O. The reverse transcript (cDNA) was synthesized from 1 μg of total RNA using the iScript cDNA Synthesis kit (Bio-Rad). Real-time PCR quantification was performed using Sybr-Green (KAPA SYBR FAST qPCR master mix) and CFX96 Thermal Cycler from Bio-Rad. The sequences of the primers used for quantitative PCR are given in Supplemental Table.
RNA-seq.
RNA-seq was performed by Q2 Solutions – EA Genomics. Briefly, total RNA was isolated from fresh frozen tissue using the Qiagen miRNeasy Mini Kit. All samples had > 100 ng of input RNA and a RNA integrity number (RIN) value ≥ 7.0 Sequencing libraries were created using the IlluminaTruSeq Stranded mRNA method, which preferentially selects for messenger RNA by taking advantage of the polyadenylated tail. Libraries were sequenced using the Illumina sequencing-by-synthesis platform, with a sequencing protocol of 50 bp paired-end sequencing and total read depth of 30 M reads per sample.
RNA-seq data generated in this study were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih gov/geo) with accession number GSE140742.
DEG analysis.
The Bioconductor packages edgeR (49) and limma (50) were used to normalize sequence count data and conduct differential gene expression analysis. FDR was calculated using the Benjamini-Hochberg method (51). GSEA (52) and EnrichR (53) were conducted to identify significantly enriched functional pathways and GO categories. DEGs specific for ASK1i-treated Nlrp3 mutant mice were subjected to validation using real-time PCR.
Statistics.
Analyses were performed with Graph Pad (version 5.03; Graph Pad Software Inc.) and Windows Excel 2016. The significance level was set at P < 0.05 for all comparisons unless otherwise stated. Gaussian distributed data were analyzed using 1-way ANOVA and Bonferroni post hoc test. Nonparametric data were compared using Kruskal-Wallis test with Dunn’s selected post hoc test. A Mann-Whitney U test was used for ordinal data (*P < 0.05; **P < 0.01; ***P < 0.001). Unless otherwise stated, data are expressed as mean ± SEM.
Study approval.
University of California San Diego IACUC approved all protocols (S92110).
Author contributions
AEF conceived the idea. SSG, MDM, CDJ, AZ, GB, HMH, and AEF designed the experiments. SSG, LJG, MDM, CJ, and AZ contributed to sample preparation. SSG, LJG, CDJ, AZ, LL, VB, and IM carried out the experiments. SSG, CDJ, AZ, LL, VB, IM, AW, LJJ, BGP, GB, HMH, and AEF contributed to the interpretation of the results. SSG and GB wrote the manuscript. AEF critically reviewed the manuscript and provided funding for the study. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Supplementary Material
Acknowledgments
This work was funded by a sponsored research grant from Gilead Sciences, NIH grants R01 DK113592 and U01AA024206 to AF, and the German Research Foundation (DFG) SCHU 3146/1-2 to SSG.
Version 1. 01/30/2020
Electronic publication
Footnotes
Conflict of interest: AZ, LL, VB, IM, and GB are employees of Gilead Sciences.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(2):e123294.https://doi.org/10.1172/jci.insight.123294.
Contributor Information
Susanne Schuster-Gaul, Email: susanne.schuster@medizin.uni-leipzig.de.
Lukas Jonathan Geisler, Email: lgeisler@ukaachen.de.
Matthew D. McGeough, Email: mmcgeough@ucsd.edu.
Casey D. Johnson, Email: caseyj2@uci.edu.
Anna Zagorska, Email: Anna.Zagorska@gilead.com.
Li Li, Email: Li.Li@gilead.com.
Alexander Wree, Email: alexander.wree@charite.de.
Igor Mikaelian, Email: Igorm@23andme.com.
Lily J. Jih, Email: Lily.Jih@va.gov.
Bettina G. Papouchado, Email: bpapouchado@ucsd.edu.
References
- 1.Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. From NAFLD to NASH to cirrhosis-new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol. 2013;10(11):627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Marengo A, Jouness RI, Bugianesi E. Progression and Natural History of Nonalcoholic Fatty Liver Disease in Adults. Clin Liver Dis. 2016;20(2):313–324. doi: 10.1016/j.cld.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Goh GB, McCullough AJ. Natural History of Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61(5):1226–1233. doi: 10.1007/s10620-016-4095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noureddin M, et al. NASH Leading Cause of Liver Transplant in Women: Updated Analysis of Indications For Liver Transplant and Ethnic and Gender Variances. Am J Gastroenterol. 2018;113(11):1649–1659. doi: 10.1038/s41395-018-0088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagström H, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. 2017;67(6):1265–1273. doi: 10.1016/j.jhep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Golabi P, et al. Non-alcoholic steatofibrosis (NASF) can independently predict mortality in patients with non-alcoholic fatty liver disease (NAFLD) BMJ Open Gastroenterol. 2018;5(1):e000198. doi: 10.1136/bmjgast-2018-000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musso G, Gambino R, Cassader M, Pagano G. Meta-analysis: natural history of non-alcoholic fatty liver disease (NAFLD) and diagnostic accuracy of non-invasive tests for liver disease severity. Ann Med. 2011;43(8):617–649. doi: 10.3109/07853890.2010.518623. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzawa A, et al. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6(6):587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 10.Shiizaki S, Naguro I, Ichijo H. Activation mechanisms of ASK1 in response to various stresses and its significance in intracellular signaling. Adv Biol Regul. 2013;53(1):135–144. doi: 10.1016/j.jbior.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Soga M, Matsuzawa A, Ichijo H. Oxidative Stress-Induced Diseases via the ASK1 Signaling Pathway. Int J Cell Biol. 2012;2012:439587. doi: 10.1155/2012/439587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tobiume K, et al. ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2001;2(3):222–228. doi: 10.1093/embo-reports/kve046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuster S, Feldstein AE. NASH: Novel therapeutic strategies targeting ASK1 in NASH. Nat Rev Gastroenterol Hepatol. 2017;14(6):329–330. doi: 10.1038/nrgastro.2017.42. [DOI] [PubMed] [Google Scholar]
- 14.Sakauchi C, Wakatsuki H, Ichijo H, Hattori K. Pleiotropic properties of ASK1. Biochim Biophys Acta Gen Subj. 2017;1861(1 Pt A):3030–3038. doi: 10.1016/j.bbagen.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Wang PX, et al. Targeting CASP8 and FADD-like apoptosis regulator ameliorates nonalcoholic steatohepatitis in mice and nonhuman primates. Nat Med. 2017;23(4):439–449. doi: 10.1038/nm.4290. [DOI] [PubMed] [Google Scholar]
- 16.Xiang M, et al. Targeting hepatic TRAF1-ASK1 signaling to improve inflammation, insulin resistance, and hepatic steatosis. J Hepatol. 2016;64(6):1365–1377. doi: 10.1016/j.jhep.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Wree A, et al. NLRP3 inflammasome activation results in hepatocyte pyroptosis, liver inflammation, and fibrosis in mice. Hepatology. 2014;59(3):898–910. doi: 10.1002/hep.26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agorio A, et al. An Arabidopsis Natural Epiallele Maintained by a Feed-Forward Silencing Loop between Histone and DNA. PLoS Genet. 2017;13(1):e1006551. doi: 10.1371/journal.pgen.1006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wree A, et al. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med. 2014;92(10):1069–1082. doi: 10.1007/s00109-014-1170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinkade JM, Pember SO, Barnes KC, Shapira R, Spitznagel JK, Martin LE. Differential distribution of distinct forms of myeloperoxidase in different azurophilic granule subpopulations from human neutrophils. Biochem Biophys Res Commun. 1983;114(1):296–303. doi: 10.1016/0006-291X(83)91627-3. [DOI] [PubMed] [Google Scholar]
- 21.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104(4):487–501. doi: 10.1016/S0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 22.Canbay A, Higuchi H, Bronk SF, Taniai M, Sebo TJ, Gores GJ. Fas enhances fibrogenesis in the bile duct ligated mouse: a link between apoptosis and fibrosis. Gastroenterology. 2002;123(4):1323–1330. doi: 10.1053/gast.2002.35953. [DOI] [PubMed] [Google Scholar]
- 23.Feldstein AE, et al. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 2003;125(2):437–443. doi: 10.1016/S0016-5085(03)00907-7. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro PS, et al. Hepatocyte apoptosis, expression of death receptors, and activation of NF-kappaB in the liver of nonalcoholic and alcoholic steatohepatitis patients. Am J Gastroenterol. 2004;99(9):1708–1717. doi: 10.1111/j.1572-0241.2004.40009.x. [DOI] [PubMed] [Google Scholar]
- 25.Takehara T, et al. Hepatocyte-specific disruption of Bcl-xL leads to continuous hepatocyte apoptosis and liver fibrotic responses. Gastroenterology. 2004;127(4):1189–1197. doi: 10.1053/j.gastro.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 26.Thomas H. NAFLD: A critical role for the NLRP3 inflammasome in NASH. Nat Rev Gastroenterol Hepatol. 2017;14(4):197. doi: 10.1038/nrgastro.2017.21. [DOI] [PubMed] [Google Scholar]
- 27.Mridha AR, et al. NLRP3 inflammasome blockade reduces liver inflammation and fibrosis in experimental NASH in mice. J Hepatol. 2017;66(5):1037–1046. doi: 10.1016/j.jhep.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ Res. 2002;90(12):1259–1266. doi: 10.1161/01.RES.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
- 29.Zhang R, He X, Liu W, Lu M, Hsieh JT, Min W. AIP1 mediates TNF-alpha-induced ASK1 activation by facilitating dissociation of ASK1 from its inhibitor 14-3-3. J Clin Invest. 2003;111(12):1933–1943. doi: 10.1172/JCI17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, Sekine S, Naguro I, Sekine Y, Ichijo H. Apoptosis Signal-regulating Kinase 1 (ASK1)-p38 Pathway-dependent Cytoplasmic Translocation of the Orphan Nuclear Receptor NR4A2 Is Required for Oxidative Stress-induced Necrosis. J Biol Chem. 2015;290(17):10791–10803. doi: 10.1074/jbc.M114.623280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta S, Campbell D, Dérijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267(5196):389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 32.Raingeaud J, Whitmarsh AJ, Barrett T, Dérijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16(3):1247–1255. doi: 10.1128/MCB.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Win S, Than TA, Zhang J, Oo C, Min RWM, Kaplowitz N. New insights into the role and mechanism of c-Jun-N-terminal kinase signaling in the pathobiology of liver diseases. Hepatology. 2018;67(5):2013–2024. doi: 10.1002/hep.29689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–252. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 35.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophys Acta. 1997;1333(2):F85–104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 36.González-Terán B, et al. p38γ and p38δ reprogram liver metabolism by modulating neutrophil infiltration. EMBO J. 2016;35(5):536–552. doi: 10.15252/embj.201591857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vallerie SN, Hotamisligil GS. The role of JNK proteins in metabolism. Sci Transl Med. 2010;2(60):60rv5. doi: 10.1126/scitranslmed.3001007. [DOI] [PubMed] [Google Scholar]
- 38.Kluwe J, et al. Modulation of hepatic fibrosis by c-Jun-N-terminal kinase inhibition. Gastroenterology. 2010;138(1):347–359. doi: 10.1053/j.gastro.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuño-Lámbarri N, Barbero-Becerra VJ, Uribe M, Chávez-Tapia NC. Mitochondrial Molecular Pathophysiology of Nonalcoholic Fatty Liver Disease: A Proteomics Approach. Int J Mol Sci. 2016;17(3):281. doi: 10.3390/ijms17030281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decaris ML, et al. Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology. 2017;65(1):78–88. doi: 10.1002/hep.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liles JT, et al. Combination of an ASK1 inhibitor and FXR agonist increases efficacy in a mouse model of non-alcoholic steatohepatitis. J Hepatol. 2017;66(1):S19–S20. [Google Scholar]
- 42.Bates J, et al. ACC inhibitor demonstrates potent anti-fibrotic activity in vitro and in vivo. J Hepatol. 2018;68:S399–S400. [Google Scholar]
- 43.Brydges SD, et al. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30(6):875–887. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 45.McGeough MD, et al. Cutting edge: IL-6 is a marker of inflammation with no direct role in inflammasome-mediated mouse models. J Immunol. 2012;189(6):2707–2711. doi: 10.4049/jimmunol.1101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mederacke I, Dapito DH, Affò S, Uchinami H, Schwabe RF. High-yield and high-purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc. 2015;10(2):305–315. doi: 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 48.Fujita M, et al. Overexpression of tumor necrosis factor-alpha produces an increase in lung volumes and pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2001;280(1):L39–L49. doi: 10.1152/ajplung.2001.280.1.L39. [DOI] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 52.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.