Abstract
In the RV144 HIV-1 phase III trial, vaccine efficacy directly correlated with the magnitude of the variable region 2–specific (V2-specific) IgG antibody response, and in the presence of low plasma IgA levels, with the magnitude of plasma antibody-dependent cellular cytotoxicity. Reenrollment of RV144 vaccinees in the RV305 trial offered the opportunity to define the function, maturation, and persistence of vaccine-induced V2-specific and other mAb responses after boosting. We show that the RV144 vaccine regimen induced persistent V2 and other HIV-1 envelope–specific memory B cell clonal lineages that could be identified throughout the approximately 11-year vaccination period. Subsequent boosts increased somatic hypermutation, a critical requirement for antibody affinity maturation. Characterization of 22 vaccine-induced V2-specific mAbs with epitope specificities distinct from previously characterized RV144 V2-specific mAbs CH58 and CH59 found increased in vitro antibody-mediated effector functions. Thus, when inducing non-neutralizing antibodies, one method by which to improve HIV-1 vaccine efficacy may be through late boosting to diversify the V2-specific response to increase the breadth of antibody-mediated anti–HIV-1 effector functions.
Keywords: AIDS/HIV
Keywords: AIDS vaccine
The HIV-1 Env vaccine-induced antibody response was characterized in RV144 clinical trial participants who were boosted 3 more times across an 11-year span.
Introduction
The canary pox vector ALVAC-HIV (vCP1521) prime followed by ALVAC-HIV plus recombinant bivalent gp120 (AIDSVAX B/E; AE.A244 and B.MN gp120s) boosting regimen used in the RV144 clinical trial (ClinicalTrials.gov NCT00223080) generated an estimated vaccine efficacy of 60% at 12 months that declined to 31% at 42 months (1, 2). HIV-1 vaccine efficacy directly correlated with the magnitude of HIV-1 envelope (Env) second variable region (V2) plasma binding IgG antibodies and inversely correlated with Env IgA antibody binding (3–6). A viral genetic sieve analysis (7) and characterization of V2-specific mAbs (8) identified Env lysine position 169 (K169) as a target site of protective antibody responses. Vaccine-induced, V2-specific mAbs CH58 and CH59 mediated antibody-dependent cellular cytotoxicity against tier 2 HIV-1 infectious molecular clone–infected target cells (8). Moreover, the predominant specificity of V2 mAbs isolated to date from PBMCs of RV144 trial participants contain a light chain complementary determining region 2 (LCDR2) glutamic acid/aspartic acid (ED) motif that is used to interact with Env K169 (8, 9).
Although V2-specific antibodies are proposed as the correlate of decreased transmission risk in RV144, to date only a limited number of linear V2-specific mAbs have been isolated from ALVAC/AIDSVAX B/E–immunized humans (8), and the effect of boosting on the quality of putative protective V2-specific antibodies is not known. The RV305 trial (10) (ClinicalTrials.gov NCT01435135) reenrolled HIV-1–uninfected RV144 vaccinees 6 to 8 years after the last RV144 immunization and boosted them twice with ALVAC/AIDSVAX B/E, AIDSVAX B/E alone, or ALVAC alone. In the clinical trial referred to as RV305a, vaccinees were boosted again 1 time with AIDSVAX B/E alone 2 to 3 years after the last RV305 immunization. Here we probed ALVAC prime, bivalent AIDSVAX B/E–induced V2-specific antibody responses from the time of RV144 vaccination over an 11-year period with the RV305 and RV305a late boosts of RV144 initial trial participants (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131437DS1). Potentially novel conformational and linear V2-specific mAbs were identified, including a class of V2-specific antibody dependent upon the N156 glycan for binding. The V2 mAbs potently inhibited α4β7 integrin binding to Env V2. Additionally, V2-specific mAbs were identified with Fc-mediated antiviral functions. Finally, persistent V2-specific and other Env-specific memory B cells were identified that spanned the 11-year immunization time period.
Results
Effect of late boosting on RV144-primed Env-reactive antibodies
This study focused specifically on 8 vaccine recipients for which serum samples were available after the RV144 clinical trial and after the RV305 clinical trial. Two weeks after the first RV305 boost (RV305 week 2), all 6 AIDSVAX B/E protein vaccine recipients studied had detectable but variable AE.A244gp120, B.MN V3, and A244 V1V2 epitope titers (Supplemental Figure 2). Five of 8 vaccine recipients had detectable antibody binding titers to the heterologous gp70 B.CaseA V1V2 antigen (expressing the full V1V2 region on a scaffold), and 6 of 8 vaccine recipients had antibody binding titers to the autologous AE.A244 V1V2 antigen (Supplemental Figure 2). The durability of the V2-specific antibody responses varied (Supplemental Figure 2). Thus, as previously reported for RV144 (11), heterogeneous V2-specific immune responses were seen following boosting.
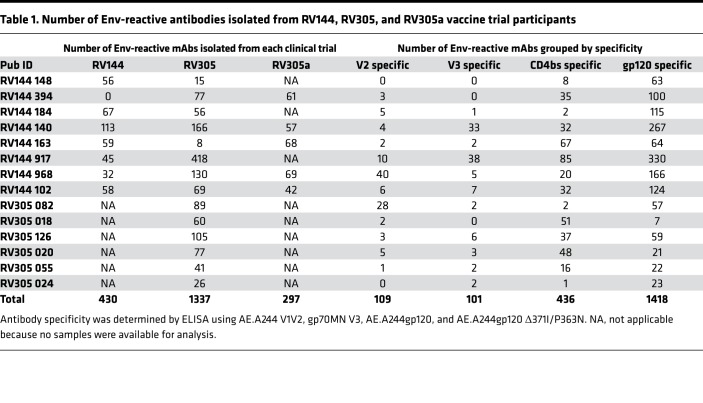
We next longitudinally analyzed at the single-mAb level vaccine-elicited responses in post-RV144, post-RV305, and post-RV305a (Supplemental Figure 1) vaccine recipients using PBMCs collected 2 weeks after the final boost given in each clinical trial. A total of 3126 mAbs were isolated by either antigen-specific B cell sorting or memory B cell cultures (8). Antibody variable heavy (VH) and light (VL) chain genes were reverse transcription PCR (RT-PCR) amplified (12). All mAbs were transiently expressed and assayed by ELISA, by which it was found that of 3126 total mAbs, 2064 (66%) bound to either AE.A244gp120 or B.MNgp120 — the protein components of AIDSVAX B/E (Table 1). Of these Env-reactive mAbs 109/2064 (5%) were V2 specific, 101/2064 (5%) were V3 specific, 436/2064 (21%) were CD4 binding site specific, and 1418/2064 (69%) were gp120 reactive with the epitope not determined (Table 1).
Table 1. Number of Env-reactive antibodies isolated from RV144, RV305, and RV305a vaccine trial participants.
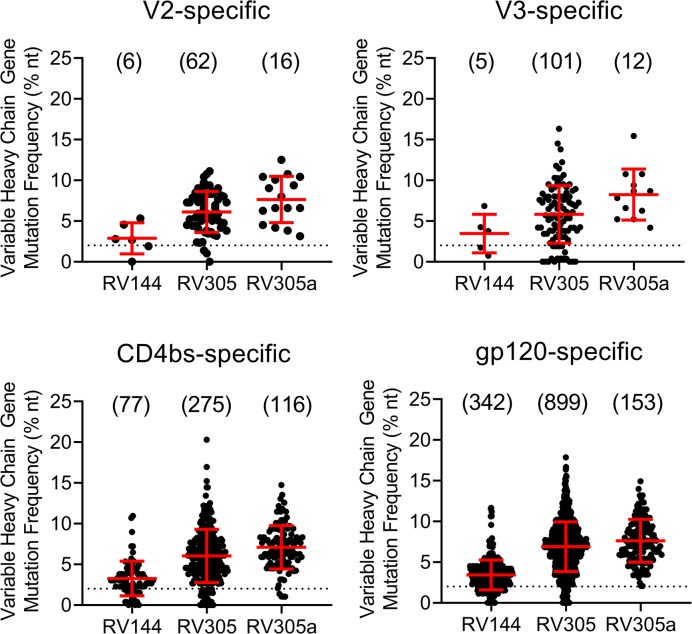
We next asked whether late boosting of RV144 vaccine recipients would either preferentially induce new Env-reactive memory B cell clonal lineages or boost preexisting memory B cells. We first assessed Env-specific mAb mutation frequencies after boosting and found that the mean VH chain gene mutation frequency for V2-specific mAbs, V3-specific mAbs, CD4bs-specific mAbs, and other gp120-specific mAbs progressively increased with boosts given in each clinical trial. VH mutation frequency in RV305a was greater than in RV305, which, in turn, was greater than in RV144 (Figure 1). Moreover, with each subsequent boost, fewer mAbs with a low level (0%–2%) of VH mutations were isolated (Figure 1, dashed line). These data suggested that the ALVAC/AIDSVAX B/E immunization regimen induced or expanded Env-reactive memory B cells that persisted and underwent continued rounds of somatic hypermutation.
Figure 1. VH chain gene mutation frequency of Env-specific mAbs isolated from the RV144, RV305, or RV305a HIV-1 clinical trials.
B cells were antigen specific single-cell sorted or isolated from memory B cell culture. The antibody VH and VL chain genes were RT-PCR amplified, Sanger sequenced, and analyzed with Cloanalyst (45). Env reactivity and epitope specificity was determined by transiently expressing mAbs and assaying by ELISA. MAbs were isolated from 8 RV144 vaccinees, 14 RV305 vaccinees, and 5 RV305a vaccinees, of which 5 were studied in all 3 clinical trials. All PBMCs used for mAb isolation were collected 2 weeks after the final boost in each clinical trial. Dashed line indicates 2% VH chain gene mutation frequency. Numbers of mAbs analyzed are in parentheses.
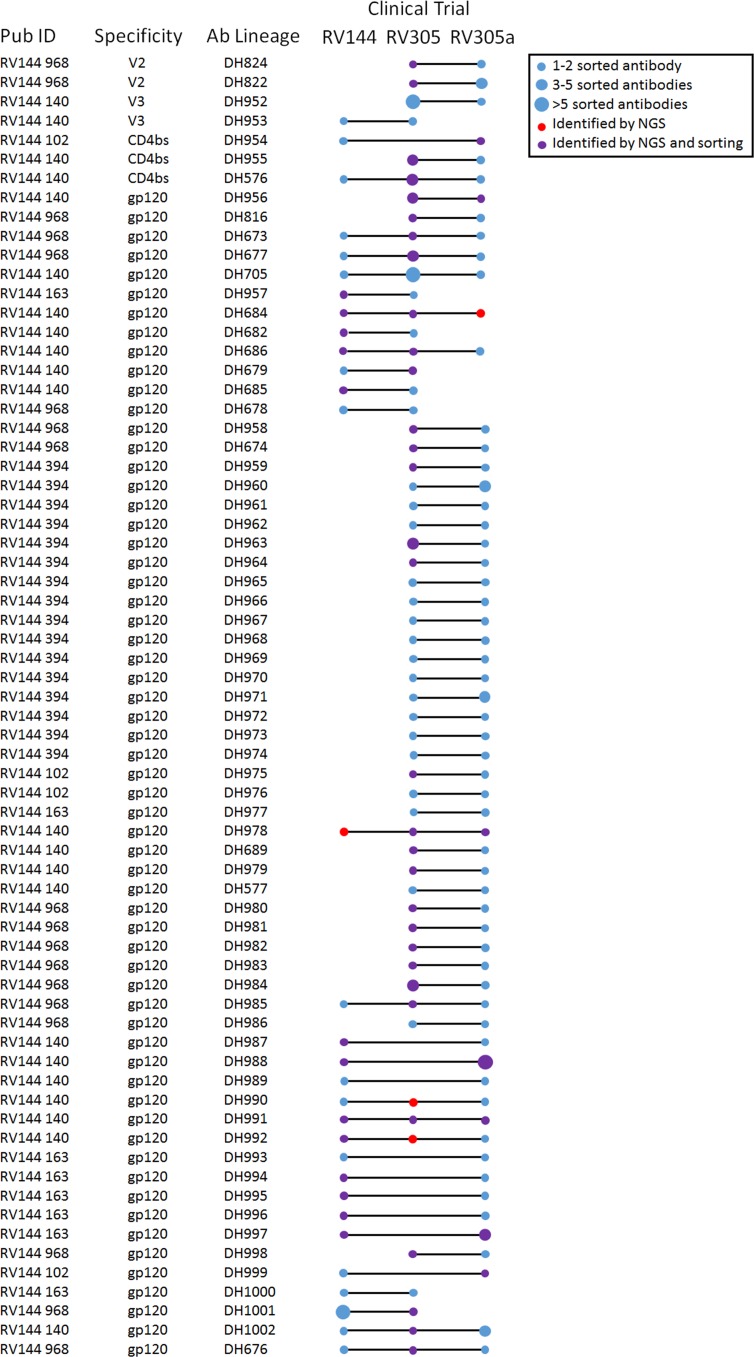
Of the 14 RV305 participants included in this study, PBMCs collected after the RV144 immunization regimen were available from 8 individuals, and 5 of these 8 individuals were boosted again in RV305a (Supplemental Figure 1 and Table 1). MAbs isolated from these vaccinees were used to identify persistent memory B cell clonal lineages. In addition, antibody VH chain next-generation sequencing (NGS) was performed on 5 of the 8 RV144, RV305, and RV305a vaccinee samples. These NGS data sets were probed for additional clonal lineage member sequences. Two out of 70 (2.86%) V2-specific mAbs were identified that persisted from RV305 to RV305a (Figure 2). In addition 2/86 (2.33%) V3-specific, 3/281 (1.07%) CD4bs-specific, and 61/1229 (4.96%) gp120-specific persistent mAb clonal lineages were identified (Figure 2). Overall 23/68 (33.82%) of the persistent lineages identified were started in RV144 and persisted for 11 years, being identified again in RV305a (Figure 2). Thus, the ALVAC/AIDSVAX B/E immunization regimen induced long-lived memory B cells against HIV-1 Env epitopes.
Figure 2. Vaccine-induced Env-reactive long-lived memory B cells isolated from the RV144, RV305, and RV305a HIV-1 clinical trials.
MAbs isolated from vaccine recipients who volunteered for 2 or more clinical trials and confirmed by direct binding ELISA to be Env reactive were assessed for clonal relatedness with Cloanalyst (45). Additional clonal lineage members were identified by VH chain gene NGS. The boosting interval between RV144 and RV305 was 6 to 8 years and between RV305 and RV305a was 2 to 3 years.
Analysis of V2-specific antibodies
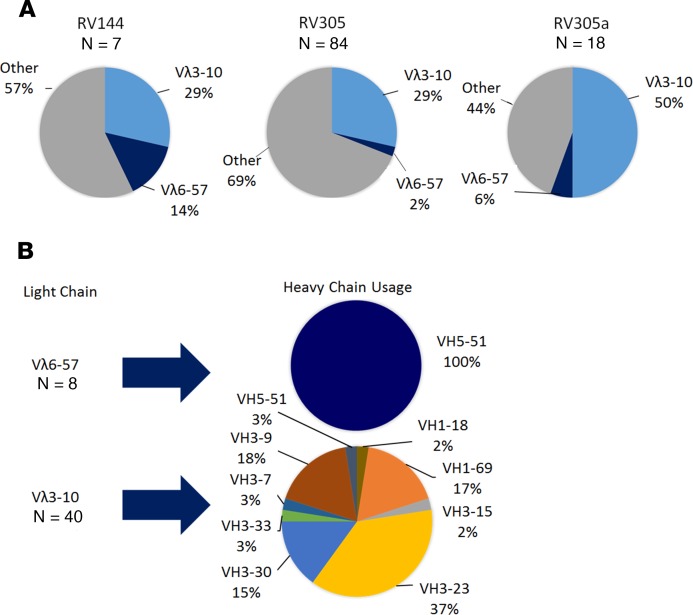
Because in RV144 HIV-1 infection risk inversely correlated with Env V1V2–specific antibody titers, here we focused specifically on V2-specific antibodies isolated after boosting. Structural analysis of RV144 V2-specific mAbs CH58 and CH59 demonstrated that these mAbs form a salt bridge with Env K169 using an ED motif located in the LCDR2 (8, 13). Here an analysis of Env V2 mAbs identified 3 potentially new CH58-like (VH5–51 / Vλ6–57) and 6 potentially new CH59-like (VH3–9 / Vλ3–10) heavy and light chain gene pairings (Supplemental Table 1). V2-specific mAbs with a Vλ3–10 light chain were more common than V2-specific mAbs with a Vλ6–57 light chain (Figure 3A). Among all V2-specific mAbs isolated to date from ALVAC/AIDSVAX B/E–immunized humans (RV144, RV305, and V2-specific mAbs unpublished from RV306 [NCT01931358] and RV328 [NCT01933685]), all that used Vλ6–57 light chains were paired with VH5–51 whereas those that used Vλ3–10 light chains paired with a variety of VH1, VH3, and VH5 family gene segments (Figure 3B). The high frequency of ALVAC/AIDSVAX B/E–induced V2-specific mAbs with Vλ3–10 light chains is related in part to the diverse number of heavy chains that can pair with Vλ3–10 while retaining epitope specificity. Thus, as in the RV144 trial, the dominant V2 response after late boosting in the RV305 trial was Env K169 targeted by mAbs with an ED motif in the light chain LCDR2.
Figure 3. V2-specific mAbs isolated from RV144 and RV305 vaccine recipients frequently utilize the Vλ3–10 light chain gene.
(A) Seven RV144, 84 RV305, and 18 RV305a mAbs were identified that bound to AE.A244 V1V2 in ELISA. VL chain gene sequences were analyzed by Cloanalyst (45). Shown is the frequency of Vλ3–10 or Vλ6–57 VL chain gene. (B) All V2-specific mAbs with Vλ6–57 or Vλ3–10 light chain genes isolated to date from ALVAC/AIDSVAX B/E humans were analyzed. The V2-specific Vλ6–57 VL chain gene sequences paired only with VH5–51 heavy chain sequences while the Vλ3–10 light chain gene paired with multiple VH chain genes.
Epitope mapping of RV144 and RV305 V2 mAbs
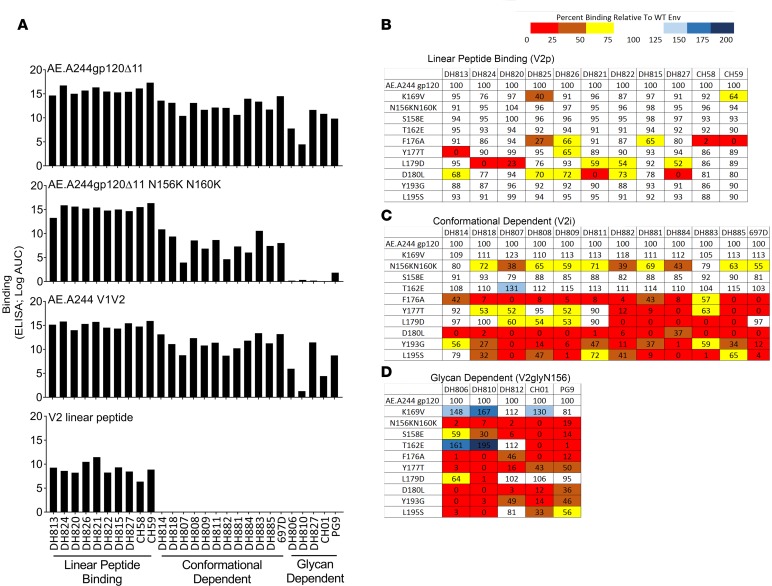
Next, we determined the epitope specificity of 22 V2-specific mAbs (DH827 from RV144 and 21 V2 mAbs from RV305) that utilized light chains with no ED motifs and represented potentially new V2-specific antibody specificities in the context of ALVAC/AIDSVAX B/E vaccination. These V2-specific mAbs were assayed by ELISA for binding to AE.A244gp120, AE.A244gp120 N156K N160K, AE.A244 V1V2, and AE.A244 linear V2 peptide (Figure 4). The non-ED motif V2-specific mAbs separated into 3 groups: V2 linear peptide–binding mAbs (V2p) and conformational dependent V2 mAbs (V2i), as previously described (14), and a third class of novel conformation-dependent V2 mAbs dependent on glycans for Env binding (V2gly156) (Figure 4A).
Figure 4. Recombinantly expressed V2-specific mAb epitope mapping by ELISA.
Recombinantly expressed mAbs were assayed by ELISA. (A) Three groups of V1V2-specific antibodies were identified: those that bound to V2 linear peptide, those that were conformational dependent, and those that were glycan dependent. The (B) linear peptide–binding, (C) conformational dependent, (D) and glycan-dependent mAbs were epitope mapped on AE.A244gp120Δ11 with point mutations in the V1V2 loop. Percentage of binding to the mutant AE.A244gp120Δ11 relative to the WT AE.A244gp120Δ11. Values are the mean of 2 independent experiments.
We compared the epitope specificity of V2p mAbs to linear peptide–binding mAbs CH58 and CH59 (8), V2i mAbs to the conformation-dependent mAb 697D (15), and V2gly156 mAbs to the V1V2-specific broadly neutralizing antibodies (bnAbs) CH01 and PG9 (16, 17). Point mutations that have previously been shown to reduce the binding of CH58 and CH59 (K169V, F176A) (8), 697D (18) (F176A, Y177T, L179D, D180L, Y193G, L195S) (19), or V1V2 bnAbs (N156K, N160K, S158E, T162E) (16, 17) were introduced singly or in combination into AE.A244gp120 and the mAbs assayed by ELISA. Unlike CH59, the newly isolated V2p mAbs were not sensitive to the AE.A244gp120 K169V mutation (Figure 4B). When epitope mapped by alanine scanning using peptides, the V2p mAbs DH824, DH820, and DH822 showed more than 50% reduced binding with a K169A mutation while 5 other V2p mAbs assayed — DH813, DH826, DH821, DH815, and DH827 — were Env K169 partially dependent or independent (Supplemental Figure 3).
The remaining V2-specific mAbs were separated into the 2 groups, V2i and V2gly156. The V2i mAbs mapped very similarly to the previously described V2-specific mAb 697D (refs. 15, 18; Figure 4C) and are likely a subset of mAbs recognizing the previously described immunodominant site in V2 that includes the α4β7 binding motif (19). In contrast, V2gly156 mAbs were sensitive to the AE.A244gp120 S158E mutation, which disrupts the glycosylation sequon for N156, but not to the AE.A244gp120 T162E mutation, which disrupts the glycosylation sequon for N160 (Figure 4D).
Glycan site occupancy of the RV144 AIDSVAX B/E gp120s used in the RV144, RV305, and RV305a trials
Because ALVAC and AIDSVAX B/E immunizations induced V2 glycan–binding mAbs, we next determined glycan site occupancy and site-specific glycosylation patterns of AIDSVAX B/E — the Env boost immunogens used in the RV144, RV305, and RV305a trials. Both AE.A244gDgp120 and B.MN gDgp120 contained predominantly processed glycans, but approximately 30% of AE.A244 gDgp120 and 15% of B.MN gDgp120 did contain a variety of high-mannose glycans at Env positions N156 and N160 (Supplemental Figure 4), including Man3, Man5, Man7, and Man8. To determine the glycan species that the newly identified V2gly156 mAbs DH806, DH810, and DH812 recognized, these mAbs were assayed for binding to free glycan. While DH810 did not bind free glycans, DH806 bound to GlcNac2 and Man8a, and DH812 bound to Man7, Man8a, and Man8b (Supplemental Figure 5).
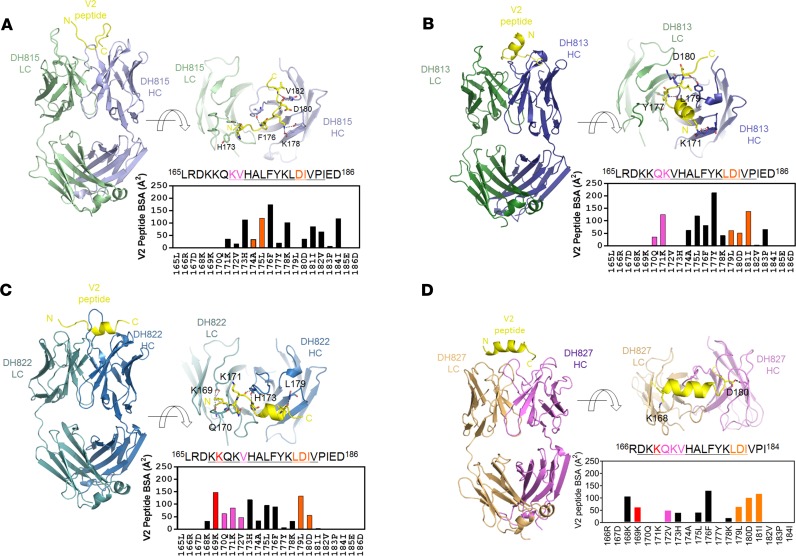
V2 antibody structures
To understand the mode of Env interaction of the new V2p mAbs, we determined the crystal structures of 4 V2p mAbs — DH813, DH815, DH822, and DH827 — in complex with linear V2 peptide. Peptide–antigen-binding fragment (peptide-Fab) co-crystals were obtained that defracted to 1.68, 2.30, 2.12, and 2.90 Å, respectively (Table 2). Structures were solved with molecular replacement and refined to R factors that ranged from 18.1 to 25.9 (Table 2) and that revealed that the 4 mAbs recognized distinct conformations of the V2 region (Figure 5, A–D). DH815 recognized a fully extended conformation of the peptide (Figure 5A), while DH822 and DH813 recognized partially helical conformations (Figure 5, B and C). DH827 recognized an extended helical form of the V2 peptide (Figure 5D). For DH815, ordered electron density was observed for peptide residues 171–184, while for DH822 and DH813, ordered density was observed for V2 residues 168–182 and 168–183, respectively. DH827 recognized residues 168–182 with antibody binding focused on 1 face of the V2 peptide helix recognizing noncontinuous amino acid residues. To determine whether the structures could explain observed differences in Env K169 dependence, we examined the mAb-V2 peptide interfaces. In total, DH822, DH815, DH813, and DH827 buried 945, 927, 1003, and 760 Å2 of surface area on the V2 peptide, respectively (Supplemental Table 2). For DH822, significant interactions between the mAb and residue K169 were observed, accounting for 148 Å2 of interactive surface on K169, consistent with binding analyses previously described (Supplemental Figure 3 and Supplemental Table 2). A salt bridge was observed between Env K169 and DH822 light chain residue D50, as well as hydrogen bonds between Env K169 and carbonyl oxygens of light chain residues L27 and Q30. In contrast, no interactions were observed between Env K169 and DH815 or DH813, consistent with binding analyses that showed their lack of dependence on this residue (Supplemental Figure 3 and Supplemental Table 2). DH827 showed 69 Å2 of surface interaction with Env K169, which matched the moderate but incomplete knockout effect observed in the binding studies (Supplemental Figure 3 and Supplemental Table 2). Env K168 did show significant binding to DH827 light chain residue D30.
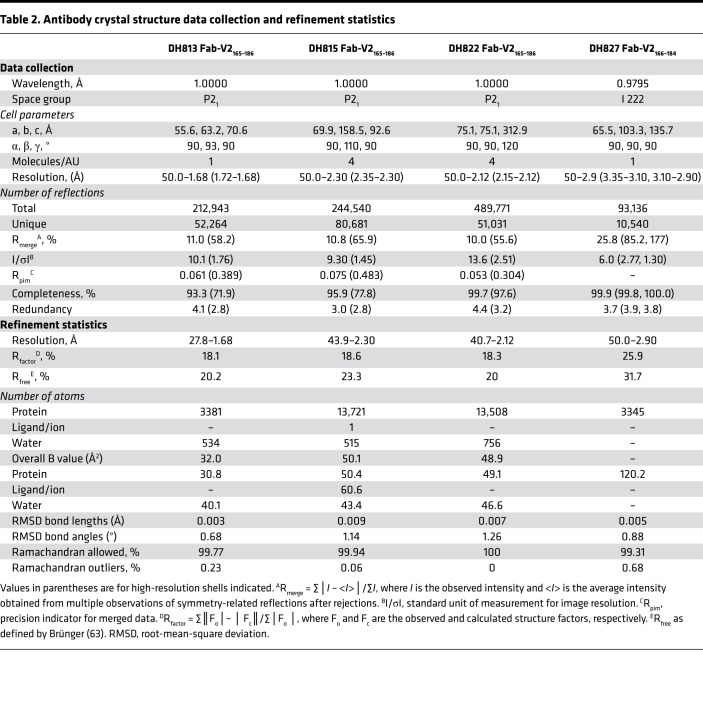
Table 2. Antibody crystal structure data collection and refinement statistics.
Figure 5. Structural analysis of ALVAC/AIDSVAX-induced V2-specific mAbs.
Crystal structures of the RV305 Fab DH815 (A, heavy chain in light blue, light chain in pale green), RV305 Fab DH813 (B, light chain in green, heavy chain in blue), RV305 Fab DH822 (C, light chain in teal, heavy chain in marine), and RV144 Fab DH827 (D, light chain in orange, heavy chain in violet), in complex with an HIV-1 V2 peptide (yellow) encompassing HIV-1 gp120 residues 165–186. Upper right: Close-up views rotated 90° relative to the orientation on the left. Residues of V2 peptide that form hydrogen bonds or salt bridges with the respective mAbs are shown in stick representation. Plots of buried interactive surface area per residue on the bound V2 peptide are shown at lower right for each mAb. Interactions with residue K169 are plotted in red and with each respective integrin binding site are plotted in magenta and orange, respectively. Ordered V2 residues in the respective structures are underlined in the shown peptide sequences.
Two patches on V2 have been reported as binding sites for integrin α4β7 — QKV and LDI — spanning gp120 V2 residues 170–172 and 179–181, respectively (20). All 4 mAbs interacted with both patches on V2, although to varying degrees (Figure 5, A–D, and Supplemental Table 2). Comparative analysis of the binding of the 4 mAbs to these integrin binding sites revealed that DH813 exhibited the most extensive interaction with the 2 sites, burying a total of 413 Å2 of surface area, while DH822 and DH815 buried 387 and 174 Å2, respectively, and DH827 buried 342.6 Å2 (Figure 5, A–D, and Supplemental Table 2).
Functional analysis of V2 mAbs
Neutralization.
No V2-specific mAbs were identified that could neutralize tier 2 isolates; only sporadic tier 1 neutralization was observed (Supplemental Table 3).
Antibody blocking of Env-α4β7 integrin binding.
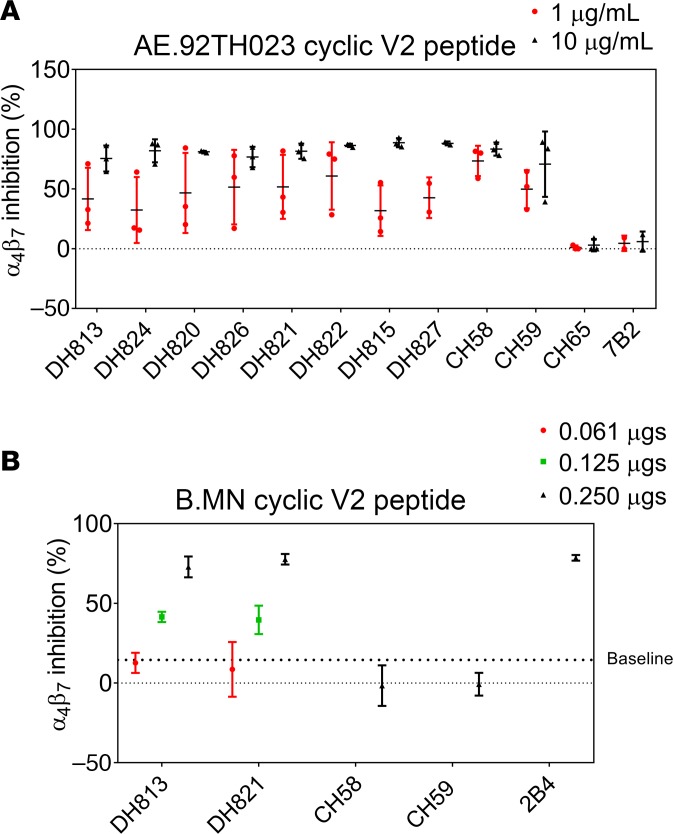
The HIV-1 Env protein has been shown to contain multiple α4β7 integrin binding motifs, including 2 patches in the V2 loop region (20–22). Purified IgG from RV144 vaccinees inhibit α4β7 integrin binding (22), and all the newly identified V2-specific mAbs were sensitive to mutations within the α4β7 integrin binding site (Figure 4, B–D, and Supplemental Figure 3). In addition to the canonical Env LDV/I amino acids (aa) 179–181 α4β7 integrin binding motif, the Env QRV/QRE aa 170–172 motif has also been shown to mediate α4β7 integrin binding (20). The AE.A244 V2 region contains a QKV aa 170–172 motif, and the linear peptide–binding mAbs DH813 and DH827 were both sensitive to mutations within this region (Supplemental Figure 3). Thus, we assessed the V2p mAbs for their ability to block α4β7 integrin binding to AE.92TH023 V2 peptide. Like the K169-dependent V2 mAbs, CH58 and CH59, all the newly isolated V2p mAbs blocked α4β7 integrin binding (Figure 6A). DH813 and DH821 bound in ELISA to B.MNgp120 and thus were also assayed for blocking of α4β7 integrin binding to B.MN cyclic V2 peptide. CH58 and CH59 did not bind to B.MNgp120 and thus were not able to block α4β7 integrin binding to B.MN cyclic V2 peptide. However, DH813 and DH821 manifested dose-dependent blocking of α4β7 integrin binding to B.MN cyclic V2 peptide (Figure 6B).
Figure 6. V2 linear peptide–binding mAbs inhibit α4β7 binding.
(A) V2 linear peptide–binding mAbs were assayed at 10 μg/mL and 1 μg/mL for blocking α4β7 binding to AE.92TH023 cyclic V2 peptide. (B) The B.MN cross-reactive mAb DH813 and DH821 were assayed at 0.061 μg, 0.125 μg, and 0.250 μg for blocking α4β7 binding to B.MN cyclic V2 peptide. Graphed are the mean and SD of 2 independent experiments.
Antibody-dependent cellular phagocytosis.
It has been reported that HIV-1 antibodies are capable of mediating virion phagocytosis, and the efficiency depends on the antibody epitope specificity, subclass usage, and isotype (23). V2-specific mAbs have been shown to mediate phagocytosis in a dose-dependent manner (24). Here we sought to determine whether our panel of V2-specific mAbs could also mediate antibody-dependent virion phagocytosis (ADCP) of AE.92TH023, AE.CM235, and AE.427299 fluorescently labeled virions. DH827, which binds an extended helical form of the V2 loop, was very effective at mediating ADCP of AE.CM235 (Supplemental Figure 6). In addition, 2 V2gly156 mAbs (DH812 and DH810) and 2 V2i mAbs (DH884 and DH807) exhibited weaker ADCP (Supplemental Figure 6) that was comparable or better than what was observed for the group reference mAbs PG9 and 697D, respectively.
Antibody-dependent cellular cytotoxicity.
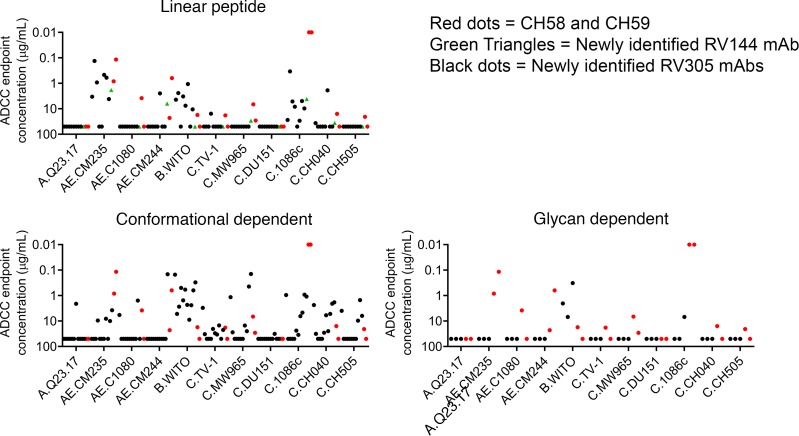
To determine whether our panel of V2-specific mAbs mediated increased antibody-dependent cellular cytotoxicity (ADCC) breadth relative to CH58 andCH59, we assayed the newly isolated mAbs against a panel of clade AE, clade B, and clade C HIV LucR infectious molecular clone–infected (IMC-infected) target cells. Infected cells were monitored for CD4 downregulation at the time of the assay, and it was determined that the percentage of CD4–p24+ was 44.3% ± 22.7% (mean ± SD). Compared with the RV144 mAbs CH58 and CH59, the newly identified V2p mAbs were less potent at mediating ADCC against C.1086c IMC–infected target cells. However, V2p mAbs gained additional breadth against B.WITO and C.CH040 IMC–infected cells (Figure 7 and Supplemental Table 4). Compared with CH58 and CH59, all V2i mAbs mediated more potent ADCC against the single clade B isolate assayed (B.WITO) and expanded clade C ADCC breadth (Figure 7 and Supplemental Table 4). V2gly156 mAbs mediated ADCC against B.WITO IMC–infected cells with 1 mAb having weak activity against C.1086c IMC–infected cells (Figure 7 and Supplemental Table 4). V2gly156 V2-specific mAbs demonstrated exceptional breadth, mediating ADCC against 10 of the 11 IMC-infected target cells assayed (Figure 7 and Supplemental Table 4).
Figure 7. Compared with CH58 and CH59, the newly identified V2-specific mAbs exhibit increased ADCC breadth against a cross-clade panel of IMC-infected target cells.
Linear peptide–binding, conformational dependent, and glycan-dependent V2-specific mAbs were assayed for ADCC against a cross-clade panel of 11 IMC-infected target cells. CH58 and CH59 responses are shown in red, a new RV144 antibody (DH827) is shown in green, and the new RV305 mAbs are shown in black. Data are expressed as mean ADCC endpoint concentration (μg/mL) per antibody from 2 independent experiments.
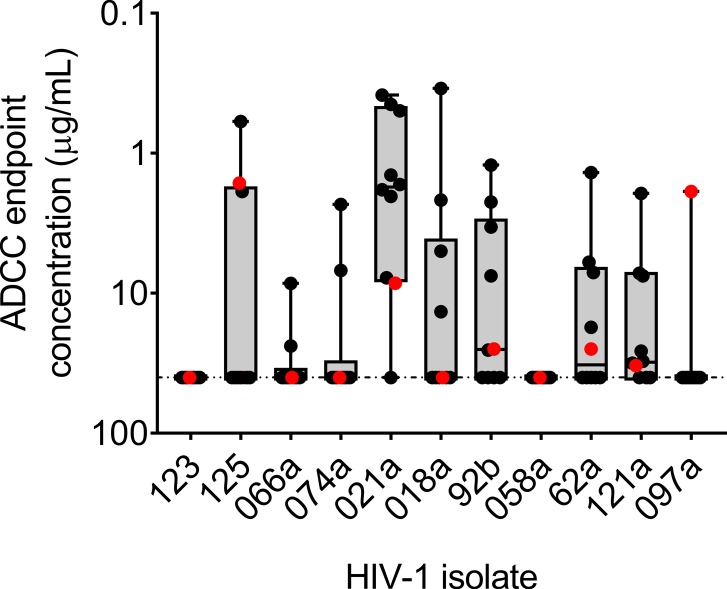
RV144 vaccinees who became infected had evidence of vaccine-induced V1V2-specific immune pressure that selected against viral strains matching the vaccine at Env V1V2 positions 169 and 181 (7). To determine whether the newly isolated V2-specific mAbs could mediate ADCC against RV144 breakthrough viruses, we assayed the V2-specific mAbs against cells infected with 11 RV144 breakthrough isolate IMCs. Infected cells were monitored for CD4 downregulation at the time of the assay, and it was determined that the percentage of CD4–p24+ was 66.7% ± 14.9% (mean ± SD). CH58 mediated ADCC against 6 of 11 isolates assayed, with an average endpoint concentration of 15.8 g/mL (range 1.6–32.9 μg/mL) (Figure 8 and Supplemental Table 5). The V2-specific mAb response that was assayed increased ADCC breadth to 10 of the 11 isolates assayed (Figure 8 and Supplemental Table 5). Moreover, the novel RV305 V2-specific mAbs had an endpoint concentration lower than that observed for CH58 against cells infected with all but 1 of the RV144 clinical HIV-1 isolates (Figure 8 and Supplemental Table 5).
Figure 8. Compared with CH58, the newly identified V2-specific mAbs exhibit increased ADCC breadth against RV144 breakthrough virus IMC-infected CD4+ T cells.
A subset of linear peptide–binding and conformational dependent V2-specific mAbs were assayed for ADCC against a panel of 11 different RV144 breakthrough virus IMC-infected target cells. CH58 and CH59 responses are shown in red, and the newly characterized RV305 V2 mAb responses are shown in black. Data are expressed as mean ADCC endpoint concentration (μg/mL) per antibody from 2 independent experiments.
Discussion
Here we studied the effects of late boosting of RV144 vaccinees with either ALVAC plus AIDSVAX B/E or AIDSVAX B/E alone. We were able to isolate persistent memory B cell clonal linages that were present after the initial RV144 immunization regimen and boosted several years later. This is in contrast to the vaccine-induced short-lived serum antibody concentrations in RV144 that waned after 12 months (2). Inducing long-lived memory B cells that can be recalled and boosted by vaccination is critical for antibody affinity maturation, but to induce durable protective immunity, it will be necessary to identify novel adjuvants or boosting methods to increase differentiation of affinity-matured long-lived memory B cell clonal lineages into long-lived plasma cells that produce persistent serum antibodies.
In RV144 V2-specific antibody responses inversely correlated with HIV-1 infection risk (3). Although these findings have been debated (25), numerous nonhuman primate immunization studies have provided additional support for the role of V2-specific antibodies in mediating protection from simian immunodeficiency virus (SIV) and simian-human immunodeficiency virus (SHIV) infection (summarized in ref. 26). In particular, the magnitude of either plasma or mucosal V2-specific antibody binding to cyclic or linear V2 peptide increased time to infection and reduced peak viral loads (27–31).
Here we found that V2-specific mAbs isolated after RV144 and after boosting were primarily those with an LCDR2 ED motif that targeted K169 and used either VL3-10 (CH59-like) or VL6-57 (CH58-like) light chains, indicating that the ALVAC/AIDSVAX-induced immune responses were stereotyped and used the same VH and VL pairs in different vaccinees. Characterization of non-ED motif boosted V2-specific mAbs showed that compared with CH58 and CH59 antibodies, the mAbs studied here had increased ADCC breadth and potency while 23% were able to mediate ADCP (Figure 4, B–D, and Supplemental Figure 3).
The capability of antibody-mediated inhibition of Env-α4β7 integrin binding to reduce HIV-1 infection risk remains uncertain. Previously it has been shown that α4β7 integrin expression levels on CD4+ T cells correlated with disease severity and progression (32). Passive infusion of the V2i mAb 830A (epitope overlapping α4β7 integrin motif) reduced viral load and decreased viral DNA in lymphoid tissue (33). These findings indicate that while the α4β7 integrin–binding motif is occluded on prefusion trimers, alternative conformations of Env V1V2 may expose the α4β7 integrin binding motif (34, 35), increasing the likelihood of α4β7 integrin binding motif–directed V2-specific antibody binding. In nonhuman primates targeting α4β7 integrin with a primatized mAb protected animals from repeated low-dose intravaginal SIV challenge (36–38), but a follow-up SIV infection study found that the same antibody had no effect on virological control (39). Blocking Env-α4β7 integrin in vitro did not inhibit CD4+ T cell infection (40). Thus the in vivo mechanism of action is more complicated than inhibiting virus-target cell interaction and is an area of ongoing research (37).
The frequency and timing of boosting will be critical considerations for future HIV-1 vaccine regimens. In RV144, the DNA prime protein-boosting strategy used was not sufficient to fully mature the vaccine-induced V2-specific antibody response. However, in other ALVAC and AIDSVAX studies, repetitive boosting led to a decline in V1V2-specific antibody responses after 4 immunizations and a skewing from the more functional IgG3 isotype to IgG4 (41, 42). In contrast, the boost given to RV144 vaccinees occurred many years later in the RV305 clinical trial. While this study demonstrates that RV144 may have benefited from a boost, the optimal timing between boosts is not known. The RV306 vaccine trial (NCT01931358) was a repeat of RV144 with a boost given at 6 months, 9 months, or 12 months. This vaccine trial will likely provide critical information on the optimal timing of the boost, which will also determine how early in childhood a vaccine regimen would need to start to affinity mature vaccine-induced immune responses prior to sexual debut.
In summary, V2-specific mAbs with light chains different from CH58 and CH59 had increased antibody effector functions with greater breadth and potency. Thus, a method to improve upon the vaccine efficacy seen in RV144 may be to use late boosting with novel vaccine regimens that induce a durable and diverse V2-specific antibody response.
Methods
Donor subjects.
In the RV305 clinical trial (NCT01435135) 162 RV144 clinical trial participants (NCT00223080) were boosted 6 to 8 years after the conclusion of RV144 in a randomized, placebo-controlled, double-blinded design. RV305 volunteers were randomized into 3 groups and boosted twice with AIDSVAX B/E plus ALVAC-HIV (vCP1521), AIDSVAX B/E alone, ALVAC-HIV (vCP1521) alone, or a placebo. In the RV305a amended trial (NCT01435135) a subset of RV305 clinical trial participants were boosted 1 time with AIDSVAX B/E in an open-label design. All PBMCs used in this study for antibody isolation were collected 2 weeks after the final boost in each clinical trial.
Antigen-specific single-cell sorting.
PBMCs from 8 RV144 vaccine recipients, 9 RV305 vaccine recipients, and 5 RV305a vaccine recipients were stained with fluorescently conjugated AE.A244gp120Δ11 proteins and a human B cell flow cytometry panel. Viable antigen-specific B cells (AqVd–CD14–CD16–CD3–CD19+IgD–) were single-cell sorted with a BD FACSAria II SORP (BD Biosciences) into 96-well PCR plates containing lysis reagent, RNAse inhibitor, and first strand buffer. The plates were immediately frozen and stored at –80°C. Antibodies used were CD14 clone M5E2 (BD Biosciences 563916), CD16 clone 3G8 (BD Biosciences 563691), CD3 clone SP34-2 (BD Biosciences 563916), CD19 clone HIB19 (BD Biosciences 348794), and IgD clone AI6-2 (BD Biosciences 562024).
Memory B cell culture.
From each of 10 vaccinees, 100 million PBMCs were stained with fluorescently conjugated AE.A244gp120Δ11 proteins and a human B cell flow cytometry panel. Viable antigen-specific B cells (AqVd–CD14–CD16–CD3–CD19+IgD–) were antigen-specific bulk sorted with a BD FACSAria II SORP and the B cells stimulated overnight with Epstein-Barr virus. Memory B cells were seeded at 2 cells per well into 96-well plates and cultured for 2 weeks as previously described (17). Ig-containing supernatants were assayed for Env binding and AE.CM244 neutralization.
Single-cell RT-PCR.
Single B cell cDNA was generated with random hexamers (Genelink) using SSIII (Thermo Fisher Scientific). Antibody VH and VL chain regions were PCR amplified using AmpliTaq360 Master Mix (Applied Biosystems, Thermo Fisher Scientific). PCR products were purified (Qiagen) and sequenced by Genewiz. Gene rearrangements, clonal relatedness, unmutated common ancestors, and intermediate ancestor inferences were made using Cloanalyst.
Antibody VH chain gene NGS.
Cryopreserved PBMCs from either RV144 visit 8 or RV305 visit 5 were rapidly thawed, and RNA was extracted from 1 × 107 PBMCs using RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. Isolated RNA was used for cDNA synthesis and to generate antibody VH chain gene amplicons for NGS as previously described (43). Briefly, reverse transcription was carried out using human heavy and light chain constant region-specific primers. The cDNA was used for PCR amplification using IGHV1–IGHV6 specific forward primers and an IgG-specific reverse primer. A second round of PCR was performed to add Nextera index sequencing adapters (Illumina). Libraries were gel purified and quantified by quantitative PCR using KAPA SYBR Fast qPCR kit (KAPA Biosystems). Sequencing was performed with a MiSeq System using more than 4 nM of amplicon and the Illumina MiSeq V3 2 × 300 bp kit. NGS reads were computationally processed as described elsewhere (43, 44). Clonal lineages were identified using Cloanalyst (45), probing the NGS data sets for sequences related to the heavy chain gene sequence recovered by single-cell sorted B cell RT-PCR. Sequences were deemed clonally related if they shared the same V and J gene segment usage and had similar CDR3 sequences. The CDR3 sequence similarity threshold was set based on the mutation frequency of the sequences such that the threshold was lowered for more highly mutated sequences.
MAb production.
PCR-amplified heavy and light chain gene sequences were transiently expressed as previously described (46). Ig-containing cell culture supernatants were used for ELISA binding assays. For large-scale expression heavy and light chain genes were synthesized (VH chain in the IgG1 4A backbone) and transformed into DH5α cells (GeneScript). Plasmids were expressed in Luria Broth, purified (Qiagen), and approximately 5 × 106 293i cells were transfected with 0.4 mg of heavy plus 0.4 mg of light chain gene using ExpiFectamine (Life Technologies, Thermo Fisher Scientific) following the manufacturer’s protocol. After 5 days of incubation at 37°C 5% CO2, the Ig-containing medium was concentrated and purified with protein A beads and the antibody buffer exchanged into PBS. All antibodies in this study not purchased from a commercial vendor were expressed by this method.
Antibody binding assays.
Direct ELISAs were performed as previously described (46). Briefly, 384-well microplates were coated overnight with 30 ng/well of protein. Antibodies were diluted and added for 1 hour. Binding was detected with an anti–IgG-HRP (polyclonal, Rockland 609-103-123) and developed with SureBlue Reserve TMB One Component (KPL). Plates were read on a plate reader (Molecular Devices) at 450 nm.
Neutralization assays.
TZM-bl neutralization assays were performed as previously described (47). Data are reported as IC50 titers.
IMCs.
The HIV-1 reporter viruses used were replication-competent IMCs designed to encode the env genes of CM235 (subtype A/E; GenBank AF259954.1), WITO (subtype B; GenBank JN944948), 1086.c (subtype C; GenBank FJ444395), TV-1 (subtype C; GenBank HM215437), MW96.5 (subtype C; GenBank), DU151 (subtype C; GenBank DQ411851), and DU422 (subtype C; GenBank DQ411854) or clinical isolates from the RV144 trial in cis within an isogenic backbone that also expresses the Renilla luciferase reporter (LucR) gene and preserves all viral open reading frames (48). The subtype AE Env-IMC-LucR viruses used were the NL-LucR.T2A-AE.CM235-ecto (IMCCM235) (plasmid provided by Jerome Kim, US Military HIV Research Program, Bethesda, Maryland, USA) and clinical env isolates from the RV144 trial that were built on the 40061-LucR virus backbone (provided by Agnes Chenine, Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, Maryland, USA). All other IMCs were built using the original NL-LucR.T2A-ENV.ecto backbone as originally described by Edmonds et al. (48). Reporter virus stocks were generated by transfecting HEK293T cells (ATCC) with proviral IMC plasmid DNA, and virus titer was determined on TZM-bl cells for quality control (48).
Infection of CEM.NKRCCR5 cell line with HIV-1 IMCs.
CEM.NKRCCR5 cells were infected with HIV-1 IMCs as previously described (49). Briefly, IMCs were titrated to achieve maximum expression within 48 to 72 hours after infection as determined by detection of Luciferase activity and intracellular p24 expression. IMC infections were performed by incubation of the optimal dilution of virus with CEM.NKRCCR5 cells for 0.5 hours at 37°C and 5% CO2 in the presence of DEAE-Dextran (MilliporeSigma) (7.5 μg/mL). The cells were subsequently resuspended at 0.5 × 106/mL and cultured for 48 to 72 hours in complete medium containing 7.5 μg/mL DEAE-Dextran. For each ADCC assay, we monitored the frequency of infected target cells by intracellular p24 staining. Assays were performed using infected target cells with at least 60% cell viability and at least 20% p24+.
Luciferase ADCC assay.
ADCC activity was determined by a luciferase-based (Luc-based) assay as previously described (8). Briefly, CEM.NKRCCR5 cells (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Alexandra Trkola) (50) were used as targets after infection with the HIV-1 IMCs. PBMCs obtained from an HIV-seronegative donor with the heterozygous 158F/V and 131H/R genotypes for Fc receptors FcR3A and FcγR2A (51, 52), respectively, were used as a source of effector cells, at an effector/target ratio of 30:1. Recombinant mAbs were tested across a range of concentrations using 5-fold serial dilutions starting at 50 μg/mL. The effector cells, target cells, and antibody dilutions were plated in opaque 96-well half area plates and were incubated for 6 hours at 37°C in 5% CO2. The final readout was the luminescence intensity (relative light units, RLU) generated by the presence of residual intact target cells that had not been lysed by the effector population (ViviRen substrate, Promega). The percentage of specific killing was calculated using this formula: percentage of specific killing = [(number of RLU of target and effector well − number of RLU of test well) / number of RLU of target and effector well] × 100. In this analysis, the RLU of the target plus effector wells represents spontaneous lysis in the absence of any source of antibody. The ADCC endpoint concentration, defined as the lowest concentration of mAb capable of mediating ADCC in our in vitro assay, was calculated by interpolation of the mAb concentration that intersected the positive cutoff of 15% specific killing. The respiratory syncytial virus–specific mAb palivizumab was used as a negative control.
ADCP.
The ADCP assay was carried out as previously described, with slight modifications (23). Briefly, human blood–derived monocytes were isolated via elutriation and cryopreserved. Monocytes were thawed and rested overnight before the assay. Fluorescently labeled HIV-192Th023-Tomato, HIV-1CM235-Tomato, and HIV-1427299-Tomato were generated as described in Shukair et al. (53–55) (provided by Thomas Hope, Northwestern University, Evanston, Illinois, USA). Virus was incubated with or without antibody at 37°C for 2 hours. Next, 1 × 104 CD4-blocked monocytes were added to a final volume of 40 μL each and spinoculated at 1200 g at 4°C. CD4 blocking was done to reduce background virus phagocytosis. This was achieved by pretreating monocytes with 20 μg/mL anti-human CD4 antibody (clone SK3, BioLegend 344602) for 15 minutes at 4°C. Following spinoculation, the immune complex–cell mixture was incubated at 37°C for 1 hour for phagocytosis to occur. Subsequently, cells were washed with 0.05% trypsin and fixed. Antibody-dependent phagocytosis was quantified using a phagocytosis score as previously described — for each sample, the percentage of cells with fluorescence above the 95th percentile of fluorescence for the no-antibody control was taken, multiplied by their MFI, and then normalized to the corresponding values for the no-antibody control.
92TH023 α4β7 blocking and adhesion assay.
The assay was performed as previously described (22). For the adhesion assay triplicate wells of a 96-well flat-bottom polypropylene plate (Greiner Bio-One) were coated overnight at 4°C with 100 μL of 2 μg/mL NeutrAvidin (Thermo Fisher Scientific). For the 92TH023 α4β7 blocking assay streptavidin-coated 96-well Immunlon 2HB round-bottom plates (Thermo Fisher Scientific) were used. In both α4β7 blocking and adhesion assays, plates were incubated with 5 μg/mL biotinylated cyclic V2 92TH023 peptide or with biotinylated cyclic V2 MN peptide (HIV isolate MN subtype B, accession M17449) (JPT Peptide Technologies; both peptides >90% purity) in bicarbonate buffer at pH 9.6 for 1 hour at 37°C. Plates were then blocked for 1 hour at 37°C in blocking buffer (25 mM Tris, 2.7mM KCl, 150 mM NaCl, 0.5% casein, 4 mM MnCl2 pH 7.2), followed by the addition of varying concentrations of V2 linear peptide–binding mAbs in sample buffer (25 mM Tris, 2.7mM KCl, 150 mM NaCl, 4 mM MnCl2, 2.5% HEPES, 1% FBS, pH 7.2) for 45 minutes. MAbs CH58 and CH59 or 2B4 served as the positive controls while CH65 and 7B2 served as the negative controls. Plates were washed 4 times with blocking buffer before the addition of 2 × 105 RPMI8866 cells/well (MilliporeSigma) that had previously been incubated in sample buffer for 1 hour at 37°C alone or in the presence of 6.7 μg/mL of positive control anti-α4β7 mAb ACT-1 (from A. A. Ansari [ref. 56]; obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) or α4 mAb (2B4). For the adhesion assay RPMI8866 cells were cultured in medium containing 1 μM retinoic acid for at least 7 days before use. Plates were then washed 5 times with blocking buffer, and bound cells were detected by the addition of Alamar Blue dye (Thermo Fisher Scientific). The fluorescence released by the reduction of Alamar Blue dye was read on an M2 plate reader (Molecular Devices) at 2-hour intervals up to 8 hours. The percentage of inhibition was calculated based on the fluorescence units obtained in the presence and in the absence of the mAb as described previously (22). For each mAb, the samples were run in triplicate in a minimum of 2 to 3 independent experiments. The data represent the average percentage of inhibition ± SD at the 6-hour time point (92TH023 α4β7 blocking assay).
Oligomannose bead immunoassay.
Binding to oligomannose and complex glycans was performed as previously described (57). In brief, LumAvidin beads (Luminex Corporation) were blocked with 3% BSA in PBS, washed with PBS, and incubated with 16 μM biotinylated glycan. Each glycan was incubated with a unique fluorescent lumavidin bead in a Mixmate (Eppendorf) for 2 hours at 2000 rpm at room temperature protected from light. Free glycan was washed away and the beads were stored in PBS-BN (1% BSA, 0.05% NaN3 in PBS). MAb binding to glycans was measured in 96-well filter plates by incubating 200 μg/mL of mAb with beads for 1.5 hours while shaking at 60 rpm. Free mAb was washed away and bound mAb was detected with mAb bound to glycan on microspheres using 100 μL of 2 μg/mL mouse anti-IgG against PE (polyclonal, Southern Biotech 1030-09S). After the secondary mAb was added beads were washed 5 times and then resuspended in 100 μL. The binding of antibody to glycan was determined with a Bio-Plex 200 HTS (Bio-Rad) and Bio-Plex Manager software (Bio-Rad). The values were represented as fluorescence minus fluorescence in wells with beads bearing no glycan but had primary and secondary mAb added. HIV-1 Env peptide-specific mAb 19B was used as a negative control antibody, and a variant of the anti–HIV-1 Env glycan-specific mAb DH501 (57) and 2G12 were used as high and low positive controls.
Binding antibody multiplex assay.
Plasma binding to AE.A244gp120Δ11, AE.A244 V1V2 tags, gp70 B.CaseA V1V2, B.MN V3 gp70, and proteins (provided by the Duke Human Vaccine Institute Protein Production Facility) was assessed by binding antibody multiplex assay (BAMA) as previously described, with modifications (3, 11, 58). Briefly, polystyrene microspheres (Luminex Corporation), covalently coupled to HIV-1 proteins (59), were incubated with plasma diluted at 1:50 in StabilGuard Immunoassay Stabilizer (BSA-Free) (SurModics) for 30 minutes. Sample wells were washed (in wash buffer containing PBS, BSA, Tween-20, sodium azide) and incubated with mouse anti-human IgG-biotin (polyclonal, Southern Biotech 2040-08), followed by washing and incubation with PE streptavidin (BD Pharmingen). Beads were washed and acquired on a Bio-Plex 200 instrument (Bio-Rad), where the magnitude of the PE-derived signal is proportional to binding, expressed MFI. RV144 and RV305 plasma samples were collected across multiple time points. Samples were tested in duplicate in the same assay under Good Clinical Laboratory Practice conditions with antigen tracking (Levey-Jennings). Purified polyclonal IgG from HIV-1–infected subjects (HIVIG) (NIH AIDS Reagent Program), CH58_4A mAb (V2-specific mAb), VRC01 mAb (CD4 binding site–specific mAb), and 2G12 mAb (glycan-specific mAb) were used as positive controls. Normal human serum (HIV-1–seronegative human serum) (MilliporeSigma), blank (uncoupled) beads, and a blank well (microspheres plus detection antibody in the absence of sample) were included as negative controls. Criteria for positivity were as follows: sample MFI value (a) greater than 3 times the matched baseline sample (RV144 visit 1 sample) MFI (or median MFI of all pooled baseline samples if the matched baseline sample was not available for testing) before and after blank bead subtraction and (b) greater than the antigen-specific cutoff (95th percentile of all baseline samples calculated for each antigen).
Fab preparation.
DH827 antibody was digested overnight at 37°C using the Pierce Fab Preparation Kit (Thermo Fisher Scientific) according to manufacturer guidelines. Antibody Fc fragment was removed using protein A resin. The Fab fragment was assessed by SDS-PAGE for purity and concentrated to 8 mg/mL.
Fab–V2 peptide complex crystallization.
Crystallizations of antibody Fabs of DH813, DH815, and DH822 in complex with peptide V2165–186 were undertaken by initial robotic screening using 576 conditions from commercially available screens (Hampton Research and Rigaku Reagents) followed by optimization. Robotic crystallization trays were set up as vapor diffusion sitting drops using a Mosquito Nanoliter Liquid Handling System (TTP Labtech). DH813 Fab-V2165–186 complex crystals were obtained in a condition made up of 30% 2-propanol, 15% PEG 8000, and 0.1 M imidazole at pH 6.5. These crystals were cryoprotected with mother liquor supplemented with ethylene glycol and cryocooled in liquid nitrogen for data collection. DH815 Fab-V2165–186 complex crystals were obtained in a condition made up of 0.2 M sodium chloride, 20% PEG 8000, and 0.1 M sodium citrate at pH 4.2. These crystals were cryoprotected with mother liquor supplemented with (2R,3R)-(−)-2,3-Butanediol and cryocooled in liquid nitrogen for data collection. DH822 Fab-V2165–186 complex crystals were obtained in a condition made up of 18% 2-propanol, 20% PEG 4000, and 0.1 M sodium citrate at pH 5.5. These crystals were flash frozen in liquid nitrogen for data collection with no additional cryoprotectant.
Following IgG digestion and Fab purification, DH827 Fab was incubated with the V2 peptide (R166DKKQKVHALFYKLDIVPI188, Genscript, >95% purity) at a ratio of 1:1.5 at 4°C for 1 hour. Excess peptide was removed by centrifugation prior to crystal condition screening using an Art Robbins Gryphon crystallization robot. A set of 1200 crystal growth conditions prepared using an Art Robbins Scorpion robot were assessed by mixing 0.2 μL of Fab-peptide complex with 0.2 μL of reservoir solution using the sitting-drop vapor diffusion method at 20°C. Initial crystallization conditions were optimized manually using 2 μL crystallization drops to produce crystals suitable for data collection. Crystals of DH827 Fab V2166–184 peptide were grown in 2.0 M ammonium sulfate and 0.1 M citric acid (pH 3.5). Crystals were transferred to a solution containing mother liquor supplemented with 18% glycerol and cryocooled in liquid nitrogen prior to x-ray diffraction data collection.
Structure determination and analysis.
X-ray diffraction data were processed with HKL-2000 (60), and all structures were solved by molecular replacement using PHASER (61). Search models for molecular replacement were PDB ID 1FVE for the DH813 complex, PDB ID 4KMT for the DH815 complex, and PDB ID 4HK0 for the DH822 complex. All structures were refined using PHENIX (62) with iterative model building in Coot (63). Interactive buried surface areas were determined using the Protein Interfaces, Surfaces and Assemblies (PISA) Web server (64). All graphical images were prepared with PyMOL Molecular Graphics System. X-ray diffraction data were collected at SER CAT ID-22 beamlines of the Advanced Photon Source at the Argonne National Laboratory.
Diffraction data for the DH827 Fab-V2166–184 peptide complex were collected at the Advanced Photon Source at the Argonne National Laboratory Northeastern Collaborative Access Team 24-ID-E beamline and measured using a DECTRIS EIGER X 16M pixel detector. Data were processed with the beamline software pipeline RAPD incorporating XDS, and the structure was solved by molecular replacement using the coordinates from the CH58 Fab-V2 peptide complex (Protein Data Bank code 4HPO) with the program PHASER and refined using REFMAC5 (65) and PHENIX (62). Refinement was coupled with manual refitting and rebuilding with Coot (63). Structure quality was assessed with MolProbity (66). Data collection and refinement statistics are shown in Table 2. PISA was used to determine antibody-antigen contact residue interactions (64). Figures showing the structure were prepared using the PyMOL Molecular Graphics System (DeLano Scientific).
AIDSVAX B/E site-specific glycan analysis.
Trizma@ hydrochloride, Trizma@ base, urea, dithiothreitol, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), iodoacetamide (IAM), and glacial acetic acid were purchased from MilliporeSigma. Other reagents used in this study included Optima liquid chromatography–mass spectrometry–grade (LC/MS-grade) formic acid and acetonitrile (Thermo Fisher Scientific), HPLC-grade water (Honeywell Burdick and Johnson), sequencing-grade trypsin and chymotrypsin (Promega), and endo-β-N-acetylglucosaminidase H (Endo H) (New England BioLabs). All reagents and buffers were prepared with deionized water purified with a Millipore Direct-Q3 water purification system. Samples containing 50 μg of the AIDSVAX B/E AE.244 gD gp120 and B. MN gD gp120 Envs were partially deglycosylated with Endo H. Partial deglycosylation was performed by incubating the samples with 2.5 μL of Endo H (≥5 units/mL) at pH 5.5 for 48 hours at 37°C. Following deglycosylation, samples were digested as described below.
AIDSVAX B/E AE.244 gD gp120 and B. MN gD gp120 Envs (50 μg) were denatured with 6 M urea in 100 mM Tris buffer (pH 8.0), reduced at room temperature (RT) for 1 hour with TCEP (5 mM), and alkylated with 20 mM IAM at RT for another hour in the dark. The reduced and alkylated samples were buffer exchanged using a 30-kDa molecular weight cutoff filter (MilliporeSigma) before protease digestion. Digestion was performed using trypsin alone and a combination of trypsin and chymotrypsin at a 30:1 protein/enzyme ratio. Samples were incubated overnight at 37°C. The resulting Env digest was either directly analyzed or stored at –20°C until further analysis. To ensure reproducibility of the method, digestion was performed at least 3 times on different days with samples obtained from the same batch and analyzed with the same experimental procedure.
High-resolution LC/MS experiments were performed using an LTQ-Orbitrap Velos Pro (Thermo Fisher Scientific) mass spectrometer equipped with electron transfer dissociation (ETD) that is coupled to an Acquity ultra-performance liquid chromatography system (Waters). Mobile phases consisted of solvent A, 99.9% deionized H2O and 0.1% formic acid, and solvent B, 99.9 % CH3CN and 0.1% formic acid. Five microliters of the sample (~2 μM) was injected onto a C18 PepMap 300 column (300 mm in diameter × 15 cm, 300 Å, Thermo Fisher Scientific) at a flow rate of 5 L/min. The following CH3CN/H2O multistep gradient was used: 3% B for 5 minutes, followed by a linear increase to 40% B in 50 minutes, then a linear increase to 90% B in 15 minutes. The column was held at 97% B for 10 minutes before re-equilibration. All mass spectrometric analysis was performed in the positive ion mode using data-dependent acquisition mode as described below. Data-dependent acquisition mode was set up to sequentially and dynamically select the 5 most intense ions in the survey scan in the mass range, 400 to 2000 m/z, for alternating collision-induced dissociation (CID) and ETD in the linear trap quadrupole ion trap using a normalized collision energy of 30% for CID and an ion-ion reaction time of 100 to 150 ms for ETD. Full MS scans were measured at a resolution (R) of 30,000 at m/z 400. Under these conditions, the measured R (full width at half maximum) in the orbitrap mass analyzer at m/z 1000 is 20,000 and 17,000 at m/z 1500.
Details of the compositional analysis have been described previously (67–69). Briefly, compositional analysis of glycopeptides was carried out by first identifying the peptide portion from tandem MS data. Once the peptide portion was determined, plausible glycopeptide compositions were obtained using the high-resolution mass spectrometry data and GlycoPep DB. The putative glycopeptide composition was confirmed manually from CID and ETD data.
Study approval.
All clinical trials were approved by Walter Reed Army Institute of Research (Silver Spring, Maryland, USA), Thai Ministry of Public Health (Mueang Nonthaburi District, Thailand), Royal Thai Army Medical Department (Bangkok, Thailand), Faculty of Tropical Medicine (Bangkok, Thailand), Mahidol University (Nakhon Pathom, Thailand), Chulalongkorn University Faculty of Medicine (Bangkok, Thailand), and Siriraj Hospital (Bangkok, Thailand). Written informed consent was obtained from all clinical trial participants. The Duke University Health System Institutional Review Board (Durham, North Carolina, USA) approved all human specimen handling.
Author contributions
DE, JP, LDW, RJO, SV, JHK, NLM, JLE, MLR, SRN, JK, PP, SN, FS, JT, SP, MGJ, GAO, GF, and BFH conceptualized the study. DE, JP, KL, LDW, MZT, AM, KP, MC, SM, PL, PZ, EPG, HD, MB, KKH, CB, MK, KW, KOS, DCM, GDT, MAM, JA, MR, MGJ, GAO, GF, and BFH developed methodology. KW developed antibody sequence analysis software. DE, JP, KL, MZT, AM, KP, MC SM, PL, PZ, EPG, HD, MB, KKH, CB, MK, KOS, DCM, GDT, MAM, JA, MR, GF, and BFH validated data. FS, JT, SP, and KOS provided critical reagents. DE, JP, MZT, AM, KP, MC, SM, PL, PZ, EPG, HD, MB, KKH, CB, MK, KOS, DCM, GDT, MAM, JA, MR, GF, and BFH engaged in formal analysis. BJ, NG, MGJ, and GAO were responsible for crystal structures. DE, JP, MZT, AM, RJO, MGJ, and GAO investigated. RJO, SV, JHK, NLM, JLE, MLR, SRN, JK, PP, and SN provided clinical trial material (PBMCs and serum). DE, JP, KL, LDW, MZT, AM, KOS, GF, and BFH curated data. DE, JP, GF, and BFH wrote the original draft of the manuscript. DE, JP, KL, BJ, NG, LDW, MZT, AM, KP, MC, SM, PL, PZ, EPG, HD, MB, KKH, CB, MK, RJO, SV, JHK, NLM, JLE, MLR, SRN, JK, PP, SN, FS, JT, SP, KW, KOS, DCM, GDT, MAM, JA, MR, MGJ, GAO, GF, and BFH reviewed and edited the manuscript. DE, JP, KP, PL, JA, MZT, AM, KOS, MGJ, GAO, GF, and BFH prepared figures. DE, JP, MB, KOS, DCM, GDT, MAM, JM, MR, MGJ, GAO, GF, and BFH supervised. BFH, KOS, HD, and GDT acquired funding.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the Duke Human Vaccine Institute Flow Cytometry Facility (Durham, North Carolina, USA) and the following individuals for their expert technical assistance: Patrice McDermott and Dawn Jones Marshall for flow cytometry; Lawrence Armand for conjugated antigens; Lashanda Cheston and Morgan Gladden for memory B cell cultures; Andrew Foulger, Erika Dunford, and Kedamawit Tilahun for transient transfections; Callie Vivian and Maggie Barr for ELISA; Giovanna Hernandez, Esther Lee, Emily Machiele, and Rachel Reed for antibody expression; Amanda Eaton, Celia C. LeBranche, Peter Gao, Kelli Greene, and Hongmei Gao for neutralization assays; and Sheetal Sawant for BAMA analysis. For project management, we thank Cynthia Nagle and Kelly Soderberg. We also thank all the RV144 and RV305 clinical trial team members and participants.
Crystal structure data were collected at Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, under a general user agreement to GAO. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38.
Crystal structure data are based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). The EIGER 16M detector on 24-ID-E beam line is funded by an NIH-ORIP High-End Instrumentation grant (S10OD021527). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract DE-AC02-06CH11357.
This work was primarily supported by a Collaboration for AIDS Vaccine Discovery Grant OPP1114721 from the Bill and Melinda Gates Foundation to BFH, the Henry M. Jackson Foundation for the Advancement of Military Medicine grant 829295 to BFH, and NIH grants R01AI120801 to KOS, R01AI125093 to HD, and P01 AI120756 and AI064518 to GDT. This work was supported by the Department of Defense through a cooperative agreement (W81XWH-18-2-0040) with the Henry M. Jackson Foundation for the Advancement of Military Medicine and the US Department of Defense.
The views expressed are those of the authors and should not be construed to represent the positions of the Uniformed Services University, US Army, Department of Defense, or Department of Health and Human Services. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70.
Version 1. 01/30/2020
Electronic publication
Version 2. 01/30/2020
The affiliations were corrected.
Footnotes
JHK and JLE’s present address is: International Vaccine Institute, Seoul, Republic of Korea.
MK’s present address is: Health Policy, New America, Washington, DC, USA.
SP’s present address is: Glaxosmithkline Discovery Performance Unit, Siena, Italy.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(2):e131437.https://doi.org/10.1172/jci.insight.131437.
Contributor Information
David Easterhoff, Email: david.easterhoff@duke.edu.
Justin Pollara, Email: justin.pollara@duke.edu.
Kan Luo, Email: kan.luo@dm.duke.edu.
Neelakshi Gohain, Email: ngohain@hivresearch.org.
LaTonya D. Williams, Email: latonya.williams@duke.edu.
Matthew Zirui Tay, Email: matthew.zirui.tay@duke.edu.
Anthony Monroe, Email: anthony.monroe@duke.edu.
Misook Choe, Email: mchoe@hivresearch.org.
Susie Min, Email: smin3642@gmail.com.
Paolo Lusso, Email: paolo.lusso@nih.gov.
Peng Zhang, Email: peng.zhang2@nih.gov.
Eden P. Go, Email: edenp@ku.edu.
Heather Desaire, Email: hdesaire@ku.edu.
Mattia Bonsignori, Email: mattia.bonsignori@duke.edu.
Kwan-Ki Hwang, Email: kwan-ki.hwang@duke.edu.
Charles Beck, Email: cebeck1221@gmail.com.
Matina Kakalis, Email: matinakakalis@gmail.com.
Sandhya Vasan, Email: SVasan@hivresearch.org.
Jerome H. Kim, Email: Jerome.Kim@ivi.int.
Jean-Louis Excler, Email: JeanLouis.Excler@ivi.int.
Supachai Rerks-Ngarm, Email: supachair@biophics.org.
Jaranit Kaewkungwal, Email: jaranitk@biophics.org.
Punnee Pitisuttithum, Email: punnee.pit@mahidol.ac.th.
Sorachai Nitayaphan, Email: SorachaiN@afrims.org.
Faruk Sinangil, Email: fsinangil@gsid.org.
James Tartaglia, Email: jim.tartaglia@sanofipasteur.com.
Sanjay Phogat, Email: sphogat@gmail.com.
Kevin Wiehe, Email: kevin.wiehe@duke.edu.
Kevin O. Saunders, Email: kevin.saunders@duke.edu.
Georgia D. Tomaras, Email: georgia.tomaras@duke.edu.
James Arthos, Email: jarthos@niaid.nih.gov.
Mangala Rao, Email: mrao@hivresearch.org.
M. Gordon Joyce, Email: gjoyce@hivresearch.org.
Guido Ferrari, Email: gflmp@dm.duke.edu.
Barton F. Haynes, Email: barton.haynes@duke.edu.
References
- 1.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Robb ML, et al. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12(7):531–537. doi: 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottardo R, et al. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS ONE. 2013;8(9):e75665. doi: 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karasavvas N, et al. The Thai Phase III HIV Type 1 Vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses. 2012;28(11):1444–1457. doi: 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zolla-Pazner S, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PLoS ONE. 2014;9(2):e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland M, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490(7420):417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao HX, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38(1):176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiehe K, et al. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41(6):909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rerks-Ngarm S, et al. Randomized, double-blind evaluation of late boost strategies for HIV-uninfected vaccine recipients in the RV144 HIV vaccine efficacy trial. J Infect Dis. 2017;215(8):1255–1263. doi: 10.1093/infdis/jix099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yates NL, et al. HIV-1 envelope glycoproteins from diverse clades differentiate antibody responses and durability among vaccinees. J Virol. 2018;92(8):e01843-17. doi: 10.1128/JVI.01843-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao HX, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158(1-2):171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicely NI, et al. Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee. EBioMedicine. 2015;2(7):713–722. doi: 10.1016/j.ebiom.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spurrier B, Sampson J, Gorny MK, Zolla-Pazner S, Kong XP. Functional implications of the binding mode of a human conformation-dependent V2 monoclonal antibody against HIV. J Virol. 2014;88(8):4100–4112. doi: 10.1128/JVI.03153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorny MK, et al. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J Virol. 1994;68(12):8312–8320. doi: 10.1128/JVI.68.12.8312-8320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Upadhyay C, et al. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J Virol. 2014;88(21):12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayr LM, Cohen S, Spurrier B, Kong XP, Zolla-Pazner S. Epitope mapping of conformational V2-specific anti-HIV human monoclonal antibodies reveals an immunodominant site in V2. PLoS ONE. 2013;8(7):e70859. doi: 10.1371/journal.pone.0070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tassaneetrithep B, et al. Cryptic determinant of α4β7 binding in the V2 loop of HIV-1 gp120. PLoS ONE. 2014;9(9):e108446. doi: 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthos J, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9(3):301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 22.Peachman KK, et al. Identification of new regions in HIV-1 gp120 variable 2 and 3 loops that bind to α4β7 integrin receptor. PLoS ONE. 2015;10(12):e0143895. doi: 10.1371/journal.pone.0143895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tay MZ, et al. Antibody-mediated internalization of infectious HIV-1 virions differs among antibody isotypes and subclasses. PLoS Pathog. 2016;12(8):e1005817. doi: 10.1371/journal.ppat.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musich T, Li L, Liu L, Zolla-Pazner S, Robert-Guroff M, Gorny MK. Monoclonal antibodies specific for the V2, V3, CD4-binding site, and gp41 of HIV-1 mediate phagocytosis in a dose-dependent manner. J Virol. 2017;91(8):e02325-16. doi: 10.1128/JVI.02325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desrosiers RC. Protection against HIV Acquisition in the RV144 Trial. J Virol. 2017;91(18):e00905-17. doi: 10.1128/JVI.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zolla-Pazner S, Alvarez R, Kong XP, Weiss S. Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV. Curr Opin HIV AIDS. 2019;14(4):309–317. doi: 10.1097/COH.0000000000000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482(7383):89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roederer M, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505(7484):502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pegu P, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol. 2013;87(3):1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon SN, et al. Antibody to the gp120 V1/V2 loops and CD4+ and CD8+ T cell responses in protection from SIVmac251 vaginal acquisition and persistent viremia. J Immunol. 2014;193(12):6172–6183. doi: 10.4049/jimmunol.1401504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh S, et al. Control of heterologous simian immunodeficiency virus SIVsmE660 infection by DNA and protein coimmunization regimens combined with different Toll-like-receptor-4-based adjuvants in macaques. J Virol. 2018;92(15):e00281-18. doi: 10.1128/JVI.00281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivro A, et al. Integrin α4β7 expression on peripheral blood CD4+ T cells predicts HIV acquisition and disease progression outcomes. Sci Transl Med. 2018;10(425):eaam6354. doi: 10.1126/scitranslmed.aam6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hessell AJ, et al. Reduced cell-associated DNA and improved viral control in macaques following passive transfer of a single anti-V2 monoclonal antibody and repeated simian/human immunodeficiency virus challenges. J Virol. 2018;92(11):e02198-17. doi: 10.1128/JVI.02198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wibmer CK, et al. Common helical V1V2 conformations of HIV-1 Envelope expose the α4β7 binding site on intact virions. Nat Commun. 2018;9(1):4489. doi: 10.1038/s41467-018-06794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Barnes CO, Yang Z, Nussenzweig MC, Bjorkman PJ. Partially open HIV-1 envelope structures exhibit conformational changes relevant for coreceptor binding and fusion. Cell Host Microbe. 2018;24(4):579–592.e4. doi: 10.1016/j.chom.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Byrareddy SN, et al. Targeting α4β7 integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nat Med. 2014;20(12):1397–1400. doi: 10.1038/nm.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arthos J, et al. The role of integrin α4β7 in HIV pathogenesis and treatment. Curr HIV/AIDS Rep. 2018;15(2):127–135. doi: 10.1007/s11904-018-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santangelo PJ, et al. Early treatment of SIV+ macaques with an α4β7 mAb alters virus distribution and preserves CD4+ T cells in later stages of infection. Mucosal Immunol. 2018;11(3):932–946. doi: 10.1038/mi.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwamoto N, et al. Blocking α4β7 integrin binding to SIV does not improve virologic control. Science. 2019;365(6457):1033–1036. doi: 10.1126/science.aaw7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parrish NF, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balasubramanian P, et al. Functional antibody response against V1V2 and V3 of HIV gp120 in the VAX003 and VAX004 vaccine trials. Sci Rep. 2018;8(1):542. doi: 10.1038/s41598-017-18863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karnasuta C, et al. Comparison of antibody responses induced by RV144, VAX003, and VAX004 vaccination regimens. AIDS Res Hum Retroviruses. 2017;33(5):410–423. doi: 10.1089/aid.2016.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams WB, et al. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349(6249):aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonsignori M, et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci Transl Med. 2017;9(381):eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kepler TB, et al. Reconstructing a B-cell clonal lineage. ii. mutation, selection, and affinity maturation. Front Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Easterhoff D, et al. Boosting of HIV envelope CD4 binding site antibodies with long variable heavy third complementarity determining region in the randomized double blind RV305 HIV-1 vaccine trial. PLoS Pathog. 2017;13(2):e1006182. doi: 10.1371/journal.ppat.1006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12:Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 48.Edmonds TG, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408(1):1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollara J, et al. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A. 2011;79(8):603–612. doi: 10.1002/cyto.a.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70(2):1100–1108. doi: 10.1128/JVI.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood. 1997;90(3):1109–1114. [PubMed] [Google Scholar]
- 52.Bruhns P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 53.Shukair SA, et al. Human cervicovaginal mucus contains an activity that hinders HIV-1 movement. Mucosal Immunol. 2013;6(2):427–434. doi: 10.1038/mi.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wills S, et al. HIV-1-Specific IgA monoclonal antibodies from an HIV-1 vaccinee mediate galactosylceramide blocking and phagocytosis. J Virol. 2018;92(7):e01552-17. doi: 10.1128/JVI.01552-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tay MZ, et al. Rare detection of antiviral functions of polyclonal IgA isolated from plasma and breast milk compartments in women chronically infected with HIV-1. J Virol. 2019;93(7):e02084-18. doi: 10.1128/JVI.02084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ansari AA, et al. Blocking of α4β7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J Immunol. 2011;186(2):1044–1059. doi: 10.4049/jimmunol.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders KO, et al. Vaccine elicitation of high mannose-dependent neutralizing antibodies against the V3-glycan broadly neutralizing epitope in nonhuman primates. Cell Rep. 2017;18(9):2175–2188. doi: 10.1016/j.celrep.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yates NL, et al. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med. 2014;6(228):228ra39. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomaras GD, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 65.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 66.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27(1):293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Go EP, et al. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res. 2008;7(4):1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Go EP, et al. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res. 2009;8(9):4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irungu J, et al. Comparison of HPLC/ESI-FTICR MS versus MALDI-TOF/TOF MS for glycopeptide analysis of a highly glycosylated HIV envelope glycoprotein. J Am Soc Mass Spectrom. 2008;19(8):1209–1220. doi: 10.1016/j.jasms.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.