The noninvasive detection of Helicobacter pylori and its resistance to clarithromycin could revolutionize the management of H. pylori-infected patients by tailoring eradication treatment without any need for endoscopy when histology is not necessary. Several real-time PCR tests performed on stools have been proposed, but their performances were either poor or they were tested on too few patients to be properly evaluated. We conducted a prospective, multicenter study including 1,200 adult patients who were addressed for gastroduodenal endoscopy with gastric biopsies and who were naive for eradication treatment in order to evaluate the performance of the Amplidiag H. pylori+ClariR assay recently developed by Mobidiag (Espoo, Finland).
KEYWORDS: diagnosis, gastric cancer, gastric ulcers, Helicobacter pylori, molecular diagnosis, noninvasive sampling, PCR, stool
ABSTRACT
The noninvasive detection of Helicobacter pylori and its resistance to clarithromycin could revolutionize the management of H. pylori-infected patients by tailoring eradication treatment without any need for endoscopy when histology is not necessary. Several real-time PCR tests performed on stools have been proposed, but their performances were either poor or they were tested on too few patients to be properly evaluated. We conducted a prospective, multicenter study including 1,200 adult patients who were addressed for gastroduodenal endoscopy with gastric biopsies and who were naive for eradication treatment in order to evaluate the performance of the Amplidiag H. pylori+ClariR assay recently developed by Mobidiag (Espoo, Finland). The results of the Amplidiag H. pylori+ClariR assay performed on DNA from stools (automatic extraction with the EasyMag system [bioMérieux]) were compared with those of culture/Etest and quadruplex real-time PCRs performed on two gastric biopsy samples (from the antrum and corpus) to detect the H. pylori glmM gene and mutations in the 23S rRNA genes conferring clarithromycin resistance. The sensitivity and specificity of the detection of H. pylori were 96.3% (95% confidence interval [CI], 92 to 98%) and 98.7% (95% CI, 97 to 99%), respectively. The positive and negative predictive values were evaluated to be 92.2% (95% CI, 92 to 98%) and 99.3% (95% CI, 98 to 99%), respectively. In this cohort, 160 patients (14.7%) were found to be infected (positive by culture and/or PCR). The sensitivity and specificity for detecting resistance to clarithromycin were 100% (95% CI, 88 to 100%) and 98.4% (95% CI, 94 to 99%), respectively.
INTRODUCTION
Helicobacter pylori is responsible for the most common chronic bacterial infection in humans (1, 2). This bacterium has been implicated in the pathogenesis of gastroduodenal diseases, such as gastritis, peptic ulcer, mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric adenocarcinoma (2, 3, 39). Eradication of H. pylori infection has been associated with a reduction of the risk of recurrent peptic ulcer diseases as well as a reduction of the risk of development of gastric adenocarcinoma and MALT lymphoma (4). Clarithromycin is a key component of the first-line treatment used to eradicate H. pylori. It is now widely accepted that clarithromycin resistance among H. pylori strains has been increasing worldwide and is the most important factor responsible for the failure of eradication treatment (5–7). Clarithromycin resistance in H. pylori was considered in 2017 by the World Health Organization (WHO) to be a high priority for antibiotic research (8). The mechanism leading to clarithromycin resistance in H. pylori is well characterized. Resistance in H. pylori is mostly due to point mutations in the peptidyltransferase region of the 23S rRNA gene. Single nucleotide variants decrease the affinity between ribosomes and clarithromycin. The mutations A2143G and A2142G/C are most commonly associated with clarithromycin resistance in natural H. pylori isolates worldwide (9, 10). In addition, other 23S rRNA gene mutations, such as T2289C, C2245T, G2224A, T2182C, T2717C, and T2243C, have occasionally been reported in clarithromycin-resistant H. pylori isolates, but their relevance is still under debate (11). Given the spread of antibiotic resistance and the subsequent failure of empirical eradication treatment, treatment guided by the results of culture and standard susceptibility tests or PCR detection of resistance has been recommended in regions or populations with high rates of clarithromycin resistance (>15%) (4, 12, 13). Up until now, only invasive methods requiring endoscopy for the collection of biopsy specimens could reliably detect clarithromycin resistance with good performances (14). Several attempts to develop methods for the detection of clarithromycin resistance with noninvasive sampling (stool sampling, for example) have been reported, but their performances were not sufficient for use in the clinical management of patients. Moreover, either the performances of these tests were poor or the tests were tested on a small cohort, limiting the quality of their evaluation (15–25).
A noninvasive test detecting H. pylori and its resistance to clarithromycin could revolutionize the management of H. pylori-infected patients by tailoring eradication treatment without any need for endoscopy when histology is not necessary.
Mobidiag (Espoo, Finland) recently developed a new multiplex real-time PCR assay (Amplidiag H. pylori+ClariR), CE-IVD marketed, detecting H. pylori infection and mutations conferring clarithromycin resistance (without the distinction of the mutations). With good performances when performed on gastric biopsy samples, this test can also be performed on stool specimens (26, 27).
The aim of this study was to evaluate the performances (sensitivity, specificity, positive predictive value [PPV], negative predictive value [NPV], accuracy) of the real-time Amplidiag H. pylori+ClariR stool PCR assay on a large cohort of adult patients.
MATERIALS AND METHODS
Ethical considerations.
The University Hospital of Poitiers was the promoter of the study. Mobidiag was the sponsor of the study but was not involved in the design, the interpretation of the results, or the decision to publish this study.
The study received ethical approval from the regional Committee for the Protection of People (CPP) and from the National Commission for Protection of Personal Data on 23 November 2015. All patients were informed before inclusion, and their consent was obtained before analysis.
Study design.
A prospective multicenter clinical study (study HepyStool 2015-A01075-44) was set up to evaluate the Amplidiag H. pylori+ClariR assay (Mobidiag) performances on stools from adult patients. To limit confusion biases, these patients were naive for H. pylori eradication treatment. The results of the test on stools were then compared with those of tests on two gastric biopsy samples, i.e., culture and/or glmM PCR, to evaluate the detection of H. pylori and Etest and/or a quadruplex real-time PCR (which detect mutations in the 23S rRNA genes) to evaluate the detection of clarithromycin resistance.
This study was carried out in four hospitals in west-central France (the University Hospital of Poitiers, Hospital of Montmorillon, Hospital of Thouars, and Hospital of Parthenay), but all of the analyses were performed in the Bacteriology Laboratory of the University Hospital of Poitiers. Consecutive patients who met the selection criteria were included from December 2015 to July 2018 (30 months). Inclusions were performed 2 days per week.
Study population.
Consecutive adult patients without diarrhea referred for endoscopy and gastric biopsy on two random days per week were screened for inclusion as representative targets for the index test. Inclusion criteria were an age of ≥18 years, referral as a hospitalized patient or an outpatient to one of the participating centers for endoscopy because of suspected H. pylori infection, a willingness to participate, and signing of an informed consent. Exclusion criteria were an age of <18 years, recent or ongoing antibiotic treatment for ≤4 weeks, previous receipt of eradication treatment, no affiliation with social insurance, receipt of legal protection, refusal to sign informed consent, inclusion in another trial, an inability to take oral medication on an ongoing basis, or the presence of severe life-threatening disease in the short term.
Gastric biopsies. (i) Collection of gastric biopsy samples.
As recommended for routine diagnosis, a single corpus and antrum biopsy specimen from each patient was collected during endoscopy and immediately placed into Portagerm pylori transport medium (Port-Pyl; bioMérieux, Marcy l’Etoile, France) before being sent to the laboratory within 48 h (14). Biopsy specimens (10 to 20 mg) were ground together with a Retsch MM400 mixer mill (Retsch, Haan, Germany) in 1 ml sterile water with two 4-mm-diameter inox beads in a 1.5-ml Eppendorf tube for 1 min 30 s at 20 Hz. The material was then split into three parts: 100 μl was used for culture, 500 μl was used for immediate molecular analysis, and 400 μl was stored at −80°C.
(ii) Culture from gastric biopsy samples.
For the culture phase, biopsy material was plated on Columbia agar supplemented with 10% (vol/vol) sheep blood and Skirrow antibiotic supplement (Oxoid, Basingstoke, Hampshire, United Kingdom). The inoculated plates were incubated at 37°C for 12 days under microaerobic conditions (by using a GENbox Microaer generator [bioMérieux] in an anaerobic jar). Identification of H. pylori was based on colony morphology, typical appearance on Gram staining, and positive urease, catalase, and oxidase activities.
For phenotypic clarithromycin susceptibility testing, a suspension of the H. pylori culture in saline buffer was adjusted to a number 3 McFarland standard (14). A Gram staining examination of a smear was then performed with each H. pylori strain to confirm the absence of coccoid forms. The suspension was plated onto Mueller-Hinton agar supplemented with 10% horse blood (Becton, Dickinson, NJ, USA). An Etest strip containing clarithromycin (bioMérieux) at concentrations ranging from 0.016 to 256 mg/liter was placed on the agar after plating. The inoculated plates were incubated at 37°C for 3 days under microaerobic conditions. The MIC was determined, and susceptibility was interpreted according to 2016 EUCAST recommendations, applying a clinical breakpoint of ≤0.5 mg/liter. To control the process, H. pylori strain CCUG 17874 was similarly analyzed.
(iii) Molecular analysis from gastric biopsy samples.
For the molecular analysis phase, DNA was extracted from 500 μl of ground material from the gastric biopsy samples by using a NucliSENS easyMAG system (bioMérieux), according to the manufacturer’s instructions, in 400 μl lysis buffer, using the specific B 2.1 protocol, 100 μl of silica, and 100 μl of eluate. The isolated DNA was stored at −20°C until use.
glmM PCR.
To detect H. pylori infection, a TaqMan real-time PCR assay targeting the H. pylori-specific gene glmM was used, optimizing published methods (27–29).
Briefly, the primer and probe sequences were as follows: forward primer, 5′-AGCGCTCTCACTTCCATAGGC-3′; reverse primer, 5′-TCTTCGGTTAAAAAAGCGAT-3′ (the primers amplified an 80-bp fragment); and TaqMan probe, 5′-6-FAM TGATCCAAATAGGGCCTATGCCTACCCC-3′-TAMRA (where 6-FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine). Real-time PCR was performed on an Applied Biosystems 7500 Fast thermocycler (Life Technologies, CA, USA). An internal control (5 μl) was included in the ground material of the gastric biopsy samples before extraction (with a Simplexa Focus Diagnostics extraction kit and an amplification control primer pair; DiaSorin, Saluggia, Italy). The amplification reaction mix (25 μl) contained Premix Ex Taq DNA polymerase (TaKaRa, Shiga, Japan), 90 nM each primer, 70 nM probe, and 5 μl of the extracted DNA. Amplification consisted of the following program: 20 s of denaturing at 95°C, followed by 45 cycles of 15 s at 95°C and 30 s at 55°C. A positive control (DNA extracted from four strains, the 23S rRNA genes of which were sequenced) and a negative control (molecular-quality distilled water) were used in each run of the PCR assay.
Scorpion PCR.
Detection of H. pylori single nucleotide variants in the 23S rRNA gene was performed by Scorpion PCR according to a previously published method in a multiplex real-time PCR assay that detects H. pylori infection and identifies four alleles of the 23S rRNA gene of H. pylori. The process detects the wild-type sequence and three mutations conferring clarithromycin resistance (A2142G, A2143G, and A2142C) (30). A positive control (DNA extracted from four strains, the 23S rRNA genes of which were sequenced) and a negative control (molecular-quality distilled water) were used in each run of the PCR assay.
PCR was performed in a final volume of 25 μl with Premix Ex Taq (TaKaRa), 5 μl of DNA extracted from the biopsy specimen, 0.1 μM oligonucleotide primer 23SF2, 0.14 μM primer 23SscA2142G, 0.18 μM primer 23SscA2143G, 0.1 μM primer 23SscA2142C, and 0.08 μM primer 23SScWT. Amplification was performed after a denaturation step (95°C for 15 s) for 50 cycles of denaturation (95°C for 15 s), annealing (55°C for 30 s), and extension (72°C for 20 s). Reading of the fluorescence was performed at the annealing step on four channels (6-carboxyfluorescein [FAM], Texas Red, Cy3, and Cy5).
Stool. (i) Stool sample collection.
Within 1 week after endoscopy and biopsies and before treatment, stools were self-collected by the patients at home using a disposable stool collection device (Daklapack, Perpignan, France) with an eNAT tube (Copan, Brescia, Italy) for transport at ambient temperature. The eNAT tubes were immediately sent to the laboratory. After receipt in the laboratory, the samples were stored at −80°C until testing. The delay between stool emission and storage in the laboratory was reported in days.
(ii) Nucleic acid extraction from stool samples.
After collection, 400 μl of the eNAT tube supernatant was extracted using the NucliSENS easyMAG system (bioMérieux), according to the manufacturer’s instructions, in 2 ml lysis buffer under the following conditions: the specific B 2.1 protocol with 140 μl of silica, after adding of 1 μl of internal control (Amplidiag Easy Process Control I; Mobidiag), for a final volume of eluate of 70 μl.
(iii) Real-time PCR on stool with the Amplidiag H. pylori+ClariR assay.
Real-time PCR was conducted according to the manufacturer’s instructions (Amplidiag H. pylori+ClariR; Mobidiag) on a Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA). Twenty-eight samples at a maximum were tested at a time with 96‐well plates, because all samples (including the negative, wild-type [WT], and mutant controls) were tested in triplicate. Note that each reaction mixture included an internal amplification control aimed at detecting PCR inhibitors (Amplidiag Easy Process Control I; Mobidiag). The raw data were analyzed by using a fully automated analysis program, the Amplidiag analyzer (Mobidiag). The test provided results for the detection of H. pylori and for the detection of mutations conferring clarithromycin resistance (without a distinction between the mutations).
(iv) Determination of analytical sensitivity of Amplidiag H. pylori+ClariR assay.
To test the analytical sensitivity of the Amplidiag H. pylori+ClariR assay, DNAs extracted from specific inocula of the four reference strains (3.5 × 106 CFU/μl of DNA for the J99 strain with the wild-type phenotype/genotype, 1 × 106 CFU/μl of DNA for clarithromycin-resistant strain HP 222 with the A2143G mutation, 2.5 × 106 CFU/μl of DNA for clarithromycin-resistant strain HP 225 with the A2142G mutation, and 1.75 × 106 CFU/μl of DNA for clarithromycin-resistant strain HP 825 with the A2142C mutation) were diluted 10-fold to 10−7 in an H. pylori-negative stool matrix. The seven 10-fold serial dilutions of each DNA were tested by PCR. To determine the specificity of H. pylori detection in stool samples, 10 Campylobacter species culture-positive stool samples were processed similarly.
Regarding the detection of multiple strains in the same biopsy sample, chromosomal DNA from the wild-type reference strain J99 and from a clarithromycin-resistant reference strain harboring either the A2143G (43G), A2142G (42G), or A2142C (42C) mutation was mixed in different proportions. To assess the performances of the PCR, geometric dilutions were tested in saline buffer and in a Helicobacter-negative stool sample. Briefly, after precise quantification of the inoculum using culture and molecular methods, both resistant and wild-type strains were diluted to the same concentration (6.25 × 107 CFU/ml, corresponding to a threshold cycle value of 22) and then geometrically diluted to determine the limit of detection of a pure WT or resistant strain. All PCRs were carried out in triplicate, and the results were considered negative when zero or one replicate was positive.
After that, each of the three mutated strains was diluted in a constant quantity of the WT strain (42G scale, 42C scale, and 43G scale). H. pylori subpopulations were obtained in the following proportions: 99%, 95%, 90%, 80%, 20%, 10%, 5%, and 1%. In this way, artificial samples (n = 24) were obtained and analyzed by PCR. To assess the performances in both a paucicellular matrix and a highly cellular matrix, volume-to-volume dilution in elution buffer or conservation buffer with Helicobacter-negative stool was carried out.
(v) External quality assessment.
Two sets of 10 samples for external quality assessment (Helicobacter pylori EQA Pilot Study 2017 and 2018; QCMD, Glasgow, Scotland, UK) were tested to control the quality of our results.
Reference tests.
Patients were tested, in parallel with stool PCR, by culture and PCR of two gastric biopsy samples collected during esophagogastroduodenoscopy, to determine a consensus patient infection status. A patient was considered infected by H. pylori when culture yielded spiral urease-, catalase-, and oxidase-positive bacteria or the glmM DNA fragment was detected by the TaqMan real-time PCR assay of gastric biopsy samples. The infecting strain of H. pylori was considered resistant to clarithromycin if the clarithromycin MIC of the cultivated strain determined by Etest was over 0.5 mg/liter or if one of the three mutations conferring clarithromycin resistance (A2142G, A2142C, A2143G) was detected by the Scorpion PCR.
The performances of the evaluated test (sensitivity, specificity, positive predictive value, negative predictive value, and accuracy) and their confidence intervals were determined for the two parameters tested: detection of H. pylori infection and detection of clarithromycin resistance (31).
Analysis of discrepant results.
The real-time PCR products of samples with discrepant results (false-positive results, i.e., a positive result with the tested method and a negative result with the reference method) were sequenced on an ABI Prism 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA) after purification (QIAquick PCR purification kit; Qiagen, Hilden, Germany). The primer sequences used were 5′-GCTAGTCTAAGGGCGTAGATTGGAGGGAAG-3′ for the forward primer and 5′-GCTTGTGCCATTACACTCAACTTGCGATTTC-3′ for the reverse primer. The sequencing program included an initial denaturation of the templates at 94°C for 10 min, followed by 40 cycles, each of which consisted of a denaturation step at 94°C for 15 s and an annealing and elongation step for 2 min at 66 to 68°C. After production, the DNA sequences were verified by BLAST analysis. Similarly, false-positive results (i.e., a positive result by the tested method and a negative result by the reference method) were verified using an Amplidiag Bacterial GE real-time multiplex PCR kit (Amplidiag), searching for Campylobacter DNA (32).
Statistical analyses.
Based on an estimated prevalence of H. pylori infection of 17% (the prevalence of the infection in the main investigation center in 2015; data not shown) and an estimated prevalence of clarithromycin resistance of 20% (the resistance prevalence in the main investigation center in 2015; data not shown), expecting a sensitivity of clarithromycin resistance detection of 0.95 ± 0.07, the minimal number of patients required to be evaluated was calculated to be 1,200 (31).
RESULTS
Analyzed population.
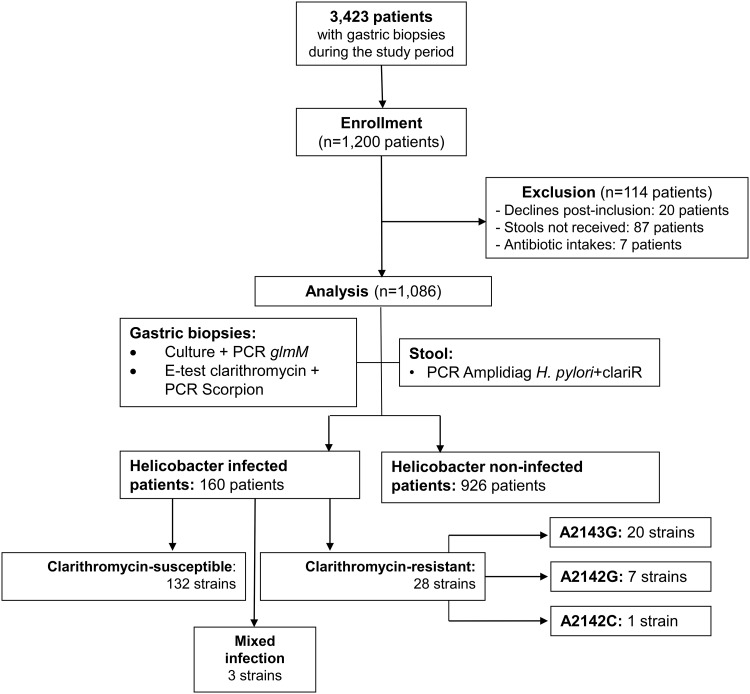
In this prospective multicenter study, 1,200 patients were included by gastroenterologists from four centers from December 2015 to July 2018 (30 months). The activities involving samples from these patients constituted 35% of the complete activity of the Bacteriology Laboratory of the University Hospital of Poitiers during the same period (1,200/3,423). A total of 114 patients (9.3%) were excluded from the study: 20 (1.7%) refused to continue the trial, 87 did not send their stool sample (7%), and 7 (<1%) took antibiotics before stool was collected (Fig. 1). This population was composed of 53% males with a median age of 56 years (range, 22 to 96 years), and two-thirds were born in France (Table 1).
FIG 1.
Study design and flowchart. Out of the 1,200 enrolled patients, after exclusion of 114 patients, the population (n = 1,086) was divided into two groups according to their H. pylori infection status (infected patients, n = 160; noninfected patients, n = 926).
TABLE 1.
Patient baseline characteristics
| Characteristic | Value for: |
|
|---|---|---|
| H. pylori-infected patients (n = 160) | All patients (n = 1,086) | |
| Age (yr) | 50 | 51.4 |
| No. (%) of patients by sex | ||
| Male | 72 (45) | 446 (41) |
| Female | 88 (55) | 640 (59) |
| No. (%) of patients with the following birth locationa : | ||
| Europe | 115 (71.9) | 988 (91.0) |
| Africa | 38 (23.7) | 76 (7.0) |
| Asia | 4 (2.5) | 11 (1.0) |
| The Americas | 3 (1.9) | 11 (1.0) |
| Median (range) delay (days) before receipt of stool | 1.47 (0–7) | 1.37 (0–10) |
Percentage of infections among patients born in each location: Europe (11.6%), Africa (50.0%), Asia (36.0%), and America (27%).
Analytical performances of Amplidiag H. pylori+ClariR assay. (i) Limit of detection of pure WT H. pylori or H. pylori strains with 23S rRNA mutations.
In saline buffer, the pure WT H. pylori strain was detected up to a concentration of 3.12 × 10−3 CFU/ml. In stool conserved in conservation medium, detection was possible up to a concentration of 3.12 CFU/ml.
In saline buffer, pure 42C, 42G, and 43G mutant H. pylori strains were detected at minimal concentrations of 3.12 × 10−4 CFU/ml, 3.12 × 10−3 CFU/ml, and 3.12 × 10−3 CFU/ml, respectively. In stool conserved in conservation medium, pure 42C, 42G and 43G mutant H. pylori strains were detected at minimal concentrations of 3.12 CFU/ml, 3.12 × 10−1 CFU/ml, and 3.12 × 10−1 CFU/ml, respectively.
(ii) Limit of detection of mixed wild-type and mutated H. pylori strains.
In saline buffer, clarithromycin-resistant subpopulations were detected when they represented ≥1% of the population. In stool sample plus conservation medium, resistant 42C, 42G, and 43G mutant subpopulations were detected when they represented ≥5% of the population. Note that in this buffer, the wild-type subpopulation was not detected when it represented less than 5% of the population in a resistant 43G mutant subpopulation.
(iii) External quality assessment.
Two sets of 9 samples for external quality assessment (Helicobacter pylori EQA Pilot Study 2017 and 2018; QCMD, Glasgow, Scotland, UK) were tested during the study to control the quality of our results. We obtained a perfect concordance with the expected results. The sets of samples consisted of 6 samples with wild-type clarithromycin-susceptible H. pylori, 10 samples with clarithromycin-resistant H. pylori, and 2 negative samples.
Detection of H. pylori and clarithromycin susceptibility testing by culture and real-time PCR.
As summarized in Table 2, among the 1,086 gastric biopsy samples tested in this study, culture was positive for 154 biopsy specimens (14.2%). Among the strains from these specimens, Etest identified 28 to be clarithromycin resistant (28/154; 18.2%). One hundred sixty gastric biopsy samples (160/1,086; 14.7%) were positive for the H. pylori-specific gene glmM. Among those samples, clarithromycin resistance-associated mutations of the 23S rRNA gene were detected in 28 gastric biopsy samples (28/160; 17.5%); i.e., the A2143G (n = 20; 71.4%), A2142G (n = 7; 25%), or A2142C (n = 1; 3.6%) mutation was detected. Rare gastric biopsy samples harbored a mixed infection (3/160; 1.9%) with both clarithromycin-susceptible and -resistant isolates. In these cases, mixed infections were not detected by phenotypic susceptibility testing with Etest, as the resistant isolate was the only one detected.
TABLE 2.
Comparison of culture and clarithromycin susceptibility testing of biopsy specimens compared to molecular testing of stoola
| No. of patients (n = 1,086) | Result for: |
||||
|---|---|---|---|---|---|
| Gastric biopsy samples |
Stool by real-time Amplidiag H. pylori+ClariR PCR | ||||
| Culture |
Real-time PCR |
||||
| Growth | Etest | glmM | 23S rRNA by Scorpion PCR | ||
| 913 | − | − | − | − | − |
| 13 | − | − | − | − | S |
| 5 | + | S | + | S | − |
| 1 | + | R | + | R | − |
| 120 | + | S | + | S | S |
| 5 | − | − | + | S | S |
| 24 | + | R | + | R | R |
| 3 | + | R | + | R + S | R |
| 1 | + | S | + | S | R |
| 1 | − | − | + | S | R + S |
Clarithromycin susceptibility testing was carried out by Etest and real-time PCR (for detection of glmM and 23S rRNA gene mutations by Scorpion PCR) of biopsy specimens. S, susceptible; R, resistant.
Finally, considering the results of both culture and PCR, 160 patients (160/1,086; 14.7%) were considered H. pylori infected.
Note that five gastric biopsy samples were negative by culture and positive by PCR (positive for a wild-type susceptible H. pylori strain), and all gastric biopsy samples in which glmM was detected by real-time PCR were positive (for a wild-type or mutated allele) by the Scorpion PCR. No discrepant results between phenotypic and genotypic methods for the detection of clarithromycin resistance were observed.
The H. pylori-infected population was not statistically significantly different from the total population in terms of demographic characteristics, except for the place of birth (Europe versus other countries at 11.6% versus 28%, P < 0.01; Table 1).
The distribution of H. pylori-related diseases (n = 147 patients for which results were available) yielded 83 cases of gastritis (83/147; 56.5%), 12 duodenal ulcers (12/147; 8.2%), 5 gastric ulcers (5/147; 3.4%), and 47 (31.2%) precancerous and cancerous lesions (23 gastric atrophies [23/147; 15.6%], 19 cases of metaplasia [19/147; 12.9%], 3 cases of dysplasia [3/147; 2.0%], and 2 cases of gastric adenocarcinoma [2/147; 1.4%]).
Clinical performances of the Amplidiag H. pylori+ClariR assay on stool samples.
For the 1,086 stool samples collected for this study, the average time between home stool delivery and sample tube freezing in the laboratory was 1.37 days (range, 0 to 6 days). All samples were tested by operators maintained blind to the culture results. Out of these samples, the results for five remained invalid due to discrepant results by the three tests (5/1,086; 0.5%), These 5 samples were run again and were then rendered valid for analysis. The PCR process did not detect any inhibition of the PCR.
The performances of the stool PCR in comparison to the results of culture and gastric biopsy were analyzed on the basis of two different parameters: (i) detection of H. pylori infection and (ii) detection of clarithromycin resistance (Table 3).
TABLE 3.
Performances of the H. pylori Amplidiag H. pylori+ClariR assay for detection and susceptibility testing of H. pylori in stoola
| Result detected | No. (%) of specimens with the following result: |
Analytical performance (%)b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | Sensitivity | Specificity | PPV | NPV | Accuracy | |
| H. pylori (n = 1,086) | 154 (14.2) | 913 (84.1) | 13 (1.2) | 6 (0.6) | 96.3 (92–98) | 98.7 (97–99) | 92.2 (92–98) | 99.3 (98–99) | 98.2 (97–99) |
| Clarithromycin resistance (n = 154) | 27 (17.5) | 125 (81.2) | 2 (1.3) | 0 (0) | 100 (88–100) | 98.4 (94–99) | 93.1 (78–98) | 100 (97–100) | 98.7 (95–99) |
True-positive specimens (TP) were defined as specimens positive by both the Amplidiag test on stool and culture or PCR of gastric biopsy samples or resistant by both the Amplidiag test on stool and Etest or Scorpion PCR of gastric biopsy samples. True-negative (TN) specimens were defined as specimens negative by both the Amplidiag test on stool and culture or PCR of gastric biopsy samples or susceptible by both the Amplidiag test on stool and Etest or Scorpion PCR of gastric biopsy samples. False-positive (FP) specimens were defined as specimens positive by the Amplidiag test on stool and negative by culture or PCR of gastric biopsy samples and susceptible by the Etest or Scorpion PCR of gastric biopsy samples and resistant by the Amplidiag test on stool. False-negative (FN) specimens were defined as specimens positive by culture or PCR of gastric biopsy samples and negative by the Amplidiag test on stool or susceptible by the Amplidiag test on stool and resistant by the Etest or Scorpion PCR of gastric biopsy samples. PPV, positive predictive value; NPV, negative predictive value.
Values in parentheses are 95% confidence intervals.
First, for the detection of H. pylori, the reference considered both culture and PCR of biopsy specimens, enabling detection of H. pylori in 15% of the total number of patients included. Among this group and even after retesting, 13 false-positive results (a positive result by the Amplidiag test of stool and a negative result by culture or PCR of gastric biopsy samples) and 6 false-negative results (a negative result by the Amplidiag test of stool and a positive result by culture or PCR of gastric biopsy samples) were observed. Considering these results, the sensitivity was 96.3% (95% confidence interval [CI], 92% to 98%), the specificity was 98.7% (95% CI, 97% to 99%), the positive predictive value (PPV) was 92.2% (95% CI, 92% to 98%), the negative predictive value (NPV) was 99.3% (95% CI, 98% to 99%), and the accuracy was 98.2% (95% CI, 97% to 99%) (Table 3). Analysis of the discrepant results demonstrated the presence of five clarithromycin-susceptible strains and one clarithromycin-resistant strain in the samples with false-negative results obtained by culture from gastric biopsy samples. For the stool samples with false-positive results, the amplified product was sequenced and was usable for six samples: two with Arcobacter spp., two with Helicobacter cinaedi strains, one with a Helicobacter canicola strain, and one with a Helicobacter pylori strain. Among the 13 samples with false-positive results, the histological status was available for 10 of them (usable histological results were not obtained for the 3 others). The gastric mucosa histology was normal for six of the samples (two with Arcobacter spp., one with an H. cinaedi strain, one with an H. canicola strain, and two with nonusable bacterial sequences), nonactive gastritis was found in three of them (two with nonusable bacterial sequences and one with an H. cinaedi strain), and active chronic gastritis with detection of an H. pylori sequence was found in one of them.
Second, for the detection of resistance to clarithromycin, the reference considered both the culture (Etest) and the Scorpion PCR result for the biopsy specimens, enabling detection of clarithromycin resistance in 17.5% of this cohort. Among the patients found to be H. pylori positive by both stool PCR test and culture or glmM PCR of biopsy specimens, no samples with false-negative results and two samples with false-positive results were detected (Table 3). Considering these results, the sensitivity was 100% (95% CI, 88% to 100%), the specificity was 98.4% (95% CI, 94% to 99%), the PPV was 93.1% (95% CI, 78% to 98%), the NPV was 100% (95% CI, 97% to 100%), and the accuracy was 98.7% (95% CI, 95% to 99%). For the two samples with false-positive results, one was found to contain a mixture of susceptible and resistant isolates with the Amplidiag test of stool, while the isolates in the other sample were not found to be susceptible at all. The isolates in both samples were found to be susceptible by the Scorpion PCR of gastric biopsy samples.
DISCUSSION
This study is the first one to evaluate the analytical performances of PCR assays detecting H. pylori infection and clarithromycin resistance in stool samples.
The main challenge in the management of H. pylori infection is linked to the direct consequences of antimicrobial resistance. Clarithromycin is a key component of the triple therapy routinely used to eradicate H. pylori. As resistance to this molecule has increased in many countries worldwide, it is the main element responsible for the failure of eradication using first-line therapy (up to 70%) (5–7, 33). Recent international guidelines have discouraged empirical clarithromycin-based triple therapy in geographical areas with a significantly high prevalence (>15%) of clarithromycin resistance (4). Antibiotic susceptibility testing was subsequently recommended and proved to be highly effective and superior to empirical treatments, thereby justifying recommendation of its use (12, 13, 34–37). This approach necessitates culture- or PCR-based tests for detection and resistance characterization. Usually performed on gastric biopsy samples (necessitating invasive endoscopic procedures) and requiring an experienced laboratory to perform culture, in routine practice this approach remains limited, rendering this guided strategy difficult to apply on a large scale and explaining why most international guidelines do not yet recommend use of the guided therapy strategy as the first-line treatment. Detection of H. pylori DNA in stool by molecular analysis is crucial to the development of the guided therapeutic strategy. Noninvasive tests using this easily obtained sample could improve patient compliance and therapeutic outcomes, optimizing the time/cost importance of disease management, especially in the context of future increases in rates of resistance. Moreover, the benefits to dysbiosis of the intestinal microbiota are clearly established, as this approach limits the number of antibiotic molecules present.
For now, few studies have focused on PCR detection in stool of both H. pylori and clarithromycin resistance. These rare studies have relied on poor criterion determination and a small number of patients, leading to very large confidence intervals for performance characteristics (15–17, 19, 20). In these studies, PCR tests performed on stool are designed for use with biopsy specimens, as described for the GenoType HelicoDR (Hain Lifescience, Nehren, Germany) and H. pylori ClariRes (Ingenetix, Wien, Austria) assays, without extensive validation on this particular matrix (15, 17–19, 21, 24). Other studies have been performed on cohorts constituted of selected H. pylori-infected patients or by a reference test with low performances (15, 16, 19, 23, 25). Finally, other studies have focused on composite reference testing to obtain a high sensitivity and specificity (18, 21, 22). In this context, the major strength of the present study resides in its prospective design and the inclusion of a large number of patients (n = 1,200) from four centers, thereby ensuring a robust representation of the target population. Moreover, this study is in accordance with the Standards for Reporting of Diagnostic Accuracy (STARD) checklist, facilitating fine reporting of important results (31). The design of the study is reinforced by the very high rate of adherence (92%) of a nonhospitalized patient population, thanks to the stool collection device proposed for use by the included subjects and the low number (0.5%) of patients excluded for other reasons. As described in the manufacturer’s recommendations, the eNAT tube sample system stabilizes microbial DNA, ensuring the good preservation of H. pylori DNA without differences between infected and noninfected patients in terms of a delay to the time of receipt in the laboratory (t test value = 1.4868; P > 0.05).
This study reports very high performances of the stool assay (with very narrow confidence intervals of performances, highlighting the reliability of these good results) for the detection of both H. pylori infection and clarithromycin resistance. We observed only five discordant results by the test (<0.5%) when done in triplicate, according to the manufacturer’s recommendations. Use of fully automated software is key to analysis of these results, as no melting curve analysis is needed, contrary to other commercialized tests (18, 21, 35). Rare false-positive results by stool testing seemed to be due mostly to nonspecific detection of phylogenetically closely related bacterial species, commensals, or pathogens of the gastrointestinal tract (Arcobacter, H. cinaedi, H. canicola). One positive case, the sequence of which allowed the identification of H. pylori, was highly suggestive of false-negative results for both reference tests. To ensure the absence of cross-reactivity with Campylobacter, infected patients (n = 10) were verified not to be positive by this molecular testing.
Moreover, for this study, patients performed stool self-sampling at home. After inclusion, they received a disposable stool collection device and a collection tube (eNAT; Copan) tolerating postal transport at ambient temperature. This procedure ensured the very good compliance by the patients, with 92% of stool specimens being received at the laboratory.
The feasibility of the molecular assay was very good and minimally time-consuming (5 min for extraction and 10 min for amplification, with a complete turnaround time of 3 h 45 min, including validation of the result). The total price per patient, including transport tube, postal transport, extraction, and amplification, was evaluated to be less than €30.
In the study population, the low prevalence of H. pylori infection (14.7%) (and a low rate of clarithromycin primary resistance of 17.5%) reflected the epidemiology of H. pylori infection in developed countries, such as France, as we observed a higher prevalence (28%) in patients born outside of Europe (1). The relatively low prevalence of resistance to clarithromycin might also be associated with the recent decrease observed in European countries, such as France (6, 7, 30, 38).
The very good performances of this noninvasive test (sensitivity, 96.3%; specificity, 98.7%) allow us to recommend its use in all cases where histological study of the gastric mucosa is not necessary. First, this test could be used for several indications for H. pylori testing and oriented treatment if the result is positive in patients <40 years of age and/or a first-degree family member of a gastric cancer patient; long-term users of aspirin and nonsteroidal anti-inflammatory drugs; long-term proton pump inhibitor users; and patients with iron deficiency anemia, idiopathic thrombocytopenic purpura, and vitamin B12 deficiency (4, 12). Second, this test could also be proposed for use for screening for H. pylori infection in patients when fibroscopy and the collection of biopsy specimens for histological study are indicated and then performed only in patients with proven infection, in which case fibroscopy could be delayed until after treatment guided by the results of the test. This approach could avoid many needless fibroscopy procedures in noninfected asymptomatic patients. Finally, use of this test can also be proposed when fibroscopy with the collection of gastric biopsy samples has been done with a histological survey only and when this examination reveals the presence of H. pylori. In this case, antimicrobial susceptibility testing by culture or PCR could be performed with the stool PCR test in order to guide treatment, without the need for a new fibroscopy with the collection of biopsy specimens. Nevertheless, several medicoeconomic studies are needed to confirm these hypothetical recommendations.
Conclusion.
This study was conducted using the best criteria to provide relevant results associating (i) the very good compliance of patients with autosampling, (ii) the good feasibility of the test (it is easy to perform), and (iii) a cost of less than €30 (including stool transport and extraction). The results demonstrated the very good performances of the noninvasive Amplidiag H. pylori+ClariR assay for the detection of H. pylori and clarithromycin resistance with stool. This stool PCR testing will clearly revolutionize the management of H. pylori-infected patients, thereby enabling guided treatment without invasive sampling.
ACKNOWLEDGMENTS
Thanks go to Emilie Pithon and Solène Tillet for their contribution as clinical research technicians. We gratefully acknowledge Jeffrey Arsham, an American translator, for his rereading and revision of the original English-language manuscript.
This study was supported by a grant from the Groupement Interrégional de Recherche Clinique et d’Innovation Sud-Ouest Outre-Mer Hospitalier (APIK-2018). This study was also supported by Mobidiag. Mobidiag is a privately owned company that made the PCR assay kit used in the study. It provided the funding and, in discussions with C.B., the basic conceptualization for the study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Burucoa C, Axon A. 2017. Epidemiology of Helicobacter pylori infection. Helicobacter 22(Suppl 1):e12403. doi: 10.1111/hel.12403. [DOI] [PubMed] [Google Scholar]
- 2.Suerbaum S, Michetti P. 2002. Helicobacter pylori infection. N Engl J Med 347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 3.Marshall BJ, Warren JR. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 4.Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM, European Helicobacter and Microbiota Study Group and Consensus Panel . 2017. Management of Helicobacter pylori infection—the Maastricht V/Florence Consensus Report. Gut 66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 5.Mégraud F. 2004. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374–1384. doi: 10.1136/gut.2003.022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savoldi A, Carrara E, Graham DY, Conti M, Tacconelli E. 2018. Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterology 155:1372–1382.e17. doi: 10.1053/j.gastro.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond J, Lamarque D, Kalach N, Chaussade S, Burucoa C. 2010. High level of antimicrobial resistance in French Helicobacter pylori isolates. Helicobacter 15:21–27. doi: 10.1111/j.1523-5378.2009.00737.x. [DOI] [PubMed] [Google Scholar]
- 8.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 9.Mégraud F, Lehours P. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev 20:280–322. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raymond J, Burucoa C, Pietrini O, Bergeret M, Decoster A, Wann A, Dupont C, Kalach N. 2007. Clarithromycin resistance in Helicobacter pylori strains isolated from French children: prevalence of the different mutations and coexistence of clones harboring two different mutations in the same biopsy. Helicobacter 12:157–163. doi: 10.1111/j.1523-5378.2007.00486.x. [DOI] [PubMed] [Google Scholar]
- 11.Burucoa C, Landron C, Garnier M, Fauchere J-L, Khan R, Rahman M. 2005. T2182C mutation is not associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother 49:868–870. doi: 10.1128/AAC.49.2.868-870.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamarque D, Burucoa C, Courillon-Mallet A, de Korwin JD, Delchier JC, Fauchère JL, Kalach N, Labigne A, Lehours P, Mégraud F, Raymond J, et le Groupe d’Etudes Français des Helicobacter . 2012. Révision des recommandations françaises sur la prise en charge de l’infection par Helicobacter pylori. Hepato-Gastro Oncol Dig 19:475–502. [Google Scholar]
- 13.Rollan A, Arab JP, Camargo MC, Candia R, Harris P, Ferreccio C, Rabkin CS, Gana JC, Cortés P, Herrero R, Durán L, García A, Toledo C, Espino A, Lustig N, Sarfatis A, Figueroa C, Torres J, Riquelme A. 2014. Management of Helicobacter pylori infection in Latin America: a Delphi technique-based consensus. World J Gastroenterol 20:10969–10983. doi: 10.3748/wjg.v20.i31.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burucoa C, Mégraud F. 2011. Helicobacter pylori, p 283–286. In Cornaglia G, Courcol R, Herrmann JL (ed), European manual of clinical microbiology, 1st ed European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 15.Brennan DE, Omorogbe J, Hussey M, Tighe D, Holleran G, O'Morain C, Smith SM, McNamara D. 2016. Molecular detection of Helicobacter pylori antibiotic resistance in stool vs biopsy samples. World J Gastroenterol 22:9214–9221. doi: 10.3748/wjg.v22.i41.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Talarico S, Yao L, He L, Self S, You Y, Zhang H, Zhang Y, Guo Y, Liu G, Salama NR, Zhang J. 2018. Droplet digital PCR-based detection of clarithromycin resistance in Helicobacter pylori isolates reveals frequent heteroresistance. J Clin Microbiol 56:e00019-18. doi: 10.1128/JCM.00019-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scaletsky IC, Aranda KR, Garcia GT, Gonçalves ME, Cardoso SR, Iriya K, Silva NP. 2011. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter 16:311–315. doi: 10.1111/j.1523-5378.2011.00845.x. [DOI] [PubMed] [Google Scholar]
- 18.Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovách Z, Rotter M, Makristathis A. 2004. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol 42:4512–4518. doi: 10.1128/JCM.42.10.4512-4518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angol DC, Ocama P, Ayazika Kirabo T, Okeng A, Najjingo I, Bwanga F. 2017. Helicobacter pylori from peptic ulcer patients in Uganda is highly resistant to clarithromycin and fluoroquinolones: results of the GenoType HelicoDR test directly applied on stool. Biomed Res Int 2017:5430723. doi: 10.1155/2017/5430723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giorgio F, Ierardi E, Sorrentino C, Principi M, Barone M, Losurdo G, Iannone A, Giangaspero A, Monno R, Di Leo A. 2016. Helicobacter pylori DNA isolation in the stool: an essential pre-requisite for bacterial noninvasive molecular analysis. Scand J Gastroenterol 51:1429–1432. doi: 10.1080/00365521.2016.1216592. [DOI] [PubMed] [Google Scholar]
- 21.Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Rüssmann H. 2007. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol 45:1718–1722. doi: 10.1128/JCM.00103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckman E, Saracino I, Fiorini G, Clark C, Slepnev V, Patel D, Gomez C, Ponaka R, Elagin V, Vaira D. 2017. A novel stool PCR test for Helicobacter pylori may predict clarithromycin resistance and eradication of infection at a high rate. J Clin Microbiol 55:2400–2405. doi: 10.1128/JCM.00506-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beer-Davidson G, Hindiyeh M, Muhsen K. 2018. Detection of Helicobacter pylori in stool samples of young children using real-time polymerase chain reaction. Helicobacter 23:e12450. doi: 10.1111/hel.12450. [DOI] [PubMed] [Google Scholar]
- 24.Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. 2010. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol 8:309–312. doi: 10.1016/j.cgh.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. 2007. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol 56:1174–1180. doi: 10.1099/jmm.0.47302-0. [DOI] [PubMed] [Google Scholar]
- 26.Bénéjat L, Ducournau A, Lehours P, Mégraud F. 2018. Real-time PCR for Helicobacter pylori diagnosis. The best tools available. Helicobacter 23:e12512. doi: 10.1111/hel.12512. [DOI] [PubMed] [Google Scholar]
- 27.Hays C, Delerue T, Lamarque D, Burucoa C, Collobert G, Billöet A, Kalach N, Raymond J. 2019. Molecular diagnosis of Helicobacter pylori infection in gastric biopsies: evaluation of the Amplidiag H. pylori+ClariR assay. Helicobacter 24:e12560. doi: 10.1111/hel.12560. [DOI] [PubMed] [Google Scholar]
- 28.Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, Braimi N, Dupont C, Labigne A. 1996. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol 147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 29.Espinoza MG, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG. 2011. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J Clin Microbiol 49:1650–1652. doi: 10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burucoa C, Garnier M, Silvain C, Fauchere JL. 2008. Quadruplex real-time PCR assay using allele-specific Scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J Clin Microbiol 46:2320–2326. doi: 10.1128/JCM.02352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, Smith PG, Sriram N, Wongsrichanalai C, Linke R, O’Brien R, Perkins M, Cunningham J, Matoso P, Nathanson CM, Olliaro P, Peeling RW, Ramsay A. 2010. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol 12:S17–S29. doi: 10.1038/nrmicro1523z. [DOI] [PubMed] [Google Scholar]
- 32.Rintala A, Munukka E, Weintraub A, Ullberg M, Eerola E. 2016. Evaluation of a multiplex real-time PCR kit Amplidiag Bacterial GE in the detection of bacterial pathogens from stool samples. J Microbiol Methods 128:61–65. doi: 10.1016/j.mimet.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y, on behalf of the Study Group Participants . 2013. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut 62:34–42. doi: 10.1136/gutjnl-2012-302254. [DOI] [PubMed] [Google Scholar]
- 34.Wenzhen Y, Yumin L, Quanlin G, Kehu Y, Lei J, Donghai W, Lijuan Y. 2010. Is antimicrobial susceptibility testing necessary before first-line treatment for Helicobacter pylori infection? Meta-analysis of randomized controlled trials. Intern Med 49:1103–1109. doi: 10.2169/internalmedicine.49.3031. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, Zhang J, Song Z, He L, Li Y, Qian J, Bai P, Xue Y, Wang Y, Lin S. 2016. Tailored versus triple plus bismuth or concomitant therapy as initial Helicobacter pylori treatment: a randomized trial. Helicobacter 21:91–99. doi: 10.1111/hel.12242. [DOI] [PubMed] [Google Scholar]
- 36.López-Góngora S, Puig I, Calvet X, Villoria A, Baylina M, Muñoz N, Sanchez-Delgado J, Suarez D, García-Hernando V, Gisbert JP. 2015. Systematic review and meta-analysis: susceptibility-guided versus empirical antibiotic treatment for Helicobacter pylori infection. J Antimicrob Chemother 70:2447–2455. doi: 10.1093/jac/dkv155. [DOI] [PubMed] [Google Scholar]
- 37.Delchier JC, Bastuji-Garin S, Raymond J, Megraud F, Amiot A, Cambau E, Burucoa C, HELICOSTIC Study Group. . 27 June 2019. Efficacy of a tailored PCR-guided triple therapy in the treatment of Helicobacter pylori infection. Med Mal Infect doi: 10.1016/j.medmal.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Ducournau A, Bénéjat L, Sifré E, Bessède E, Lehours P, Mégraud F. 2016. Helicobacter pylori resistance to antibiotics in 2014 in France detected by phenotypic and genotypic methods. Clin Microbiol Infect 22:715–718. doi: 10.1016/j.cmi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Pichon M, Burucoa C. 2019. Impact of the gastro-intestinal bacterial microbiome on Helicobacter-associated diseases. Healthcare (Basel) 7(1):34. doi: 10.3390/healthcare7010034. [DOI] [PMC free article] [PubMed] [Google Scholar]