U.S. gonorrhea rates are rising, and antibiotic-resistant Neisseria gonorrhoeae (AR-Ng) is an urgent public health threat. Since implementation of nucleic acid amplification tests for N. gonorrhoeae identification, the capacity for culturing N. gonorrhoeae in the United States has declined, along with the ability to perform culture-based antimicrobial susceptibility testing (AST). Yet AST is critical for detecting and monitoring AR-Ng.
KEYWORDS: antimicrobial resistance, antibiotic resistance, Neisseria gonorrhoeae, gonorrhea, antimicrobial susceptibility testing, whole-genome sequencing
ABSTRACT
U.S. gonorrhea rates are rising, and antibiotic-resistant Neisseria gonorrhoeae (AR-Ng) is an urgent public health threat. Since implementation of nucleic acid amplification tests for N. gonorrhoeae identification, the capacity for culturing N. gonorrhoeae in the United States has declined, along with the ability to perform culture-based antimicrobial susceptibility testing (AST). Yet AST is critical for detecting and monitoring AR-Ng. In 2016, the CDC established the Antibiotic Resistance Laboratory Network (AR Lab Network) to shore up the national capacity for detecting several resistance threats including N. gonorrhoeae. AR-Ng testing, a subactivity of the CDC’s AR Lab Network, is performed in a tiered network of approximately 35 local laboratories, four regional laboratories (state public health laboratories in Maryland, Tennessee, Texas, and Washington), and the CDC’s national reference laboratory. Local laboratories receive specimens from approximately 60 clinics associated with the Gonococcal Isolate Surveillance Project (GISP), enhanced GISP (eGISP), and the program Strengthening the U.S. Response to Resistant Gonorrhea (SURRG). They isolate and ship up to 20,000 isolates to regional laboratories for culture-based agar dilution AST with seven antibiotics and for whole-genome sequencing of up to 5,000 isolates. The CDC further examines concerning isolates and monitors genetic AR markers. During 2017 and 2018, the network tested 8,214 and 8,628 N. gonorrhoeae isolates, respectively, and the CDC received 531 and 646 concerning isolates and 605 and 3,159 sequences, respectively. In summary, the AR Lab Network supported the laboratory capacity for N. gonorrhoeae AST and associated genetic marker detection, expanding preexisting notification and analysis systems for resistance detection. Continued, robust AST and genomic capacity can help inform national public health monitoring and intervention.
INTRODUCTION
THE NEED FOR NEISSERIA GONORRHOEAE AR DETECTION CAPACITY
In the United States, rates of reported gonorrhea have been rising since 2009. In 2018, the U.S. Centers for Disease Control and Prevention (CDC) received 583,405 case reports (1), and gonorrhea remained the second most commonly reported nationally notifiable condition. Neisseria gonorrhoeae can rapidly develop resistance due to its genomic plasticity (2), and, indeed, N. gonorrhoeae has developed resistance to several classes of antibiotics. Currently, dual therapy with ceftriaxone and azithromycin is recommended for treatment of uncomplicated infections (3). This recommendation was made in 2015 based on the rationale that only ceftriaxone is still considered fully effective (3), while azithromycin could serve as a shield and preserve ceftriaxone efficacy. The CDC considers rising antibiotic resistance (AR) among N. gonorrhoeae isolates an urgent threat, initially along with two other AR threats, i.e., Clostridioides difficile and carbapenem-resistant Enterobacteriaceae (CRE), according to the CDC’s 2013 antibiotic resistance threat report (4). Laboratory preparedness is one of several public health strategies to combat these threats.
N. gonorrhoeae antimicrobial susceptibility testing (AST) is currently not part of routine clinical diagnostics for uncomplicated gonorrhea in the United States (3, 5). Persons with symptomatic gonorrhea are often empirically treated with antibiotics (3) before definitive laboratory results from a nucleic acid amplification test (NAAT) are available. Similarly, when asymptomatic persons are screened for N. gonorrhoeae, testing is nearly always done by NAAT, and test results typically come back after a few hours or days, depending on laboratory setup (6). Rapid and presumptive administration of antibiotics is often the next step. It has the advantage of interrupting transmission chains quickly but is imperfect due to unknown antibiotic resistance patterns of the patient’s infection. In cases of suspected treatment failure and for disseminated gonococcal infections (DGI), culture collection and phenotypic AST are recommended (5) and can aid in the selection of an alternative treatment regimen. In the future, AST may take on a more important role for individual patients should no drugs remain universally effective.
AR-N. gonorrhoeae (AR-Ng) data are also needed for surveillance and other public health efforts to respond to the spread of concerning strains. Since such data are not automatically generated from patient diagnoses, there has been a longstanding public health effort to obtain them. Prior to the launch of the AR Lab Network, there was already a CDC-directed effort to support primarily academic laboratories to conduct AST for CDC’s national AR-Ng surveillance. This article describes how these preexisting activities were expanded into collaborative efforts to develop and expand a robust infrastructure for N. gonorrhoeae AST and other activities in U.S. public health laboratories.
N. gonorrhoeae culture and phenotypic AST are cumbersome. N. gonorrhoeae growth is fastidious; i.e., the organism does not grow well in broth medium in suspension, and it requires a CO2-enriched environment. Better broth-based culture systems would enhance automation of testing, reduce turnaround time for results, and allow for more objective interpretation of results. However, the Clinical and Laboratory Standards Institute (CLSI) describes agar dilution or disk diffusion methods as standard methods (7), both with limited opportunity for scale-up and efficiency. Commercial gradient strip diffusion systems can be used with incremental increases in speed but still require bacterial isolation and days of bacterial growth. Molecular assays that detect resistance markers in lieu of phenotypic AST assays have not yet advanced to U.S. Food and Drug Administration (FDA) clearance. NAATs for ciprofloxacin or other antibiotic drug susceptibility markers are emerging (8) as laboratory-developed tests, and diagnostic whole-genome sequence (WGS) analyses are also in developmental stages (9). Development and broad implementation of these methods will be challenging due to multiple factors, including rapidly changing resistance patterns among N. gonorrhoeae strains.
OVERVIEW AND GOALS OF CDC’S AR LAB NETWORK FOR MULTIPLE PATHOGENS, INCLUDING N. GONORRHOEAE
The CDC gonorrhea program has managed a network of local laboratories and primarily academic reference laboratories associated with its national surveillance project (Gonococcal Isolate Surveillance Project [GISP], described below) since 1986 (10). In 2016, when the CDC established its AR Lab Network, N. gonorrhoeae activities became part of this network. The overall CDC AR Lab Network supports a nationwide laboratory capacity to rapidly detect resistance and inform local responses to prevent spread and protect people (details are available on the CDC’s website [https://www.cdc.gov/drugresistance/solutions-initiative/ar-lab-network.html]). The network tracks changes in resistance and helps identify and respond to outbreaks. The CDC gonorrhea program is part of this effort to share resources, using CDC’s nationwide investment to enhance preexisting public health laboratory activities. The network in its entirety includes laboratories in 50 states, four major cities, and Puerto Rico, including seven regional labs and the National Tuberculosis Molecular Surveillance Center. It is funded through CDC’s Epidemiology and Laboratory Capacity for Infectious Diseases (ELC) Cooperative Agreement (11, 12). The AR Lab Network is not a research activity. It is centrally managed by CDC’s Antibiotic Resistance Coordination and Strategy (ARX) Unit, in close collaboration with CDC divisions that focus on individual pathogens. For N. gonorrhoeae, this is CDC’s Division of STD Prevention (DSTDP). This article describes in depth only activities for AR-Ng.
IMPLEMENTATION OF THE AR LAB NETWORK PORTFOLIO FOR AR-Ng
In August 2016, four state public health laboratories received funding for regional laboratory N. gonorrhoeae activities, for a combined capacity of testing 20,000 isolates annually: the Maryland Public Health Laboratory (MD) for the Mid-Atlantic region, the Texas Department of State Health Services Laboratory (TX) for the Mountain region, the Tennessee State Public Health Laboratory (TN) for the Southeast region, and the Washington State Public Health Laboratories (WA) for the West region. The WA laboratory subcontracts part of its work to the University of Washington. Three additional AR Lab Network state laboratories (in Minnesota [MN], Wisconsin [WI], and New York [NY]) do not perform N. gonorrhoeae testing.
CLINICS AND N. GONORRHOEAE SPECIMENS
In its initial concept, the AR Lab Network’s AR-Ng program received specimens from three CDC-managed sexually transmitted disease (STD) public health projects for surveillance or rapid public health action. Of these, the Gonococcal Isolate Surveillance Project (GISP) is the largest and longest-running project as it has been in operation since 1986. GISP is a national sentinel surveillance project and is described in depth elsewhere (10). In brief, STD clinics in 25 to 30 jurisdictions (the exact number varies each year) collect specimens from the first 25 male patients presenting with symptomatic gonococcal urethritis at their sentinel sites each month. Here, the terms “jurisdiction” (a funded location) and “sentinel site” (a location that submits isolates) are used interchangeably, with the recognition that some jurisdictions have more than one sentinel site and that some sentinel sites have more than one clinic in order to meet the required number of isolates each month. The creation of the AR Lab Network has introduced new regional laboratories that perform AST for the project. However, GISP isolate-submitting local laboratories have largely remained unchanged, as have the receiving CDC national reference laboratory and CDC data analysis activities. National GISP N. gonorrhoeae susceptibility data are published in CDC’s STD surveillance report every year (1). They are used to inform clinical treatment guidelines and are not further discussed in this review of laboratory capacity. The enhanced GISP project (eGISP) is a newer project and was initiated in August 2017. In addition to collecting urethral specimens, eGISP clinics also collect extragenital specimens and specimens from women. In 2017, 12 sites participated, followed by nine participating sites in 2018. The third project is the Strengthening the U.S. Response to Resistant Gonorrhea (SURRG) project. It is designed to enhance local capacity to rapidly detect and respond to emerging resistance. SURRG is also a newer project. Sites participating in SURRG (nine in 2017 and eight in 2018) collect specimens from persons of all genders at exposed anatomic sites and who are attending STD clinics and other non-STD clinic health care facilities (such as family planning clinics and emergency departments). Unlike the GISP and eGISP facilities, local laboratories participating in SURRG perform gradient strip AST (Etest, bioMérieux, France) for local actionable data generation before submitting N. gonorrhoeae isolates to the AR Lab Network (details below). The three projects are further described in depth elsewhere (10, 13, 14) and briefly summarized here only to describe submitted specimens. Each regional member of the AR Lab Network may also accept patient specimens for N. gonorrhoeae testing, as decided by each participating regional laboratory. This activity is not specifically CDC directed or enhanced. It is possible that in the future other specimen streams may be added to the portfolio, particularly since the current specimen sources have not yielded 20,000 isolates per year (see also below).
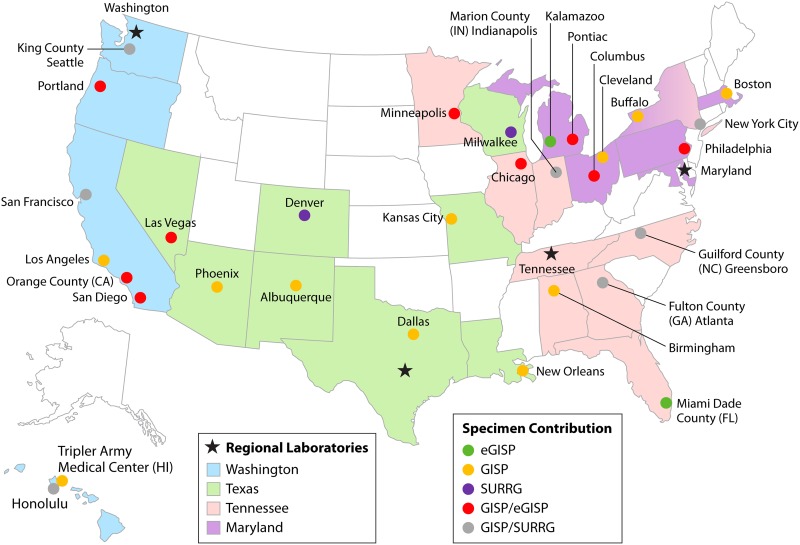
Figure 1 shows a map of the four AR Lab Network regional laboratories performing AR-Ng testing, with 27 GISP, 12 eGISP, and 9 SURRG sites. Funding decisions and awards for laboratories and specimen-collecting projects typically arrive in August each year; therefore, sites can vary each year and are shown here as of 31 August 2017. Of note, male urethral isolates and AST results from the first 25 symptomatic men collected by eGISP or SURRG sites each month are also included in GISP data; thus, many sites participate in more than one project, e.g., in both SURRG and GISP or in eGISP and GISP. There is typically one main local laboratory associated with these sites; overall, there are approximately 35 local laboratories. Some participating sites have multiple submitting clinics; for example, New York City has 10. Over 60 clinics contribute specimens (Table 1). In 2018, the number of GISP sites increased to 35, and 8 sites participated in SURRG.
FIG 1.
Map of antibiotic-resistant Neisseria gonorrhoeae isolate collection and laboratory testing in the AR Lab Network as of 31 August 2017. States submitting isolates to the network are color coded as indicated, as are the approximately 35 specimen-collecting sites from GISP, eGISP, and SURRG. States shown in white do not submit specimens to the network. GISP, Gonococcal Isolate Surveillance Project; eGISP, enhanced GISP; SURRG, Strengthening the U.S. Response to Resistant Gonorrhea project.
TABLE 1.
AR-Ng testing in the CDC’s AR Lab Networka
| Network parameter | Value for the parameter |
|
|---|---|---|
| 2017 | 2018 | |
| Sites (no.)b | ||
| GISP | 27 | 35 |
| eGISP | 12 | 9 |
| SURRG | 9 | 8 |
| Participating clinics | 63 | 63 |
| N. gonorrhoeae isolate testing (no. of isolates)c | ||
| AST | 8,214 | 8,628 |
| Alert | 531 | 646 |
| Quick-send alert | 47 | 55 |
| WGS | 605 | 3159 |
The table summarizes information about sites and their associated laboratories participating in the AR Lab Network, as well as gonococcal isolates managed by the AR Lab Network.
Project funding cycles are August 1 to July 31 of the following year. The number of sites here refers to contribution status as of 31 August 2017 or 31 August 2018. All other numbers refer to calendar years 2017 and 2018. The number of reported sites can vary slightly between different publications, depending on whether sites funded by carryover funds are counted and whether sites submitting isolates without funding are included. GISP, Gonococcal Isolate Surveillance Project; eGISP, enhanced GISP; SURRG Strengthening the US. Response to Resistant Gonorrhea.
Isolate numbers are as reported by the regional laboratories and may vary slightly from final data reconciliation of each project. The number of alerts may slightly overrepresent a final, reconciled count since some isolates (<10) may be identified as an alert in both the cefixime and ceftriaxone categories. AST, antimicrobial susceptibility test; WGS, whole-genome sequencing.
A TIERED LABORATORY NETWORK
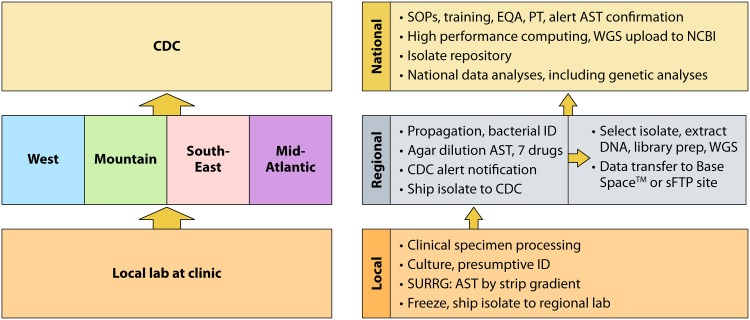
The various laboratory responsibilities, including work and specimen flow, are shown in Fig. 2. It is a tiered laboratory system. Providers at local clinics collect specimens, and microbiological staff at associated laboratories initiate bacterial culture (further described below, including exceptions) and then send isolates to the next level reference laboratory, i.e., a regional laboratory which performs reference AST and WGS. Thereafter, some isolates are sent to the third tier, i.e., the national reference laboratory at the CDC, for further characterization and/or archiving. The work flow for bacterial isolation and AST is based on preexisting GISP experiences (15), while WGS was newly implemented. What follows is an in-depth description of microbiologic procedure portfolios.
FIG 2.
Tiered laboratory responsibilities for Neisseria gonorrhoeae testing in the AR Lab Network. Specimens enter the tiered system at the local clinical laboratory as soon as possible after being collected from the patient. Recovered isolates are then sent to the appropriate regional laboratory (West, Mountain, Southeast, or Mid-Atlantic) where AST and genome sequencing are conducted. Confirmatory AST testing and genomic sequence analysis are performed at the CDC. Arrows indicate the flow of isolates within the laboratory network. SOP, standard operating protocol; SURRG, Strengthening the U.S. Response to Resistant Gonorrhea project; AST, antimicrobial susceptibility testing; WGS, whole-genome sequencing; ID, identification; EQA, external quality assurance; PT, proficiency testing; NCBI, National Center for Biotechnology Information. BaseSpace is a cloud-based genomics sequence hub built by Illumina (San Diego, CA), sFTP, secure file transfer protocol site.
THE FIRST LABORATORY TIER: PATIENT SPECIMENS, BACTERIAL VIABILITY, AND ISOLATION
In the first laboratory tier, a variety of preexisting microbiologic protocols and methods for bacterial isolate generation are in operation, in accordance with national or international laboratory guidance (5, 16). This allows the participation of laboratories with differing local conditions, e.g., previously obtained assay validation data, previously purchased equipment, and differing geographic distances between clinic and an associated laboratory. Local laboratories are mostly public health laboratories associated with participating clinics. However, laboratories associated with academic or other hospital-affiliated health centers also participate. Laboratories work with different N. gonorrhoeae growth-supporting, nutrient-based selective growth media (e.g., modified Thayer-Martin) in a CO2 environment or with commercial transporting media (e.g., including, but not limited to InTray GC [Biomed Diagnostics, Inc., White City, OR] and Copan Liquid Amies Elution Swab [Eswab, BD] Collection and Transport systems [Copan Diagnostics Inc., Murrieta, CA]) (17) to maintain bacterial viability. Most GISP and eGISP sites presumptively identify N. gonorrhoeae, typically by (i) growth of typically appearing colonies with characteristic morphologies (e.g., small and transparent) on a selective medium such as modified Thayer-Martin, (ii) a positive oxidase test, and (iii) the observation of Gram-negative diplococci in stained smears. SURRG laboratories perform definitive identification which can include additional biochemical or enzymatic testing or matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF), if available. SURRG laboratories have clinical laboratory improvement amendments (CLIA) certification for gradient strip AST for azithromycin, ceftriaxone, and cefixime (Etest, bioMérieux, France), with the goal of producing actionable patient results within a 5-day turnaround time. They follow CDC notification procedures as outlined below for AR Lab Network laboratories when potential resistance is encountered. The SURRG sites use the AST results for local, rapid public health follow-up; e.g., they prioritize contact, interview, and testing of sexual partners in cases of concerning AST results.
All local laboratories make frozen isolate stocks and ship them to the AR Lab Network monthly. Personal identifiable information is removed locally before a limited amount of data are transmitted from local laboratories to the four regional laboratories. Data transmission occurs with electronic manifests that include several variables (e.g., specimen collection date, specimen source, patient age, and a unique specimen identifier) using data error check mechanisms and standardized result reports. A secure file transfer protocol (FTP) site is used to transmit data.
THE SECOND LABORATORY TIER: AR LAB NETWORK REGIONAL REFERENCE LABORATORIES PERFORM AST AND WGS
Regional laboratories contact submitting local laboratories if problems are noted upon specimen receipt, e.g., nonviability, contamination, or inappropriate transport. They propagate N. gonorrhoeae from frozen stocks and perform definitive identification as described above for SURRG laboratories. Using GISP protocols (15) and CLSI methods (18), the regional laboratories conduct AST by agar dilution for seven antibiotics (azithromycin, ceftriaxone, cefixime, gentamicin, penicillin, tetracycline, and ciprofloxacin) and β-lactamase testing. CDC selected these drugs due to treatment recommendations in effect when the AR Lab Network was created (3) and to maintain long-standing susceptibility monitoring. Unlike the SURRG AST turnaround time of five working days, agar dilution AST in regional laboratories takes up to 1 month, and its current main purpose is monitoring of national susceptibility trends.
Laboratories notify the CDC of elevated MIC results for azithromycin, ceftriaxone, and cefixime to alert the CDC to potential isolates of concern within 24 h of result generation. While concerning elevated MICs do not trigger the same rapid public health follow-up as that described for the SURRG project, the notification system nevertheless gives CDC an opportunity for public health follow-up in all three projects, i.e., GISP, eGISP, and SURRG. The selection of drugs is based on preexisting GISP protocols (15) and consists of drugs recommended for uncomplicated gonorrhea and expedited partner treatment at the inception of the AR Lab Network (3). The selection can change should recommended drugs change. “Quick-send” alert values were newly introduced for highest urgency cases due to a higher likelihood of associated treatment failure, while the alert system was adopted from GISP (15). Table 2 shows the respective MIC values, in comparison to available CLSI interpretive criteria (19), which consist only of susceptible (S) breakpoints for these drugs. This is mostly due to a scarcity of clinical resistance data for monotherapy (20) and uncertainty about the MIC at which treatment failures generally occur. All quick-send MICs are well above CLSI susceptibility breakpoint MICs. Of note, the ceftriaxone alert MIC is lower than the CLSI susceptibility breakpoint MIC due to particular concern about potential resistance development to this last remaining, fully effective drug. This allows the CDC to conduct sensitive surveillance for shifts toward higher MICs. In contrast, alert values for azithromycin and cefixime are set higher than and equal to CLSI susceptibility breakpoints, respectively; their higher U.S. prevalence is already documented (1). Quick-send isolates are sent to the CDC for confirmation immediately, while alert isolates are sent quarterly. Regular AST data are transmitted to the CDC monthly using the secure Association of Public Health Laboratories (APHL) Informatics Messaging System (AIMS) portal, which is managed by APHL.
TABLE 2.
2018 Antibiotic-resistant Neisseria gonorrhoeae MIC values for CDC notification and WGS selection in the AR Lab Network in comparison to CLSI breakpointsa
| Antibiotic | MIC (μg/ml) for the category |
|||
|---|---|---|---|---|
| Quick-send alert | Alert | WGS selectionb | CLSI S breakpointc | |
| Ceftriaxone | ≥0.5 | ≥0.125 | ≥0.125 | ≤0.25 |
| Cefixime | ≥1.0 | ≥0.25 | ≥0.25 | ≤0.25 |
| Azithromycin | ≥16 | ≥2.0 | ≥4.0 | ≤1.0 |
The AR Lab Network selects additional isolates for sequencing independent of susceptibility to the indicated antibiotics.
WGS, whole-genome sequencing.
Susceptibility breakpoint as determined by the Clinical and Laboratory Standards Institute (CLSI).
Two regional laboratories began whole-genome sequencing (WGS) as a supplemental activity in 2017. In 2018, all four laboratories received funding to sequence up to 1,250 isolates per year, i.e., 5,000 isolates nationally. To streamline activities, laboratories use sequencing protocols developed by the CDC’s PulseNet program for foodborne pathogen surveillance (21) and utilize already available Illumina MiSeq equipment (San Diego, CA). The CDC shared protocols for DNA extraction, Nextera XT library preparation, and sequencing, minimally adapted to N. gonorrhoeae from the manufacturer’s instructions (Illumina, San Diego, CA). The CDC developed an isolate selection algorithm for continuous sequencing as opposed to selecting isolates after end-of-year AST data analysis for specific projects. In 2018, all alert isolates were sequenced, with the exception of those for azithromycin (sequenced only if isolates had MICs of ≥4) (Table 2), allowing monitoring of AR markers. Laboratories sequence a selection of isolates from all geographic locations, regardless of MIC values, to allow description of a subset of all strains. In addition, they sequence isolates from extragenital anatomical sites and from women to increase knowledge of associated strain characteristics. GISP, eGISP, SURRG project officers, and other partners have an opportunity to request additional sequencing based on specific local or project data needs outside the routine sequencing algorithm as long as 5,000 sequences are not exceeded annually.
To manage sequence data, the CDC assigns unique sequence identifiers. Regional laboratory staff prepare data for a National Center for Biotechnology Information (NCBI) metadata file, including collection date, geographical region, patient age range, gender, and anatomical site, but are not responsible for its upload. After quality check, raw sequence data are transmitted to the CDC through BaseSpace, a cloud-based genomics sequence hub built by Illumina (San Diego, CA), or via an FTP site.
THE THIRD TIER: NATIONAL CDC REFERENCE LABORATORY CONFIRMS ALERT AST RESULTS AND CONDUCTS ADDITIONAL ANALYSES
CDC staff develop and distribute standard operation protocols (SOPs) for laboratories working with AR-Ng. The CDC conducts laboratory training at the CDC, site visits, and conference calls to direct activities. To ensure quality, reproducibility, and comparability for AST, the CDC national reference laboratory, i.e., the STD Laboratory Reference and Research Branch, provides N. gonorrhoeae strain ATCC 49226 and two additional well-defined strains for quality control. The CDC requires external quality assurance testing with panels of 15 isolates twice per year, with a requirement of ≥80% concordance with the modal MIC ±1 dilution. The CDC performs confirmatory AST to further ensure comparability. In 2017 to 2018, the CDC retested all alert isolates and confirmed all quick-send isolates shortly after receipt.
Upon receipt of raw read sequencing files, the CDC uses its high-performance computing infrastructure for data management and bioinformatics analysis (22). The CDC assembles and analyzes reads to detect and characterize isolates with unique or concerning antimicrobial susceptibility patterns and genetic resistance markers. These data allow greater resolution of the relationship and potential transmission parameters of concerning strains than can be deduced from phenotypic AST alone. The CDC is also responsible for upload of all raw sequences to the Sequence Read Archive (SRA/NCBI).
The CDC maintains an extensive isolate repository to make residual isolates available for research and development to combat antibiotic resistance. A small subset of isolates from this permanent collection is deposited in the CDC & FDA AR Isolate Bank (23). The CDC & FDA AR Isolate Bank is centrally managed at the CDC and currently contains two N. gonorrhoeae panels. The CDC repository is separately managed; custom-curated isolate collections can be developed in collaborative efforts and requested directly from the CDC (stdlaboratoryspecim@cdc.gov). In 2017 to 2018, the CDC fulfilled approximately 60 requests, not counting routine shipments for external quality control in associated laboratories.
AR LAB NETWORK AR-Ng ACCOMPLISHMENTS BY THE NUMBERS
After initial laboratory training, some regional laboratories started testing in early 2017. The AR Lab Network produced 8,214 and 8,628 N. gonorrhoeae AST results in 2017 and 2018, respectively (Table 1). This constitutes an increase over the approximately 5,000 to 6,000 GISP isolates (numbers varied by year) submitted to and tested annually in regional laboratories prior to launch of the AR Lab Network (10). The 2017 and 2018 isolates included 531 and 646 alerts and 47 and 55 quick-send alerts, respectively. Two laboratories sequenced 605 isolates through supplemental funds in 2017, and all four sequenced 3,159 in 2018; WGS had not been a funded activity in GISP and its regional laboratory partners prior to AR Lab Network launch. As of 14 August 2019, the CDC has contributed approximately 5,000 N. gonorrhoeae sequences to NCBI, including sequences predating the AR Lab Network and uploaded by personnel associated with the AR Lab Network project.
AR LAB NETWORK DATA ANALYSES
AR Lab Network N. gonorrhoeae AST laboratory data are primarily reported in the national surveillance report (GISP data [1]) and eGISP and SURRG project analyses (13, 14). Importantly, in 2017, the national surveillance report partially contained AR Lab Network AST data for the first time, in addition to data still generated by previous regional partner laboratories. In 2018, reported national levels of isolates with alert MICs (alert values are listed in Table 2) were from N. gonorrhoeae testing in the AR Lab Network and were as follows: azithromycin, 4.6%; ceftriaxone, 0.2%; cefixime, 0.3% (1). National levels of GISP isolates with resistance (CLSI criteria [19]) were as follows for these drugs: penicillin, 13.7%; tetracycline, 25.6%; ciprofloxacin, 31.2% (1). These data contribute to treatment guideline review, laboratory and clinical interpretive criteria decisions (20), and other activities that promote gonorrhea prevention and control.
The value of N. gonorrhoeae WGS data for public health purposes is increasingly being recognized (24). AR Lab Network WGS data have readily enhanced monitoring of concerning strains or resistance markers (22). They may continue to become part of an integral surveillance effort to monitor spread of resistance in greater depth than is possible if only AST data are available. Comprehensive SURRG and eGISP project analyses are in progress. When they are completed, it will be possible to evaluate whether the described isolate sequence selection algorithm resulted in a representative sample of isolates according to antimicrobial susceptibilities, geographic distribution, and sex.
Residual AR Lab Network isolates and NCBI-deposited sequences have been used for supplemental studies when regulations and resources allow. For example, sequences are used for research on new resistance mechanisms, vaccines, diagnostic test development, and other uses. Newer molecular technologies are clearly the way forward for future N. gonorrhoeae antimicrobial resistance (AMR) diagnostics. The increase in sequence generation and sharing, together with quality-controlled AST data, has the potential to make critical contributions to the development and validation of such technologies.
Last, monitoring international resistance is in the national interest to prepare for potential importation of associated strains and their transmission and spread in the United States. AR Lab Network laboratory protocols and GISP surveillance results have been shared with international partners (25) for efforts to curb AR-Ng worldwide.
POTENTIAL FUTURE DIRECTIONS OF THE AR LAB NETWORK
Currently, GISP, eGISP, and SURRG isolate submissions have not resulted in the full capacity of antimicrobial susceptibility tests for 20,000 isolates. If more specimens were submitted to the network, a greater proportion of reported U.S. cases could be tested for antibiotic susceptibility. In 2018, 8,628 isolates from 583,405 reported U.S. cases were tested in the AR Lab Network, i.e., approximately 1 in 68 cases. If 20,000 isolates were tested, it would be roughly 1 in 28 cases, similar to what the Australian Gonococcal Surveillance Program reported in 2017 (26). This estimate of the extent of laboratory AST has limitations because not all gonorrhea cases are diagnosed and reported to the CDC, and other laboratories may offer AST which is not captured in CDC data. The estimate may also be inaccurate because a few SURRG and eGISP participants had multiple isolates from different anatomical sites from individual patients submitted to the AR Lab Network; those could not be identified or removed for this analysis.
Since capacity has not yet been reached, additional testing could be completed if an outbreak of AR-Ng occurs. Another future possibility is that the network could accept isolates or clinical samples from unaffiliated STD service providers and/or associated local clinical laboratories. This could allow all states to connect to and benefit from the network. As shown in Fig. 1, several states have no submitting clinics, albeit mainly states with lower gonorrhea case rates. The challenge for such laboratories has been that many are not equipped to obtain culture isolates from clinical specimens. There has not been technical support capacity available to help them through a substantial, directed effort outside the GISP, eGISP, or SURRG projects. Priority cases could be persons with suspected treatment failure, including pharyngeal persistent N. gonorrhoeae, international travel, frequent reinfection and/or need for retreatment, or other clinical concerns, e.g., DGI. Collectively, this could improve the availability of AST for patient care and maximize the likelihood of detecting very concerning cases of multidrug or very high-level resistance. If such an expansion were to occur, the respective roles of local and regional laboratories would need to be determined. Regional laboratories could aid submitters in their region in culture recovery by providing transport media to clinics and/or by training others to perform limited gradient-strip AST for rapid local follow-up. There is also a need for technical assistance to improve culture yields since many isolate collection attempts fail due to poor viability, growth, or transport problems (data not shown). This applies particularly to nonurethral specimens. They are included in new eGISP and SURRG projects. In contrast, GISP historically collected only male urethral specimens, which are easier to culture. This has raised questions as to whether GISP isolates represent gonorrhea in women or other potentially undersampled populations. Obtaining isolates from a higher proportion of cases has been a substantial hurdle in reaching the network capacity of 20,000 isolates. The CDC is currently designing laboratory projects to aid sites by developing best-practices documents for bacterial isolation, transport, and culture.
In addition, regional laboratories could implement local genomic sequence data analysis for concerning resistance markers to rapidly identify concerning resistance or strains, perhaps using forthcoming CDC-developed, freely available N. gonorrhoeae genome analysis tools. Should PCR-based AR marker detection in clinical samples become available in validated tests, such approaches could also be implemented at local or regional laboratories.
Recent international “supergonorrhea” cases of multidrug resistance received much attention (27, 28). Patients originally had urogenital symptoms which resolved after treatment; however, test of cure and collection of pharyngeal specimens revealed persisting strains with combined azithromycin and ceftriaxone resistance (27, 28). Such rare cases may not be detected through existing sentinel surveillance or SURRG activities but, rather, by observation of cases submitted by vigilant medical providers. With the dual-therapy regimen of azithromycin and ceftriaxone that was recommended in 2015 (3), no confirmed treatment failures have been reported to the CDC. In cases of suspected treatment failure, the CDC currently advises contacting the CDC for specimen submission and assistance (instructions are online [29]). Regional or local laboratories could provide additional capacity should an increase in suspected treatment failures occur. Thus, the newly built network presents an opportunity to expand national capacity to address treatment failures and could become a leading partner in strengthening the local capacity for rapid AST in every state in the United States.
SUMMARY AND SIGNIFICANCE
The CDC has partnered with four regional state public health laboratories, i.e., a subset of the seven regional laboratories in the CDC’s AR Lab Network and an extensive network of local laboratories associated with STD service providers, to strengthen AR-Ng detection capacity in the United States. This has increased N. gonorrhoeae AST and culture capacity, with the goal of reaching 20,000 cultured isolates per year. While this capacity has not yet been reached, the current work nevertheless facilitates monitoring of antibiotic susceptibility patterns and genetic markers of AR and prepares the nation for the potential of emerging multidrug resistance and dwindling drug options for gonorrhea treatment.
ACKNOWLEDGMENTS
Many individuals contribute to the success of the AR Lab Network AR-Ng program, but not all can be named. We thank former GISP laboratories for their fundamental work developing N. gonorrhoeae testing protocols. We gratefully acknowledge GISP, eGISP, and SURRG projects and the clinics and their associated local laboratories that submit data and specimens to these projects.
This study received funding from the CDC and/or CDC’s Combating AR Bacteria (CARB) initiative.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
AR-Ng Working Group members are the following: Jillian Loomis, Liore Klein, Rebecca Abelman, and Ami Patel (all, Maryland Department of Health); Christina Moore (Tennessee Department of Health); John Leavitt (Texas Department of State Health Services); Sopheay Hun (Washington State Department of Health); Jennifer Ludovic, Shacara Johnson, Tremeka Sanders, Jesse Thomas, Alesia Harvey, Stephanie Gumbis, and Jennifer Reimche (all, CDC).
REFERENCES
- 1.CDC. 2019. Sexually transmitted disease surveillance 2018. https://www.cdc.gov/std/stats18/default.htm.
- 2.Unemo M, Shafer WM. 2014. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Workowski KA. 2015. Centers for Disease Control and Prevention sexually transmitted diseases treatment guidelines. Clin Infect Dis 61(Suppl 8):S759–S762. doi: 10.1093/cid/civ771. [DOI] [PubMed] [Google Scholar]
- 4.CDC. 2013. Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 5.Centers for Disease Control and Prevention. 2014. Recommendations for the laboratory-based detection of Chlamydia trachomatis and Neisseria gonorrhoeae—2014. MMWR Recomm Rep 63:1–19. [PMC free article] [PubMed] [Google Scholar]
- 6.Buono SA, Watson TD, Borenstein LA, Klausner JD, Pandori MW, Godwin HA. 2015. Stemming the tide of drug-resistant Neisseria gonorrhoeae: the need for an individualized approach to treatment. J Antimicrob Chemother 70:374–381. doi: 10.1093/jac/dku396. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Taylor TH Jr, Pettus K, Johnson S, Papp JR, Trees D. 2016. Comparing the disk-diffusion and agar dilution tests for Neisseria gonorrhoeae antimicrobial susceptibility testing. Antimicrob Resist Infect Control 5:46. doi: 10.1186/s13756-016-0148-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis O, Hemarajata P, Shahkolahi A, Masinde G, Buchs K, Humphries RM, Klausner JD. 2019. A multisite implementation of a real-time polymerase chain reaction assay to predict ciprofloxacin susceptibility in Neisseria gonorrhoeae. Diagn Microbiol Infect Dis 94:213–317. doi: 10.1016/j.diagmicrobio.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eyre DW, Golparian D, Unemo M. 2019. Prediction of minimum inhibitory concentrations of antimicrobials for Neisseria gonorrhoeae using whole-genome sequencing. Methods Mol Biol 1997:59–76. doi: 10.1007/978-1-4939-9496-0_4. [DOI] [PubMed] [Google Scholar]
- 10.Kirkcaldy RD, Harvey A, Papp JR, Del Rio C, Soge OO, Holmes KK, Hook EW III, Kubin G, Riedel S, Zenilman J, Pettus K, Sanders T, Sharpe S, Torrone E. 2016. Neisseria gonorrhoeae antimicrobial susceptibility surveillance–The Gonococcal Isolate Surveillance Project, 27 Sites, United States, 2014. MMWR Surveill Summ 65:1–19. doi: 10.15585/mmwr.ss6507a1. [DOI] [PubMed] [Google Scholar]
- 11.CDC. 2019. Epidemiology and laboratory capacity for prevention and control of emerging infectious diseases (ELC). https://www.cdc.gov/ncezid/dpei/epidemiology-laboratory-capacity.html. Accessed 1 May 2019.
- 12.Association of Public Health Laboratories. 2019. ELC program: essential funding for public health lab response. https://www.aphl.org/policy/Pages/ELC.aspx. Accessed 1 May 2019.
- 13.Bizune DJ, Johnson SD, Bachmann L, Bhattacharyya S, Pathela P, Garrett-Cherry T, Golden M, Holderman J, Nguyen TQ, Riley C, Tripplett LR, Kersh EN, Nash E, Pettus K, Sharpe S, Papp J, Pham CD, Bernstein K, Kirkcaldy RD, Schlanger K. 2018. Launching a U.S. Public Health Response to Resistant Gonorrhea: implementation of Strengthening U.S. Response to Resistant Gonorrhea (SURRG), 2017, poster 360-T. 2018 STD Prevention Conference, Washington, DC, 27 to 30 August 2018. [Google Scholar]
- 14.St Cyr S, Quilter L, Pham CD, Torrone E, Weinstock H. 2019. Concurrent gonococcal infections with differing susceptibility results enhanced Gonococcal Isolate Surveillance Project (eGISP). Open Forum Infect Dis 6(Suppl 2):S216. doi: 10.1093/ofid/ofz360.507. [DOI] [Google Scholar]
- 15.CDC. 2016. Gonococcal Isolate Surveillance Project (GISP) protocol. https://www.cdc.gov/std/gisp/GISP-Protocol-May-2016.pdf.
- 16.Unemo M, Ballard R, Ison C, Lewis D, Ndowa F, Peeling R. (ed). 2013. Laboratory diagnosis of sexually transmitted infections, including human immunodeficiency virus. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 17.Papp JR, Henning T, Khubbar M, Kalve V, Bhattacharyya S, Travanty E, Xavier K, Jones K, Rudrik JT, Gaynor A, Hagan C. 2016. Recovery of Neisseria gonorrhoeae from 4 commercially available transport systems. Diagn Microbiol Infect Dis 86:144–147. doi: 10.1016/j.diagmicrobio.2016.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed CLSI standard M07 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.CLSI. 2019. performance standards for antimicrobial susceptibility testing, 29th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 20.Kersh EN, Allen V, Ransom E, Schmerer M, St Cyr S, Workowski K, Weinstock H, Patel J, Ferraro MJ. 9 April 2019. Rationale for a Neisseria gonorrhoeae susceptible only interpretive breakpoint for azithromycin. Clin Infect Dis doi: 10.1093/cid/ciz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribot EM, Hise KB. 2016. Future challenges for tracking foodborne diseases: PulseNet, a 20-year-old US surveillance system for foodborne diseases, is expanding both globally and technologically. EMBO Rep 17:1499–1505. doi: 10.15252/embr.201643128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas JC, Seby S, Abrams AJ, Cartee J, Lucking S, Vidyaprakash E, Schmerer M, Pham CD, Hong J, Torrone E, St Cyr S, Shafer WM, Bernstein K, Kersh EN, Gernert KM, Antimicrobial-Resistant Neisseria gonorrhoeae Working G. 2019. Evidence of recent genomic evolution in gonococcal strains with decreased susceptibility to cephalosporins or azithromycin in the United States, 2014–2016. J Infect Dis 220:294–305. doi: 10.1093/infdis/jiz079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutgring JD, Machado MJ, Benahmed FH, Conville P, Shawar RM, Patel J, Brown AC. 2018. FDA-CDC Antimicrobial Resistance Isolate Bank: a publicly available resource to support research, development, and regulatory requirements. J Clin Microbiol 56:e01415-17. doi: 10.1128/JCM.01415-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golparian D, Unemo M. 2019. Now is the time to implement whole genome sequencing in the global antimicrobial resistance surveillance for Neisseria gonorrhoeae? EClinicalMedicine 7:11–12. doi: 10.1016/j.eclinm.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wi T, Lahra MM, Ndowa F, Bala M, Dillon JR, Ramon-Pardo P, Eremin SR, Bolan G, Unemo M. 2017. Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLoS Med 14:e1002344. doi: 10.1371/journal.pmed.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahra MM, Enriquez R, George C. 2019. Australian Gonococcal Surveillance Programme annual report, 2017. Commun Dis Intell 2019:43. doi: 10.33321/cdi.2019.43.13. [DOI] [PubMed] [Google Scholar]
- 27.Eyre DW, Sanderson ND, Lord E, Regisford-Reimmer N, Chau K, Barker L, Morgan M, Newnham R, Golparian D, Unemo M, Crook DW, Peto TE, Hughes G, Cole MJ, Fifer H, Edwards A, Andersson MI. 2018. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Euro Surveill 23(27):pii=1800323. doi: 10.2807/1560-7917.ES.2018.23.27.1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiley DM, Jennison A, Pearson J, Lahra MM. 2018. Genetic characterisation of Neisseria gonorrhoeae resistant to both ceftriaxone and azithromycin. Lancet Infect Dis 18:717–718. doi: 10.1016/S1473-3099(18)30340-2. [DOI] [PubMed] [Google Scholar]
- 29.CDC. 2019. Antibiotic-resistant gonorrhea basic information. https://www.cdc.gov/std/gonorrhea/arg/basic.htm. Accessed 11 November 2019.