LETTER
The positive impact that matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has had on clinical microbiology over the past decade has been well established (1). However, unfortunately, the reliable identification to the species level of Mycobacterium tuberculosis complex (MTC) isolates has not to date been achievable by MALDI-TOF MS. Nevertheless, some researchers have shown that MALDI-TOF MS coupled with in silico analysis has been useful as a typing tool for other mycobacterial groupings that were not previously possible, including Mycobacterium kansasii (2), differentiating Mycobacterium chimaera from Mycobacterium intracellulare (3), and distinguishing Mycobacterium avium subsp. paratuberculosis from other M. avium subspecies (4). In this study, we set out to examine whether MALDI-TOF MS could be used as a typing tool for outbreak investigations.
We used 32 MTC isolates that had undergone 24-digit mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing by the Irish Mycobacterial Reference Laboratory. All isolates in the present study were cultured in Cork University Hospital from patient respiratory specimens. The 32 isolates encompassed the following three lineages: European-American (EA) (8 sublineages: Haarlem [9], H37Rv [5], Cameroon [2], X [3], Ural [1], NEW-1 [1], S [1], and EA with no subtype [4]), Oceanic (East African-Indian [EAI] subtype [3]) and Mycobacterium bovis (Bovis BCG subtype [3]). There were 9 isolates which had identical MIRU-VNTR numbers; 3 were Haarlem subtypes, 2 were H37Rv subtypes, 2 were EAI subtypes and 2 were M. bovis BCG subtypes. Two of the three identical Haarlem subtypes and both of the identical EAI and H37rv subtype pairs were confirmed as cluster cases through contact tracing.
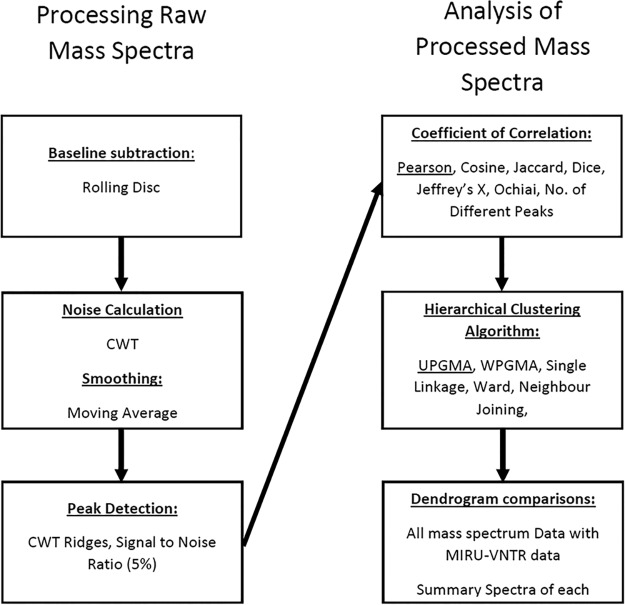
The isolates were recovered, extracted and identified using a Bruker Biotyper Microflex LT system with version 1.0 of the Mycobacteria Library to attain a minimum of 5 biological replicates, as previously described (5). All identifications by MALDI-TOF MS were correct (MTC), with log scores greater than 2.0. The mass spectrum data were exported to BioNumerics version 7.6.2 (bioMérieux). The data initially underwent processing, over 120 permutations were examined, and the parameters outlined in Fig. 1 were visually determined to be optimal. This was done by zooming into the baseline of each mass spectrum and visually assessing the whole mass spectrum to ensure that the processing algorithms utilized did not eliminate spectral peaks (there are no quantitative evaluation criteria available [6]). The coefficient of correlation and hierarchical clustering algorithm (HCA) that were most appropriate for these data were then analyzed (itemized in Fig. 1); it was found that Pearson’s correlation coefficient coupled with the unweighted pair group method with arithmetic mean (UPGMA) was most suitable, having the highest global cophenetic correlation (91%) of the algorithms analyzed. Next, an examination of the HCA of individual mass spectra for each isolate and also for combined summary spectra for each isolate was compared to HCA of the MIRU-VNTR data (Pearson’s correlation coefficient with UPGMA, global cophenetic correlation of 84%) to ascertain whether the data type was suitable as a typing tool. Summary spectra allow for the highlighting of peaks that are unique to an isolate while eliminating those that may be erroneous on a single mass spectrum.
FIG 1.
Summary of processing protocol for raw mass spectra and subsequent analysis, including creation of dendrograms. CWT, continuous wavelet transformation; UPGMA, unweighted pair group method with arithmetic mean; WPGMA, weighted pair group method with arithmetic mean.
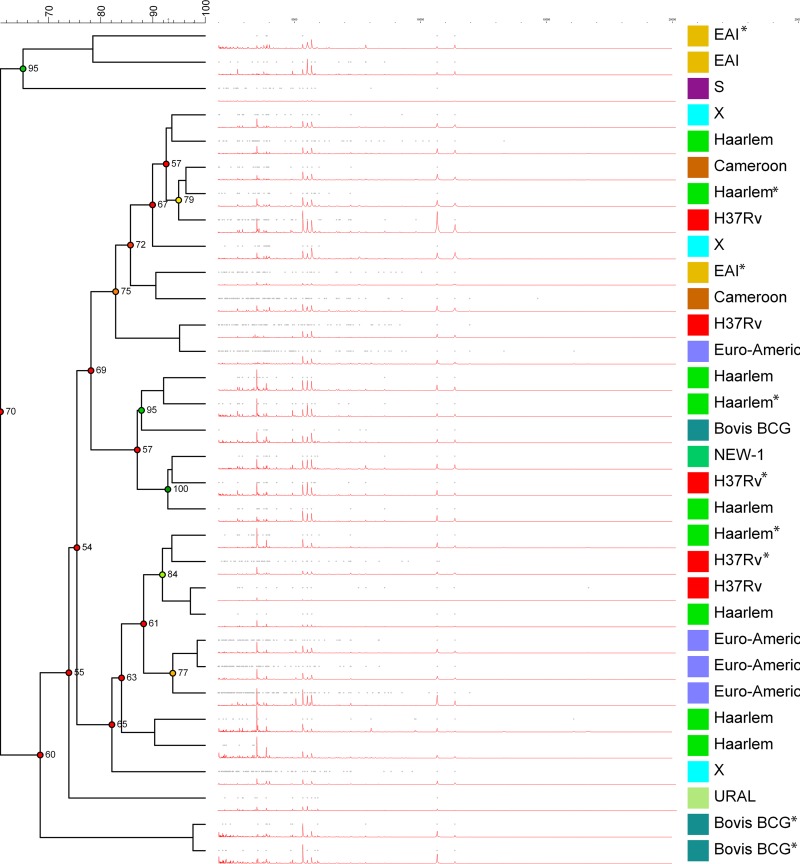
As highlighted in Fig. 2, there was no coherent clustering of MIRU-VNTR lineages/sublineages for the summary spectra. The only isolates that had identical MIRU-VNTR numbers that grouped together were two Bovis-BCG isolates with identical MIRU-VNTR numbers. The dendrograms of individual spectra performed worse than the summary spectra dendrogram, with mass spectra from the same isolate not grouping together. This may be due to the high clonality of MTC isolates or due to the low resolution of MALDI-TOF MS. The data presented demonstrates that the combination of a previously published method to improve identification and enhanced processing of mass spectra do not currently facilitate the typing of clinical MTC isolates.
FIG 2.
Dendrogram of summary spectra (pictured in red) calculated following comparison by Pearson’s coefficient of correlation and determined by the unweighted pair group method with arithmetic mean (UPGMA). Isolates with identical MIRU-VNTR digits are marked with an asterisk.
ACKNOWLEDGMENTS
J.A.O. was funded by the Irish Research Council (grant GOIPG/2014/72), J.A.O., B.L., and G.D.C. were also funded by the Strategy for the Control of Antimicrobial Resistance in Ireland (SARI). Neither agency had any role in study design, data collection and interpretation, or in the decision to submit the work for publication.
We declare no conflicts of interest or funding.
We thank all staff in the Microbiology Department, Cork University Hospital (CUH), Wilton, Cork, Ireland, for their help, especially Mary Lynch Healy.
REFERENCES
- 1.Rodríguez-Sánchez B, Cercenado E, Coste AT, Greub G. 2019. Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Euro Surveill 24:pii=1800193 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2019.24.4.1800193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murugaiyan J, Lewin A, Kamal E, Bakuła Z, van Ingen J, Ulmann V, Unzaga Baranano MJ, Humiecka J, Safianowska A, Roesler UH, Jagielksi T. 2018. MALDI spectra database for rapid discrimination and subtyping of Mycobacterium kansasii. Front Microbiol 9:587. doi: 10.3389/fmicb.2018.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epperson LE, Timke M, Hasan NA, Godo P, Durbin D, Helstrom NK, Shi G, Kostrzewa M, Strong M, Salfinger M. 2018. Evaluation of a Novel MALDI Biotyper algorithm to distinguish Mycobacterium intracellulare from Mycobacterium chimaera. Front Microbiol 9:3140. doi: 10.3389/fmicb.2018.03140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricchi M, Mazzarelli A, Piscini A, Di Caro A, Cannas A, Leo S, Russo S, Arrigoni N. 2017. Exploring MALDI-TOF MS approach for a rapid identification of Mycobacterium avium ssp. paratuberculosis field isolates. J Appl Microbiol 122:568–577. doi: 10.1111/jam.13357. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor JA, Lynch-Healy M, Corcoran D, O’Reilly B, O’Mahony J, Lucey B. 2016. Improved matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based identification of Mycobacterium spp. by use of a novel two-step cell disruption preparatory technique. J Clin Microbiol 54:495–496. doi: 10.1128/JCM.02998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM. 2007. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom 260:212–221. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]