Candida auris is a multidrug-resistant yeast which has emerged in health care facilities worldwide; however, little is known about identification methods, patient colonization, environmental survival, spread, and drug resistance. Colonization on both biotic (patients) and abiotic (health care objects) surfaces, along with travel, appear to be the major factors for the spread of this pathogen across the globe. In this investigation, we present laboratory findings from an ongoing C. auris outbreak in New York (NY) from August 2016 through 2018.
KEYWORDS: Candida auris, mycology, antifungals, molecular biology, phylogenetics
ABSTRACT
Candida auris is a multidrug-resistant yeast which has emerged in health care facilities worldwide; however, little is known about identification methods, patient colonization, environmental survival, spread, and drug resistance. Colonization on both biotic (patients) and abiotic (health care objects) surfaces, along with travel, appear to be the major factors for the spread of this pathogen across the globe. In this investigation, we present laboratory findings from an ongoing C. auris outbreak in New York (NY) from August 2016 through 2018. A total of 540 clinical isolates, 11,035 patient surveillance specimens, and 3,672 environmental surveillance samples were analyzed. Laboratory methods included matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for yeast isolate identification, real-time PCR for rapid surveillance sample screening, culture on selective/nonselective media for recovery of C. auris and other yeasts from surveillance samples, antifungal susceptibility testing to determine the C. auris resistance profile, and Sanger sequencing of the internal transcribed spacer (ITS) and D1/D2 regions of the ribosomal gene for C. auris genotyping. Results included (a) identification and confirmation of C. auris in 413 clinical isolates and 931 patient surveillance isolates as well as identification of 277 clinical cases and 350 colonized cases from 151 health care facilities, including 59 hospitals, 92 nursing homes, 1 long-term acute care hospital (LTACH), and 2 hospices, (b) successful utilization of an in-house developed C. auris real-time PCR assay for the rapid screening of patient and environmental surveillance samples, (c) demonstration of relatively heavier colonization of C. auris in nares than in the axilla/groin, and (d) predominance of the South Asia clade I with intrinsic resistance to fluconazole and elevated MIC to voriconazole (81%), amphotericin B (61%), flucytosine (5FC) (3%), and echinocandins (1%). These findings reflect greater regional prevalence and incidence of C. auris and the deployment of better detection tools in an unprecedented outbreak.
INTRODUCTION
Candida auris, a multidrug-resistant pathogenic yeast has emerged in health care facilities across the globe. Candida auris was first described as a new species in 2009 from the ear discharge of a hospitalized patient in Japan (1). In the last decade, cases of C. auris have been reported from more than 35 countries on five continents, including Asia, Africa, Europe, South America, and North America (2–13). Fungemia caused by C. auris is associated with a therapeutic failure and high mortality rate (14). Ecological niches of C. auris remain unknown. However, successful colonization of human body sites and survival and persistence on surfaces within health care environments may be contributing to the outbreak and prevalence of C. auris worldwide. Whole-genome sequencing (WGS) of C. auris isolates revealed four major populations, which emerged independently and spread locally within each region in Asia, South Africa, and South America (7, 15, 16). These populations were grouped into South Asian (I), East Asian (II), African (III), and South American (IV) clades. Recently, a fifth clade of C. auris was identified from a patient in Iran who never traveled outside that country (17). The WGS revealed that the Iran C. auris isolate was genetically distinct and separated by >200,000 single nucleotide polymorphisms (SNPs) from the other four known C. auris clades (18).
The identification of C. auris has been challenging for most clinical laboratories because of reliance on biochemical-based identification systems such as Vitek 2 and API 20C AUX. Because C. auris is not present in the databases of these systems, it is misidentified as Candida haemulonii (5, 7, 9, 11, 15, 19). Other less commonly used biochemical platforms also misidentified C. auris as Rhodotorula glutinis, Candida famata, Candida sake, and Saccharomyces cerevisiae (19–21). Accurate identification of C. auris requires matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (22) or sequencing of the 18S ribosomal gene (1). These reliable techniques are not readily available in most clinical laboratories. In the United States, the Food and Drug Administration (FDA) approved the use of C. auris on the MALDI-TOF MS platform such as Bruker Daltonik, Bremen, Germany, as recently as 24 April 2018 (http://www.cidrap.umn.edu/news-perspective/2018/04/fda-approves-rapid-diagnostic-test-candida-auris) and bioMérieux Vitek MS, Marcy-l’Etoile, France, on 21 December 2018 (https://www.rapidmicrobiology.com/news/new-fda-clearance-for-vitek-ms-expanded-id-for-challenging-pathogens).
Apart from the difficult identification of C. auris, antifungal resistance testing has also been challenging. Approximately 90% of C. auris isolates are resistant to fluconazole (15). Moreover, elevated MICs for voriconazole have been reported in 50% of isolates and higher MICs for amphotericin B in 30% of isolates (16, 19, 23). Resistance to echinocandins, although low, has also been reported (23).
Adams and colleagues described the first 51 clinical cases and 61 colonized cases in the outbreak in New York (NY) health care facilities (19). As part of an ongoing response to this C. auris outbreak, 540 clinical isolates and 11,035 patient and 3,762 environmental surveillance samples have been collected in New York from 2016 through 2018. Here, we provide the laboratory analysis of these specimens which highlights the application of rapid molecular screening to outbreak control, the unique characteristics of C. auris isolates, the spectrum of antifungal resistance, and the prevalence of other pathogenic yeasts.
MATERIALS AND METHODS
Sample collection and case definitions.
Clinical yeast isolates suspected of C. auris received from various health care facilities in NY from August 2016 to December 2018 were part of this investigation. However, surveillance samples collected from August 2016 to October 2018 from patients and their environment in various C. auris-affected health care facilities in New York were the major focus of these studies. Clinical cases were defined as the identification of a first C. auris isolate recovered from a specimen obtained to diagnose or treat disease (19). These C. auris isolates and specimens recovered were grouped as clinical samples. Colonized cases were defined as the recovery of the first C. auris isolate from a sample for surveillance purposes (16). The C. auris isolate recovered from colonized or clinical cases or from environmental samples for surveillance purposes were all grouped as surveillance samples. The environmental samples processed in this study were classified as porous (e.g., linen, carpet, etc.) or nonporous (e.g., metal knob, phone, TV monitor, bed rail, etc.) based on surface texture. Environmental samples which did not fall within these two categories were excluded from this investigation (see Table S1 in the supplemental material).
Candida identification.
The species of Candida isolates were determined by MALDI-TOF MS (Bruker, Bremen, Germany) using both the manufacturer’s and in-house validated library databases. The in-house library database was enriched by adding spectra of several C. auris isolates from the current NY outbreak and from the Centers for Disease Control and Prevention Antibiotic Resistance (CDC AR) bank (https://www.cdc.gov/drugresistance/resistance-bank/index.html). Some closely related species from the CDC AR panel, including C. haemulonii (2), and Candida duobushaemulonii (3), were also added to the in-house library. These efforts led to the successful identification of C. auris and other closely related species (19).
Colonization study.
(i) Sample types. Axilla, groin, and nare swabs were collected individually or as a composite swab (axilla/groin or nares/axilla/groin) using the BD ESwab Liquid Amies Collection and Transport System (Becton, Dickinson, Franklin Lakes, NJ, USA) from patients in various health care facilities. Occasionally, rectal or other body site swabs or body fluids were also collected for surveillance purposes. For environmental sampling, various objects and surfaces in health care facilities were swabbed with 3M Sponge Sticks (3M Health Care, St. Paul, MN, USA), and after an area was sampled, the sponge was removed from the collection stick and placed in a zip-top bag. All surveillance swabs, other body fluids, and environmental sponges were transported to the laboratory within 24 to 48 h at ambient temperature. Based on the number of samples received, they were processed immediately or stored at 4°C before processing.
(ii) Culture of swabs and sponges. Each ESwab containing 1 ml modified liquid Amies medium was vortexed for 30 s, and 50 μl was inoculated onto nonselective (Sabouraud dextrose agar containing antibacterials [SDA-A], including chloramphenicol, 25 mg/liter; gentamicin, 40 mg/liter; penicillin, 20,000 U/liter; streptomycin; 40 mg/liter) and selective (Sabouraud dulcitol agar containing antibacterials [SDulA-AS; see above for composition] and 10% salt) media as described previously (19). Two hundred microliters of the ESwab liquid were also inoculated in selective broth medium minus agar (SDulB-AS). Sputum and bronchial aspirates were streaked on all the media as described above. Urine samples, if received in large volume (10 to 20 ml), were concentrated by centrifugation at 4,000 rpm for 5 min and the supernatant was decanted, leaving about 3 ml; 100 μl was inoculated onto both SDA-A and SDulA-AS plates, and 1 ml was inoculated in 5 ml of SDulB-AS broth.
Each environmental sponge sample was placed in a Whirl-Pak Homogenizer Blender Filter bag containing 45 ml of phosphate-buffered saline (PBS) with 0.02% Tween 80. The bags were gently mixed in the Stomacher 400 Circulator (Laboratory Supply Network, Inc., Atkinson, NH, USA) at 260 rpm for 1 min, and the suspension was transferred into a 50-ml conical tube and centrifuged at 4,000 rpm for 5 min; the supernatant was decanted, leaving about 3 ml of liquid at the bottom of the tube. The 3 ml liquid was vortexed briefly, and of these, 1 ml was removed, centrifuged, washed, and resuspended in 50 μl of PBS-bovine serum albumin (BSA) for DNA extraction as described below for swabs. From the remaining 2 ml of liquid, 100 μl each was inoculated on SDA-A and SDulA-AS plates, and 1 ml was inoculated in 5 ml of SDulB-AS broth. Agar plates and broth tubes were incubated at 40°C for a maximum of 2 weeks.
Enumeration of CFU.
To determine the extent of colonization on skin by C. auris, the colonies recovered on selective agar medium were counted. If colonies were numerous, a 10-fold dilution series of the swab was prepared and plated for colony counts. Recovered colonies were identified by MALDI-TOF MS and results were expressed as CFU per swab. For other Candida species, colonies recovered on nonselective medium were randomly picked and identified by MALDI-TOF MS.
Real-time PCR.
A real-time PCR assay developed in the laboratory (24) for the rapid screening of surveillance samples for C. auris was deployed effective 17 May 2017 following approval from the New York State Clinical Laboratory Evaluation Program (CLEP). In brief, 200 μl of ESwab liquid Amies and 1 ml of concentrated liquid following sponge processing were washed twice with phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA). The pellet was resuspended in 50 μl of PBS-BSA followed by freezing, heating, bead-beating, and centrifugation at 13,000 rpm for 5 min; 5 μl of the extracted DNA was tested in duplicates on the real-time PCR assay. A cycle threshold value of ≤37 was reported as positive, and >37 was reported as negative for C. auris. If PCR inhibition was observed, the results was reported as inconclusive (24).
Antifungal susceptibility testing of C. auris.
C. auris isolates were tested according to Clinical and Laboratory Standards Institute reference methodology M27-A4 (25). The MICs of azoles and echinocandins were determined by custom TREK frozen broth microdilution panels (catalog number CML2FCAN; Thermo Fisher Scientific, Marietta, OH, USA), and MICs of amphotericin B and flucytosine (5FC) were determined by Etest as recommended by the manufacturer (AB Biodisk; bioMérieux, Solna, Sweden) except that MICs were read at 24 h postincubation or until a confluent lawn of growth was seen. There are currently no established C. auris-specific susceptibility breakpoints. Therefore, CDC-defined breakpoints were used for azoles, amphotericin B, and echinocandins, and also, the MIC value of 1.5 for amphotericin B was rounded up to 2.0 (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html).
Phylogenetic analysis of Candida auris.
Genomic DNA of C. auris from the current outbreak and reference strains procured from the CDC AR bank was extracted using a QIAcube DNA extractor with the QIAamp DNA Minikit (Qiagen, Hilden Germany). The extracted DNA was amplified for the internal transcribed spacer (ITS) and D1/D2 regions of the ribosomal gene with primer sets ITS1-ITS4 and NL1-NL4, respectively (26). Conventional PCR was performed using proofreading AccuTaq LA DNA polymerase (Sigma-Aldrich, St. Louis, MO, USA) with initial denaturation at 95°C for 3 min followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 3 min and the final extension at 68°C for 10 min. The PCR amplicons were sequenced, assembled, and edited using Sequencher 5.0 software (Gene Codes Corp., Ann Arbor, MI, USA) and BLAST searched against two databases: GenBank (www.ncbi.nlm.nih.gov/) and CBS-KNAW (www.cbs.knaw.nl/). Multiple alignments of ITS and D1/D2 sequences of C. auris from the current outbreak and reference strains were conducted using the Geneious R9 version 9.1.6 (Biomatters, Inc., Newark, NJ) and the phylogenetic analysis of the aligned sequences was performed using the neighbor-joining (NJ) method with 2,000 bootstrap replicates on the same software.
Statistical analysis.
GraphPad Prism 8 software for Mac (GraphPad Software, Inc., La Jolla, CA) was used for statistical analysis of the results. Two-by-two tables and the nonparametric Mann Whitney test were used to compare samples.
Results communication.
Any positive real-time PCR result was immediately communicated to New York State Department of Health (NYSDOH) epidemiologists and the requesting facilities, allowing facilities to rapidly implement an infection control response. Following real-time PCR, culture results and then antifungal susceptibility testing results were communicated. Unusual findings, such as resistance to echinocandins, were also communicated to the CDC as an alert.
Data availability.
All nucleotide sequences of C. auris were deposited in GenBank under the following accession numbers: ITS sequences, MN338097 to MN338196; D1/D2 sequences, MN337432 to MN337531.
RESULTS
New York state-wide Candida auris identification.
From various clinical, public, and commercial laboratories, 540 isolates of yeasts suspected of being C. auris were received. Of these, 413 were confirmed as C. auris, 12 as C. duobushaemulonii, and 7 as C. haemulonii (see Table S2 in the supplemental material). The remaining 108 isolates were confirmed as other yeasts (data not shown). In most cases, there was no correlation seen between presumptive identification (ID) by the requesting laboratory and the method they used for identification. Candida auris was correctly identified 100% (42/42) of the time by laboratories using ITS sequencing, 72% (106/147) of the time by laboratories using Bruker MALDI, and 21% (9/43) of the time by laboratories using Vitek MS. Interestingly, some clinical laboratories using the biochemical-based system, Vitek 2, provided a presumptive C. auris identification. Whether these laboratories presumed identification of C. haemulonii as C. auris or used a “research-use-only” database of MALDI-TOF from Bruker or bioMérieux is unclear.
Clinical cases.
A total of 277 clinical cases were confirmed by culture during this period. The majority of C. auris isolates from clinical cases were recovered from blood (51%) followed by urine (23%) and then other body sites (Table 1). Based on these results, a bloodstream infection (BSI) was by far the most common clinical infection. An additional 136 C. auris isolates were recovered from some of these 277 clinical case patients, resulting in a total recovery of 413 C. auris isolates.
TABLE 1.
Source of first C. auris isolate to define a clinical case
| Source | No. of samples culture positive | % of total first-positive isolates |
|---|---|---|
| Blood | 140 | 51 |
| Urine | 64 | 23 |
| Wound | 37 | 13 |
| Lung | 23 | 8 |
| Bile | 4 | 1.5 |
| Corneal/eye | 2 | <1.0 |
| Ear | 1 | <1.0 |
| Bone | 1 | <1.0 |
| Stool | 1 | <1.0 |
| Unspecified | 4 | 1.5 |
| Total | 277 |
Colonized cases.
Nine hundred thirty-one (8.4%) of 11,035 patient surveillance samples tested positive for C. auris by culture (Table 2). These included 450 first-positive patient surveillance samples representing 350 colonized cases, 183 subsequent-positive patient surveillance samples from some of the 350 colonized cases, and 298 positive patient surveillance samples from known clinical cases. A total of 151 health care facilities in which the patients received care in the 90 days prior to their C. auris diagnosis were affected by the outbreak, and these included 56 hospitals, 92 nursing homes, 1 long-term acute care hospital (LTACH), and 2 hospices.
TABLE 2.
Testing of various surveillance samples for recovery of C. auris in culture from August 2016 to October 2018
| Source | No. of samples culture positive/no. of samples tested (%) |
|---|---|
| Axilla/groin | 231/3,928 (6.0) |
| Nares | 275/4,058 (6.8) |
| Nares/axilla/groin | 121/1,846 (6.6) |
| Axilla | 68/340 (20.0) |
| Groin | 70/311 (22.5) |
| Rectal | 37/165 (22.4) |
| Wound | 69/187 (36.9) |
| Urine | 20/57 (35.1) |
| Respiratory | 18/51 (35.3) |
| Ear | 6/15 (40.0) |
| Skin | 5/47 (8.8) |
| Unspecified | 11/30 (36.7) |
| Total | 931/11,035 (8.4) |
Of 624 surveillance samples cultured, 450 were positive, defining 350 colonized cases (Table 3). Although first positive surveillance samples defining 350 colonized cases originated from different body sites, we chose axilla/groin and nares to understand the probability and the extent of C. auris colonization as they made up the bulk of surveillance samples. Of 222 axilla/groin samples tested, 178 (80%) were positive for C. auris, while of 215 nares samples tested, 125 (58%) were positive for C. auris (Table 3). When the extent of C. auris colonization in 178 axilla/groin and 125 nare positive sites were analyzed randomly; nares harbored 2 logs (P < 0.0001) higher C. auris than the axilla/groin (Fig. 1A). When 74 of axilla/groin and nares in parallel (from the same patient) were analyzed; nares harbored 2 logs higher C. auris than the axilla/groin (Fig. 1B). These results indicated that axilla/groin is the preferred site of C. auris colonization compared to nares in the colonized patients, but if nares are colonized, they harbor a relatively higher burden of C. auris than axilla/groin. These results plus practical logistics and resource issues prompted us to use one composite swab of nares/axilla/groin for determining colonized cases, effective January 2018 for all point prevalence studies. Of 350 colonized cases, 106 were indeed identified using one composite swab of nares/axilla/groin in the present investigation.
TABLE 3.
Source of first C. auris isolate to define a colonized case
| Source | No. of samples positive/no. of samples tested | % of total first-positive samples |
|---|---|---|
| Axilla/groin (bilateral) | 178/222 | 80 |
| Nares (bilateral) | 125/215 | 58 |
| Nares/axilla/groin (bilateral) | 106/106 | 100 |
| Axilla (unilateral) | 10/20 | 50 |
| Groin (unilateral) | 10/20 | 50 |
| Nares (unilateral) | 6/14 | 43 |
| Wound | 4/11 | 36 |
| Rectal | 4/7 | 57 |
| Ear | 4/6 | 67 |
| Skin | 1/1 | 100 |
| Not specified | 2/2 | 100 |
| Total | 450/624 | 72 |
FIG 1.
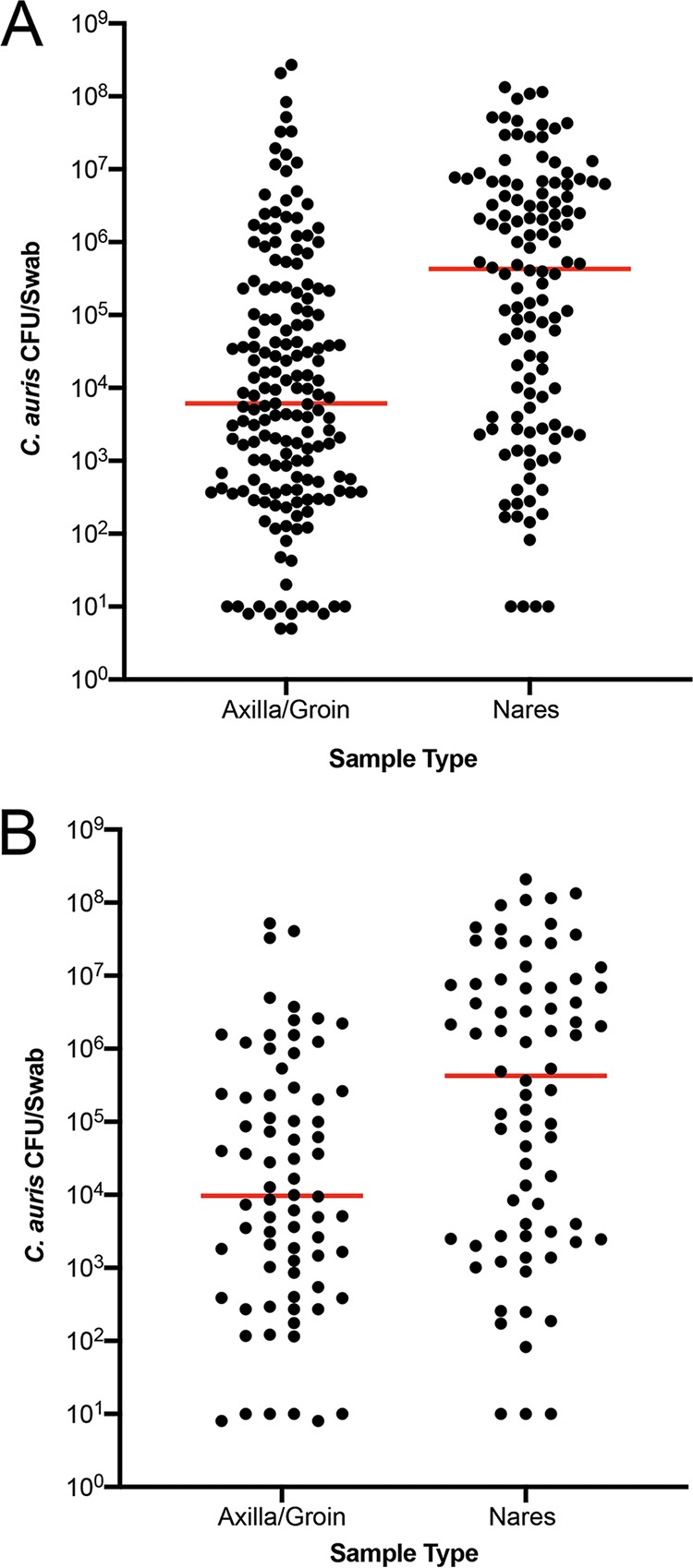
Colonization of the axilla/groin and nares. Axilla/groin and nares were swabbed and processed for culture. Recovered colonies were counted, and results were expressed as CFU/swab. Each dot represents total CFU/swab/patient, and the horizontal bar within each group represents the median. (A) Unpaired samples from patients colonized with C. auris. (B) Paired samples from patients colonized with C. auris. In both cases, the median C. auris CFU was 2-log higher in nares than in axilla/groin (P < 0.0001).
Utility of C. auris real-time PCR assay in patient surveillance screening.
Following NYSDOH CLEP approval of C. auris real-time PCR assay for patient surveillance samples effective May 2017, a total of 9,982 patient samples were tested from May 2017 to October 2018, and 6,834 of those samples were cultured as part of point prevalence studies. In comparison to culture as a gold standard, the diagnostic accuracy, sensitivity, and specificity of the real-time PCR assay were 98.36%, 93.32%, and 98.38%, respectively (Table 4). The performance was consistent with findings in our earlier investigation (24).
TABLE 4.
Efficacy of the C. auris real-time PCR assay for surveillance samples
| PCR result | No. of cultures testing: |
Accuracy (% or 95% CIa) | Sensitivity (% or 95% CI) | Specificity (% or 95% CI) | PPVb (%) | NPVc (%) | |
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| Patient samples | |||||||
| Positive | 545 | 73 | 98.36 | 93.32 | 98.83 | 88 | 99 |
| Negative | 39 | 6177 | 98.03–98.65 | 90.98–95.21 | 98.53–99.08 | ||
| Environmental samples | |||||||
| Positive | 72 | 362 | 69.42 | 98.63 | 67.5 | 16.67 | 99.87 |
| Negative | 1 | 752 | 66.72–72.09 | 92.60–99.97 | 64.67–70.25 | ||
CI, confidence interval.
PPV, positive predictive value.
NPV, negative predictive value.
Utility of C. auris real-time PCR assay in environmental surveillance screening.
Four hundred thirty-four of 3,672 environmental samples were positive by real-time PCR (11.8%). Candida auris was cultured from 109 of 434 PCR-positive samples (25%). The diagnostic accuracy, sensitivity, and specificity of the real-time PCR assay for the environmental samples were 69.42%, 98.63%, and 67.5%, respectively (Table 4). C. auris colonization of culture-positive environmental surfaces as quantified by CFU revealed a range of concentrations from a low of <50 CFU/surface to a high of >105 CFU/surface; degree of colonization was significantly higher (P < 0.001) on nonporous than on porous surfaces (Fig. 2). However, these results should be interpreted with caution, as many more nonporous samples were analyzed than porous samples.
FIG 2.
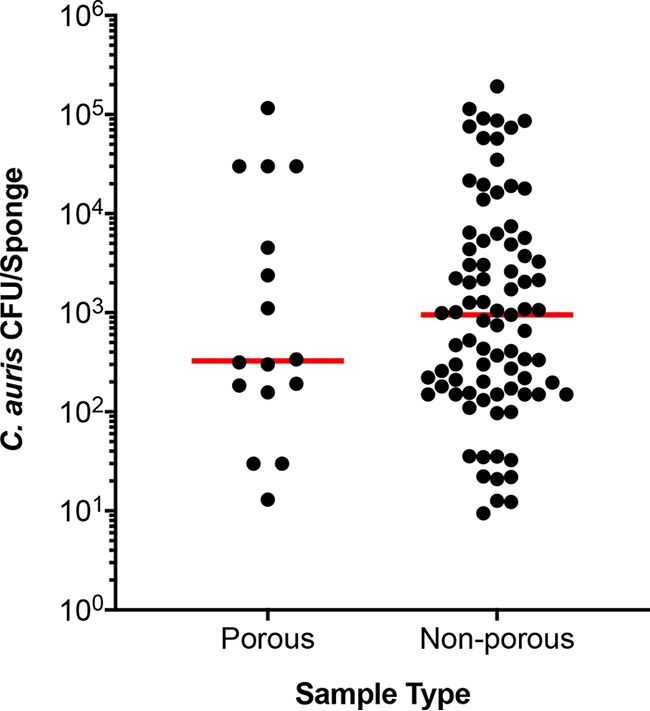
Colonization of environmental surfaces with C. auris. Environmental surfaces of various health care facilities were sponge swabbed and processed for culture. Recovered colonies were counted, and results were expressed as total CFU/sponge. The environmental surfaces were divided into porous (i.e., linen and carpet) and nonporous (i.e., plastic and metal devices) for data analysis. Each dot represents total CFU recovered/per area swabbed, and the horizontal bar within each group represents the median. The median C. auris CFU was approximately 3-fold higher on nonporous surfaces than on the porous surfaces (P < 0.001).
Multiple alignment and phylogenetic analysis of ribosomal genes of C. auris.
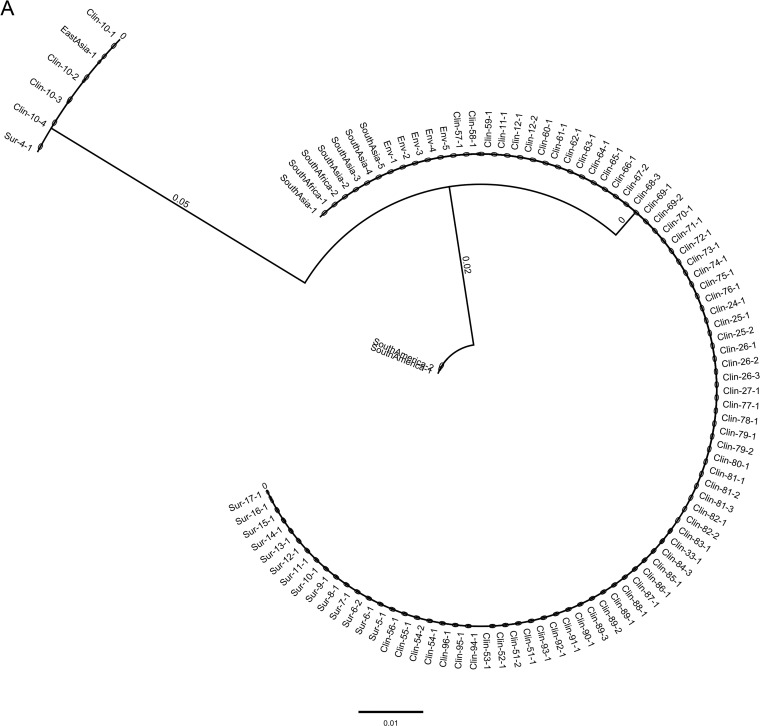
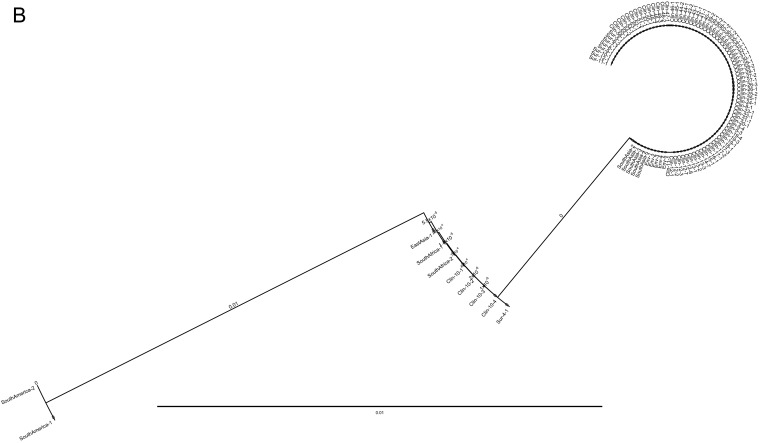
To determine the genetic makeup of C. auris causing the current NY outbreak, ITS and D1/D2 regions of the ribosomal gene were used. The ITS conventional PCR yielded a 400-bp amplicon, and following sequencing and trimming of 5′ and 3′ prime ends of the ITS gene, a 311-bp sequence was used for the multiple alignment. Between the South Asia clade I and East Asia clade II isolates, there were four mutations at the 5′ end of the gene at positions 67 (A to C), 68 (C to T), 70 (A to T), and 74 (C to G) and three mutations at the 3′ prime end of the gene at positions 307 (T to C), 308 (C to G), and 309 (G to T). Additionally, two nucleotide insertions, A and T at 75 and 76, respectively, were present in the South Asia clade I but absent in East Asia clade II. The phylogenetic analyses using a neighbor-joining method revealed these two clades were well separated from each other (Fig. 3A).
FIG 3.
(A) Phylogenetic analysis of C. auris isolates from New York using ITS and D1/D2 sequences. Nucleotide sequences of 622 isolates of C. auris recovered from clinical patients (Clin), colonized patients (Sur), environmental surfaces (Env), and 10 standard isolates from the CDC AR bank were aligned. The neighbor-joining method was used to construct the phylogenetic tree. The bootstrap scores are based on 2,000 reiterations. The outbreak was predominately the South Asia clade I, with a minor population of the East Asia clade II. Trees representing a few C. auris isolates from each group are shown. (A) Analysis of ITS sequences. Note, ITS could not distinguish South Africa clade III (CDC AR) from South Asia clade I. (B) Analysis of D1/D2 sequences. Note, D1/D2 did not differentiate South Africa clade III from East Asia clade II.
Amplification of the D1/D2 region revealed a 500-bp amplicon, and following sequencing and trimming of 5′ and 3′ primed ends, a 426-bp sequence was used for the multiple alignment. Present in the South Asia clade I, but not in the East Asia clade II, were T to C mutations at position 372 and 389 and T insertions at positions 392 and 393. The topology of the neighbor-joining tree was similar to that of ITS (Fig. 3B).
Analysis of 622 clinical, surveillance, and environmental isolates revealed that the South Asia clade I was the major genotype, while the East Asia clade II was the minor genotype of the outbreak in NY. The other two well-known genotypes of C. auris, including South Africa clade III and South America clade IV, were not found in New York.
Antifungal susceptibility testing.
Antifungal susceptibility testing was conducted for 966 C. auris isolates. These included 277 first clinical isolates from 277 clinical patients, 116 subsequent isolates from 74 of those 277 clinical patients, 215 patient surveillance isolates from 48 of the 277 clinical patients, 240 randomly selected patient surveillance isolates from colonized patients, and 95 environmental isolates (Table 5). Of 277 first clinical isolates, one isolate was susceptible to all the antifungals tested, while the rest of the 276 isolates were resistant to fluconazole (≥32.0 μg/ml). Of the fluconazole resistant isolates, 224 (81%) had an elevated MIC to voriconazole (MIC ≥ 2.0 μg/ml), 170 (61%) were resistant to amphotericin B (MIC ≥ 2.0 μg/ml), and 2 (0.7%) were resistant to 5FC (≥32.0 μg/ml). None of the isolates were resistant to echinocandins.
TABLE 5.
Antifungal susceptibility data of C. auris isolates recovered from clinical cases, colonized cases, and the environmenta
| Antifungalb | Tentative resistance breakpoint (μg/ml) | First clinical isolate (no. 277) from 277 clinical patients |
Subsequent clinical isolate (no. 116) from 74 clinical patients |
Surveillance isolates (no. 215) from 48 clinical patients |
Surveillance isolates (no. 240) from colonized patients |
Surveillance isolates (no. 95) from environment |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) |
No (%) resistant | MIC (μg/ml) |
No. (%) resistant | MIC (μg/ml) |
No. (%) resistant | MIC (μg/ml) |
No. (%) resistant | MIC (μg/ml) |
No. (%) resistant | |||||||||||||||||
| 50% | 90% | Range | Mode | 50% | 90% | Range | Mode | 50% | 90% | Range | Mode | 50% | 90% | Range | Mode | 50% | 90% | Range | Mode | |||||||
| FLC | 32 | >256 | >256 | 8 to ≥ 256 | >256 | 276 (99) | >256 | >256 | 128 to ≥256 | >256 | 116 (100) | ≥256 | ≥256 | 8 to ≥256 | >256 | 214 (99) | >256 | >256 | 4 to ≥256 | >256 | 239 (99) | >256 | >256 | 8 to ≥256 | >256 | 94 (99) |
| ITC | NAc | 0.5 | 1 | 0.12 to 1 | 0.5 | NA | 0.5 | 1 | 0.12 to 1 | 0.5 | NA | 0.5 | 0.5 | 0.05 to 1 | 0.5 | NA | 0.5 | 1 | 0.015 to 1 | 0.5 | NA | 0.5 | 1 | 0.25 to 1 | 1 | NA |
| VRC | NA | 2 | 2 | 0.25 to 4 | 2 | 224 (81) | 2 | 2 | 0.5 to 4 | 2 | 96 (83) | 2 | 4 | 0.125 to 4 | 2 | 175 (81) | 2 | 4 | 0.015 to 8 | 2 | 199 (83) | 2 | 4 | 0.06 to 4 | 2 | NA |
| POS | NA | 0.25 | 0.5 | 0.03 to 1 | 0.25 | NA | 0.25 | 0.5 | 0.03 to 0.5 | 0.25 | NA | 0.25 | 0.25 | 0.015 to 1 | 0.25 | NA | 0.25 | 0.5 | 0.008 to 1 | 0.25 | NA | 0.5 | 0.5 | 0.03 to 1 | 0.5 | NA |
| ISA | NA | 0.5 | 1 | 0.05 to 2 | 1 | NA | 0.5 | 1 | 0.12 to 2 | 0.5 | NA | 1 | 2 | 0.12 to 2 | 1 | NA | 0.5 | 1 | 0.008 to 2 | 1 | NA | 1 | 2 | 0.03 to 4 | 1 | NA |
| CAS | 2.0 | 0.12 | 0.25 | 0.008 to >16.0 | 0.12 | 0 | 0.12 | 0.25 | 0.008 to >16.0 | 0.25 | 3 (2.6) | 0.06 | 0.25 | 0.008 to 2 | 0.25 | 6 (2.8) | 0.06 | 0.25 | 0.015 to 0.5 | 0.25 | 0 (0) | 0.25 | 0.5 | 0.015 to 1 | 0.25 | 0 (0) |
| MFG | 4 | 0.12 | 0.25 | 0.03 to 0.5 | 0.12 | 0 | 0.12 | 0.25 | 0.03 to 4 | 0.12 | 4 (3.4) | 0.12 | 1 | 0.03 to 4 | 0.12 | 6 (2.8) | 0.12 | 0.25 | 0.03 to 0.5 | 0.12 | 0 (0) | 0.25 | 0.25 | 0.06 to 0.5 | 0.25 | 0 (0) |
| AFG | 4 | 0.25 | 0.5 | 0.12 to 0.5 | 0.25 | 0 | 0.25 | 0.5 | 0.12 to 0.5 | 0.25 | 3 (2.6) | 0.25 | 1 | 0.03 to 4 | 0.25 | 6 (2.8) | 0.25 | 0.5 | 0.03 to 0.5 | 0.25 | 0 (0) | 0.25 | 1 | 0.08 to 1 | 0.25 | 0 (0) |
| AMB | 2.0 | 2 | 2 | 0.125 to 3 | 2 | 170 (61) | 2 | 2 | 0.125 to 3 | 2 | 67 (58) | 2 | 2 | 0.19 to 4 | 2 | 143 (67) | 2 | 2 | 0.5 to 3.0 | 2 | 160 (67) | 2 | 2 | 0.25 to 3.0 | 2 | 59 (62) |
| 5FC | NA | 0.094 | 0.125 | 0.023 to >32 | 0.094 | 2 (0.7) | 0.094 | 0.125 | 0.023 to >32 | 0.094 | 6 (5.1) | 0.064 | 0.094 | 0.016 to >32 | 0.064 | 1 (0.5) | 0.064 | 0.19 | 0.016 to >32 | 0.047 | 5 (2.1) | 0.064 | 0.125 | 0.032 to 0.19 | 0.047 | 0 (0) |
MICs for azoles and echinocandins are defined as the lowest drug concentration that caused 50% growth inhibition compared to growth of the drug-free controls; MICs for amphotericin B and flucytosine are defined as the lowest concentration at which there was 100% and 90% growth inhibition, respectively.
FLC, fluconazole; ITC, itraconazole; VRC, voriconazole; POS, posaconazole; ISA, isavuconazole; CAS, caspofungin; MFG, micafungin; AFG, anidulafungin; AMB, amphotericin B; 5FC, flucytosine.
NA, not available.
Of the 116 subsequent clinical isolates analyzed from 74 clinical patients, four (3.4%) were resistant to micafungin (≥4.0 μg/ml); two were recovered from urine, and one each was recovered from blood and ascites from four individual patients. Of two urine isolates with micafungin resistance, one also showed resistance against caspofungin (≥2.0 μg/ml) and anidulafungin (≥4.0 μg/ml). Of the echinocandin-resistant isolates, two isolates from two patients exhibited borderline resistance to amphotericin B (MIC = 1.5 μg/ml), but CDC’s Mycotic Diseases Branch found these isolates to be susceptible to amphotericin B (≤1.0 μg/ml). Of the follow-up surveillance isolates analyzed from some of the clinical cases, one patient’s six isolates were resistant to echinocandins, and of these, one isolate recovered from a rectal swab collected to assess ongoing colonization after resolution of infection was pan-resistant, as it also showed resistance to amphotericin B (MIC = 3.0 μg/ml), which was retrospectively confirmed in 2019 by CDC’s Mycotic Diseases Branch (MIC =1.5 μg/ml). Interestingly, all urine isolates from this patient collected on different days were resistant to echinocandins, but isolates recovered from other body sites were variably resistant or susceptible (Table 6), raising the possibility of the presence of heterogenous populations. None of the isolates recovered either from colonized patients or from the environment showed any resistance to echinocandins (i.e., echinocandin resistance was only identified in patients meeting the clinical case definition).
TABLE 6.
List of serial C. auris patient isolates with resistance to various antifungals and/or pan-resistance
| Patient no. | Collection date (mo/day/yr) | Isolate type | Specimen | Antifungal susceptibility test results (μg/ml)a
|
||||
|---|---|---|---|---|---|---|---|---|
| CAS | MFG | AFG | 5FC | AMB | ||||
| Clin-1 | 12/29/2016 | Clinical | Blood | 0.25 | 0.12 | 0.25 | 0.094 | 1 |
| Clin-1 | 3/1/2017 | Clinical | Urine | >16 (R) | 4 (R) | 4 (R) | 0.064 | 1.5 (2.0, R)b |
| Clin-2 | 2/22/2017 | Clinical | Bile | 0.25 | 0.12 | 0.25 | 0.094 | 0.75 |
| Clin-2 | 3/10/2017 | Clinical | Blood | 4 (R) | 4 (R) | 4 (R) | 0.094 | 1 |
| Clin-2 | 3/17/2017 | Clinical | Abdominal fluid | 0.03 | 0.06 | 0.12 | 0.094 | 1.5 (2.0, R) |
| Clin-3 | 2/19/2017 | Clinical | Blood | 0.12 | 0.12 | 0.25 | 0.094 | 1.5 (2.0, R) |
| Clin-3 | 3/9/2017 | Surveillance | Axilla/groin | 0.06 | 0.06 | 0.12 | 0.023 | 0.75 |
| Clin-3 | 3/9/2017 | Surveillance | Nasal | 0.03 | 0.06 | 0.12 | 0.023 | 1.5 (2.0, R) |
| Clin-3 | 3/9/2017 | Surveillance | Skin rash | 0.03 | 0.06 | 0.25 | 0.023 | 0.75 |
| Clin-3 | 3/20/2017 | Surveillance | Urine | 2 (R) | 4 (R) | 4 (R) | 0.023 | 1 |
| Clin-3 | 3/20/2017 | Surveillance | Respiratory | 0.06 | 0.06 | 0.25 | 0.023 | 1.5 (2.0, R) |
| Clin-3 | 3/20/2017 | Surveillance | Axilla | 0.12 | 0.12 | 0.25 | 0.023 | 2 (R) |
| Clin-3 | 3/20/2017 | Surveillance | Groin | 2 (R) | 4 (R) | 4 (R) | 0.032 | 0.75 |
| Clin-3 | 3/20/2017 | Surveillance | Skin rash | 2 (R) | 4 (R) | 4 (R) | 0.032 | 0.75 |
| Clin-3 | 3/20/2019 | Surveillance | Rectum | 0.12 | 0.12 | 0.25 | 0.023 | 1 |
| Clin-3 | 4/5/2017 | Surveillance | Skin rash | 0.015 | 0.06 | 0.25 | 0.023 | 2 (R) |
| Clin-3 | 4/5/2017 | Surveillance | Groin | 0.06 | 0.06 | 0.25 | 0.032 | 1.5 (2.0, R) |
| Clin-3 | 4/5/2017 | Surveillance | Urine | 2 (R) | 4 (R) | 4 (R) | 0.023 | 1 |
| Clin-3 | 4/5/2017 | Surveillance | Rectum | 0.06 | 0.12 | 0.25 | 0.023 | 1.5 (2.0, R) |
| Clin-3 | 4/20/2017 | Surveillance | Urine | 2 (R) | 4 (R) | 4 (R) | 0.023 | 1 |
| Clin-3 | 4/20/2017 | Surveillance | Groin | 0.03 | 0.06 | 0.25 | 0.023 | 0.75 |
| Clin-3 | 4/20/2017 | Surveillance | Respiratory | 0.12 | 0.12 | 0.25 | 0.023 | 1.5 (2.0, R) |
| Clin-3 | 4/20/2017 | Surveillance | Rectum | 2 (R) | 4 (R) | 4 (R) | 0.023 | 3.0 (R)c |
| Clin-4 | 4/19/2017 | Clinical | Urine | 0.06 | 0.12 | 0.25 | 0.094 | 2 (R) |
| Clin-4 | 5/11/2017 | Surveillance | Urine | 0.12 | 0.25 | 0.5 | 0.094 | 2 (R) |
| Clin-4 | 5/11/2017 | Surveillance | Groin | 0.25 | 0.5 | 0.5 | >32 (R) | 2 (R) |
| Clin-5 | 4/11/2018 | Clinical | Abdomen | 0.06 | 0.12 | 0.25 | 0.094 | 2 (R) |
| Clin-5 | 4/12/2018 | Clinical | Ascites fluid | 0.5 | 0.25 | 1 | 0.064 | 2 (R) |
| Clin-5 | 5/9/2018 | Clinical | Blood | 0.06 | 0.06 | 0.25 | 0.064 | 2 (R) |
| Clin-5 | 5/10/2018 | Clinical | Ascites fluid | 2 (R) | 4 (R) | 8 (R) | 0.064 | 1.5 (2.0, R)b |
| Clin-6 | 9/1/2018 | Clinical | Urine | 0.25 | 0.06 | 0.5 | 0.094 | 1.5 (2.0, R) |
| Clin-6 | 9/9/2018 | Clinical | Urine | 0.25 | 0.12 | 0.25 | 0.064 | 1 |
| Clin-6 | 9/19/2018 | Clinical | Urine | 2 (R) | 4 (R) | 1 | ND | 1 |
| Clin-7 | 3/11/2018 | Clinical | Blood | 0.03 | 0.12 | 0.06 | >32 (R) | 2 (R) |
| Clin-7 | 3/9/2018 | Clinical | Right arm wounds | 0.03 | 0.12 | 0.12 | >32 (R) | 1 |
| Clin-7 | 3/9/2018 | Clinical | Left arm wound | 0.03 | 0.06 | 0.12 | >32 (R) | 1 |
| Clin-7 | 3/9/2018 | Clinical | Leg wound | 0.03 | 0.12 | 0.12 | >32 (R) | 2 (R) |
| Clin-7 | 3/9/2018 | Clinical | Buttock wound | 0.03 | 0.12 | 0.12 | >32 (R) | 1 |
| Clin-8 | 5/7/2018 | Clinical | Urine | 0.12 | 0.12 | 0.25 | >32 (R) | 2 (R) |
| Clin-8 | 6/19/2018 | Clinical | Blood | 0.06 | 0.06 | 0.25 | >32 (R) | 1 |
| Clin-9 | 3/19/2017 | Clinical | Urine | 0.03 | 0.06 | 0.25 | 0.094 | 2 (R) |
| Clin-9 | 3/23/2017 | Clinical | Urine | 0.12 | 0.12 | 0.25 | 0.125 | 2 (R) |
| Clin-9 | 4/18/2018 | Clinical | Urine | 0.12 | 0.12 | 0.25 | >32 (R) | 3.0 (R) |
| Clin-9 | 4/13/2017 | Surveillance | Nares | 0.5 | 0.25 | 1 | 0.125 | 3.0 (R) |
| Clin-9 | 6/20/2017 | Surveillance | Groin | 0.06 | 0.12 | 0.12 | 0.047 | 2 (R) |
| Clin-9 | 6/20/2017 | Surveillance | Nares | 0.06 | 0.12 | 0.12 | 0.047 | 2 (R) |
| Clin-9 | 9/12/2017 | Surveillance | Nares | 0.25 | 0.12 | 0.5 | 0.125 | 2 (R) |
| Clin-9 | 9/12/2017 | Surveillance | Rectal | 0.015 | 0.03 | 0.06 | 0.064 | 2 (R) |
| Clin-9 | 1/17/2018 | Surveillance | Nares | 0.06 | 0.25 | 0.25 | 0.094 | 1 |
| Sur-1 | 10/10/2017 | Surveillance | Axilla/groin | 0.03 | 0.06 | 0.06 | >32 (R) | 2 (R) |
| Sur-1 | 2/22/2018 | Surveillance | Right arm wound | 0.03 | 0.06 | 0.06 | >32 (R) | 1 |
| Sur-1 | 2/22/2018 | Surveillance | Left axilla | 0.015 | 0.06 | 0.06 | >32 (R) | 1.5 (2.0, R) |
| Sur-1 | 2/22/2018 | Surveillance | Left groin | 0.015 | 0.03 | 0.06 | >32 (R) | 1 |
| Sur-1 | 2/22/2018 | Surveillance | Sacral pressure injury | 0.03 | 0.06 | 0.06 | >32 (R) | 1 |
| Sur-1 | 2/22/2018 | Surveillance | Rectal | 0.015 | 0.03 | 0.06 | >32 (R) | 1.5 (2.0, R) |
| Sur-2 | 1/11/2018 | Surveillance | Axilla/groin | 0.06 | 0.12 | 0.12 | >32 (R) | 2 (R) |
| Sur-3 | 1/17/2018 | Surveillance | Nares | 0.06 | 0.25 | 0.12 | >32 (R) | 1 |
| Sur-3 | 1/17/2018 | Surveillance | Axilla/groin | 0.03 | 0.12 | 0.12 | >32 (R) | 2 (R) |
| Sur-3 | 2/21/2018 | Surveillance | Axilla/groin | 0.03 | 0.03 | 0.03 | >32 (R) | 0.38 |
R, resistant; CAS, caspofungin; MFG, micafungin; AFG, anidulafungin; 5FC, flucytosine; AMB, amphotericin B.
Isolate suspected of pan-resistant.
Isolate confirmed as pan-resistant.
Isolation of other fungal species from patient and environmental surveillance samples.
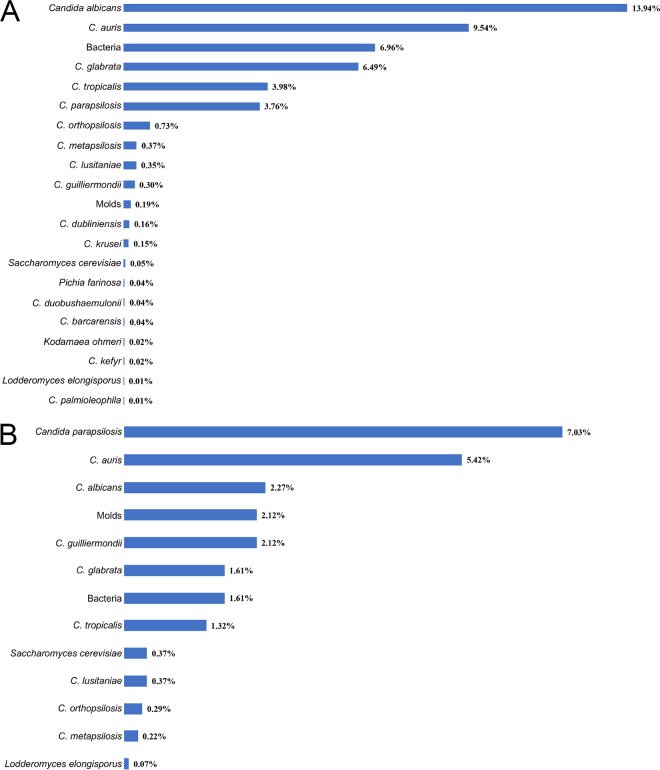
Other organisms present in the patient and environmental surveillance samples were determined. Candida albicans (13.94%) was by far the dominant pathogen in patient surveillance samples followed by C. auris (9.54%), C. glabrata (6.49%), C. tropicalis (3.98%), and C. parapsilosis (3.76%). Several other rare Candida spp., other yeasts, molds, and bacterial species were also isolated (Fig. 4 A). The environmental samples were predominantly positive for C. parapsilosis (7.03%) followed by C. auris (5.42%), C. albicans (2.27%), C. guilliermondii (2.1%), and C. glabrata (1.61%). Other Candida species, yeast, molds, and bacteria were also isolated (Fig. 4 B).
FIG 4.
Prevalence of Candida species in surveillance samples. (A) The bar diagram represents different Candida species and their frequency of isolation (%) from patient surveillance samples. Candida albicans was the dominant pathogen followed by C. auris, C. glabrata, C. tropicalis, and C. parapsilosis. Additional Candida species, and other yeasts, were minor components. (B) Bar diagram represents different Candida species and their frequency of isolation (%) from environmental surveillance samples. Candida parapsilosis was the dominant pathogen followed by C. auris, C. albicans, C. guilliermondii, C. glabrata, and C. tropicalis. Additional Candida species, molds, and other yeasts were minor components.
DISCUSSION
Laboratory investigations of an outbreak of C. auris in New York with 277 clinical and 350 colonized cases from August 2016 through 2018 are summarized. The New York State and New York City metropolitan areas have borne the brunt of this unprecedented epidemic. As of 22 November 2019, the outbreak has impacted 192 health care facilities in which the patients received care in the 90 days prior to their C. auris diagnosis. Facilities included 70 hospitals, 118 nursing homes, two hospices, and one each of long-term acute care hospital (LTACH) and Veterans Administration (VA) hospital. Despite coordinated efforts of public health authorities, C. auris colonization and infections continue to increase across New York State. The C. auris epidemic is especially alarming considering Candida species are major causes of health care-associated infections in the United States (27). Infections by these pathogens result in mortality rates of >30% to 40% and are responsible for the highest total annual hospitalization costs of any invasive fungal disease (N = 26,735; total cost $1.4 billion) (27–29). The recent emergence of multidrug-resistant (MDR) C. auris has further exacerbated this crisis (9, 19). A recent announcement from the Centers for Disease Control and Prevention (CDC) recognizes C. auris as among the five bacterial and fungal pathogens that pose urgent threats to U.S. public health.
Detecting an epidemic of infectious disease at an early stage is crucial for the timely implementation of infection control measures and for minimizing morbidity and mortality. During this epidemic, our laboratory developed and deployed a novel real-time PCR assay for the rapid detection of C. auris (24). This assay, with high sensitivity and specificity, proved invaluable for confirmation of C. auris from patient surveillance samples. Compared to those for patient surveillance samples, the calculated diagnostic accuracy and specificity of the real-time PCR assay for environmental samples were lower. However, these calculations rely on culture as the gold standard, but this is not an appropriate comparator for environmental samples. For example, we did not know how long C. auris was present in the health care environment prior to sample collection. Also, several environmental samples received for testing were often collected after the site had been cleaned and disinfected. The ability of the real-time PCR assay to pick up DNA from live, dead, and growth-defective C. auris, as well as its ability to pick up leftover DNA stuck to the objects, likely impacted the calculated diagnostic accuracy. Nevertheless, environmental testing contributed substantially to the institution of infection control practices in the affected health care facilities. We would recommend that while PCR is an excellent screening tool for the testing of environmental samples, culture should be used as the basis for follow-up remediation efforts.
In the present laboratory-based aggregate analysis, we quantified C. auris in colonized patients and their surrounding environment to determine the extent of colonization. Our results revealed that the colonized patients harbored large numbers of live C. auris cells on skin (axilla/groin) and mucosal (nares) surfaces. Likewise, the patient’s contact points, including several health care objects, were also contaminated with the large numbers of live C. auris cells. These results indicate that patients who are heavily colonized with C. auris on their skin or mucosal surfaces can contaminate their surroundings, which might be a key to the successful transmission of C. auris in the health care facilities. Identifying reservoirs, prompt notification of C. auris identification, and implementation of effective infection control practices are key to controlling the spread of C. auris.
Among colonized patients, the nares appeared to serve as an excellent site for C. auris growth, as they harbored approximately 2-log higher CFU than the axilla/groin. The precise mechanism(s) leading to the extensive colonization of nares is currently unclear, but results of this investigation led us to use one combination swab of nares/axilla/groin rather than a separate swab for colonization screening, an approach necessary to handle an outbreak of this magnitude. Further research is needed to understand the mechanism(s) of colonization. Although C. auris recovery from the patient’s skin and health care environment was established previously (8, 9, 19), in this report, we have used a quantitative approach to determine the extent of C. auris colonization of skin and mucosal surfaces of the colonized patients and their surrounding environment.
Identification of C. auris has been challenging for clinical and public health microbiology laboratories, as common biochemical-based platforms could not accurately identify this newly emerged fungal pathogen. Diagnostic devices based on matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) can differentiate C. auris from other Candida species, but not all the reference databases included in MALDI-TOF devices allowed for such detection earlier in the epidemic. In the beginning of the outbreak (August to December 2016), our MALDI score for C. auris was in the lower range of 1.7 to 1.9, which could be attributed to fewer C. auris isolates in the Bruker library database. Enrichment of the database led to the higher scores ranging from 2.2 to 2.6. These results agreed with other investigators who used either a Bruker or bioMérieux MALDI platform (23, 30). Recently, the FDA approved Bruker and bioMérieux MALDI-TOF databases for C. auris identification, an important step to facilitate testing by the hospital laboratories.
Candida auris isolates in the New York epidemic were intrinsically resistant to fluconazole, and approximately 81% were resistant to voriconazole and 61% to amphotericin B. The resistance observed in the current outbreak was higher than in the two large studies previously published (16, 23). This discrepancy could be due to the fact that the C. auris outbreak in New York was dominated by South Asia clade I. The echinocandin resistance was noted in fewer isolates, which was consistent with other studies (16, 23). We also found three isolates suspected to have pan-resistance (concurrent resistance to azoles, echinocandins, and amphotericin B); one of these isolates was confirmed as pan-resistant at the CDC Mycotic Diseases Branch Laboratory. Our MIC values for the two suspected pan-resistant isolates were 1.5 μg/ml, while the CDC’s Mycotic Diseases Branch reported them to be lower than 1.5 μg/ml. A recent publication suggests C. auris amphotericin B resistance is inducible and transient, and MIC values of some isolates decrease following subcultures in the laboratory (31). Amphotericin B resistance is rare in most of the Candida species and it is thought to be associated with a fitness cost (32). Interestingly, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii, closely related to C. auris, have high-level intrinsic resistance to amphotericin B (23). It is not clear if the associated fitness cost leads to rare incidences of their infections worldwide. In contrast, the borderline amphotericin B resistance observed in C. auris might suggest no loss of fitness, which could allow for higher prevalence. More research is needed to understand the mechanisms leading to amphotericin B resistance in C. auris. A small percentage of NY C. auris isolates showed resistance to flucytosine. It is not clear if affected patients received amphotericin B and flucytosine combination therapy. To overcome the therapeutic challenges posed by multidrug resistance in C. auris, both a new drug(s) and old drugs with combination therapy are needed. A few new drugs in the pipeline have shown excellent in vitro activity against C. auris (33, 34). Similarly, in vitro antifungal combination testing revealed synergy between echinocandins and azoles (34–36). However, in vivo studies with suitable animal models of C. auris infection, and pharmacodynamic/pharmacokinetic studies, are needed to confirm in vitro findings.
The South Asia clade I was the major genotype, and the East Asia clade II was a minor genotype of the C. auris outbreak in New York. These results are consistent with the WGS results reported for NY strains (37). Our ability to genotype South and East Asia clades by Sanger sequencing of the ribosomal genes (ITS and D1/D2) is encouraging, as fungal WGS is not readily available in public health and diagnostic laboratories. Interestingly, we did not find any isolate of the South Africa clade III or the South America clade IV in New York despite it being a large international port of entry. The ribosomal gene sequencing allows reliable separation of South Asia clade I, East Asia clade II, and South America clade III with the exception of South Africa clade IV. Using ITS sequences, South Africa clade IV clustered with South Asia clade I, consistent with an earlier study (5), but it clustered with East Asia clade I using D1/D2 sequences. Recently, a fifth clade of C. auris was identified from a patient in Iran who never travelled outside that country (17). The WGS revealed that the Iran C. auris isolate was >200,000 SNPs apart from the other four clades (18). It is yet to be determined if ITS and D1/D2 genes can differentiate the fifth clade from other clades of C. auris. Thus, a combination of ITS and D1/D2 genes served as an excellent marker for the separation of predominant clades of C. auris in the NY epidemic. Although ITS and D1/D2 phylogenetic analyses did not reveal any SNPs in isolates within the clades, limited WGS of NY isolates in a published study revealed low genetic variations (37). There is an urgent need to perform more extensive whole-genome analysis of these isolates to investigate the extent of genetic variations, if any, and the presence of clonal strains.
Despite extensive efforts, the NY C. auris epidemic is still ongoing. This could be due to the complexity of the health care systems in the metropolitan area, including the density and proximity of health care facilities in the region, and/or the lack of rapid testing within these facilities. The Wadsworth Center Mycology Laboratory has carried out nearly all testing since the beginning of the epidemic, and the test volume by itself has become a limiting factor in the comprehensive assessment of this epidemic. It is imperative that more clinical, public health, and commercial laboratories introduce C. auris testing. Laboratories must determine the species of all Candida cultures recovered from blood, urine, and other body parts using appropriate methods for early recognition of any incidences of C. auris. In the absence of such laboratory capacities, the suspect C. auris isolate ought to be promptly submitted to the reference or public health laboratories. At a minimum, diagnostic laboratories in the affected areas ought to provide rapid molecular testing. Overall, the need for the development of high-throughput assays, as well as point-of-care tests, cannot be overemphasized in view of threats posed by C. auris epidemic (38, 39).
In summary, use of a real-time PCR assay as a rapid detection tool, quantification of the extent of colonization of C. auris on biotic (patient) and abiotic (environment) surfaces, successful use of one combination swab of nares/axilla/groin for surveillance studies, and the use of ribosomal genes (ITS and D1/D2) to determine C. auris clades were among some of the highlights of this investigation and provided further insight into C. auris outbreak in New York.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Healthcare Epidemiology and Infection Control Program and staff members from various hospitals and long-term-care facilities for assistance with surveillance samples. We thank Wadsworth Center for its Applied Genomic Technologies and Media & Tissue Cultures Cores for DNA sequencing and culture media, respectively. We are also thankful to Lisa Biega for editorial comments.
This publication was supported by funding from the Wadsworth Center and cooperative agreement number NU50CK000516, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services.
YanChun Zhu was responsible for the processing of the surveillance sample, culture identification of all yeasts and data analyses. Lynn Leach was responsible for the real-time PCR assay of patient surveillance samples and data analysis. Brittany O’Brien performed antifungal susceptibility testing of all C. auris isolates. Alexandra Clarke and Marian Bates were responsible for the analysis of environmental samples using real-time PCR assay. Debra Blog, Eleanor Adams, Belinda Ostrowsky, Karen Southwick, Richard Erazo, Elizabeth Dufort, Valerie B. Haley, Coralie Bucher, and Monica Quinn assisted in epidemiology investigation of positive results, including advising and arranging for onsite point prevalence surveys, performing onsite infection control assessments, and contact tracing. Eleanor Adams, Belinda Ostrowsky, and Monica Quinn also critically read the manuscript. Ronald J. Limberger coordinated the lab-epi meeting and critically read the manuscript. Emily Lutterloh coordinated the epidemiologic C. auris response and critically reviewed the manuscript. Vishnu Chaturvedi provided expertise in antifungal susceptibility testing and critically reviewed the manuscript. Sudha Chaturvedi conceived and designed the study, interpreted data, and wrote the manuscript.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.02083-19.
Supplemental material is available online only.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Bing J, Zheng Q, Zhang F, Liu J, Yue H, Tao L, Du H, Wang Y, Wang H, Huang G. 2018. The first isolate of Candida auris in China: clinical and biological aspects. Emerg Microbes Infect 7:93. doi: 10.1038/s41426-018-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R. 2015. Candida auris candidemia in Kuwait. Emerg Infect Dis 21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, MSD, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus - United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-López SE, Parra-Giraldo CM, Ceballos-Garzón A, Martínez HP, Rodríguez GJ, Álvarez-Moreno CA, Rodríguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz Gaitán AC, Moret A, López Hontangas JL, Molina JM, Aleixandre López AI, Cabezas AH, Mollar Maseres J, Arcas RC, Gómez Ruiz MD, Chiveli MÁ, Cantón E, Pemán J. 2017. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34:23–27. doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Saris K, Meis JF, Voss A. 2018. Candida auris. Curr Opin Infect Dis 31:334–340. doi: 10.1097/QCO.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 14.Spivak ES, Hanson KE. 2018. Candida auris: an emerging fungal pathogen. J Clin Microbiol 56:e01588-17. doi: 10.1128/JCM.01588-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abastabar M, Haghani I, Ahangarkani F, Rezai MS, Taghizadeh Armaki M, Roodgari S, Kiakojuri K, Al-Hatmi AMS, Meis JF, Badali H. 2019. Candida auris otomycosis in Iran and review of recent literature. Mycoses 62:101–105. doi: 10.1111/myc.12886. [DOI] [PubMed] [Google Scholar]
- 18.Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. 2019. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis 25:1780–1781. doi: 10.3201/eid2509.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams E, Candida auris Investigation Workgroup, Quinn M, Tsay S, Poirot E, Chaturvedi S, Southwick K, Greenko J, Fernandez R, Kallen A, Vallabhaneni S, Haley V, Hutton B, Blog D, Lutterloh E, Zucker H. 2018. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis 24:1816–1824. doi: 10.3201/eid2410.180649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Manuel R, Brown CS. 2018. Candida auris: a review of the literature. Clin Microbiol Rev 31:e00029-17. doi: 10.1128/CMR.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizusawa M, Miller H, Green R, Lee R, Durante M, Perkins R, Hewitt C, Simner PJ, Carroll KC, Hayden RT, Zhang SX. 2017. Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J Clin Microbiol 55:638–640. doi: 10.1128/JCM.02202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leach L, Zhu Y, Chaturvedi S. 2017. Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J Clin Microbiol 56:e01223-17. doi: 10.1128/JCM.01223-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts—4th ed CLSI document M27 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 26.Zhang T, Chaturvedi V, Chaturvedi S. 2015. Novel Trichoderma polysporum strain for the biocontrol of Pseudogymnoascus destructans, the fungal etiologic agent of bat white nose syndrome. PLoS One 10:e0141316. doi: 10.1371/journal.pone.0141316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strollo S, Lionakis MS, Adjemian J, Steiner CA, Prevots DR. 2016. Epidemiology of hospitalizations associated with invasive candidiasis, United States, 2002–2012. Emerg Infect Dis 23:7–13. doi: 10.3201/eid2301.161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magill SS, O'Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle J, Wilson LE, Kainer MA, Lynfield R, Greissman S, Ray SM, Beldavs Z, Gross C, Bamberg W, Sievers M, Concannon C, Buhr N, Warnke L, Maloney M, Ocampo V, Brooks J, Oyewumi T, Sharmin S, Richards K, Rainbow J, Samper M, Hancock EB, Leaptrot D, Scalise E, Badrun F, Phelps R, Edwards JR, Emerging Infections Program Hospital Prevalence Survey Team . 2018. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med 379:1732–1744. doi: 10.1056/NEJMoa1801550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benedict K, Jackson BR, Chiller T, Beer KD. 2018. Estimation of direct healthcare costs of fungal diseases in the United States. Clin Infect Dis 68:1791–1797. doi: 10.1093/cid/ciy776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girard V, Mailler S, Chetry M, Vidal C, Durand G, van Belkum A, Colombo AL, Hagen F, Meis JF, Chowdhary A. 2016. Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption ionisation time of flight mass spectrometry. Mycoses 59:535–538. doi: 10.1111/myc.12519. [DOI] [PubMed] [Google Scholar]
- 31.Lockhar SR. 2019. Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol 131:103243. doi: 10.1016/j.fgb.2019.103243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. 2013. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol 11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berkow EL, Lockhart SR. 2018. Activity of novel antifungal compound APX001A against a large collection of Candida auris. J Antimicrob Chemother 73:3060–3062. doi: 10.1093/jac/dky302. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien B, Chaturvedi S, Chaturvedi V. 13 January 2020. In vitro evaluation of antifungal drug combinations against multidrug-resistant Candida auris isolates from New York outbreak. Antimicrob Agents Chemother doi: 10.1128/AAC.02195-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fakhim H, Chowdhary A, Prakash A, Vaezi A, Dannaoui E, Meis JF, Badali H. 2017. In vitro interactions of echinocandins with triazoles against multidrug-resistant Candida auris. Antimicrob Agents Chemother 61:e01056-17. doi: 10.1128/AAC.01056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Barat SA, Borroto-Esoda K, Angulo D, Chaturvedi S, Chaturvedi V. 2019. In vitro efficacy of novel glucan synthase inhibitor, ibrexafungerp (SCY-078), against multidrug- and pan-resistant Candida auris isolates from the outbreak in New York. bioRxiv doi: 10.1101/811182. [DOI] [PubMed]
- 37.Chow NA, US Candida auris Investigation Team, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, Barrett PM, Kerins JL, Lockhart SR, Chiller TM, Litvintseva AP, US Candida auris Investigation Team . 2018. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis 18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leach L, Russell A, Zhu Y, Chaturvedi S, Chaturvedi V. 2019. A rapid and automated sample-to-result Candida auris real-time PCR assay for high-throughput testing of surveillance samples with the BD Max open system. J Clin Microbiol 57:e00630-19. doi: 10.1128/JCM.00630-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulberry G, Chaturvedi S, Chaturvedi V, Kim B. 2020. Toward point-of-care diagnostics of Candida auris. medRxiv doi: 10.1101/2020.02.10.20021683. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All nucleotide sequences of C. auris were deposited in GenBank under the following accession numbers: ITS sequences, MN338097 to MN338196; D1/D2 sequences, MN337432 to MN337531.