Abstract
Idiopathic inflammatory myopathies (IIM) are characterized by muscle inflammation and weakness, myositis-specific autoantibodies (MSAs), and extramuscular organ damage. The role of neutrophil dysregulation and neutrophil extracellular traps (NETs) in IIM is unclear. We assessed whether pathogenic neutrophil subsets (low-density granulocytes [LDGs]) and NETs were elevated in IIM, associated with clinical presentation and MSAs, and their effect on skeletal myoblasts and myotubes. Circulating NETs and LDGs were quantified and correlated with clinical measures. Specific MSAs were tested for their ability to induce NETs. NETs and neutrophil gene expression were measured in IIM biopsies. Whether NETs damage skeletal myoblasts and myotubes was tested. Circulating LDGs and NETs were increased in IIM. IIM LDGs had an enhanced ability to form NETs. LDGs and NETs correlated with IIM disease activity and muscle damage. The serum MSA anti-MDA5 correlated with circulating and tissue NETs and directly enhanced NET formation. An enhanced neutrophil gene signature was present in IIM muscle and associated with muscle injury and tissue IFN gene signatures. IIM NETs decreased the viability of myotubes in a citrullinated histone-dependent manner. Dysregulated neutrophil pathways may play pathogenic roles in IIM through their ability to directly injure muscle cells and other affected tissues.
Keywords: Autoimmunity, Muscle Biology
Keywords: Innate immunity, Neutrophils
Neutrophil dysregulation and enhanced NET formation promote skeletal muscle damage and immune dysregulation in idiopathic inflammatory myopathies.
Introduction
Idiopathic inflammatory myopathies (IIM) are heterogeneous autoimmune conditions that affect adults and children and are characterized by muscle inflammation and frequent development of extramuscular manifestations in lung, skin, and joints (1). IIM include several clinicopathologic subtypes: dermatomyositis (DM), polymyositis (PM), immune-mediated necrotizing myopathy, sporadic inclusion body myositis, and myositis in association with other autoimmune conditions. The discovery of autoantibodies associated with specific clinical phenotypes has been instrumental in the understanding of IIM. Myositis-specific autoantibodies (MSAs) target cytoplasmic or nuclear antigens. Abs against histidyl tRNA synthetase (Jo-1) are the most common in adult IIM and are associated with the anti-synthetase syndrome, a distinct clinical entity (2, 3). Autoantibodies against the IFN-inducible antigen melanoma differentiation-associated protein 5 (MDA5) have been described in DM, in association with skin ulceration, arthritis, and rapidly progressive interstitial lung disease (ILD) (4).
Although significant work has focused on the role of the adaptive immune system and lymphocyte subsets in IIM, the role of cellular components of the innate immune system is less well understood. In particular, how neutrophils may contribute to this group of diseases has only started to be investigated. In other systemic autoimmune diseases, including systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and antineutrophil cytoplasmic antibody-associated vasculitis (AAV), neutrophils have recently been proposed to play important pathogenic roles, at least in part, through their enhanced ability to form neutrophil extracellular traps (NETs) (5–7). NETs are extracellular lattices that are extruded by neutrophils in response to microbial and sterile danger signals, and they contain nucleic acids, histones, and neutrophil granule proteins. Studies from our group and others have identified NETs as important sources of modified autoantigens and immunostimulatory molecules as well as drivers of enhanced type I IFN response (5–8). NETs also induce tissue damage through deleterious effects on endothelial cells, fibroblasts, and macrophages, among various cell types (8–10). In particular, a subset of neutrophils called low-density granulocytes (LDGs), previously identified in increased levels in SLE, AAV, psoriasis, and others inflammatory diseases, has an enhanced capacity to form immunogenic NETs that can cause more tissue damage and are more immunostimulatory and interferogenic than NETs synthesized by other neutrophil subsets (5, 7, 11–13).
A previous study in the Chinese population reported that NETs are associated with ILD in IIM, particularly in subjects who are positive for anti-MDA5 autoantibodies (14), indicating that neutrophils potentially may be involved in the pathogenesis of this group of conditions. Furthermore, a recent association among circulating NETs, neutrophil activation, and calcinosis in juvenile DM (JDM) has been described, implicating calcium deposits characteristic of this disease with the induction of tissue and peripheral NET formation (15).However, whether aberrant neutrophil responses and NET formation play an important role in IIM and associated muscle damage remains to be systematically studied. Here, we investigated whether neutrophil dysregulation is prevalent in IIM and examined its associations with clinical manifestations and immune perturbances as well as its potential role in muscle damage.
Results
LDGs and NET complexes are present in IIM and associate with clinical manifestations.
The demographic and clinical characteristics of the different IIM cohorts studied are included in the Methods and in Supplemental Tables 1–4 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.134189DS1). LDGs were detected in various forms of IIM at levels comparable to those observed in SLE, except for in adult DM, in which levels were found to be significantly higher than in SLE (Supplemental Figure 1). Similar to what has been described in SLE, IIM LDGs displayed enhanced spontaneous ability to form NETs in vitro when compared with healthy control (HC) neutrophils or normal-dense IIM neutrophils (Figure 1, A and B). Circulating NET remnants were measured with 2 assays: human neutrophil elastase–DNA (HNE-DNA) complexes and myeloperoxidase-DNA (MPO-DNA) complexes. HNE-DNA NET complexes were significantly elevated in IIM as a whole, as well as in adult DM and JDM subgroups when compared with age-matched controls, but not in the PM group (Figure 1C). MPO-DNA complexes were significantly elevated in IIM group as a whole when compared with HCs (Figure 1D).
Figure 1. IIM LDGs display enhanced spontaneous NET formation, and circulating NET complexes are increased in adult and juvenile DM.
(A) Representative microphotographs display spontaneous NET formation at 2 hours in healthy control neutrophils, IIM normal dense neutrophils, and autologous IIM LDGs. Blue represents DNA (DAPI) and red represents MPO. Original magnification, ×10. (B) The percentage of neutrophils (mean ± SEM) undergoing NET formation in healthy control normal-density granulocytes (Ctrl-NDG), IIM NDGs, and IIM LDGs. Results are for 5 healthy control, 16 IIM-NDG, and 5 LDG samples. (C and D) HNE-DNA (C) and MPO-DNA (C) plasma NET complexes quantified by ELISA in adult or juvenile (Juv) healthy controls (HC), IIM (all IIM), adult DM (ADM), juvenile DM (JDM), or polymyositis (PM). Dots represent individual subjects. HC = 30; ADM = 46; PM = 20; JDM = 86. Bars represent median values ± IQR. Statistical analysis was performed using Mann-Whitney for B and Kruskal-Wallis for C and D. *P < 0.05; **P < 0.01; ****P < 0.0001.
When assessing associations of circulating LDGs and/or NETs with various markers of disease activity and damage, there were specific associations depending on myositis subtype, which are reported in Supplemental Table 5. In the adult DM and PM groups, NET levels correlated with serum muscle enzymes, which is indicative of skeletal muscle injury. In the JDM group, NET levels correlated with lung, vascular, and muscular components of the Myositis Disease Activity Assessment Tool (MDAAT), a validated assessment of disease activity of extramuscular organ systems and muscle, while LDG levels correlated with severity of skin disease and negatively correlated with muscle strength. In the PM group, levels of circulating NETs also significantly correlated with the cardiovascular and muscle components of the MDAAT. No correlation analysis was performed for LDGs in the PM group, given the small sample size. These results indicate that the presence and levels of LDGs and NETs significantly correlate with IIM disease activity, including muscle and skin activity and extramuscular manifestations of IIM. Abnormalities in small blood vessels are a hallmark of JDM/DM and are likely associated with tissue damage (16). LDGs negatively correlated with periungual capillary density in JDM (r = –0.58, P < 0.05), supporting previous observations that lupus LDGs damage endothelial cells (9). No associations between LDG or NETs were observed with calcinosis in the DM or JDM group. Overall, neutrophil subsets and NETs correlated with disease activity in IIM and with the microvascular abnormalities characteristic of these conditions. In general, LDGs showed correlations with clinical disease parameters in JDM but not as strongly in adult DM, while correlations of these parameters with NETs were present in both adult and pediatric forms of the disease.
There were no associations between use of specific immunosuppressive therapies and LDG or NET complexes levels, except for a correlation between circulating HNE-DNA NET complexes in the circulation with steroid dose in the JDM group (r = 0.29, P < 0.05) but not with other IIM. No associations were observed with levels of circulating LDGs and steroid use in any form of IIM.
MSAs are associated with NET levels and directly induce NET formation.
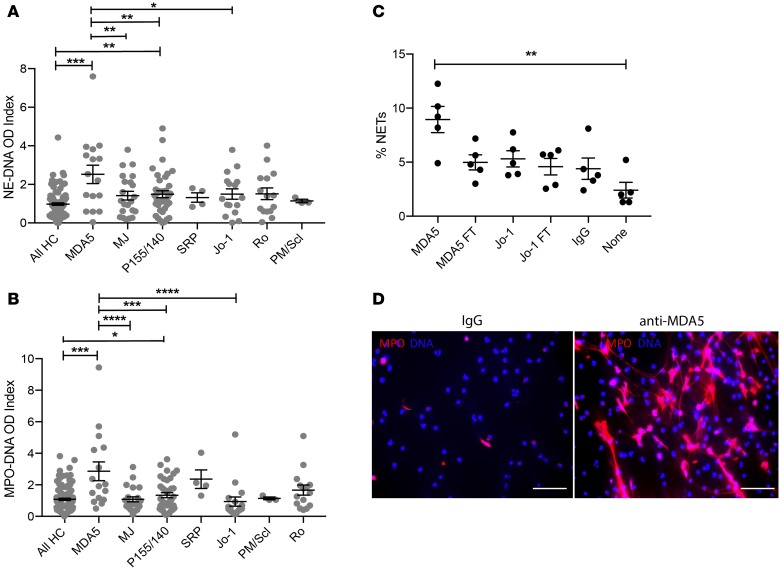
When assessing associations with specific MSA profiles, circulating NET levels (both HNE-DNA and MPO-DNA complexes) were significantly higher in IIM subjects that tested positive for anti-MDA5 MSAs. In addition, NET levels were higher in those subjects that had anti-transcriptional intermediary factor 1 (TIF1, also known as p155/140) autoantibodies, a MSA associated with DM and JDM (17). In contrast, other MSAs (including anti-Jo1) were not associated with elevated circulating levels of NETs, while LDG numbers did not correlate with any specific MSA (Figure 2, A and B; P > 0.05).
Figure 2. Anti-MDA5 MSAs are associated with higher levels of circulating NETs and directly induce NET formation.
Graphs represent levels of circulating NETs quantified as either plasma HNE-DNA complexes (A) or MPO-DNA complexes (B) in IIM subjects with presence of specific serum MSAs. HC = 30; ADM = 46; PM = 20; JDM = 86. (C) Healthy control neutrophils (n = 5 HC) were incubated with purified anti-MDA5 Ab (MDA5) or its corresponding flow through (MDA5 FT, see Methods), purified anti-Jo-1 (Jo-1) or corresponding flow through (Jo-1 FT), control IgG (IgG), or no treatment (none), and the percentage of netting neutrophils was quantified by fluorescence microscopy. Dots represent individual subjects, and data are expressed as median ± IQR. HC, healthy controls. Kruskal-Wallis was performed for nonparametric comparisons, while 1-way ANOVA was used for parametric comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (D) Representative microphotographs display HC neutrophils incubated in the presence or absence of purified anti-MDA5. Images depict cells stained with DAPI and MPO. Original magnification, ×10. p155/140 also known as TIF-1 alpha; Mj also known as NXP-2.
Given that enhanced NET formation was preferentially observed in IIM subjects with anti-MDA5, we assessed whether this Ab had preferential abilities to induce NET formation in HC neutrophils. Indeed, purified anti-MDA5 isolated from an adult subject with IIM, significantly enhanced NET formation in HC neutrophils when compared with control IgG (Figure 2, C and D), while purified anti-Jo1 MSA (also isolated from an adult IIM subject) did not (Figure 2C). Overall, anti-MDA5 MSAs enhanced NET formation and were associated with in vivo neutrophil dysregulation in IIM.
NETs and infiltrating neutrophils are detected in IIM tissues.
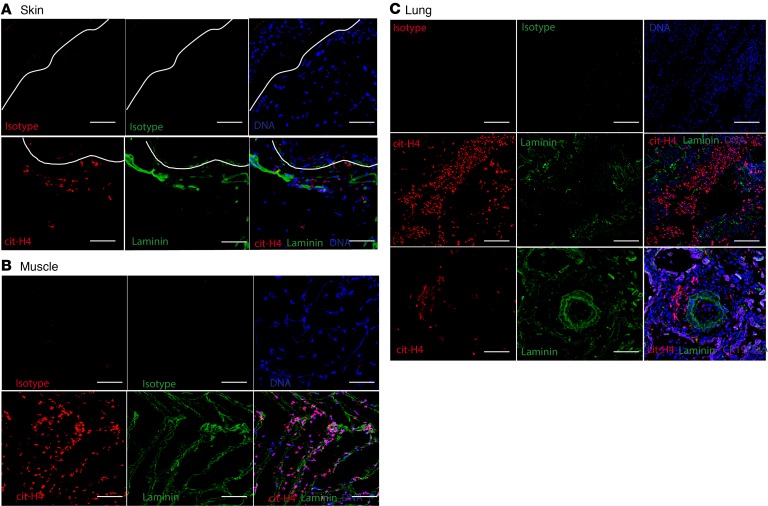
To further assess whether enhanced NET formation occurs in vivo in affected tissues in IIM, skin, muscle, and lung biopsies were analyzed. In skin, we studied 2 cohorts. The first cohort consisted of samples selected based on the presence or absence of an identified neutrophil mRNA gene signature in skin; these participants were selected so that we could identify those patients whose neutrophils were infiltrating this organ (n = 9) versus those who did not (n = 3). None of the patients in this cohort were positive for MSAs (Supplemental Table 2). The second cohort comprised DM subjects with anti-MDA5 (n = 4) or anti-Mi2 (n = 1) MSAs (Supplemental Table 3). In the MSA-positive cohort, all skin biopsies displayed evidence of infiltrating netting neutrophils, while this was not observed in the MSA-negative cohort, even if intact neutrophils were present in the skin.(Figure 3A and data not shown). NETs were also detected in muscle biopsies in the adult DM group (n = 5 of 5; all MSAs positive for anti-MDA5, anti-nuclear matrix protein-2 [NXP2, also known as Mj], or anti-TIF1) but not in unaffected muscle (Figure 3B and data not shown). Furthermore, NETs were detected in lung biopsies from subjects with DM and associated ILD (n = 2 of 2; 1 with anti-MDA5 and 1 with anti-Ro52 and anti-Pm/Scl75 autoantibodies), particularly surrounding blood vessels in pulmonary parenchyma (Figure 3C). Overall, these results indicate that netting neutrophils infiltrate clinically affected tissues in MSA-positive IIM, particularly in association with anti-MDA5. Given the association between anti-MDA5 and the presence of NETs in tissues, and previous reports of high MDA5 mRNA expression during muscle regeneration (18), we assessed whether the MDA5 protein could be detected in these biopsies. Indeed, MDA5 antigen was present in DM muscle and skin tissue biopsies (Supplemental Figure 2A). Purified NETs did not express MDA5, indicating that they were not the source of this antigen in skin (Supplemental Figure 2B).
Figure 3. NETs are detected in IIM skin, muscle, and lung in anti-MDA5–positive subjects.
Representative immunofluorescence images of (A) skin, (B) muscle, and (C) lung biopsies of subjects with anti-MDA5–positive adult DM. Red represents citrullinated histone H4, green represents laminin, and blue represents DNA. Isotype controls are shown on top, and right lower images depict merged images for each tissue. Original magnification, ×10. Details of the subject population are provided in Supplemental Table 3.
Neutrophil-associated gene expression in skeletal muscle correlate with markers of IIM disease activity and the type I and II IFN gene signatures.
RNA-sequencing was performed in muscle biopsy specimens from 39 subjects with adult DM (11 anti-Mi2-, 12 anti-NXP2-, 11 anti-TIF1-, and 5 anti-MDA5–positive MSAs; Supplemental Table 4). In addition, 20 normal muscle biopsy specimens were used as comparators. A neutrophil gene signature was evaluated using 5 neutrophil genes: ELANE, MPO, CTSG, S100A8, and S100A9, as previously described (19). Relative to HC muscle, neutrophil-associated gene expression was significantly upregulated in the DM group (Table 1). S100A8, S100A9, and MPO showed a positive correlation with type I IFN-regulated genes (ISG15, IFI6, and MX1) and type II IFN-regulated genes (PSMB8, GPB1, and GBP2; all q values < 0.001), while CSTG showed a positive correlation only with type II IFN-regulated genes (all q values < 0.006; Figure 4A). Muscle strength measurements and creatine kinase (CK) levels obtained near the time of the muscle biopsy were available in 17 DM subjects. S100A8, S100A9, and MPO positively correlated with CK levels and negatively correlated with muscle strength (Figure 4B). Overall, these results indicated that a skeletal muscle neutrophil gene signature was present in DM, in association with muscle damage and with enhanced muscle type I and II IFN responses, pathways considered pathogenic in DM.
Table 1. Gene expression (fold change) and q values of 5 neutrophil genes in DM skeletal muscle compared with healthy skeletal muscle.
Figure 4. A neutrophil gene signature is increased in DM skeletal muscle and correlates with type I and II IFN gene signatures and with clinical markers of muscle damage.
(A) Heatmap representing correlations between individual neutrophil genes and type I and II IFN-inducible genes in skeletal muscle from DM subjects, calculated by Spearman’s correlation. (B) Heatmap representing correlations between among neutrophil genes and serum CK and mean muscle strength (MEAN_STR) in DM subjects, calculated by Spearman’s correlation. Results are from 39 DM and 20 control biopsies.
NETs hamper myotube viability.
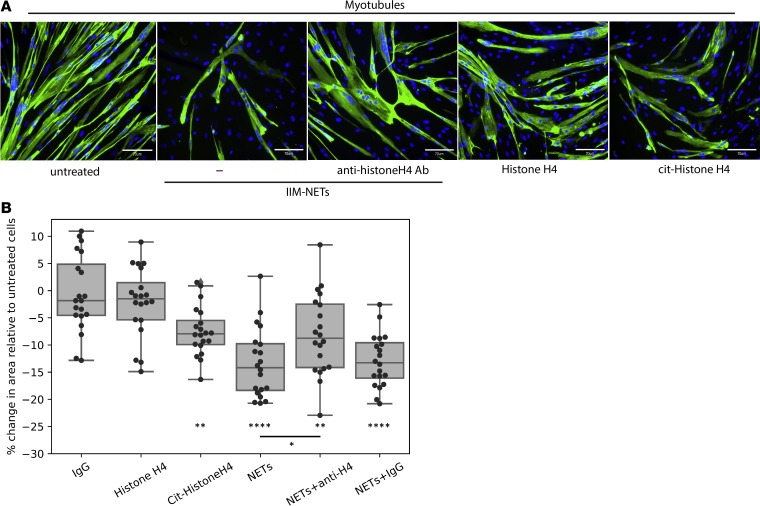
Given that NETs were detected in IIM muscle tissue, we determined whether they could play a local pathogenic role. Skeletal muscle fibers form as a result of the fusion of the precursors myoblasts into multinucleated fibers called myotubes. To assess whether NETs could directly harm muscle cells, we grew skeletal muscle myoblasts in the presence or absence of spontaneously formed NETs isolated from IIM subjects. We assessed both the ability of myoblasts to proliferate as well as the viability of skeletal myotubes. NETs did not disrupt the ability of myoblasts to proliferate after 72 hours in culture (Supplemental Figure 3A). In contrast, already formed myotubes exposed to NETs showed enhanced cell death that was evident as early as 1 hour after exposure. Significantly fewer myotubes were present by 24–48 hours when cells were exposed to NETs compared with untreated cells (Figure 5 and Supplemental Figure 3, B and C). To assess potential mechanisms of muscle cell damage by NETs, we blocked HNE, a component of NETs previously associated with cell injury. However, adding the HNE inhibitor Sivelestat did not abrogate the damaging effect of NETs on myotube viability (Supplemental Figure 3C). Histones externalized in NETs have recently been implicated in vascular smooth muscle cell damage in the context of atherosclerosis by forming pores in the target cells and inducing cell death (20). During NET formation, histones can undergo citrullination due to the activation of the enzyme peptidylarginine deiminase-4 (PAD4), and these citrullinated histones can be extruded as part of the NETs (21). Thus, we assessed whether native or citrullinated histones could cause skeletal myotube cell death. Indeed, while recombinant histone H4 was not toxic to these myotubes, citrullinated histone H4 significantly decreased myotube viability, similar to the effect observed with NETs. Furthermore, a blocking antibody targeting histone H4 partially and significantly reversed the toxic effect of the NETs on the skeletal myotubes, while control IgG did not (Figure 5B). Overall, these observations indicated that NETs have the ability to directly disrupt skeletal myotubes, at least in part, through a toxic effect mediated by citrullinated histones present in NETs. These results indicated that NETs in IIM muscle may contribute to the direct tissue damage that represents the hallmark of these diseases.
Figure 5. NETs interfere with myotube viability in a citrullinated histone-dependent manner.
(A) Representative microphotographs display myotube fibers stained with DAPI and MF-20. Images were obtained at 48 hours of culture. Original magnification, ×20. (B) Percentage change in skeletal myotube cell area relative to that in untreated cells. Results represent individual measurements of 5 fields over 4 replicates of each experiment. Each condition included 4 independent wells and 5 images/well. Comparisons between groups were performed using a 2-sided Student’s t test. *P < 0.05; **P < 0.01; ****P < 0.0001. The central line of the box plot represents the median, while the limits of the box correspond to quartiles 1 and 3. The whiskers show the minimum and maximum values of the distribution within the quartile 1 or 3 ± 1.5 × IQR range.
Discussion
The role of neutrophils in inflammatory myopathies remains poorly characterized, although recent data suggest that neutrophil numbers are associated with decreased patient survival (22). We now report that IIM is characterized by the presence of a pathogenic subset of neutrophils, LDGs, previously described in other autoimmune diseases. As is the case in SLE and AAV (5, 7), LDGs present in IIM subjects are characterized by their enhanced ability to form NETs. The levels of circulating LDGs and/or NETs significantly associate with disease activity in the muscle and skin and with the presence of specific MSAs (prevalently with anti-MDA5). Indeed, anti-MDA5 MSAs directly enhance NET formation. Further supporting that specific MSAs induce NET formation, netting neutrophils infiltrate organs affected in DM, including muscle, lung, and skin, in those IIM subjects who are positive for MSAs but not in MSA-negative IIM. Furthermore, the presence of an upregulated neutrophil gene signature in DM skeletal muscle biopsies associates with clinical features of the disease and with the type I–II IFN signatures. All these observations appear to have important pathogenic implications, as NETs purified from IIM neutrophils directly cause damage to myotubes, at least in part, via a citrullinated histone-dependent manner.
It is now generally accepted that inflammatory responses, if tightly regulated, are integral to muscle repair and regeneration (23). However, when inflammation becomes chronic and dysregulated, it can have profound effects on muscle injury and regeneration. Skeletal muscle has a remarkable capacity for repair, even following severe damage. The regenerative capacity of skeletal muscle relies on the presence of satellite cells/myoblasts that retain their ability to proliferate and differentiate to either fuse with existing muscle fibers or with other myogenic cells to generate new fibers (24). Furthermore, chronic inflammatory damage to already formed muscle fibers can significantly affect the function of muscles and their ability to recover from injury. The response of the immune system to muscle injury and disease is a complex process. Although myeloid cells, including neutrophils, participate in chronic striated muscle injury and regeneration, the role of NETs in this process had not been reported.
It is known that neutrophils can cause muscle fiber damage in vitro and in vivo (25) and that oxidants released by these cells can cause severe cellular damage. Recent studies also implicate a role for HNE in muscular dystrophies by impairing myoblast cell survival, proliferation, and differentiation (26). We found that, while NETs do not impair myoblast proliferation, they significantly decrease skeletal myotube viability. This is relevant, as biopsies from IIM subjects, but not healthy muscle, displayed evidence of infiltrating netting neutrophils and NET structures. Given the high levels of HNE present in NETs, it is possible that it and other proteases play prominent roles in the induction of myotube cell death (26). However, blocking the effects of HNE did not improve myotube viability, while citrullinated histones recapitulated the toxic effect of NETs on myotube viability. This is consistent with a recent report that histone H4 can bind and lyse vascular smooth muscle cells and destabilize atherosclerotic plaques (20). Although that study did not address the role of specific posttranslational modifications of histones in their ability to damage cells, we observed that citrullinated histone H4 has an enhanced ability to damage skeletal myotubes compared with native histone H4. Future studies should address the mechanisms by which specific protein modifications may dictate cell injury in various disease states. Of note, a recent study reported evidence of tissue citrullination in IIM-affected muscles, while was absent in control muscle tissue (27). This supports our observations implicating PAD activation and histone citrullination in skeletal muscle damage.
In other diseases, specific autoantibodies were found to induce enhanced NET formation, including anti-ribonucleoprotein autoantibodies in SLE (13), anti-citrullinated protein antibodies in RA (6), and antineutrophil cytoplasmic antibodies in AAV (28). We now report that anti-MDA5 MSAs can have similar effects in IIM and that their presence is associated with enhanced NET formation in circulation and in tissues. The mechanisms of this distinct association need to be further investigated and may have therapeutic implications. Future studies should also test how other MSAs, besides anti-MDA5 and anti-Jo, may modulate NET formation and the role of the NET-autoantibody interactions in the degree of induced skeletal myotube damage. It will also be important to enhance understanding of why some disease-specific autoantibodies, but not others, induce NET formation in this and other autoimmune conditions.
IIM subtypes are characterized by significant vasculopathy, which may contribute to muscle injury and dysfunction. Of interest, NETs were often found close to vascular structures, and their presence closely correlated with in vivo evidence of vasculopathy. We previously described a significant vasculopathic effect of LDG NETs, with endothelial cell apoptosis ensuing after NET exposure primarily triggered by the presence of matrix metalloproteinase-9 (9). Furthermore, the levels of LDGs were previously found to significantly correlate with vascular injury in other autoimmune diseases, such as SLE, and with poor treatment response in AAV (7, 19). It is therefore possible that NETs in muscle tissue promote injury and disruptions in regeneration by dual effects on muscle cell and on vascular structures. NETs were also found in other tissues affected by the inflammatory response characteristic of IIM, including the lungs and the skin in anti-MDA5–positive DM, suggesting that neutrophils and their NETs are also implicated in extramuscular involvement characteristic of this group of diseases. This finding is supported by the clinical correlations we detected between levels of NETs and/or LDGs and clinical scores of myositis disease activity and extramuscular disease as well as the inverse correlation with periungual nailfold capillary density as a sign of vasculopathy in JDM. Neutrophil infiltration was observed in IIM skin, while the presence of netting neutrophils was particularly evident in the anti-MDA5–positive group. The presence of infiltrating neutrophils in IIM skin supports the findings of a previous publication reporting that the cathelicidin LL-37, derived from these infiltrating neutrophils, can promote type I IFN responses in IIM (29). Furthermore, levels of circulating NETs significantly correlated with skin disease activity, further implicating these cells in the pathogenesis of DM. With regards to lung involvement, our observations are supported by previous reports of enhanced neutrophil peptides in ILD lungs in IIM (30) and that NET levels are increased in ILD in anti-MDA5–positive DM (14). Indeed, previous observations have suggested that impaired NET degradation may also contribute to ILD in IIM (31). Similarly, a recent study reported a link between calcium deposition in tissues, the calcinosis characteristic of JDM, and the development of neutrophil activation and NETs in muscle tissue (15). This may be an additional mechanism that induces NET formation in these individuals. However, we also observed enhanced NET formation and tissue infiltration by NETs in the absence of clinical calcinosis and in subsets of IIM not characterized by calcium deposits. Indeed, we found no correlations between circulating NET or LDG levels and calcinosis in DM and JDM. This suggests that other mechanisms in addition to crystals are implicated in enhanced neutrophil activation and NET formation in IIM and may include specific MSAs. In previous studies, numbers of LDGs and levels of NET formation were not linked to specific medications used to treat autoimmune diseases. In this study, however, some associations were present with the use of corticosteroids. It remains to be determined whether this represents the true effects of the drugs on neutrophil dysregulation in IIM or merely that individuals who require these medications tend to have more severe disease.
Overall, our results indicate that neutrophil dysregulation in IIM may play important pathogenic roles in autoimmune responses and tissue damage and that investigating strategies that modulate aberrant neutrophil responses in IIM may lead to changes in the outcome of these potentially devastating conditions.
Methods
Patient recruitment for peripheral blood studies.
Patients fulfilled probable or definite criteria for IIM from Bohan, Peter, and colleagues (32) or revised criteria for SLE from the American College of Rheumatology (33) and were enrolled at the NIH Clinical Center in the NIAMS/National Institute of Diabetes and Digestive and Kidney Diseases IRB-approved myositis (94-E0165) or SLE (94-AR-0066) natural history studies, respectively. HCs were enrolled from the NIH Blood Bank or the NIH healthy volunteer cohort. MSAs were detected by standardized immunoprecipitation and immunoprecipitation-immunoblot methods (34) at the NIH Clinical Center Central Laboratory. Clinical characterization and measurements of IIM disease activity and damage were obtained as previously described (35, 36). Videocapillaroscopy was quantified as described previously (37).
Quantification of circulating NET complexes.
HNE-DNA and MPO-DNA complexes were measured in plasma as previously described (19). In brief, high-binding 96-well microplates were incubated overnight at 4°C with either rabbit anti-HNE (Calbiochem, 481001) or mouse anti-human MPO (Bio-Rad, clone 4A4, 0400-0002). At 4°C, the plates were washed and blocked with 1% BSA overnight, and then plasma samples diluted in blocking buffer were added and incubated overnight. Plates were washed; they were then incubated for 1 hour with anti-ds-DNA (MilliporeSigma, clone BV16-13, MAB030) for the HNE-DNA plate or HRP-conjugated anti-DNA POD (Roche, 11774425001) for the MPO-DNA plate. The HNE-DNA plate was washed and incubated for 1 hour with anti–mouse IgG-HRP conjugate (Bio-Rad, 1721012). ELISAs were visualized with tetramethylbenzidine and stop solution.
Quantification of circulating LDGs.
Quantification of circulating LDGs was performed as previously described (19). In brief, human PBMCs were isolated by Ficoll-Paque Plus density gradient (GE Healthcare, GE17-1440-03), RBCs were lysed with hypertonic solution, and cells were resuspended in FACs buffer. Cells were blocked for 15 minutes with Human TruStain Fc Receptor Blocking Solution (BioLegend, 422301); stained with Abs recognizing human CD10 (BioLegend, clone H10A, 312209), CD15 (BioLegend, clone HI98, 301906), and CD14 (BioLegend, clone HCD14, catalog 325610) or isotype control for 15 minutes at 4°C in the dark; and fixed with 2% paraformaldehyde overnight. Data were collected using a BD FACSCanto RUO and analyzed using FlowJo Software version 10. Cutoff values for positive staining were determined using compensation controls for each fluorophore. LDGs were classified as CD10+CD15+CD14lo, as previously described (11).
Isolation of MSAs.
MSAs were isolated as previously reported (38). Plasma from 1 anti-Jo1-positive IIM subject (39-year-old female) and from 1 anti-MDA5–positive IIM subject (58-year-old male) were used to purify MSAs. Human MDA5 (amino acids 298–1025, Uniprot ID Q9BYX4) and HisRS (amino acids 1–509, Uniprot ID P12081) were subcloned separately into the expression vector pNIC-Bio3 (GenBank accession no. JN792439), which contains an N-terminal His6-tag and a C-terminal Avi-tag. Expression and purification were performed as described previously (39, 40). Biotinylated MDA5 and HisRS were produced by coexpressing the generated constructs with the bacterial BirA biotin ligase in E. coli strain BL21(DE3) R3. The proteins were purified from soluble cell extracts by a 2-step procedure, including immobilized metal affinity chromatography, followed by size exclusion chromatography. The final protein batches were assessed for quality and protein identity by SDS-PAGE and mass spectrometry and subsequently flash frozen in liquid nitrogen and stored at –80°C until use. N-hydroxysuccinimide–activated prepacked sepharose columns (NHS columns, GE Healthcare) were coupled with 5 or 1 mg recombinant MDA5 and Jo1, respectively, and according to the manufacturer’s instructions. Anti-MDA5 and anti-Jo1 IgG were isolated as previously reported (38). Briefly, plasma was centrifuged at 3000 g for 5 minutes, diluted 1:5 in PBS, and filtered using a 0.45-μM filter, before loading in the HiTrap Protein G HP columns (GE Healthcare) for IgG enrichment. Total IgGs eluted from the column using 0.1 M glycine-HCl, pH 2.7, were immediately neutralized to approximately 7.4 with 1 M Tris, pH 9, and buffer exchanged to PBS by dialysis. Anti-MDA5 IgG and anti-Jo1 IgG were enriched by applying total IgGs to MDA5 or Jo1 affinity columns, as described for the protein G column. Non-MDA5 and non-Jo1 reactive IgGs were collected and used as control Abs (flow through [FT]). No endotoxin contamination was detected in the Ab preparations after buffer exchange to PBS and the preparations were sterile filtered (<0.05 EU/mL). HC IgG was isolated from serum using the Melon Gel IgG Spin Purification Kit (Thermo Fisher Scientific, 45206) and quantified with a BCA assay. The efficiency of anti-MDA5 and anti-Jo1 IgG enrichment from the protein G–derived total IgG was confirmed by ELISA. Streptavidin-coated 96-well plates (50 ng, overnight at 4°C) were first blocked for 1 hour at room temperature with PBST 0.1% BSA, followed by a 1-hour incubation with either recombinant biotinylated MDA5 or Jo1 (1 μg/well). Anti-MDA5 IgG, anti-Jo1 IgG and corresponding FT fractions were added to the plates in serial dilutions (ranging from 0.05 ng/mL to 1000 ng/mL per well) and incubated for 90 minutes. Reactivity was recorded at 450 nm absorbance after adding Peroxidase F(ab′)2 Fragment Goat Anti-Human IgG, Fcγ fragment specific antibody (dilution 1:10,000, ref. 109-036-008, Jackson ImmunoResearch) for 1 hour at room temperature, followed by a 15-minute incubation with 3,3′,5,5′-tetramethylbenzidine substrate (reaction stopped with 1 M H2SO4).
Assessment of NET formation by immunofluorescence microscopy.
HC neutrophils and IIM neutrophils and LDGs were isolated as previously described (13). Cells were incubated on poly-L-lysine–coated coverslips for 15 minutes, blocked with Fc-blocking reagent (Miltenyi, 130-059-901), and then treated for 1 hour with anti-Jo-1 IgG, anti-MDA5 IgG, FT of anti-Jo-1 IgG, FT of anti-MDA5 IgG, or HC IgG. Coverslips were gently fixed with 4% paraformaldehyde overnight at 4°C, then stained with rabbit anti-MPO (Agilent Technologies, A039829-2) for 1 hour, followed with Alexa Fluor 555–conjugated donkey anti-rabbit IgG (Life Technologies, A-31572) and Hoechst (Life Technologies, H3570). After mounting with Prolong Gold (Life Technologies, P36930), slides were visualized by microscopy. Three images were taken per coverslip at ×40, and NET and total cell counts were quantified using ImageJ (NIH).
NET collection.
NETs were collected as previously described (11). Neutrophils or LDGs were seeded at 5 million cells per well and incubated in 5% CO2 at 37°C. After 4 hours, NETs were harvested using micrococcal nuclease and centrifuged, and the supernatant was collected and stored at –20°C. Protein content was quantified by BCA assay before use.
Preparation of skin, muscle, and lung biopsies.
Only cases that met clinical and histologic criteria for DM were included. DM skin lesion biopsies were identified through the University of Michigan Pathology Archives (IRBMED HUM00072843). RNA was extracted from 10-micron sections of formalin-fixed, paraffin-embedded tissue using E.N.Z.A FFPE RNA Kit (Omega Bio-tek), following the manufacturer’s instructions. Neutrophil-associated gene expression was assessed via human ST 2.1 Affymetrix array. Cases were separated into “neutrophil high” (n = 9) and “neutrophil low” (n = 3) based on expression of 7 genes, ELANE, MPO, CTSG, S100A8, S100A9, CRAMP1, and LTF, as previously described (19). Additional skin, subcutaneous tissue, muscle, and lung samples used for immunofluorescence microscopy analysis were obtained at the Instituto Nacional de Ciencias Médicas y de la Nutrición Salvador Zubiran (IRB protocol 2152), for anti-MSA–positive DM subjects. MSAs were assessed by the EUROIMMUNE immunoblot method. Paraffin-embedded tissue sections were deparaffinized with 2 sequential washes in xylene and 100% ethanol for 10 minutes and 1 wash for 5 minutes in 95%, 70%, and 50% ethanol. Epitope retrieval was carried out by immersing the slides in IHC-TEK epitope retrieval solution (IHC WORLD Life Science Products & Services) for 40 minutes in a steamer. After blocking for 30 minutes with 10% BSA, rabbit anti–human citrullinated histone H4 (Merck Millipore;07-596; dilution 1:250), mouse anti–human laminin-γ (R&D Systems; MAB2139; 1: 250), goat anti–human cytokeratin-19 (Santa Cruz Biotechnology; sc-376126; 1: 100), and rabbit anti–human MDA5 (Thermo Fisher Scientific, 33H12L34;1:100) were added and incubated overnight at 4°C. After washing 3 times with PBS, secondary Abs were added (donkey anti–rabbit Alexa Fluor 555, donkey anti–mouse Alexa Fluor 488, and donkey anti–goat Alexa fluor 647 [1:200 in 5% BSA], all from Thermo Fisher Scientific). After washes, slides were incubated for 10 minutes at room temperature with 1:1000 Hoechst 33342 (Thermo Fisher Scientific) and mounted with ProLong Gold Antifade (Thermo Fisher Scientific). For the frozen sections, similar steps were followed starting with the blocking step. The samples were acquired in an Eclipse Ti-E Nikon confocal microscope (Minato).
RNA-sequencing analysis of skeletal muscle.
Muscle biopsies used for the RNA-sequencing analysis were obtained from DM subjects enrolled in IRB-approved longitudinal cohorts from the NIH (IRB 91-AR-0196), the Johns Hopkins Myositis Center (IRB NA_00007454), the Clinic Hospital (Barcelona; IRB HCB/2015/0479), and the Vall d’Hebron Hospital (Barcelona; IRB PR[AG] 68/2008). CK levels and strength assessments obtained closest to the time of muscle biopsy were used to measure the clinical activity. Muscle strength was evaluated by the examining physician using the Medical Research Council scale. This scale was transformed to Kendall’s 0–10 scale; the right- and left-side measurements for arm abduction and hip flexion strength were combined and the average was used for calculations (possible range 0–10), as previously described (36). Normal muscle biopsies were obtained from the Johns Hopkins Neuromuscular Pathology Laboratory (n = 10) and the Skeletal Muscle Biobank of the University of Kentucky (n = 10).
RNA-sequencing was performed as previously described (41). Briefly, RNA was prepared using TRIzol (Invitrogen). Libraries were prepared using the NeoPrep system according to the TruSeq Stranded mRNA Library Prep protocol (Illumina) and sequenced using the Illumina HiSeq 2500 or 3000. Reads were aligned using STAR v.2. (42), the abundance of each gene was quantified using StringTie v.1.3.3. (43), and differential gene expression analysis was performed using DESeq2 v.1.20.0. (44) The Benjamini-Hochberg correction was used to adjust for multiple comparisons, and a corrected P value (q value) of 0.05 or less was considered statistically significant. A neutrophil gene signature was calculated based on the expression of ELANE, MPO, CTSG, S100A8, and S100A9, as mentioned above. IFN pathway genes were collected from the Reactome biorepository (https://reactome.org/). General IFN-related genes and genes from the type I and II IFN pathways were merged in a single list. The 13 genes included in the previously proposed IFN scoring system in myositis were also added to the list (45). The expression of the genes of on list was analyzed in the different MSA and clinical myositis subsets. Gene expression (fragments per kilobase of transcript per million mapped reads [FPKM]) values were log transformed (logFPKM: log2[FPKM+1]) for visualization purposes using the Python programming language and the packages Numpy, Pandas, and Seaborn. Correlation among continuous variables was measured using Spearman’s rho. Skin microarray and muscle RNA-sequencing data were deposited in GEO (accession GSE142807 and GSE143323, respectively).
Skeletal myoblast cell cultures.
Healthy human skeletal muscle myoblasts (Lonza, CC2580) were grown following the manufacturer’s recommendations. Myoblasts were seeded at 3500 cells/cm2 and maintained in growth medium (Lonza, CC-3245) at 37°C, 5% CO2, until 50% or 60% confluence, changing growth medium every other day. Myoblasts were then treated with HC NETs, IIM NETs, ionophore, or media in duplicates. Within 10 minutes of treatment, plates were imaged on the IncuCyte ZOOM platform as previously described (46), using phase contrast. Nine areas per well were imaged every 30 minutes for the first 6 hours and every 2 hours for the next 18 hours for 3 days, using a ×20 dry objective. Videos were created using the IncuCyte Basic Software and ImageJ (NIH).
Preparation of citrullinated histone H4.
Recombinant human histone H4 (New England Biolabs) and recombinant PAD4 (Signalchem) were suspended in RPMI containing 2 mM CaCl2 and 1 mM DTT. The reaction was performed at 37°C overnight and stored at –20°C until further use.
Skeletal myotube experiments.
Healthy human skeletal muscle myoblasts at passage 4 were cultured in 24-well plates (3500 cells/ cm2) in SkBM-2 Basal Medium (Lonza, CC-3245). Differentiation started when the cells were about 50%–60% confluent, replacing growth medium with DMEM/F12 (Lonza), 2% horse serum, 1% L-glutamine, and 1% Insulin (Invitrogen, 1:100). On day 4, when myotubes were fully elongated and fused, NETs (50 μg), recombinant histone H4 (New England Biolabs; 1 μg), or citrullinated H4 (prepared as above; 1 μg) were added to myotubes. In specific experiments, NETs were pretreated with the HNE inhibitor Sivelestat (MilliporeSigma, 20 μM), rabbit polyclonal anti-histone H4 (Abcam, ab10158; 1 μg), or rabbit IgG isotype control (Invitrogen, 1 μg) before adding to the myotube cultures. Studies were performed in duplicate or quadruplicate. After 8 hours, 24 hours, and 48 hours of adding the NETs, cells were washed with PBS 3 times, fixed with ice-cold methanol for 10 minutes at –20°C, washed, and blocked with 5% BSA/PBS. The monoclonal Ab MF 20 (MYH1; DSHB, University of Iowa; deposited to DSHB by D.A. Fischman) was used for staining myosin heavy chain protein. The Ab was added at 1:10 dilution, followed by incubation for 1 hour at 37°C. Cells were then washed and secondary antibody Alexa goat anti–mouse IgG 488 (Invitrogen; 1:200 dilution) was added for 1 hour at 37°C. After a wash, cells were stained with DAPI (Vector Laboratories; Vectashield Antifade Mounting Medium DAPI, catalog H-1200). Five pictures were taken per well (×10 magnification), using a Revolve microscope (Echo Laboratories Inc). ImageJ (NIH) was used to count DAPI positivity (nuclei) and to calculate the muscle fiber area.
Statistics.
All graphs depict the mean ± SEM or median and IQR range unless otherwise stated. Associations of measurements with clinical characteristics and disease activity indices were assessed with Spearman’s correlation using IBM SPSS software (IBM Analytics). Student’s 2-tailed t test, Kruskal-Wallis, Mann-Whitney were used as appropriate. Multiple comparisons were analyzed by 1-way ANOVA using GraphPad Prism. P values of less than 0.05 were considered significant.
Study approval.
All human studies were reviewed and approved by appropriate institutional review boards (NIAMS/National Institute of Diabetes and Digestive and Kidney Diseases 94-AR-0066 and 91-AR-0196; NIEHS 94E0165; University of Michigan HUM00072843; Instituto Nacional de Ciencias Medicas y de la Nutrición Salvador Zubiran 2152; Johns Hopkins University NA_00007454; Vall d’Hebron Hospital PR(AG)68/2008; Hospital Clinic de Barcelona HCB/2015/0479 (Barcelona, Spain). All subjects provided informed consent before their participation in the study according to the standards of the Declaration of Helsinki.
Author contributions
NS, JJTR, CCR, IPF, KP, MMP, YH, CFC, PG, and NA performed experiments and analyzed data. AG, OB, DGM, FWM, IEL, AIS, AM, and LGR supervised data and performed critical analysis of data. ASO, JCM, JMGJ, LCS, and JMK obtained and provided tissues and clinical analyses. AIS, AM, and LGR evaluated clinical subjects and provided clinical scores and statistical analyses. All authors reviewed the paper. MJK conceived the study, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was funded by the Intramural Research Program of the NIH, NIAMS (ZIAAR041199) and the NIEHS (Z01ES101074). This work was also funded in part by the Consejo Nacional de Ciencia y Tecnología (CONACYT SEP-CB2017-2018, A1-S23262). We thank Ira Targoff for MSA testing, Stephen Hewitt for technical support, and Edvard Wigren and Susanne Graslund for recombinant protein preparation.
Version 1. 01/16/2020
In-Press Preview
Version 2. 02/13/2020
Electronic publication
Footnotes
DGM’s present address is: Tecnológico de Monterrey, Escuela de Medicina y Ciencias de la Salud, Monterrey, Mexico.
OB’s present address is: Department of Internal Medicine and Clinical Immunology, Pitié Salpêtrière Hospital, AP-HP, Sorbonne Université, Paris, France.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(3):e134189.https://doi.org/10.1172/jci.insight.134189.
Contributor Information
Jose Jiram Torres-Ruiz, Email: jiram_torres@hotmail.com.
Carmelo Carmona-Rivera, Email: carmelo.carmona-rivera@nih.gov.
Iago Pinal-Fernandez, Email: iago.pinalfernandez@nih.gov.
Katherine Pak, Email: katherine.pak@nih.gov.
Monica M. Purmalek, Email: monica.purmalek@gmail.com.
Yuji Hosono, Email: yuji.hosono@tsc.u-tokai.ac.jp.
Catia Fernandes-Cerqueira, Email: catiafcerqueira@gmail.com.
Nathan Arnett, Email: arnett.nathan.gl@gmail.com.
Alexander Gorbach, Email: gorbacha@mail.nih.gov.
Olivier Benveniste, Email: olivier.benveniste@psl.aphp.fr.
Diana Gómez-Martín, Email: dgomar79@gmail.com.
Josep M. Grau-Junyent, Email: jmgrau@clinic.cat.
Lisa Christopher-Stine, Email: lchrist4@jhmi.edu.
Frederick W. Miller, Email: millerf@mail.nih.gov.
Ingrid E. Lundberg, Email: ingrid.lundberg@ki.se.
J. Michelle Kahlenberg, Email: mkahlenb@umich.edu.
Adam I. Schiffenbauer, Email: schiffenbauera2@niehs.nih.gov.
Lisa G. Rider, Email: riderl@mail.nih.gov.
Mariana J. Kaplan, Email: mjkaplansch@hotmail.com.
References
- 1.Selva-O’Callaghan A, Pinal-Fernandez I, Trallero-Araguás E, Milisenda JC, Grau-Junyent JM, Mammen AL. Classification and management of adult inflammatory myopathies. Lancet Neurol. 2018;17(9):816–828. doi: 10.1016/S1474-4422(18)30254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tournadre A, Miossec P. A critical role for immature muscle precursors in myositis. Nat Rev Rheumatol. 2013;9(7):438–442. doi: 10.1038/nrrheum.2013.26. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida S, Akizuki M, Mimori T, Yamagata H, Inada S, Homma M. The precipitating antibody to an acidic nuclear protein antigen, the Jo-1, in connective tissue diseases. A marker for a subset of polymyositis with interstitial pulmonary fibrosis. Arthritis Rheum. 1983;26(5):604–611. doi: 10.1002/art.1780260505. [DOI] [PubMed] [Google Scholar]
- 4.Moghadam-Kia S, Oddis CV, Sato S, Kuwana M, Aggarwal R. Antimelanoma differentiation-associated gene 5 antibody: expanding the clinical spectrum in North American patients with dermatomyositis. J Rheumatol. 2017;44(3):319–325. doi: 10.3899/jrheum.160682. [DOI] [PubMed] [Google Scholar]
- 5.Villanueva E, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. 2011;187(1):538–552. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khandpur R, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5(178):178ra40. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grayson PC, et al. Neutrophil-related gene expression and low-density granulocytes associated with disease activity and response to treatment in antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol. 2015;67(7):1922–1932. doi: 10.1002/art.39153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carmona-Rivera C, et al. Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Sci Immunol. 2017;2(10):eaag3358. doi: 10.1126/sciimmunol.aag3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmona-Rivera C, Zhao W, Yalavarthi S, Kaplan MJ. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Ann Rheum Dis. 2015;74(7):1417–1424. doi: 10.1136/annrheumdis-2013-204837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahlenberg JM, Carmona-Rivera C, Smith CK, Kaplan MJ. Neutrophil extracellular trap-associated protein activation of the NLRP3 inflammasome is enhanced in lupus macrophages. J Immunol. 2013;190(3):1217–1226. doi: 10.4049/jimmunol.1202388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny MF, et al. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J Immunol. 2010;184(6):3284–3297. doi: 10.4049/jimmunol.0902199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin AM, et al. Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J Immunol. 2011;187(1):490–500. doi: 10.4049/jimmunol.1100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med. 2016;22(2):146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Y, Zhang S, Zhao Y, Liu Y, Yan B. Neutrophil extracellular traps may contribute to interstitial lung disease associated with anti-MDA5 autoantibody positive dermatomyositis. Clin Rheumatol. 2018;37(1):107–115. doi: 10.1007/s10067-017-3799-y. [DOI] [PubMed] [Google Scholar]
- 15.Duvvuri B, et al. Neutrophil extracellular traps in tissue periphery in juvenile dermatomyositis. Arthritis Rheumatol. doi: 10.1002/art.41078. doi: 10.1002/art.41078. [published online ahead of print August 12, 2019]. [DOI] [PubMed] [Google Scholar]
- 16.Bertolazzi C, Cutolo M, Smith V, Gutierrez M. State of the art on nailfold capillaroscopy in dermatomyositis and polymyositis. Semin Arthritis Rheum. 2017;47(3):432–444. doi: 10.1016/j.semarthrit.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Fiorentino D, Casciola-Rosen L. Autoantibodies to transcription intermediary factor 1 in dermatomyositis shed insight into the cancer-myositis connection. Arthritis Rheum. 2012;64(2):346–349. doi: 10.1002/art.33402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinal-Fernandez I, et al. Myositis autoantigen expression correlates with muscle regeneration but not autoantibody specificity. Arthritis Rheumatol. 2019;71(8):1371–1376. doi: 10.1002/art.40883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlucci PM, et al. Neutrophil subsets and their gene signature associate with vascular inflammation and coronary atherosclerosis in lupus. JCI Insight. 2018;3(8):99276. doi: 10.1172/jci.insight.99276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silvestre-Roig C, et al. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature. 2019;569(7755):236–240. doi: 10.1038/s41586-019-1167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grayson PC, Kaplan MJ. At the Bench: Neutrophil extracellular traps (NETs) highlight novel aspects of innate immune system involvement in autoimmune diseases. J Leukoc Biol. 2016;99(2):253–264. doi: 10.1189/jlb.5BT0615-247R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha YJ, et al. Baseline peripheral blood neutrophil-to-lymphocyte ratio could predict survival in patients with adult polymyositis and dermatomyositis: A retrospective observational study. PLoS One. 2018;13(1):e0190411. doi: 10.1371/journal.pone.0190411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peake JM, Neubauer O, Della Gatta PA, Nosaka K. Muscle damage and inflammation during recovery from exercise. J Appl Physiol. 2017;122(3):559–570. doi: 10.1152/japplphysiol.00971.2016. [DOI] [PubMed] [Google Scholar]
- 24.Robinson DCL, Dilworth FJ. Epigenetic regulation of adult myogenesis. Curr Top Dev Biol. 2018;126:235–284. doi: 10.1016/bs.ctdb.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Tidball JG, Villalta SA. Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol. 2010;298(5):R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arecco N, et al. Elastase levels and activity are increased in dystrophic muscle and impair myoblast cell survival, proliferation and differentiation. Sci Rep. 2016;6:24708. doi: 10.1038/srep24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makrygiannakis D, et al. Citrullination is an inflammation-dependent process. Ann Rheum Dis. 2006;65(9):1219–1222. doi: 10.1136/ard.2005.049403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessenbrock K, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. 2009;15(6):623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, et al. The host defense peptide LL-37 a possible inducer of the type I interferon system in patients with polymyositis and dermatomyositis. J Autoimmun. 2017;78:46–56. doi: 10.1016/j.jaut.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Sakamoto N, et al. Elevated α-defensin levels in plasma and bronchoalveolar lavage fluid from patients with myositis-associated interstitial lung disease. BMC Pulm Med. 2018;18(1):44. doi: 10.1186/s12890-018-0609-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Shu X, Tian X, Chen F, Lu X, Wang G. Enhanced formation and impaired degradation of neutrophil extracellular traps in dermatomyositis and polymyositis: a potential contributor to interstitial lung disease complications. Clin Exp Immunol. 2014;177(1):134–141. doi: 10.1111/cei.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohan A, Peter JB, Bowman RL, Pearson CM. Computer-assisted analysis of 153 patients with polymyositis and dermatomyositis. Medicine (Baltimore) 1977;56(4):255–286. doi: 10.1097/00005792-197707000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 34.Targoff IN, et al. A novel autoantibody to a 155-kd protein is associated with dermatomyositis. Arthritis Rheum. 2006;54(11):3682–3689. doi: 10.1002/art.22164. [DOI] [PubMed] [Google Scholar]
- 35.Rider LG, et al. Update on outcome assessment in myositis. Nat Rev Rheumatol. 2018;14(5):303–318. doi: 10.1038/nrrheum.2018.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rider LG, et al. Measures of adult and juvenile dermatomyositis, polymyositis, and inclusion body myositis: Physician and Patient/Parent Global Activity, Manual Muscle Testing (MMT), Health Assessment Questionnaire (HAQ)/Childhood Health Assessment Questionnaire (C-HAQ), Childhood Myositis Assessment Scale (CMAS), Myositis Disease Activity Assessment Tool (MDAAT), Disease Activity Score (DAS), Short Form 36 (SF-36), Child Health Questionnaire (CHQ), physician global damage, Myositis Damage Index (MDI), Quantitative Muscle Testing (QMT), Myositis Functional Index-2 (FI-2), Myositis Activities Profile (MAP), Inclusion Body Myositis Functional Rating Scale (IBMFRS), Cutaneous Dermatomyositis Disease Area and Severity Index (CDASI), Cutaneous Assessment Tool (CAT), Dermatomyositis Skin Severity Index (DSSI), Skindex, and Dermatology Life Quality Index (DLQI) Arthritis Care Res (Hoboken) 2011;63 Suppl 11:S118–S157. doi: 10.1002/acr.20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minniti CP, et al. Vasculopathy, inflammation, and blood flow in leg ulcers of patients with sickle cell anemia. Am J Hematol. 2014;89(1):1–6. doi: 10.1002/ajh.23571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandes-Cerqueira C, et al. Patients with anti-Jo1 antibodies display a characteristic IgG Fc-glycan profile which is further enhanced in anti-Jo1 autoantibodies. Sci Rep. 2018;8(1):17958. doi: 10.1038/s41598-018-36395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keates T, et al. Expressing the human proteome for affinity proteomics: optimising expression of soluble protein domains and in vivo biotinylation. N Biotechnol. 2012;29(5):515–525. doi: 10.1016/j.nbt.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong N, et al. Optimizing production of antigens and Fabs in the context of generating recombinant antibodies to human proteins. PLoS One. 2015;10(10):e0139695. doi: 10.1371/journal.pone.0139695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amici DR, et al. Calcium dysregulation, functional calpainopathy, and endoplasmic reticulum stress in sporadic inclusion body myositis. Acta Neuropathol Commun. 2017;5(1):24. doi: 10.1186/s40478-017-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg SA, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57(5):664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, Chan DW, Zaal KJ, Kaplan MJ. A high-throughput real-time imaging technique to quantify NETosis and distinguish mechanisms of cell death in human neutrophils. J Immunol. 2018;200(2):869–879. doi: 10.4049/jimmunol.1700905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.