Abstract
Few therapeutic methods exist for preventing preterm birth (PTB), or delivery before completing 37 weeks of gestation. In the US, progesterone (P4) supplementation is the only FDA-approved drug for use in preventing recurrent spontaneous PTB. However, P4 has limited effectiveness, working in only approximately one-third of cases. Computational drug repositioning leverages data on existing drugs to discover novel therapeutic uses. We used a rank-based pattern-matching strategy to compare the differential gene expression signature for PTB to differential gene expression drug profiles in the Connectivity Map database and assigned a reversal score to each PTB-drug pair. Eighty-three drugs, including P4, had significantly reversed differential gene expression compared with that found for PTB. Many of these compounds have been evaluated in the context of pregnancy, with 13 belonging to pregnancy category A or B — indicating no known risk in human pregnancy. We focused our validation efforts on lansoprazole, a proton-pump inhibitor, which has a strong reversal score and a good safety profile. We tested lansoprazole in an animal inflammation model using LPS, which showed a significant increase in fetal viability compared with LPS treatment alone. These promising results demonstrate the effectiveness of the computational drug repositioning pipeline to identify compounds that could be effective in preventing PTB.
Keywords: Reproductive Biology, Therapeutics
Keywords: Bioinformatics
Computational drug repositioning method discovers novel uses for existing drugs for the prevention of preterm birth.
Introduction
Preterm birth (PTB), or birth before 37 weeks of gestation, is the leading cause of infant mortality worldwide. According to global estimates in 2015, complications arising from PTB were responsible for 35.5% of neonatal deaths (and 17.8% of deaths in children under 5) (1). Estimated rates of PTB vary by country, with rates as low as 5% and as high as 18% and a worldwide estimate of 11.1% (2). Even in nonfatal cases, prematurely born children are at a higher risk for life-long neurodevelopmental sequelae, such as motor and cognitive impairments (3, 4). Approximately two-thirds of PTB cases are spontaneous PTBs (sPTBs), an umbrella term for both spontaneous preterm labor (early onset of regular contractions and cervical changes) and preterm premature rupture of the membranes (early water breaking) (5). sPTB is difficult to predict, with ongoing studies focused on identifying biological and demographic risk factors as well as potential warning signs during pregnancy. Some studies have suggested that sPTB may be caused by changes in the maternal immune system or in the interplay between the fetal and maternal immune systems (6–8).
Although early diagnosis and identification of patients at risk for sPTB is an important goal, there are also very few therapeutic options available to prevent sPTB. Cervical cerclage, a surgical procedure to close the cervix, has been shown to have some success in preventing sPTB (9, 10); however, it is an invasive procedure. For pharmacological measures, vaginally applied progesterone (P4) supplementation has shown some success in preventing PTB in women with short cervixes (10–12). In 2011, the US FDA approved injections of hydroxyprogesterone caproate, a synthetic progestin or P4, to prevent recurrent sPTB in the US (13). However, these treatments have limited success, reducing the risk of PTB only by approximately one-third (11–14). As such, a high degree of emphasis should be placed on finding potential new therapies and other preventative medicines.
Traditional development of novel pharmaceuticals is both expensive and time consuming (15). Drug repositioning, or drug “repurposing,” is the process of discovering new indications for existing drugs (16). By searching for new applications of drugs that are already FDA-approved for use in humans, development time and costs can be substantially reduced. Existing drugs have already undergone extensive clinical testing and development and have well-established safety and pharmacokinetic profiles. Some examples of successful repositioning include atomoxetine for attention-deficit hyperactivity disorder, sildenafil for erectile dysfunction, and finasteride for hair loss (16). Safety is an especially important consideration within the context of pregnancies; therefore, by working with existing FDA-approved drugs, we can focus on potential drugs for sPTB with no known adverse effects to the mother or the fetus.
With the increasing availability of large public data sets, computational drug repositioning has emerged as an efficient approach to identify novel indications for existing drugs (17–23). Gene expression microarrays have been used extensively to study genome-wide effects of perturbagens (e.g., drugs) as well as disease states. Differential gene expression profiles can be generated by comparing microarray data from drug-treated samples with those of unperturbed samples. These profiles consist of genes that are upregulated or downregulated in treated cells relative to untreated cells. Similarly, disease gene expression signatures can be generated by comparing disease samples with healthy controls. One computational approach uses a pattern-matching strategy to find drugs and diseases with opposite differential gene expression profiles — that is, combinations in which genes upregulated or downregulated in a disease are downregulated or upregulated, respectively, by drug treatment. The hypothesis behind this strategy is that if a disease with a set of gene changes is matched with a drug with the opposite set of gene changes, then that drug could potentially be used therapeutically for the disease. This pattern-matching strategy has been used to identify existing and novel treatments for various indications (18), including inflammatory bowel disease (19), dermatomyositis (20), and various cancers (21–23).
This transcriptomics-based drug repositioning approach has been successful primarily for diseases with a strong immune system component. During pregnancy, the immune system maintains a tolerogenic antiinflammatory state by proliferating regulatory T cells until late gestation (24, 25). At the end of pregnancy, a switch to a proinflammatory state promotes labor (6, 25, 26). To sustain pregnancy, the balance between innate and adaptive immune cells must be maintained, with a premature shift in the balance potentially leading to preterm labor (27). In a prior study by our group, we performed a cross-study meta-analysis to identify genome-wide differences associated with sPTB (8). Pathway analyses of these genes identified enrichment in immune-related pathways, with sPTB associated with the upregulation of innate immunity and downregulation of adaptive immunity. In particular, genes that were downregulated were involved in biological processes, including T cell receptor signaling and transduction, B cell receptor signaling, leukocyte activation, and lymphocyte activation. Upregulated innate immunity processes included IL-1 signaling and neutrophil degranulation.
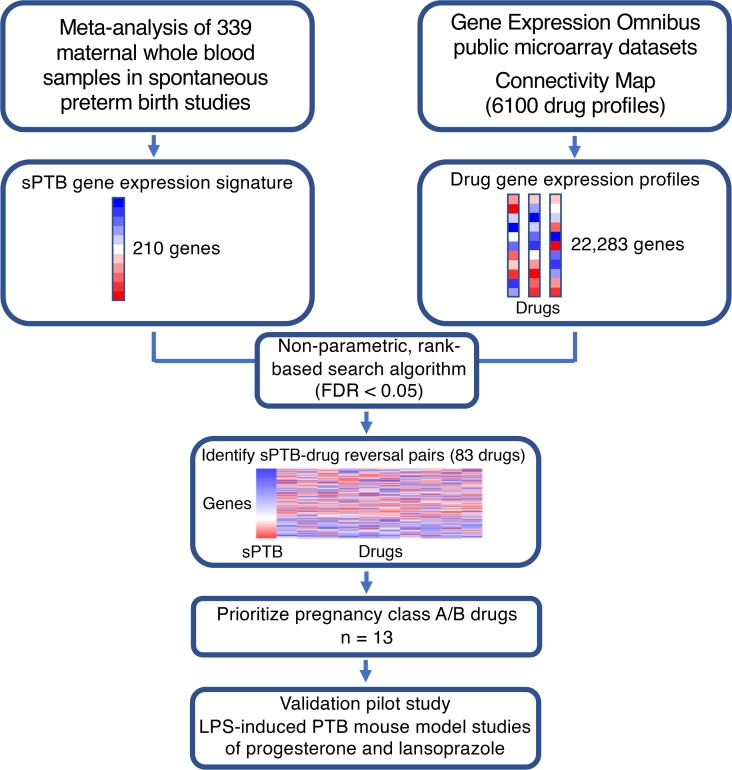
With the immune system implicated in sPTB, we applied the computational drug repositioning approach, leveraging publicly available drug and disease gene expression profiles to identify potential new treatments for sPTB. Figure 1 illustrates the general workflow of our repositioning pipeline. The Gene Expression Omnibus (GEO), which was created in an effort to make high-throughput gene expression data more accessible, is a public genomics data repository administered by the National Center for Biotechnology Information; it currently contains over 103,000 gene expression studies across 2.6 million microarray samples (28). The existence of a central data hub allows a high degree of flexibility for potential new discoveries. We leveraged the publicly available data in GEO to first identify a human gene expression profile of sPTB (8) and then query existing drug gene expression profiles (derived from human cell lines) from the Connectivity Map (CMap) (17) and the Library of Integrated Network-Based Cellular Signatures (LINCS) L1000 data sets (29), resulting in the identification a set of drugs with reversed differential gene expression profiles from that of sPTB. These drugs are medications that are all currently used in humans for different indications and may be potentially useful in the prevention of sPTB.
Figure 1. Workflow of drug repositioning pipeline.
The gene expression signature of sPTB was extracted from a meta-analysis of maternal whole blood samples (8). By comparing the signature with drug gene expression profiles from publicly available data sets on gene responses to drug exposure (17, 29), we identified sPTB/drug pairs with reversed gene expressions as candidate therapeutics.
P4, the existing standard for preventing sPTB, is 1 of the 83 compounds we identified; 81 of these have a higher reversal score than P4. We further narrowed down the list by focusing on those with plausible safety profiles that fall within the pregnancy category A or B designation, which then yielded a total of 13 potential drugs. We focused our validation efforts on lansoprazole, a proton pump inhibitor (PPI), which has one of the strongest reversal scores and a good safety profile. Here, we tested lansoprazole in an animal inflammation model of fetal wastage using LPS (30), which showed a significant increase in fetal viability compared with treatment with LPS alone. These promising results demonstrate the potential effectiveness of the computational drug repurposing pipeline for identifying compounds that might be effective in preventing sPTB.
Results
Computational identification of drug repositioning candidates.
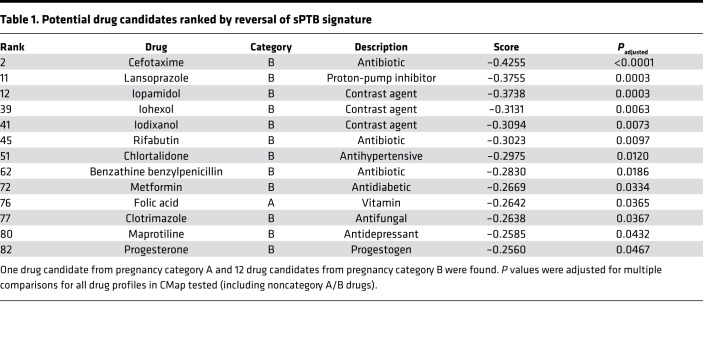
By comparing the drug signatures from CMap with the differential gene expression for sPTB, we identified 83 potential candidate drugs, including P4, which significantly reversed the disease gene expression signature. We further narrowed down potential candidates by their safety profiles during pregnancy. In the past, the FDA applied broad labels known as pregnancy categories to drugs based on results from both human and animal studies. Letter grades of categories were broadly based upon risk to the pregnant mother and her fetus. Pregnancy category B corresponds to drugs that have failed to demonstrate risk in animal studies with no adequate studies in pregnant women, while pregnancy category A is reserved for drugs for which human studies have failed to demonstrate a risk. By ruling out drugs with no known pregnancy category and drugs with pregnancy categories with positive evidence of risk, we further reduced our list to 13 category A or category B compounds (Table 1). Notably, P4 is 1 of the 13 drug hits identified. Only 1 pregnancy category A drug was identified: folic acid. Belonging to pregnancy category B are P4 and 11 of the other drugs: 3 antibiotics (benzathine benzylpenicillin, cefotaxime, and rifabutin), an antifungal (clotrimazole), a PPI (lansoprazole), an antidepressant (maprotiline), an antidiabetic (metformin), an antihypertensive (chlortalidone), and 3 contrast agents (iopamidol, iohexol, and iodixanol). We focused on the therapeutic agents, excluding the 3 contrast agents. A heatmap of the differential gene expressions of the sPTB signature and the 10 remaining drugs is shown in Figure 2A. Each of the compounds exhibited a clear reversal of the sPTB gene expression signature.
Table 1. Potential drug candidates ranked by reversal of sPTB signature.
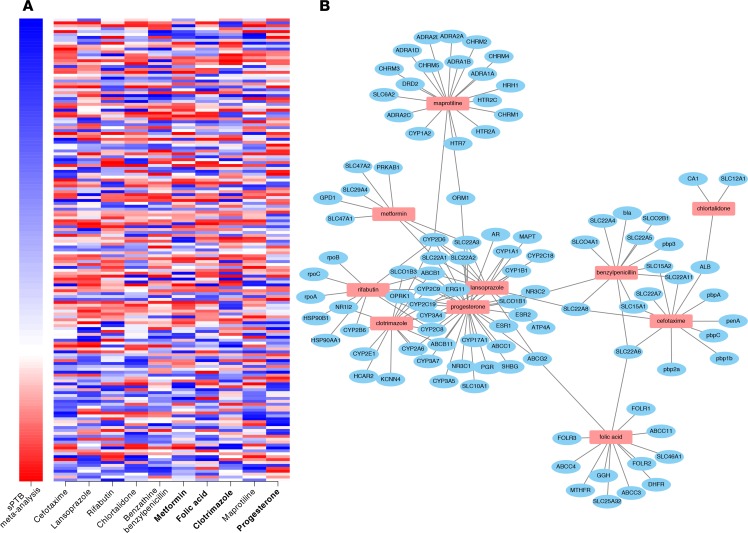
Figure 2. Gene expression heatmap and protein-interaction connectivity of category A/B therapeutic drug hits.
(A) Differential gene expression heatmap of sPTB meta-analysis and 10 computationally identified drugs in pregnancy categories A or B. Each row corresponds to a gene (sorted top to bottom from most downregulated to most upregulated in sPTB relative to term births). A rank-based coloring scheme was used. The sPTB meta-analysis differential gene expression signature ranked the 115 downregulated genes (blue) and the 44 upregulated genes (red) separately. The 10-drug gene expression profiles were ordered from left to right by reversal score (most negative to least negative), with all 159 genes ranked from 1–159 (blue-red). Bluer color corresponds to more downregulated genes, and redder color corresponds to more upregulated genes. Bolded drugs (metformin, folic acid, clotrimazole, and progesterone) have been observed in past studies to have some efficacy in the prevention of PTB. (B) Drug-protein interactions for the 10 drug hits according to DrugBank.
Common protein interactions among 13 pregnancy category A/B drug hits.
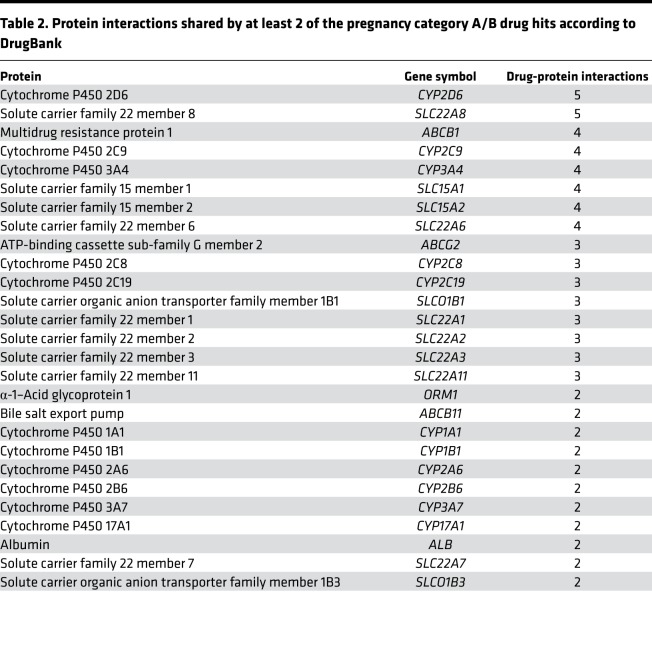
Using DrugBank (31) as a reference, we identified 27 proteins that interact with at least 2 of the drug hits (Table 2). By using these interactions to construct a network (Figure 2B), we can visualize the shared interactions between the drug hits (with no interactions recorded for the 3 contrast agents). Common proteins include members of the cytochrome P450 family (most commonly CYP2D6), members of solute carrier family 22, and multidrug resistance protein 1 (ABCB1). Cytochrome P450 1B1 (CYP1B1) is the only common protein that is also shared by the PTB signature.
Table 2. Protein interactions shared by at least 2 of the pregnancy category A/B drug hits according to DrugBank.
Inflammation-induced mouse model validation of lansoprazole.
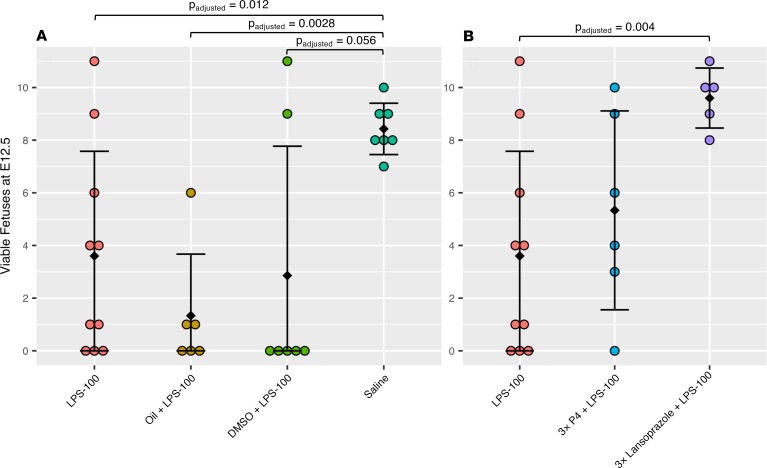
Despite having the highest reversal score, cefotaxime shares few protein interactions with the other drug hits. Lansoprazole has the next highest reversal score. Moreover, lansoprazole is available over the counter, whereas cefotaxime requires a prescription. Lansoprazole has also been connected to the stress-response protein heme oxygenase-1 (HO-1) (32), and deficiencies in HO-1 have been linked with pregnancy disorders (33). Thus, we focused our validation efforts on lansoprazole. We used a mouse model of inflammation using LPS to induce fetal wastage in pregnant mice (30). In addition to lansoprazole, we chose to test P4 as a positive control, because it is currently indicated for the prevention of sPTB. The counts of viable mouse fetuses at E12.5 for each treatment group are summarized in Figure 3. The number of viable mouse fetuses at E12.5 for each treatment group is summarized in Figure 3. Pregnant mice treated with 100 µg/kg LPS (LPS-100) had significantly (P < 0.05) fewer viable fetuses compared with the saline group, demonstrating the effectiveness of the LPS-induced model. Treatment with oil (oil + LPS-100) alone (P4 vehicle) had no effect on fetal viability. The positive control group, which received 3 consecutive daily doses of P4 in addition to the LPS treatment (3xP4 + LPS-100), showed some effectiveness at recovering the number of viable fetuses at E12.5. Finally, in our lansoprazole trials, we compared LPS-100 alone to the lansoprazole-treated group, which also received 3 consecutive daily doses of lansoprazole alongside the LPS treatment (3xlansoprazole + LPS-100). The group treated with lansoprazole showed a significantly (P < 0.05) increased number of viable fetuses compared with the group treated with LPS-100 alone. Treatment with 5% DMSO (DMSO + LPS-100) alone (lansoprazole vehicle) had no effect on fetal viability.
Figure 3. Results from LPS-induced inflammation mouse model of fetal wastage.
Colored circles represent results from independent mouse pregnancies. Error bars represent mean ± SD. Comparisons were made using the Student’s 2-sided t test. (A) Comparisons between pregnancies receiving LPS-100 (n = 10), oil + LPS-100 (n = 6), DMSO + LPS-100 (n = 7), and saline (n = 7) showed a significantly reduced number of viable fetuses at E12.5 in the LPS-100 group compared with the saline group and no significant differences between the LPS-100 group and either of the vehicle groups (oil + LPS-100 and DMSO + LPS-100). (B) The LPS-100 group compared with the P4-positive control group (3xP4 + LPS-100) and the lansoprazole treatment group (3xlansoprazole + LPS-100). The P4-positive control group showed some effectiveness, while the lansoprazole treatment was significantly effective in increasing the number of viable fetuses compared with the LPS-100 group.
Discussion
Considering both the prevalence of PTB and the sparsity of preventative approaches, finding new treatments should be a priority. This study identified existing drugs for repositioning by applying a computational approach leveraging transcriptomics data. By comparing the differential gene expression signature for sPTB (derived from maternal blood samples) with the differential gene expression profiles of drug experiments (derived from human cell lines), we found 83 drugs whose profiles were significantly (FDR < 0.05) reversed compared with sPTB. As the data were derived from human gene expression, the physiology of sPTB should be especially represented in our drug predictions. Thirteen of these drugs — benzathine benzylpenicillin, cefotaxime, chlortalidone, clotrimazole, folic acid, iodixanol, iohexol, iopamidol, lansoprazole, maprotiline, metformin, P4, and rifabutin — belong to pregnancy category A or B, indicating no known risk in pregnant women.
P4, clotrimazole, metformin, and folic acid have shown some efficacy against PTB in past studies. P4 treatment has been shown to be effective in reducing the rate of sPTB in cases where pregnant women have short cervixes (11). Clotrimazole, an antifungal with no teratogenic effects (34), is used to treat yeast and fungal infections. An analysis of 17 years of Hungarian births found that mothers receiving clotrimazole treatment during pregnancy tended to have longer pregnancies, with a significant reduction in the rate of PTB, suggesting a protective effect that could not be explained by other factors (35). Metformin, an antidiabetic, has been found to reduce the rate of PTB compared with placebo in women with polycystic ovary syndrome (36). Folic acid, commonly taken during early pregnancy, was observed to have a possible reduction of PTB in another Hungarian population study (37).
We have previously shown that the administration of lansoprazole can induce the stress-response protein HO-1, which may account for its gastroprotective effects (32). Recently, we have demonstrated that administration of pravastatin can induce the expression of HO-1 and reduce pregnancy loss in HO-1–deficient mice (38). In HO-1–deficient pregnancies, there is abnormal development of the placental vasculature (39), with small litter sizes, increased maternal diastolic pressures, and increased plasma sFlt-1 levels (33), and we speculated that targeting HO-1 expression may have potential in preventing recurrent pregnancy loss. Therefore, we performed preliminary studies to evaluate the protective effects of lansoprazole using an LPS-induced inflammation mouse model of fetal wastage. We found an increase in the number of viable fetuses in LPS-treated pregnant dams given lansoprazole. These promising results show the potential of lansoprazole to prevent inflammation and have potential for the prevention of sPTB.
Lansoprazole and P4 both interact with CYP1B1, a cytochrome that is upregulated in the PTB signature (8). The encoding gene CYP1B1 is involved in estrogen hormone synthesis, and 3 genotypes (CYP1B1*2, CYP1B1*3, and CYP1B1*7) have been found to be significantly more expressed in preterm labor patients in a prior study (40). Further investigation of the interactions of the drug hits presented here could provide insight into the etiology of PTB. Given the existing success of P4 and our initial validation trials on lansoprazole, the rest of the drug hits identified should be treated as repositioning candidates. In particular, clotrimazole, maprotiline, and metformin interact with several of the same proteins as lansoprazole and P4 and thus could have a similar mechanism of action in the treatment of sPTB.
Our study has several limitations. As noted by Vora et al., the sPTB gene expression signature was the product of a meta-analysis of 3 studies with varying designs and may have been influenced by confounding factors (8). The PTB gene expression signature that is used in the analysis is based primarily on mRNA differences from maternal blood samples collected at the time of threatened preterm labor and during preterm labor, with only few gene differences being contributed from data collected from asymptomatic women. Therefore, the signature represents, in large, after the fact effects in maternal circulation. Ideally, the signature would be derived from asymptomatic women only, which can be obtained from GSE59491. Despite the fact that this signature is very limited (26 genes, only 17 of which are in the drug database), through a sensitivity analysis we do observe a nominally significant reversal for lansoprazole (P = 0.021). A differential gene expression signature for sPTB assembled from studies with larger better phenotyped cohorts could result in a better prediction of therapeutics. Another limitation is the overlap between the gene sets of the sPTB signature and the genomics databases. Although there was a relatively large overlap between the sPTB signature and CMap, there was only a small overlap (16 genes) between the sPTB signature and genes measured in LINCS. The LINCS database measured approximately 66,000 profiles, 10 times more than CMap, but only measured 978 genes. By not using LINCS, we may be excluding other promising candidates for drug repositioning. Additionally, the data from CMap and LINCS were derived from cancer cell lines; it would be informative to see drug effects in relevant reproductive tissues. Finally, our validation model was an inflammation-induced fetal wastage mouse model demonstrating the similar therapeutic effects of P4 and lansoprazole. Although LPS elicits a strong maternal inflammatory response that results in preterm delivery and fetal death in a murine model, it is only one model, with considerable differences from human pregnancy. Additional testing in different animal models of PTB is needed to further characterize the potential to prevent sPTB.
Ideally, lansoprazole or any other potential therapeutics could be evaluated in women at high risk for PTB. However, there is still an open question in the field for predicting PTB. Identifying women who could be at risk would be an important step before deciding on any preventative measures. Although there are known risk factors of PTB (including history of a prior PTB, a short cervical length, or a raised fetal fibronectin concentration) (5), no definitive diagnostic test exists to our knowledge. Therefore, in addition to improving therapeutic strategies for PTB, diagnostics need to be improved.
In summary, we applied a computational drug repurposing pipeline to identify potential novel therapeutics for the prevention of PTB. In addition to P4, the only drug currently used to prevent recurrent sPTB, we identified 12 other plausible candidates with no known safety risk in pregnancy. We validated lansoprazole in pilot studies on an LPS-induced inflammation mouse model of fetal wastage. These results demonstrate the therapeutic potential of the PPI lansoprazole and warrant further investigation into the other drug hits identified by our analysis.
Methods
Study design
We developed and applied a bioinformatics approach, leveraging publicly available transcriptomic data to identify potential new treatments for sPTB. This computational pipeline scored the reversal between 2 lists of differentially expressed genes, with the hypothesis that an sPTB-drug pair with a negative reversal score indicated the drug could be therapeutic for sPTB. We scored the gene expression signature of sPTB with each drug in CMap, resulting in a list of 83 sPTB-drug pairs with a significant (FDR < 0.05) reversal score. We further narrowed down the list by focusing on 13 drugs belonging to pregnancy categories A or B and then selected lansoprazole for validation. We performed initial mouse model studies using an LPS-induced inflammation model (30).
Gene expression signature of sPTB
Recently, our group published a cross-study meta-analysis to identify genome-wide differential gene expression signals in sPTB (8). We found 3 studies (GSE46510, GSE59491, and GSE73685) relating to sPTB from GEO. These studies analyzed a total of 339 whole blood samples from women delivering prematurely (n = 134) or at term (n = 205). The 3 studies were aggregated together and then corrected for batch effects, resulting in a normalized data set of 4,648 genes after applying a FDR of 0.1. After applying a threshold of a 1.3-fold increase or decrease in differential gene expression, the gene list was further reduced to a final set containing 210 genes (65 upregulated and 145 downregulated; Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133761DS1).
Gene expression profiles of drugs
Differential gene expression profiles for drugs were collected from 2 complementary sources: CMap (GSE70138) (17) and the LINCS L1000 data set (29). Both are collections of differential gene expression profiles of cultured human cells treated with a variety of small-molecule perturbagens. Using DNA microarrays to assay mRNA expression, the creators of CMap generated a broad database cataloging the effects of drugs on genes (17). CMap is a deep, genome-wide profiling of the effects of approximately 1300 drugs on a variety of cell lines. The LINCS L1000 data set can be characterized as a shallow profiling (978 genes) of over 10,000 drugs. Although the depth of the CMap data set and the breadth of the LINCS L1000 data set provide 2 complementary views into the effects of drugs on gene expression, the genome-wide aspect of the CMap was crucial to our study. Of the 210 differentially expressed genes identified by the sPTB meta-analysis study, 159 were also profiled in CMap, while only 16 were profiled in LINCS. As a result, we focused our study on combining the sPTB gene expression signature with the drug gene expression profiles from CMap.
Computational gene expression pattern matching
To identify potential therapeutic drugs, we used a nonparametric rank-based method to identify candidates based on differential gene expression profiles (18, 19, 21, 23). We hypothesized that drugs with gene changes that are the reverse of those observed in sPTB can have a therapeutic effect in preventing sPTB. Based on the Kolmogorov-Smirnov statistic, this type of analysis was originally suggested by Lamb et al. in their original publication of the CMap database (17). We extended and applied the methodology to specifically identify new uses for existing drugs. Several successful applications of this approach were used in the context of inflammatory bowel disease (18, 19), dermatomyositis (20), and liver cancer (21, 23).
We used the build 02 version of CMap to obtain genome-wide gene expression profiles (relative to controls) from a variety of cell lines treated with small-molecule perturbagens (17). This database provided differential gene expression profiles (~22,000 genes) for 1309 drugs/compounds cultured in up to 5 different cell lines. A total of 6100 drugs were tested with variations in both concentrations and durations of treatment. We applied a prefiltering step for the profiles similar to a previous computational drug repurposing pipeline (21).
Of the 210 differentially expressed genes identified by the cross-study meta-analysis, 159 (44 upregulated and 115 downregulated; Supplemental Table 2) were present in the drug gene expression profiles in CMap. Drugs were given scores based on the relationship between their differential gene expression profiles relative to the sPTB gene expression profile. A negative score indicated a reverse profile between the drug and sPTB, where the upregulated genes in sPTB were lower ranked (more downregulated) and the downregulated genes in sPTB were higher ranked (more upregulated) in the drug profiles. Conversely, a positive score indicated a similar profile between the drug and sPTB. Significances of scores were obtained by a comparison to a distribution of scores generated from random permutations and then adjusted for multiple comparisons; only scores with FDR < 0.05 were kept. For drugs with multiple gene expression profiles (from testing on different cell lines and dosages), we kept the profile with the lowest negative (most reversed) score.
LPS-induced inflammation mouse model
Animals.
Adult male (6- to 8-week-old) and female (8- to 10-week-old) FVB/n wild-type mice were obtained from Charles River Laboratories. Mice were allowed food (Teklad Rodent Diet 2018, Envigo) and prefilled acidified drinking water (Aquavive, Innovive) ad libitum. Twenty-four hours after breeding, gestational ages were determined by the presence of a vaginal plug representing E0.5.
Reagents
LPS.
A stock solution of 1 μg/μL LPS (E. coli O55:B5 LPS [Sigma-Aldrich]) was prepared in saline. 500 μL or 100 μg/kg BW was injected i.p. into each pregnant dam.
P4.
A stock solution of 20 mg/mL P4 (Sigma-Aldrich) was prepared in mineral oil. 100 μL or 2 mg P4 was injected i.m. into each pregnant dam.
Lansoprazole.
A stock solution of 80 mg/mL lansoprazole (Sigma-Aldrich) was prepared in 100% DMSO. A working solution of 4 mg/mL in 5% DMSO (or 24 mg/kg BW) was then made by serial dilutions with saline, and 150 μL of this solution was injected i.p. into each pregnant dam. From prior experience, we used 5% DMSO as the vehicle at the doses used, because lansoprazole is not soluble in mineral oil (32). The dose of lansoprazole used for this study was equivalent to the typical doses of 15–30 mg per day used in humans (41).
Mouse model study design.
For 3 consecutive days starting at E6.5, P4, oil, lansoprazole, or DMSO was administered as described above. At E7.5, LPS was administered i.p. 2 hours after treatment with P4, oil, lansoprazole, or 5% DMSO.
Assessment of viable fetuses and resorbed concepti.
At E12.5, the number of viable fetuses and resorbed concepti was recorded for each dam. As described by Zenclussen et al., resorbed concepti were identified by their hemorrhagic/necrotic appearances and absence of fetuses (42).
Data and materials availability
All of the data used for analysis of this manuscript is publicly available and referenced in the Methods section. The code for the computational pipeline was adapted from ref. 21.
Statistics
Reversal scores were generated for sPTB-drug pairs using a nonparametric, rank-based method based on the Kolmogorov-Smirnov test. More details on this method can be found in previous references (17, 18, 21). Reversal score P values were obtained by comparing with a distribution of 100,000 random scores and then adjusted via FDR for multiple comparisons. An individual random score was generated by selecting a random subset of genes matching the disease signature (44 ranked upregulated genes, 115 ranked downregulated genes) from the entire CMap gene list to act as a proxy signature and then compared with a random CMap drug for score calculation. To ensure that the significance of the score was specific to the drug in question, for our candidate drug, lansoprazole, we also compared 1000 random input signatures to the lansoprazole profile. The PTB reversal score for lansoprazole was more extreme than each of the 1000 generated random scores for lansoprazole, highlighting the significance of the findings. For testing the distributions in the mouse model studies, comparisons were made using the 2-tailed Student’s t test and adjusted via FDR for multiple comparisons. A P value of less than 0.05 was considered significant.
Study approval
All mouse model studies were approved by Stanford University’s Institutional Animal Care and Use Committee and conducted with strict adherence to the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
BLL and MS conceived the study. BLL and MS implemented the computational pipeline and performed data analysis. SI, RJW, and DKS carried out the animal studies. All authors wrote and edited the manuscript.
Supplementary Material
Acknowledgments
The authors thank Aolin Wang, Idit Kosti, and Bianca Vora for useful discussions. This project was in part supported by the March of Dimes and NIH grant NLM K01LM012381. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the March of Dimes.
Version 1. 02/13/2020
Electronic publication
Footnotes
Conflict of interest: MS is a scientific advisor to twoXAR. A US provisional patent (application serial number 62/821,408) has been filed based on this work.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(3):e133761.https://doi.org/10.1172/jci.insight.133761.
Contributor Information
Brian L. Le, Email: Brian.Le@ucsf.edu.
Sota Iwatani, Email: sotaiwa@stanford.edu.
Ronald J. Wong, Email: rjwong@stanford.edu.
David K. Stevenson, Email: dstevenson@stanford.edu.
Marina Sirota, Email: marina.sirota@ucsf.edu.
References
- 1.Liu L, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet. 2016;388(10063):3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe H, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379(9832):2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 3.Mwaniki MK, Atieno M, Lawn JE, Newton CR. Long-term neurodevelopmental outcomes after intrauterine and neonatal insults: a systematic review. Lancet. 2012;379(9814):445–452. doi: 10.1016/S0140-6736(11)61577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spittle A, Orton J, Anderson P, Boyd R, Doyle LW. Early developmental intervention programmes post-hospital discharge to prevent motor and cognitive impairments in preterm infants. Cochrane Database Syst Rev. 2012;12:CD005495. doi: 10.1002/14651858.CD005495.pub3. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero R, Espinoza J, Gonçalves LF, Kusanovic JP, Friel LA, Nien JK. Inflammation in preterm and term labour and delivery. Semin Fetal Neonatal Med. 2006;11(5):317–326. doi: 10.1016/j.siny.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frascoli M, et al. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci Transl Med. 2018;10(438):eaan2263. doi: 10.1126/scitranslmed.aan2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vora B, et al. Meta-analysis of maternal and fetal transcriptomic data elucidates the role of adaptive and innate immunity in preterm birth. Front Immunol. 2018;9:993. doi: 10.3389/fimmu.2018.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berghella V, Rafael TJ, Szychowski JM, Rust OA, Owen J. Cerclage for short cervix on ultrasonography in women with singleton gestations and previous preterm birth: a meta-analysis. Obstet Gynecol. 2011;117(3):663–671. doi: 10.1097/AOG.0b013e31820ca847. [DOI] [PubMed] [Google Scholar]
- 10.Conde-Agudelo A, et al. Vaginal progesterone is as effective as cervical cerclage to prevent preterm birth in women with a singleton gestation, previous spontaneous preterm birth, and a short cervix: updated indirect comparison meta-analysis. Am J Obstet Gynecol. 2018;219(1):10–25. doi: 10.1016/j.ajog.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca EB, Celik E, Parra M, Singh M, Nicolaides KH, Fetal Medicine Foundation Second Trimester Screening Group Progesterone and the risk of preterm birth among women with a short cervix. N Engl J Med. 2007;357(5):462–469. doi: 10.1056/NEJMoa067815. [DOI] [PubMed] [Google Scholar]
- 12.Romero R, et al. Vaginal progesterone for preventing preterm birth and adverse perinatal outcomes in singleton gestations with a short cervix: a meta-analysis of individual patient data. Am J Obstet Gynecol. 2018;218(2):161–180. doi: 10.1016/j.ajog.2017.11.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norwitz ER, Caughey AB. Progesterone supplementation and the prevention of preterm birth. Rev Obstet Gynecol. 2011;4(2):60–72. [PMC free article] [PubMed] [Google Scholar]
- 14.Meis PJ, et al. Prevention of recurrent preterm delivery by 17 alpha-hydroxyprogesterone caproate. N Engl J Med. 2003;348(24):2379–2385. doi: 10.1056/NEJMoa035140. [DOI] [PubMed] [Google Scholar]
- 15.Reichert JM. Trends in development and approval times for new therapeutics in the United States. Nat Rev Drug Discov. 2003;2(9):695–702. doi: 10.1038/nrd1178. [DOI] [PubMed] [Google Scholar]
- 16.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 17.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 18.Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3(96):96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley JT, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3(96):96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho HG, Fiorentino D, Lewis M, Sirota M, Sarin KY. Identification of alpha-adrenergic agonists as potential therapeutic agents for dermatomyositis through drug-repurposing using public expression datasets. J Invest Dermatol. 2016;136(7):1517–1520. doi: 10.1016/j.jid.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, et al. Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling. Gastroenterology. 2017;152(8):2022–2036. doi: 10.1053/j.gastro.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahchan NS, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3(12):1364–1377. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, et al. Reversal of cancer gene expression correlates with drug efficacy and reveals therapeutic targets. Nat Commun. 2017;8:16022. doi: 10.1038/ncomms16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe JH, Ertelt JM, Xin L, Way SS. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature. 2012;490(7418):102–106. doi: 10.1038/nature11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mor G, Aldo P, Alvero AB. The unique immunological and microbial aspects of pregnancy. Nat Rev Immunol. 2017;17(8):469–482. doi: 10.1038/nri.2017.64. [DOI] [PubMed] [Google Scholar]
- 26.Mor G, Cardenas I, Abrahams V, Guller S. Inflammation and pregnancy: the role of the immune system at the implantation site. Ann N Y Acad Sci. 2011;1221:80–87. doi: 10.1111/j.1749-6632.2010.05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gomez-Lopez N, StLouis D, Lehr MA, Sanchez-Rodriguez EN, Arenas-Hernandez M. Immune cells in term and preterm labor. Cell Mol Immunol. 2014;11(6):571–581. doi: 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017;171(6):1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aisemberg J, Vercelli CA, Bariani MV, Billi SC, Wolfson ML, Franchi AM. Progesterone is essential for protecting against LPS-induced pregnancy loss. LIF as a potential mediator of the anti-inflammatory effect of progesterone. PLoS One. 2013;8(2):e56161. doi: 10.1371/journal.pone.0056161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz-Geske S, Erdmann K, Wong RJ, Stevenson DK, Schröder H, Grosser N. Molecular mechanism and functional consequences of lansoprazole-mediated heme oxygenase-1 induction. World J Gastroenterol. 2009;15(35):4392–4401. doi: 10.3748/wjg.15.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Wong RJ, Kalish FS, Nayak NR, Stevenson DK. Effect of heme oxygenase-1 deficiency on placental development. Placenta. 2009;30(10):861–868. doi: 10.1016/j.placenta.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czeizel AE, Tóth M, Rockenbauer M. No teratogenic effect after clotrimazole therapy during pregnancy. Epidemiology. 1999;10(4):437–440. doi: 10.1097/00001648-199907000-00016. [DOI] [PubMed] [Google Scholar]
- 35.Czeizel AE, Fladung B, Vargha P. Preterm birth reduction after clotrimazole treatment during pregnancy. Eur J Obstet Gynecol Reprod Biol. 2004;116(2):157–163. doi: 10.1016/j.ejogrb.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Vanky E, DE Zegher F, Díaz M, Ibáñez L, Carlsen SM. On the potential of metformin to prevent preterm delivery in women with polycystic ovary syndrome - an epi-analysis. Acta Obstet Gynecol Scand. 2012;91(12):1460–1464. doi: 10.1111/aogs.12015. [DOI] [PubMed] [Google Scholar]
- 37.Czeizel AE, Puhó EH, Langmar Z, Acs N, Bánhidy F. Possible association of folic acid supplementation during pregnancy with reduction of preterm birth: a population-based study. Eur J Obstet Gynecol Reprod Biol. 2010;148(2):135–140. doi: 10.1016/j.ejogrb.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Tsur A, et al. Pravastatin improves fetal survival in mice with a partial deficiency of heme oxygenase-1. Placenta. 2019;75:1–8. doi: 10.1016/j.placenta.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Azuma J, Kalish F, Wong RJ, Stevenson DK. Maternal heme oxygenase 1 regulates placental vasculature development via angiogenic factors in mice. Biol Reprod. 2011;85(5):1005–1012. doi: 10.1095/biolreprod.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mustafa M, et al. Genetic polymorphisms in Cytochrome P 4501B1 and susceptibility to idiopathic preterm labor in North Indian population. Clin Biochem. 2013;46(18):1812–1815. doi: 10.1016/j.clinbiochem.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 41.Andersson T, Holmberg J, Röhss K, Walan A. Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br J Clin Pharmacol. 1998;45(4):369–375. doi: 10.1046/j.1365-2125.1998.t01-1-00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zenclussen ML, et al. Haem oxygenase-1 dictates intrauterine fetal survival in mice via carbon monoxide. J Pathol. 2011;225(2):293–304. doi: 10.1002/path.2946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.