Abstract
Acute kidney injury (AKI) and chronic kidney diseases are associated with high mortality and morbidity. Although the underlying mechanisms determining the transition from acute to chronic injury are not completely understood, immune-mediated processes are critical in renal injury. We have performed a comparison of 2 mouse models leading to either kidney regeneration or fibrosis. Using global gene expression profiling we could identify immune-related pathways accounting for the majority of the observed transcriptional changes during fibrosis. Unbiased examination of the immune cell composition, using single-cell RNA sequencing, revealed major changes in tissue-resident macrophages and T cells. Following injury, there was a marked increase in tissue-resident IL-33R+ and IL-2Ra+ regulatory T cells (Tregs). Expansion of this population before injury protected the kidney from injury and fibrosis. Transcriptional profiling of Tregs showed a differential upregulation of regenerative and proangiogenic pathways during regeneration, whereas in the fibrotic environment they expressed markers of hyperactivation and fibrosis. Our data point to a hitherto underappreciated plasticity in Treg function within the same tissue, dictated by environmental cues. Overall, we provide a detailed cellular and molecular characterization of the immunological changes during kidney injury, regeneration, and fibrosis.
Keywords: Immunology, Nephrology
Keywords: Adaptive immunity, Chronic kidney disease, Fibrosis
Acute kidney injury unbiased immune cell landscape reveals a protective role for regulatory T cells during kidney injury, regeneration, and fibrosis.
Introduction
Acute kidney injury (AKI) is clinically characterized by an abrupt loss of renal function, usually occurring in the first 7 days after an initial insult (1). Hospitalized patients, especially those in the intensive care unit, are at a high risk of developing AKI. Despite advances in medical care, AKI patients continue to exhibit high morbidity and mortality (2). Furthermore, several comprehensive metastudies have confirmed that patients suffering from AKI have significantly higher risk of progression to chronic kidney diseases (CKDs) and end-stage renal diseases (3, 4). One of the main causes of AKI is renal ischemia/reperfusion injury (IRI), which can result from a variety of conditions, including major surgery, sepsis, trauma, dehydration and toxic drug damage (5). Reduced renal blood flow ultimately leads to acute cellular injury, particularly the renal proximal tubular epithelial cells. Tubular cell death and associated damage to the endothelium initiates an inflammatory response through the release of cytokines and chemokines followed by recruitment of leukocytes. In addition, disruption of peritubular capillary endothelial cells and loss of interaction with pericytes largely contributes to the propagation of the pathology (6, 7). Depending on the severity and duration of the ischemic injury, surviving tubular epithelial cells begin an adaptive repair process that can result in full recovery of renal function. However, for reasons that remain largely unclear, incomplete repair and persistent damage result in tubular cell loss, vascular rarefaction, chronic inflammation, fibroblast proliferation, and fibrosis (8, 9). Such maladaptive fibrosis is a common feature of many kidney diseases, driving progressive deterioration of renal function and culminating in end-stage renal failure.
A number of the advances in our understanding of the mechanisms of fibrosis have been achieved through the use of mouse models of IRI. These include the critical role of tubular epithelial cell cycle arrest and fibroblast activation as early triggers of fibrosis (8–10). However, the cellular and molecular mechanisms that determine whether an injury proceeds to repair or fibrotic disease remain elusive. Increasing experimental evidence suggests that inflammatory processes play both beneficial and deleterious roles in the initiation, maintenance, and resolution phases following renal injury. Several tissue-resident and -infiltrating cells, from both innate and adaptive immune compartments, have been implicated as important determinants of disease outcomes (11–13). Thus, it is important to understand the immune cell composition of injured kidneys, which factors and cell types are involved in the late phase after ischemia, and how they contribute to fibrosis.
Here, we have performed extensive transcriptional characterization of 2 distinct yet functionally related mouse models of AKI leading to either full regeneration or resulting in chronic disease and fibrosis. Temporal gene expression patterns identified inflammation-associated pathways as the predominant feature distinguishing fibrosis development. Further unbiased cell-specific characterization of isolated kidney immune cells, using single-cell RNA sequencing (scRNA-Seq), resulted in a detailed profile of the kidney immune cell composition during acute injury, repair and fibrotic phases. Surprisingly, we observed marked and unexpectedly diverging roles for kidney-resident regulatory T cells (Tregs) in repair and fibrosis following injury. Although Tregs isolated from regenerating kidneys expressed proangiogenesis- and tissue repair–associated genes, Tregs in fibrotic kidneys expressed proinflammatory- and apoptosis-related transcripts. The functional capacity of Tregs to confer protection in acute injury was experimentally demonstrated through in vivo Treg expansion by IL-2 and IL-33. Altogether, we have defined the immunological landscape spanning acute injury, regeneration, and fibrosis, delineated hitherto unappreciated plasticity and functional relevance of Tregs, and provide molecular targets for the prevention and treatment of kidney fibrosis.
Results
Two murine models of AKI to study kidney regeneration or fibrosis development after IRI.
In order to study the mechanisms that lead to the progression from acute to chronic renal injury, we investigated 2 established murine models of IRI. Warm IRI is a widely used model of AKI in mice, which results in differential outcomes depending upon other experimental variables (14, 15). To study kidney regeneration, we adopted the established unilateral IRI with immediate contralateral nephrectomy (9, 14). Alternatively, to study kidney fibrosis, we also used the unilateral IRI without contralateral nephrectomy. Because the primary goal of our study was to investigate regeneration versus fibrosis development induced by the same ischemic injury, which by definition is a temporal process, we performed pilot studies to determine the appropriate conditions of renal ischemia that generated high reproducibly of outcome without compromising long-term survival. Mice were subjected to 25, 30, or 45 minutes of unilateral ischemia by renal vessel clamping, followed by reperfusion. The best survival rate (>90%) in the unilateral IRI with immediate contralateral nephrectomy was observed in female mice following 30 minutes of ischemia. However, as previously reported (16), the same ischemic time and model lead to survival of only 50% in male mice (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130651DS1). We therefore adopted 30 minutes of unilateral IRI with immediate contralateral nephrectomy in C57BL/6 female mice as a model to study kidney repair and regeneration after injury, hereafter named the “regeneration model” (Supplemental Figure 1B). To study fibrosis development, males and females were subjected to 30 minutes of unilateral renal pedicle clamping, without contralateral kidney nephrectomy. However, only males developed marked fibrosis (data not shown). Survival was over 95% in both cases, presumably due to compensation from the normal contralateral kidney (17). Therefore, we adopted 30 minutes of ischemia without contralateral kidney nephrectomy in C57BL/6 male mice as a model to study kidney fibrosis after IRI injury, hereafter referred to as the “fibrosis model.” We used appropriate age- and sex-matched naive and sham-operated mice as controls to remove gender bias (Supplemental Figure 2, E and F). To characterize the different phases following kidney injury, regeneration, and fibrosis development, we performed a time-course study, starting from day 3 up to day 42 in both models. We evaluated kidney function and histopathology and performed a comprehensive bulk RNA-Seq of whole kidneys, from injured or sex-matched controls. We also measured serum creatinine (S-cre) levels as the standard method to evaluate the decay of kidney function (18). In the regeneration model, S-cre was transiently elevated during the first 7 days after injury and then returned to normal levels. In the fibrosis model, S-cre levels stayed constantly low, throughout all time points analyzed (Supplemental Figure 1C), an expected observation due to the contralateral kidney compensation.
Histological examination of kidney sections revealed that 30 minutes of ischemia induced early renal damage in both models. The damage was most pronounced in the corticomedullary junction, evidenced by disruption of tubular architecture, presence of tubular casts, and marked tubular necrosis, which were not seen in control mice (Supplemental Figure 1D). Importantly, and in agreement with previous reports (14, 15), in the regeneration model, renal damage and alterations in kidney structure returned to basal levels by day 14 and normal kidney architecture was seen on days 28 and 42 after injury (Supplemental Figure 1D). On the other hand, in the fibrosis model, the damage persisted throughout all time points analyzed. Persistent damage and apoptotic cell death in the kidney are some of the factors that can lead to the development of tissue interstitial fibrosis (19). Indeed, in the fibrosis model, we observed increased collagen deposition in the inner and outer medullary regions compared with sham controls (Supplemental Figure 1, E and F). We confirmed these findings by morphometric quantification (Supplemental Figure 1G). Notably, we did not observe any increase in collagen deposition in the regeneration model, for all time points analyzed, compared to sham and naive controls (data not shown). We further confirmed fibrosis evolution by α–smooth muscle actin (α-SMA) immunostaining (Supplemental Figure 1H). Morphometric quantification revealed a significant increase in positive staining, at all time points analyzed in the fibrosis model, compared with sham controls. In contrast, in the regeneration model, only a transient increase in α-SMA staining was seen, which returned to basal levels after day 14 (Figure 1, A and B). When interpreting these data is important to be aware that both gender and nephrectomy are influencing the fate decision toward regeneration or fibrosis. These confounders could influence the findings that are reported here; nevertheless, the main goal of the study was to independently study these 2 distinct disease outcomes.
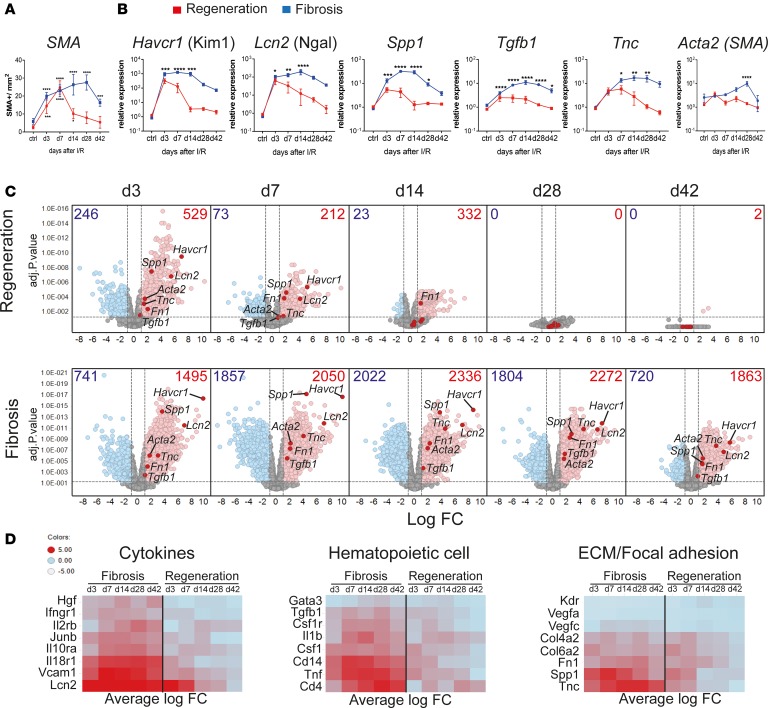
Figure 1. Kidney regeneration and fibrosis characterization after ischemia/reperfusion injury (IRI) reveal sustained dysregulation in immune system–related pathways during kidney fibrosis.
Time course after kidney injury in regeneration and fibrosis models. Mice were subjected to 30 minutes of unilateral left renal IRI, with (regeneration) or without contralateral kidney nephrectomy (fibrosis). (A) Morphometric quantification of α-smooth muscle actin (SMA) staining on kidney sections at indicated time points from regeneration (red line) and fibrosis (blue line). (B) qPCR analysis of the indicated genes, on regeneration (red line) and fibrosis (blue line) samples for the indicated time points, as fold change relative to control samples. (C) Volcano plots showing the fold change (FC) versus P value of differentially expressed (DE) genes in regeneration (top) and fibrosis (bottom) for the indicated time points (in days), compared with age- and sex-matched naive and sham control mice. Numbers indicate upregulated (red) FC > 2, or downregulated genes (blue) FC < 2; P < 0.05. Selected kidney injury and fibrosis biomarker genes are highlighted. (D) Heatmaps showing the average FC of selected upregulated genes from key regulated pathways. Results are representative of 1 (regeneration model) or 2 (fibrosis model) independent experiments, with 3 to 5 mice per time point/condition. Mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 compared with controls by 2-way ANOVA followed by Dunnett’s post hoc test for multiple comparisons (A and B).
Overall, we demonstrated that a single acute ischemic injury could induce acute tubular damage with distinct long-term outcomes. We therefore decided to use these 2 murine models of IRI to study kidney regeneration versus long-term development of fibrosis throughout the rest of the study.
In order to identify the transcriptional changes that correlate with kidney regeneration versus fibrosis development, we performed time-course bulk RNA-Seq from whole kidneys. Principle component analysis (PCA) segregated regenerating and fibrosing samples into 2 distinct groups, while sham and naive controls grouped together (Supplemental Figure 2A). Early after injury (day 3), regeneration and fibrosis were very similar, as indicated by close proximity in the PCA plot. However, starting from day 7, differences between the 2 became more apparent and the samples started to segregate. The clearest separation between regeneration and fibrosis samples occurred on day 14. By day 42, regeneration samples had returned to the basal state (indicated by close proximity to naive and sham controls), whereas the fibrosis samples were distantly located from them, revealing their differences (Supplemental Figure 2A). Differentially expressed (DE) gene analysis revealed that in regeneration most of the genes peaked at early time points (day 3 and 7), and returned to basal levels 14 days after IRI (Figure 1C), in agreement with the histological findings. Conversely, in the fibrosis model, we observed not only a higher number, but also maintained expression of DE genes throughout the time points analyzed. This suggested that constant and ongoing processes were happening in the fibrotic but not in the regenerating kidney (Figure 1C). Indeed, known kidney injury biomarkers (Havcr1, Lcn2, and Spp1) (20, 21) were highly upregulated at early time points, in both models, but only remained constantly upregulated at later time points in the fibrosis model, consistent with the presence of persistent injury. Of note, only at later time points in the fibrosis model did we observe upregulation of key extracellular matrix and profibrotic genes such as Tgfb1, Tnc, Acta2 (α-SMA), and Fn1 by RNA-Seq and quantitative real-time PCR (qPCR) (Figure 1, B and C, and refs. 9, 22).
To gain insight into the changes in cellular composition within regenerating and fibrosing kidneys, we assessed the changes in expression of genes known to be expressed in specific kidney cell types, thereby enabling an estimate of their relative abundance over time in both models. As a reference, we used the previously published mouse kidney-specific scRNA-Seq signatures from Park et al. (9, 23). Using this method, we observed decreased expression of proximal tubule epithelial cell genes early after injury, consistent with these cells being the most abundant and affected cell type during ischemic kidney injury. Interestingly, in the regeneration model, this cell type recovered over time, while in fibrosis we saw a progressive decline. On the other hand, the fibroblast cell signature showed an early and transient increase during regeneration, whereas during fibrosis it remained constantly elevated (Supplemental Figure 2B). This specific-cell-type deconvolution analysis further supports kidney regeneration versus fibrosis development in our mouse models.
To characterize major changes related to the biological process between regeneration and fibrosis over all time points, we performed gene enrichment KEGG pathway analysis. Unsupervised hierarchical clustering analysis revealed that the majority of the upregulated pathways were related to the immune system (KEGG category 5, organismal systems) (Supplemental Figure 2C). They showed enhanced and persistent expression during fibrosis, until late time points. Among those, we found that cytokine–cytokine receptor interaction, TNF signaling pathway, chemokine signaling pathway, hematopoietic cell lineage, and IL-17 signaling pathway were most significantly upregulated when ranked by P value (Supplemental Figure 2D). During fibrosis, some key genes from these selected pathways showed a substantially higher fold change (Figure 1D), supporting the notion that persistent inflammation contributes to kidney fibrosis. Overall, this extensive in situ and molecular characterization of regenerating and fibrotic kidneys suggests that the immune system plays a major role in the decision to regenerate or progress to kidney fibrosis.
scRNA-Seq reveals major changes in tissue-resident myeloid and T cells after kidney injury.
To obtain an unbiased characterization of the resident and infiltrating immune cell populations within mouse kidneys in healthy and diseased states, we performed scRNA-Seq of FACS-isolated CD45+ immune cells from naive and injured kidneys. To filter out poor quality cells, we performed several stringent quality control steps, based on the number of detected genes and features per cell, number of unique reads (unique molecular identifiers [UMIs]), and the ratio of mitochondria gene content per cell (Supplemental Figure 3, A and B). The final data set contained 28,767 cells, which passed all quality control steps, with a median of 1,500 unique genes detected per cell (Supplemental Figure 3C). All batches had similar coverage and no batch effects have been observed.
We used the single-cell clustering algorithm SC3 (24) to perform normalization and unsupervised clustering of cells. To determine meaningful clusters, informative genes were selected from a mean dispersion scatter plot using Ndisp/M3Drop (24). The top 10% genes with a significantly increased dispersion were retained. SC3 was then used to determine clusters and marker genes. From the total 11,065 expressed genes, 1,956 were selected as informative. Using this approach, SC3 identified an optimal number of 9 clusters in a t-distributed stochastic neighbor embedding (t-SNE) plot (Figure 2A). However, from those, only 7 clusters demonstrated clear marker genes and a minimum number of cells that allowed a cell type to be identified (Supplemental Figure 3D). In addition, by testing different clustering methods (e.g., Seurat) (25), we found that most methods identified similar cell groups, expressing the same group of marker genes with limited variations in cell separation (data not shown). To define the identity of each cell cluster, we generated an SC3-based cluster-specific marker gene list (Supplemental Table 1) and a heatmap (Supplemental Figure 3D), showing the top 10 marker genes per cluster (area under the receiver operating characteristic curve [AUROC] > 0.8). Each subset expressed a unique set of markers, receptors, and signaling pathway components that were either previously described (26–28) or de novo defined in our data set. Top marker genes (AUROC > 0.8) with high cluster specificity, combined with existing biological knowledge were used to classify cell clusters into cell types. Using this approach the 7 main clusters were identified as: 1, tissue resident macrophages; 2, inflammatory macrophages; 5, B cells; 6, T cells; 7, dendritic cells (DCs)/monocytes; 8, natural killer (NK) cells; and 9, neutrophils/monocytes (Figure 2, A and B). Clusters 3 and 4 contained too few cells to which no specific marker genes could be assigned. To confirm the cluster assignment identity, we compared the marker gene list from each cluster to each of the 249 immune cell bulk RNA-Seq results, from sorted cell population data profiled by the Immgen consortium (http://www.immgen.org), using a slight modification of the algorithm described by Zemmour et al. (ref. 29 and Supplemental Figure 3, E and F). Using this approach, we identified over 20 different immune cell types. However, once related cell types were combined, we observed a high correlation to our annotated cells types. The B, T, and NK cell clusters strongly corresponded with the Immgen cell annotation. The myeloid clusters contained a mixed population of DCs, macrophages, and monocytes. A small population of stromal cells mapped to a minor distant cluster and probably represented contaminating cells. Only a few cells representing neutrophils or eosinophils were identified (Supplemental Figure 3, E and F).
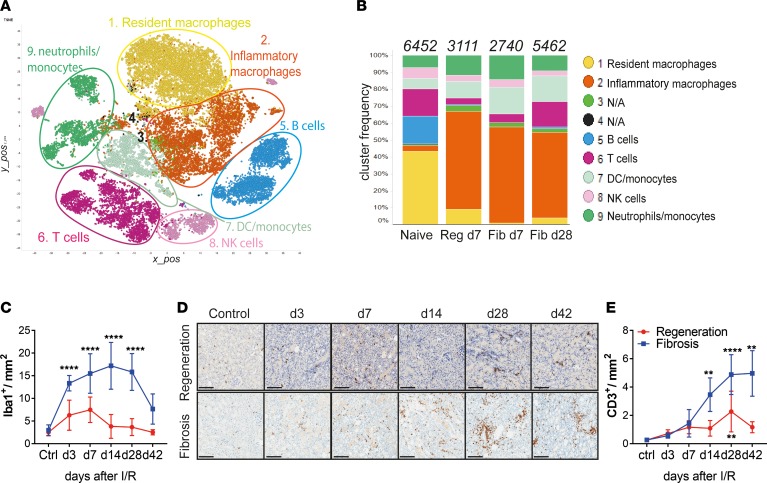
Figure 2. Single-cell RNA-Seq reveals major alterations in tissue-resident myeloid and T cells after kidney injury.
(A) Unsupervised clustering showing 9 major clusters in 2-dimensional t-SNE plots. A total of 28,767 combined CD45+ single cells from naive, regeneration day 7, and fibrosis days 7 and 28, derived from 2 independent biological replicates each. (B) Bar graphs showing the relative frequencies of the respective clusters from A, in each disease model from indicated time points. Numbers on top indicate the total number of cells per condition analyzed. (C) Morphometric quantification of Iba-1+ macrophage immunostaining on kidney sections from regeneration and fibrosis. (D) Representative CD3 immunostaining on kidney sections from regeneration (top) and fibrosis (bottom) for the indicated time points and (E) morphometric quantification. Scale bars: 100 μm. Results are representative of 1 (regeneration model) or 2 (fibrosis model) independent experiments, with 3 to 5 mice per time point/condition. Mean ± SEM. **P < 0.01; ****P < 0.0001 compared with controls by 2-way ANOVA followed by Dunnett’s post hoc test for multiple comparisons (E and F). Reg, regeneration; Fib, fibrosis.
When analyzing the relative frequencies of each population, we observed a dynamic change in the composition of the inflammatory infiltrate after injury. The healthy kidney was dominated by tissue-resident macrophages (43%), followed by B (16%) and T (16%) cells and a relatively equal proportion of DCs, monocytes/neutrophils, and NK cells (Figure 2B). Early after injury (day 7), irrespective of the model (regeneration or fibrosis), we observed a complete depletion of B cells and a partial depletion of T cells. The myeloid compartment predominated after injury in all time points analyzed. Inflammatory macrophages (expressing Apoe, Lyz, Ctsb, Trem2, Cx3cr1, Lamp1, Lyz2, Fcgr3, and others), almost nonexistent in naive kidneys, expanded markedly after injury, regardless of the condition and time point. In contrast, tissue-resident macrophages (expressing Cd14, Cd68, Csf1r, Fcgr4, and Cst3, among others), were present mostly in naive mice and disappeared after injury. We confirmed the macrophage expansion after injury and accumulation during kidney fibrosis by immunohistochemical staining for the macrophage marker Iba-1 (Supplemental Figure 3G). Quantification of Iba-1+ staining showed a significant and persistent increase in kidney macrophages in the fibrosis model (Figure 2C). Importantly, the expression of the inflammatory macrophage marker genes Apoe and Lyz2, in addition to classical macrophage markers, Cd68 and Csf1r, could be detected by qPCR in whole-kidney extracts in the fibrosis model. The expression of those genes was higher at later time points after injury, further supporting the accumulation of inflammatory macrophages in the fibrotic kidney (Supplemental Figure 3H).
Although the inflammatory macrophage population consistently predominated both early and late after injury, the T cell compartment was the only population that was initially depleted and had a marked increased at the late time point, pointing to a potential role for T cells during fibrosis development. To confirm our findings on the T cell expansion, we applied a set of complementary approaches. Immunohistochemical staining of CD3 revealed T cell accumulation in the corticomedullary junction and cortex area, places of concentrated initial damage in injured kidneys (Figure 2D). Morphometric quantifications confirmed a significant increase in CD3+ T cells at later time points in fibrosing kidneys (Figure 2E). This marked alteration in the T cell compartment prompted us to investigate further the role of T cells, specifically the CD4+ compartment, during kidney repair and fibrosis development.
scRNA-Seq of kidney CD4+ T cells reveals the expansion of Th17 T cells and Tregs during fibrosis.
To better understand the contribution of the various CD4+ T cell subsets, we performed scRNA-Seq experiments on CD4+ T cells isolated from kidneys of naive and injured mice by FACS, at selected time points. Following the same quality control steps outlined in Figure 2, the combined data set contained 22,851 cells. t-SNE and unsupervised clustering analysis revealed an optimal number of 5 clusters (Figure 3A). The limited number of marker genes identified per cluster (and lack of kidney-resident CD4+ T cell signatures in the public domain) prevented the cell-type profiling based on the Immgen algorithm. However, based on canonical markers, we could identify the following cell clusters: clusters 1 and 2, Th1/effector cells (Ifng, Nkg7, Ccl4, and Ccl5); cluster 3, naive T cells (Ccr7, Lef1, Satb1, Sell, and others); cluster 4, Th17 cells (Il17a, Tmem176a, and Tmem176b); and cluster 5, Tregs (Ikzf2, Klrg1, Foxp3, IL2ra, and others) (Figure 3B). Extended marker genes are in Supplemental Figure 4A (30). Notably, the Treg cluster was the only population expressing a clear set of marker genes (Supplemental Figure 4A). Among those, classical core Treg genes (Foxp3, IL2ra, and Ikzf2) in addition to tissue-resident Treg genes (Il1rl1, Areg, Ctla4, Tnfrsf9, Tnfrsf4, and Tnfrsf18) were expressed at higher levels in cluster 5, and were absent in other cell types. These genes have been previously assigned to Tregs based on bulk and scRNA-Seq data (29–31). Once more, we could observe major changes in the composition of the CD4+ T cell subsets following kidney injury. The population of naive T cells (cluster 3) was markedly reduced after injury, especially during fibrosis on day 28. Clusters 1 and 2 shared similar markers, suggesting that the same cell type was present in naive and injured kidneys. In addition, 2 new clusters (cluster 4, Th17; and cluster 5, Tregs) expanded substantially after injury, with the highest frequencies during fibrosis (Figure 3C). To further validate the kinetics of T cell accumulation, we applied these CD4+ single-cell-derived signatures (Supplemental Figure 4A) to deconvolute the different T cell populations in the bulk RNA-Seq data. Using the same deconvolution approach as in Supplemental Figure 2B, we could observe the upregulation of some signature genes from Th1 (Ccl4, Tcf7, and Itgb1), Th17 (Rora, Tmem176a, and Tmem176b), and Tregs (Ikzf2, Il1rl1, Gata3, Itgae, Cd83, Rgs2, and Rgs16) in whole-kidney bulk RNA-Seq (Supplemental Figure 4B). Interestingly, we could confirm that the Treg genes had the highest upregulation at later time points during fibrosis. To further elucidate the role of Th17 T cells we performed experiments in the fibrosis model, using an anti–IL-17A neutralizing antibody. However, and in agreement with previously published results (32), anti–IL-17A neutralization did not influence kidney injury or fibrosis (data not shown). Therefore, we decided to further focus our investigation on the role of Tregs in kidney fibrosis.
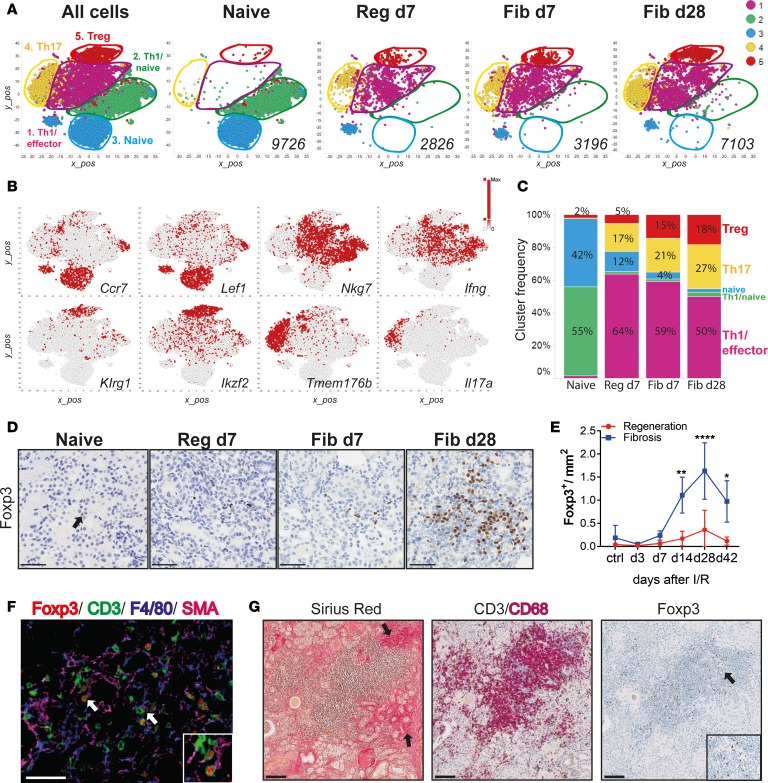
Figure 3. Single-cell RNA-Seq of kidney CD4+ T cells identifies a population of regulatory T cells that expands during fibrosis.
(A) Unsupervised cell clustering colored by the 5 major populations from sorted kidney CD4+ T cells in 2-dimensional t-SNE plots. A total of 22,851 combined kidney single cells (left) and separated by indicated conditions and time points (right). Numbers indicate the total number of cells per condition. (B) t-SNE plots showing selected marker genes for each cluster. Colors indicate the gene expression level. (C) Bar graphs showing the frequency of each cluster (as in A) per condition and time point. (D) Representative Foxp3 immunostaining on kidney sections from naive, regeneration day 7, and fibrosis days 7 and 28, and (E) morphometric quantification. (F) Multicolor immunofluorescence of kidney sections from fibrosis day 28. Arrows indicate Tregs interacting with SMA+ (zoomed area in the square insert at the bottom right) and F4/80+ cells. (G) Kidney biopsies from a patient after transplant with fibrosis, stained with Sirius Red and for CD3/CD68 and Foxp3. Arrows on the left picture indicate fibrotic areas and the right picture shows Treg accumulation (zoomed areas in the small square). Scale bars: 50 μm (D and F) and 100 μm (G). Results are representative of 1 (regeneration model) or 2 (fibrosis model) independent experiments, with 3 to 5 mice per time point/condition. Mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001 compared with controls by 2-way ANOVA followed by Dunnett’s post hoc test for multiple comparisons (E). Reg, regeneration; Fib, fibrosis.
To confirm the Treg expansion during fibrosis, we performed immunohistochemical analysis on kidney sections stained for Foxp3 from both models, at indicated time points after injury (Figure 3D). Morphometric quantification confirmed that Foxp3+ Tregs were significantly increased at later time points, only during fibrosis development (Figure 3E). To further characterize Tregs in the fibrotic kidneys, we performed multiplexed immunofluorescence (IF). IF revealed that Tregs could be found in SMA+ areas in close proximity to F4/80+ macrophages, suggesting a possible interaction. Importantly, F4/80+ macrophages were mostly located in areas of SMA+ staining (Figure 3F). Interestingly, when analyzing human kidney sections from transplanted patients showing significant levels of interstitial fibrosis, we could observe areas of similar immune infiltrates (Figure 3G). Specifically, in the fibrotic areas, we observed intense CD3+ T cells and CD68+ macrophage infiltration. Importantly, in the same areas we also identified Foxp3+ cells (Figure 3G). These results suggest that similar immune infiltrates are present in fibrotic kidneys derived from human renal transplant recipients.
Altogether, these results provide a comprehensive characterization of the immune cell populations infiltrating the kidney at distinct phases after injury and highlight Tregs as a prominent population that accumulated during fibrosis development.
IL-2 complex– and IL-33–mediated Treg expansion confers protection from kidney injury and fibrosis.
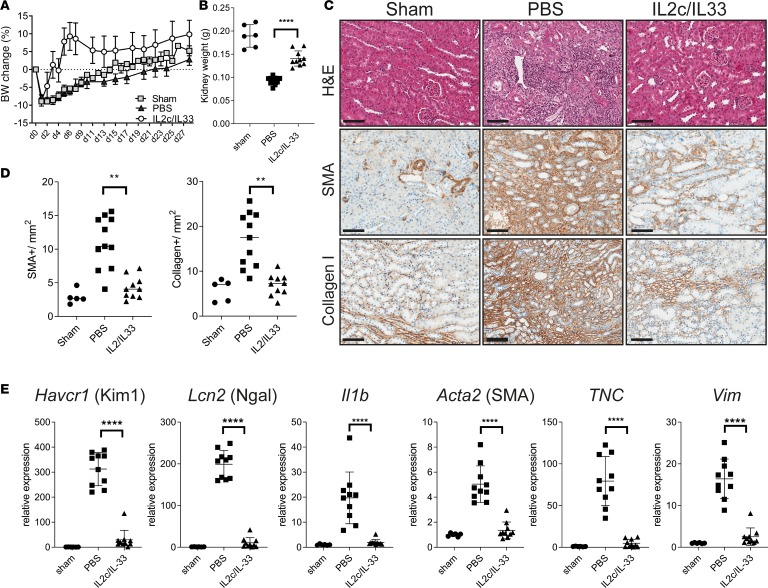
Given that we saw a late Treg accumulation during kidney fibrosis, which apparently was not sufficient to resolve the disease, we reasoned that early Treg expansion may protect from injury and fibrosis. To this end, we first trialed treatment with IL-2 complexed with JES6-1A12 (IL-2c) to expand tissue Tregs in vivo (33–35). In our hands, IL-2c treatment induced an approximately 4-fold Treg expansion in the kidney (Supplemental Figure 5A and ref. 33). In addition, we observed that expression of the IL-33 receptor, ST2 (encoded by Il1rl1), was restricted to Tregs within the CD4+ T cell populations in the kidney (Supplemental Figure 4A) and that ST2 expression was increased in kidney Tregs after injury compared with naive mice or Tregs in the spleen (Supplemental Figure 5B). In line with this, IL-33 treatment of naive mice induced an approximately 3-fold Treg expansion in kidney (Supplemental Figure 5C). Therefore, we adopted a modified version of a previously established protocol to expand kidney Tregs (33, 34), specifically using a combination of IL-2c and IL-33 treatment before fibrosis-inducing kidney injury. Treg-expanded mice had markedly lower body weight loss and significantly lower kidney weight loss following IRI (Figure 4, A and B). Histological analysis and morphometric quantification revealed protection from tissue damage (H&E staining) and absence of marked fibrosis (α-SMA and collagen I staining) in mice treated with Treg-expanding IL-2c/IL-33, compared with PBS-treated mice (Figure 4, C and D). In addition, qPCR analysis confirmed that the expression of markers of kidney injury (Ngal and Kim1), inflammation (Il1b), and fibrosis (Acta2, Tnc, and Vim) had returned to the same levels as those seen in control mice (Figure 4E). Taken together, our results demonstrate that prophylactic Treg expansion confers protection to kidney injury and fibrosis development.
Figure 4. In vivo Treg expansion ameliorates kidney fibrosis.
Male mice were subjected to IRI, fibrosis model. Mice were treated with a mixture of IL-2–IL-2 mAb (IL-2c) and IL-33 (IL-2c/IL-33) for 5 consecutive days, starting from day –3. Samples were harvested and analyzed 28 days after injury. (A) Body weight change graph for the indicated groups. (B) Left kidney weight at termination. (C) Representative images showing H&E, α-SMA, and collagen I staining on kidney sections from sham, PBS, or IL-2c/IL-33–treated mice. Scale bars: 100 μm. (D) Morphometric quantification of α-SMA and collagen I positive staining, from C. (E) qPCR analysis of whole kidneys for the indicated markers of kidney injury Kim1 and Ngal, inflammation (IL-1b) and fibrosis (α-SMA; TNC, tenascin; Vim, vimetin), shown as fold change relative to control samples. Results are a pool of 2 independent experiments; n = 5 for sham; n = 11 for PBS; n = 10 for IL-2c/IL-33. Mean ± SEM. **P < 0.01; ****P < 0.0001 compared with PBS-treated group by 2-tailed Student’s t test.
Markers of kidney-resident Tregs after injury and fibrosis.
To elucidate the mechanisms by which Tregs protected the kidney from injury and fibrosis, we first generated a kidney scRNA-Seq–derived Treg signature, by analyzing the genes upregulated in Foxp3+ cells versus all other Foxp3–cells (Supplemental Figure 6A and Supplemental Table 2). Despite the low coverage inherent in scRNA-Seq experiments, we could still observe genes common to Tregs from naive, regeneration, and fibrosis (such as Foxp3, IL2ra, Klrg1, Areg, Ikzf2, Tnfrsf4, and Tnfrsf18), which thus constituted the “core kidney Treg single-cell signature” (Supplemental Figure 6A). The Treg single-cell signature was dominated by activation markers, which were more highly expressed after injury (Supplemental Figure 6B). Furthermore, when comparing the DE genes between Tregs from naive, regenerating, and fibrosing kidneys, we observed that fibrotic Tregs had higher expression of transcription factors previously linked to fibrosis (Id2, Nfkb, Rgs2, and Junb) (Supplemental Figure 6C and refs. 36, 37).
Next, we performed bulk RNA-Seq analysis on FACS-isolated Tregs and conventional CD4+ T cells (Tconvs) from kidneys and spleens of Foxp3-EGFP reporter mice, at different phases after injury. First, to determine the specific tissue signatures, we compared expression changes between Treg and Tconv genes, for spleen and each kidney disease model (Figure 5, A and B). We could validate the presence of genes from the single-cell–derived Tconv signature of naive T cells (Ccr7, Tcf7, Lef1, and Satb1), Th1 (Ifng, Nkg7, and Ccl5), and Th17 cells (Il17a, Temem176a, and Temem176b) (Figure 5B, genes highlighted in blue). Moreover, previously described Tconv genes (Cd40lg, Fasl, Tnf, Tbx21, IL2, and Bach2) (29, 30) were also upregulated in fibrotic kidneys.
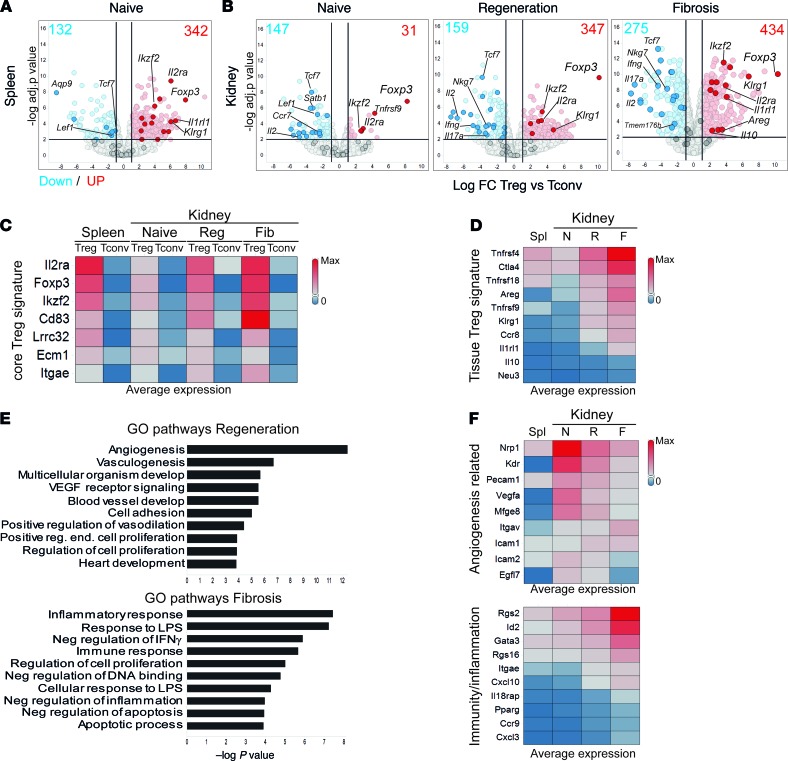
Figure 5. New markers of tissue-resident regulatory T cells during kidney injury and fibrosis.
Bulk RNA-Seq analysis showing detected transcriptional differences. (A and B) Volcano plots with the log fold change (FC) and P value for the comparison between Treg versus Tconv genes for the indicated conditions and tissues. Selected Tconv and core Treg genes are highlighted in blue and red, respectively. Numbers indicate the amount of upregulated (red, FC > 2) and downregulated (blue, FC < –2) genes. (C and D) Heatmap representations showing the average expression (FPKM, fragments per kilobase of exon model per million reads mapped) of core Treg signature genes (C) and tissue Treg genes (D) at the indicated conditions. (E) Gene Ontology (GO) pathway analysis showing the top 10 upregulated pathways in Tregs from regeneration (red, top) and fibrosis (blue, bottom), ranked by P value. (F) Heatmaps showing the expression of selected genes from the pathways in E. N, naive; R, regeneration; F, fibrosis; Spl, spleen.
We then focused on the Treg signatures from spleen and kidneys. We could confirm the expression of genes previously assigned to a core Treg signature (Foxp3, IL2ra, Ikzf2, Cd83, and Itgae), based on the genes upregulated under all conditions (fold change > 2, P < 0.05) in Tregs compared with Tconvs (Figure 5C and refs. 29–31). In addition, when comparing kidney versus lymphoid Treg signatures, we observed that genes previously assigned to a tissue Treg signature (Areg, Il1rl1, Tnfrsf4, Tnfrsf9, Ctla4, and Klrg1) (Figure 5D refs. 29, 30) had indeed higher expression in kidney Tregs compared with splenic Tregs. Furthermore, most genes were also present in our core kidney Treg single-cell signature (Supplemental Figure 6A). We could validate the differential expression of CD25 and KLRG1 between Tregs and Tconvs in spleen and kidney and between naive and injury by flow cytometry (Supplemental Figure 7). These results suggest that a “pan” tissue Treg signature is maintained, irrespective of inflammatory alterations and tissue localization, in agreement with previous reports (29, 30).
To identify changes in Treg signatures after kidney injury, we performed pathway analysis on the upregulated genes in Tregs compared to Tconvs (from Figure 5B), from kidney regeneration and fibrosis (Figure 5E). The fibrotic Treg signature was enriched in pathways related to inflammation and apoptosis (Gata3, Cxcl3, Pparg, Ccr9, Rgs2, and Rgs16), whereas Tregs from regeneration were enriched for angiogenesis-related pathways (Nrp1, Kdr, Mfge8, Vegfa, and Icam1) (Figure 5, E and F), highlighting their differences. Importantly, the transcription factors Id2 and Rgs2, identified in fibrotic Tregs in the scRNA-Seq analysis, were exclusively upregulated in the fibrotic Treg bulk RNA-Seq signature (Figure 5F and Supplemental Figure 6D). These observations indicated that kidney Tregs differentially express a set of response genes, depending on the inflammatory environment.
Discussion
Previous organ-wide studies combined with recent gene expression profiling of mouse kidneys have helped to discover several clinical AKI biomarkers, derived from epithelial and kidney stromal cell genes (38). However, until now, an unbiased and comprehensive characterization of kidney immune cell-type-specific signatures and their contribution to the intrinsic repair and fibrotic mechanisms had not been performed to the best of our knowledge. Here, we combined complementary multiomics approaches to generate a detailed cellular and molecular comparison between kidney regeneration and fibrosis, after ischemic injury in mice. Recent studies have identified early-injury biomarkers with sustained expression during the chronic phase (39–41). Our work complements and extends these findings to the level of global pathway changes. By comparing DE genes upregulated at different phases after initial injury, we have identified key immune-related pathways driving differences between kidney regeneration and fibrosis.
The immune response during IRI is complex, and different immune cells have been shown to play varying roles in acute and chronic pathogenesis following kidney injury (13, 42–44). So far, the composition of the immune cell infiltrates of kidneys has been investigated mainly using conventional approaches, such as histology and immunohistochemistry, and mostly focused on the roles of immune cell populations during the early phase after injury (11, 43). Recent studies using scRNA-Seq have identified marker genes for the majority of renal epithelial cell types, but most tissue-resident and -infiltrating immune cells remained poorly characterized (28, 45, 46), mainly because CD45+ immune cells account for a small fraction (2%–17%) of the total kidney cell population (47). Our study has revealed the complexity and the importance of the immune cell compartment in restoring kidney function following injury. These included major early and late injury-induced changes in kidney-resident macrophages and T cell populations. The changes in macrophage subtypes from a tissue-resident to monocyte-derived population following AKI has also been described by others (48, 49). Interestingly, we saw a major contribution of the T cell response and a highly dynamic adaptation of this population during injury, regeneration, and fibrosis. This response is surprising, as T cell responses are generally thought to be triggered by cognate antigens like self-antigens in autoimmune diseases, such as lupus nephritis or alloantigens in kidney transplantation. In the kidney injury setting, intense levels of danger signals may overcome the requirement for T cell receptor–antigen interaction.
In this regard, it is of high interest that our detailed single-cell analysis and in situ characterization of the CD4+ T cell populations revealed that Tregs were an important population preferentially accumulating in fibrotic mouse kidneys. This special population of tissue-resident Tregs are critically dependent on danger signaling like IL-33 or IL-18 for tissue homing and expansion (50, 51). Interestingly, Tregs have been shown to have a protective role in limiting initial inflammation and cellular injury in different models of renal diseases (33–35, 52, 53). Importantly, our study could extend this observation and showed that early Treg expansion was not only sufficient to protect the kidney from initial damage, but also to prevent late fibrosis development. The mechanisms used by Tregs to induce tissue protection are not fully understood; however, studies in other models of organ injury have elucidated several factors like amphiregulin (AREG) to be involved (51, 54). In agreement with these studies, we could also identify AREG and ST2 upregulation in Tregs in the kidney during injury and fibrosis. Our analysis extends this current picture by showing that Tregs present during kidney regeneration were transcriptionally clearly different from fibrotic Tregs. Whereas the first had a “proangiogenic” and preregenerative signature, fibrotic Tregs showed upregulation of proinflammatory pathways. This hitherto unappreciated plasticity in Treg function indicates that resident Tregs exhibit more complex and likely tissue-specific functions that require further investigation. Beyond the distinct transcripts identified in our study, we speculate that close proximity of Tregs to macrophages following kidney injury holds functional significance, since it has been shown that Tregs can control macrophage phenotype and function to prevent inflammation and promote repair (35, 55, 56). Further studies, focused on targeting fibrotic Tregs and their interactions with macrophages or endothelial cells could shed light on the precise mechanism of Treg-mediated protection during kidney regeneration and fibrosis.
Finally, the presence of large numbers of CD3+CD4+ T cells in human fibrotic kidneys, up to 1 year after the initial injury (39), combined with our own human data, suggests that T cells contribute to the transition from AKI to CKD in humans. Further, in a recent study describing single-cell analysis of immune cells from lupus nephritis patients (57) the authors have identified similar immune cell subsets, with overlapping marker genes, to those our study. These results not only support the relevance of our murine study to humans, but also suggest that the immune composition of autoimmune kidneys shares features with ischemic mouse kidneys, especially in the chronic phase of the disease. This is remarkable, as it suggests that the same immune cell types could be targeted in different kidney diseases. Given that renal transplants undergo an episode of IRI at the time of surgery, they provide a good case to study the response after AKI in conditions in which tissue material is limited (41). Further studies evaluating the presence of Tregs in fibrotic lesions from AKI patients would further confirm these findings and reveal them as an important cell type to target to prevent the progression from AKI to CKD in patients.
Our study, for the first time to our knowledge, comprehensively characterizes gene expression in immune cells isolated from injured kidneys and identifies Treg heterogeneity during the acute and chronic phases after kidney injury. Taken together, the kidney immune cell atlas presented in this study enhances the understanding of the kidney cellular composition and provides information that will help to design targeted therapeutic strategies to promote adaptive renal recovery and minimize progressive fibrosis and CKDs.
Methods
Patients.
The kidney tissue biopsy of a renal transplant recipient was collected at the Cantonal Hospital Baselland, Liestal, Switzerland, in order to determine if there were signs of acute or chronic rejection. The kidney tissue biopsy did not show any signs of acute rejection, but marked interstitial fibrosis and was selected for further immunostaining for CD3, CD68, and Foxp3.
Animals and surgical protocol.
C57BL/6N mice were purchased from Charles River and B6.Cg-Foxp3tm2(EGFP)Tch/J (58) (Foxp3-EGFP) were purchased from The Jackson Laboratory (stock 006772) and bred in house. Male and female mice aged 8 to 12 weeks (weighing 25–35 g) underwent warm IRI, as previously described (14). Two established models of warm IRI were used to induce kidney injury (9, 14). In the first model, female C57BL/6 or B6-Foxp3-EGFP underwent unilateral 30 minutes of ischemia (left kidney) by renal pedicle clamping, with immediate contralateral (right) kidney nephrectomy, defined in this study as the regeneration model. Female, sex-matched, sham-operated mice, without ischemia but with contralateral kidney nephrectomy, and naive nonoperated mice were used as controls. In the second model, to induce kidney fibrosis, male C57BL/6 or B6-Foxp3-EGFP underwent unilateral 30 minutes of ischemia (left kidney) by renal pedicle clamping, without contralateral (right) kidney nephrectomy, defined as the fibrosis model. Sham animals in this model were only subjected to laparotomy, without renal vessel clamping. Male, sex-matched animals were used as sham and naive controls. The animals were kept at a constant body temperature of 37°C using a temperature-adjustable pad. Mice were weighed and scored daily for the first 7 days, and then twice a week, until the end of the experiment (and a percentage weight loss or gain was calculated). Up to 5 mice were used per group in each experimental condition, and 3 sham or naive controls. Following surgery, mice were sacrificed at indicated times per experiment, transcardially perfused with PBS, and biological specimens were retrieved and processed as further described.
IL-33 and IL-2c treatment.
For Treg expansion experiments, mice were injected intraperitoneally with 1 μg of recombinant murine IL-33 (aa 109–266, Novartis) on days 1, 3, and 5 (59). Alternatively, mice were also treated with a combination of 1 μg of recombinant mouse IL-2 (R&D Systems) and 5 μg of anti–mouse IL-2 mAb (JES6-1A12) (BioXCell) at a 1:5 ratio, as previously described (33). For kidney fibrosis experiments, mice received a mixture of IL-33/IL-2/JES6-1A12 (1:1:5 ratio) for 5 consecutive days, starting 3 days before IRI. Kidney tissues were harvested 28 days after injury for histology and qPCR analysis.
Tissue dissociation and cell isolation.
Leukocytes from murine kidneys were isolated as previously described (60). Briefly, kidneys were cut into small pieces and digested in a mixture of 1 mg/mL collagenase IV (Worthington) and 100 μg/mL DNase I (Roche) in DMEM at 37°C for 36 minutes, using an octoMACS tissue dissociator (Miltenyi Biotec). Single-cell suspensions were filtered through a 40-μm cell strainer to remove debris and further washed with PBS. Leukocytes were enriched through 40%–80% Percoll (GE Healthcare) density gradient centrifugation before FACS analysis. Alternatively, kidney CD45+ or CD4+ immune cells were enriched using microbeads (Miltenyi Biotec) before FACS isolation.
Histology, immunohistochemistry, and morphometric evaluation.
Half or a quarter of a kidney from each animal was fixed in 10% normal buffered formalin, processed, and embedded in paraffin. Three-micrometer-thick sections were cut and stained with H&E and Gomori’s Trichrome Green (kit 38016SS1, Leica Biosystems). Automated enzymatic immunohistochemical staining for CD3, CD31, collagen I, Iba-1, Ki-67, and α-SMA) were performed on a Ventana Discovery XT immunostainer (Roche Diagnostics) and Leica Bond RX stainer (Leica Biosystems). Multicolor immunofluorescence staining was performed using a sequential multiplex assay with Opal 4-Color anti-Rabbit Automation IHC Kit and Opal 7-Color Automation IHC Kit, according to manufacturer’s instructions (NEL830001KT and NEL821001KT, PerkinElmer). Slides were mounted with Invitrogen ProLong Gold Antifade (P36930, Thermo Fischer Scientific). Details of the antibodies used are provided in Supplemental Table 3.
All mouse biopsy samples were digitized using a ScanScope XT (Aperio, Leica Biosystems) and VERSA (Leica Biosystems) slide scanners at ×20 and ×40 magnification. Digital images were viewed using Aperio Imagescope v12.1.0.5029 software. Quantitative image analyses were performed using the HALO 2.2 software and the Area Quantification v1.0 and Cytonuclear v1.6 algorithms (Indica Labs).
Flow cytometry and cell sorting.
Single-cell suspensions were stained in FACS buffer (PBS, 2% FBS, 2 mM EDTA) with fluorochrome-labeled antibodies against CD45 (30-F11), CD4 (GK-1.5), CD3 (17A2), CD8 (53-6.7), CD25 (PC61), TCRβ (H57-597), Foxp3 (FJK-16s), and KLRG1 (2F1) from BD Biosciences, BioLegend, and eBioscience and acquired with an LSRFortessa flow cytometer or sorted with a FACSAria Fusion cell sorter (BD Biosciences). Data were analyzed using FlowJo software (Tree Star). For scRNA-Seq, CD45+ cells were sorted as CD45+ live singlets and CD4+ T cells as CD45+CD4+CD8–TCRβ+ live singlets. For Treg bulk RNA-Seq, Tregs were sorted as CD45+CD4+TCRβ+Foxp3–GFP+ live singlets and Tconvs were isolated as CD45+CD4+TCRβ+Foxp3–GFP– live singlets. In all cases, purity exceeded 90%.
RNA isolation and qPCR.
For whole-kidney bulk RNA-Seq and qPCR analysis, whole kidneys were weighed and preserved in RNAlater (Qiagen) solution for further processing. RNA was extracted using an RNeasy micro kit (Qiagen) according to the manufacturer’s instructions. Quality and quantity of RNA was tested with the Agilent 2100 Bioanalyzer using the RNA 6000 Pico Chip and reagents (Agilent). For Treg bulk RNA-Seq, 3 to 5 biological replicates of 3000–5000 Tregs or Tconvs, consisting of a pool of 5 to 10 mice, were FACS isolated in lysis buffer (Qiagen) containing 1% 2-mercaptoethanol (MilliporeSigma) and preserved at –80°C until RNA extraction. cDNA was synthesized using a High-Capacity RNA-to-cDNA kit (Applied Biosystems). Gene expression analysis was determined by qPCR using TaqMan-based assays (Applied Biosystems). Probes used are listed in Supplemental Table 4. Transcript values were normalized to the mouse Gapdh or Tbp housekeeping genes. Data are expressed as fold change ± SEM, relative to sham and naive controls.
Bulk RNA-Seq library preparation and data analysis.
For low-yield RNA samples (primary tissue-resident lymphocytes), SMARTer Stranded RNA-Seq libraries were prepared from 1.6 to 2.5 ng of total RNA, following the manufacturer’s instructions (Takara). Whole-kidney RNA extracts were processed using 250 ng of total RNA as input for library construction with the TruSeq Stranded Total RNA Sample Preparation kit with Ribo-Zero Gold, following the manufacturer’s instructions (Illumina). Libraries were pooled and sequenced to a minimum depth of 30 million on a HiSeq 2500 or 4000 (Illumina). Reads were aligned to the mm10 genome with STAR aligner (version 2.5.2a) (61). Gene-level count data were normalized with the voom R package and further analyzed for differential expression of relevant contrasts with the limma R package (significance cutoffs were FDR < 0.01 and absolute log2 fold change > 1) (62). Before statistical assessment, genes having expression values below a given threshold in a majority of experimental group samples were removed from the subsequent analysis.
Pathway analysis was performed using the output from the differential expression analysis with the GAGE methodology, as implemented in the gage R package (63). The GAGE analysis was run on the KEGG pathway database. The KEGG pathway list was first downloaded from http://www.genome.jp/kegg/pathway.html on April 25, 2018. To be able to assess pathway gene enrichment in each KEGG subgroup independently from one another, a GAGE analysis was run per pathway family with local FDR controls of the enrichment P values. Human Diseases and Drug Development categories were not considered. The pathway to gene identifier mappings was retrieved from the KEGG API REST server (http://www.kegg.jp/kegg/rest/keggapi.html). GO pathway analysis of Treg signatures was done using DAVID (64). Spotfire (TIBCO) was used to generate volcano plots and heatmaps.
scRNA-Seq: 10× Genomics.
FACS-isolated kidney CD45+ and kidney CD4+ cells were resuspended in PBS with 0.05% BSA. Viability was accessed by Trypan blue cell counting and exceeded 90%. Cells (2000–10,000) were loaded on a 10× Genomics Chromium instrument to generate single-cell GEMs. scRNA-Seq libraries were prepared using a GemCode Single Cell 3′ Gel Bead Kit according to the manufacturer’s instructions (10× Genomics). Amplified cDNA products were cleaned up with the SPRIselect Reagent Kit (0.6× SPRI). Indexed sequencing libraries were constructed using the reagents in the Chromium Single Cell 3′ library kit V2 (10× Genomics). The barcoded sequencing libraries were quantified using a Qubit 2.0 dsDNA HS Assay Kit (Invitrogen). The quality of the libraries was accessed on an Agilent 2100 Bioanalyzer using an Agilent High Sensitivity DNA kit. Sequencing libraries were loaded at 10 to 12 pM on an Illumina HiSeq 2500 platform.
scRNA-Seq data analysis.
Raw gene counts obtained from Cell Ranger were processed with an R (3.5.2) pipeline based on scater v1.8.4 (https://bioconductor.org/packages/release/bioc/html/scater.html). The initial total cell numbers were 29,044 CD45+ and 23,704 CD4+ cells. CD45+ and CD4+ sorted cells were processed separately. The data sets comprised 2 to 4 biological replicates for each time point, from a pool of 2 mice each. Low quality cells were initially filtered out based on the following. For CD45+: fewer than 512 expressed genes, fewer than 1024 UMIs, and more than 12% UMIs from mitochondrial genes. For CD4+: fewer than 1024 expressed genes, fewer than 512 UMIs, and more than 10% UMIs from mitochondrial genes. This resulted in 28,767 cells for CD45+ and 22,851 cells for CD4+. Normalization was performed using scran v1.8.4. Informative genes were selected using M3Drop (v3.10.3) (24). CD45+ selection was based on dispersion and the top 10% genes were selected. CD4+ selection was based on dropout rate with cutoff of 0.1 for the significance level. Clustering was performed on the set of informative genes using SC3 (v1.8.0) (24) in the range of 2 to 12 clusters. Results are shown with k = 9 clusters for CD45+ and k = 5 clusters for CD4+. An initial PCA representation on the set of informative genes with d = 50 was obtained using irlba (v2.3.2; https://cran.r-project.org/web/packages/irlba/index.html). Based on this PCA, a t-SNE representation was obtained using Rtsne function (v0.15; https://cran.r-project.org/web/packages/Rtsne/index.html).
Accession number.
The data reported in this paper have been deposited in ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) with accession numbers E-MTAB-7957, E-MTAB-7961, and E-MTAB-8002.
Statistics.
Graphs in the figures represent mean values ± SEM. Statistical tests include Student’s t test and ordinary 1-way and 2-way ANOVA (Turkey’s or Dunnett’s post hoc test for multiple comparisons, respectively), using GraphPad Prism. Significance is indicated in the graphs as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Nonsignificant differences were not indicated.
Study approval.
Human kidney tissue biopsy was obtained from the Institute of Pathology, Cantonal Hospital Baselland, Liestal, Switzerland, and initially collected under the approval of the Ethics Committee Basel (KEK-EKBB-No.2016-01499). Animal handling and all surgical procedures were performed according the animal experimentation laws and guidelines approved by the Swiss Cantonal Veterinary Authority, under license number BS-2785.
Author contributions
FVD designed studies, conducted experiments, analyzed data, and wrote the manuscript. KM, KD, RC, AW, and JL conducted experiments and analyzed data. AL, UN, MB, SG, and SP performed data analysis. GR and GW analyzed data and edited the manuscript. KDM provided the images from kidney tissue biopsy from the transplant patient. MW designed and supervised the study and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Tobias Junt, Ben Roediger, and Till Roehn for carefully reading and editing the manuscript; Sandro Bruno for bioinformatics analysis; Cyril Allard and the ATI flow cytometry core facility for outstanding sorting support; and Claudine Maurer, Alexander Cattini, Corinne Debon, Patrick Gfeller, and Vincent Boehler for excellent technical assistance.
Version 1. 02/13/2020
Electronic publication
Footnotes
Conflict of interest: All authors except KDM are employees of Novartis Pharma AG.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(3):e130651.https://doi.org/10.1172/jci.insight.130651.
Contributor Information
Armelle Lafont, Email: Armelle.Lafont@novartis.com.
Martin Beibel, Email: martin.beibel@novartis.com.
Kea Martin, Email: keamartin@gmail.com.
Katy Darribat, Email: katy.Darribat@novartis.com.
Annick Waldt, Email: annick.waldt@novartis.com.
Ulrike Naumann, Email: ulrike.naumann@novartis.com.
Grazyna Wieczorek, Email: grazyna.wieczorek@novartis.com.
Swann Gaulis, Email: swann.gaulis@novartis.com.
Sabina Pfister, Email: Sabina.Pfister@novartis.com.
Kirsten D. Mertz, Email: Kirsten.Mertz@ksbl.ch.
Jianping Li, Email: jianping.li@novartis.com.
Guglielmo Roma, Email: guglielmo.roma@novartis.com.
Max Warncke, Email: max.warncke@novartis.com.
References
- 1.Chawla LS, et al. Acute kidney disease and renal recovery: consensus report of the Acute Disease Quality Initiative (ADQI) 16 Workgroup. Nat Rev Nephrol. 2017;13(4):241–257. doi: 10.1038/nrneph.2017.2. [DOI] [PubMed] [Google Scholar]
- 2.Hoste EA, Schurgers M. Epidemiology of acute kidney injury: how big is the problem? Crit Care Med. 2008;36(4 suppl):S146–S151. doi: 10.1097/CCM.0b013e318168c590. [DOI] [PubMed] [Google Scholar]
- 3.See EJ, et al. Long-term risk of adverse outcomes after acute kidney injury: a systematic review and meta-analysis of cohort studies using consensus definitions of exposure. Kidney Int. 2019;95(1):160–172. doi: 10.1016/j.kint.2018.08.036. [DOI] [PubMed] [Google Scholar]
- 4.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371(1):58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk A, Bonventre JV. Acute kidney injury. Annu Rev Med. 2016;67:293–307. doi: 10.1146/annurev-med-050214-013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest. 2011;121(11):4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devarajan P. Update on mechanisms of ischemic acute kidney injury. J Am Soc Nephrol. 2006;17(6):1503–1520. doi: 10.1681/ASN.2006010017. [DOI] [PubMed] [Google Scholar]
- 8.Ferenbach DA, Bonventre JV. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol. 2015;11(5):264–276. doi: 10.1038/nrneph.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med. 2010;16(5):535–543. doi: 10.1038/nm.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bechtel W, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. 2010;16(5):544–550. doi: 10.1038/nm.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinsey GR, Okusa MD. Role of leukocytes in the pathogenesis of acute kidney injury. Crit Care. 2012;16(2):214. doi: 10.1186/cc11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandolfo MT, et al. Foxp3+ regulatory T cells participate in repair of ischemic acute kidney injury. Kidney Int. 2009;76(7):717–729. doi: 10.1038/ki.2009.259. [DOI] [PubMed] [Google Scholar]
- 13.Jang HR, Rabb H. The innate immune response in ischemic acute kidney injury. Clin Immunol. 2009;130(1):41–50. doi: 10.1016/j.clim.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Clef N, Verhulst A, D’Haese PC, Vervaet BA. Unilateral renal ischemia-reperfusion as a robust model for acute to chronic kidney injury in mice. PLoS One. 2016;11(3):e0152153. doi: 10.1371/journal.pone.0152153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Q, Dong Z. Mouse model of ischemic acute kidney injury: technical notes and tricks. Am J Physiol Renal Physiol. 2012;303(11):F1487–F1494. doi: 10.1152/ajprenal.00352.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park KM, Kim JI, Ahn Y, Bonventre AJ, Bonventre JV. Testosterone is responsible for enhanced susceptibility of males to ischemic renal injury. J Biol Chem. 2004;279(50):52282–52292. doi: 10.1074/jbc.M407629200. [DOI] [PubMed] [Google Scholar]
- 17.Skrypnyk NI, Harris RC, de Caestecker MP. Ischemia-reperfusion model of acute kidney injury and post injury fibrosis in mice. J Vis Exp. 2013;78 doi: 10.3791/50495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Donnell MP, Burne M, Daniels F, Rabb H. Utility and limitations of serum creatinine as a measure of renal function in experimental renal ischemia-reperfusion injury. Transplantation. 2002;73(11):1841–1844. doi: 10.1097/00007890-200206150-00025. [DOI] [PubMed] [Google Scholar]
- 19.Bonventre JV. Primary proximal tubule injury leads to epithelial cell cycle arrest, fibrosis, vascular rarefaction, and glomerulosclerosis. Kidney Int Suppl (2011) 2014;4(1):39–44. doi: 10.1038/kisup.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alge JL, Arthur JM. Biomarkers of AKI: a review of mechanistic relevance and potential therapeutic implications. Clin J Am Soc Nephrol. 2015;10(1):147–155. doi: 10.2215/CJN.12191213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlton JR, Portilla D, Okusa MD. A basic science view of acute kidney injury biomarkers. Nephrol Dial Transplant. 2014;29(7):1301–1311. doi: 10.1093/ndt/gft510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H, et al. Tenascin-C is a major component of the fibrogenic niche in kidney fibrosis. J Am Soc Nephrol. 2017;28(3):785–801. doi: 10.1681/ASN.2016020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Park J, Susztak K, Zhang NR, Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat Commun. 2019;10(1):380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiselev VY, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14(5):483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 27.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemmour D, Zilionis R, Kiner E, Klein AM, Mathis D, Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19(3):291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiSpirito JR, et al. Molecular diversification of regulatory T cells in nonlymphoid tissues. Sci Immunol. 2018;3(27):eaat5861. doi: 10.1126/sciimmunol.aat5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27(5):786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Thorenz A, et al. IL-17A blockade or deficiency does not affect progressive renal fibrosis following renal ischaemia reperfusion injury in mice. J Pharm Pharmacol. 2017;69(9):1125–1135. doi: 10.1111/jphp.12747. [DOI] [PubMed] [Google Scholar]
- 33.Kim MG, et al. IL-2/anti-IL-2 complex attenuates renal ischemia-reperfusion injury through expansion of regulatory T cells. J Am Soc Nephrol. 2013;24(10):1529–1536. doi: 10.1681/ASN.2012080784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stremska ME, et al. IL233, A novel IL-2 and IL-33 hybrid cytokine, ameliorates renal injury. J Am Soc Nephrol. 2017;28(9):2681–2693. doi: 10.1681/ASN.2016121272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuswanto W, et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity. 2016;44(2):355–367. doi: 10.1016/j.immuni.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gervasi M, et al. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-β. J Cell Biol. 2012;196(5):589–603. doi: 10.1083/jcb.201109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang HS, Kim JI, Noh M, Rhee MH, Park KM. Regulator of G protein signaling 2 (RGS2) deficiency accelerates the progression of kidney fibrosis. Biochim Biophys Acta. 2014;1842(9):1733–1741. doi: 10.1016/j.bbadis.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Liu J, McMahon AP. Defining the acute kidney injury and repair transcriptome. Semin Nephrol. 2014;34(4):404–417. doi: 10.1016/j.semnephrol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, et al. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight. 2017;2(18):94716. doi: 10.1172/jci.insight.94716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharifian R, et al. Distinct patterns of transcriptional and epigenetic alterations characterize acute and chronic kidney injury. Sci Rep. 2018;8(1):17870. doi: 10.1038/s41598-018-35943-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cippà PE, Sun B, Liu J, Chen L, Naesens M, McMahon AP. Transcriptional trajectories of human kidney injury progression. JCI Insight. 2018;3(22):123151. doi: 10.1172/jci.insight.123151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams TM, Wise AF, Layton DS, Ricardo SD. Phenotype and influx kinetics of leukocytes and inflammatory cytokine production in kidney ischemia/reperfusion injury. Nephrology (Carlton) 2018;23(1):75–85. doi: 10.1111/nep.12941. [DOI] [PubMed] [Google Scholar]
- 43.Jang HR, Rabb H. Immune cells in experimental acute kidney injury. Nat Rev Nephrol. 2015;11(2):88–101. doi: 10.1038/nrneph.2014.180. [DOI] [PubMed] [Google Scholar]
- 44.Alikhan MA, Huynh M, Kitching AR, Ooi JD. Regulatory T cells in renal disease. Clin Transl Immunology. 2018;7(1):e1004. doi: 10.1002/cti2.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Combes AN, Zappia L, Er PX, Oshlack A, Little MH. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019;11(1):3. doi: 10.1186/s13073-019-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karaiskos N, et al. A single-cell transcriptome atlas of the mouse glomerulus. J Am Soc Nephrol. 2018;29(8):2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nistala R, Meuth A, Smith C, Annayya A. Reliable and high efficiency extraction of kidney immune cells. J Vis Exp. 2016;114 doi: 10.3791/54368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao Q, Harris DC, Wang Y. Macrophages in kidney injury, inflammation, and fibrosis. Physiology (Bethesda) 2015;30(3):183–194. doi: 10.1152/physiol.00046.2014. [DOI] [PubMed] [Google Scholar]
- 49.Puranik AS, et al. Kidney-resident macrophages promote a proangiogenic environment in the normal and chronically ischemic mouse kidney. Sci Rep. 2018;8(1):13948. doi: 10.1038/s41598-018-31887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-Treg phenotype. Cell. 2018;174(2):285–299.e12. doi: 10.1016/j.cell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162(5):1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sabapathy V, Cheru NT, Corey R, Mohammad S, Sharma R. A novel hybrid cytokine IL233 mediates regeneration following doxorubicin-induced nephrotoxic injury. Sci Rep. 2019;9(1):3215. doi: 10.1038/s41598-019-39886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gousopoulos E, et al. Regulatory T cell transfer ameliorates lymphedema and promotes lymphatic vessel function. JCI Insight. 2016;1(16):e89081. doi: 10.1172/jci.insight.89081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155(6):1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panduro M, Benoist C, Mathis D. Treg cells limit IFN-γ production to control macrophage accrual and phenotype during skeletal muscle regeneration. Proc Natl Acad Sci U S A. 2018;115(11):E2585–E2593. doi: 10.1073/pnas.1800618115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q, et al. IL-33-mediated IL-13 secretion by ST2+ Tregs controls inflammation after lung injury. JCI Insight. 2019;4(6):123919. doi: 10.1172/jci.insight.123919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arazi A, et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol. 2019;20(7):902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haribhai D, Lin W, Relland LM, Truong N, Williams CB, Chatila TA. Regulatory T cells dynamically control the primary immune response to foreign antigen. J Immunol. 2007;178(5):2961–2972. doi: 10.4049/jimmunol.178.5.2961. [DOI] [PubMed] [Google Scholar]
- 59.Sedhom MA, et al. Neutralisation of the interleukin-33/ST2 pathway ameliorates experimental colitis through enhancement of mucosal healing in mice. Gut. 2013;62(12):1714–1723. doi: 10.1136/gutjnl-2011-301785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.