Abstract
Alternative polyadenylation (APA) is a widespread and important mechanism in regulation of gene expression. Dysregulation of the 3′ UTR cleavage and polyadenylation represents a common characteristic among many disease states, including lung fibrosis. In this study, we investigated the role of mammalian cleavage factor I–mediated (CFIm-mediated) APA in regulating extracellular matrix production in response to mechanical stimuli from stiffened matrix simulating the fibrotic lungs. We found that stiff matrix downregulated expression of CFIm68, CFIm59 and CFIm25 subunits and promoted APA in favor of the proximal poly(A) site usage in the 3′ UTRs of type I collagen (COL1A1) and fibronectin (FN1) in primary human lung fibroblasts. Knockdown and overexpression of each individual CFIm subunit demonstrated that CFIm68 and CFIm25 are indispensable attributes of stiff matrix–induced APA and overproduction of COL1A1, whereas CFIm did not appear to mediate stiffness-regulated FN1 APA. Furthermore, expression of the CFIm subunits was associated with matrix stiffness in vivo in a bleomycin-induced mouse model of pulmonary fibrosis. These data suggest that stiff matrix instigates type I collagen biogenesis by selectively targeting mRNA transcripts for 3′ UTR shortening. The current study uncovered a potential mechanism for regulation of the CFIm complex by mechanical cues under fibrotic conditions.
Keywords: Pulmonology
Keywords: Collagens, Extracellular matrix, Fibrosis
The mammalian cleavage factor I complex is a key mechanosensing mediator of matrix stiffness-regulated collagen I expression in lung fibroblasts.
Introduction
Excessive synthesis relative to breakdown of the extracellular matrix (ECM) is an important cause of fibrotic diseases, as is evident in human idiopathic pulmonary fibrosis (IPF), a lethal fibrotic lung disease contributing to significant morbidity and mortality (1). Crosslinking of the ECM by enzyme- and/or nonenzyme-dependent mechanisms stiffens the ECM (2). Matrix stiffening in the lung has been identified as a critical factor that perpetuates/amplifies pulmonary fibrosis (3–11). Understanding the mechanisms by which matrix stiffness regulates fibrotic lung progression will provide new interventions and therapeutic approaches to prevent, treat, and potentially reverse pulmonary fibrosis.
Polyadenylation involves endonucleolytic cleavage and addition of a poly(A) tail at the 3′ end of pre-mRNAs. It is an essential step in processing nascent eukaryotic transcripts into mature mRNAs (12). Constitutive polyadenylation occurs in pre-mRNAs with a single encoded cleavage and polyadenylation site. In contrast, nascent mRNAs harboring multiple potential polyadenylation sites may have alternative polyadenylation (APA), mostly within the 3′ UTR (13, 14). Since the 3′ UTR of pre-mRNAs often contains many binding sites for regulatory RNA-binding proteins and microRNAs, APA can considerably affect mRNA stability, cellular localization, and protein translation efficiency (13). Owing to the tendency of transcripts to harbor destabilizing elements and their size, transcripts with proximal poly(A) sites and therefore shorter 3′ UTRs are generally thought to produce increased protein levels due to the exclusion of regulatory sequences that mediate degradation or translational efficiency of mRNAs (15). However, this concept has been challenged by more recent studies (16–18).
The positioning and efficiency of 3′ end cleavage and polyadenylation are largely deter-mined by a complex machinery of cleavage and polyadenylation factors, including mammalian cleavage factor I (CFIm) complex and other RNA-binding proteins, such as cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), and poly(A) polymerase. These factors bind specific cis-acting sequence elements surrounding the potential poly(A) site and coordinately regulate the cleavage of pre-mRNAs and polyadenylation (19). The CFIm complex is a heterodimer consisting of CFIm25 (also known as CPSF5) and either of 2 closely related subunits, CFIm68 (also known as CPSF6) or CFIm59 (also known as CPSF7). The CFIm heterodimers selectively bind an upstream U-rich element (USE) containing UGUA motifs located within 40–100 nucleotides of the cleavage site (20). Transcriptome-wide studies have shown that the association of CFIm complex with USEs prevents the access of other 3′-end processing factors (21). As such, knockdown or loss of function of CFIm exposes the USEs to the 3′-end processing complexes, leading to significant shortening of 3′ UTRs and a global usage of the proximal poly(A) site (21–24).
APA is tightly controlled by the level and activity of cleavage and polyadenylation factors as well as by the availability and strength of cis elements in the 3′ UTR (13). Dysregulation of APA has been found in multiple disease states, including oncological, immunological, and neurological diseases (25). A recent study has focused on the potential role of the CFIm25 subunit in lung fibrogenesis (26). The authors showed that CFIm25 expression was selectively reduced in α-smooth muscle actin+ (α-SMA+) fibroblasts in the lungs of patients with IPF as well as in bleomycin injury–induced experimental lung fibrosis in mice (26). Overexpression of CFIm25 inhibited expression of key profibrotic factors in human IPF fibroblasts, whereas selective knockout of CFIm25 in (myo)fibroblasts augmented bleomycin-induced mouse lung fibrosis (26). This study suggests that CFIm25 plays a functional role in the development of pulmonary fibrosis. However, the roles of CFIm68 and CFIm59, essential components of the CFIm complex, in lung fibrogenesis remain to be determined. The mechanisms involved in dysregulation of the CFIm expression in lung fibrosis remain incompletely understood.
In the current study, we sought to determine whether mechanical stimuli derived from the stiffened matrix substrates simulating fibrotic lungs regulate expression of CFIm68, CFIm59, and CFIm25 in human lung fibroblasts. We identified stiff matrix as a negative regulator of the CFIm subunits. We demonstrated that stiff matrix promotes type I collagen (COL1A1) production by CFIm68/CFIm25 complex-dependent APA. Our study identified a potential mechanism for regulation of the CFIm complex by mechanical cues under fibrotic conditions.
Results
Stiff matrix inhibits expression of CFIm68, CFIm59, and CFIm25 subunits at the mRNA and/or protein level
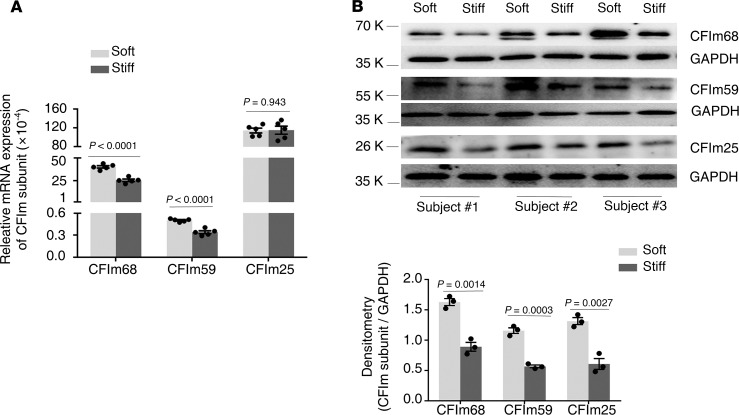
To determine effects of matrix stiffness on expression of the CFIm subunits, primary human lung fibroblasts isolated from failed donors were cultured on 1 kPa (soft) and 20 kPa (stiff) polyacrylamide hydrogels, mimicking the stiffness grades of normal and fibrotic lungs (8, 27). Lung fibroblasts cultured on stiff matrix expressed significantly lower levels of CFIm68 and CFIm59 at both the mRNA and protein level than cells cultured on soft matrix (Figure 1). Stiff matrix conditions significantly decreased CFIm25 protein expression as compared with soft matrix conditions (Figure 1B), whereas mRNA expression of CFIm25 was equivalent between different stiffness conditions (Figure 1A). These data suggest that matrix stiffening negatively regulates expression of the CFIm subunits. Stiff matrix downregulation of CFIm25 likely occurs through a posttranscriptional mechanism.
Figure 1. Stiff matrix inhibits expression of the CFIm subunits at the mRNA and/or protein level.
Primary lung fibroblasts were isolated from failed donor lungs of 3 human subjects (subj). Cells were cultured on soft and stiff polyacrylamide gels for 48 hours. (A) Relative levels of CFIm68, CFIm59, and CFIm25 mRNA were determined by real-time RT-PCR. GAPDH was used as internal reference control. Bar graphs represent (mean ± SD) 5 separate experiments. (B) Protein levels of CFIm68, CFIm59, and CFIm25 were determined by immunoblot. GAPDH was used as loading control. Densitometry was performed using ImageJ (NIH). Relative density of CFIm subunits was normalized to GAPDH. Bar graphs represent (mean ± SD) 3 separate experiments. A 2-tailed Student’s t test was used for comparison between groups.
Stiff matrix promotes expression of COL1A1 and FN1 expression at both the mRNA and protein level
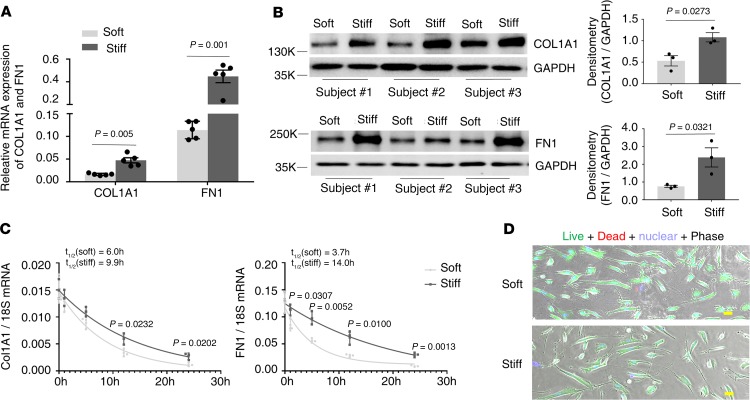
Pulmonary fibrosis is characterized by excessive synthesis and deposition of the ECM proteins, including collagens and fibronectin (FN1), in the lung. Human lung fibroblasts cultured on stiff matrix expressed significantly higher levels of COL1A1 and FN1 at both the mRNA and protein level than cells cultured on soft matrix (Figure 2, A and B), indicating that stiff matrix promotes expression of COL1A1 and FN1. To determine the stability of COL1A1 and FN1 mRNAs under soft versus stiff matrix conditions, we treated human lung fibroblasts with actinomycin D, a transcription inhibitor that interferes with transcription by intercalating into DNA, and then cultured the cells on soft or stiff matrix for 24 hours. The mRNA kinetics were measured by quantitative PCR (qPCR) at 0, 1, 5, 12, and 24 hours. Cells cultured on stiff matrix had a slower mRNA decay rate than those cultured on soft matrix (Figure 2C). The half-lives of COL1A1 mRNA were 6.0 hours under soft matrix conditions and 9.9 hours under stiff matrix conditions. The half-lives of FN1 mRNA were 3.7 hours under soft matrix conditions and 14.0 hours under stiff matrix conditions. Live/dead cell viability assays demonstrated no significant cell death after actinomycin D treatment for 24 hours under either soft or stiff matrix conditions (Figure 2D). Together, these data suggest that lung fibroblast expression of the CFIm complex is inversely correlated with the ECM protein expression (COL1A1 and FN1) under different matrix stiffness conditions. Stiff matrix–induced increases in COL1A1 and FN1 expression are associated with decreased degradation of mRNA.
Figure 2. Stiff matrix promotes expression of COL1A1 and FN1 at both the mRNA and protein level.
Human lung fibroblast populations (n = 3) were cultured on soft and stiff polyacrylamide gels for 48 hours. (A) Relative levels of COL1A1 and FN1 mRNA were determined by real-time RT-PCR. GAPDH was used as internal reference control. Bar graphs represent (mean ± SD) 5 separate experiments. (B) Protein levels of COL1A1 and FN1 were determined by immunoblot. GAPDH was used as loading control. Relative density of COL1A1 and FN1 bands was normalized to GAPDH. Bar graphs represent (mean ± SD) 3 separate experiments. (C) Human lung fibroblasts were treated with 10 μg/mL actinomycin D, followed by culturing on soft and stiff polyacrylamide gels for 24 hours. Relative levels of COL1A1 and FN1 mRNA were determined by real-time RT-PCR at the indicated time points. 18S rRNA was used as internal reference control. Graphs represent mRNA decay and half-lives of COL1A1 and FN1 under soft and stiff matrix conditions after actinomycin D treatment and represent (mean ± SD) 3 separate experiments. (D) Cell viability after actinomycin D treatment for 24 hours under both soft and stiff matrix conditions was evaluated by live/dead cell viability assays. Scale bar: 20 μm. A 2-tailed Student’s t test was used for comparison between groups.
The 3′ UTRs of COL1A1 and FN1 harbor functional cis-regulatory elements that mediate the cleavage of the 3′ UTR and APA
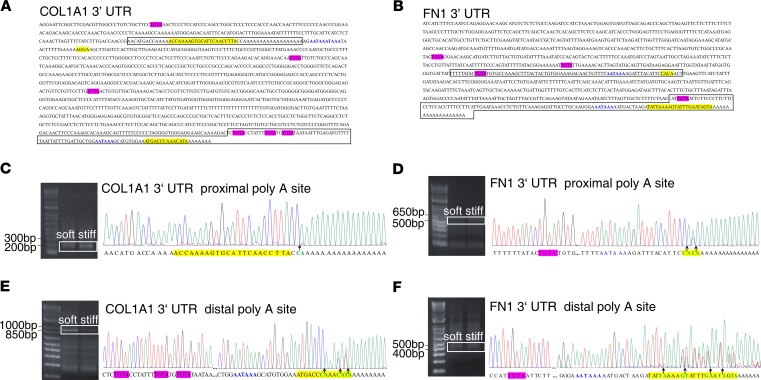
To test the hypothesis that stiff matrix promotes the production of COL1A1 and FN1 by CFIm-dependent APA, we first performed sequence-based computational prediction analysis to identify putative cis elements for CFIm complex–dependent APA in the 3′ UTRs of COL1A1 and FN1 (28, 29). A total of 6 USEs (UGUA or TGTA), 3 polyadenylation signals (AATAAA), and 3 potential cleavage sites were identified in the 3′ UTR of COL1A1 (Figure 3A); a total of 4 USEs, 2 polyadenylation signals, and 2 potential cleavage sites were identified in the 3′ UTR of FN1 (Figure 3B). Next, we determined CFIm-mediated APA in the 3′ UTRs of COL1A1 and FN1 experimentally by rapid amplification of 3′ cDNA ends (3′ rapid amplification of cDNA ends [3′ RACE]). DNA sequencing of the 3′-end PCR products demonstrated the existence of both short and long 3′-end cDNA fragments cleaved at or near predicted proximal and distal cleavage sites (Figure 3, C–F), suggesting that CFIm-dependent cleavage and APA indeed occurred at the 3′ UTRs of both COL1A1 and FN1. Furthermore, DNA sequencing data showed a single peak before the proximal COL1A1 poly(A) tail (Figure 3C), indicating that the cleavage (Figure 3C, arrow) occurred at a single site (between C and A), which was only 2 nucleotides downstream the predicted cleavage region. In contrast, additional double peak(s) were observed before the distal COL1A1 poly(A) tail (Figure 3E) and both the proximal and distal FN1 poly(A) tails (Figure 3, D and F), indicating that the cleavage occurred at multiple sites (between C and A, T and A, G and T, or G and A, as indicate by arrows in Figure 3, C–F), rather than a single site. Notably, the cleavage sites determined by the 3′ RACE experiments were consistent with the prediction analysis (Figure 3, D–F, highlighted yellow). Together, the 3′ RACE analysis demonstrated the cleavage and APA in the 3′ UTRs of COL1A1 and FN1.
Figure 3. The 3′ UTRs of COL1A1 and FN1 harbor functional cis elements that mediate the cleavage and APA of COL1A1 and FN1 transcripts.
(A and B) Prediction analysis of the cleavage sites (yellow), UGUA-containing (or TGTA-containing) USEs (hot pink), and polyadenylation signals (blue) in the 3′ UTRs of COL1A1 and FN1 transcripts. (C–F) 3′ RACE products were separated on agarose gels, purified, and sequenced. Graphs show DNA sequences flanking the cleavage sites, as indicated by the boxed areas in A and B. Arrows indicate the cleavage sites.
Stiff matrix promotes APA of COL1A1 and FN1 transcripts in favor of the proximal poly(A) site usage
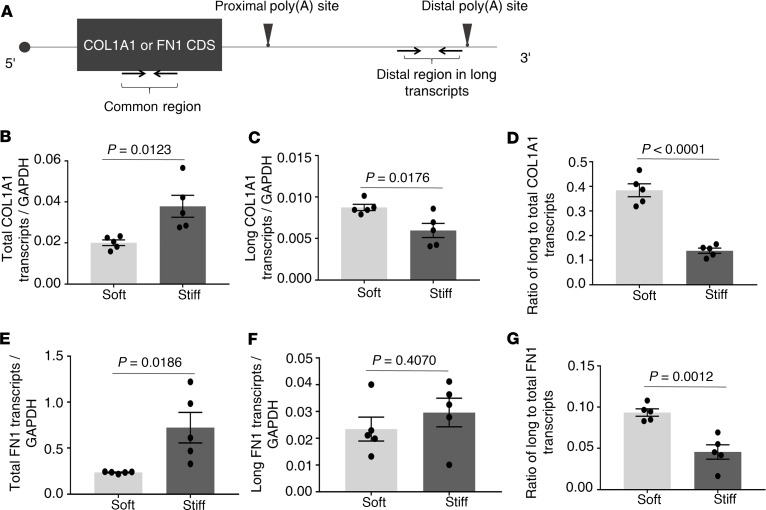
To determine whether matrix stiffness regulates APA in the 3′ UTRs of COL1A1 and FN1, we compared the distal and proximal poly(A) site usage under stiff versus soft matrix conditions. The distal poly(A) site usage can be measured directly by qPCR (Figure 4A). However, since both long and short transcripts contain the proximal mRNA sequences, it would be challenging to directly measure the proximal poly(A) site usage by qPCR. In this study, we evaluated the proximal poly(A) site usage indirectly by the ratio of long transcripts/total transcripts. The amount of total transcripts were measured by a common region shared by both long and short transcripts (Figure 4A). Real-time qPCR analysis showed that stiff matrix increased the level of total COL1A1 mRNAs as compared with soft matrix (Figure 4B). The result is consistent with the observation that stiff matrix promotes COL1A1 transcription (Figure 2A). Stiff matrix decreased the relative level of long COL1A1 transcripts as compared with soft matrix (Figure 4C), suggesting that the distal poly(A) usage is decreased under stiff matrix conditions. As expected, the ratio of long COL1A1 transcripts to total COL1A1 transcripts was decreased under stiff matrix conditions as compared with soft matrix conditions (Figure 4D). Together, these results suggest that stiff matrix increases the proximal poly(A) usage in the 3′ UTR of COL1A1.
Figure 4. Stiff matrix promotes APA of COL1A1 and FN1 in favor of the proximal poly(A) site usage.
(A) A schematic diagram of the COL1A1 or FN1 transcripts, in which the relative positions of complete coding sequence (CDS), proximal and distal poly(A) sites, and primer pairs for amplification of a common region and a distal region in the long transcripts are shown. (B–G) Lung fibroblasts were cultured on soft and stiff hydrogels for 48 hours. Relative levels of total/common COL1A1 transcripts (B), long/distal COL1A1 transcripts (C), total/common FN1 transcripts (E), and long/distal FN1 transcripts (F) were determined by real-time RT-PCR. GAPDH was used as internal reference control. Ratios of long-to-total COL1A1 (D) and FN1 (G) transcripts were used to indirectly evaluate the proximal poly(A) site usage. Bar graphs represent (mean ± SD) 5 separate experiments. A 2-tailed Student’s t test was used for comparison between groups.
Analysis of FN1 by similar approaches showed that stiff matrix increased the level of total FN1 mRNAs as compared with soft matrix (Figure 4E). This is, again, in agreement with the previous observations (Figure 2A). Stiff matrix had no significant effects on the level of long FN1 transcripts as compared with soft matrix (Figure 4F). The ratio of long FN1 transcripts to total FN1 transcripts was decreased under stiff matrix conditions as compared with soft matrix conditions (Figure 4G), suggesting that stiff matrix increases the proximal poly(A) site usage in the 3′ UTR of FN1.
Effects of specific CFIm subunits on matrix stiffness–regulated APA and expression of COL1A1 and FN1
To investigate the role of specific CFIm subunits in matrix stiffness–regulated APA and expression of COL1A1 and FN1, we knocked down and overexpressed CFIm68, CFIm59, or CFIm25 and determined the effects of knockdown and overexpression of each individual CFIm subunit on APA and expression of COL1A1 and FN1 under different matrix stiffness conditions.
CFIm68.
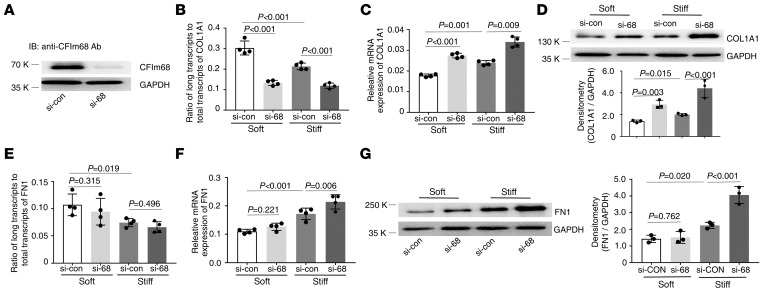
Knockdown of CFIm68 (Figure 5A) decreased the distal COL1A1 poly(A) site usage (Figure 5B) and increased expression of COL1A1 mRNA (Figure 5C) and protein (Figure 5D) under both soft and stiff matrix conditions. Knockdown of CFIm68 had no significant effects on the distal FN1 poly(A) site usage under either soft or stiff matrix conditions (Figure 5E). Knockdown of CFIm68 increased expression of FN1 mRNA and protein under stiff matrix conditions but not under soft matrix conditions (Figure 5, F and G).
Figure 5. Effects of knockdown of CFIm68 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) CFIm68-specific siRNA (si-68) and the control siRNA (si-con) were transfected into human lung fibroblasts. Knockdown of CFIm68 expression was determined by immunoblot. GAPDH was used as loading control. (B–G) Lung fibroblasts transfected with si-68 and si-con were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
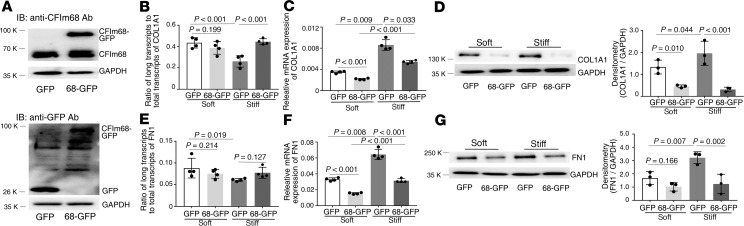
Overexpression of CFIm68 (Figure 6A) increased the distal COL1A1 poly(A) site usage under stiff matrix conditions but not under soft matrix conditions (Figure 6B). Overexpression of CFIm68 decreased expression of COL1A1 mRNA (Figure 6C) and protein (Figure 6D) under both soft and stiff matrix conditions. Overexpression of CFIm68 had no significant effects on the distal FN1 poly(A) site usage under either soft or stiff matrix conditions (Figure 6E). Overexpression of CFIm68 significantly decreased FN1 mRNA expression under both soft and stiff matrix conditions (Figure 6F) and significantly decreased FN1 protein expression under stiff matrix conditions (Figure 6G). Overexpression of CFIm68 appeared to decrease FN1 protein expression on soft matrix conditions (Figure 6G). However, the difference did not reach to statistical significance.
Figure 6. Effects of overexpression of CFIm68 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) Human lung fibroblasts were infected by recombinant lentiviral vectors expressing CFIm68-GFP or GFP alone. Ectopic expression of CFIm68-GFP fusion protein and GFP protein was determined by immunoblot with both anti-CFIm68 and anti-GFP antibodies. GAPDH was used as loading control. (B–G) Lung fibroblasts infected by CFIm68-GFP or GFP viruses were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
CFIm59.
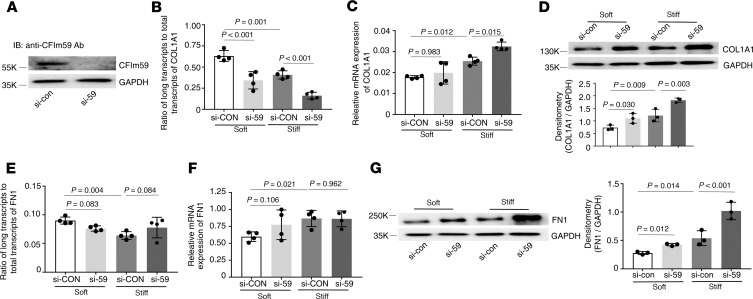
Knockdown of CFIm59 (Figure 7A) decreased the distal COL1A1 poly(A) site usage under both soft and stiff matrix conditions (Figure 7B), increased COL1A1 mRNA expression under stiff matrix conditions (Figure 7C), and increased COL1A1 protein expression under both soft and stiff matrix conditions (Figure 7D). Knockdown of CFIm59 had no significant effects on the distal FN1 poly(A) site usage (Figure 7E) and FN1 mRNA expression (Figure 7F) under either soft or stiff matrix conditions. In contrast, knockdown of CFIm59 significantly increased FN1 protein expression under both soft and stiff matrix conditions (Figure 7G).
Figure 7. Effects of knockdown of CFIm59 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) CFIm59-specific siRNA (si-59) and the control siRNA (si-con) were transfected into human lung fibroblasts. Knockdown of CFIm59 expression was determined by immunoblot. GAPDH was used as loading control. (B–G) Lung fibroblasts transfected with si-59 and si-con were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
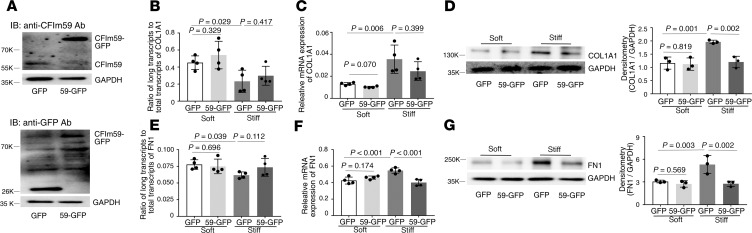
Overexpression of CFIm59 (Figure 8A) had no significant effects on the distal COL1A1 poly(A) site usage (Figure 8B) and expression of COL1A1 mRNA (Figure 8C) and protein, except a decrease in COL1A1 protein under stiff matrix conditions (Figure 8D). Overexpression of CFIm59 did not alter the distal FN1 poly(A) site usage (Figure 8E). However, overexpression of CFIm59 significantly decreased expression of FN1 mRNA (Figure 8F) and protein (Figure 8G) under stiff matrix conditions.
Figure 8. Effects of overexpression of CFIm59 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) Human lung fibroblasts were infected by recombinant lentiviral vectors expressing CFIm59-GFP or GFP alone. Ectopic expression of CFIm59-GFP fusion protein and GFP protein was determined by immunoblot with both anti-CFIm59 and anti-GFP antibodies. GAPDH was used as loading control. (B–G) Lung fibroblasts infected by CFIm59-GFP or GFP viruses were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
CFIm25.
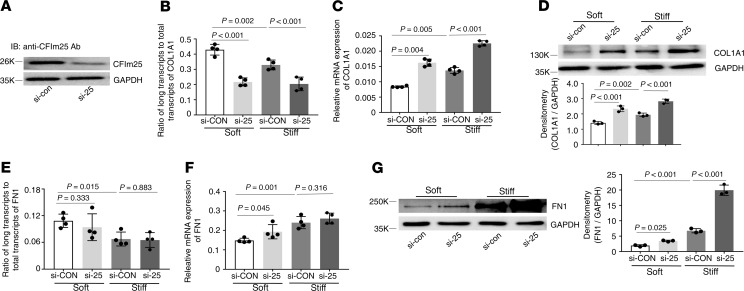
Knockdown of CFIm25 (Figure 9A) decreased the distal COL1A1 poly(A) site usage (Figure 9B) and increased expression of COL1A1 mRNA (Figure 9C) and protein (Figure 9D) under both soft and stiff matrix conditions. Knockdown of CFIm25 had no effects on the distal FN1 poly(A) site usage (Figure 9E). Knockdown of CFIm25 increased FN1 mRNA expression under soft matrix conditions (Figure 9F) and increased FN1 protein expression under both soft and stiff matrix conditions (Figure 9G).
Figure 9. Effects of knockdown of CFIm25 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) CFIm25-specific siRNA (si-25) and the control siRNA (si-con) were transfected into human lung fibroblasts. Knockdown of CFIm25 expression was determined by immunoblot. GAPDH was used as loading control. (B–G) Lung fibroblasts transfected with si-25 and si-con were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
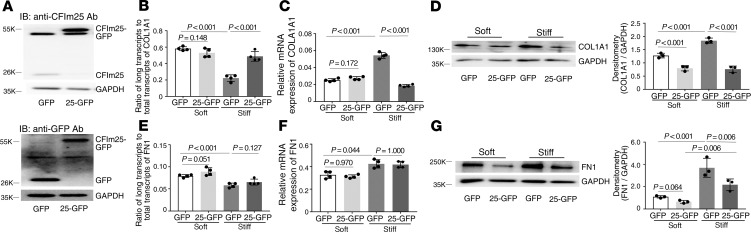
Overexpression of CFIm25 (Figure 10A) increased the distal COL1A1 poly(A) site usage (Figure 10B) and decreased expression of COL1A1 mRNA (Figure 10C) under stiff matrix conditions and decreased expression of COL1A1 protein under both soft and stiff matrix conditions (Figure 10D). Overexpression of CFIm25 has no effects on the distal COL1A1 poly(A) site usage (Figure 10B) and COL1A1 mRNA expression (Figure 10C) under soft matrix conditions. Overexpression of CFIm25 had no effects on the distal FN1 poly(A) site usage and FN1 mRNA expression (Figure 10, E and F) under either soft or stiff matrix conditions. Overexpression of CFIm25 decreased FN1 protein expression under stiff matrix conditions (Figure 10G).
Figure 10. Effects of overexpression of CFIm25 on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
(A) Human lung fibroblasts were infected by recombinant lentiviral vectors expressing CFIm25-GFP or GFP alone. Ectopic expression of CFIm25-GFP fusion protein and GFP protein was determined by immunoblot with both anti-CFIm25 and anti-GFP antibodies. GAPDH was used as loading control. (B–G) Lung fibroblasts infected by CFIm25-GFP or GFP viruses were cultured on soft and stiff hydrogels for 48 hours. Ratios of long-to-total COL1A1 (B) and FN1 (E) transcripts were used to evaluate the proximal poly(A) site usage. Relative levels of COL1A1 (C) and FN1 (F) mRNA were determined by real-time RT-PCR. Protein levels of COL1A1 (D) and FN1 (G) were determined by immunoblot. GAPDH was used as internal reference or loading control. Bar graphs represent (mean ± SD) 3 or 4 separate experiments. Statistical analysis was performed by 1-way ANOVA.
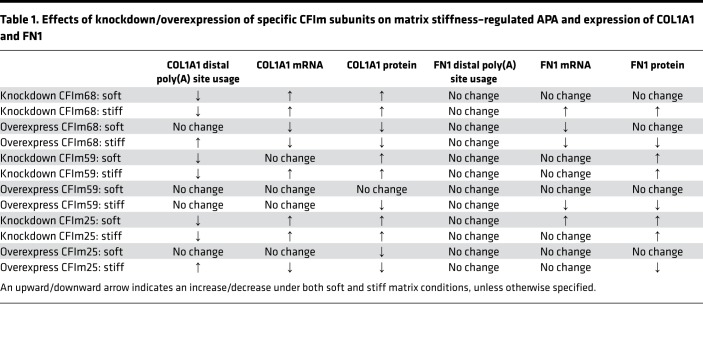
Results from the gain- and loss-of-function studies are summarized in Table 1. Collectively, these data suggest that CFIm68 and CFIm25 are required for matrix stiffness–regulated APA and expression of COL1A1. Although knockdown/overexpression of CFIm59 altered COL1A1 expression at the mRNA and/or protein level, there was no consistent correlation between COL1A1 expression and preferential usage of poly(A) sites under different matrix stiffness conditions, suggesting that CFIm59-regulated COL1A1 expression may occur through an APA-independent mechanism. Likewise, despite the fact that knockdown/overexpression of specific CFIm subunits altered FN1 expression, this does not appear to occur by CFIm-mediated APA.
Table 1. Effects of knockdown/overexpression of specific CFIm subunits on matrix stiffness–regulated APA and expression of COL1A1 and FN1.
Stiffened fibrotic lungs demonstrate decreased expression of the CFIm subunits in a bleomycin-induced mouse lung fibrosis model
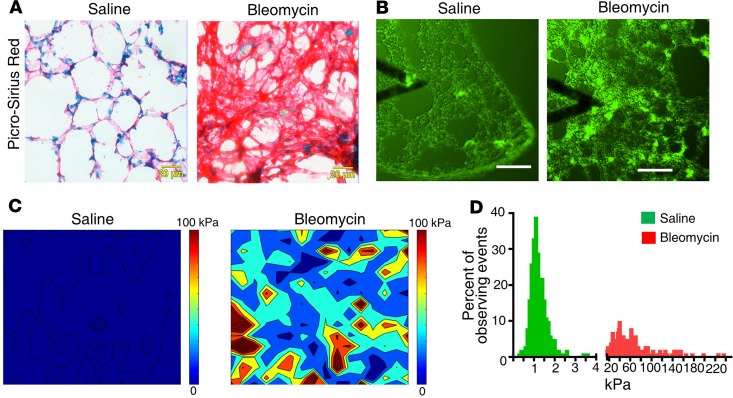
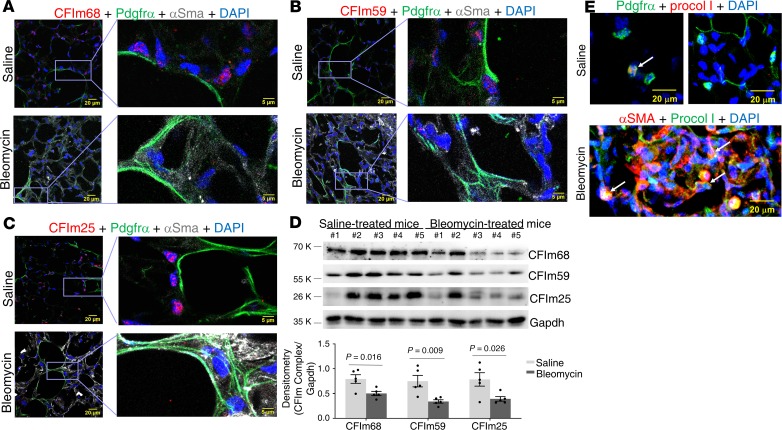
To evaluate whether expression of CFIm subunits is associated with lung matrix stiffness in vivo, we analyzed expression of CFIm68, CFIm59, and CFIm25 in stiffened fibrotic lungs and the control lungs in a bleomycin injury–induced murine model of lung fibrosis. Histological analysis demonstrated a marked increase in the collagen deposition in bleomycin-treated mouse lungs as compared with saline-treated control lungs (Figure 11A), indicative of lung fibrosis. The stiffness levels of normal and fibrotic mouse lungs were measured by atomic force microscopy (AFM) microindentation. To facilitate localization of the fibrotic area and positioning of the cantilever tip for lung mechanical testing, we stained unfixed, native mouse lungs with a collagen-binding green fluorescence probe (Col-F collagen-binding reagent) (Figure 11B). The fibrotic area in bleomycin-treated mouse lungs demonstrated increased elastic moduli (Young’s moduli) as compared with the alveolar wall in the saline-treated control lungs (Figure 11C). The average stiffness of the alveolar wall in saline-treated mice was 1.19 ± 0.42 kPa (median = 1.10 kPa), whereas the fibrotic area in bleomycin-treated mice had an average stiffness of 50.28 ± 36.49 kPa (median = 40.89 kPa) (Figure 11D). To evaluate expression of the CFIm subunits by (myo)fibroblasts in normal versus fibrotic mouse lungs, we costained the CFIm subunits with Pdgfra (a marker of lung fibroblast subpopulation) or α-SMA (a marker of myofibroblasts). Confocal immunofluorescent microscopy showed that saline-treated mouse lungs displayed positive signals for CFIm68, CFIm59, and CFIm25 in Pdgfrα+ fibroblasts, whereas the CFIm signals were reduced in α-SMA–expressing myofibroblasts in the fibrotic area (Figure 12, A–C). Consistent with these results, immunoblot analysis showed that the levels of CFIm68, CFIm59, and CFIm25 protein were significantly reduced in bleomycin-treated mouse lungs as compared with saline-treated mouse lungs (Figure 12D). Finally, α-SMA–expressing myofibroblasts in the stiffened fibrotic area actively synthesized a high level of procollagen I (Figure 12E). In contrast, only a small portion of Pdgfrα-expressing fibroblasts in saline-treated control lungs expressed procollagen I at a relatively low level. Collectively, these data suggest that expression of the CFIm subunits is suppressed in stiffened fibrotic lungs and is associated with increased production of procollagen I.
Figure 11. Matrix stiffness increases in lung fibrosis in a bleomycin injury mouse model.
C57BL/6 mice were administered bleomycin intratracheally at a dose of 3 U/kg. Control mice were given an equal volume of saline. Lung tissues were harvested at 28 days. (A) Frozen tissue sections were subjected to picrosirius red staining for collagen. (B–D) Mechanical testing of unfixed native mouse lung tissues was performed by AFM microindentation. Tissue sections were stained by a collagen Col-F collagen-binding reagent to facilitate localization of specific lung areas (e.g., the alveolar wall and collagen-enriched fibrotic area) and positioning of the cantilever tip (B). Heatmaps show the elastic moduli (Young’s moduli) of the alveolar wall in the saline-treated lungs and the fibrotic area in bleomycin-treated lungs (C). Bar graphs show the distribution of stiffness in saline- and bleomycin-treated mouse lungs (D). Scale bars: 20 μm (A); 100 μm (B).
Figure 12. Stiffened fibrotic mouse lungs demonstrate decreased expression of CFIm68, CFIm59, and CFIm25.
C57BL/6 mice were administered bleomycin intratracheally at a dose of 3 U/kg. Control mice were given an equal volume of saline. Lung tissues were harvested at 28 days. (A–C) Expression of CFIm68 (A), CFIm59 (B), and CFIm25 (C) in Pdgfrα+ fibroblasts and α-SMA+ myofibroblasts in saline- and bleomycin-treated mouse lungs was evaluated by confocal immunofluorescent microscopy. (D) Protein levels of CFIm68, CFIm59, and CFIm25 in saline-treated (n = 5) and bleomycin-treated (n = 5) mouse lungs were determined by immunoblot. GAPDH was used as loading control. Relative density of CFIm subunits was normalized to GAPDH. Bar graphs represent (mean ± SD); n = 5. A 2-tailed Student’s t test was used for comparison between groups. (E) Expression of procollagen I (Procol I) in Pdgfrα+ fibroblasts and α-SMA+ myofibroblasts in saline- and bleomycin-treated mouse lungs was evaluated by confocal immunofluorescent microscopy. Arrows indicate procollagen I–expressing (myo)fibroblasts. Scale bars: 20 μm (A–C, leftmost images, and E); 5 μm (A–C, rightmost images).
Discussion
APA is an important and widespread mechanism for modulating gene expression. Dysregulation of the 3′ UTR cleavage and polyadenylation can cause abnormal gene expression and represents a common characteristic among many diseases (25, 30). In this study, we investigated the role of CFIm complex–mediated APA in the regulation of synthesis of the ECM proteins (COL1A1 and FN1) in response to mechanical stimuli derived from stiffened matrix substrates simulating fibrotic lungs. We demonstrated that (a) stiff matrix downregulates expression of both the large (CFIm68 and CFIm59) and small (CFIm25) CFIm subunits in human lung fibroblasts; (b) the 3′ UTRs of COL1A1 and FN1 harbor functional cis elements that mediate CFIm complex–dependent APA; (c) stiff matrix promotes APA in favor of the proximal poly(A) site usage in the 3′ UTRs of both COL1A1 and FN1; (d) both the CFIm68 and CFIm25 subunits are required for stiff matrix–induced APA of COL1A1pre-mRNAs and overproduction of the protein; (e) the CFIm complex mediates stiffness-regulated FN1 expression likely by APA-independent mechanisms; and (f) expression of the CFIm subunits is associated with matrix stiffness in vivo in a bleomycin-induced mouse lung fibrosis. Our findings indicate that mechanical cues derived from the stiffened fibrotic matrix instigate type I collagen biogenesis by selectively targeting mRNA transcripts for 3′ UTR shortening. The present study expanded the findings of a recent report on CFIm25 in the regulation of lung fibrogenesis (26). It highlighted a pivotal role of the CFIm68/CFIm25 complex in stiff matrix–induced APA and augmentation of COL1A1 expression, uncovering a potentially novel mechanism for the regulation of CFIm expression by mechanical signals derived from the ECM under fibrotic conditions. As demonstrated in this and other studies (26), the APA events can most likely play significant roles in regulating the pathogenesis of lung fibrosis.
The CFIm complex is a heterotetramer composed of a small subunit (CFIm25) and a larger subunit (CFIm59 or CFIm68) (31). In this study, although stiff matrix downregulates expression of both CFIm68 and CFIm59 in addition to CFIm25, the knockdown and overexpression experiments strongly suggest that the CFIm68/CFIm25 complex is required for stiff matrix–induced APA in the 3′ UTR and overproduction of COL1A1. Knockdown of CFIm59 decreased the distal poly(A) site usage in the 3′ UTR of COL1A1 and increased the level of COL1A1 mRNA under stiff matrix conditions. However, unlike CFIm68, overexpression of CFIm59 had no effects on the distal poly(A) site usage and mRNA expression of COL1A1. These data suggest that, despite their structural similarity, CFIm68 and CFIm59 may have different roles in matrix stiffness–dependent 3′-end processing of the COL1A1 transcripts. Compared with CFIm59, CFIm68 contains an additional glycine-arginine–rich motif (32), which could potentially confer a different function for the 2 larger subunits. The crystal structures of the CFIm25/CFIm59 and CFIm25/CFIm68 complexes also display differences in which CFIm59 has a narrower RNA exit cleft, resulting in a different mode of RNA binding, which may explain its lesser effect on APA compared with CFIm68 (32, 33). Additionally, CFIm68 and CFIm59 have distinct methylation patterns (34, 35), but the potential effect of posttranslational modifications on 3′ UTR processing will require further investigation.
In contrast to COL1A1, neither knockdown nor overexpression of CFIm68, CFIm59 or CFIm25 altered the usage of poly(A) sites in the 3′ UTR of FN1 pre-mRNAs. However, knockdown or overexpression of the CFIm subunits did alter FN1 mRNA and/or protein expression under different stiffness conditions. These findings suggest that the CFIm complex may mediate matrix stiffness–dependent FN1 expression by an APA-independent mechanism(s). Besides the function in alternative poly(A) site selection, the CFIm complex has been shown to regulate mRNA splicing and export (36, 37), molecular events known to regulate gene expression. Additionally, as aforementioned, the CFIm subunits are subjected to posttranslational modifications, including phosphorylation, acetylation, and ubiquitination, in addition to methylation (38). Thus, the posttranslational modifications, rather than alterations of CFIm expression per se may alter the affinity of the CFIm complex for binding to the cleavage sites or its interaction with other cleavage factors, resulting in functional APA.
Upon cis element–directed cooperative binding of sets of CPSF, CstF, CFIm, and other 3′-end processing factors, mRNA cleavage occurs downstream of the highly conserved hexanucleotide AAUAAA (29). The nuclease responsible for this specific cleavage has been reported to be CPSF73 (39, 40). An intriguing finding in our studies is that the mRNA cleavage can occur simultaneously at multiple adjacent poly(A) sites. The biological consequences of coexistence of mRNA isoforms with only a few different nucleotides immediate upstream of the poly(A) signal currently remain unknown and warrant further investigation.
It has been shown that APA is regulated by extracellular cues, including epidermal growth factor, IL-2, and coapplication of anti-IgM and CpG in various cell types (41). The CFIm subunits are coded by 3 separate genes, whereas CFIm72 is an alternatively spliced form of CFIm68 (31). In this study, we identified for the first time to our knowledge that the mechanical properties of the ECM regulate expression of all 3 CFIm subunits. We observed that stiff matrix downregulates expression of the 2 large CFIm subunits at both the mRNA and protein level, whereas stiffness-dependent downregulation of CFIm25 was observed only at the protein level. These findings suggest that matrix stiffness regulates the large and small CFIm subunits, likely through distinct mechanisms. A vast majority of mechanosensing studies, including the current study, have used the PA hydrogels as 2D matrix substrates to study the effects of ECM stiffness on a range of cellular processes across various cell types (42, 43). PA gels have a number of advantages, including cost effectiveness, ease of preparation, constant surface chemistry, reproducibility, and availability in a wide range of stiffness (44–47). However, due to the cytotoxicity of precursor components, PA hydrogels cannot be used to encapsulate cells in 3D, in contrast to those grown in vivo. Future studies will need to confirm the regulation of CFIm expression by matrix stiffness in the context of a cell-laden 3D ECM. Additionally, more detailed investigations are needed to delineate the mechanotransductive mechanisms involved in mechanical signal-regulated CFIm expression.
While we were preparing this manuscript, Ko and colleagues reported that TGF-β1 treatment reduced mRNA expression of CFIm25 at 24 hours and protein expression after day 3 in human lung fibroblasts, suggesting that CFIm25 expression is downregulated by the fibrogenic cytokine (48). Furthermore, the authors demonstrated that TGF-β1 downregulates CFIm25 expression by a miR203-dependent mechanism. Our study and that by Ko et al. demonstrated that CFIm25 is regulated by both the biochemical and mechanical signals associated with lung fibrogenesis. It remains to be determined whether TGF-β1 regulates expression of the 2 large subunits, CFIm68 and CFIm59.
Unraveling the functional consequences of dysregulated APA in lung fibrosis is promising for biomarker discovery and development of novel therapies. In this and other recent studies, we and others demonstrated that suppression of expression of the CFIm complex is associated with increased production of the ECM proteins and lung fibrosis progression. These findings raise the possibilities of predicting the progression of IPF by expression of the CFIm complex and/or targeting the CFIm complex for halting the progression of this devastating fibrotic lung disease. Clearly, more research needs to be done to better understand the functional role and significance of the CFIm complex–mediated APA events in lung fibrogenesis and to explore the potential of targeting these APA events as therapeutic means.
Methods
Lung fibroblast isolation, culture, and transfection.
Human lung fibroblasts were established from failed donors undergoing lung transplantation. Lung fibroblast isolation, culture, and transfection are described in the Supplemental Methods (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.133972DS1).
Preparation of polyacrylamide hydrogels with varying stiffness.
PA gels with tunable stiffness were fabricated using a published protocol (44). Gel surfaces were coated with 0.1 mg/mL rat tail collagen I (Thermo Fisher Scientific).
3′ RACE and DNA sequencing.
3′ RACE was performed with a 3′ RACE System for Rapid Amplification of cDNA Ends (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Gene-specific primers for amplification of the proximal and distal poly(A) tails were as follows: COL1A1 proximal, 5′-CCAACCCGGAAACAGACAAG-3′; COL1A1 distal, 5′-GCAACATCTCCCCTTCGTTT-3′; FN1 proximal, 5′-GCACATTGCCTGTTCTGCTT-3′; and FN1 distal, 5′-ACACCTTCGGGGGAAATAAT-3′. 3′ RACE PCR products were visualized on a 1% agarose gel. RACE products were subjected to fluorescent DAN sequencing on an ABI 3730 DNA Analyzer (Applied Biosystems) at the Genomics Core Laboratory of the Heflin Center, University of Alabama at Birmingham.
Lung tissue preparation and mechanical testing.
Mouse lungs were inflated with and imbedded in Tissue-Plus Optimal Cutting Temperature (O.C.T.) Compound (Thermo Fisher Scientific). Lung tissues were frozen on dry ice and stored at –80°C. 60-μm-thickness tissue slices were cut from lung tissue samples by a Leica Cryostat. Cryosections were placed on poly-l-lysine–coated coverslips. Samples were mechanically characterized using an MFP-3D-BIO AFM (Asylum Research), mounted on a Nikon eclipse Ti2 microscope (Nikon Inc.), in contact mode. To facilitate localization of the specific lung area (e.g., alveoli and fibrotic regions) and positioning of the cantilever tip, lung tissue sections were stained by a Col-F collagen-binding reagent (ImmunoChemistry Technologies) at 37°C for 1 hour followed by 2 PBS washes at room temperature. Detailed AFM methods were provided the Supplemental Methods.
Confocal laser scanning microscopy.
Fluorescent signals were detected using a confocal laser-scanning microscope (Zeiss LSM510) equipped with a digital color camera (Zeiss Axiocam). All fluorescent images were generated using sequential laser scanning, with only the corresponding single wavelength laser line, activated using acousto-optical tunable filters to avoid cross-detection of either one of the fluorescence channels.
Animals and experimental protocol.
Six- to eight-week-old pathogen-free female mice (The Jackson Laboratory) were used in this study. Bleomycin sulphate (Almirall) was dissolved in 50 μL sterile saline solution and intratracheally instilled into mice as a single dose of 3 U/kg bodyweight. Control mice received an equal volume of 50 μL saline. Mice were sacrificed at 21 days. Lung tissues were excised and inflated with O.C.T. for immunofluorescent or immunohistochemical analyses.
To prepare for lung homogenates, fresh lung tissues were transferred to prechilled tubes containing T-PER and Complete Mini Protease Inhibitor Cocktail (Thermo Fisher Scientific) at a proportion of 1 tablet/10 mL T-PER and were homogenized at 4°C. Homogenates were centrifuged at 9,000 g for 10 minutes at 4°C. Supernatants were transferred to clean microcentrifuge tubes. Total protein concentrations in the lung tissue homogenates were determined using a BCA Protein Assay kit (Thermo Fisher Scientific).
Statistics.
Statistical differences among treatment conditions were determined by 1-way ANOVA (Newman-Keuls method for multiple comparisons) or 2-tailed Student’s t test using GraphPad Prism 7.02 software. Values of P < 0.05 were considered significant.
Study approval.
The animal studies were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and approved by Institutional Animal Care and Use Committee at the University of Alabama at Birmingham. The studies involving human subjects were approved by Institutional Review Board at the University of Alabama at Birmingham. Participants have been provided with written informed consent.
Additional details on the methods are provided in the Supplemental Methods.
Author contributions
Y. Zhou and ZZ designed the study. ZZ, JQ, LH, Y. Zhu, SZY, FZ, and TG performed the experiments. ZZ, Y. Zhu, and Y. Zhou analyzed the data. HP and PC participated in the discussion. ZZ and Y. Zhou wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants R01HL124076, R01HL139584, and R01EY027924 (to Y. Zhou). The authors would like to thank the Clinical Core for support from NIH grant P01HL114470 (PI: Victor J. Thannickal) for providing primary human lung fibroblasts and the Molecular Core for support from NIH grant P30EY003039 (PI: Steven J. Pittler) for providing Cryostats.
Version 1. 01/14/2020
In-Press Preview
Version 2. 02/13/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(3):e133972.https://doi.org/10.1172/jci.insight.133972.
Contributor Information
Zijing Zhou, Email: zhouzijing1114@126.com.
Jing Qu, Email: jingqu@uabmc.edu.
Li He, Email: heli8415@uab.edu.
Yi Zhu, Email: yzhu1@uab.edu.
Feng Zhang, Email: fengz@uab.edu.
Ting Guo, Email: tingg925@uab.edu.
Hong Peng, Email: phdiana@126.com.
Ping Chen, Email: pingchen0731@csu.edu.cn.
Yong Zhou, Email: yongzhou@uabmc.edu.
References
- 1.Zhou Y, et al. Extracellular matrix in lung development, homeostasis and disease. Matrix Biol. 2018;73:77–104. doi: 10.1016/j.matbio.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells RG. The role of matrix stiffness in regulating cell behavior. Hepatology. 2008;47(4):1394–1400. doi: 10.1002/hep.22193. [DOI] [PubMed] [Google Scholar]
- 3.Liu F, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190(4):693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y, et al. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J Clin Invest. 2013;123(3):1096–1108. doi: 10.1172/JCI66700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahaman SO, et al. TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice. J Clin Invest. 2014;124(12):5225–5238. doi: 10.1172/JCI75331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J Cell Biol. 2007;179(6):1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiore VF, Strane PW, Bryksin AV, White ES, Hagood JS, Barker TH. Conformational coupling of integrin and Thy-1 regulates Fyn priming and fibroblast mechanotransduction. J Cell Biol. 2015;211(1):173–190. doi: 10.1083/jcb.201505007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, et al. Mechanosensing by the α6-integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat Commun. 2016;7:12564. doi: 10.1038/ncomms12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, et al. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol. 2015;308(4):L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Southern BD, et al. Matrix-driven myosin II mediates the pro-fibrotic fibroblast phenotype. J Biol Chem. 2016;291(12):6083–6095. doi: 10.1074/jbc.M115.712380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker MW, et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest. 2014;124(4):1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis JD, Gunderson SI, Mattaj IW. The influence of 5’ and 3’ end structures on pre-mRNA metabolism. J Cell Sci Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 13.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18(1):18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 15.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber AR, et al. Global 3’ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun. 2014;5:5465. doi: 10.1038/ncomms6465. [DOI] [PubMed] [Google Scholar]
- 17.Spies N, Burge CB, Bartel DP. 3’ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res. 2013;23(12):2078–2090. doi: 10.1101/gr.156919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spangenberg L, et al. Polysome profiling shows extensive posttranscriptional regulation during human adipocyte stem cell differentiation into adipocytes. Stem Cell Res. 2013;11(2):902–912. doi: 10.1016/j.scr.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11(21):2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 20.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11(10):1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3’ end processing reveals a decisive role of human cleavage factor I in the regulation of 3’ UTR length. Cell Rep. 2012;1(6):753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Li W, et al. Systematic profiling of poly(A)+ transcripts modulated by core 3’ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 2015;11(4):e1005166. doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber AR, Martin G, Keller W, Zavolan M. Cleavage factor Im is a key regulator of 3’ UTR length. RNA Biol. 2012;9(12):1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- 24.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang JW, Yeh HS, Yong J. Alternative polyadenylation in human diseases. Endocrinol Metab (Seoul) 2017;32(4):413–421. doi: 10.3803/EnM.2017.32.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weng T, et al. Cleavage factor 25 deregulation contributes to pulmonary fibrosis through alternative polyadenylation. J Clin Invest. 2019;129(5):1984–1999. doi: 10.1172/JCI122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu J, et al. Reversing Mechanoinductive DSP Expression by CRISPR/dCas9-mediated Epigenome Editing. Am J Respir Crit Care Med. 2018;198(5):599–609. doi: 10.1164/rccm.201711-2242OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19(11):1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Q, Gilmartin GM, Doublié S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3’ processing. Proc Natl Acad Sci USA. 2010;107(22):10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Implications of polyadenylation in health and disease. Nucleus. 2014;5(6):508–519. doi: 10.4161/nucl.36360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rüegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1(2):243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Gilmartin GM, Doublié S. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5’ capping and splicing. RNA Biol. 2011;8(5):748–753. doi: 10.4161/rna.8.5.16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y, et al. Molecular Mechanisms for CFIm-mediated regulation of mRNA alternative polyadenylation. Mol Cell. 2018;69(1):62–74.e4. doi: 10.1016/j.molcel.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin G, et al. Arginine methylation in subunits of mammalian pre-mRNA cleavage factor I. RNA. 2010;16(8):1646–1659. doi: 10.1261/rna.2164210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 36.Ruepp MD, et al. Mammalian pre-mRNA 3’ end processing factor CF I m 68 functions in mRNA export. Mol Biol Cell. 2009;20(24):5211–5223. doi: 10.1091/mbc.e09-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millevoi S, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3’ end processing machineries. EMBO J. 2006;25(20):4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardy JG, Norbury CJ. Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation. Biochem Soc Trans. 2016;44(4):1051–1057. doi: 10.1042/BST20160078. [DOI] [PubMed] [Google Scholar]
- 39.Mandel CR, et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3’-end-processing endonuclease. Nature. 2006;444(7121):953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3’ processing endonuclease. RNA. 2004;10(4):565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 43.Charest JM, Califano JP, Carey SP, Reinhart-King CA. Fabrication of substrates with defined mechanical properties and topographical features for the study of cell migration. Macromol Biosci. 2012;12(1):12–20. doi: 10.1002/mabi.201100264. [DOI] [PubMed] [Google Scholar]
- 44.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;Chapter 10:Unit 10.16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 45.Cretu A, Castagnino P, Assoian R. Studying the effects of matrix stiffness on cellular function using acrylamide-based hydrogels. J Vis Exp. 2010;(42):2089. doi: 10.3791/2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelham RJ, Wang Yl Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khatiwala CB, Peyton SR, Putnam AJ. Intrinsic mechanical properties of the extracellular matrix affect the behavior of pre-osteoblastic MC3T3-E1 cells. Am J Physiol, Cell Physiol. 2006;290(6):C1640–C1650. doi: 10.1152/ajpcell.00455.2005. [DOI] [PubMed] [Google Scholar]
- 48.Ko J, et al. Transforming growth factor β1 alters the 3’-UTR of mRNA to promote lung fibrosis. J Biol Chem. 2019;294(43):15781–15794. doi: 10.1074/jbc.RA119.009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.