Abstract
The Arp2/3 complex has so far been considered to be a single seven-subunit protein complex required for actin nucleation and actin filament polymerization in diverse critical cellular functions including phagocytosis, vesicular trafficking and lamellipodia extension. The Arp2/3 complex is also exploited by bacterial pathogens and viruses during cellular infectious processes. Recent studies suggest that some subunits of the complex are dispensable in specific cellular contexts, pointing to the existence of alternative ‘hybrid Arp2/3 complexes’ containing other components such as vinculin or α-actinin, as well as different isoforms or phosphorylation variants of canonical Arp2/3 subunits. Therefore, this diversity should be now considered when assigning specific Arp2/3 assemblies to different actin-dependent cellular processes.
Introduction
The actin cytoskeleton is a main component of eukaryotic cells, that not only provides the molecular basis for cellular morphogenesis and migration, but also participates dynamically in mechanical resistance to deformation, uptake of extracellular material, intracellular vesicular transport and cell adhesion. The actin cytoskeleton also participates in the organization of complex cellular structures such as filopodia, lamellipodia and podosomes [1,2].
Polymerization of actin monomers into actin filaments requires the activity of cellular actin nucleators. The Arp2/3 complex, the first nucleator identified in eukaryotic cells, plays a central role in many cellular processes and is highly conserved from trypanosomes to the fission yeast and humans [3–5]. Other nucleators include formins, Spire, Cordon-bleu (COBL) and Leiomodins [6].
The Arp2/3 complex is composed of seven subunits [7] and it has been traditionally considered as a single entity, associated with the vast majority of cellular processes in which its function is required and has been studied. Recent studies [8–10] in the same mammalian cell system reveal that diverse Arp2/3 complexes may regulate different cellular and pathogen-associated functions, raising the possibility that Arp2/3 complex compositions may have been overlooked, and paving the way for the identification of novel complexes associated to different actin polymerization-mediated processes.
Discovery and Functions of the Classical Arp2/3 Complex
The Arp2/3 complex was first isolated from Acanthamoeba castellanii during a search for ligands of the actin-binding protein profilin [11]. It contained seven proteins: the actin-related proteins Arp2 (44-kD) and Arp3 (47-kD) considered as ‘unconventional actins’, together with a 40 kD protein similar to a WD40 β-propeller protein from Dictyostelium discoideum, and four additional proteins of 35 kD, 19 kD, 18 kD and 13 kD [11]. Subsequently, the Arp2/3 complex was also identified and associated with actin-rich structures in the fission yeast Schizosaccharomyces pombe [12] and in the budding yeast Saccharomyces cerevisiae [13]. In human cells, the Arp2/3 complex consists of Arp2 and Arp3, together with the Arp complex subunits ARPC1, ARPC2, ARPC3, ARPC4 and ARPC5 [14]. While consensus exists concerning the Arp2 and Arp3 nomenclature, different names have been used in the literature concerning other Arp2/3 complex subunits: a nomenclature update across species is presented in Table 1.
Table 1. Arp2/3 Complex Nomenclature.
| Common Vertebrate Name | Homo sapiens | Mus musculus | Danio rerio | Drosophila melanogaster | Caenorhabditis elegans | Leishmania major | Saccharomyces cerevisiae | Arabidopsis thaliana | Acanthamoeba castellanii | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arp2 | Arp2 | Arp2 | Arp2 | Arp2A Arp2B | Arp2 | Arp2 | Arp2 | Arp2 | Arp2 | Arp2 | ||||
| Arp3 | Arp3 | Arp3A Arp3B | Arp3 | Arp3A Arp3B | Arp3 | Arp3A Arp3B | Arp3 | Arp3 | Arp3 | Arp3 | Arp3 | Arp3 | ||
| p41 | ARPC1 | ARPC1A ARPC1B | ARPC1 | ARPC1A ARPC1B | ARPC1 | ARPC1A ARPC1B | ARPC1 | ARPC1 | ARPC1 | Arc40/Sop2 | ARPC1 | ARPC1A ARPC1B | p40 | |
| p34 | ARPC2 | ARPC2 | ARPC2 | ARPC2 | ARPC2 | ARPC2 | ARC35 | ARPC2 | ARPC2A ARPC2B | p35 | ||||
| p21 | ARPC3 | ARPC3 | ARPC3 | ARPC3 | ARPC3A ARPC3B | ARPC3 | ARC18 | ARPC3 | p19 | |||||
| p20 | ARPC4 | ARPC4 | ARPC4 | ARPC4 | ARPC4 | ARPC4 | ARC19 | ARPC4 | p18 | |||||
| p16 | ARPC5 | ARPC5A ARPC5B | ARPC5 | ARPC5 | ARPC5A ARPC5B | ARPC5 | ARPC5 | ARC15 | ARPC5 | ARPC5A ARPC5B | p14 | |||
The function of the Arp2/3 complex was shown for the first time to be critical in triggering actin polymerization when it was isolated from a subcellular fraction of human platelets that sustained actin assembly by the bacterial pathogen Listeria monocytogenes (see Glossary) [15]. The L. monocytogenes surface protein ActA activates the Arp2/3 complex to initiate actin polymerization, and was the first actin nucleation promoting factor (NPF) to be identified [16,17]. Several mammalian NPFs were subsequently identified, including WASP [18], N-WASP [19], and Scar/WAVE [20] (see Box 1). The purified A. castellanii Arp2/3 complex was shown to nucleate the formation of actin filaments at 70° from other filaments [21]. A series of elegant microscopy and biochemistry investigations then definitively established the key role of Arp2/3 in actin polymerization and the formation of branched structures [21–23] (see Box 2).
Glossary.
Focal adhesions: large multiprotein complexes that act as transmembrane links between the extracellular matrix and the actin cytoskeleton. The protein network associated to integrin receptor that bridges between the extracellular matrix and the termini of actin stress fibers is called ‘the adhesome’.
Listeria monocytogenes: a Gram-positive bacterium responsible for a food-borne disease named listeriosis, which can lead to meningitis in newborns and abortions in pregnant women. L. monocytogenes is the prototype intracellular pathogen, inducing its internalization within nonphagocytic cells, lysing its phagosome and using an actin-based motility system to promote cell-to-cell spread.
Nucleation promoting factors (NPFs): factors that activate the Arp2/3 complex to polymerize actin above the basal nucleation rate threshold displayed by the native complex. The activities of the NPFs are regulated by signal transduction pathways that coordinate actin cytoskeleton polymerization in time and space.
Vaccinia virus: a prototypic poxvirus closely related to variola virus, the causative agent of smallpox. Poxviruses are enveloped DNA viruses that have the particular ability to exist in two infectious forms: mature virions are viral particles contained within a single membrane, while extracellular virions are contained within two concentric membranes. Vaccinia virus particles are propelled on the tip of actin tails at the surface of infected host cells.
Box 1. Activation of the Arp2/3 Complex by NPFs.
The Arp2/3 complex is by itself an inefficient actin nucleator, and requires the binding of nucleation promoting factors (NPFs) to stimulate its nucleation activity. While preformed actin filaments are also Arp2/3 complex activators [20], the most efficient NPFs include the bacterial surface proteins ActA and RickA from L. monocytogenes and R. conorii respectively [15,33,61] as well as the eukaryotic WASP (Wiskott–Aldrich syndrome protein) [62], SCAR/WAVE (suppressor of cyclic AMP repressor/WASp-family verprolin-homologous protein) [20], WASH (WASP and Scar homologue) [63], WHAMM (WASP homolog associated with actin, membranes and microtubules) [64], and JMY (junction mediating and regulatory protein, p53 cofactor) [65]. These NPFs characteristically display a WCA domain (W: WASP-homology 2 domain or WH2, C: Connector, A: Acidic; this domain is also known as VCA domain: Verprolin-Connector and Acidic-rich) that increases the affinity of Arp2/3 complex for the mother filament, activating the complex [4,66]. NPFs like ActA and WASP dimerize to deliver actin monomers to the Arp2/3 complex [67]. NPFs dissociate from the Arp2/3 complex and may participate in multiple rounds of activation [68].
Box 2. Actin Branching by the Arp2/3 Complex.
The Arp2/3 complex generates new actin filaments that branch-off from the side of pre-existing filaments at a 70° angle to form a Y-branched network [23]. The use of electron tomography to reconstruct the branch junction suggested that the Arp2 and Arp3 subunits reorganize into a dimer, providing a template for elongation of the daughter filament [52]. The remaining subunits, in particular ARPC2 and ARPC4, were reported to make substantial contacts with the mother filament and allow the anchoring of Arp3 as the first subunit of the daugther filament [52]. Subsequent mutagenesis studies have confirmed a role for ARPC2 and ARPC4 in providing an actin-filament-binding interface which is critical for nucleation and branch stability [69] and suggest for the yeast Arc40 (ARPC1 in mammalian cells, see nomenclature in Table 1) multiple essential roles, including suppression of spontaneous nucleation by the Arp2/3 complex and propagation of NPF activation signals [70].
As mentioned above, the function of the Arp2/3 complex is subverted by bacterial pathogens at different stages of their infectious processes [24]. The Gram-positive pathogen L. monocytogenes uses Arp2/3 not only to mediate intracellular and intercellular movements but also to trigger cellular invasion [25–27]. The Gram-negative pathogen Shigella flexneri also requires Arp2/3 function for actin-based motility [28] and for bacterial internalization within host cells [29]. Interestingly, S. flexneri does not express an ActA-like protein but instead recruits on its surface, via the protein IcsA/VirG, the NPF N-WASP which in turn activates Arp2/3 to mediate actin-based motility [28,30]. The Gram-negative bacteria Rickettsia conorii and Rickettsia parkerii activate Arp2/3 during early stages of bacterial intracellular motility via a protein called RickA [31–33]. Moreover, R. conorii and R. parkerii require Arp2/3 activity to invade diverse host cells [34,35]. Vaccinia virus is able to move at the surface of cells on actin-based structures that resembles pedestals [36], which requires the function of the Arp2/3 complex [37]. Other bacteria including Mycobacterium marinum [38] and Burkholderia thailandensis also move via an actin-based motility requiring Arp2/3 functions [39–42].
In S. pombe and S. cerevisiae, deletions of genes encoding each of the subunits of the Arp2/3 complex cause severe growth defects or lethality [43,44], suggesting a major role for all subunits in vivo. In particular, Arp2/3 had been shown to be important for the formation and function of cortical actin patches where clathrin-mediated endocytosis takes place [13]. Mammalian Arp2/3 complex was localized to regions of lamellipodial protrusion [14,45] and together with cofilin and other actin-binding proteins, was shown to control the organization and treadmilling of actin filaments in lamellipodia [22]. The Arp2/3 complex has been associated to other cellular functions requiring actin polymerization including phagocytosis [46], trafficking within and from the Golgi apparatus [47] as well as formation of focal adhesions [48]. The critical role of Arp2/3 in humans is highlighted by the Wiskott–Aldrich syndrome (WAS), a recessive X-linked genetic disorder characterized by mutations in the WAS protein (WASP), which is characterized by defects in the actin-rich immunological synapse between T cells and antigen presenting cells, leading to severe defects in immunological responses [49,50].
Initial cryo-electron microscopy [51], X-ray crystallography [52] and biochemical reconstitution [53] studies identified major roles played by each subunit of the Arp2/3 complex, which have been complemented by electron tomography of the branch junction [54]. Arp2 and Arp3 are folded like actin and form the first two subunits of the daughter filament [51,52]. A dimer of ARPC2 and ARPC4 forms the structural backbone of the complex [52,54] which provides the main surface for interaction of the complex with the mother actin filament [53]. ARPC3 is proposed to form a bridge between Arp3 and the mother actin filament [54] and increases the efficiency of nucleation [53], but complexes lacking ARPC3 display minor functional defects [44,53]. While ARPC1 is supposed to make only minor contacts with the mother actin filament [54], complexes lacking this subunit are far less effective in actin nucleation, suggesting additional roles for ARPC1 including binding of NPFs [55]. ARPC5 was proposed to tether Arp2 to the rest of the complex [54].
Several reports also suggest a functional role played by phosphorylation of different subunits of the Arp2/3 complex. ARPC1 phosphorylation by p21-activated kinase (Pak1) was reported to be crucial for mammalian cell motility [56]. It has been suggested that Arp2 phosphorylation is required and critical for Arp2/3 complex binding to the pointed end of actin filaments and actin nucleation in cultured Drosophila cells [57], but mutation of the phosphorylated residues had only subtle effects on motility in Dictyostelium [58]. As shown recently, phosphorylation of Arp3 by the Legionella pneumophila kinase LegK2 inhibits actin polymerization at the surface of bacterial-containing phagosomes [59].
Several subunits of the Arp2/3 complex (i.e. Arp3, ARPC1 and ARPC5) display more than one isoform [14], but the functional significance of these variants has not been investigated in detail. While the major subunit Arp3 is detected in all tissues, a gene encoding the isoform ARP3β has been detected predominantly in brain neuronal cells and was proposed to play a role in the development and/or maintenance of nerve cells [60]. Two variants of ARPC1 presenting 70% homology had been known for long [12,45] and a mutation in the gene ARPC1A was shown to impact cell migration and invasion in pancreatic cancer [61]. ARPC5 was also found to display a second isoform, named ARPC5B, expressed constitutively in many tissues but with the highest levels in the brain, while the original ARPC5A was found highly enriched in the spleen and thymus [62].
Diversity of Arp2/3 Complexes
Focal Adhesions
Association of the Arp2/3 complex to focal adhesions in human skin cells had previously been shown to require interactions with vinculin [48]. A recent native mass spectrometry analysis of proteins extracted from the dense plaques (focal adhesion homologous structures) of chicken smooth muscle detected Arp2/3 complexes consisting of a core composed of Arp2, Arp3 and ARPC2, together with α-actinin and vinculin, or Arp2, Arp3, ARPC2, ARPC3 and vinculin, that is, ‘hybrid complexes’ [8]. This study therefore supported, for the first time, the notion that alternative Arp2/3 complexes that do not consist of the seven classical subunits are involved in specific cellular processes. Notably, these alternative complexes contain vinculin, which can mediate the recruitment of the complex to focal adhesions and compete with ARPC1B in HeLa cells. Accordingly, knock-down of ARPC1B had a positive effect on focal adhesion and stress fiber formation, in line with an equilibrium shifted towards formation of Arp2/3-vinculin hybrid complexes [8].
Listeria monocytogenes Infection
L. monocytogenes, as mentioned above, uses the Arp2/3 complex for entry into cells and for actin-based motility. Specific roles for ARPC1A and ARPC1B were recently identified in human genome-wide RNA interference (RNAi) screens investigating HeLa cell infection by L. monocytogenes [9]. Knockdown of ARPC1B but not of ARPC1A significantly diminished bacterial entry, while inactivation of ARPC1A had a more profound impact on actin tail formation than inactivation of ARPC1B. The use of small interfering RNAs targeting the subunits ARPC5A (the product of the ARPC5 gene) and ARPC5B (the product of the ARPC5L gene) did not alter bacterial entry nor actin tail formation, suggesting that the ARPC5 subunits are dispensable for both cellular processes. ArpC4 was also shown to be dispensable for L. monocytogenes early invasion of host cells [9]. At this stage, taking into account the central place of the ARPC4 subunit in the Arp2/3 complex function according to previous functional and structural results [53,54], it is not possible to exclude that residual ARPC4 levels may explain the absence of phenotype upon anti-ARPC4 RNAi treatment during bacterial entry.
Vaccinia Virus Mobility
In a recent study of actin polymerization by vaccinia virus, specific roles for ARPC1B and ARPC5B have been found [10]. Indeed, it has been observed that Arp2/3 complexes containing ARPC1B and ARPC5B are significantly more efficient at promoting actin assembly than those containing ARPC1A and ARPC5A. Interestingly, actin networks induced by complexes containing the subunits ARPC1B and ARPC5B were found more stable since in the presence of these specific subunits, cortactin stabilizes the Arp2/3 complexes against coronin-mediated disassembly [10].
Overall, these three reports confirm that the subunits Arp2 and Arp3 play a critical role as an actin nucleation core module, whereas the role of the other subunits is regulatory, determining the efficiency of actin nucleation as well as localization of the complex [8,53,54]. In the case of the L. monocytogenes model, it is interesting to mention that vinculin inactivation by siRNA did not perturb bacterial cellular invasion nor actin-based motility [9], raising the possibility that other cellular molecule(s) not yet identified participate in the localization/modulation of the Arp2/3 complex during L. monocytogenes infection-related processes.
Concluding Remarks
While the Arp2/3 complex has been classically considered as a single molecular entity for 20 years since its discovery, an emerging possibility from recent research suggests that multiple versions of the Arp2/3 complex may coexist in cells (see Outstanding Questions; Figure 1, Key Figure, presents a summary of currently described complexes and their mode of regulation). Differential expression of the Arp2/3 complex subunits in various cells may drive this diversity.
Outstanding Questions.
The regulation of the Arp2/3 complex diversity in cells is unclear at present. Available subunits could determine the composition of the complexes in some cellular contexts. No evidence exists for subunit exchange in already formed Arp2/3 complexes, but this possibility can not be formally excluded.
Which other molecules may be associated with canonical subunits of the Arp2/3 complex? A future challenge will be to isolate and characterize the various Arp2/3 complexes, and assess their activity in vitro.
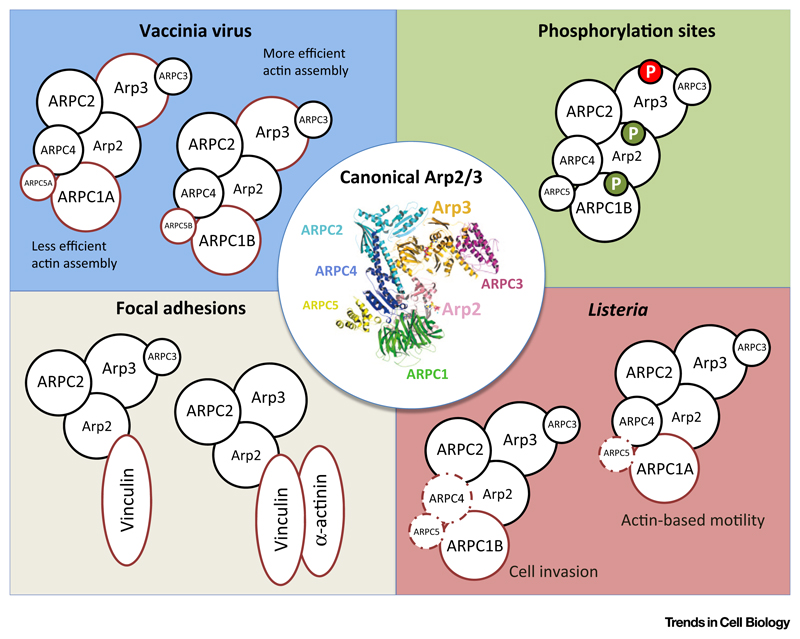
Figure 1. Key Figure: Diversity of Arp2/3 Complexes.
Central circle (white): ribbon diagram of the canonical 7-subunit Arp2/3 complex [63]; copyright (2004) National Academy of Sciences, U.S.A. Top left (blue): Arp2/3 complex variants used by Vaccina virus (alternative subunits are enclosed by a red line). Many combinations of Arp2/3 subunits are recruited by the virus. Bottom left: two ‘hybrid complexes’, containing the actin nucleation core and vinculin, or vinculin plus α-actinin, which presumably localize the complex to focal adhesions. Bottom right: Arp2/3 complexes hijacked by L. monocytogenes during cellular infection (alternative subunits are enclosed by a red bold line, dispensable subunits are enclosed by a red pointed line). Top right: variations in Arp2/3 complexes caused by phosphorylation of specific subunits. The effect of the subunit substitution on the actin nucleation activity is color coded (red: reduce; green: enhance).
In the case of the L. monocytogenes system, in wild-type cells the subunit ARPC5 can be detected at both bacterial entry sites and actin comet tails by fluorescence microscopy, indicating that although ARPC5 is dispensable, Arp2/3 complexes containing this subunit may still be functional during both processes [9]. It is possible that Arp2/3 complexes of different composition have overlapping functions during L. monocytogenes infection, but current data suggests that the precise composition of different Arp2/3 complexes plays a role in fine-tuning actin rearrangements in both cases. This hypothesis is supported by the observation that ARPC4 can be found predominantly early during bacterial actin comet tail formation and that knockdown of ARPC4 affects initial actin polymerization at the bacterial surface rather than actin tail elongation, indicating that different Arp2/3 complexes may be required in a sequential manner. The fine-tuning of Arp2/3 complex actin polymerization activity depending on the subunit composition is also supported by results on the vaccinia virus system [10]. It is highly anticipated that other examples of diverse Arp2/3 complexes in other systems will soon be reported.
Trends.
Mass spectrometry analysis of proteins extracted from focal adhesions identified two Arp2/3 hybrid complexes: the first is composed of Arp2, Arp3, ARPC2 and ARPC3 together with vinculin; the second is composed of Arp2, Arp3, ARPC2 and vinculin together with α-actinin.
Functional studies of Arp2/3 complex subunits during Listeria monocytogenes cell invasion and actin-based motility suggest that diverse complexes participate at each infection stage. The subunit ARPC1B is predominantly required for cellular entry while the subunit ARPC1A is predominantly required for intracellular actin polymerization. Moreover, the subunit ARPC5 is dispensable for both processes.
Arp2/3 complexes containing the subunits ARPC1B and ARPC5B are more efficient at promoting actin assembly by vaccinia virus than complexes containing ARPC1A and ARPC5A subunits.
Post-translational modifications of different subunits affect the efficiency of Arp2/3 complex activity. Phosphorylation of Arp2 and ARPC1B enhances actin nucleation, while phosphorylation of Arp3 attenuates it.
Acknowledgments
This work was supported by the Institut Pasteur, the Institut National de la Santé et de la Recherche Médicale (INSERM Unité 604), the Institut National de la Recherche Agronomique (INRA Unité Sous Contrat 2020), the Institut Pasteur Programmes Transversaux de Recherche’ (PTR 460 and PTR521 to JPC), L’Agence Nationale de la Recherché (ANR-15-CE15-0017 StopBugEntry to JPC), Fondation Le Roch Les Mousquetaires, European Research Council Advanced Grant (670823 BacCellEpi to PC), and under grant agreement n° 294852-SynAd (to BG). P.C. is an International Senior Research Scholar of the Howard Hughes Medical Institute. B.G. holds the Erwin Neter Chair in Tumor and Cell Biology.
Footnotes
The authors declare no conflict of interest.
References
- 1.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 4.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 5.Krause M, Gautreau A. Steering cell migration: lamel-lipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 6.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard TD, Beltzner CC. Structure and function of the Arp2/3 complex. Curr Opin Struct Biol. 2002;12:768–774. doi: 10.1016/s0959-440x(02)00396-2. [DOI] [PubMed] [Google Scholar]
- 8.Chorev DS, et al. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms4758. [DOI] [PubMed] [Google Scholar]
- 9.Kühbacher A, et al. Genome-wide siRNA screen identifies complementary signaling pathways involved in Listeria infection and reveals different actin nucleation mechanisms during Listeria cell invasion and actin comet tail formation. mBio. 2015;6:e00598–e615. doi: 10.1128/mBio.00598-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abella JVG, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2015;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 11.Machesky LM, et al. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balasubramanian MK, et al. Fission yeast Sop2p: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- 13.Winter D, et al. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- 14.Welch MD, et al. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welch MD, et al. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 16.Kocks C, et al. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 17.Welch MD, et al. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 18.Yarar D, et al. The Wiskott–Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- 19.Rohatgi R, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 20.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullins RD, et al. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and tread-milling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchoin L, et al. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 24.Cossart P. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2000;2:195–205. doi: 10.1046/j.1462-5822.2000.00053.x. [DOI] [PubMed] [Google Scholar]
- 25.Pizarro-Cerdá J, et al. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a010009. a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bierne H, et al. A role for cofilin and LIM kinase in Listeria-induced phagocytosis. J Cell Biol. 2001;155:101–112. doi: 10.1083/jcb.200104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sousa S, et al. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeria into cells. Cell Microbiol. 2007;9:2629–2643. doi: 10.1111/j.1462-5822.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 28.Egile C, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bougnères L, et al. Cortactin and Crk cooperate to trigger actin polymerization during Shigella invasion of epithelial cells. J Cell Biol. 2004;166:225–235. doi: 10.1083/jcb.200402073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, et al. Neural Wiskott–Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 1998;17:2767–2776. doi: 10.1093/emboj/17.10.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeng RL, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- 32.Gouin E, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- 33.Reed SC, et al. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2013;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez JJ. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117:5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- 35.Reed SCO, et al. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell Microbiol. 2012;14:529–545. doi: 10.1111/j.1462-5822.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cudmore S, et al. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 37.Frischknecht F, et al. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- 38.Stamm LM, et al. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med. 2003;198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sitthidet C, et al. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J Bacteriol. 2010;192:5249–5252. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouin E, et al. Actin-based motility of intracellular pathogens. Curr Opin Microbiol. 2005;8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Benanti EL, et al. Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell. 2015;161:348–360. doi: 10.1016/j.cell.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouin E, et al. Intracellular bacteria find the right motion. Cell. 2015;161:199–200. doi: 10.1016/j.cell.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 43.Lees-Miller JP, et al. Identification of act2, an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc Natl Acad Sci USA. 1992;89:80–83. doi: 10.1073/pnas.89.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter DC, et al. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA. 1999;96:7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machesky LM, et al. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.May RC, et al. Involvement of the Arp2/3 complex in phagocytosis mediated by FcγR or CR3. Nat Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- 47.Luna A, et al. Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell. 2002;13:866–879. doi: 10.1091/mbc.01-12-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demali KA, et al. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolluri R, et al. Direct interaction of the Wiskott–Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci USA. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirchhausen T, Rosen FS. Disease mechanism: unravelling Wiskott–Aldrich syndrome. Curr Biol. 1996;6:676–678. doi: 10.1016/s0960-9822(09)00447-3. [DOI] [PubMed] [Google Scholar]
- 51.Volkmann N, et al. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 52.Robinson RC, et al. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 53.Gournier H, et al. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8:1041–1052. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 54.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly AE, et al. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vadlamudi RK, et al. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.LeClaire LL, et al. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182:647–654. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi CH, et al. Phosphorylation of actin-related protein 2 (Arp2) is required for normal development and cAMP chemotaxis in Dictyostelium. J Biol Chem. 2013;288:2464–2474. doi: 10.1074/jbc.M112.435313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michard C, et al. The Legionella kinase LegK2 targets the ARP2/3 complex to inhibit actin nucleation on phagosomes and allow bacterial evasion of the late endocytic pathway. mBio. 2015;6:e00354–e415. doi: 10.1128/mBio.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jay P, et al. ARP3β, the gene encoding a new human actin-related protein, is alternatively spliced and predominantly expressed in brain neuronal cells. Eur J Biochem. 2000;267:2921–2928. doi: 10.1046/j.1432-1327.2000.01306.x. [DOI] [PubMed] [Google Scholar]
- 61.Laurila E, et al. Characterization of the 7q21-q22 amplicon identifies ARPC1A, a subunit of the Arp2/3 complex, as a regulator of cell migration and invasion in pancreatic cancer. Genes Chromosom Cancer. 2009;48:330–339. doi: 10.1002/gcc.20643. [DOI] [PubMed] [Google Scholar]
- 62.Millard TH, et al. Identification and characterisation of a novel human isoform of Arp2/3 complex subunit p16-ARC/ARPC5. Cell Motil Cytoskeleton. 2003;54:81–90. doi: 10.1002/cm.10087. [DOI] [PubMed] [Google Scholar]
- 63.Nolen BJ, et al. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campellone KG, et al. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuchero JB, et al. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rotty JD, et al. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2012;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 67.Padrick SB, et al. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Egile C, et al. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goley ED, et al. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci USA. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balcer HI, et al. The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation. J Biol Chem. 2010;285:8481–8491. doi: 10.1074/jbc.M109.054957. [DOI] [PMC free article] [PubMed] [Google Scholar]