Abstract
With its large proportion of endemic taxa, complex geological past, and location at the confluence of the highly diverse Malesian and Australian floristic regions, Papuasia – the floristic region comprising the Bismarck Archipelago, New Guinea, and the Solomon Islands – represents an ideal natural experiment in plant biogeography. However, scattered knowledge of its flora and limited representation in herbaria have hindered our understanding of the drivers of its diversity. Focusing on the woody angiosperm genus Schefflera (Araliaceae), we ask whether its morphologically defined infrageneric groupings are monophyletic, when these lineages diverged, and where (within Papuasia or elsewhere) they diversified. To address these questions, we use a high-throughput sequencing approach (Hyb-Seq) which combines target capture (with an angiosperm-wide bait kit targeting 353 single-copy nuclear loci) and genome shotgun sequencing (which allows retrieval of regions in high-copy number, e.g., organellar DNA) of historical herbarium collections. To reconstruct the evolutionary history of the genus we reconstruct molecular phylogenies with Bayesian inference, maximum likelihood, and pseudo-coalescent approaches, and co-estimate divergence times and ancestral areas in a Bayesian framework. We find strong support for most infrageneric morphological groupings, as currently circumscribed, and we show the efficacy of the Angiosperms-353 probe kit in resolving both deep and shallow phylogenetic relationships. We infer a sequence of colonization to explain the present-day distribution of Schefflera in Papuasia: from the Sunda Shelf, Schefflera arrived to the Woodlark plate (present-day eastern New Guinea) in the late Oligocene (when most of New Guinea was submerged) and, subsequently (throughout the Miocene), it migrated westwards (to the Maoke and Bird’s Head Plates and thereon) and further diversified, in agreement with previous reconstructions.
Keywords: sequence capture, target enrichment, herbariomics, historical biogeography, Papuasia, New Guinea, Araliaceae, Schefflera
Introduction
Situated at the crossroads between Asia and Australia (Lohman et al., 2011), Papuasia has inspired biogeographic research since the time of Wallace (1869). Plate tectonic processes, from volcanism and deformation to ophiolite obduction and island-arc accretion (Baldwin et al., 2012), have created a plethora of terrestrial ecosystems – from mangroves to subalpine grasslands, through tropical forests (Paijmans, 1976; Wikramanayake, 2002; Marshall, 2007) – that support some of the richest diversity on Earth (Williams, 2011). The Papuasian floristic region comprises the main island of New Guinea, the Bismarck Archipelago, and the Solomon Islands (Warburg, 1891; Brummitt, 2001). Its 54% endemic plant taxa (van Welzen et al., 2011) is attributed to its high environmental heterogeneity and isolation (Beehler, 2007; Mutke et al., 2011). Indeed, elevation and terrain ruggedness (i.e., elevational heterogeneity) have been shown to strongly correlate with orchid diversity (Vollering et al., 2016) and even terrestrial-plant genus richness (Hoover et al., 2017), hinting toward orogenetic (i.e., mountain building) processes as catalysts of plant radiation in the region. The spatial distribution of morphological clades in families Sapindaceae (van Welzen et al., 2001) and Ericaceae (Heads, 2003) broadly correspond to geological terranes (i.e., crust fragments sutured to a plate other than that of origin) of various ages leading the authors of both studies to ascribe cladogenesis to vicariance events. Despite these apparent associations, the evolutionary links between past geological events and present-day distributions remain largely unexplored in the region.
With over 600 species, Schefflera J.R. Forst & G. Forst s.l. is one of the largest angiosperm genera and the most speciose genus in Araliaceae (Frodin, 1975; Frodin and Govaerts, 2003; Frodin, 2004). In Papuasia, the genus has around 200 estimated species and exhibits a wide environmental tolerance (Figure 1) and plasticity of growth forms. Schefflera s.l. attains its greatest diversity in Papuasia as trees in montane forests between 1,000 and 2,500 m a.s.l., but other growth forms also include prominent epiphytes or shrubs in lower-montane (650–1,500 m) to mid- and upper-montane forests (1,500–3,200 m; Johns et al., 2007b) or even canopy-emergent trees in sub-alpine ecosystems (3,200–4,200 m; Brass, 1941; van Royen, 1979; Johns et al., 2007a), making Schefflera an ideal case study to investigate woody angiosperm diversification in Papuasia. Tectonic models and stratigraphic evidence indicate that New Guinea’s mountains attained their present height by rapid uplift from the late Miocene to the early Pliocene (van Ufford and Cloos, 2005; Hall, 2009). While this suggests that Papuasian Schefflera rapidly diversified within the last 10–5 Myr, studies that test this hypothesis using a representative sample of Papuasian Schefflera have so far been wanting (i.e., nine accessions in Li and Wen, 2014).
FIGURE 1.
Distribution of Schefflera s.l. collections in Papuasia. Over 2,000 collections (colored diamonds) are known from the region, across an elevation gradient from 0 to 4025 m a.s.l. Collections are color-coded according to morphologically defined groups.
Morphologically, most Papuasian Schefflera are classified into three largely endemic infrageneric groups: Brassaia, Papuoschefflera, and Pagiophyllae (Plunkett et al., 2005). The first two have overlapping distributions, while the third one is restricted to montane forests and grasslands above 2000 m in the Maoke Mountains (Papua prov., Indonesia). Papuoschefflera is further classified into six provisional morphological groupings which we here name “Bougainvilleanae,” “Ischnoacrae,” “Versteegiae,” “Schumannianae,” “Ischyrocephalae,” and “Morobeae” (Papuoschefflera one through six, respectively, in Frodin et al., 2010), most of which are geographically restricted within Papuasia. Other morphogroups in the region include Malesian Heptapleurum, Pacific Plerandra, and Philippine-Papuasian Cephaloschefflera. Schefflera elliptica (Heptapleurum group) is widespread in Southeast Asia, while Plerandra and Cephaloschefflera mostly occur around south-eastern New Guinea.
Unraveling the evolutionary history of Papuasian Schefflera requires time-calibrated molecular phylogenies both to ascertain the monophyly of these morphologically defined groupings and to shed light on their observed geographic ranges (Wen et al., 2013).
Phylogenies reconstructed from a nuclear region (ITS) and a plastid locus (trnL-trnF) indicate that Schefflera s.l. is polyphyletic, comprising five geographically distinct clades (Plunkett et al., 2005). Using five additional plastid regions, Li and Wen (2014) established the monophyly of nine accessions from western New Guinea and dated their divergence from Heptapleurum to have taken place ∼22 Ma. However, the Papuan clade recovered by Li and Wen (2014) included S. lucescens and S. polybotrya, which have been recorded from Java, Sumatra, and Borneo but not Papuasia. Frodin et al. (2010) placed S. lucescens in the mainly Sundan Paratropia group, together with S. “zollingeriana” sp. ined., which Plunkett et al. (2005) had resolved as the sister group to Papuasian Schefflera. As a result, two competing hypotheses on the origins of Papuasian Schefflera exist: either they originated within or outside of Papuasia.
Hyb-Seq (Weitemier et al., 2014) is a high-throughput sequence capture approach that combines target enrichment of, e.g., low-copy nuclear orthologs with genome skimming (additionally yielding high copy number regions, like organellar ones). Whereas previous molecular phylogenies of Papuasian taxa relied almost exclusively on silica-dried samples, Hyb-Seq can be implemented on DNA of varying quality, including that of range-restricted and under-collected species only accessible as herbarium specimens (Hart et al., 2016). In this way, Hyb-Seq has been successfully implemented at the population level and above, even with degraded DNA from centuries-old specimens (Villaverde et al., 2018; Brewer et al., 2019). Still, Hyb-Seq, and other target enrichment techniques, have yet to be widely adopted due to the high cost, prior knowledge needed (e.g., available transcriptomes or genomes), and expertise required (e.g., computational skills) at the initial design and optimization stages of taxon-specific probes (Lemmon and Lemmon, 2013; Dodsworth et al., 2019). To overcome this design and optimization hurdle, the Angiosperms-353 probe set was developed from available transcriptome and genome data (One Thousand Plant Transcriptomes Initiative, 2019) – including twelve Apiales representatives, three of them (Hedera, Hydrocotyle, and Polyscias) in Araliaceae – with universal probes that capture 353 nuclear single-copy loci shared across all angiosperms (Johnson et al., 2018). By using the Angiosperms-353 sequence capture probes, our study is among the first to test the efficacy of this probe set to resolve species-level relationships from either historical herbarium specimens (Brewer et al., 2019; Larridon et al., 2020; Murphy et al., 2020) or fresh tissue (Van Andel et al., 2019).
Here, we aim to reconstruct the evolutionary history of Papuasian Schefflera. To do so, we ask whether its morphologically defined infrageneric groupings are monophyletic, when these lineages diverged, and where (within Papuasia or elsewhere) they diversified.
Materials and Methods
Taxon Sampling
We sampled foliar tissue (i.e., lamina, petioles) from 195 herbarium specimens collected between 1850 and 2018 (including four type specimens). These were selected from a georeferenced database of Schefflera s.l. compiled by D.G. Frodin, with the addition of four samples from the Royal Botanic Gardens, Kew (RBGK) DNA Bank1, and four silica-preserved tissue samples from the RBGK Living Collection2 and the Raja Ampat Islands, West Papua, Indonesia (J. Schrader, GAU Göttingen). Of the 203 sampled specimens, only 74 could be successfully sequenced (due to funding constraints), though these are representative of all Papuasian morphogroups (Frodin et al., 2010) and cover the entire geographic range of the genus in Papuasia (Figure 1 and Supplementary Data Sheet S1A). We selected outgroup taxa from the four primary clades of Asian Schefflera (Li and Wen, 2014), plus its closest Araliad relatives: Heteropanax fragrans (Roxb.) Seem and Tetrapanax papyrifer (Hook.) K.Koch. We also sequenced geographic clades of Schefflera that diverged earlier than Asian Schefflera (Nicolas and Plunkett, 2014; Supplementary Data Sheet S1B) and included genomic sequences of taxa from other major clades in Araliaceae, available through the Plant and Fungal Tree of Life (PAFTOL) Research Programme (Johnson et al., 2018) and the 1000 Plants (1KP) Initiative (Matasci et al., 2014; Supplementary Data Sheet S1C).
Laboratory Protocols
DNA Extraction
Samples were washed with 70% ethanol, then kept at −80°C for 12 h (to facilitate cell wall breakage) and milled in an MM400 (Retsch Inc.) grinder. Genomic DNA was extracted using a modified-CTAB protocol (Doyle and Doyle, 1987), further adjusted to improve yield from herbarium samples (Supplementary Data Sheet S2A). Key changes include incubating samples in CTAB at 65°C for 12 h (to optimize DNA isolation) and in isopropanol at −20°C for 48 h (to improve precipitation of fragmented DNA). Precipitated DNA pellets were washed twice with 70% ethanol and resuspended in Milli-Q ultrapure (Type 1) water (Merck KGaA). We measured relative DNA concentration (ng/μL) with the QuantiFluor® dsDNA System (Promega Corp.). Samples were purified using Agencourt AMPure XP beads (Beckman Coulter Life Sciences) (Supplementary Data Sheet S2B). Extractions from pre-1970 collections or with concentrations <10 ng/μL were treated as having highly fragmented DNA and were cleaned using AMPure beads and isopropanol following Lee (2014) to reduce loss of small (<300 bp) DNA fragments (Särkinen et al., 2012). Where material was available, we repeated extractions to obtain at least 200 ng of DNA from each sample. DNA fragment size distribution was determined by gel electrophoresis (Supplementary Data Sheet S3A). Extractions with fragments predominantly above the target insert size (≥ 500 bp) were sonicated with an ME220 Focused-ultrasonicatorTM (Covaris Inc.).
Genomic Library Preparation
We prepared genomic libraries using the NEBNext® UltraTM II DNA Library Prep Kit and Multiplex Oligos (Dual Index Primers, sets 1 and 2) for Illumina® (New England Biolabs) at half-volumes to reduce per sample costs. Target insert size was 350 bp and size selection was not required where DNA template was highly degraded (<300 bp). Size-selected libraries were amplified with 13 PCR cycles and all others with 14 cycles. Libraries were re-amplified, where required, using KAPA HiFi HotStart ReadyMix (Roche) with i5 and i7 forward and reverse primers (as described in Meyer and Kircher, 2010) to obtain at least 75 ng/library.
Hybridization and Sequencing
Libraries were multiplexed (11–12 per pool) for hybridization. Equimolar library pools were made homogenizing phylogenetic distances (if known) and avoiding combinations of libraries originating from different quality DNA to reduce uneven bait competition within hybridization pools. We considered the following criteria: (i) whether re-amplification was required, (ii) whether sonication was required, (iii) whether size selection was required, and (iv) whether there was sufficient library template (Supplementary Table S3B). Outgroup taxa were pooled separately from Papuasian taxa where possible to even out occupation of bait binding sites. Each pool contained 500–1,000 ng DNA in total.
Pools were enriched using the Angiosperms-353 myBaits® Expert Panel target capture kit (Arbor Biosciences). They were hybridized at 65°C for 24 h, then amplified with i5 and i7 forward and reverse primers (Meyer and Kircher, 2010) for 14–18 PCR cycles to obtain pools at least 1 nM. Each pool was quality-controlled with a TapeStation 4200 (Agilent Technologies) (Supplementary Data Sheet S3C). Due to funding constraints, we only sequenced the eight library pools with the highest quality, which covered the broadest diversity range and consisted of 90 accessions in total (though only 74 passed quality filters). These were denatured and diluted following manufacturer’s specifications (Illumina® protocol # 15039740) and loaded at 16 pM for sequencing in an Illumina® MiSeq using two v2 (300-cycles) reagent kits (Illumina®, Inc.) at the Jodrell Laboratory (Royal Botanic Gardens, Kew, Richmond, United Kingdom).
Bioinformatic Analyses
Sequence Rescue and Alignment
Sequences were trimmed using Trimmomatic 0.38 (Bolger et al., 2014), employing “palindrome mode” adapter removal and Maximum Information quality filter settings to favor longer reads (ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10:2:TRUE MAXINFO:40:0.2 LEADING:3 TRAILING:3 MINLEN:36). We examined sequence quality with FastQC 0.11.7 (Andrews, 2018) before and after trimming to ensure complete adapter removal and to identify surviving artifacts that could affect downstream analyses.
HybPiper 1.3.1 (Johnson et al., 2016) was used to retrieve target sequences of nuclear genes (exonerate script) and flanking off-target regions (intronerate.py script, which reruns exonerate but, instead of removing flanking regions, it keeps them), with the BWA mapper (Li, 2013) and the SPAdes assembler (Bankevich et al., 2012) (–bwa -cov-cutoff = 3). We investigated polymorphic sequences for paralogy using exploratory trees built from MAFFT 7.215 (Katoh and Standley, 2013) (–auto) alignments in FastTree 2 (Price et al., 2010) (-nt -gtr). We discarded any sequence that may have resulted from gene duplication and retained the most common allele when sequences were homologous (Kates et al., 2018). We used our own custom script, max_overlap3 (Supplementary Data Sheet S4A), to calculate a coverage score for each sequence that is proportional to representativeness (proportion of accessions/loci with sequences), data matrix completeness (percent sequence recovered against overall length of target), and evenness of distribution (adapted from Pielou, 1966) across accessions per locus and so, to reduce noise in our data matrixes by filtering out underrepresented, incomplete, and unevenly distributed sequences.
Internal Transcribed Spacer (ITS1-5.8S-ITS2) nuclear ribosomal DNA sequences (hereafter ITS) were also retrieved with HybPiper using a target file we made from aligned ITS sequences – from Li and Wen (2014) and Plunkett et al. (2004, 2005) – deposited in GenBank (Benson et al., 2018). Off-target sequences corresponding to 142 plastid loci (genes and intergenic spacers) were similarly rescued with HybPiper using a plastid-target file we generated from the complete plastid genomes of S. heptaphylla (L.) Frodin (Zong et al., 2016: KT748629), Aralia elata (Miq.) Seem. (Kim and Yang, 2016: KT153023), S. delavayi (Franch.) Harms, and Metapanax delavayi (Franch.) J.Wen & Frodin (Li et al., 2013: KC456166, KC456165).
All sequences were aligned with UPP (Nguyen N.-P.D. et al., 2015) to produce accurate alignments from fragmentary datasets (i.e., historical herbarium DNA template), using only those with >95% of the longest available sequence length for the backbone dataset. UPP uses hidden Markov models (HMM) for multiple sequence alignment (MSA) and it relies on PASTA 1.8.4 (Mirarab et al., 2015) (a divide-and-conquer MSA method) to generate initial backbone alignments. In turn, PASTA relies on FastTree 2 (Price et al., 2010) for tree estimation, MAFFT 7.215 (Katoh and Standley, 2013) for alignment, and OPAL 2.1.3 (Wheeler and Kececioglu, 2007) for merging. We trimmed alignments using our own custom script, optrimAl4 (Supplementary Data Sheet S4B), which optimizes the gap threshold value in trimAl 1.2 (Capella-Gutiérrez et al., 2009), to obtain the highest proportion of parsimony-informative characters (PPIC) while retaining adequate sequence length (Shen et al., 2016). Alignments where trimming resulted in data loss exceeding 30% were interpreted to contain low ratios of phylogenetic signal to noise and were discarded. Alignment statistics were calculated in AMAS 0.98 (Borowiec, 2016) using the summary function. Sequence capture statistics were calculated using R 3.5.3 (R Core Team, 2019).
Gene and Species Tree Inference
Gene trees were inferred with IQ-TREE 1.6.10 (Nguyen L.-T. et al., 2015) after selecting substitution models with ModelFinder (-m TEST) (Kalyaanamoorthy et al., 2017). Outlier branches that increased the diameter of each gene tree by more than 20% were identified using TreeShrink 1.3.1 (Mai and Mirarab, 2018) with centroid re-rooting (-b 20 -c) and removed. Each locus was then realigned without the outlier sequences. We estimated bipartition support with 1000 UFBoot2 (Hoang et al., 2018) bootstrap replicates using IQ-TREE (-bb 1000) and contracted branches with support values below 10% (Mirarab, 2019) using Newick Utilities 1.6 (nw_ed “i & b ≤ 10”) (Junier and Zdobnov, 2010).
Pseudo-coalescent species trees were inferred using ASTRAL III v5.6.3 (Zhang et al., 2018). Species trees were inferred separately for the nuclear and chloroplast genomes as they represent different evolutionary pathways (Ravi et al., 2008). We used the local posterior probabilities (PPlocal) calculated in ASTRAL to estimate quartet support for the recovered topology at each node. Conflict, concordance, and phylogenetic signal were assessed with phyparts (Smith et al., 2015) and displayed with the PhyPartsPieCharts script5, which depicts the number of gene trees that support, oppose or provide no information with respect to the dominant species tree topology. Unresolved polytomies in the final species tree were tested in ASTRAL (-t 10) to determine if they are due to insufficient data or could possibly reflect a true polytomy (Sayyari and Mirarab, 2018).
To verify placement of our sampled Papuasian and outgroup accessions within family Araliaceae, we inferred the ITS gene tree for these together with sequences of other Araliaceae accessions from Li and Wen (2014) and Plunkett et al. (2004, 2005). We used two accessions from Myodocarpaceae (Delarbrea paradoxa Vieill. and Myodocarpus fraxinifolius Brongn. & Gris.) as the outgroup. The gene tree was estimated in MrBayes 3.2.6 (Ronquist et al., 2012) under a GTR + Γ substitution model (with settings: nchains = 4, nruns = 4, and MCMC ngen = 100M).
Divergence Time Estimation and Ancestral Area Reconstruction
To limit variation in substitution rates and minimize overestimation of recent divergence times (Ho et al., 2005), we selected nuclear genes that (i) were at least 10% concordant with the species tree (bipartition support > 0.1), (ii) likely evolved according to a strict molecular clock (we set root-tip variation to <0.024), (iii) contained the most information (tree length > 0.1), and (iv) represented the most accessions (at least 25) with SortaDate (Smith et al., 2018). To reconstruct the biogeographic history of Papuasian Schefflera (Crisp et al., 2011), we included in the data matrix ITS sequences from other Asian Schefflera clades sampled by Li and Wen (2014). All other loci for these Asian accessions were coded as missing data. This resulted in a 51-taxa data matrix, partitioned by locus (we used independent substitution models for each locus, as selected by ModelFinder), consisting of only Asian and Papuasian Schefflera sequences. This data matrix comprised 31,496 bp across 10 nuclear exonic regions (6,469 bp), 15 nuclear flanking regions (24,177 bp), and nuclear rDNA ITS. The data matrix had 11.6% parsimony-informative sites and, as 16 taxa (32% of the total sampling) are represented only by ITS sequences, 55.5% missing data.
Divergence times were estimated in BEAST 1.10.4 (Suchard et al., 2018) on the CIPRES Science Gateway v3.3 online platform (Miller et al., 2011). Since known Araliaceae fossils lay outside our focus group, three secondary time constraints – drawn from Li and Wen (2014) – were imposed on: (i) the root node (normal prior distribution with mean = 42.0 and st.dev. = 8.0); (ii) the Heptapleurum crown node (normal, mean = 36.0, st.dev. = 7.0); and (iii) the Papuasian Schefflera crown node (normal, mean = 22.0, st.dev. = 5.0). To reduce search-space and avoid miss-rooting problems with BEAST analyses, we enforced monophyly on the Agalma, Brassaia, S. elliptica alliance, Heptapleurum, Heptaphylla + Hypoleuca, Ischyrocephalae, and Papuoschefflera s.s. clades (fully supported in our species tree), as well as the root node, to prevent inverted ingroup-outgroup topologies (for further details see Springer et al., 2018).
Concurrently, ancestral areas were reconstructed using a fully probabilistic approach – first described by Sanmartín et al. (2008) and implemented in BEAST by Lemey et al. (2009) – that infers diffusion processes among discrete locations in timed evolutionary histories under Bayesian stochastic search variable selection (BSSVS). Continuous-time Markov chains (CTMC) are used to model instantaneous geographic locations of any given sequence, together with the transition and migration rates between these locations. Since this Bayesian CTMC phylogeographic model assumes ancestral ranges are limited to single regions, it requires discretizing the entire distribution of any given taxon (i.e., a given taxon can be represented by multiple accessions), while tolerating incomplete sampling (Drummond et al., 2012). Moreover, unlike other approaches (e.g., dispersal-vicariance parsimony or dispersal-extinction cladogenesis), it can be implemented in scenarios where the number of areas is large (>10 areas), allowing for fine-scale area explorations (e.g., Mairal et al., 2015, 2017). Thus, we included a partition with collection localities – coded according to tectonic plate boundaries (Bird, 2003) – in our BEAST input file (generated in BEAUTi 1.10.4; Suchard et al., 2018) and we tested six models, comprising all possible combinations of three clock priors – strict, random local, and uncorrelated relaxed lognormal – and two species-tree priors robust to incomplete sampling (our case) – Yule process (Yule, 1925) and birth-death incomplete sampling (Stadler, 2009). We selected the best-supported model by estimating marginal likelihoods (MLEs, path steps = 100, chain length = 1M), using path sampling (PS) and stepping-stone sampling (SS) (Baele et al., 2013), from runs that converged after 100M iterations. BEAST log files were loaded into Tracer 1.7.1 (Rambaut et al., 2018) and visually inspected to check that the chains had converged, and that mixing and Effective Sample Sizes (ESS > 200) were adequate for all parameters (after 100M iterations). After discarding burn-in iterations, trees were annotated and posterior probabilities (PP) summarized in TreeAnnotator 1.10.4 (Suchard et al., 2018) on the tree in the posterior sample with the maximum sum of the posterior clade probabilities (MCC tree), rescaling to reflect median node heights for clades contained in said tree. The resulting MCC tree was visualized in FigTree 1.4.4 (Rambaut, 2018).
Results
Sequence Capture Success and Bioinformatic Analyses
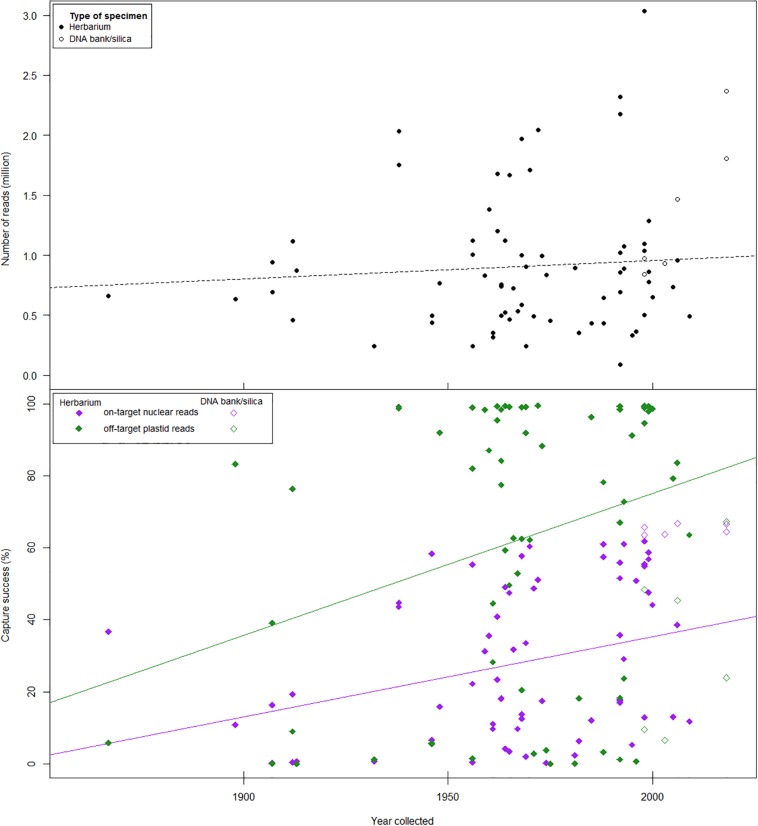
Of the 90 accessions sequenced, only 74 had sufficient reads after quality filtering for target retrieval with HybPiper (median = 737,691 reads/sample; Supplementary Table S5A). We find that specimen age had no significant effect on the number of reads and that read yield did not differ between herbarium specimens and other material (i.e., Kew DNA bank and silica-dried samples; Figure 2). For nuclear loci in the Angiosperms-353 enrichment panel, on average 11.5% reads/sample were on-target. Capture success (defined as the proportion of total reference sequence recovered) varied widely (range = 0.1 – 64.6%) across samples for herbarium material (median = 28% reads/sample) and was weakly correlated with specimen age (F = 7.01, DF = 66, p < 0.01, R2 = 0.08). DNA bank and silica samples yielded higher capture success (t = 14.1, DF = 71.6, p < 0.001). For off-target plastid regions (including both coding loci and intergenic spacers), on average 16.9% of reads/sample mapped to our “plastid-target” custom file, with 58% median “capture success” for herbarium material. Plastid “capture success” was nearly complete for 16 samples (Supplementary Table S5B) and was weakly correlated with specimen age (F = 6.34, DF = 66, p < 0.01, R2 = 0.07). In total, we obtained sequences for 352 coding (on-target) and 349 flanking (off-target) regions from the nucleus, and 73 coding and 64 intergenic spacers from the chloroplast (off-target), as well as the nuclear rDNA ITS region (recovered with “ITS-target” custom file). Sixty potential paralogs were detected based on gene tree topology (Supplementary Table S5C), of which 23 nuclear and two plastid genes were probable duplications and excluded from downstream analyses.
FIGURE 2.
Reads mapped and capture success with respect to specimen age and template type (DNA bank/silica vs. herbarium). (Top) The number of reads obtained did not differ between herbarium specimens (filled circles) and DNA bank/silica samples (empty circles). There was no correlation between the number of reads and specimen age (dotted black line). (Bottom) Capture success (proportion of total reference sequence recovered) of nuclear genes (empty purple diamonds) was significantly higher in DNA bank/silica samples than in herbarium specimens (filled purple diamonds). Capture success in herbarium samples was weakly correlated with specimen age for both nuclear genes (purple line) and plastid loci (green line).
The proportion of parsimony-informative characters (PPIC) was highly variable between multiple sequence alignments (MSA) for nuclear coding loci and flanking regions (Table 1). PPIC was generally low for off-target plastid genes and intergenic spacers. Non-coding off-target regions (nuclear flanking regions and plastid spacers) had higher PPIC than their respective coding counterparts (nuclear targets and off-target plastid loci). Missing data accounted for 27.1% of nuclear targets, 42.3% of nuclear flanking regions (off-target), 7.2% of plastid loci, and 8.0% of plastid spacers. No phylogenetic bias was observed in the distribution of missing data. We included only nuclear regions (both coding and flanking) for phylogenetic inference as the low levels of informativeness in the plastid sequences were more likely to lead to gene tree estimation errors (Meiklejohn et al., 2016). Only 39 accessions had a coverage score of at least 0.5 for nuclear sequence capture; this coverage score is proportional to representativeness x completeness x evenness (see Methods above). These 39 accessions, together with three sequences from 1KP (One Thousand Plant Transcriptomes Initiative, 2019) and one sequence from PAFTOL (Johnson et al., 2018), were used for downstream phylogenomic analyses (pseudo-coalescent framework). The final data set included 33 Papuasian Schefflera accessions and 12 outgroup accessions. After trimming, the data set to be used in pseudo-coalescent analyses comprised 354,057 bases (Supplementary Table S5D) across 141 nuclear coding regions (93,325 bp) and 163 nuclear flanking regions (260,732 bp).
TABLE 1.
Alignment statistics across retrieved regions.
| Genomic compartment | Region | Alignment length |
PPIC* |
Missing data |
|||
| Mean (bp) | SD | Mean (%) | SD | Mean (%) | SD | ||
| Nuclear | On-target coding | 613 | 368 | 17.0 | 10.4 | 27.1 | 11.8 |
| Off-target flanking | 886 | 484 | 28.2 | 14.3 | 42.3 | 10.5 | |
| Plastid | Off-target coding | 873 | 830 | 1.7 | 1.7 | 7.2 | 5.2 |
| Off-target non-coding | 534 | 531 | 2.7 | 2.2 | 8.0 | 5.5 | |
| All | 696 | 506 | 20.3 | 13.6 | 47.6 | 15.6 | |
* PPIC, Proportion of Parsimony-informative characters.
Phylogenetic Relationships in Schefflera
The monophyly of most of the currently accepted genera in Araliaceae was strongly supported in the Bayesian ITS tree (Figure 3 left), save Polyscias (sensu Lowry and Plunkett, 2010), which had relatively low support. Panax was nested within Aralia and Chengiopanax was nested in Gamblea. As expected, Schefflera was highly polyphyletic, following the geographical clades of Plunkett et al. (2005). Schefflera’s “Asian Palmate clade” (Plunkett et al., 2005) was strongly supported (with Neotropical Schefflera nested within it) and had Tetrapanax papyrifer and Heteropanax fragrans gradually leading to the “Asian Schefflera clade” with maximum support. Within this latter “Asian Schefflera clade,” all major clades (sensu Li and Wen, 2014) were also strongly supported including Heptapleurum, which comprises the Schefflera elliptica alliance, the Philippine Schefflera, and the Papuasian clade, which has Schefflera tristis as sister lineage. The pseudo-coalescent species tree also recovered monophyletic Papuasian Schefflera nested within Heptapleurum, which was itself nested in the “Asian Schefflera clade” (Figure 4 left) with maximum quartet support.
FIGURE 3.
Bayesian ITS gene tree (MrBayes). Dashed lines represent low confidence (PPITS < 0.8). Solid lines have maximum support (PPITS = 1) unless otherwise stated (percentages by branches). (Left) Araliaceae genera. All generic clades have been collapsed in this tree, including Papuasian Schefflera. (Right) Papuasian Schefflera. Accession labels are color-coded according to infrageneric morphogroups as in Frodin et al. (2010).
FIGURE 4.
Pseudo-coalescent species tree of Papuasian Schefflera (ASTRAL). Pie charts show gene tree concordance with coalescent species tree. Dashed lines represent low confidence (PPlocal < 0.8). Solid lines have maximum support (PPlocal = 1) unless otherwise stated (percentages by branches). (Left) Outgroup taxa and Papuasian subgeneric groupings. Papuoschefflera has been collapsed in this tree. (Right) Papuoschefflera infrageneric groupings. Accessions are color-coded according to infrageneric morphogroups as in Frodin et al. (2010).
Within Papuasian Schefflera (Figure 3 right, Figure 4 right, and Figure 5), the monophyly of the Brassaia morphogroup was strongly supported, regardless of the inference approach taken. However, phylogenetic relationships within Brassaia were poorly resolved. The Papuoschefflera morphogroup was paraphyletic with regard to Brassaia in the ITS tree, it was supported as sister to Brassaia in the pseudo-coalescent tree and, although the latter topology was also retrieved in the chronogram, it had low support. All three trees disagreed on the placement of Cephaloschefflera (S. eriocephala) but found Pagiophyllae (S. “frigidariorum” sp. ined.) to be nested within Papuoschefflera. Hereafter, we refer to the taxa that are not part of the Brassaia clade as Papuoschefflera s.l. and restrict the circumscription of Papuoschefflera s.s. to Pagiophyllae plus Bougainvilleanae, Schumannianae, and Versteegiae. These morphogroups were reconstructed in a highly supported clade in all trees and are generally distributed across the western half of Papuasia (Figure 5 bottom left). Morphogroup Ischyrocephalae had maximum support both in the ITS tree and the chronogram but had S. “goodenoughiana” sp. ined. (Oreopolae, D.G. Frodin pers. comm.) nested in the pseudo-coalescent tree, which resulted in a paraphyletic Ischyrocephalae (and a polyphyletic Oreopolae). Monophyly of morphogroups Ischnoacrae and Morobeae could not be tested as sequenced representative samples did not yield sufficient reads on target.
FIGURE 5.
(Bottom left) Distribution of Schefflera clades across Papuasia. Accession symbols are coded according to clade. Areas are color-coded according to tectonic plates. The background map is a hillshade of the Digital Elevation Model. (Right) Bayesian ancestral area reconstruction for Papuasian Schefflera (BEAST). Branches are color-coded according to reconstructed ancestral areas. Symbols next to accession labels indicate collection elevation.
At the species level, all morphological taxa sampled (except for S. actinophylla) were reconstructed as monophyletic, though support was variable. S. “aeruginosa” sp. ined., S. “frigidariorum” sp. ined., S. pilematophora, and S. pueckleri had high support in all trees. Support for S. simbuensis was high in the chronogram and the ITS tree but low in the pseudo-coalescent tree. Similarly, S. eriocephala was well supported in the chronogram but not so in the ITS tree. Schefflera actinophylla had two non-Papuasian accessions group together to form a well-supported clade in the ITS tree and a third Papuasian accession highly supported as sister to also Papuasian S. thaumasiantha.
Divergence Time and Ancestral Area Co-estimation
Speciation under a non-coalescent Yule process and an uncorrelated relaxed molecular clock (lognormal distribution) was selected as the best-fit model (Table 2). Heptapleurum, with an early Oligocene crown age of ∼33.4 Ma, was inferred to have originated in the Sunda plate (Figure 5 right). Heptapleurum transitioned from Sunda into the Woodlark plate sometime in the late Oligocene, between ∼29 and 26.3 Ma, giving rise to the Papuasian Schefflera clade.
TABLE 2.
Model selection for divergence time estimation and ancestral area reconstruction in BEAST 1.10.4 (Suchard et al., 2018): marginal likelihood estimates (MLEs) for six tree and clock model comparisons.
| Tree model | Clock model | Log marginal likelihood |
|
| Path sampling | Stepping-stone sampling | ||
| Yule process | Strict | −127,161.38 | −127,159.31 |
| Random local* | N/A | N/A | |
| Uncorrelated relaxed lognormal | −126,125.37 | −126,125.16 | |
| Birth-death incomplete sampling | Strict | −127,149.93 | −127,149.49 |
| Random local* | N/A | N/A | |
| Uncorrelated relaxed lognormal | −126,130.93 | −126,130.65 | |
* Random local clock models exhibited inadequate chain mixing and did not converge after 100M iterations. Italics indicate best-supported model and associated log MLEs.
Within the Papuasian clade, Brassaia was reconstructed as having originated in the Sahul shelf, having arrived from the Woodlark plate in the early Miocene, between ∼23.8 and 18.1 Ma. Paratropia, Bordenia, and Tupidanthus (S. rigida, S. macgregorii, and S. subintegra, respectively) are here monophyletic and sister to Brassaia, though with low support. These three morphogroups were reconstructed to have transitioned back to Sunda, from Woodlark, in the Early Miocene (between ∼23.8 and 16 Ma) and, from there, onward to the Philippines sometime between ∼13.4 Ma and the present. An additional dispersal event to the Philippines, this time from Sunda, took place sometime between ∼33.5 Ma and the present and resulted in the Philippine Schefflera clade (S. blancoi). Morphogroup Ischyrocephalae was inferred as sister to S. boridiana plus S. stolleana. Crown age for Ischyrocephalae is ∼17.6 Ma and it reached the South Bismarck plate, from the Woodlark plate, twice between ∼11.6 Ma and the present. Morphogroup Oreopolae (S. oreopola and S. “goodenoughiana” sp. ined.) was reconstructed in a clade with maximum support and as sister to Barbatae (S. polyclada). This well-supported Oreopolae + Barbatae clade transitioned into the Sahul shelf from Woodlark in the Middle Miocene, between ∼17.3 and 12.6 Ma, returning to Woodlark between ∼7.43 Ma and the present. Additionally, the Oreopolae + Barbatae clade converged with an also well-supported Cephaloschefflera clade (S. eriocephala and S. eriocephala f. centralis) ∼17.6 Ma to form an Eastern clade. Papuoschefflera s.s. moved from the Woodlark plate to the Maoke plate in the Late Miocene, between ∼23.1 Ma and 20.0 Ma. From there, it colonized the Bird’s Head plate multiple times, expanded into the Sahul shelf and re-entered the Woodlark plate.
Discussion
Efficacy of Universal Probes
To date, other than in Schefflera, the Angiosperms-353 enrichment panel (Johnson et al., 2018) has only been tested at the species level in sedges (Cyperus; Larridon et al., 2020) and Old World pitcher plants (Nepenthes; Murphy et al., 2020) and at the population level (SNPs) in rice (Oryza; Van Andel et al., 2019). When comparing data matrices containing on-target loci only, both Larridon et al. (2020) and Murphy et al. (2020) had a higher capture success (Table 3); an expected result since they worked with a greater proportion of silica-dried tissue (Brewer et al., 2019). However, their mean PPIC was lower than ours (Table 3), probably as a result of our filtering approach, which combined two custom scripts – max_overlap (representativeness x completeness x evenness) and optrimAl (per locus gap threshold optimization) – to increase signal while reducing missing data in our data matrixes. The relatively high informativeness of our final data set suggests that this adaptive trimming may help strike a balance between retaining sequence length and improving phylogenetic informativeness (Hartmann and Vision, 2008).
TABLE 3.
Comparison of nuclear exon target capture and alignment statistics* across comparable data matrixes (on-target coding only) in herbariomic studies.
| Study | Probe set | Target loci | Sample type (Herb/Other) | Collection year | Mean reads per sample | Reads on target (%) | Capture success (%) | Alignment length | Mean PPIC (%) | Missing data (%) |
| Hart et al., 2016 | Leguminosae (specific) | 214 | 11/2 | 1835–2009 | 1,241,592 | 79.9 | 89 | 229,995 | 6.7 | 22.4 |
| Vatanparast et al., 2018 | Leguminosae (specific) | 507 | 12/13 | 1985–2014 | 6,356,207 | 32 | 81.9 | 737,309 | 18.3 | 19.7 |
| Villaverde et al., 2018 | Euphorbia (specific) | 431 | 56/88 | 1891–2014 | 1,036,822 | 48.6 | 26.8 | 486,878 | 8 | 23.3 |
| Soto Gomez et al., 2019 | Dioscoreaceae (specific) | 260 | 22/3 | 1994–2007 | 922,847 | 31.6 | 93.8 | 276,920 | 24 | 5.2 |
| Larridon et al., 2020 | Angiosperms (universal) | 353 | 30/8 | 1948–2017 | 5,155,095 | 8.5 | 33.2 | 233,429 | 9.9 | 40.4 |
| Murphy et al., 2020 | Angiosperms (universal) | 353 | 31/194 | 1835–2019 | 1,633,509 | 5.6 | 59.3 | 160,320 | 13.6 | 9.6 |
| Shee et al., this study | Angiosperms (universal) | 353 | 68/6 | 1850–2018 | 946,130 | 11.5 | 28.2 | 194,215 | 17 | 27.1 |
∗Calculated from published results, including Supplementary Material.
The low specificity of the Angiosperms-353 baits (probes are <30% divergent; Johnson et al., 2018) would explain why Larridon et al. (2020) and Murphy et al. (2020), and this study retrieve a lower mean percent of on-target reads per sample than other herbariomic studies (Hart et al., 2016; Vatanparast et al., 2018; Villaverde et al., 2018) relying on taxon-specific probes (Table 3). Mean capture success of target loci from herbarium specimens in Schefflera was comparable to that obtained by Villaverde et al. (2018), which used custom probes, designed for genus Euphorbia, rather than universal ones. Increased sequencing depth has been shown to correlate with capture success (Johnson et al., 2018) which, combined with kit specificity and sample age, may explain why mean capture success is higher (than ours) in two legume studies (>80%) using different family-specific bait kits (Hart et al., 2016; Vatanparast et al., 2018). Like Villaverde et al. (2018), we also found that specimen age affected capture success (Figure 2), probably due to accumulated DNA damage and its effect on genomic library preparation (Der Sarkissian et al., 2015). The large variance in capture success of post-1940s specimens could be explained in terms of variability in collection, preservation, and storage techniques (Brewer et al., 2019; Forrest et al., 2019).
Other recent studies have also demonstrated the efficacy of taxon-specific probe sets in resolving species-level relationships from herbarium DNA (Finch et al., 2019; Soto Gomez et al., 2019; White et al., 2019). Whereas Kadlec et al. (2017) argued that high sequence variability across angiosperm orders precluded the usefulness of universal probes in resolving species-level relationships, Chau et al. (2018) found that general purpose probes can be as effective as taxon-specific ones. While we do not compare these alternative probes, the results from our study, Larridon et al. (2020) and Murphy et al. (2020) suggest that an appropriately designed universal probe set can capture adequate phylogenomic information to resolve relationships at the species-level and even at the population-level (Van Andel et al., 2019). Liu et al. (2019) showed experimentally that when probes are <30% divergent from regions targeted, enrichment worked adequately (see Figure 6 and Supplementary Figure S6 in Liu et al., 2019). Johnson et al. (2018) took this threshold into account when designing the Angiosperms-353 probe set and included sufficient probes to account for the diversity the panel encompasses (i.e., all angiosperms). Thus, if universal probe sets can indeed be as informative at shallower phylogenetic levels as lineage-specific ones, this would considerably reduce the cost and effort associated with designing and optimizing taxon-specific probes for phylogenomic studies (McKain et al., 2018; Dodsworth et al., 2019).
Phylogenomic Support for Morphological Groupings
The paraphyly of the Papuasian accessions in our study corroborates the previous results of Li and Wen (2014). The Papuasian clade we recovered included accessions from three non-Papuasian lineages: Sundan Paratropia, Philippine Bordenia and mainly Indochinese Tupidanthus (Figure 3 right). Given our divergence time estimates for the Papuasian clade (Figure 5 right) and considering the timing of the Sunda-Sahul floristic exchange between ∼34 and 12 Ma (Crayn et al., 2015), Asian Schefflera appears to have crossed Wallacea – the floristic province within the Malesia biogeographic region connecting the Sunda and Sahul shelves – at least twice before both these shelves finally merged, supporting the observation made by van Welzen et al. (2011) that “there is no sharp E-W boundary for plant distributions in Malesia” (though see Bacon et al., 2013 for a counter-example in palms).
All our topologies support the current circumscription of Brassaia proposed by Frodin et al. (2010). Brassaia and Papuoschefflera s.l. primarily differ in floral morphology (Table 4). An earlier treatment (Harms, 1921) classified Papuasian Schefflera into two sections: (i) Cephaloschefflera, with flowers arranged in heads and (ii) Euschefflera, with flowers in umbellules. Our molecular analysis supports the proposal of Frodin (1975) that this character is plesiomorphic in Cephaloschefflera sensu Harms (1921). Brassaia was historically treated as a separate genus (Bentham, 1867) until it was incorporated into sect. Cephaloschefflera (Harms, 1921). Later, the four conspicuous floral bracts present in the clade were proposed as an apomorphic character (Frodin, 1975), an observation which appears to be validated by all our topologies (Figures 3–5).
TABLE 4.
Morphological characters distinguishing Brassaia and Papuoschefflera s.l.
| Morphological character | Brassaia | Papuoschefflera s.l. |
| Ovary position | 1/2–2/3 superior | <1/6 superior |
| Floral bracts, ligules | Glabrous | Setulose |
| Main inflorescence axis | Sessile/short | Up to 60 cm long |
| Flower color | Red to pink | Pale green to white, some purple to red |
Within Brassaia, phylogenetic relationships better match geography than morphology. This is exemplified by the strongly supported clade comprising S. macrostachya ssp. “australis” ssp. ined. and S. “ovalis.narrow” sp. ined. which was recovered in all three trees (Figures 3–5). Members of the S. “ovalis” alliance have never been formally described, although their affinity with S. macrostachya in leaf venation has been noted (Frodin, 1975). Since the two collections were made within 30 km of each other along the Aikwa River, in Mimika Regency, they may well represent variation within a single species. The same seems to be happening with regards to the polyphyly of S. actinophylla in the ITS tree (Figure 3 right). Our New Guinean S. actinophylla accession and S. thaumasiantha were from the same locality and formed a well-supported clade. The other clade consisted of a Queensland collection and a cultivated plant from New York Botanical Garden (NYBG) of unknown origin. It is worth noting that S. actinophylla is widespread across the world as an ornamental primarily from Australian stock, which suggests this NYBG collection might be Australian in origin. Interestingly, S. thaumasiantha is also cultivated locally in SE New Guinea (Frodin, 1975), which could help explain the observed morphological similarities. Previous work on domestication points to possible multiple origins in a number of crops, with parallelism and convergence being the norm (Fuller et al., 2014; Purugganan, 2019).
Papuoschefflera s.l. was reconstructed as either paraphyletic with respect to Brassaia (Figure 3 right) or as reciprocally monophyletic and sister to Brassaia with some (Figure 4 left) or no support (Figure 5 right). Although we could not place Cephaloschefflera (represented by S. eriocephala) with confidence, we found that morphogroup Pagiophyllae (represented by S. “frigidariorum” sp. ined.) belonged in Papuoschefflera s.s. (see section Phylogenetic Relationships in Schefflera in Results), together with morphogroups Bougainvilleanae, Schumannianae, and Versteegiae (Figures 3–5). Other Papuoschefflera s.l. morphogroups (e.g., Ischyrocephalae or Oreopolae) were monophyletic in some but not all of our trees, which should be further explored.
Evolutionary History of Papuasian Lineages
The sampled accessions of Papuasian Schefflera tended to form ecological and morphological clades within broader geographical clades. Analogous patterns have been observed in other Araliaceae clades, such as Polyscias (Plunkett and Lowry, 2010), Neotropical Schefflera (Fiaschi and Plunkett, 2011) and Plerandra, which is the Melanesian clade of Schefflera s.l. (Plunkett and Lowry, 2012). The dominance of Brassaia and Papuoschefflera s.l., on either side of the New Guinea Highlands also recalls a similar arrangement in the two clades of Afro-Malagasy Schefflera s.l. on either side of the Mozambique Channel, which are now recognized as genera Astropanax and Neocussonia (Gostel et al., 2017). These distribution patterns may prove to be fertile ground for the testing of biogeographic hypotheses.
The Woodlark Plate: A Source Area for Papuasian Schefflera
Though our estimated crown age of ∼26.3 Ma for the Papuasian Schefflera clade is older than the crown age of ∼21.9 Ma estimated by Li and Wen (2014) – which is the source of our secondary time constraints –, as expected, the highest posterior density intervals for both these estimates overlap. These divergence dates may seem to be at odds with the assertion by Lohman et al. (2011) that most of New Guinea was submerged prior to 20 Ma. Yet, several studies have also inferred the origin of Papuasian taxa back to the Oligocene (Cibois et al., 2014; Kodandaramaiah et al., 2018) or even as early as the late Eocene (Jønsson et al., 2011; Cozzarolo et al., 2019). During the Oligocene, there may have been an island archipelago, located where present-day New Guinea is, formed by the collision of both the Philippine-Sea and the Pacific plates with the Australia plate (Hill and Hall, 2003). The largest landmass would probably have been where the present-day Papuan Peninsula is and have resulted from the docking of the East Papua Composite Terrane (EPCT; part of the Woodlark plate) onto the Australia plate (Davies, 2012). Stratigraphic examination of sediments deposited in the Aure Trough provides evidence for mountain building on the Woodlark plate during this period (van Ufford and Cloos, 2005), indicating that the terranes forming the Papuan Peninsula were already emergent. Our reconstruction of the Woodlark plate as the best-supported ancestral area for Papuasian Schefflera (Figure 5) is consistent with the above scenario.
Hall (2012) estimated that the Sahul and Sunda shelves first collided ∼25 Ma (see Figure 32 in Hall, 2012). Phylogenies reconstructed by Li and Wen (2014) and Plunkett et al. (2005) and this study support Papuasian Schefflera as nested within Sundan Heptapleurum. It is thus within reason that our ancestral area reconstruction suggests that the common ancestor of Papuasian Schefflera dispersed from Sunda to colonize the Woodlark plate right before this contact took place (Figure 5). In this scenario, we hypothesize that a light-loving and pioneer ancestral Papuasian Schefflera would have rapidly colonized areas of the New Guinea land mass as they gradually emerged above sea level. The Woodlark plate could, therefore, have functioned as a source area for the colonization of New Guinea along the predominantly west-to-east axis of the Sunda-Sahul floristic exchange (Crayn et al., 2015).
Vicariance in Brassaia
Our reconstruction of the Sahul shelf as the ancestral area for Brassaia points to a vicariance scenario for the early evolutionary history of this clade (Figure 5). The Brassaia crown is dated at ∼18.1 Ma, which is earlier than the ∼5 Ma date estimated for the emergence of the Sahul shelf to form the southern half of New Guinea (Hall, 2009). As Brassaia also occurs on the southern fall of the Owen Stanley Range (near present-day Port Moresby, in the “tail” of Papua New Guinea) and is partly formed by the Sahul shelf, it is possible that ancestral Brassaia evolved in isolation from ancestral Papuoschefflera s.l. on either side of the proto-Owen Stanley Range during the early Miocene.
The placement of S. kraemeri in Brassaia supports the morphological circumscription of the group, despite this species’ disjunct distribution with regard to the rest of the group. Schefflera kraemeri is found only in the Truk Islands, more than 800 km away from Papuasia. It is most likely to have arrived via long distance dispersal as there are no intervening landmasses in the Pacific Ocean to serve as stepping-stones. Human-mediated dispersal is unlikely, not only because of the inferred timing (which predates humanity), but also because Schefflera has limited uses in Papuasia and S. kraemeri has not been recorded among the region’s indigenous people as a useful species (Cámara-Leret and Dennehy, 2019a,b). The estimated divergence time of ∼6.8 Ma is consistent with the geological age of these islands, which were determined to have been the result of volcanism ∼11 Ma (Keating et al., 1984).
Papuoschefflera s.l. Speciated on Geographic and “Sky” Islands
The divergence of the major clades of Papuoschefflera s.l. between ∼24.6 and 16.7 Ma overlaps with the period in the early Miocene when most of New Guinea was submerged (Figure 5). While Ischyrocephalae and the Eastern clade are inferred to have remained on the Woodlark plate at this time, Papuoschefflera s.s. may have arisen from an early dispersal to then-emerged islands in the Maoke plate, corresponding to the present-day Maoke Mountains. The splitting of the Eastern clade into Cephaloschefflera and the Oreopolae + Barbatae clade probably resulted from another dispersal to Sunda shelf terranes between ∼17.3 and 12.6 Ma. Indeed, zoochorous dispersal across narrow water barriers has been found to play an important role in the intercontinental floristic exchange of the Malesian flora (Crayn et al., 2015). The Papuasian Schefflera, with their fleshy drupaceous fruits, could have been widely dispersed (for example by birds; Snow, 1981) across the proto-Papuasian archipelago.
Accessions from islands located in the Bird’s Head plate illustrate how Schefflera species may have colonized islands in geological times. The shallowest seafloor connecting the islands of Biak and Waigeo to mainland New Guinea is about 200 m deep, precluding an overland connection even during the Pleistocene, when sea level was up to 120 m shallower than nowadays (Voris, 2000). The divergence of S. “bougainvill.biak” sp. ined. ∼5.6 Ma coincides with the end of a 6-Myr upper Oligocene hiatus in deposition in the Biak Basin near Biak Island (Gold et al., 2014), indicating that sea level was shallower at that point in time. A larger area of land would have been exposed on both the island and the mainland, facilitating dispersal over a narrower water barrier. Similarly, the divergence of Schefflera sp. (Raja Ampat) ∼4.7 Ma coincided with an active period of Pliocene deformation, resulting in more land emerging above sea level (Charlton et al., 1991). The placement of this collection as sister to S. “KB22” (Sorong) sp. ined. suggests descent from a common ancestor that occupied the NW Bird’s Head peninsula.
Ischyrocephalae is restricted to upper montane forests and subalpine grasslands, which could be explained in terms of phylogenetic biome conservatism – phenomenon that has been observed in vascular plants from the Malesian island of Borneo (Merckx et al., 2015) and also worldwide (Crisp et al., 2009). Schefflera ischyrocephala and S. pilematophora are found in the Saruwaged Range on the NE coast of New Guinea, which is interpreted to be a terrane of the South Bismarck plate. These taxa are inferred to have arrived separately from the Woodlark plate sometime between ∼11.6 and 10.7 Ma to the present, respectively, concurrent with the Finisterre volcanic arc accretion to the EPCT (Davies, 2012). This pattern could suggest that adaptation to montane regions in ancestral Papuoschefflera s.l. may have a role in promoting the highly diverse “sky island” flora of New Guinea (Sklenáø et al., 2014). Given the prevalence of high- and mid-elevation taxa both within the clade and its most recently diverging sister lineages, as well as a putative temperate origin for Asian Schefflera (Valcárcel and Wen, 2019), Papuasian Schefflera may have been pre-adapted to these environments (Antonelli, 2015). Pre-adaptation has also been invoked in a broad range of high-elevation taxa on Gunung Kinabalu (Merckx et al., 2015), a Malesian mountain located in Borneo, with uplift timing similar to that of the New Guinea Highlands. In both cases, however, this hypothesis remains to be tested by ancestral trait reconstruction, while accounting for diversification rate shifts.
Based on morphological characters, Philipson (1978) mused that Schefflera may be “vigorously diversifying at the present time.” In Papuasia, the species richness of various plant and animal taxa has been shown to peak at mid-elevations (Colwell et al., 2016; Vollering et al., 2016). Papuasian Schefflera exhibits a similar distribution in species richness, which may have arisen from rapid adaptive radiation into new ecological niches created by mountain-building. This evolutionary mechanism has also been observed in the genus Pseuduvaria (Annonaceae) on New Guinea (Su and Saunders, 2009), various genera of Andean alpine plants (Nürk et al., 2018), and Ericaceae in montane regions worldwide (Schwery et al., 2015). Many biotic and environmental factors, including its presumed temperate ancestry and the ready availability of new uncolonized habitats resulting from the uplift of the New Guinea Highlands, would have predisposed Papuasian Schefflera to speciation via biome shifts (Donoghue and Edwards, 2014). Our chronogram hints to a speciation burst in the last 12–5 Myr and we hypothesize recent adaptive radiation, facilitated by mountain building, could be the primary driver for the present-day diversity of Schefflera. Further insights into this phenomenon could be achieved by examining an expanded sample of taxa for trait shifts and diversification rates along an elevational gradient.
Conclusion
Several phylogeographical studies on Papuasian fauna have produced comparable results that point to major themes in Papuasian biogeography at different points in geological time that we recapitulate below.
Woodlark plate: source area for the colonization of New Guinea in the late Oligocene. Our inference that the Woodlark plate acted as the source area for the colonization of Papuasia by Schefflera in the late Oligocene is supported by faunistic studies on the endemic frog genus Mantophryne (Oliver et al., 2013). A similar pattern of colonization, albeit from Australia, would also explain the present-day distribution of Gondwana-derived Melanotaeniid rainbowfish (Unmack et al., 2013). However, more studies involving divergence time estimation and ancestral area reconstruction are required to establish the role of the Woodlark plate in the diversification of Papuasian taxa.
New Guinea Highlands: a barrier since the late Oligocene. The divergence of Papuasian Schefflera into northern Papuoschefflera and southern Brassaia – along a topographical barrier running east to west across the length of Papuasia – lends support to the role played by the New Guinea Highlands with respect to north-south disjunctions and has also been observed in lineages of freshwater organisms such as Melanotaeniid rainbowfish (Unmack et al., 2013), Mantophryne frogs (Oliver et al., 2013), and the turtle Elseya novoguinae (Georges et al., 2014). The more recent rapid uplift of the New Guinea Highlands in the late Miocene and Pliocene epochs coincides with the divergence of Sericulus bowerbird species (Zwiers et al., 2008) and populations of the passerine bird-species Colluricincla megarhyncha (Deiner et al., 2011). Thus, it is possible that initial mountain-building created physical barriers by compartmentalizing the Papuasian terranes into separate basins. Subsequently, the Pliocene uplift would then have raised the New Guinean Highlands to the point that new ecological barriers (i.e., alpine and subalpine zones) effectively isolated populations adapted to lower elevations.
Geographic and ecological (“sky”) islands shaped evolutionary relationships, both deep and shallow. In the Miocene, when Papuasia existed only as a chain of islands, major clades nested in Papuoschefflera s.l. originated and diversified into high- and mid-elevation clades across New Guinea’s mountain ranges. Speciation on geographic islands is evident in the Miocene divergence of freshwater taxa on the Bird’s Head Peninsula and Maoke terranes (Unmack et al., 2013; Georges et al., 2014), which probably existed as isolated landmasses prior to docking with the Maoke plate. These putative geographic islands have also been cited as cradles of diversity for corvoid birds (Jønsson et al., 2011) and Ptilinopus fruit doves (Cibois et al., 2014). Similarly, Orthonyx logrunners in the Bird’s Head Peninsula have been shown to be genetically distinct from those in the rest of New Guinea (Joseph et al., 2001). Additionally, a more recent genetic divergence (late Miocene onward) has been found on an unnamed Petaurus glider species on Normanby Island (Malekian et al., 2010). Isolation on an ecological island in the Cromwell Range during the Pleistocene may also explain the observed genetic differentiation in an isolated population of the pademelon Thylogale browni browni, which is restricted to the edges of subalpine forests (Macqueen et al., 2011).
New ecological niches followed the New Guinea Highlands uplift and could have driven rapid recent radiations. The divergence dates of most Papuasian Schefflera taxa in our chronogram coincide with the uplift of the New Guinea Highlands in the late Miocene. This geological event created new ecological niches and has been invoked as the primary driver for the diversity of several groups of Papuasian mammals and birds, such as Australasian rats (Rowe et al., 2011), Dendrolagus tree kangaroos (Eldridge et al., 2018), Exocelina diving beetles (Toussaint et al., 2014), Meliphaga honeyeaters (Norman et al., 2007), Prenolepis ants (Matos-Maraví et al., 2018), Pseudocheiridae ringtail possums (Meredith et al., 2010), Syma kingfishers (Linck et al., 2019), Thraulus mayflies (Cozzarolo et al., 2019), and Thylogale pademelons (Macqueen et al., 2011). Together, these studies strongly suggest that rapid adaptive radiation into newly created ecological niches resulting from recent mountain-building would best explain the richness of Papuasia’s biodiversity.
In closing, we have found Asian Schefflera to be among the first plant genera to have crossed from Sunda toward the Australian plate at the start of the Sunda-Sahul floristic exchange in the late Oligocene. The widespread distribution of this lineage and its existence in Papuasia since its known geologic origin suggest that its evolutionary history will prove instructive in understanding the region’s plant diversity. Our study demonstrates that Hyb-Seq with universal probes on a sample set comprising mostly herbarium specimens can resolve both deep and shallow phylogenetic relationships to elucidate the drivers of this diversity. Our results suggest an important role for (1) the Woodlark plate (present-day Papuan Peninsula), (2) the New Guinea Highlands, (3) isolation on geographic and “sky” islands, and (4) the late Miocene New Guinea Highlands uplift in explaining plant biogeography in Papuasia.
Data Availability Statement
The raw reads (FASTQ files) are available from the NCBI BioProject database (ID: PRJNA604390). The datasets generated and analyzed are available from Zenodo (doi: 10.5281/zenodo.3534088).
Author Contributions
The study and sampling scheme were jointly conceived by all authors based on taxonomic expertise and collection data provided by DF. ZS and LP sampled the specimens and carried out the molecular work and data analysis. ZS wrote the manuscript, with contributions from all authors.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our colleagues at Royal Botanic Gardens, Kew for their invaluable assistance: Edith Kapinos, Isabel Fairlie, Juan Viruel, Laszlo Csiba, Olivier Maurin, Oscar Pérez-Escobar, Marcelo Sellaro, Miranda Janatka, Penny Malakasi, Robyn Cowan, Sally Dawson, Sidonie Bellot, Tim Utteridge, Bill Baker, Félix Forest, and Ilia Leitch. We extend our gratitude to the herbaria of K, L, BM, SING and to Julian Schrader (GAU Göttingen) for providing samples, and to GenBank, 1KP, and PAFTOL for making sequences publicly available. We thank the Sackler Phylogenomic Laboratory and the Jodrell Laboratory at RBG Kew for facilitating molecular lab resources and computing resources.
Funding. LP had funding from the Garfield Weston Foundation (Global Tree Seed Bank Project) and EU-SYNTHESYS (NL-TAF-6894). RC-L had funding from EU-SYNTHESYS (GB-TAF-6305). This article was based on a final manuscript by ZS for his MSc in Plant and Fungal Taxonomy, Diversity and Conservation at Queen Mary, University of London and at RBG Kew, funded by a scholarship from the National Parks Board, Singapore.
Dedication
We dedicate this manuscript to the memory of our friend and mentor David Gamman Frodin, who passed away during the course of this study (1940–2019). His legacy lives on in the “Schefflera Team” and the scientists he inspired during his fruitful career.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00258/full#supplementary-material
References
- Andrews S. (2018). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed August 9, 2019). [Google Scholar]
- Antonelli A. (2015). Biodiversity: multiple origins of mountain life. Nature 524 300–301. 10.1038/nature14645 [DOI] [PubMed] [Google Scholar]
- Bacon C. D., Michonneau F., Henderson A. J., McKenna M. J., Milroy A. M., Simmons M. P. (2013). Geographic and taxonomic disparities in species diversity: dispersal and diversification rates across Wallace’s line. Evolution 67 2058–2071. 10.1111/evo.12084 [DOI] [PubMed] [Google Scholar]
- Baele G., Lemey P., Vansteelandt S. (2013). Make the most of your samples: Bayes factor estimators for high-dimensional models of sequence evolution. BMC Bioinformatics 14:85. 10.1186/1471-2105-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S. L., Fitzgerald P. G., Webb L. E. (2012). Tectonics of the New Guinea Region. Annu. Rev. Earth Planet. Sci. 40 495–520. 10.1146/annurev-earth-040809-152540 [DOI] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Computat. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beehler B. M. (2007). “Papuan terrestrial biogeography, with special reference to birds,” in The Ecology of Papua: Part One, eds Marshall A. J., Beehler B. M. (Singapore: Periplus Editions (HK) Ltd; ), 196–206. [Google Scholar]
- Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K. D., et al. (2018). GenBank. Nucleic Acids Res. 46 D41–D47. 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentham G. (1867). “Araliaceae,” in Genera Plantarum, eds Bentham G., Hooker J. D. (London: William Pamplin; ), 931–941. [Google Scholar]
- Bird P. (2003). An updated digital model of plate boundaries. Geochem. Geophys. Geosyst. 4 13–50. 10.1029/2001GC000252 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec M. L. (2016). AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4:e1660. 10.7717/peerj.1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass L. J. (1941). The 1938-39 expedition to the snow mountains, Netherlands New Guinea. J. Arnold Arbor. 22 271–295. [Google Scholar]
- Brewer G. E., Clarkson J. J., Maurin O., Zuntini A. R., Barber V., Bellot S., et al. (2019). Factors affecting targeted sequencing of 353 nuclear genes from herbarium specimens spanning the diversity of angiosperms. Front. Plant Sci. 10:1102. 10.3389/fpls.2019.01102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummitt R. K. (ed.) (2001). “World geographical scheme for recording plant distributions,” in Hunt Institute for Botanical Documentation, 2nd Edn (Pittsburgh: Carnegie Mellon University; ). [Google Scholar]
- Cámara-Leret R., Dennehy Z. (2019a). Indigenous knowledge of New Guinea’s useful plants: a review. Econ. Bot. 73 405–415. 10.1007/s12231-019-09464-1 [DOI] [Google Scholar]
- Cámara-Leret R., Dennehy Z. (2019b). Information gaps in indigenous and local knowledge for science-policy assessments. Nat. Sustain. 2 736–741. 10.1038/s41893-019-0324-0 [DOI] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton T. R., Hall R., Partoyo R. (1991). The geology and tectonic evolution of Waigeo Island, NE Indonesia. J. Southeast Asian Earth Sci. 6 289–297. 10.1016/0743-9547(91)90074-8 [DOI] [Google Scholar]
- Chau J. H., Rahfeldt W. A., Olmstead R. G. (2018). Comparison of taxon-specific versus general locus sets for targeted sequence capture in plant phylogenomics. Appl. Plant Sci. 6:e1032. 10.1002/aps3.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibois A., Thibault J.-C., Bonillo C., Filardi C. E., Watling D., Pasquet E. (2014). Phylogeny and biogeography of the fruit doves (Aves: Columbidae). Mol. Phylogenet. Evol. 70 442–453. 10.1016/j.ympev.2013.08.019 [DOI] [PubMed] [Google Scholar]
- Colwell R. K., Gotelli N. J., Ashton L. A., Beck J., Brehm G., Fayle T. M., et al. (2016). Midpoint attractors and species richness: modelling the interaction between environmental drivers and geometric constraints. Ecol. Lett. 19 1009–1022. 10.1111/ele.12640 [DOI] [PubMed] [Google Scholar]
- Cozzarolo C.-S., Balke M., Buerki S., Arrigo N., Pitteloud C., Gueuning M., et al. (2019). Biogeography and ecological diversification of a Mayfly Clade in New Guinea. Front. Ecol. Evol. 7:233 10.3389/fevo.2019.00233 [DOI] [Google Scholar]
- Crayn D. M., Costion C., Harrington M. G. (2015). The Sahul-Sunda floristic exchange: dated molecular phylogenies document cenozoic intercontinental dispersal dynamics. J. Biogeogr. 42 11–24. 10.1111/jbi.12405 [DOI] [Google Scholar]
- Crisp M. D., Arroyo M. T. K., Cook L. G., Gandolfo M. A., Jordan G. J., McGlone M. S., et al. (2009). Phylogenetic Biome Conservatism on a Global Scale. Nature 458 754–756. 10.1038/nature07764 [DOI] [PubMed] [Google Scholar]
- Crisp M. D., Trewick S. A., Cook L. G. (2011). Hypothesis testing in biogeography. Trends Ecol. Evol. 26 66–72. 10.1016/j.tree.2010.11.005 [DOI] [PubMed] [Google Scholar]
- Davies H. L. (2012). The geology of New Guinea - the Cordilleran Margin of the Australian Continent. Episodes 35 1–16. [Google Scholar]
- Deiner K., Lemmon A. R., Mack A. L., Fleischer R. C., Dumbacher J. P. (2011). A passerine Bird’s evolution corroborates the geologic history of the Island of New Guinea. PLoS One 6:e19479. 10.1371/journal.pone.0019479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der Sarkissian C., Allentoft M. E., Ávila-Arcos M. C., Barnett R., Campos P. F., Cappellini E., et al. (2015). Ancient Genomics. Philos. Trans. R. Soc. B Biol. Sci. 370:20130387. 10.1098/rstb.2013.0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S., Pokorny L., Johnson M. G., Kim J. T., Maurin O., Wickett N. J., et al. (2019). Hyb-Seq for flowering plant systematics. Trends Plant Sci. 24 887–891. 10.1016/j.tplants.2019.07.011 [DOI] [PubMed] [Google Scholar]
- Donoghue M. J., Edwards E. J. (2014). Biome shifts and niche evolution in plants. Annu. Rev. Ecol. Evol. Syst. 45 547–572. 10.1146/annurev-ecolsys-120213-091905 [DOI] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19 11–15. [Google Scholar]
- Drummond C. S., Eastwood R. J., Miotto S. T. S., Hughes C. E. (2012). Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Syst. Biol. 61 443–460. 10.1093/sysbio/syr126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge M. D. B., Potter S., Helgen K. M., Sinaga M. H., Aplin K. P., Flannery T. F., et al. (2018). Phylogenetic analysis of the tree-kangaroos (Dendrolagus) reveals multiple divergent lineages within New Guinea. Mol. Phylogenet. Evol. 127 589–599. 10.1016/j.ympev.2018.05.030 [DOI] [PubMed] [Google Scholar]
- Fiaschi P., Plunkett G. M. (2011). Monophyly and phylogenetic relationships of neotropical Schefflera (Araliaceae) based on plastid and nuclear markers. Syst. Bot. 36 806–817. 10.1600/036364411X583754 [DOI] [Google Scholar]
- Finch K. N., Jones F. A., Cronn R. C. (2019). Genomic resources for the neotropical tree genus Cedrela (Meliaceae) and its relatives. BMC Genomics 20:58. 10.1186/s12864-018-5382-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest L. L., Hart M. L., Hughes M., Wilson H. P., Chung K.-F., Tseng Y. H., et al. (2019). The limits of Hyb-Seq for herbarium specimens: impact of preservation techniques. Front. Ecol. Evol. 7:439 10.3389/fevo.2019.00439 [DOI] [Google Scholar]
- Frodin D. G. (1975). Studies in Schefflera (Araliaceae): the Cephaloschefflera complex. J. Arnold Arbor. 56 427–448. [Google Scholar]
- Frodin D. G. (2004). History and concepts of big plant genera. Taxon 53 753–776. 10.2307/4135449 [DOI] [Google Scholar]
- Frodin D. G., Govaerts R. (2003). World Checklist and Bibliography of Araliaceae. Kew: Royal Botanic Gardens. [Google Scholar]
- Frodin D. G., Lowry P. P., II, Plunkett G. M. (2010). Schefflera (Araliaceae): taxonomic history, overview and progress. Plant Divers. Evol. 128 561–595. 10.1127/1869-6155/2010/0128-0028 [DOI] [Google Scholar]
- Fuller D. Q., Denham T., Arroyo-Kalin M., Lucas L., Stevens C. J., Qin L., et al. (2014). Convergent evolution and parallelism in plant domestication revealed by an expanding archaeological record. Proc. Natl. Acad. Sci. U.S.A. 111 6147–6152. 10.1073/pnas.1308937110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A., Zhang X., Unmack P., Reid B. N., Le M., McCord W. P. (2014). Contemporary genetic structure of an endemic freshwater turtle reflects miocene Orogenesis of New Guinea. Biol. J. Linn. Soc. 11 192–208. 10.1111/bij.12176 [DOI] [Google Scholar]
- Gold D., Hall R., Burgess P., BouDagher-Fadel M. (2014). “The Biak Basin and its setting in the bird’s head region of West Papua,” in Proceedings of the Thirty-Eighth Annual Convention & Exhibition on Indonesian Petroleum Association, IPA14–G–298, Jakarta. [Google Scholar]
- Gostel M. R., Plunkett G. M., Lowry P. P., II (2017). Straddling the Mozambique Channel: molecular evidence for two major clades of Afro-Malagasy Schefflera (Araliaceae) co-occurring in Africa and Madagascar. Plant Ecol. Evol. 150 87–108. 10.5091/plecevo.2017.1193 [DOI] [Google Scholar]
- Hall R. (2009). Southeast Asia’s changing Palaeogeography. Blumea 54 148–161. 10.3767/000651909X475941 [DOI] [Google Scholar]
- Hall R. (2012). Late Jurassic–Cenozoic reconstructions of the Indonesian Region and the Indian Ocean. Tectonophysics 57 1–41. 10.1016/j.tecto.2012.04.021 [DOI] [Google Scholar]
- Harms H. (1921). Die Araliaceae Papuasiens. Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 56 374–414. [Google Scholar]
- Hart M. L., Forrest L. L., Nicholls J. A., Kidner C. A. (2016). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon 65 1081–1092. 10.12705/655.9 [DOI] [Google Scholar]
- Hartmann S., Vision T. J. (2008). Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol. Biol. 8:95. 10.1186/1471-2148-8-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heads M. (2003). Ericaceae in Malesia: vicariance biogeography, terrane tectonics and ecology. Telopea 10 311–449. 10.3732/ajb.0900109 [DOI] [PubMed] [Google Scholar]
- Hill K. C., Hall R. (2003). “Mesozoic-cenozoic evolution of Australia’s New Guinea margin in a West Pacific context,” in Evolution and Dynamics of the Australian Plate, eds Hillis R. R., Müller R. D. (Sydney: Geological Society of Australia; ), 265–290. [Google Scholar]
- Ho S. Y. W., Phillips M. J., Cooper A., Drummond A. J. (2005). Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22 1561–1568. 10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover J. D., Kumar S., James S. A., Leisz S. J., Laituri M. (2017). Modeling hotspots of plant diversity in New Guinea. Trop. Ecol. 58 623–640. [Google Scholar]
- Johns R. J., Shea G. A., Puradyatmika P. (2007a). “Subalpine and Alpine Vegetation of Papua,” in The Ecology of Papua: Part Two, eds Marshall A. J., Beehler B. M. (Singapore: Periplus Editions (HK) Ltd; ), 1025–1053. [Google Scholar]
- Johns R. J., Shea G. A., Vink W., Puradyatmika P. (2007b). “Montane vegetation of Papua,” in The Ecology of Papua: Part Two, eds Marshall A. J., Beehler B. M. (Singapore: Periplus Editions (HK) Ltd; ), 977–1024. [Google Scholar]
- Johnson M. G., Gardner E. M., Liu Y., Medina R., Goffinet B., Shaw A. J., et al. (2016). HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 4:1600016. 10.3732/apps.1600016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. G., Pokorny L., Dodsworth S., Botigué L. R., Cowan R. S., Devault A., et al. (2018). A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using K-medoids clustering. Syst. Biol. 68 594–606. 10.1093/sysbio/syy086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson K. A., Fabre P.-H., Ricklefs R. E., Fjeldsa J. (2011). Major global radiation of corvoid birds originated in the proto-papuan archipelago. Proc. Natl. Acad. Sci. U.S.A. 108 2328–2333. 10.1073/pnas.1018956108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph L., Slikas B., Alpers D., Schodde R. (2001). Molecular systematics and phylogeography of New Guinean Logrunners (Orthonychidae). Emu 101 273–280. 10.1071/MU01008 [DOI] [Google Scholar]
- Junier T., Zdobnov E. M. (2010). The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26 1669–1670. 10.1093/bioinformatics/btq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec M., Bellstedt D. U., Le Maitre N. C., Pirie M. D. (2017). Targeted NGS for species level phylogenomics: “made to measure” or “one size fits all”? PeerJ 5:e3569. 10.7717/peerj.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kates H. R., Johnson M. G., Gardner E. M., Zerega N. J. C., Wickett N. J. (2018). Allele phasing has minimal impact on phylogenetic reconstruction from targeted nuclear gene sequences in a case study of Artocarpus. Am. J. Bot. 105 404–416. 10.1002/ajb2.1068 [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating B. H., Mattey D. P., Helsley C. E., Naughton J. J., Epp D., Lazarewicz A., et al. (1984). Evidence for a hot spot origin of the Caroline Islands. J. Geophysical Res. Solid Earth 89 9937–9948. 10.1029/JB089iB12p09937 [DOI] [Google Scholar]
- Kim K., Yang T. -J. (2016). Data from: The Complete Chloroplast Genome of Aralia elata. GenBank. Available at: https://www.ncbi.nlm.nih.gov/nuccore/KT153023.1 (accessed August 9, 2019). [Google Scholar]
- Kodandaramaiah U., Braby M. F., Grund R., Müller C. J., Wahlberg N. (2018). Phylogenetic relationships, biogeography and diversification of Coenonymphina butterflies (Nymphalidae: Satyrinae): intercontinental dispersal of a Southern Gondwanan Group? Syst. Entomol. 43 798–809. 10.1111/syen.12303 [DOI] [Google Scholar]
- Larridon I., Villaverde T., Zuntini A. R., Pokorny L., Brewer G. E., Epitawalage N., et al. (2020). Tackling rapid radiations with targeted sequencing. Front. Plant Sci. 10:1655. 10.3389/fpls.2019.01655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. N. (2014). Solid Phase Reverse Immobilization (SPRI) Bead Technology for Micro RNA Clean up Using the Agencourt RNAclean XP Kit. Ph.D. thesis, Beckman-Coulter, Brea, CA. [Google Scholar]
- Lemey P., Rambaut A., Drummond A. J., Suchard M. A. (2009). Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5:e1000520. 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon E. M., Lemmon A. R. (2013). High-throughput genomic data in systematics and phylogenetics. Ann. Rev. Ecol. Evol. Syst. 44 99–121. 10.1146/annurev-ecolsys-110512-135822 [DOI] [Google Scholar]
- Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] Available at: https://arxiv.org/abs/1303.3997 [Google Scholar]
- Li R., Ma P.-F., Wen J., Yi T.-S. (2013). Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS One 8:e78568. 10.1371/journal.pone.0078568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Wen J. (2014). Phylogeny and biogeography of Asian Schefflera (Araliaceae) based on nuclear and plastid DNA sequence data. J. Syst. Evol. 52 431–449. 10.1111/jse.12052 [DOI] [Google Scholar]
- Linck E., Freeman B. G., Dumbacher J. P. (2019). Speciation with gene flow across an elevational gradient in New Guinea Kingfishers. bioRxiv [Preprint] Available at: https://www.biorxiv.org/content/10.1101/589044v1 [DOI] [PubMed] [Google Scholar]
- Liu Y., Johnson M. G., Cox C. J., Medina R., Devos N., Vanderpoorten A., et al. (2019). Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat. Commun. 10:1485. 10.1038/s41467-019-09454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman D. J., de Bruyn M., Page T., von Rintelen K., Hall R., Ng P. K. L., et al. (2011). Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42 205–226. 10.1146/annurev-ecolsys-102710-145001 [DOI] [Google Scholar]
- Lowry P. P., II, Plunkett G. M. (2010). Recircumscription of Polyscias (Araliaceae) to include six related genera, with a new infrageneric classification and a synopsis of species. Plant Divers. Evol. 128 55–84. 10.1127/1869-6155/2010/0128-0003 [DOI] [Google Scholar]
- Macqueen P., Goldizen A. W., Austin J. J., Seddon J. M. (2011). Phylogeography of the Pademelons (Marsupialia: Macropodidae: Thylogale) in New Guinea Reflects Both Geological and Climatic Events during the Plio-Pleistocene. J. Biogeogr. 38 1732–1747. 10.1111/j.1365-2699.2011.02522.x [DOI] [Google Scholar]
- Mai U., Mirarab S. (2018). TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics 19:272. 10.1186/s12864-018-4620-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairal M., Pokorny L., Aldasoro J. J., Alarcón M., Sanmartín S. (2015). Ancient vicariance and climate-driven extinction explain continental-wide disjunctions in Africa: the case of the Rand Flora genus Canarina (Campanulaceae). Mol. Ecol. 24 1335–1354. 10.1111/mec.13114 [DOI] [PubMed] [Google Scholar]
- Mairal M., Sanmartín I., Herrero A., Pokorny L., Vargas P., Aldasoro J. J., et al. (2017). Geographic barriers and Pleistocene climate change shaped patterns of genetic variation in the Eastern Afromontane biodiversity hotspot. Sci. Rep. 7:45749. 10.1038/srep45749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekian M., Cooper S. J. B., Norman J. A., Christidis L., Carthew S. M. (2010). Molecular systematics and evolutionary origins of the genus Petaurus (Marsupialia: Petauridae) in Australia and New Guinea. Mol. Phylogenet. Evol. 54 122–135. 10.1016/j.ympev.2009.07.026 [DOI] [PubMed] [Google Scholar]
- Marshall A. J. (2007). “The diversity and conservation of Papua’s ecosystems,” in The Ecology of Papua: Part Two, eds Marshall A. J., Beehler B. M. (Singapore: Periplus Editions (HK) Ltd; ), 753–770. [Google Scholar]
- Matasci N., Hung L.-H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., et al. (2014). Data access for the 1,000 Plants (1KP) Project. Gigascience 3:17. 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos-Maraví P., Clouse R. M., Sarnat E. M., Economo E. P., LaPolla J. S., Borovanska M., et al. (2018). An Ant Genus-Group (Prenolepis) Illuminates the Biogeography and Drivers of Insect Diversification in the Indo-Pacific. Mol. Phylogenet. Evol. 123 16–25. 10.1016/j.ympev.2018.02.007 [DOI] [PubMed] [Google Scholar]
- McKain M. R., Johnson M. G., Uribe-Convers S., Eaton D., Yang Y. (2018). Practical considerations for plant phylogenomics. Appl. Plant Sci. 6:e1038. 10.1002/aps3.1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn K. A., Faircloth B. C., Glenn T. C., Kimball R. T., Braun E. L. (2016). Analysis of a rapid evolutionary radiation using ultraconserved elements: evidence for a bias in some multispecies coalescent methods. Syst. Biol. 65 612–627. 10.1093/sysbio/syw014 [DOI] [PubMed] [Google Scholar]
- Merckx V. S. F. T., Hendriks K. P., Beentjes K. K., Mennes C. B., Becking L. E., Peijnenburg K. T. C. A., et al. (2015). Evolution of endemism on a young tropical mountain. Nature 524 347–350. 10.1038/nature14949 [DOI] [PubMed] [Google Scholar]
- Meredith R. W., Mendoza M. A., Roberts K. K., Westerman M., Springer M. S. (2010). A Phylogeny and timescale for the evolution of Pseudocheiridae (Marsupialia: Diprotodontia) in Australia and New Guinea. J. Mamm. Evol. 17 75–99. 10.1007/s10914-010-9129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Kircher M. (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010:pdb.prot5448. 10.1101/pdb.prot5448 [DOI] [PubMed] [Google Scholar]
- Miller M. A., Pfeiffer W., Schwartz T. (2011). “The CIPRES science gateway: a community resource for phylogenetic analyses,” in Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Vol. 41 (New York, NY: ACM; ). 10.1145/2016741.2016785 [DOI] [Google Scholar]
- Mirarab S. (2019). Species tree estimation using ASTRAL: practical considerations. arXiv [Preprint] Available at: https://arxiv.org/abs/1904.03826v2 [Google Scholar]
- Mirarab S., Nguyen N., Guo S., Wang L.-S., Kim J., Warnow T. (2015). PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J. Comput. Biol. 22 377–386. 10.1089/cmb.2014.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy B., Forest F., Barraclough T., Rosindell J., Bellot S., Cowan R., et al. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Mol. Phylogenet. Evol. 144:1066682. 10.1016/j.ympev.2019.106668 [DOI] [PubMed] [Google Scholar]
- Mutke J., Sommer J. H., Kreft H., Kier G., Barthlott W. (2011). “Vascular plant diversity in a changing world: global centres and biome-specific patterns,” in Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas, eds Zachos F. E., Habel J. C. (Heidelberg: Springer; ), 83–96. [Google Scholar]
- Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.-P. D., Mirarab S., Kumar K., Warnow T. (2015). Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 16:124. 10.1186/s13059-015-0688-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A. N., Plunkett G. M. (2014). Diversification times and biogeographic patterns in Apiales. Bot. Rev. 80 30–58. 10.1007/s12229-014-9132-4 [DOI] [Google Scholar]
- Norman J. A., Rheindt F. E., Rowe D. L., Christidis L. (2007). Speciation dynamics in the Australo-Papuan Meliphaga honeyeaters. Mol. Phylogenet. Evol. 42 80–91. 10.1016/j.ympev.2006.05.032 [DOI] [PubMed] [Google Scholar]
- Nürk N. M., Michling F., Linder H. P. (2018). Are the radiations of temperate lineages in tropical alpine ecosystems pre-adapted? Glob. Ecol. Biogeogr. 27 334–345. 10.1111/geb.12699 [DOI] [Google Scholar]
- Oliver L. A., Rittmeyer E. N., Kraus F., Richards S. J., Austin C. C. (2013). Phylogeny and phylogeography of Mantophryne (Anura: Microhylidae) reveals cryptic diversity in New Guinea. Mol. Phylogenet. Evol. 67 600–607. 10.1016/j.ympev.2013.02.023 [DOI] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574 679–685. 10.1038/s41586-019-1693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijmans K. (1976). “Vegetation,” in New Guinea Vegetation, ed. Paijmans K. (Canberra: Australian National University Press; ), 23–105. [Google Scholar]
- Philipson W. R. (1978). “Araliaceae: growth forms and shoot morphology,” in Tropical Trees as Living Systems, eds Tomlinson P. B., Zimmermann M. H. (Cambridge: Cambridge University Press; ), 269–284. [Google Scholar]
- Pielou E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13 131–144. 10.1016/0022-5193(66)90013-0 [DOI] [Google Scholar]
- Plunkett G. M., Lowry P. P., II (2010). Paraphyly and Polyphyly in Polyscias Sensu Lato: molecular evidence and the case for Recircumscribing the “Pinnate Genera” of Araliaceae. Plant Divers. Evol. 128 23–54. 10.1127/1869-6155/2010/0128-0002 [DOI] [Google Scholar]
- Plunkett G. M., Lowry P. P., II (2012). Phylogeny and diversification in the melanesian Schefflera Clade (Araliaceae) based on evidence from nuclear rDNA spacers’. Syst. Bot. 37 279–291. 10.1600/036364412X616837 [DOI] [Google Scholar]
- Plunkett G. M., Lowry P. P., II, Frodin D. G., Wen J. (2005). Phylogeny and geography of Schefflera: pervasive polyphyly in the largest genus of Araliaceae. Ann. Mo. Bot. Gard. 92 202–204. [Google Scholar]
- Plunkett G. M., Wen J., Lowry P. P., II (2004). Infrafamilial classifications and characters in Araliaceae: insights from the phylogenetic analysis of nuclear (ITS) and Plastid (trnL-trnF) sequence data. Plant Syst. Evol. 245 1–39. 10.1007/s00606-003-0101-3 [DOI] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan M. D. (2019). evolutionary insights into the nature of plant domestication. Curr. Biol. 29 R705–R714. 10.1016/j.cub.2019.05.053 [DOI] [PubMed] [Google Scholar]
- R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rambaut A. (2018). FigTree v1.4.4. Available at: https://github.com/rambaut/figtree/releases/tag/v1.4.4 (accessed August 9, 2019). [Google Scholar]
- Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V., Khurana J. P., Tyagi A. K., Khurana P. (2008). An update on chloroplast genomes. Plant Syst. Evol. 271 101–122. 10.1007/s00606-007-0608-0 [DOI] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe K. C., Aplin K. P., Baverstock P. R., Moritz C. (2011). Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst. Biol. 60 188–203. 10.1093/sysbio/syq092 [DOI] [PubMed] [Google Scholar]
- Sanmartín I., van der Mark P., Ronquist F. (2008). Inferring dispersal: a Bayesian approach to phylogeny-based island biogeography, with special reference to the Canary Islands. J. Biogeogr. 35 428–449. 10.1111/j.1365-2699.2008.01885.x [DOI] [Google Scholar]
- Särkinen T., Staats M., Richardson J. E., Cowan R. S., Bakker F. T. (2012). How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One 7:e43808. 10.1371/journal.pone.0043808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari E., Mirarab S. (2018). Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes 9:E132. 10.3390/genes9030132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwery O., Onstein R. E., Bouchenak-Khelladi Y., Xing Y., Carter R. J., Linder H. P. (2015). As old as the mountains: the radiations of the Ericaceae. New Phytol. 207 355–367. 10.1111/nph.13234 [DOI] [PubMed] [Google Scholar]
- Shen X.-X., Salichos L., Rokas A. (2016). A genome-scale investigation of how sequence, function, and tree-based gene properties influence phylogenetic inference. Genome Biol. Evol. 2565–2580. 10.1093/gbe/evw179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenáø P., Hedberg I., Cleef A. M. (2014). Island biogeography of tropical Alpine floras. J. Biogeogr. 41 287–297. 10.1111/jbi.12212 27555436 [DOI] [Google Scholar]
- Smith S. A., Brown J. W., Walker J. F. (2018). So many genes, so little time: a practical approach to divergence-time estimation in the genomic Era. PLoS One 13:e0197433. 10.1371/journal.pone.0197433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. A., Moore M. J., Brown J. W., Yang Y. (2015). Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol. 15:150. 10.1186/s12862-015-0423-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow D. W. (1981). Tropical frugivorous birds and their food plants: a world survey. Biotropica 13 1–14. 10.2307/2387865 [DOI] [Google Scholar]
- Soto Gomez M., Pokorny L., Kantar M. B., Forest F., Leitch I. J., Gravendeel B., et al. (2019). A customized nuclear target enrichment approach for developing a phylogenomic Baseline for Yams (Dioscoreaceae). Appl. Plant Sci. 7:e11254. 10.1002/aps3.11254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Murphy W. J., Roca A. L. (2018). Appropriate fossil calibrations and tree constraints uphold the Mesozoic divergence of Solenodons from other extant mammals. Mol. Phylogenet. Evol. 121 158–165. 10.1016/j.ympev.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Stadler T. (2009). On incomplete sampling under birth-death models and connections to the sampling-based coalescent. J. Theor. Biol. 261 58–66. 10.1016/j.jtbi.2009.07.018 [DOI] [PubMed] [Google Scholar]
- Su Y. C. F., Saunders R. M. K. (2009). Evolutionary divergence times in the Annonaceae: evidence of a late Miocene origin of Pseuduvaria in Sundaland with subsequent diversification in New Guinea. BMC Evol. Biol. 9:153. 10.1186/1471-2148-9-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4:vey016. 10.1093/ve/vey016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint E. F., Hall R., Monaghan M. T., Sagata K., Ibalim S., Shaverdo H. V., et al. (2014). The Towering Orogeny of New Guinea as a Trigger for Arthropod Megadiversity. Nat. Commun. 5:4001. 10.1038/ncomms5001 [DOI] [PubMed] [Google Scholar]
- Unmack P. J., Allen G. R., Johnson J. B. (2013). Phylogeny and Biogeography of Rainbowfishes (Melanotaeniidae) from Australia and New Guinea. Mol. Phylogenet. Evol. 67 15–27. 10.1016/j.ympev.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Valcárcel V., Wen J. (2019). Chloroplast phylogenomic data support Paleocene - Eocene Amphi-pacific early radiation for the Asian Palmate Core Araliaceae. J. Syst. Evol. 57 547–560. 10.1111/jse.12522 [DOI] [Google Scholar]
- Van Andel T., Veltman M. A., Bertin A., Maat H., Polime T., Lambers D., et al. (2019). Hidden rice diversity in the Guianas. Front. Plant Sci. 10:1161. 10.3389/fpls.2019.01161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Royen P. (1979). The Alpine Flora of New Guinea. Vaduz: A. R. Gantner Verlag K. G. [Google Scholar]
- van Ufford A. Q., Cloos M. (2005). Cenozoic Tectonics of New Guinea. AAPG Bull. 89 119–140. 10.1306/08300403073 [DOI] [Google Scholar]
- van Welzen P. C., Parnell J. A. N., Ferry Slik J. W. (2011). Wallace’s line and plant distributions: Two or three phytogeographical areas and where to group java? Biol. J. Linn. Soc. 103 531–545. 10.1111/j.1095-8312.2011.01647.x [DOI] [Google Scholar]
- van Welzen P. C., Turner H., Roos M. (2001). New Guinea: a correlation between accreting areas and dispersing Sapindaceae. Cladistics 17 242–247. 10.1006/clad.2001.0173 [DOI] [PubMed] [Google Scholar]
- Vatanparast M., Powell A., Doyle J. J., Egan A. N. (2018). Targeting Legume Loci: a comparison of three methods for target enrichment bait design in Leguminosae phylogenomics. Appl. Plant Sci. 6:e1036. 10.1002/aps3.1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaverde T., Pokorny L., Olsson S., Rincón-Barrado M., Johnson M. G., Gardner E. M., et al. (2018). Bridging the Micro- and Macroevolutionary levels in Phylogenomics: Hyb-Seq solves relationships from populations to species and above. New Phytol. 220 636–650. 10.1111/nph.15312 [DOI] [PubMed] [Google Scholar]
- Vollering J., Schuiteman A., de Vogel E., van Vugt R., Raes N. (2016). Phytogeography of New Guinean Orchids: patterns of species richness and turnover. J. Biogeography 43 204–214. 10.1111/jbi.12612 [DOI] [Google Scholar]
- Voris H. K. (2000). Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27 1153–1167. 10.1046/j.1365-2699.2000.00489.x [DOI] [Google Scholar]
- Wallace A. R. (1869). The Malay Archipelago: The Land of the Orang-Utan and the Bird of Paradise: A Narrative of Travel, with Studies of Man and Nature. New York, NY: Harper & Brothers. [Google Scholar]
- Warburg O. (1891). Beiträge zur Kenntnis der papuanischen Flora. Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 13 230–455. [Google Scholar]
- Weitemier K., Straub S. C. K., Cronn R. C., Fishbein M., Schmickl R., McDonnell A., et al. (2014). Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2:1400042. 10.3732/apps.1400042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Ree R. H., Ickert-Bond S. M., Nie Z., Funk V. (2013). Biogeography: Where do we go from here? Taxon 62 912–927. 10.12705/625.15 [DOI] [Google Scholar]
- Wheeler T. J., Kececioglu J. D. (2007). Multiple alignment by aligning alignments. Bioinformatics 23 i559–i568. 10.1093/bioinformatics/btm226 [DOI] [PubMed] [Google Scholar]
- White D. M., Islam M. B., Mason-Gamer R. J. (2019). Phylogenetic inference in section Archerythroxylum informs taxonomy, biogeography, and the domestication of coca (Erythroxylum Species). Am. J. Bot. 106 154–165. 10.1002/ajb2.1224 [DOI] [PubMed] [Google Scholar]
- Wikramanayake E. D. (2002). Terrestrial Ecoregions of the Indo-Pacific: A Conservation Assessment. Washington, DC: Island Press. [Google Scholar]
- Williams J. N. (2011). “Human population and the hotspots revisited: a 2010 assessment,” in Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas, eds Zachos F. E., Habel J. C. (Heidelberg: Springer; ), 61–81. [Google Scholar]
- Yule G. U. (1925). A mathematical theory of evolution based on the conclusions of Dr. J. C. Willis, F.R.S. Philos. Trans. Roy. Soc. Lond. B 213 21–87. [Google Scholar]
- Zhang C., Rabiee M., Sayyari E., Mirarab S. (2018). ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19:153. 10.1186/s12859-018-2129-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong X., Song J., Lv J., Wang S. (2016). The complete chloroplast genome sequence of Schefflera octophylla. Mitochondrial DNA 27 4685–4686. 10.3109/19401736.2015.1106502 [DOI] [PubMed] [Google Scholar]
- Zwiers P. B., Borgia G., Fleischer R. C. (2008). Plumage based classification of the bowerbird genus Sericulus evaluated using a multi-gene, multi-genome analysis. Mol. Phylogenet. Evol. 46 923–931. 10.1016/j.ympev.2007.11.019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw reads (FASTQ files) are available from the NCBI BioProject database (ID: PRJNA604390). The datasets generated and analyzed are available from Zenodo (doi: 10.5281/zenodo.3534088).