Abstract
Epidemics of coffee leaf rust (CLR) leads to great yield losses and huge depreciation of coffee marketing values, if no control measures are applied. Societal expectations of a more sustainable coffee production are increasingly imposing the replacement of fungicide treatments by alternative solutions. A protection strategy is to take advantage of the plant immune system by eliciting constitutive defenses. Based on such concept, plant resistance inducers (PRIs) have been developed. The Greenforce CuCa formulation, similarly to acibenzolar-S-methyl (ASM), shows promising results in the control of CLR (Hemileia vastatrix) in Coffea arabica cv. Mundo Novo. The molecular mechanisms of PRIs action are poorly understood. In order to contribute to its elucidation a proteomic, physiological (leaf gas-exchange) and biochemical (enzymatic) analyses were performed. Coffee leaves treated with Greenforce CuCa and ASM and inoculation with H. vastatrix were considered. Proteomics revealed that both PRIs lead to metabolic adjustments but, inducing distinct proteins. These proteins were related with photosynthesis, protein metabolism and stress responses. Greenforce CuCa increased photosynthesis and stomatal conductance, while ASM caused a decrease in these parameters. It was further observed that Greenforce CuCa reinforces the redox homeostasis of the leaf, while ASM seems to affect preferentially the secondary metabolism and the stress-related proteins. So, the PRIs prepare the plant to resist CLR but, inducing different defense mechanisms upon pathogen infection. The existence of a link between the primary metabolism and defense responses was evidenced. The identification of components of the plant primary metabolism, essential for plant growth and development that, simultaneously, participate in the plant defense responses can open new perspectives for plant breeding programs.
Keywords: coffee leaf rust (CLR), Coffea arabica cv. Mundo Novo, proteomics, enzymatic activities, physiological parameters, 2DE-MALDI/TOF/TOF-MS/MS, greenforce CuCa, acibenzolar-S-methyl (ASM)
Graphical Abstract
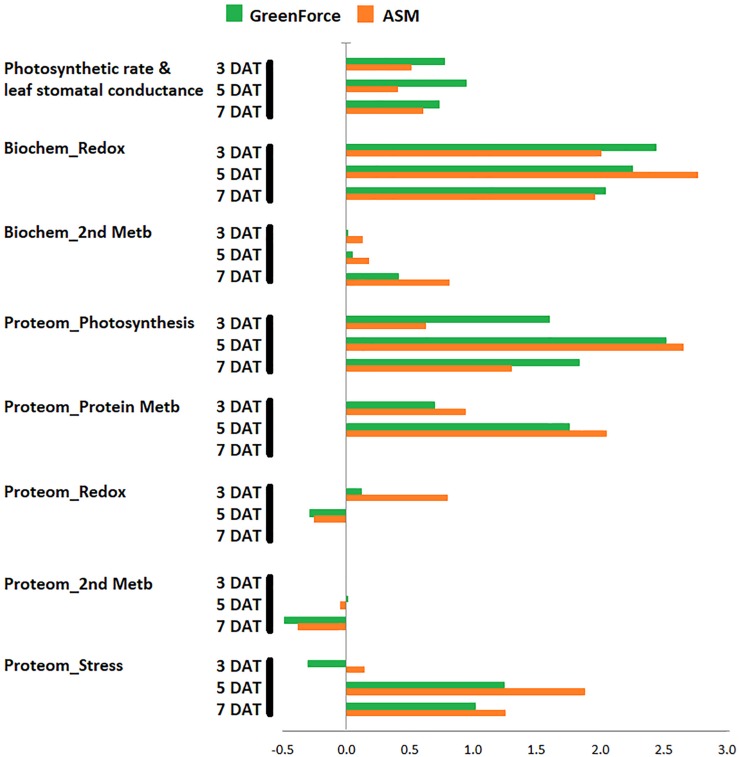
Main metabolic classes affected by the resistance inducers in response to coffee leaf rust.
Introduction
Coffee, one of the most important beverage crops in the world (with billions of cups consumed per day), is cultivated across Africa, Asia, and the Americas (Läderach et al., 2017). Crucial for the economy of more than 60 countries, coffee is the main source of income for more than 100 million people (Hoffmann, 2014; Läderach et al., 2017; ICO, 2020). Brazil is the first world coffee producer (mostly Coffea arabica L.), Minas Gerais being the state responsible for more than 50% of the Brazilian coffee production. In 2019, and considering the planted area (1.81 Mha) the productivity of coffee was 1.632 tons per hectare (CONAB, 2019).
Coffea arabica has the best quality/aroma, but most of its commercial varieties are highly susceptible to several pathogens, namely Hemileia vastatrix. This biotrophic fungus, that is spread to all coffee growing regions causes coffee leaf rust (CLR), a devastating disease characterized by large orange colonies of urediniospores in the lower surface of the leaves (Bettencourt and Rodrigues, 1988; Várzea and Marques, 2005; Talhinhas et al., 2017). In susceptible coffee leaves, after the urediniospores germination and appressorium differentiation over stomata, the fungus penetrates leaf tissues and grows into the substomatal chamber. Fungal growth continues with the formation of more intercellular hyphae and a large number of haustoria (highly specialized intracellular hyphae) in the spongy and palisade parenchyma cells. The host metabolism is modified to serve the fungus nutrients uptake allowing the completion of its life cycle (which take about 30 days) (Silva et al., 1999, 2006). The CLR causes the premature leaf fall as result of direct damages, weakening and favoring dieback of branches, decreasing the photosynthetic capacity and vigor of the infected coffee plants (Silva et al., 2006; Talhinhas et al., 2017). A recent intense epidemic of CLR in Colombia and Central America was responsible for estimated losses of several hundred million dollars (Avelino et al., 2015). In Brazil, the disease threatens coffee production, losses ranging from 30 to 50%, if no chemical control is undertaken. CLR damage is prevented by the use of protective (copper-based) and/or systemic fungicides (triazoles and strobilurin) (Zambolim, 2016). However, increasing societal expectations for sustainable coffee production demands the replacement of fungicide treatments by alternative strategies of plant protection, such as the use of coffee resistant varieties and plant resistance inducers (PRIs) (Resende et al., 2002; Várzea and Marques, 2005; Avelino et al., 2015).
The application of PRIs mimics a pathogen infection, and thus, activates a sort of unspecific systemic immunity, known as systemic acquired resistance (SAR). SAR is considered as one of the players in a multifaceted inducible defense system in plants, characterized by various signaling pathways, and metabolic responses (Cavalcanti et al., 2006; Gozzo and Faoro, 2013; Balmer et al., 2015). The PRIs application results in a stronger and faster defense response when biotic or abiotic stresses occur. Different PRI treatments, either biotic (viable or inactivated microorganisms) or chemical [e.g., acibenzolar-S-methyl (ASM), ethylene, plant natural formulations], have been used (Conrath et al., 2015).
Acibenzolar-S-methyl is a salicylic acid functional analog belonging to the benzothiadiazole (BTH) family, which is rapidly absorbed by the leaves and activating SAR (Medeiros et al., 2009; Furtado et al., 2010). Genes encoding pathogenesis-related proteins (PR proteins) are expressed and regulate secondary metabolic pathways and defense responses (Iriti and Faoro, 2003; Glazebrook, 2005; Gozzo and Faoro, 2013). ASM treatment of coffee leaves showed some protection against CLR, but without affecting H. vastatrix germination (Guzzo et al., 2009). Instead, what was observed was the induction by ASM of some SAR-related genes, such as those involved in: signal perception and transduction, oxidative burst and cell death, synthesis and transport of antimicrobial metabolites, synthesis of PR proteins and lipid metabolism (Guzzo et al., 2009). Under field conditions ASM application also satisfactorily controlled rust and other coffee diseases (Fernandes et al., 2013).
Formulations based on natural products have also been intensively studied, and it was found that they also activate the plant defense responses (Barguil et al., 2005; Medeiros et al., 2009; Conrath et al., 2015). Coffee industry by-products were effective in the control of Xanthomonas vesicatoria infection in tomato through the up-regulation of PR and antioxidant proteins (Medeiros et al., 2009). Theses formulations are being used as a control measure of plant diseases in coffee and other crops (e.g., tomato and eucalyptus) (Barguil et al., 2005; Jackson et al., 2000; Cavalcanti et al., 2006; Medeiros et al., 2009). Greenforce CuCa formulation is a plant based extract prepared with coffee industry by-products supplemented with calcium and copper salts displaying antioxidant properties. These properties derived from the high content of chlorogenic acids and caffeine, and other compounds like nicotinic acid, trigonelline, tocopherols, cafestol, and heterocyclic compounds (Esquivel and Jiménez, 2012; Murthy and Naidu, 2012). In field works, this formulation reduced CLR by about 50% (Costa et al., 2014; Silva et al., 2019).
Although the involvement of secondary metabolism in plant defense is fully documented, the relationship between defense and primary metabolism is less studied. Upon pathogen infection several genes associated with primary metabolic pathways are induced, namely, those involved in the synthesis or degradation of carbohydrates, amino acids, and lipids (Rojas et al., 2014). Links between photosynthesis and immunity in plants has been proposed (Göhre, 2015), but the role of primary metabolism in SAR has not been fully understood/analyzed. The present study aims to obtain an overview of the protein changes occurring in the C. arabica cv. Mundo Novo leaves upon treatment with the resistance inducers Greenforce Cuca and ASM and, subsequently, challenged by the obligate biotrophic fungus H. vastatrix. This proteomic phenotyping was complemented by physiological and biochemical analyses (leaf gas-exchange and enzymatic assays).
Materials and Methods
Biological Material and Treatments
Six month old seedlings (with five leaf pairs) of C. arabica cv. Mundo Novo IAC 376/4 (susceptible to CLR) were used. The experiment was conducted in a growth chamber (Eletrolab) at 24°C and 12 h photoperiod with fluorescent light (600 μmoles m–2 s–1). Plants were acclimated in the growth chamber, 30 days prior to the beginning of the experiment.
The upper surface of young fully expanded leaves of C. arabica cv. Mundo Novo were sprayed with two resistance inducers (PRIs), ASM (Bion 500 WG, Syngenta) and Greenforce CuCa (formulation prepared from products of coffee industry supplemented with copper and calcium salts; Universidade Federal de Lavras – patent pending PI063575-2, National Institute of Industrial Property, Brazil). The doses of the products were set according to the manufacturer’s recommendations: 0.2 g L–1 for ASM and 5 mL L–1 for Greenforce CuCa. Approximately 1 mL of the PRI solutions per leaf, using a manual sprayer was used. Leaves sprayed with water were used as mock-treated control.
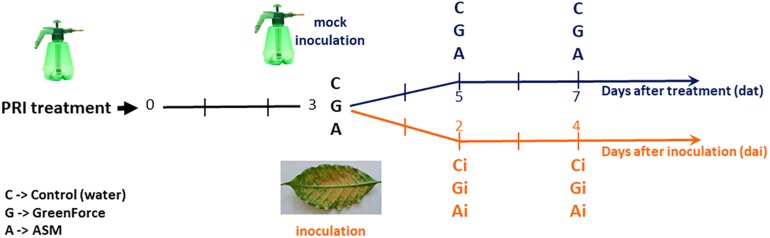
Urediniospores of H. vastatrix obtained from diseased field grown coffee (C. arabica cv. Mundo Novo) were used as inoculum source. Inoculation was performed at 3 days after PRIs treatment by spraying the undersurfaces of the leaves with a urediniospore suspension in 0.2% agar distilled water (v/v) with 0.05% Tween to a final concentration of 106 urediniospore mL–1. After inoculation all plants were kept for 24 h in a dark moist chamber (Guzzo et al., 2009). Leaves sprayed with water and kept in the same conditions of the inoculated leaves were used as mocked-inoculated control. Samples were collected at 3, 5, and 7 days after PRIs treatment, which correspond to 0, 2, and 4 days after H. vastatrix inoculation, respectively (Figure 1).
FIGURE 1.
Experimental design – Coffea arabica cv. Mundo Novo leaves were either treated with the PRIs, ASM (A) and Greenforce CuCa (G) or subsequently inoculated with Hemileia vastatrix (Ai, Gi). Leaves were collected at 3, 5, and 7 days after PRIs treatment (black timeline) and at 2 and 4 days after H. vastatrix inoculation (orange timeline). Leaves treated with water were used as control: mock-treated/inoculated (C) or inoculated (Ci).
A complete randomized block design with six treatments and three blocks (replicates) per time-point was performed. Three plants (one leaf pair per plant) were used as experimental unit for either proteomic, physiological or biochemical studies.
The rust disease severity assessment (percentage of affected area) was done by visual evaluation of the inoculated leaves (four leaves for plant) after the appearance of the first symptoms in a total of six evaluations (every 7 days) according to a diagrammatic scale (Capucho et al., 2011). The observed indexes of severity were transformed in the area under the corresponding disease severity progress curve (AUDsPC) as proposed by Shaner and Finney (1977) (Supplementary Tables S1A,B).
Proteomic Analysis
Coffee leaves (1 g) were ground with liquid nitrogen and proteins extracted using the trichloroacetic acid (TCA) precipitation method (Damerval et al., 1986) and recovered in 30 mM Tris-HCl pH 8.8 buffer solution containing 2% SDS and 50 mM DTT. After 2 h of agitation the samples were centrifuged at 12000 g, during 15 min and the clear supernatants were purified using the 2D clean-up kit following the manufacturer instructions (GE Healthcare). The precipitates were resuspended in 30 mM Tris-HCl pH 8.5 buffer containing 7M urea, 2M thiourea and 4% CHAPS; protein content was measured using a modified Bradford assay method (Ramagli, 1999).
Protein samples (300 μg) were run in 18 cm long IPG strips, pH 4–7 L (GE Healthcare). IEF was performed using the Ettan IPGphor (GE Healthcare) under the following conditions: a total of 32000 Vhrs at 20°C; Step-n-hold 100 V–2 h; Step-n-hold 30 V–10 h; Step-n-hold 150 V–3 h; Step-n-hold 300 V–3 h; Gradient 1000 V–6 h; Step-n-hold 1000 V–1 h; Gradient 8000 V–4 h; Step-n-hold 8000 V–3200 V/h; Step-n-hold 100 V–18 h; maximum current setting of 50 μA per strip. After IEF, the proteins in the IPG strip were equilibrated for 15 min in a buffer (100 mM Tris–HCl pH 8.8, 6M urea, 2% SDS, 30% glycerol and 0.2 mg mL–1 bromophenol blue) containing 5 mg mL–1 DTT (to reduce proteins), followed by another 15 min equilibration in the same buffer but which had DTT replaced by 25 mg mL–1 iodoacetamide (to alkylate proteins). The second dimension SDS-PAGE was performed at 25°C with 12% resolving gels using the 2-D Ettan Dalt II Gel apparatus (GE Healthcare) at 10 mA for 15 min and then at 20 mA until the bromophenol blue dye front had run off the gel (around 22 h). The molecular mass markers used were the “Precision Plus Protein All Blue Standards” (Bio-Rad, Hercules, CA, United States).
Gels were stained in Colloidal Coomassie Blue (Neuhoff et al., 1985) and the protein profiles were scanned using an ImageScanner II (Amersham Biosciences). The image gel analysis was carried out using the Progenesis SameSpots 2D software v. 4.5 (Non-linear Dynamics, Ltd.). The spot volumes were normalized using the mean value of the replicates (Grove et al., 2008). One-way ANOVA analyses were performed using a p-value of 0.05. For the proteins with statistically significant changes (and a fold change > 1.5) a principal component analysis (PCA) was carried out and a hierarchical clustering was performed applying a Pearson correlation using the MeV5 v. 4.8 (Supplementary Table S2).
Polypeptide spots visually detected in Colloidal Coomassie Blue stained gels were excised from the gels and processed using the Tecan freedom EVO200 (Tecan, Männedorf, CH) as previously described (Guerra-Guimarães et al., 2015). The ProteinPilotTM software 4.0.8085 was used for database searches with an in-house MASCOT platform (version 2.3, Matrix Science1, London, United Kingdom). All proteins were identified by search against a Coffea database downloaded from NCBI2 on January 17, 2019 with the taxID 13442 and containing 133 773 sequences. All searches (combined MS and 10 MS/MS spectra) were carried out using a mass window of 100 ppm for the precursor and 0.5 Da for the fragments. During the different searches the following parameters were defined: two missed cleavages, carbamidomethylation of cysteine as fixed modification, and as variable modifications we selected; oxidation of methionine, single or double oxidation of tryptophan and tryptophan to kynurenine. The proteins identified without clear annotation were BLAST analyzed and those proteins with the highest homology (when significant) were added to the Supplementary Table S3.
All identifications were manually validated and extra precursors were selected for fragmentation if the obtained data were judged as insufficient. When high quality spectra were not matched to sequences, a sequence was determined manually and, so, in the current data set they could be linked to the identified protein by allowing for more missed cleavages, semi-tryptic peptides, specific modifications or broader taxonomy search. All the spots identified in the 5 day sample control, containing one or more proteins were considered for the leaf proteome establishment. For the comparative analysis only spots having unique and significant protein identification were considered.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE partner repository (Perez-Riverol et al., 2019) with the dataset identifier PXD016012.
The identified proteins were also subjected to InterPro, UniProt, and NCBI databases analyses. The conserved domains of each protein, as well as, the superfamily were determined using the NCBI tools2. Assignment for functional annotation and protein location was based on MapMan ‘Bin’ ontology3 using Mercator Automated Sequence Annotation Pipeline4 (Lohse et al., 2014) and on Gene Ontology Annotation (GO5) using Blast2GO software (version 5 basic6) (Conesa and Götz, 2008). In addition, protein subcellular location was also assigned using the LocTree37 (Goldberg et al., 2014). Default parameters were used for all the programs.
Physiological Measurements
Gas-exchange characteristics were evaluated in fully expanded leaves using a LI-6400XT Portable Photosynthesis System (LI-COR, Lincoln, United Kingdom), at a photosynthetic photon flux density (PPFD) of 1000 μmol m–2 s–1, from a red/blue light source (6400-02B LI-COR, Lincoln, United Kingdom LED). Measurements took place between 8:30 and 11:30 h (solar time). Two leaves per plant (3rd leaf pair), in a total of six leaves were used per experimental unit. Leaf net photosynthetic rate (A; μmol CO2 m–2 s–1), stomatal conductance (gs; mol H2O m–2 s–1), mesophyll intercellular CO2 concentration (ci), the ratio of intercellular to ambient CO2 concentrations (ci/ca), water use efficiency (WUE, A/transpiration) and carboxylation efficiency of photosynthesis (A/ci) were estimated. Data related to each time-point was analyzed by one-way analysis of variance (ANOVA). When treatments were significant by the F-test, a pairwise multiple comparison was performed using Tukey test (p ≤ 0.05).
Biochemical Assays
The activities of peroxidase (POX), ascorbate peroxidase (APX), superoxide dismutase (SOD), phenylalanine ammonia lyase (PAL), and polyphenol oxidase (PPO) were quantified. Protein content was determined using bovine serum albumin (BSA) as standard (Bradford, 1976). Data related to each time-point was analyzed by one-way analysis of variance (ANOVA). When treatments were significant by the F-test, a pairwise multiple comparison was performed using Tukey test (p ≤ 0.05).
POX, APX, and SOD measurements were made according to Biemelt et al. (1998) with some modifications. Leaves (0.2 mg) were ground with liquid nitrogen in the presence of 1% polyvinylpolypyrrolidone (PVPP) (w/w) and extracted with 1.5 ml of potassium phosphate buffer (100 mM, pH 7.8) containing 0.1 mM ethylenedinitrilotetraacetic acid (EDTA) and 10 mM ascorbic acid. The extract was centrifuged at 13,000 g, 4°C for 15 min and supernatants used for the enzymatic analyses.
POX activity was performed according to Urbanek et al. (1991) at 30°C, and recording the absorbance at 480 nm. POX specific activity (μmol/min/mg protein) was calculated using the molar absorption coefficient (ε) of 1.235 mM–1 cm–1 (Chance and Maehly, 1955).
APX activity was performed according to Nakano and Asada (1981) at 25°C, and recording the absorbance at 290 nm. APX specific activity (μmol/min/mg protein) was calculated using the molar absorption coefficient (ε) of 1.4 mM–1 cm–1.
SOD activity was performed according to Giannopolitis and Ries (1977) following the nitro blue tetrazolium chloride (NBT) photoreduction, by measuring the absorbance at 560 nm. Data was presented as SOD specific activity (U/min/mg protein). One unit of SOD activity was defined as the amount of enzyme able to reduce NBT photoreduction by 50%.
For PAL quantification, and after liquid nitrogen grinding, 1 g of leaves were extracted with 3 mL of sodium phosphate buffer (50 mM, pH 6.5) containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 1% PVPP (w/v). The extract was centrifuged at 13,000 g, 4°C for 25 min, using the supernatant for enzymatic analysis. PAL activity was performed according to Zucker (1965) at 37°C, and recording absorbance at 280 nm. PAL specific activity (μmol/min/mg protein) was calculated by the phenylalanine decrease using the molar absorption coefficient (ε) of 10000 mM–1 cm–1.
For PPO quantification, after liquid nitrogen grinding, 1 g of leaves were extracted with 4 mL of potassium phosphate buffer (30 mM, pH 7.0) containing 0.1 mM EDTA. The extract was centrifuged at 13,000 g, 4°C for 25 min and using the supernatant for enzymatic analysis. PPO activity was performed according to Kar and Mishra (1976) at 30°C, recording absorbance at 410 nm. PPO specific activity (μmol/min/mg protein) was calculated through catechol degradation using the molar absorption coefficient (ε) of 1.235 mM–1 cm–1.
Hydrogen peroxide (H2O2) content was estimated in 0.2 g of leaves ground in liquid nitrogen, according to Velikova et al. (2000). The extract was centrifuged (12000 g, 4°C for 15 min), and the supernatant was used for H2O2 estimation at 390 nm, using a standard curve prepared with the following H2O2 concentrations: 0, 5, 15, 25, 35 and 45 μmol/ml.
Lipid peroxidation was determined in 0.2 g of leaves ground in liquid nitrogen, according to Buege and Aust (1978). The extract was centrifuged (10,000 g, 4°C for 10 min) and the supernatant was incubated at 95°C for 30 min. The reaction was stopped by lowering the temperature 0°C. The absorbance was recorded at 535 and 600 nm and the malondialdehyde (MDA) content was calculated through the formula: [MDA] = (A535–A600) using the molar absorption coefficient (ε) of 1.56 × 10–5 mM–1 cm–1.
Results
Coffee Leaf Proteome
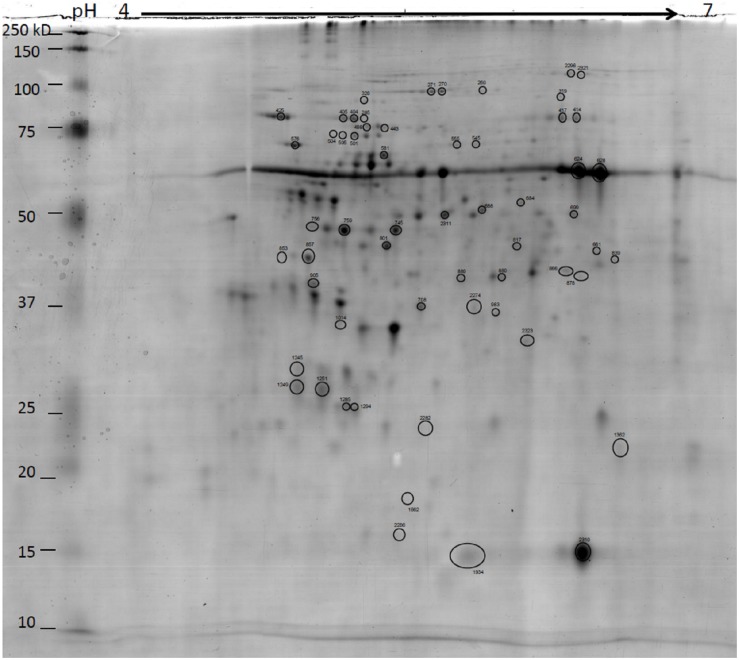
Three hundred and fifty polypeptide spots were detected by 2-DE (linear pH gradient of 4–7) in extracts of the control water-treated leaves. Proteins could be successfully identified in 179 of these spots, 43 of them containing more than one protein (Supplementary Figure S1). All the protein identifications were achieved within the Coffea genomes, mainly C. arabica (107 proteins) and C. eugenioides (97 proteins). The 219 identified proteins belong to 66 superfamilies according to their conserved domains (Supplementary Table S4). The five most represented superfamilies were: FliI (25 spots); RuBisCO_large (14 spots), P-loop_NTPase (13 spots); GH18-Chitinase (9 spots) and Chloroa_b-bind superfamily (7 spots).
The 25 FliI superfamily spots are ATP synthases, one is a mitochondrial ATP synthase related with oxidative phosphorylation; five are V-ATPases related with transport across tonoplast; and nine are ATP synthase CF1 related with photochemical reactions. Also related with the photochemical pathway are the 10 chlorophyll a-b binding proteins from the Chloro a_b-bind and PLN00048 superfamilies (photosystem light harvesting chlorophyll a/b binding proteins from both LHCI and LHCII). Largely represented is the RuBisCO_large subunit superfamily (14 spots), but RuBisCO related proteins represent a much larger proportion since RuBisCO activase (P-loop_NTPase superfamily, 13 spots), RuBisCO small subunit (4 spots) and RuBisCO chaperonins (Chaperonin_like superfamily, 7 spots) were also detected. Glycosyl hydrolases were also found, namely acidic endochitinase-like proteins (GH18-Chitinase superfamily, 9 spots) and one alpha-mannosidase (GH38-57_N_LamB_YdjC_SF superfamily).
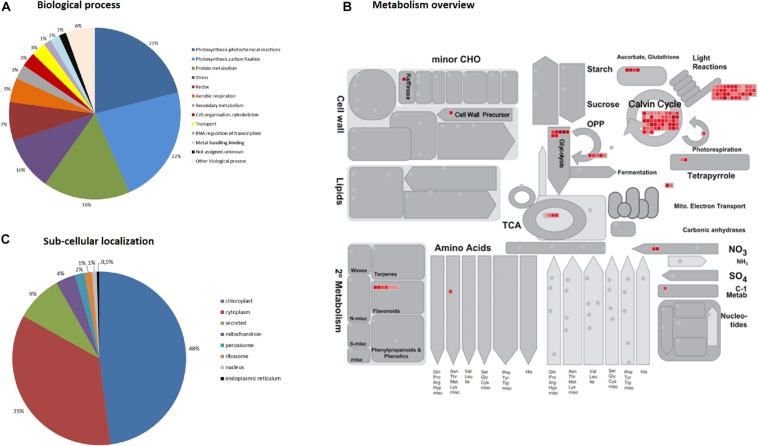
Functional categorization of the 219 identified proteins, based on MapMan “Bin” and GO annotation, indicated that they are mostly involved in: photosynthesis (43%), protein metabolism (16%), stress (10%), redox (7%), aerobic respiration (5%), and secondary metabolism (3%) (Figures 2A,B). Their predicted localizations are: chloroplast (48%), cytoplasm (35%), mitochondrion (4%), peroxisome (2%), ribosome (1%), nucleus (1%), endoplasmic reticulum (0.5%), and secreted (9%) (Figure 2C). Considering that for 124 out of the 219 proteins (56%) EC numbers were assigned (Blast2GO analysis), it was found that 53 spots were hydrolases (ATPases and NTPases), 26 were oxidoreductases, 26 were lyases (carboxy-lyases) and 11 were transferases (Supplementary Table S4). Blast2GO did not assign EC numbers to nine spots identified as acidic endochitinase-like proteins, but they will eventually have hydrolytic activity. Furthermore, distinct isoforms of the same protein (with different pI) were detected for several spots; such as: sedoheptulose-bisphosphatase (spots #853, #857), RuBisCO activase (spots #759, #756), phosphoglycerate kinase (spots #684, #688), RuBisCO large subunit (spots #628, #624) and Glyceraldehyde-3-phosphate dehydrogenase (spots #699, #700, #1500).
FIGURE 2.
Functional annotation of the 219 identified coffee leaf proteins based on MapMan “Bin” and GO ontology. (A) Biological process; (B) Metabolism overview; (C) Sub-cellular localization.
Changes in the Leaf Proteome Due to PRI Treatments and H. vastatrix Infection
The 2-DE protein patterns of coffee leaves treated with the PRIs (Greenforce CuCa and ASM) at 3, 5 and 7 days were analyzed. The simultaneous effect of PRI treatments and H. vastatrix infection at 2 and 4 days after inoculation (dai) were also studied. Overall, the number of spots that changed in abundance for all the comparisons were 165, of which 112 have an unique protein identification and were, thus, considered for further analysis (Table 1). These 112 spots represent 58 distinct proteins, since some proteins were present in more than one sample/comparison/time-point (Table 2 and Figure 3). These 58 proteins represent 26% of the 219 reproducibly identified proteins of the leaf proteome shown in Supplementary Table S4.
TABLE 1.
Proteomic analysis of coffee leaves treated with PRIs and infected with Hemileia vastatrix.
| Assessment | Comparisons | Days | Number of spots* | Spots with unique ID |
| PRI treatments | C x G x A | 3 | 16 | 12 |
| 5 | 44 | 24 | ||
| 7 | 13 | 9 | ||
| PRI + Hv treatments | Ci x Gi x Ai | 5 (2) | 20 | 14 |
| 7 (4) | 13 | 8 | ||
| Multiple interactions | C x G x A x Ci x Gi x Ai | 5 (2) | 44 | 34 |
| 7 (4) | 15 | 11 |
Number of spots that significantly changed in abundance due to PRI treatments Greenforce CuCa (G) and ASM (A) and Control leaves (C) at 3, 5, and 7 days after treatments, and at 2 and 4 days after inoculation (i) with H. vastatrix (Hv).*ANOVA (p-value < 0.05) and Fold Change > 1.5.
TABLE 2.
Annotation of the coffee leaf proteins that changed in abundance after PRI treatments and H. vastatrix infection.
| Biological processa | Spot no.b | Protein identificationc | Accession no.d | Dayse |
| Aerobic respiration | 555 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase | 1531848506 | 5 |
| 2323 | Triosephosphate isomerase, cytosolic | 1527522406 | 5 | |
| 319 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial | 1527604080 | 3 | |
| 2321 | Aconitate hydratase, cytoplasmic | 1527562521 | 5 | |
| C1-metabolism | 2274 | S-Formylglutathione hydrolase-like isoform X1 | 1527584126 | 5 |
| Hormone metabolism | 878 | Auxin-induced protein PCNT115-like | 1531837410 | 5 |
| Minor CHO metabolism | 260 | Galactinol–sucrose galactosyltransferase | 1531812739 | 5 |
| Misc.enzymes.oxidases | 817 | 2-Methylene-furan-3-one reductase-like | 1527480760 | 7 |
| N-metabolism | 2311 | Glutamine synthetase leaf isozyme, chloroplastic | 1531836679 | 5 |
| Photosynthesis | 414 | Transketolase, chloroplastic | 1527513560 | 5 |
| 417 | Transketolase, chloroplastic | 1531823890 | 5,7 | |
| 699 | Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic | 1527478725 | 3,5,7 | |
| 684 | Phosphoglycerate kinase, chloroplastic | 1527563431 | 5 | |
| 688 | Phosphoglycerate kinase, chloroplastic | 1527557669 | 5 | |
| 801 | Phosphoribulokinase, chloroplastic-like | 1527610096 | 5 | |
| 745 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic-like isoform X2 | 1527509480 | 7 | |
| 756 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic-like isoform X2 | 1531823432 | 3, 5 | |
| 759 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic-like isoform X2 | 1531823432 | 5 | |
| 545, 624 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) | 11230404 | 7 | |
| 628 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (plastid) | 1184802834 | 5,7 | |
| 1934, 2310 | Plastid ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit, partial | 83375926 | 5 | |
| 853, 857 | Sedoheptulose-1,7-bisphosphatase, chloroplastic | 1527522116 | 5 | |
| 501 | ATP synthase CF1 alpha subunit (plastid) | 1184802828 | 5,7 | |
| 270 | ATP synthase CF1 alpha subunit (plastid) | 1184802831 | 5 | |
| 866 | ATP synthase gamma chain, chloroplastic | 1527619509 | 3, 5 | |
| 1285 | Chlorophyll a-b binding protein 8, chloroplastic | 1527556260 | 5 | |
| 1362 | Photosystem I reaction center subunit II, chloroplastic-like | 1531835629 | 5 | |
| 1245 | Chlorophyll a-b binding protein of LHCII type 1 | 1531876544 | 7 | |
| 1249 | Chlorophyll a-b binding protein of LHCII type 1-like | 1527620709 | 3 | |
| 1251 | Chlorophyll a-b binding protein of LHCII type 1-like | 1531879126 | 3 | |
| 1294 | Chlorophyll a-b binding protein 36, chloroplastic | 1531849907 | 3, 5 | |
| 1014 | Oxygen-evolving enhancer protein 1, chloroplastic | 1531816163 | 5 | |
| 2282 | Oxygen-evolving enhancer protein 2, chloroplastic-like | 1531849935 | 5, 7 | |
| Protein.degradation | 498, 504 | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | 1531812697 | 5 |
| 505 | ATP-dependent zinc metalloprotease FTSH 2, chloroplastic | 1531812697 | 3,5 | |
| 271 | ATP-dependent Clp protease ATP-binding subunit ClpA homolog CD4B, chloroplastic | 1531841929 | 5 | |
| Protein.folding | 576 | LOW QUALITY PROTEIN: RuBisCO large subunit-binding protein subunit alpha-like | 1527493650 | 5 |
| 581 | RuBisCO large subunit-binding protein subunit beta, chloroplastic | 1531799225 | 3,5 | |
| Protein.synthesis | 328 | Elongation factor G-2, chloroplastic-like | 1527609809 | 5 |
| 2296 | Elongation factor 2 | 1531868107 | 3 | |
| Redox | 983 | L-Ascorbate peroxidase, cytosolic-like | 1527502141 | 5 |
| 1662 | Thioredoxin H-type-like | 1527479082 | 3 | |
| 2286 | Thioredoxin M4, chloroplastic-like | 1531852444 | 7 | |
| RNA | 661 | Chloroplast stem-loop binding protein of 41 kDa a, chloroplastic-like | 1527504431 | 7 |
| Secondary metabolism | 880 | Isoflavone reductase homolog PCBER-like | 1531818231 | 5 |
| 889 | Isoflavone reductase homolog PCBER-like | 1531818231 | 7 | |
| Stress | 285 | Heat shock cognate 70 kDa protein 2-like | 1531823375 | 7 |
| 404 | Heat shock cognate 70 kDa protein 2 | 1531799105 | 3, 5 | |
| 405 | Heat shock cognate 70 kDa protein 2 | 1531799105 | 5,7 | |
| 425 | Stromal 70 kDa heat shock-related protein, chloroplastic-like | 1527478027 | 5 | |
| 443 | Heat shock 70 kDa protein, mitochondrial | 1527512911 | 5,7 | |
| 839 | Auxin-binding protein ABP20-like | 1527581463 | 5, 7 | |
| 905 | Acidic endochitinase-like | 1527489669 | 5 | |
| Unknown.not assigned | 708 | Abscisic stress-ripening protein 5-like | 1531806318 | 7 |
aFunctional characterization of the proteins based on MapMan ‘Bin’ and Go ontology.bThe number that identified protein spots on 2-DE gel (Figure 3).cThe proteins were identified by MALDI-TOF/TOF-MS/MS followed by homology search in Coffea NCBI database.dThe accession number from GenBank assigned to the polypeptide after MS/MS analysis.eDays after PRIs treatments.
FIGURE 3.
Representative 2DE gels of coffee leaf proteins. Circled spots that significantly changed in abundance between Greenforce CuCa (G), ASM (A) and Control (C) at 3, 5, and 7 dat and 2 and 4 days after inoculation (i) with H. vastatrix (Hv) [one way ANOVA analysis was performed using a p-value < 0.05 and Fold Change > 1.5.] The proteins were successfully identified by MALDI-TOF/TOF-MS (detailed information on Table 2). Gels were stained in Colloidal Coomassie Blue.
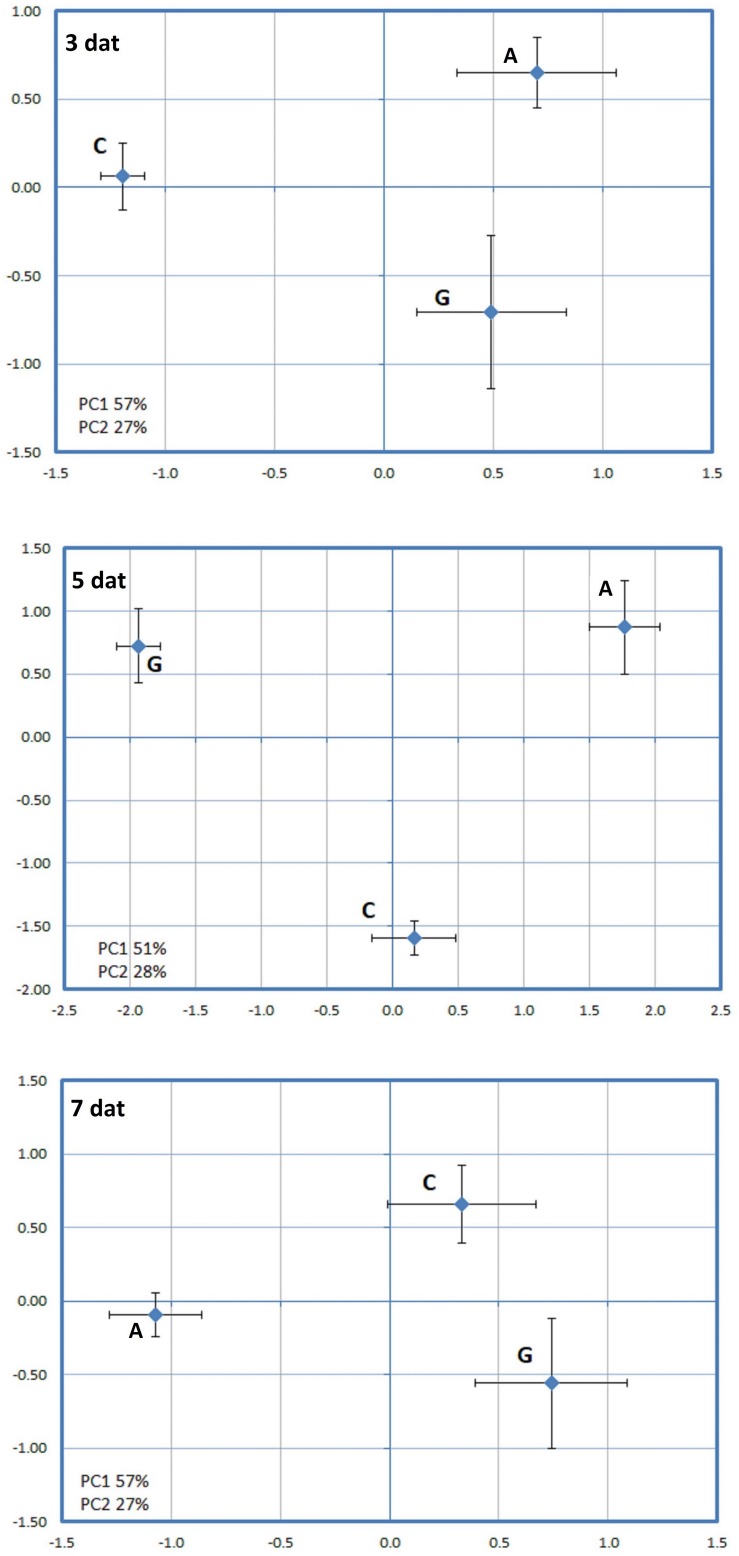
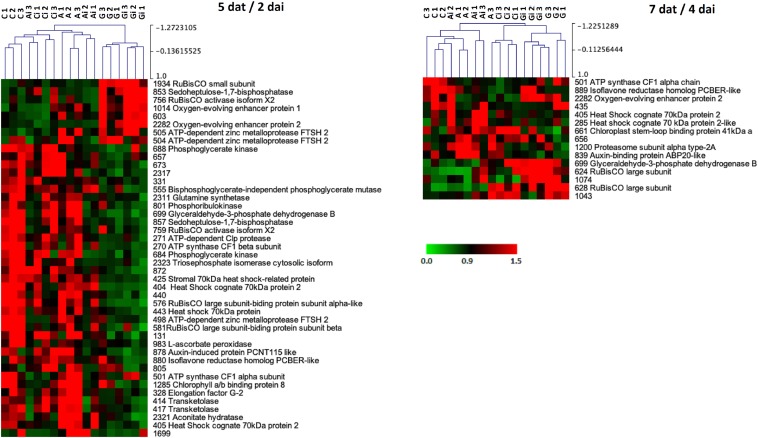
A principal components analysis (PCA) applied to the proteins that changed in abundance due to the PRI treatments [Greenforce CuCa (G), ASM (A), and Control (C)] at 3, 5, and 7 days (Figure 4) revealed a clear separation of the samples. The hierarchical cluster analysis additionally showed the variation in abundance of the proteins in the samples (Figure 5). At 3 days after the treatments (dat), 16 spots significantly differed between samples. This number increased to 44 spots at 5dat and decreased to 13 spots at 7dat. Therefore, at day 5 the protein profiles were more diverse.
FIGURE 4.
Principal component analysis (PCA) performed for the spots whose volume significantly changed in abundance (p-value < 0.05) between Greenforce CuCa (G), ASM (A), and Control (C) at 3, 5 and 7 days after treatments.
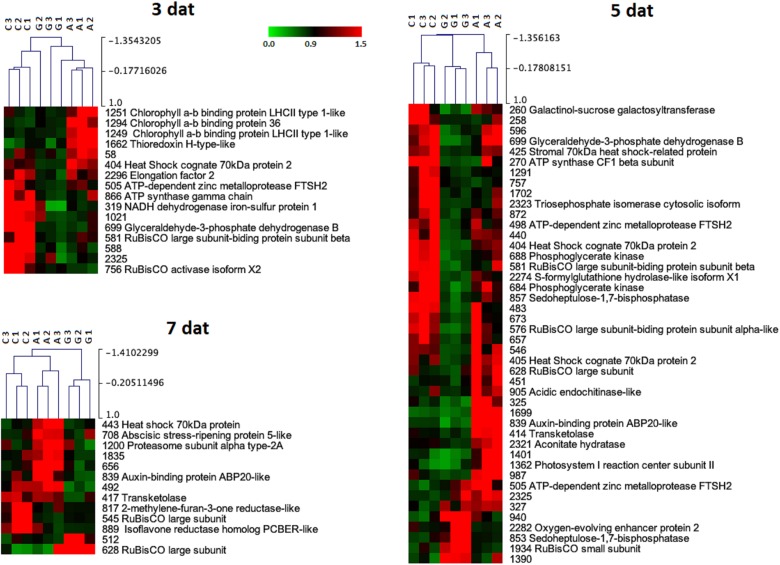
FIGURE 5.
Hierarchical cluster analysis of the proteins that significantly changed in abundance (p-value < 0.05) between Greenforce CuCa (G), ASM (A), and Control (C) at 3, 5, and 7 days after treatments. The signals are shown in a red-green color scale, from a gradient of red (higher expression) to green (lower expression).
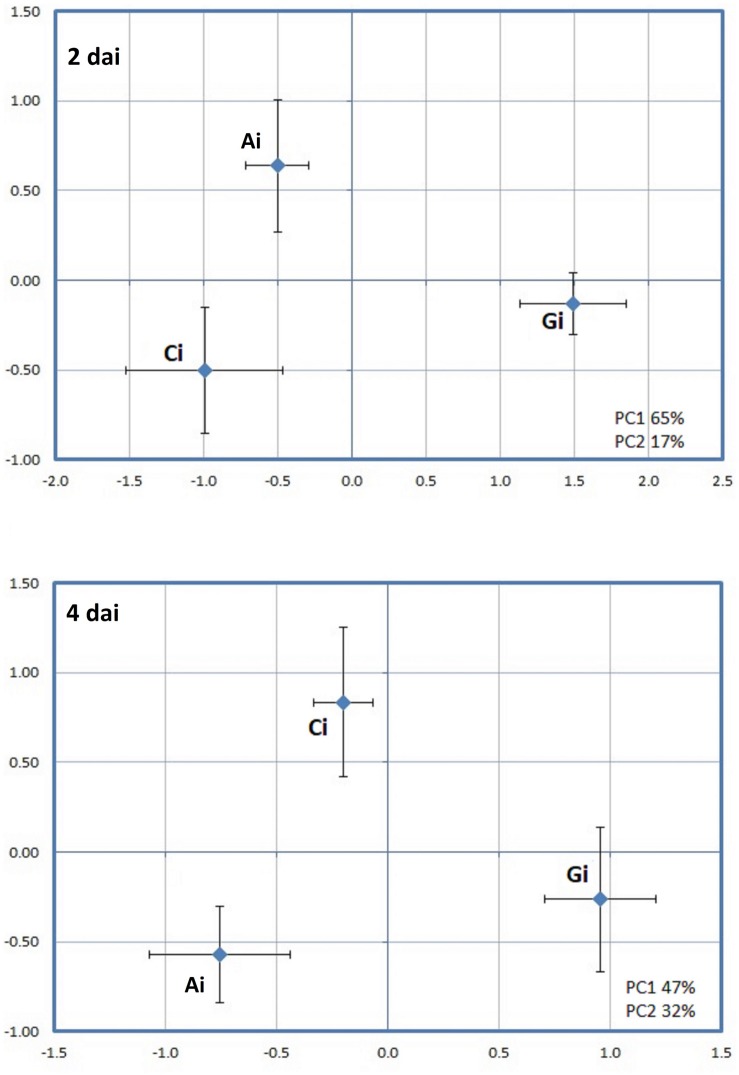
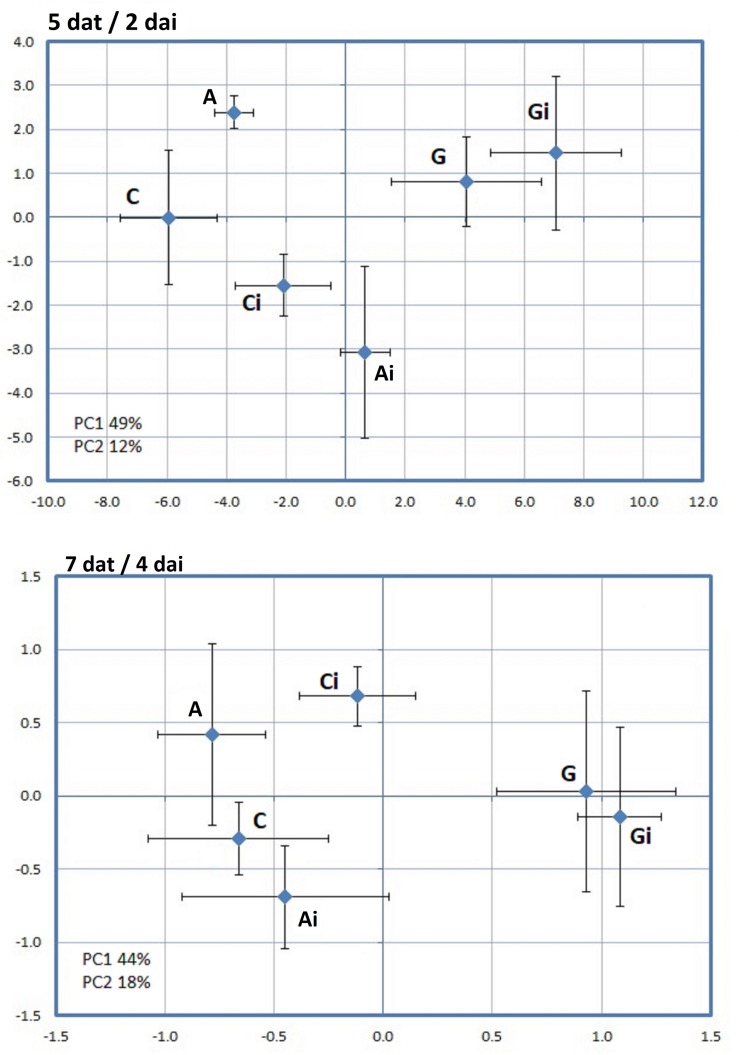
When a PCA was applied to the proteins that were effect by PRI treatments followed by the H. vastatrix infection at 2 or 4 days (Figure 6), it was also observed a clear separation of the three conditions [Greenforce CuCa infected (Gi), ASM infected (Ai), and Control infected (Ci)]. The hierarchical cluster analysis showed the variation of protein abundance in the samples (Figure 7). At 2dai, 20 spots differed significantly between samples, while at 4dai this number decreased to 13 spots. ASM and Greenforce CuCa had a quite distinct protein profile at both time-points, being the ASM protein pattern more similar to that of the Control, particularly at 2dai.
FIGURE 6.
Principal component analysis (PCA) performed for the spots whose volume significantly changed in abundance (p-value < 0.05) between PRI treatments followed by H. vastatrix infection, at 2 and 4 days after inoculation (i): Greenforce CuCa (Gi), ASM (Ai), and Control (Ci).
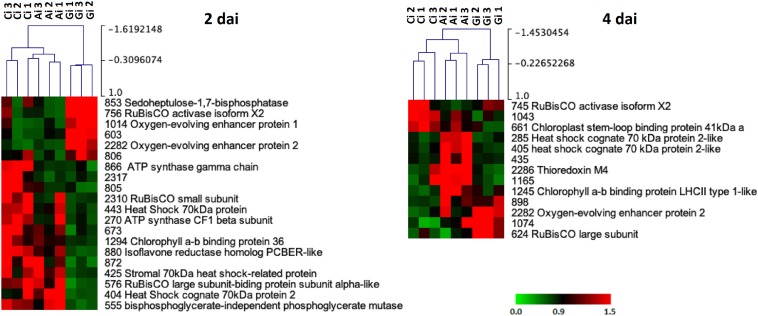
FIGURE 7.
Hierarchical cluster analysis of the proteins that significantly changed in abundance (p-value < 0.05) between PRI treatments followed by H. vastatrix infection, at 2 and 4 days after inoculation (i): Greenforce CuCa (Gi), ASM (Ai), and Control (Ci). The signals are shown in a red-green color scale, from a gradient of red (higher expression) to green (lower expression).
A PCA analysis was further applied to the effects of all PRIs treatments (C x G x A x Ci x Gi x Ai) on the leaf proteome, aiming to reveal the factor that has the major influence on the dynamics of the leaf proteome, i.e., PRI treatments or H. vastatrix infection. Greenforce CuCa treatments (G and Gi) were clearly separated from Control and ASM (C, Ci, A, Ai) at 5dat/2dai and at 7dat/4dai (Figure 8). A hierarchical cluster analysis reinforced the PCA results and evidenced that: at 5dat/2dai 44 spots changed in abundance, while at 7dat/4dai this number decreased to 15 spots (Figure 9); the ASM treatment seems to induce fewer changes in the plant than the Greenforce CuCa treatments (at 5dat), exhibiting a profile more similar to Control. The protein profiles of the ASM treated and inoculated leaves (Ai) cluster together with those of inoculated Control leaves (Ci), while profiles of Greenforce CuCa treated and/or inoculated leaves cluster together (G and Gi).
FIGURE 8.
Principal component analysis (PCA) performed for the spots whose volume significantly changed in abundance (p-value < 0.05) between Greenforce CuCa (G), ASM (A), and Control (C) and treated and inoculation (i) with H. vastatrix, (Ci, Gi, Ai), at 5dat/2dai and 7dat/4dai.
FIGURE 9.
Hierarchical cluster analysis of the proteins that significantly changed in abundance (p-value < 0.05) between Greenforce CuCa (G), ASM (A), and Control (C) and treated and inoculation (i) with H. vastatrix, (Ci, Gi, Ai), at 5dat/2dai and 7dat/4dai. The signals are shown in a red-green color scale, from a gradient of red (higher expression) to green (lower expression).
Proteins Differentially Expressed
The 58 proteins that changed in abundance due to the PRIs treatments and infection were mainly involved in: photosynthesis (27 spots, 47%), protein metabolism (8 spots, 14%), stress (7 spots, 12%), aerobic respiration (4 spots, 7%), and redox (4 spots, 7%) (Table 2). Considering the most discriminating time-point, 5dat/2dai (Figure 9), several proteins related to photosynthesis and to protein degradation positively contributed for the Greenforce CuCa cluster (G and Gi); oxygen-evolving enhancer proteins, OEE1 and OEE2 (#1014 and #2282 spots) and RuBisCO small subunit (#1934 spot) were only responsive to this treatment. Regarding the ASM treatment, other distinct proteins were induced, namely; RuBisCO chaperones (#576 and #581 spots), transketolase (#414 and #417 spots), phosphoglycerate kinase (#684 spots), HSP70 (#404 and #405 spots), acidic endochitinase-like protein (#905 spot) and one auxin-binding protein ABP20-like (#839 spot). Sedoheptulose-1,7-bisphosphatase (#853 and #857 spots) and ATP-dependent zinc metalloproteases (#498, #504 and #505 spots) changed with both PRIs treatments, but different putative isoforms were affected. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, spot #699) was also significantly increased by both treatments, but in a time-dependent way, earlier in ASM (5dat) than in Greenforce CuCa treatments (7dat).
Physiological Analysis
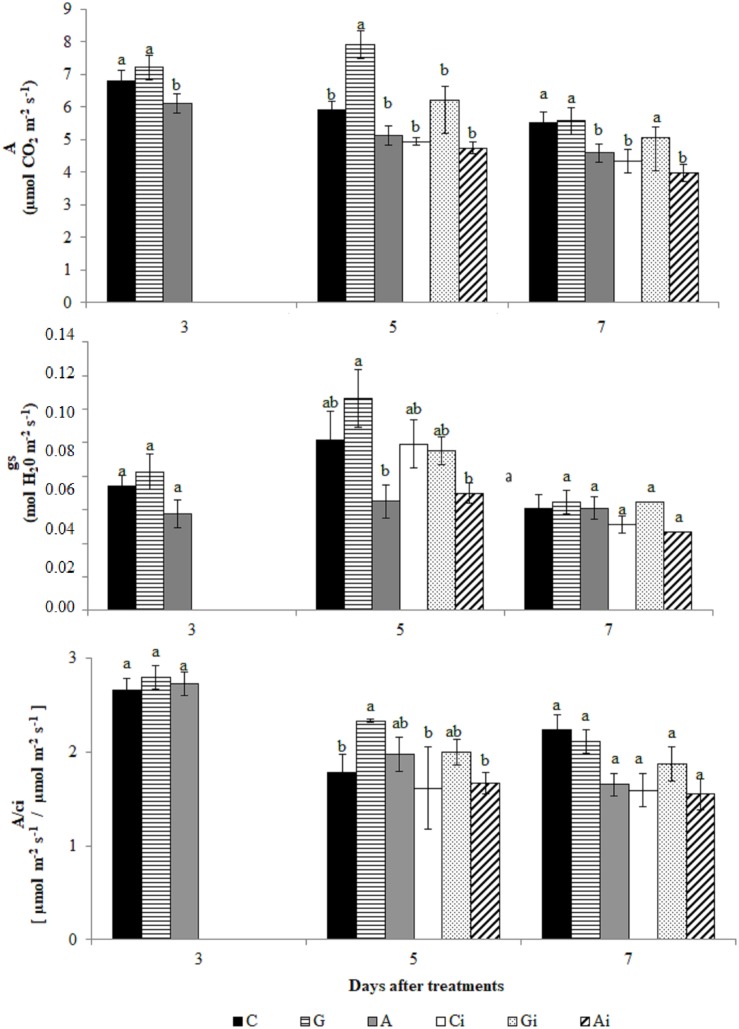
The physiological alterations observed in the coffee leaves due to the PRI treatments and H. vastatrix inoculation, were also studied. Greenforce CuCa treated leaves exhibited an increase in the photosynthetic rate (A) and stomatal conductance (gs) (at 5dat) when compared to ASM treatments (Figure 10). However, when comparing with Control, Greenforce CuCa showed a higher A/ci ratio. PRI treatments followed by H. vastatrix infection either decreased values or had no effect on the physiological parameters studied (Figure 10 and Supplementary Figure S2). Estimated values of water use efficiency (WUE, estimated as A/transpiration), mesophyll intercellular CO2 concentration (ci) and the ratio of intercellular to ambient CO2 concentrations (ci/ca) did not change significantly between treatments/inoculation (Supplementary Figure S2).
FIGURE 10.
Evaluation of photosynthesis rate (A), stomatal conductance (gs) and A/ci in coffee leaves treated with the plant resistance inducers (PRIs) Greenforce CuCa (G), ASM (A), and control (C) and inoculated with H. vastatrix (i). Inoculation occurred 3 days after treatment with PRIs. The means from the treatments, at each evaluation time, followed by different letters were significantly different (p ≤ 0.05) according to Tukey test. Bars represent the standard error.
Biochemical Analyses
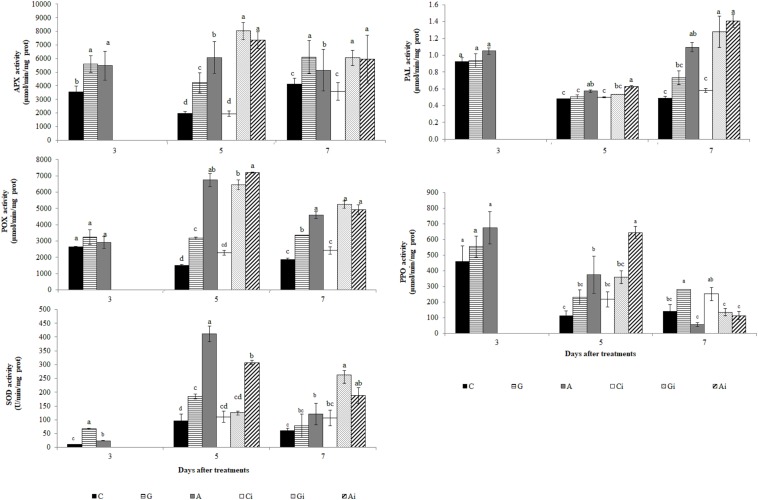
At the biochemical level the PRI treatments increased the activity of all the enzymes studied (APX, POX, SOD, PPO, and PAL) when comparing to a mock-treated control (Figure 11). An increase in H2O2 accumulation was only observed with the Greenforce CuCa treatment, at 5dat/2dai (Supplementary Figure S3). Upon H. vastatrix infection of PRI treated leaves the same pattern of response for all enzymes was observed (relatively to infected control, Ci) except for PPO, whose activity decreased. However, no significant differences were observed between the infected and non-infected controls (Ci and C). The highest values for the PAL activity were obtained at 7dat/4dai, contrary to what was observed for all the other enzymes, whose activity increase at 5dat/2dai.
FIGURE 11.
Evaluation of APX, POX, SOD, PAL and PPO activities in coffee leaves treated with the PRIs Greenforce CuCa (G), ASM (A), and control (C) and inoculated with H. vastatrix (i). Inoculation occurred 3 days after treatment with PRIs. The means from the treatments, at each evaluation time, followed by different letters were significantly different (p ≤ 0.05) according to Tukey test. Bars represent the standard error.
Discussion
Coffee Leaf Proteome
When analyzing the whole coffee leaf proteome the proteins mostly represented were annotated as related to energy production and carbon fixation (“photosynthesis”) followed by “protein metabolism” (essentially protein synthesis) and “stress” response, similarly to what was found in the leaf proteomes of several other plants (Supplementary Table S5). These results contrast with those of the coffee leaf apoplast proteome previously studied (Guerra-Guimarães et al., 2014, 2015) which evidence protein degradation as the main biological process. It is notorious that among the proteins identified in the whole coffee leaf proteome, circa 9% were annotated as extracellular proteins, namely chitinase-like and cupin-like proteins that had already been referred for the coffee leaf apoplast proteome (Guerra-Guimarães et al., 2014, 2015).
Effects of the PRI Treatments and H. vastatrix Infection
As far as we know, this is the first report on the proteomic analysis of the effect of PRIs on the leaves of a woody plant (coffee) before and after infection with a biotrophic pathogen (H. vastatrix). The PRIs ASM and Greenforce CuCa had been shown to reduce the incidence of CLR in the fields (Costa et al., 2014; Silva et al., 2019). We now confirmed their protective effect under controlled conditions, Greenforce CuCa showing a higher protective effect than ASM (Supplementary Table S1A). When analyzing the molecular mechanisms involved in such induced resistance, we verified that both PRIs modulate the primary metabolism (photosynthesis) and protein degradation, although through different proteins (Figure 9 and Graphical Abstract). This is in line with results of other authors, either in coffee (De Nardi et al., 2006) or in other plants (Barilli et al., 2012; Arasimowicz-Jelonek et al., 2013; Anup et al., 2015; Balmer et al., 2015). PRIs cause minimal changes in primary metabolism switching the plant defense mechanism to a standby state, which according to these authors is important in the defense against pathogens. We observed that at early stages of the H. vastatrix infection fewer alterations were induced in the leaf proteome of Greenforce CuCa treated leaves than by ASM. On the other hand, Greenforce CuCa also showed a better physiological performance than ASM, as expressed by an increase of about 20% in stomatal conductance (gs) and in the photosynthetic rate (A). These results might suggest that the differences in the carbon assimilation metabolism may have implications in the defense mechanisms induced by the two PRIs against H. vastatrix.
Our proteomic data did not show major changes in proteins of redox and secondary metabolism, but we did observed an increase in the activity of several related enzymes namely, APX, POX, SOD, PPO, and PAL. The integration of proteomic and biochemical data, point out for an additional level of regulation since different activities could be related with post-translation regulation of proteins and not with changes in protein abundance (Yin et al., 2019). Furthermore, 2-DE capacity to distinguish small changes of low abundant proteins is limited, which make harder the correlation of both data sets. The increases in activity of APX, POX, SOD, PPO, and PAL due to the PRIs treatments and to infection can be related with the defense responses. It is known that the redox enzymes (e.g., APX, POX, and SOD) have a role in eliminating excessive ROS caused by pathogen infection, while the increases in PPO and PAL activities might point out for modifications in secondary metabolism, eventually leading to lignin formation. In fact, increase in POX, SOD and PAL activities, accumulation of phenolics and cell wall modification/lignification at the infection sites were previously observed during the coffee resistance to H. vastatrix (Silva et al., 2002, 2008; De Nardi et al., 2006; Guerra-Guimarães et al., 2009, 2015; Leitão et al., 2011).
Relevance of Proteins Differentially Affected by PRI Treatments
It is notorious the lack of similarity in the protein profiles induced in the coffee leaves by Greenforce CuCa and ASM, what suggests the modulation of different metabolic pathways by these PRIs. The exceptions were glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and sedoheptulose-1,7-bisphosphatase (SBPase). GAPDH respond to both PRIs in a time-dependent way, while putative SBPase isoforms were distinctly induced.
The annotation of GAPDH indicated its localization in the chloroplast, with a probable participation in the reductive pentose phosphate pathway (Calvin-Benson cycle). It is known that GAPDH is a component of the triose phosphate/pentose phosphate pool, important in several biosynthetic pathways that require carbon skeletons (Lim et al., 2013). It has also been considered that this enzyme has a role in defense responses, namely, as a regulator of ROS accumulation and cell death in Arabidopsis infected with Pseudomonas syringae (Henry et al., 2015). GAPDH respond positively to pathogen infection and to PRIs treatments (McGregor et al., 2009; Mutuku and Nose, 2012; Lemaître-Guillier et al., 2017), indicating a possible involvement of this enzyme in the resistance induction in coffee. SBPase has been classified as a key enzyme in photosynthetic carbon fixation and growth (Lefebvre et al., 2005; Nilo-Poyanco et al., 2013). It was shown to be positively related with resistance of peanuts to Phaeoisariopsis personata (Kumar and Kirti, 2015) and to be decreased in susceptible mulberry when infected with mulberry dwarf phytoplasma (Ji et al., 2009). SBPase seems to be also important during coffee resistance induction, but further studies are needed in order to reveal the role of the distinct putative isoforms detected.
One group of the proteins specifically affected by the ASM treatment were the stress-related proteins HSP70 probably involved in housekeeping folding metabolism. Indeed, it was described in leaves of V. vinifera the increase in abundance of HSPs proteins due to a PRI treatment (Lemaître-Guillier et al., 2017). HSP70 proteins are ATP-dependent chaperones considered to maintain protein homeostasis by their involvement in: the proper folding of nascent synthesized proteins, the prevention of protein aggregation, the translocation of proteins across membranes, and the protein targeting to degradation (Sarkar et al., 2013). As a consequence of their important role in protein homeostasis they also participate in plant immunity through the quality control of the pattern recognition receptors (PRRs) (Park and Seo, 2015). These PRRs are essential in the detection of pathogen molecules (PAMPs-pathogen associated molecular patterns), as for instance, in the resistance of Nicotiana tabacum against Ralstonia solanacearum (Maimbo et al., 2007). Chitinase-like protein and an auxin-binding protein ABP20-like (germin-like protein) were also induced by ASM. Chitinases (PR-3, 4, 8, 11) are well-characterized glycosyl hydrolases (GH18) with potential to degrade PAMPs, limiting pathogen growth and functioning as signals for resistance responses (Silva et al., 2006; Guerra-Guimarães et al., 2009, 2015). The germin-like protein family is a considerably heterogeneous group exhibiting three different enzymatic activities, oxalate oxidase, ADP-glucose pyrophosphatase or phosphodiesterase and superoxide dismutase (SOD) (Dunwell et al., 2008). The induction of the germin-like proteins may contribute to the significantly higher SOD activity detected due to the ASM treatment. The germin-like proteins can have a role in the oxidative crosslinking of cell wall proteins around the site of infection (Bradley et al., 1992; Silva et al., 2008). Crosslink between phenolic compounds, plant cell wall polysaccharides and proteins enhance the protection of the cell wall to digestion by microbial degrading enzymes and, thus, increase the global resistance to fungi (Bily et al., 2003). The apoplastic localization of these proteins, in combination with the H2O2 generating SOD activity, offers a role in cell-wall fortification (Rietz et al., 2012). Indeed, the modulation of apoplastic chitinases, germin-like proteins and SOD during the early stages of C. arabica-H. vastatrix interactions have been reported (Guerra-Guimarães et al., 2015).
Greenforce CuCa treatment consistently induces changes in oxygen-evolving enhancer proteins (OEE1 and OEE2), RuBisCO activase and RuBisCO small subunit, which might improve the coffee response to rust. Potato treated with different PRIs, also shown relevant variations in OEE2 protein abundance (Arasimowicz-Jelonek et al., 2013). It was proposed that OEE1 and OEE2 may have a role in defense (Heide et al., 2004; Coppola et al., 2013). In Arabidopsis OEE1 and OEE2 exhibited properties of thioredoxins, which are positive regulators of plant-induced defense responses (Tada et al., 2008). The involvement of OEEs in ROS detoxification as a response to biotic stress (Heide et al., 2004; Coppola et al., 2013) is in line with our findings. These results, together with the increased activity of APX, POX, SOD and H2O2 production, strongly suggests a role of the Greenforce CuCa treatment in the redox homeostasis during the H. vastatrix infection.
The RuBisCO activase and the RuBisCO small subunit, regulators of RuBisCO activity and stability (Buchanan et al., 2015), were described as involved in the resistance of grapevine to Plasmopora viticola (Lemaître-Guillier et al., 2017). Differences in both RuBisCO activase and RuBisCO small subunit had been used to distinguish between two genetically close inbred tomato lines with opposite responses to TYLCV virus infection (one resistant and one susceptible) (Moshe et al., 2012). Moshe et al. (2012) further highlighted the decrease in abundance of ATP-dependent zinc metalloproteases (FtsH, Filamentation Temperature-Sensitive protein H) in susceptible inbred tomato lines. The effect of Greenforce CuCa on protein degradation was evidenced by the increase in abundance of FtsH, proteases known to be involved in different biological processes namely: thylakoid membrane organization, proteolysis and ROS metabolism (Kato et al., 2009; Tibiletti et al., 2016). In addition, it was also suggested that these proteins might act as chaperones (Adam and Clarke, 2002). Greenforce CuCa treatment is outstanding in inducing the increase in abundance of the above referred proteins what, together with an increase in the photosynthetic rate, is important for the coffee plant to deal with CLR.
Conclusion
We used a proteomic approach in order to shed light on the effect of the PRIs, Greenforce CuCa and ASM, on the coffee leaf metabolism. The aim was to improve knowledge on the resistance mechanisms induced by the PRIs against CLR. Proteomic adjustments mainly related to photosynthesis, protein metabolism and stress responses were shown. However, the proteins affected by Greenforce CuCa were different from those affected by ASM. It was further observed that Greenforce CuCa reinforces the redox homeostasis of the leaf, while ASM seems to affect preferentially the secondary metabolism and the stress-related proteins. A link between the both PRI treatments, the primary metabolism and the defense responses was also evidenced. While both PRIs prepare the plant to resist CLR, they induce distinct defense mechanisms. The identification of components of the plant primary metabolism (essential for plant growth and development) that, simultaneously, participate in the plant defense responses (e.g., SAR) could open new perspectives for a plant breeding program.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016012.
Author Contributions
MR and LG-G conceived and designed the research and were involved in funding acquisition. KP and JS conducted all experiments, and collected the biological material. KP and RT conducted the 2-DE electrophoresis and image analysis. SP and JR carried out the MS-based spot identification. CP, IC, and LG-G conducted the statistical analyses and functional annotation of the identified proteins. JS and MC extracted and analyzed all the physiological parameters. JS and AM extracted and analyzed the data from the biochemical assays. CP, CR, and LG-G wrote the manuscript. All authors have read and approved the final version of the manuscript.
Conflict of Interest
The formulation “Greenforce CuCa” is patent pending. There is no commercial interest regarding this patent since the purpose of INCT coffee (the intellectual property owner) is to make it freely available to coffee growers. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Deila Botelho (Pos-Doc, UFLA) for the statistical analysis assistance during the revision of the manuscript.
Funding. This work was supported by the Portuguese Funds through Fundação para a Ciência e a Tecnologia, under the project PTDC/AGR-GPL/109990/2009, LEAF/UID/AGR/04129/2019, PEst-OE/EQB/LA0004/2011, and by Brazilian Funds through CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), INCT-Café (Instituto Nacional de Ciência e Tecnologia do Café), and FAPEMIG (Fundação de Amparo à Pesquisa no Estado de Minas Gerais). RT acknowledges COST FA1306 for granting funds for a “Short Term Scientific Mission.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00309/full#supplementary-material
References
- Adam Z., Clarke C. K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7 451–456. 10.1016/S1360-1385(02)02326-9 [DOI] [PubMed] [Google Scholar]
- Anup C. P., Melvin P., Shilpa N., Gandhi M. N., Jadhav M., Ali H., et al. (2015). Proteomic analysis of elicitation of downy mildew disease resistance in pearl millet by seed priming with b-aminobutyric acid and Pseudomonas fluorescens. J. Proteomics 120 58–74. 10.1016/j.jprot.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Arasimowicz-Jelonek M., Kosmala A., Janus L., Abramowski D., Floryszak-Wieczorek J. (2013). The proteome response of potato leaves to priming agents and S-nitrosoglutathione. Plant Sci. 198 83–90. 10.1016/j.plantsci.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Avelino J., Cristancho M., Georgiou S., Imbach P., Aguilar L., Bornemann G., et al. (2015). The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Sec. 7 303–321. 10.1007/s12571-015-0446-9 [DOI] [Google Scholar]
- Balmer A., Pastor V., Gamir J., Flors V., Mauch-Mani B. (2015). The ‘prime-ome’: towards a holistic approach to priming. Trends Plant Sci. 20 443–452. 10.1016/j.tplants.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Barguil B. M., Resende M. L. V., Resende R. S., Beserra Júnior J. E. A., Salgado S. M. L. (2005). Effect of extracts from citric biomass, rusted coffee leaves and coffee berry husks on Phoma costarricensis of coffee plants. Fitopal. Bras. 30 535–537. 10.1590/s0100-41582005000500014 [DOI] [Google Scholar]
- Barilli E., Rubiales D., Castillejo M. A. (2012). Comparative proteomic analysis of BTH and BABA-induced resistance in pea (Pisum sativum) toward infection with pea rust (Uromyces pisi). J. Proteomics 75 5189–5205. 10.1016/j.jprot.2012.06.033 [DOI] [PubMed] [Google Scholar]
- Bettencourt A. J., Rodrigues C. J., Jr. (1988). “Principles and practice of coffee breeding for resistance to rust and other diseases,” in Coffee Agronomy, Vol. 4 eds Clarke R. J., Macrae R. (New York, NY: Elsevier Applied Science Publishers LTD; ), 199–234. [Google Scholar]
- Biemelt S., Keetman U., Albrecht G. (1998). Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiol. 116 651–658. 10.1104/pp.116.2.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bily A. C., Reid L. M., Taylor J. H., Johnston D., Malouin C., Burt A. J., et al. (2003). Dehydrodimers of ferulic acid in maize grain pericarp and aleurone: Resistance factors to Fusarium graminearum. Phytopathology 93 712–719. 10.1094/PHYTO.2003.93.6.712 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Bradley D. J., Kjellbom P., Lamb C. J. (1992). Elicitor and wound-induced oxidative cross linking of a plant cell wall proline-rich protein: a novel rapid defense response. Cell 70 21–30. 10.1016/0092-8674(92)90530-P [DOI] [PubMed] [Google Scholar]
- Buchanan B. B., Gruissem W., Jones R. L. (2015). Biochemistry and Molecular Biology of Plants, 2nd Edn Hoboken, NJ: John Wiley & Sons Ltd, 1280. [Google Scholar]
- Buege J. A., Aust S. D. (1978). Microsomal lipid peroxidation. Methods Enzymol. 52 302–310. 10.1016/S0076-6879(78)52032-6 [DOI] [PubMed] [Google Scholar]
- Capucho A. S., Zambolim L., Duarte H. S., Vaz G. R. (2011). Development and validation of a standard area diagram set to estimate severity of leaf rust in Coffea arabica and C. canephora. Plant Pathol. 60 1144–1150. 10.1111/j.1365-3059.2011.02472.x [DOI] [Google Scholar]
- Cavalcanti F. R., Resende M. L. V., Lima J. P. M. S., Silveira J. A. G., Oliveira J. T. A. (2006). Activities of antioxidant enzymes and photosynthetic responses in tomato pre-treated by plant activators and inoculated by Xanthomonas vesicatoria. Physiol. Mol. Plant Pathol. 68 198–208. 10.1016/j.pmpp.2006.11.001 [DOI] [Google Scholar]
- Chance B., Maehly A. C. (1955). Assay of catalase and peroxidase. Methods Enzymol. 2 764–775. 10.1016/S0076-6879(55)02300-8 [DOI] [Google Scholar]
- CONAB (2019). Acompanhamento da Safra Brasileira de Café, v. 5 – Safra 2019, n. 4 - Quarto levantamento - Dezembro 2019. Brasília: Companhia Nacional de Abastecimento, 1–44. [Google Scholar]
- Conesa A., Götz S. (2008). Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008:619832. 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J. M., Langenbach C. J. G., Jaskiewicz M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53 97–119. 10.1146/annurev-phyto-080614-120132 [DOI] [PubMed] [Google Scholar]
- Coppola V., Coppola M., Rocco M., Digilio M. C., D’Ambrosio C., Renzone G., et al. (2013). Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genomics 14:515. 10.1186/1471-2164-14-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa B. H. G., Resende M. L. V., Ribeiro Júnior P. M., Mathioni S. M., Pádua M. A., Silva Júnior M. B. (2014). Suppression of rust and brown eye spot diseases on coffee by phosphites and by-products of coffee and citrus industries. J. Phytopathol. 162 635–642. 10.1111/jph.12237 [DOI] [Google Scholar]
- Damerval C., Vienne D., Zivy M., Thiellement H. (1986). Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7 52–54. 10.1002/elps.1150070108 [DOI] [Google Scholar]
- De Nardi B., Dreos R., Del Terra L., Martellossi C., Asquini E., Tornincasa P., et al. (2006). Differential responses of Coffea arabica L. leaves and roots to chemically induced systemic acquired resistance. Genome 49 1594–1605. 10.1139/g06-125 [DOI] [PubMed] [Google Scholar]
- Deutsch E. W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., et al. (2017). The proteomexchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 54 D1100–D1106. 10.1093/nar/gkw936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell J. M., Gibbings J. G., Mahmood T., Saqlan Naqvi S. M. (2008). Germin and germin-like proteins: evolution. Struct. Funct. Plant Sci. 27 342–375. 10.1080/07352680802333938 [DOI] [Google Scholar]
- Esquivel P., Jiménez V. M. (2012). Functional properties of coffee and coffee by-products. Food Res. Int. 46 488–495. 10.1016/j.foodres.2011.05.028 [DOI] [Google Scholar]
- Fernandes L. H. M., Resende M. L. V., Pereira R. B., Costa B. H. G., Monteiro A. C. A., Junior P. M. R. (2013). Acibenzolar-S-metil in rust and blotch control in field conditions. Coffee Sci. 8 21–29. 10.25186/cs.v8i1.309 [DOI] [Google Scholar]
- Furtado L. M., Rodrigues A. A. C., Araújo V. S., Silva L. L. S., Catarino A. M. (2010). Utilização de Ecolife§e Acibenzolar–S-metil (ASM) no controle da Antracnose da banana em pós-colheita. Summa Phytopathol. 36 237–239. 10.1590/S0100-54052010000300009 [DOI] [Google Scholar]
- Giannopolitis N., Ries S. K. (1977). Superoxide dismutase. I. Occurrence in higher plants. Plant Physiol. 59 309–314. 10.1104/pp.59.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Ann. Rev. Phytopathol. 43 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Göhre V. (2015). Immune responses: photosynthetic defense. Nat. Plants 1:15079. 10.1038/nplants.2015.79 [DOI] [PubMed] [Google Scholar]
- Goldberg T., Hecht M., Hamp T., Karl T., Yachdav G., Ahmed N., et al. (2014). LocTree3 prediction of localization. Nuclic Acids Res. 42 350–355. 10.1093/nar/gku396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozzo F., Faoro F. (2013). Systemic acquired resistance (50 years after discovery): moving from the lab to the field. J. Agric. Food Chem. 61 12473–12491. 10.1021/jf404156x [DOI] [PubMed] [Google Scholar]
- Grove H., Jorgensen B. M., Jessen F., Sondergaard I., Jacobsen S., Hollung K., et al. (2008). Combination of statistical approaches for analysis of 2-DE data gives complementary results. J. Proteome Res. 7 5119–5124. 10.1021/pr800424c [DOI] [PubMed] [Google Scholar]
- Guerra-Guimarães L., Cardoso S., Martins I., Loureiro A., Bernardes A. S., Varzea V., et al. (2009). “Differential induction of superoxide dismutase in Coffea arabica–Hemileia vastatrix interactions,” in Proceedings of the 22th International Conference on Coffee Science (ASIC2008), Campinas. [Google Scholar]
- Guerra-Guimarães L., Tenente R., Pinheiro C., Chaves I., Silva M. C., Cardoso F. M. H., et al. (2015). Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 6:478. 10.3389/fpls.2015.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Guimarães L., Vieira A., Chaves I., Pinheiro C., Queiroz V., Renaut J., et al. (2014). Effect of greenhouse conditions on the leaf apoplastic proteome of Coffea arabica plants. J. Proteomics 104 128–139. 10.1016/j.jprot.2014.03.024 [DOI] [PubMed] [Google Scholar]
- Guzzo S. D., Harakava R., Tsai S. M. (2009). Identification of coffee genes expressed during systemic acquired resistance and incompatible interaction with Hemileia vastatrix. J. Phytopathol. 157 625–638. 10.1111/j.1439-0434.2008.01538.x [DOI] [Google Scholar]
- Heide H., Kalisz H. M., Follmann H. (2004). The oxygen evolving enhancer protein 1 (OEE) of photosystem II in green algae exhibits thioredoxin activity. J. Plant Physiol. 161 139–149. 10.1078/0176-1617-01033 [DOI] [PubMed] [Google Scholar]
- Henry E., Fung N., Liu J., Drakakaki G., Coaker G. (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 11:e1005199. 10.1371/journal.pgen.1005199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann J. (2014). The World Atlas of Coffee: From Beans to Brewing – Coffees Explored, Explained and Enjoyed. London: Octopus Publishing Group. [Google Scholar]
- ICO (2020). World Coffee Production. Available online at: http://www.ico.org (accessed January 30, 2020). [Google Scholar]
- Iriti M., Faoro F. (2003). Benzothiadiazole (BTH) Induces Cell-Death Independent Resistance in Phaseolus vulgaris against Uromyces appendiculatus. J. Phytopathol. 151 171–180. 10.1046/j.1439-0434.2003.00700.x [DOI] [Google Scholar]
- Jackson T. J., Burgess T., Colquhoun I., Hardy G. E. S. (2000). Action of the fungicide phosphite on Eucalyptus marginata inoculated with Phytophthora cinnamomi. Plant Pathol. 49 147–154. 10.1046/j.1365-3059.2000.00422.x [DOI] [Google Scholar]
- Ji X., Gai Y., Zheng C., Mu Z. (2009). Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.). Proteomics 9 5328–5339. 10.1002/pmic.200900012 [DOI] [PubMed] [Google Scholar]
- Kar M., Mishra D. (1976). Catalase, peroxidase, and polyphenolase activities during rice leaf senescence. Plant Physiol. 57 315–319. 10.1104/pp.57.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151 1790–1801. 10.1104/pp.109.146589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kirti P. B. (2015). Transcriptomic and proteomic analyses of resistant host responses in Arachis diogoi challenged with late leaf spot pathogen, Phaeoisariopsis personata. PLoS One 10:e0117559. 10.1371/journal.pone.0117559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läderach P., Ramirez-Villegas J., Navarro-Racines C., Zelaya C., Martinez-Valle A., Jarvis A. (2017). Climate change adaptation of coffee production in space and time. Clim. Change 141 47–62. 10.1007/s10584-016-1788-9 [DOI] [Google Scholar]
- Lefebvre S., Lawson T., Zakhleniuk O. V., Lloyd J. C., Raines C. A. (2005). Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 138 451–460. 10.1104/pp.104.055046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão S., Guerra-Guimarães L., Bronze M. R., Vilas-Boas L., Sá M., Almeida M. H., et al. (2011). “Chlorogenic acid content in coffee leaves: possible role in coffee leaf rust resistance,” in Proceedings of the 24th International Conference on Coffee Science (ASIC2010), Bali. [Google Scholar]
- Lemaître-Guillier C., Hovasse A., Schaeffer-Reiss C., Recorbet G., Poinssot B., Trouvelot S., et al. (2017). Proteomics towards the understanding of elicitor induced resistance of grapevine against downy mildew. J. Proteomics 156 113–125. 10.1016/j.jprot.2017.01.016 [DOI] [PubMed] [Google Scholar]
- Lim S., Borza T., Peters R. D., Coffin R. H., Al-Mughrabi K. I., Pinto D. M., et al. (2013). Proteomics analysis suggests broad functional changes in potato leaves triggered by phosphites and a complex indirect mode of action against Phytophthora infestans. J. Proteomics 93 207–223. 10.1016/j.jprot.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Lohse M., Nagel A., Herter T., May P., Schroda M., Zrenner R., et al. (2014). Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37 1250–1258. 10.1111/pce.12231 [DOI] [PubMed] [Google Scholar]
- Maimbo M., Ohnishi K., Hikichi Y., Yoshioka H., Kiba A. (2007). Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 145 1588–1599. 10.1104/pp.107.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor C. E., Miano D. W., LaBonte D. R., Hoy M., Clark C. A., Rosa G. J. M. (2009). Differential gene expression of resistant and susceptible sweetpotato plants after infection with the causal agents of sweet potato virus disease. J. Am. Soc. Hort. Sci. 134 658–666. 10.21273/JASHS.134.6.658 [DOI] [Google Scholar]
- Medeiros F. C. L., Resende M. L. V., Medeiros F. H. V., Zhang H. M., Paré P. W. (2009). Defense gene expression induced by a coffee-leaf extract formulation in tomato. Physiol. Mol. Plant Pathol. 74 175–183. 10.1016/j.pmpp.2009.11.004 [DOI] [Google Scholar]
- Moshe A., Pfannstiel J., Brotman Y., Kolot M., Sobol I., Henryk C., et al. (2012). Stress responses to tomato yellow leaf curl virus (TYLCV) infection of resistant and susceptible tomato plants are different. Metabolomics 1:006 10.4172/2153-0769.S1-006 [DOI] [Google Scholar]
- Murthy P. S., Naidu M. M. (2012). Sustainable management of coffee industry by-products and value addition—A review. Resour. Conservat. Recycl. 66 45–58. 10.1016/j.resconrec.2012.06.005 [DOI] [Google Scholar]
- Mutuku J. M., Nose A. (2012). Changes in the contents of metabolites and enzyme activities in rice plants responding to Rhizoctonia solani kuhn infection: activation of glycolysis and connection to phenylpropanoid pathway. Plant Cell Physiol. 53 1017–1032. 10.1093/pcp/pcs047 [DOI] [PubMed] [Google Scholar]
- Nakano Y., Asada K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22 867–880. 10.1093/oxfordjournals.pcp.a076232 [DOI] [Google Scholar]
- Neuhoff V., Stamm R., Eibl H. (1985). Clear background and highly sensitive protein staining with coomassie blue dyes in polyacrylamide gels – a systematic analysis. Electrophoresis 6 427–448. 10.1002/elps.1150060905 [DOI] [Google Scholar]
- Nilo-Poyanco R., Olivares D., Orellana A., Hinrichsen P., Pinto M. (2013). Proteomic analysis of grapevine (Vitis vinifera L.) leaf changes induced by transition to autotrophy and exposure to high light irradiance. J. Proteomics 91 309–330. 10.1016/j.jprot.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Park C. J., Seo Y. S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31 323–333. 10.5423/PPJ.RW.08.2015.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagli L. S. (1999). Quantifying protein in 2-D PAGE solubilization buffers. 2-D proteome analysis protocols. Methods Mol. Biol. 112 99–103. [DOI] [PubMed] [Google Scholar]
- Resende M. L. V., Nojosa G. B. A., Cavalcanti L. S., Aguilar M. A. G., Silva L. H. C. P., Perez J. O., et al. (2002). Induction of resistance in cocoa against Crinipellis perniciosa and Verticillium dahliae by acibenzolar-S-methyl (ASM). Plant Pathol. 51 621–628. 10.1046/j.1365-3059.2002.00754.x [DOI] [Google Scholar]
- Rietz S., Bernsdorff F. E. M., Cai D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63 5507–5519. 10.1093/jxb/ers203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C., Senthil-Kumar M., TzinVered M. K. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 10:17. 10.3389/fpls.2014.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar N. K., Kundnani P., Grover A. (2013). Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaper. 18 427–437. 10.1007/s12192-012-0395-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner G., Finney R. F. (1977). The effect of nitrogen fertilization on the expression of slow−mildewing resistance in knox wheat. Phytopathology 67 1051–1056. 10.1094/Phyto-67-105 [DOI] [Google Scholar]
- Silva J. A. G., Resende M. L. V., Monteiro A. C. A., Pádua M. A., Guerra-Guimarães L., Medeiros F. L., et al. (2019). Resistance inducers applied alone or in association with fungicide for the management of leaf rust and brown eye spot of coffee under field conditions. J. Phytopathol. 167 430–439. 10.1111/jph.12814 [DOI] [Google Scholar]
- Silva M. C., Guerra-Guimarães L., Loureiro A., Nicole M. (2008). Involvement of peroxidases in the hypersensitive reaction of coffee (Coffea arabica) plants to orange rust (Hemileia vastatrix). Physiol. Mol. Plant Pathol. 72 29–38. 10.1016/j.pmpp.2008.04.004 [DOI] [Google Scholar]
- Silva M. C., Nicole M., Guerra-Guimarães L., Rodrigues C. J., Jr. (2002). Hypersensitive cell death and post-haustorial defense responses arrest the orange rust (Hemileia vastatrix) growth in resistant coffee leaves. Physiol. Mol. Plant Pathol. 60 169–183. 10.1006/pmpp.2002.0389 [DOI] [Google Scholar]
- Silva M. C., Nicole M., Rijo L., Geiger J. P., Rodrigues C. J., Jr. (1999). Cytochemistry of plant-rust fungus interface during the compatible interaction Coffea arabica (cv. Caturra) – Hemileia vastatrix (race III). Int. J. Plant Sci. 160 79–91. 10.1086/314113 [DOI] [Google Scholar]
- Silva M. C., Várzea V., Guerra-Guimarães L., Azinheira H. G., Fernandez D., Petitot A.-S., et al. (2006). Coffee resistance to the main diseases: leaf rust and coffee berry disease. Brazilian J. Plant Physiol. 18 119–147. 10.1590/S1677-04202006000100010 22611856 [DOI] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956. 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhinhas P., Batista D., Diniz I., Vieira A., Silva D. N., Loureiro A., et al. (2017). The coffee leaf rust pathogen Hemileia vastatrix: one and a half centuries around the tropics. Mol. Plant Pathol. 18 1039–1051. 10.1111/mpp.12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibiletti T., Hernández-Prieto M. A., Semeniuk T. A., Funk C. (2016). “Assembly and degradation of pigment-binding proteins,” in Chloroplasts – Current Research and Future Trends, ed. Kirchhoff H. (Norfolk: Caister Academic Press; ), 25–57. [Google Scholar]
- Urbanek H., Kuzniak-Gebarowska E., Herka K. (1991). Elicitation of defense responses in bean leaves by Botrytis cinerea polygalacturonase. Acta Physiol. Plant. 13 43–50. [Google Scholar]
- Várzea V. M. P., Marques D. V. (2005). “Population variability of Hemileia vastatrix vs coffee durable resistance,” in Proceedings of Durable Resistance to Coffee Leaf Rust, eds Zambolim L., Zambolim E., Várzea V. M. P. (Viçosa: Universidade Federal de Viçosa; ). [Google Scholar]
- Velikova V., Yordanov I., Edreva A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci. 151 59–66. 10.1016/S0168-9452(99)00197-1 [DOI] [Google Scholar]
- Yin J., Yi H., Chen X., Wang J. (2019). Post-translational modifications of proteins have versatile roles in regulating plant immune responses. Int. J. Mol. Sci. 20:2807. 10.3390/ijms20112807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambolim L. (2016). Current status and management of coffee leaf rust in Brazil. Trop. Plant Pathol. 41 1–8. 10.1007/s40858-016-0065-9 [DOI] [Google Scholar]
- Zucker M. (1965). Induction of phenylalanine deaminase by light and its relation to chlorogenic acid synthesis in potato tuber tissue. Plant Physiol. 40 779–784. 10.1104/pp.40.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016012.