Abstract
Introduction
Acetaminophen toxicity has been associated with elevation of microRNAs. The present study was to evaluate overall microRNA profiles and previously identified microRNAs to differentiate acetaminophen (APAP) toxicity from other causes of transaminase elevation.
Methods
This was an observational study of adults with presumed acetaminophen toxicity at presentation. Serum samples were collected every 12 hours during hospitalization. Total miRNAs were extracted from plasma and levels of 327 microRNAs were quantified using real-time polymerase chain reaction. A standard measure of miRNA expression (delta-delta cycle threshold) was calculated for each microRNAs. A two-level cluster analysis was performed using a random k-means algorithm. Demographic and clinical characteristics of each cluster were compared using ANOVA, Wilcoxon rank sum, Kruskal-Wallis, and chi-square tests. Performance of specific miRNAs of interest was also evaluated.
Results
Twenty-seven subjects were enrolled (21 with a final diagnosis of acetaminophen toxicity), and a total of 61 samples were analyzed. Five clusters were identified, two of which demonstrated clear clinical patterns and included specific elevated miRNAs previously reported to be elevated in APAP toxicity patients. Features associated with clusters 1 and 5 included confirmed acetaminophen toxicity, high peak alanine aminotransferase, and late presentation. Clusters 2–4 contained lower peak microRNAs, lower peak alanine aminotransferase, and heterogeneous clinical characteristics.
Conclusions
Severe cases of acetaminophen toxicity showed two distinct patterns of microRNA elevation which were similar to previous work, while less severe cases were difficult to distinguish from non-acetaminophen-associated cases. Further work is needed to incorporate microRNA profiles into the diagnostic algorithm of acetaminophen toxicity.
Electronic supplementary material
The online version of this article (10.1007/s13181-019-00739-6) contains supplementary material, which is available to authorized users.
Keywords: Acetaminophen, Drug-induced liver injury, MicroRNAs, Biomarkers
Background
Acetaminophen (APAP) toxicity is a leading cause of drug-induced liver injury, accounting for more than 70,000 emergency department visits annually in the USA [1]. Approximately 570 cases with a major adverse outcome or fatality were reported to the national poison data system in 2016 [2]. Current diagnostic standards are based on a history of ingestion, which is often unreliable, and a rise in serum transaminases due to hepatocellular injury, which is typically delayed by 24–72 hours from the time of ingestion. There is an urgent unmet need for earlier biomarkers of injury to improve both diagnostic sensitivity and specificity and to identify patients who are appropriate candidates for treatment with the antidote, N-acetylcysteine (NAC). An ideal assay would reduce costs by decreasing both unnecessary hospitalizations and cases of delayed diagnosis and treatment. Proposed candidates for early biomarkers of acetaminophen hepatotoxicity include proteins, cytokines, messenger RNAs, and microRNAs (miRNAs) [3].
First described in 1993 [4], miRNAs are short, noncoding RNA sequences which influence a wide array of physiologic processes via effects on gene regulation and protein synthesis [5, 6]. To date, more than 2000 human miRNAs have been identified [7], and this list is growing. MicroRNAs are known to be stable in circulation and, unlike protein biomarkers, are not susceptible to post-translational modifications [6]. MicroRNAs are ordinarily intracellular and often tissue-specific. They are released upon cell stress, making extracellular miRNA levels an excellent marker for tissue injury. Serum and tissue profiles of these biomarkers are currently under investigation for detection and treatment of multiple cancer subtypes, neurologic diseases, and cardiovascular diseases [8, 9].
MicroRNA profiles have tremendous potential in the diagnosis and mechanistic exploration of acetaminophen hepatotoxicity. Significant changes in circulating miRNA profiles of various biologic matrices (e.g., blood, urine) have been demonstrated in mouse and rat models of acetaminophen toxicity [10–12]. In humans, key miRNAs have been notably upregulated in urine [13], hepatocytes [14, 15], and blood [15–20]. Human studies have largely focused on liver-associated miRNAs, specifically miR-122, which has repeatedly been found to be elevated in acetaminophen hepatotoxicity [21, 22]. More recent studies have focused on expanded panels of miRNAs as markers, facilitators, and inhibitors of acetaminophen-induced hepatotoxicity [17, 23].
In a preliminary work from this laboratory [20], samples from 42 acetaminophen toxic patients and seven patients with ischemic hepatitis were analyzed for sequential miRNA profiles. These data showed that circulating miRNAs rise before other serum biomarkers, can distinguish acetaminophen-induced hepatotoxicity from ischemic hepatitis, and may be predictive of response to treatment. The objective of the present study was to evaluate the previously identified candidate panel of miRNAs in a clinically diverse sample of subjects with suspected acetaminophen toxicity. The objectives of this study were to explore whether these markers can differentiate acetaminophen toxicity from other causes of hepatic injury, and to identify miRNA profile patterns associated with various clinical phenotypes.
Methods
Setting
This protocol was reviewed and approved by the University of Massachusetts Medical School Institutional Review Board. A convenience sample of subjects was recruited from a large, tertiary care emergency department with greater than 130,000 patient visits per year. Patients whose clinical care included consultation to the medical toxicology service for suspected acetaminophen toxicity during business hours when study staff were available to enroll were screened for participation. Inclusion criteria were as follows: (1) 18 years of age or greater; (2) known or suspected acetaminophen poisoning based on a clinical history compatible with the diagnosis; and (3) either detectable serum acetaminophen concentration (> 10 mcg/mL) or elevated alanine aminotransferase (ALT, > 40 U/L) at presentation. Individuals were excluded if they were pregnant and/or unable to provide informed consent.
Data Acquisition and Management
Demographic data, laboratory values, and clinical data were obtained from the electronic medical record. All study data were collected and managed using REDCap (Research Electronic Data Capture), a secure, web-based application designed to support data capture for research studies [24].
Variable Definitions
Laboratory values (including liver enzyme profiles, serum chemistry profiles, coagulation profiles, blood gas analyses, and acetaminophen concentrations) were considered as continuous variables. Peak ALT was considered as a continuous variable, and a categorical ALT variable was created for clustering purposes, using previously defined criteria [20]: low ALT < 50 IU/L, medium ALT 51–1000 IU/L, and high ALT > 1000 IU/L.
Length of NAC treatment and hospitalization were analyzed as continuous clinical variables, while need for transplant, survival to hospital discharge, concomitant ingestions, history of liver disease, heavy alcohol use (daily use or > 10 drinks per week), and illicit drug use were analyzed as dichotomous variables. Type of APAP ingestion was classified as acute (occurred over a period of < 1 hour, with defined time of ingestion), chronic repeated doses (occurred over a period of > 1 hour), or chronic with unknown timing. Time to presentation was defined as time from reported ingestion to hospital presentation and was assigned a binary classifier of early (≤ 12 hours) or late (> 12 hours). Ingestions with unknown timing were classified as late. Dichotomous variables were created for a variety of other clinical and historical categories of interest. Upon hospital discharge, laboratory and clinical data (including clinical course and history provided by patient and/or collateral sources) from each subject’s encounter were reviewed by a board-certified medical toxicologist (SC). Each case was assigned a binary classifier based on the final diagnosis of APAP toxicity (APAP toxicity or non-APAP toxicity).
Sample Acquisition and Storage
Whole blood was collected in an ethylenediaminetetraacetic acid (EDTA) tube upon hospital presentation and every 12 hours thereafter until the serum acetaminophen concentration was undetectable and ALT returned to baseline. Tubes were immediately placed on ice and sent to a central lab where plasma was isolated by centrifugation at 1500×g for 20 minutes at 4 °C. Extracted serum was stored at − 80 °C within 60 minutes of the original draw and remained frozen until microRNA analysis was performed.
Sample Preparation and Quantitative Real-Time PCR
Samples were prepared and processed according to previously published methods [20]. Briefly, after thawing, plasma was spun to remove debris and deplete platelets, mixed with 100 μL of lysis buffer, and digested with Proteinase K at 65 °C while shaking for 15 minutes. After cooling to room temperature, 250 μL of phenol:chloroform 5:1 (pH 8.0) and 250 μL of nuclease-free water were added. This mixture was shaken for 5 minutes at room temperature and spun at 16,000× for 5 minutes, and the aqueous layer (∼ 450–500 μL) was removed. Three volumes of 100% ethanol were added. The RNA was recovered using an Enzymax Tini spin column, dissolved in nuclease-free water, and stored at − 80 °C. Preamplified complementary DNA (cDNA) prepared from the RNA equivalent of 7.5 μL of plasma was assayed using the miScript 372 miRNA PCR Array for Human Serum and Plasma (QUIAGEN, Germantown, MD). QIAGEN reagents were used according to the manufacturer’s instructions in the reverse transcription, preamplification, and quantitative real-time PCR (qRT-PCR). Reverse transcription control (miRTC) primer assay and positive PCR control (PPC) were used to evaluate as a measure of RNA sample purity and qRT-PCR performance. Quantitative real-time PCR reactions were run on Viia 7 (ThermoFisher).
Sample Size
Our sample size was estimated by employing false discovery rate methods to correct for the number of statistical comparisons conducted. Based upon previously described methods for microarray sample size calculations [25], we estimated that 25 samples (with an average of 4 time point samples per subject) were required to maintain a false discovery rate of 5% to detect a 2-fold change in expression with 80% power assuming that the rate of differentially expressed miRNAs is less than < 10% and that miRNA expression levels may not be independent (i.e., are correlated).
Statistical Analysis
Standard measures of miRNA expression cycle threshold (CT) and delta-delta cycle threshold (ΔΔCT or ddCT) were used. Analysis of the miRNA data set was performed as previously described [20] except as follows: Samples with miRTC CT > 25 were excluded from analysis, the normalization set included those miRNAs with the lowest score calculated by mean times standard deviation of CT values, and a set of 149 miRNAs with ddCT ≥ 3 in at least three subjects was used for further analysis. The ten miRNAs with the lowest scores were selected in order to obtain a normalization set that is both present at a relatively high concentration (more likely to be accurately measured) and relatively invariable across samples. Peak point (defined as the sample from each subject that has the most miRNAs at a level at least eightfold higher than the normal control mean) clustering was performed by calculating the total number of miRNAs with ddCT ≥ 3 in each sample and selecting from each subject the sample with the most miRNAs with ddCT ≥ 3. Two-step clustering was performed using unsupervised machine learning with a random k-means algorithm. Clustering was first performed on all subjects; then, a second cluster analysis was performed on the subset of subjects classified as APAP toxicity. Receiver operator characteristic (ROC) curve analysis was conducted using the ROCR package in R to evaluate the sum of ddCTs of the seven APAP-associated microRNAs as a metric to distinguish clinical phenotypes [26].
Demographic and clinical characteristics were compared between APAP and non-APAP group and by clusters. Continuous normally distributed variables were compared using ANOVA, and continuous skew variables were compared using the Wilcoxon rank sum (2 groups) or Kruskal-Wallis t (> 2 groups) test, and categorical variables were compared using Pearson’s chi-square. Analysis was performed in STATA (Version 15.1, College Station, TX).
Results
Sample Characteristics
Subject characteristics are summarized in Table 1. Twenty-seven subjects were enrolled over a 27-month period with a mean age of 37.9 years (SD = 17.5). Females comprised 63% of the sample (N = 17). Sixty-three percent (N = 17) had a detectable APAP concentration on presentation, and 26% (N = 7) were considered early presenters. Seventy-eight percent (N = 21) were categorized as APAP toxicity; the remaining 22% (N = 6) were categorized as non-APAP toxicity at the time of discharge. There was one death (a non-APAP subject) and no subjects required a liver transplant.
Table 1.
Sample characteristics, by final diagnosis.
| APAP toxicity | Non-APAP toxicity | Total sample | ||
|---|---|---|---|---|
| N | 21 (78%) | 6 (22%) | 27 | |
| Subject characteristics | ||||
| Age in years, mean (SD) | 36.0 (16.6) | 44.6 (20.8) | 37.9 (17.5) | |
| Female sex | 12 (57%) | 5 (83%) | 17 (63%) | |
| History of liver disease | 4 (19%) | 0 (0%) | 4 (15%) | |
| History of heavy EtOH use | 4 (19%) | 3 (50%) | 7 (26%) | |
| History of illicit drug use | 10 (48%) | 1 (17%) | 11 (41%) | |
| History of IV drug use | 4 (19%) | 0 (0%) | 4 (15%) | |
| Presentation characteristics | ||||
| Time to presentation | Early (< 12 hours) | 4 (19%) | 3 (50%) | 7 (26%) |
| Late (> 12 hours) | 17 (81%) | 3 (50%) | 20 (74%) | |
| Type of ingestion | Acute | 7 (33%) | 2 (33%) | 9 (33%) |
| Chronic, repeated doses | 9 (43%) | 1 (17%) | 10 (37%) | |
| Chronic, time unknown | 5 (24%) | 3 (50%) | 8 (30%) | |
| Detectable APAP concentration at presentation | 13 (62%) | 4 (67%) | 17 (63%) | |
| Concomitant anticholinergic ingestion | 4 (19%) | 2 (33%) | 6 (22%) | |
| Concomitant EtOH ingestion | 4 (19%) | 1 (17%) | 5 (19%) | |
| Concomitant opioid ingestion | 2 (10%) | 2 (33%) | 4 (15%) | |
| Outcome characteristics | ||||
| Duration of NAC therapy in hours, mean (SD) | 56.9 (33.5) | 31.3 (36.3) | 51.2 (35.1) | |
| Length of hospitalization in days, mean (SD) | 5.6 (4.0) | 4.8(3.9) | 5.4 (3.9) | |
| Liver transplant requirement | 0 | 0 | 0 | |
| Death | 0 | 1 | 3% | |
| Group | Non-APAP | 0 (0%) | 6 (100%) | 6 (22%) |
| APAP-low ALT | 6 (29%) | 0 (0%) | 6 (22%) | |
| APAP-med ALT | 3 (14%) | 0 (0%) | 3 (11%) | |
| APAP-high ALT | 12 (57%) | 0 (0%) | 12 (44%) | |
Normalization Set
The normalization set used for this study included the following: miR-486-5p, miR-92a-3p, miR-126-5p, miR-1280, miR-16-5p, miR-25-3p, miR-126-3p, miR-451a, miR-21-5p, and miR-125a-5p obtained from the plasma of healthy control subjects.
Cluster Analysis A—All Subjects
Two-level k-means clustering of the peak samples were initially performed on all 27 subjects, with the best balance of separation and number of subjects in each cluster at k = 5 (maximum number of iterations = 2000; number of different random start seeds = 10). Given that heterogeneity in timing of initial sample procurement relative to the reported APAP ingestion event confounded comparison of initial samples among subjects, peak samples were used for clustering purpose to provide more consistent comparisons.
The resulting five clusters (clusters A1–A5) contained between three and eight subjects each (Table 2, Fig. 1, Supplemental Fig. 1). These clusters were described based on their miRNA profiles, corresponding ALT and APAP concentrations, and diagnostic data. Two clusters (clusters A1 and A5) demonstrated the most clinical homogeneity, while clusters A2, A3, and A4 were more heterogeneous.
Table 2.
All subjects, characteristics by cluster.
| Factor | Level | Cluster A1 | Cluster A2 | Cluster A3 | Cluster A4 | Cluster A5 | p |
|---|---|---|---|---|---|---|---|
| N | 8 | 6 | 3 | 4 | 6 | ||
| Participant characteristics | |||||||
| Age in years, mean (SD) | 34.0 (13.6) | 53.8 (23.5) | 28 (14.7) | 29.5 (10.7) | 37.8 (13.8) | 0.11 | |
| Sex, female | 5 (62%) | 3 (50%) | 2 (67%) | 3 (75%) | 4 (67%) | 0.95 | |
| Duration of NAC in hours, mean (SD) | 55.6 (36.4) | 48 (35.7) | 27.7 (20.6) | 29.5 (22.4) | 74.8 (37.3) | 0.22 | |
| Days hospitalized, mean (SD) | 6.8 (6.1) | 5 (3.8) | 4.3 (2.3) | 4 (1.4) | 5.7 (2.0) | 0.80 | |
| Peak sample characteristics | |||||||
| Time from admission to peak sample in hours, mean (SD) | 50.7 (46.7) | 44.2 (36.9) | 22 (14.8) | 14.6 (16.2) | 16.7 (17.3) | 0.25 | |
| Peak occurred after NAC treatment complete | 3 (38%) | 1 (17%) | 0 (0%) | 0 (0%) | 1 (17%) | 0.47 | |
| Serum APAP concentration at peak, mean (SD) | 0 (0) | 10.3 (25.3) | 8.3 (14.4) | 15 (13.2) | 4.33 (10.6) | 0.49 | |
| Serum creatinine in mg/dL at peak, mean (SD) | 1.9 (2.3) | 1.7 (0.7) | 0.77 (0.1) | 0.66 (0.3) | 0.7 (0.2) | 0.45 | |
| Serum bicarbonate in mEq/L at peak, mean (SD) | 23.3 (4.9) | 19.3 (5) | 26 (0.1) | 26 (0) | 24 (5.4) | 0.59 | |
| Serum ALT in IU/L at peak, mean (SD) | 683 (964) | 359 (692) | 135 (182) | 317 (565) | 2107 (1742) | 0.04 | |
| INR at peak, mean (SD) | 1.2 (0.1) | 1.4 (0.1) | 1 (0.1) | 1.2 (0.1) | 1.4 (.4) | 0.71 | |
| Ingestion history characteristics | |||||||
| Concomitant anticholinergic use | 1 (12%) | 0 (0%) | 2 (67%) | 2 (50%) | 1 (17%) | 0.11 | |
| Concomitant EtOH | 1 (12%) | 1 (17%) | 1 (33%) | 0 (0%) | 2 (33%) | 0.66 | |
| Concomitant opioid use | 0 (0%) | 2 (33%) | 0 (0%) | 2 (50%) | 1 (17%) | 0.42 | |
| Time to initial presentation | Early, < 12 hours | 1 (12%) | 3 (50%) | 0 (0%) | 2 (50%) | 1 (17%) | 0.28 |
| Late, > 12 hours | 7 (88%) | 3 (50%) | 3 (100%) | 2 (50%) | 5 (83%) | ||
| Detectable APAP at presentation | 2 (25%) | 5 (83%) | 2 (67%) | 4 (100%) | 4 (67%) | 0.07 | |
| History of heavy EtOH use | 2 (25%) | 3 (50%) | 0 (0%) | 1 (25%) | 1 (17%) | 0.54 | |
| History of illicit drug use | 4 (50%) | 0 (0%) | 0 (0%) | 3 (75%) | 4 (67%) | 0.04 | |
| History of IV drug use | 2 (29%) | 0 (0%) | 0 (0%) | 1 (25%) | 1 (17%) | 0.58 | |
| History of liver disease | 2 (25%) | 0 (0%) | 0 (0%) | 1 (25%) | 1 (17%) | 0.63 | |
| Type of ingestion | Acute, known time of ingestion | 1 (12%) | 2 (33%) | 1 (33%) | 2 (50%) | 3 (50%) | 0.31 |
| Chronic, repeated doses | 5 (62%) | 3 (50%) | 0 (0%) | 1 (25%) | 1 (17%) | ||
| Chronic, unknown timeline | 2 (25%) | 1 (17%) | 2 (66%) | 1 (25%) | 2 (33%) | ||
| Diagnosis at discharge | Non-APAP toxicity | 1 (12%) | 3 (50%) | 1 (33%) | 1 (25%) | 0 (0%) | 0.65 |
| APAP toxicity | 7 (88%) | 3 (50%) | 2 (67%) | 3 (75%) | 6 (100%) | ||
| Low ALT | 1 (12%) | 1 (17%) | 1 (33%) | 2 (50%) | 1 (17%) | ||
| Med ALT | 1 (12%) | 1 (17%) | 1 (33%) | 0 (0%) | 0 (0%) | ||
| High ALT | 5 (62%) | 1 (17%) | 0 (0%) | 1 (25%) | 5 (83%) | ||
Continuous normally distributed variables were compared using ANOVA. Continuous skew variables were compared using the Kruskal-Wallis test
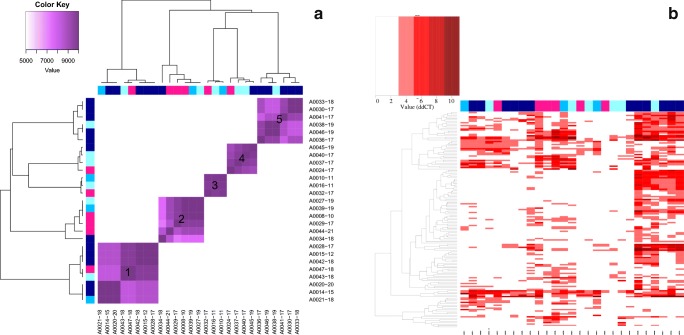
Fig. 1.
MicroRNA results for all subjects. a Cluster analysis A. X- and Y-axes represent individual cases with blue squares indicating APAP toxicity (darker blue indicates higher peak ALT concentration) and pink squares indicating non-APAP subjects. b Heat map. X-axis represents individual case. Y-axis represents individual microRNAs.
Cluster A1 had eight subjects, seven of which were classified as APAP toxicity. The peak samples in this set had a modest number of miRNAs elevated above our established threshold, the maximum being 55 miRNAs. Membership in cluster A1 was significantly associated with undetectable serum APAP on presentation and high ALT. Most subjects were from the APAP toxicity group and were late presenters.
Clusters A2, A3, and A4 included all but one of the of the non-APAP subjects and showed the least homogeneity in clinical characteristics. Cluster A2 consisted of six subjects: three APAP subjects that had a detectable APAP concentration at presentation and three non-APAP subjects. Of the three APAP subjects, only one had an elevated ALT, and this subject also reported significant alcohol abuse. Cluster A3 consisted of three subjects: two APAP subjects and one non-APAP subject. All were late presenters, reported acetaminophen ingestions in self-harm attempts, and had modest peak ALTs (mean 132 IU/L, max 344 IU/L). Cluster A4 consisted of four subjects: a non-APAP subject and three subjects that initially had a detectable APAP concentration but never demonstrated notable miRNA elevations (none having more than 25 miRNAs elevated at any time). Clinically, this group consisted of subjects with reported low-dose ingestions of acetaminophen-containing combination products either co-formulated with diphenhydramine (N = 2) or with an opioid (N = 2). The only subject in this cluster to have elevated transaminases also had chronic hepatitis C and alcohol use disorder.
Cluster A5 had six subjects, five of which were categorized as late presenters and high ALT; all were APAP toxicity subjects. Membership in cluster A5 was significantly associated with high ALT and detectable APAP concentration on presentation (p < 0.05).
Cluster Analysis B—APAP Subjects Only
A second k-means clustering analysis was subsequently performed on only the subjects categorized as APAP toxicity (Fig. 2, Supplemental Table 1), and resulted in three clusters (clusters B1, B2, and B3) with improved consistency with regard to clinical characteristics within each cluster. Even when the number of k-centers is adjusted (for 3, 4, or 5 centers), three k-centers provide the best balance of separation and number of subjects in each cluster when the non-APAP subjects are excluded.
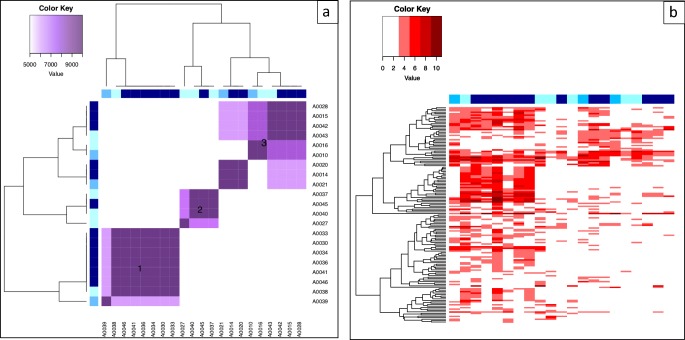
Fig. 2.
MicroRNA results for APAP toxicity subjects. a Cluster analysis B. X- and Y-axes represent individual cases with blue squares indicating APAP toxicity (darker blue indicates higher peak ALT concentration). b Heat map. X-axis represents individual case. Y-axis represents individual microRNAs.
Cluster B1 had eight subjects: all subjects from the original cluster A5, plus two additional subjects (including a subject with APAP toxicity and high ALT from the original cluster A3). Again, characteristics associated with membership in this cluster were high serum ALT at peak and a detectable serum APAP concentration at presentation.
Cluster B2 had four subjects, all from original clusters A2 and A4. Most had low ALT, and interestingly, all four subjects presented with an ingestion of a combination APAP product or a polypharmacy overdose.
Cluster B3 had nine subjects: all but one subject from the original cluster A1 plus two additional subjects from the original cluster A3. Characteristics associated with membership in this cluster were high serum ALT at peak and an undetectable serum APAP at presentation. Although ALT was significantly elevated in this cluster, it was notably lower than cluster B1 (with mean of 641 IU/L and standard deviation of 900 IU/L, compared to 2058 and 1596 respectively).
Notably, in this re-clustering, three subjects (A0021, A0014, and A0020) clustered most closely together, and also demonstrated peak miRNA elevation after NAC treatment was complete, suggesting that there may be a rebound phenomenon where miRNAs increase late in these subject’s post-treatment.
Performance of Specific miRNAs
We evaluated the miRNAs in each cluster demonstrating the largest magnitude increase compared with normalization sets (Supplemental Table 2). Two sets of miRNAs of particular interest based on previously published [20] work from this laboratory were evaluated. The first set (miRNA Set Seven contained seven miRNAs (miR-194-5b, miR-125b-5b, miR-193a-5b, miR-122-5p, miR-21-5p, miR-27b-3p, and miR-1290) that were identified to differentiate APAP toxicity from other forms of liver injury (ischemic hepatitis). The second set (miRNA Set Twelve) contained 12 miRNAs associated with the most severely APAP-poisoned subjects (miR-3646-3p, miR-412, miR-2467-3p, miR-1207-5p, miR-138-1-3p, miR-605, miR-4258, miR-372, miR-4524a-3p, miR-19b-1-5p, miR-122-5p, and miR-483-5p).
Trends in APAP-Associated microRNAs (miRNA Set Seven)
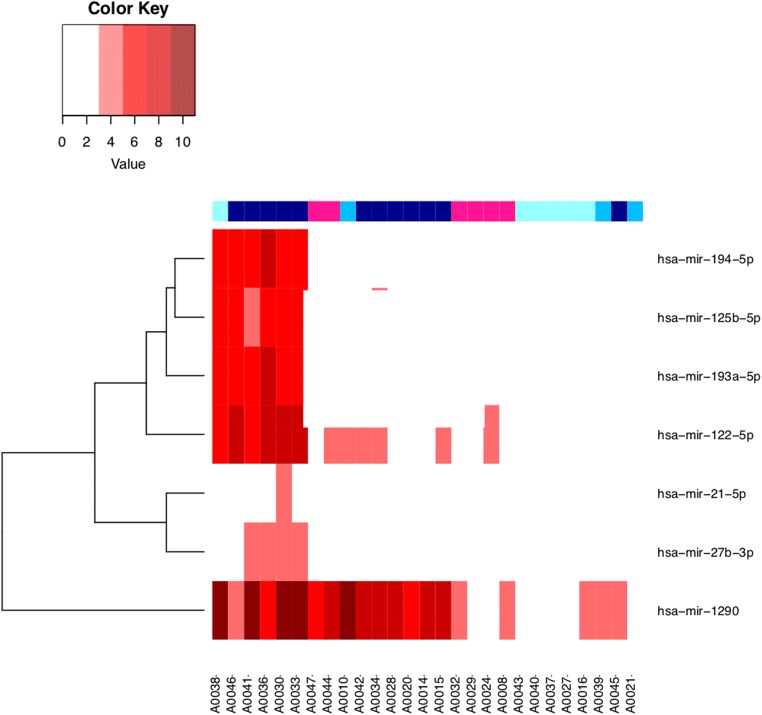
Figure 3 shows a heat map of all subjects based on similarities in expression of miRNA Set Seven. All of the subjects from the original cluster A5 were grouped on the left, having the highest levels of the seven APAP-associated microRNAs. These subjects were also all categorized as APAP toxicity with high ALT (plus one APAP toxicity with low ALT). Five of the seven miRNAs (miR-1290, miR-122-5p, miR-194-5p, miR-193a-5p, and miR-125b-5p) were among the most elevated miRNAs in this cluster (Supplemental Table 2), being elevated more than eightfold above the normal mean. The remaining two miRNAs in this set of seven (miR-21-5p and miR-27b-3p) were both significantly elevated, but with lower magnitude. Notably, only subjects in cluster A5 had five or more elevated miRNAs from miRNA Set Seven. In cluster A1, which contained the majority of the other APAP toxicity subjects, no more than two of the seven miRNAs in miRNA Set Seven were elevated in any sample in this cluster. Only one of the seven (miR-1290) was in the top twenty list (Supplemental Table 2) for cluster A1; however, two others (miR-193a-5b and miR-122-5p) were also significantly elevated. miR-1290 was notably seen in the top 20 of clusters A1, A2, A3, and A5.
Fig. 3.
Heat map of seven APAP-associated microRNAs, all subjects. X-axis represents individual cases with blue squares indicating APAP toxicity (darker blue indicates higher peak ALT concentration) and pink squares indicating non-APAP subjects. Y-axis indicates microRNA of interest.
To evaluate whether these seven APAP-associated miRNAs could be used to distinguish diagnostic categories in the present study, we employed the sum of the ddCTs of those seven microRNAs (ddCT Sum 7) as a measure of their aggregate elevation in a given sample. We tested whether the ddCT Sum 7 could differentiate cases in each category: non-APAP, APAP with low ALT (APAP-low ALT), APAP with medium ALT (APAP-medium ALT), and APAP with high ALT (APAP-high ALT) (Supplemental Figs. 2 and 3). Receiver operating characteristic (ROC) analysis demonstrated that ddCT Sum 7 could differentiate APAP-high ALT subjects from both non-APAP subjects and from all others (including APAP-medium ALT and APAP-low ALT subjects). This analysis indicates that the ddCT Sum 7 could be effective at distinguishing the most severely APAP toxic subjects.
Trends in Severe APAP-Associated miRNAs (miRNA Set Twelve)
Cluster A5 again showed the most consistency with prior data, with five of the 12 miRNAs in miRNA Set Twelve (miR-2467, miR-4524a-3p, miR-122-5p, miR-483-5p, and miR-1207-5p) being among the top 20 elevated miRNAs in this cluster (Supplemental Table 2). Four additional miRNAs in the set (miR-3646-3p, miR-138-1-3p, miR-605, and miR-4258) were significantly elevated, but not among the top 20 in this cluster. Cluster A1 showed elevation of only one of the 12 miRNAs (miR-2467-3p); 3 additional miRNAs (miR-605, miR-122-5p, and miR-483-5p) showed significant elevation but did not rank among the top 20 elevated in this cluster. miR-2467 was seen in the top 20 in clusters A1, A2, A4, and A5.
Discussion
In this clinically diverse population of patients presenting with suspected APAP toxicity, miRNAs in peak samples tended to identify the more severely ill subjects (i.e., those with significant elevations in transaminases), while the less severely APAP-poisoned subjects were more difficult to differentiate from the non-APAP subjects. In the initial clustering analysis, clusters A2–A4 were more difficult to explain, though repeating the clustering analysis without the non-APAP subjects did provide some insight.
Several interesting trends within the clusters raise the possibility that miRNA profiles can differentiate clinical phenotypes, which warrant further exploration. We noted a delayed peak and rebound in miRNA elevations after completion of NAC treatment in a subset of subjects in cluster A1. This was described in prior work as well [20]. Interestingly, all three of these subjects had similar clinical presentations: accidental acetaminophen overdoses in the setting of supratherapeutic ingestions in an attempt to treat unrelated conditions over a period of several days to a week (e.g., viral syndromes, musculoskeletal pain, dental pain). A rebound in key miRNA may indicate incomplete treatment; however, this seems unlikely give that the subjects continued to improve clinically. The miRNAs involved in this rebound may hold clues to the mechanisms by which these biomarkers are involved in the pathophysiology of APAP-induced hepatotoxicity; e.g., they may be involved in reparative or regenerative processes. Further exploration of this phenomenon is warranted.
A second interesting subgroup appeared in the cluster analysis of APAP toxicity only subjects (cluster analysis B). In cluster B2, all subjects (N = 4) presented with a polysubstance overdose or a co-formulated product that contained acetaminophen plus another class of drug (e.g., an antihistamine or opioid). Many co-ingestants are known to change the expected pharmacokinetics (and potentially pharmacodynamics) of acetaminophen toxicity due to alterations in absorption, metabolism, and susceptibility to liver injury. Both of these clinical scenarios (chronic and polysubstance ingestions) challenge our routine management paradigms of acetaminophen toxicity; diagnostic pathways guided by miRNA elevations would provide a useful tool for risk stratification.
In evaluating both initial and peak samples, we found initial samples were less useful than peak samples to produce meaningful clusters of subjects. This may be due to the variation in time of presentation to care, which created significant variation in the time the initial sample was taken with respect to time of reported ingestion. This heterogeneity, although complicating the analytic picture, is a more reflective clinical practice where patients present at various times throughout the course of ingestion. In a previous study, initial samples provided useful information and were successfully used to cluster subjects as APAP toxicity or ischemic hepatitis [20]. However, the previous cohort represented a much more severely ill population and overall may have presented to the hospital later in the course of poisoning. Peak miRNA elevations occurred at various points throughout hospitalization course; however, miRNA trajectories showed that the same miRNAs tend to rise and fall over the same time course as opposed to different groups at different times. A single predictive test at the time of presentation would represent the ideal assay; however, we did not find this in our data. This suggests the best approach may not be a single miRNA assay, but that serial assays to evaluate trajectory may be warranted to predict outcome and/or guide therapy.
Prior literature on human miRNA profiling in acetaminophen toxicity reports a range of miRNA candidates associated with APAP-induced liver injury. Among them, miR-122 is frequently implicated [21, 22]. In our sample, miR-122 was most elevated in cluster A5 (> sevenfold from control) and was also significantly elevated in cluster A1 although to a lesser degree (1.5 fold). miR-122 was not significantly elevated in clusters A2-A4. Other miRNAs associated with APAP-induced liver injury, including miR-194-5p and miR-483-5 [17, 27], were most significantly elevated in cluster A5 (> sixfold), but not the others. Overall, miRNA profiles in cluster A5 (which contained the most severely ill subjects with the highest ALT) were most consistent with prior literature (both from this laboratory and others), with cluster A1 following.
For a successful clinical assay, a consistent normalization set is necessary. The normalization set for the present study has partial overlap with the set used in previous work from this group, with four of the 10 microRNAs from the current study appearing in the prior set [20] (Supplemental Table 3). With future additional similar studies of diverse clinical populations and sites, we anticipate a common normalization set will emerge that could eventually be accepted as a normalization set for a clinical assay.
The goals of this study were to gain deeper understanding of the pathology of APAP toxicity and to identify circulating biomarkers with utility in diagnostic and prognostic clinical assays. A previous study identified a panel of microRNAs dramatically upregulated in the plasma of suspected APAP patients that could distinguish APAP toxicity from ischemic hepatitis [20]. In the present study, we found that many of those same microRNAs were elevated in APAP toxicity subject. Therefore, this study, which utilized a separate patient population at a distinct geographic location, helped confirm the utility of a subset of the previously identified panel of elevated microRNAs. However, this current patient population is more diverse clinically as acetaminophen toxicity is a clinically heterogeneous disease that presents in a variety of phenotypes due a multitude of factors (e.g., delayed of onset of symptoms, frequent coformulation with other xenobiotics). Further study with a larger number of subjects will be required to identify miRNA profiles that correspond to the various clinical phenotypes of APAP toxicity in a way that will enhance clinical decision-making.
This study also corroborates the previous finding that the set of microRNAs dramatically elevated in blood plasma in association with APAP liver toxicity does not simply reflect the set of microRNAs that would be predicted to reflect known microRNA profiles of hepatocyte injury. Further study will be required to determine the basis for this discrepancy, for example, whether certain liver microRNAs are selectively released from hepatocytes in response to APAP toxicity, whether liver-released microRNAs are differentially stable in circulation, and/or whether tissues other than liver may release microRNAs, perhaps as a secondary response to liver pathology.
The present findings move us one step closer to the utilization of miRNAs in the clinical care of acetaminophen-poisoned patients. Recent advances in microarray technology have spurred efforts to develop rapid point-of-care testing for miRNA. Ultimately, a point-of-care miRNA assay, based on the profiles described, has the potential to detect APAP toxicity earlier than current biomarkers, identify patients at risk for morbidity and mortality, and allow for earlier resource mobilization (e.g., transfer to a tertiary care facility or involvement of transplant teams). Conversely, this strategy may also be used to identify those who are unlikely to develop APAP toxicity, providing substantial cost savings by preventing unnecessary treatments and hospitalizations. Finally, miRNA profiles can provide mechanistic data and identify potential novel therapeutic targets.
Limitations
Our sample consisted of a heterogeneous group of all-comers with presumed acetaminophen toxicity, making our results more clinically relevant. However, this was a small convenience sample, and none of the subjects demonstrated the most severe consequences of acetaminophen toxicity (need for transplant or death). In addition, the subjects varied in their time to presentation, and the time to peak sample, which may have impacted the comparisons. Some of the non-APAP subjects had a detectable acetaminophen serum concentration on presentation, but were later determined to be non-APAP toxicity, either after history of ingestion was clarified to be sub-toxic, an alternative etiology for transaminase elevation was discovered, or both. This raises the possibility that APAP may have played a role, however small, in those cases, influencing the results. Similarly, a portion of our sample had concomitant alternative sources of liver injury (alcoholic liver disease, viral hepatitis), the clinical significance of which we cannot determine in this small sample.
Conclusions
The most severe cases of APAP toxicity showed two distinct patterns of miRNA elevation, while less severe cases were more difficult to distinguish from non-APAP subjects. The profiles of circulating microRNAs in APAP hepatotoxicity are partially consistent with prior studies of distinct patient populations. Further study is needed to understand the diversity and dynamics of circulating microRNA profiles associated with APAP toxicity and to determine the clinical utility of miRNAs in the diagnostic algorithm of APAP toxicity.
Electronic Supplementary Material
(DOCX 139 kb)
Sources of Funding
This work was generously sponsored by an investigator-initiated grant from McNeil Consumer Healthcare. Specimen collection, storage, and processing was provided by the University of Massachusetts Medical School/UMass Memorial Healthcare biorepository (National Center for Advancing Translational Sciences/National Institutes of Health grant number UL1TR000161).
Compliance with Ethical Standards
This protocol was reviewed and approved by the University of Massachusetts Medical School Institutional Review Board.
Conflicts of Interest
None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Budnitz DS, Lovegrove MC, Crosby AE. Emergency department visits for overdoses of acetaminophen-containing products. Am J Prev Med. 2011;40(6):585–592. doi: 10.1016/j.amepre.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Gummin DD, Mowry JB, Spyker DA, Brooks DE, Fraser MO, Banner W. 2016 Annual report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 34th annual report. Clin Toxicol (Philadelphia, Pa) 2017;55(10):1072–1252. doi: 10.1080/15563650.2017.1388087. [DOI] [PubMed] [Google Scholar]
- 3.Amacher DE, Schomaker SJ, Aubrecht J. Development of blood biomarkers for drug-induced liver injury: an evaluation of their potential for risk assessment and diagnostics. Mol Diagn Ther. 2013;17(6):343–354. doi: 10.1007/s40291-013-0049-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 5.Ward J, Szabo G, McManus D, Boyer E. Advanced molecular biologic techniques in toxicologic disease. J Med Toxicol. 2011;7(4):288–294. doi: 10.1007/s13181-011-0189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osaki M, Kosaka N, Okada F, Ochiya T. Circulating microRNAs in drug safety assessment for hepatic and cardiovascular toxicity: the latest biomarker frontier? Mol Diagn Ther. 2014;18(2):121–126. doi: 10.1007/s40291-013-0065-0. [DOI] [PubMed] [Google Scholar]
- 7.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. doi: 10.12688/f1000research.2-136.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 10.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology (Baltimore, Md) 2012;56(5):1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward J, Bala S, Petrasek J, Szabo G. Plasma microRNA profiles distinguish lethal injury in acetaminophen toxicity: a research study. World J Gastroenterol. 2012;18(22):2798–2804. doi: 10.3748/wjg.v18.i22.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su YW, Chen X, Jiang ZZ, Wang T, Wang C, Zhang Y, Wen J, Xue M, Zhu D, Zhang Y, Su YJ, Xing TY, Zhang CY, Zhang LY. A panel of serum microRNAs as specific biomarkers for diagnosis of compound- and herb-induced liver injury in rats. PLoS One. 2012;7(5):e37395. doi: 10.1371/journal.pone.0037395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Greenhaw J, Shi Q, Su Z, Qian F, Davis K, Mendrick DL, Salminen WF. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol Sci. 2012;125(2):335–344. doi: 10.1093/toxsci/kfr321. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima T, Hamada Y, Yamada H, Horii I. Changes of micro-RNA expression in rat liver treated by acetaminophen or carbon tetrachloride--regulating role of micro-RNA for RNA expression. J Toxicol Sci. 2007;32(4):401–409. doi: 10.2131/jts.32.401. [DOI] [PubMed] [Google Scholar]
- 15.Yu D, Wu L, Gill P, Tolleson WH, Chen S, Sun J, Knox B, Jin Y, Xiao W, Hong H, Wang Y, Ren Z, Guo L, Mei N, Guo Y, Yang X, Shi L, Chen Y, Zeng L, Dreval K, Tryndyak V, Pogribny I, Fang H, Shi T, McCullough S, Bhattacharyya S, Schnackenberg L, Mattes W, Beger RD, James L, Tong W, Ning B. Multiple microRNAs function as self-protective modules in acetaminophen-induced hepatotoxicity in humans. Arch Toxicol. 2018;92(2):845–858. doi: 10.1007/s00204-017-2090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krauskopf J, de Kok TM, Schomaker SJ, Gosink M, Burt DA, Chandler P, Warner RL, Johnson KJ, Caiment F, Kleinjans JC, Aubrecht J. Serum microRNA signatures as “liquid biopsies” for interrogating hepatotoxic mechanisms and liver pathogenesis in human. PLoS One. 2017;12(5):e0177928. doi: 10.1371/journal.pone.0177928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong A, Cheung B, Nejad C, Gantier M, Graudins A. Hepatotoxicity after paracetamol overdose in a patient with cystic fibrosis despite early acetylcysteine and utility of microRNA to predict hepatotoxicity. Clin Toxicol (Philadelphia, PA) 2018;56(10):904–906. doi: 10.1080/15563650.2018.1454596. [DOI] [PubMed] [Google Scholar]
- 19.Wong A, Nejad C, Gantier M, Choy KW, Doery J, Graudins A. MicroRNA from a 12-h versus 20-h acetylcysteine infusion for paracetamol overdose. Hum Exp Toxicol. 2019;38:646–654. doi: 10.1177/0960327119833740. [DOI] [PubMed] [Google Scholar]
- 20.Ward J, Kanchagar C, Veksler-Lublinsky I, Lee RC, McGill MR, Jaeschke H, et al. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci U S A. 2014;111(33):12169–12174. doi: 10.1073/pnas.1412608111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology (Baltimore, Md) 2011;54(5):1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 22.Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, Bateman DN, Goldring CE, Park BK. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology (Baltimore, Md) 2013;58(2):777–787. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munakata C, Fuchigami Y, Hiroishi S, Haraguchi A, Hagimori M, Enomoto H, Tachiki H, Kodama Y, Sasaki H, Kawakami S. Evaluation of miR-122 to predict high dose acetaminophen-induced liver injury in mice: the combination uses of 5-fluorouracil. Biol Pharm Bull. 2018;41(11):1732–1735. doi: 10.1248/bpb.b18-00504. [DOI] [PubMed] [Google Scholar]
- 24.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Hwang JTG. Quick calculation for sample size while controlling false discovery rate with application to microarray analysis. Bioinformatics (Oxford, England) 2007;23(6):739–746. doi: 10.1093/bioinformatics/btl664. [DOI] [PubMed] [Google Scholar]
- 26.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics (Oxford, England) 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 27.Vliegenthart ADB, Shaffer JM, Clarke JI, Peeters LEJ, Caporali A, Bateman DN, Wood DM, Dargan PI, Craig DG, Moore JK, Thompson AI, Henderson NC, Webb DJ, Sharkey J, Antoine DJ, Park BK, Bailey MA, Lader E, Simpson KJ, Dear JW. Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury. Sci Rep. 2015;5:15501. doi: 10.1038/srep15501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 139 kb)