Expression of genes controlling the virulence activities of a bacterial pathogen is expected to occur preferentially at host sites vulnerable to that pathogen. Host-derived molecules that induce such activities in the plant pathogen Agrobacterium tumefaciens are found in the soil, as well as in the plant. Here, we tested the hypothesis that mechanisms exist to suppress the sensitivity of Agrobacterium species to a virulence gene-inducing molecule by selecting for mutant bacteria that are hypersensitive to its inducing activity. The mutant genes identified encode an efflux pump whose proposed activity increases the concentration of the inducer necessary for vir gene expression; this pump is also involved in antibiotic resistance, demonstrating a relationship between cellular defense activities and the control of virulence in Agrobacterium.
KEYWORDS: Agrobacterium, virulence gene expression, RND efflux pump
ABSTRACT
Expression of the tumor-inducing (Ti) plasmid virulence genes of Agrobacterium tumefaciens is required for the transfer of DNA from the bacterium into plant cells, ultimately resulting in the initiation of plant tumors. The vir genes are induced as a result of exposure to certain phenol derivatives, monosaccharides, and low pH in the extracellular milieu. The soil, as well as wound sites on a plant—the usual site of the virulence activity of this bacterium—can contain these signals, but vir gene expression in the soil would be a wasteful utilization of energy. This suggests that mechanisms may exist to ensure that vir gene expression occurs only at the higher concentrations of inducers typically found at a plant wound site. In a search for transposon-mediated mutations that affect sensitivity for the virulence gene-inducing activity of the phenol, 3,5-dimethoxy-4-hydroxyacetophenone (acetosyringone [AS]), an RND-type efflux pump homologous to the MexE/MexF/OprN pump of Pseudomonas aeruginosa was identified. Phenotypes of mutants carrying an insertion or deletion of pump components included hypersensitivity to the vir-inducing effects of AS, hypervirulence in the tobacco leaf explant virulence assay, and hypersensitivity to the toxic effects of chloramphenicol. Furthermore, the methoxy substituents on the phenol ring of AS appear to be critical for recognition as a pump substrate. These results support the hypothesis that the regulation of virulence gene expression is integrated with cellular activities that elevate the level of plant-derived inducers required for induction so that this occurs preferentially, if not exclusively, in a plant environment.
IMPORTANCE Expression of genes controlling the virulence activities of a bacterial pathogen is expected to occur preferentially at host sites vulnerable to that pathogen. Host-derived molecules that induce such activities in the plant pathogen Agrobacterium tumefaciens are found in the soil, as well as in the plant. Here, we tested the hypothesis that mechanisms exist to suppress the sensitivity of Agrobacterium species to a virulence gene-inducing molecule by selecting for mutant bacteria that are hypersensitive to its inducing activity. The mutant genes identified encode an efflux pump whose proposed activity increases the concentration of the inducer necessary for vir gene expression; this pump is also involved in antibiotic resistance, demonstrating a relationship between cellular defense activities and the control of virulence in Agrobacterium.
INTRODUCTION
Agrobacterium strains thrive in many types of soils around the globe, predominantly as nonpathogenic forms lacking a Ti (tumor-inducing) plasmid (1, 2). Like many such microbes, it has evolved a variety of mechanisms to survive in the highly competitive soil environment, including, for example, a very large number of ABC transporters used to acquire a variety of nutrients (3). Upon acquisition of a Ti plasmid—generally via conjugative transfer from a Ti-containing cell (4)—Agrobacterium gains the capacity to induce tumors at infection sites, generally wound sites, in a broad range of plant hosts (1, 5). This process requires the activation of Ti plasmid virulence (vir) gene expression and the utilization of the vir gene products to process and transfer proteins and a single-stranded DNA (the T-strand) into the host, where the DNA ultimately integrates into the host genome (for reviews, see references 6, to ,9). There, this transferred DNA (the T-DNA) expresses proteins that (i) cause uncontrolled cell proliferation and tumor formation and (ii) catalyze the formation of sugar derivatives—the opines—that can be utilized by the Ti-containing inciting strain as a carbon, nitrogen, phosphate, and/or sulfur source.
The transition to the pathogenic state is tightly controlled (10, 11), as expected given the energetic expense of processing and transferring proteins and DNA into the plant cell (12). Exposure of virulent strains of Agrobacterium to a wide variety of phenol derivatives, certain monosaccharides, and low pH—all found at a plant wound site (13–18)—is required to activate expression of the vir genes (for reviews, see references 6, 19, 20). These ligands accomplish this via the Ti plasmid-encoded VirA and VirG two-component regulatory system, in which the phosphorylated form of the VirA histidine kinase transfers its phosphoryl group to the response regulator VirG. Phospho-VirG can then activate transcription of all of the vir genes on the Ti plasmid, including virA and virG. Recognition of the host-derived ligands by VirA is either direct, in the case of the phenols (21, 22), or indirect, in the case of sugars and low pH (18, 23–25). Critical to the sugar/pH recognition is the activity of the chromosomally encoded periplasmic sugar binding protein ChvE (18, 23, 25). In the presence of certain “inducing” sugars, the ChvE-sugar complex is thought to interact with VirA—likely in a low-pH-dependent manner (24)—to cause VirA to be sensitive to lower concentrations of the essential phenol derivatives than would otherwise be the case (23, 26, 27).
One unexpected finding in the analysis of ChvE was that the KD (equilibrium dissociation constant) for the neutral inducing sugar-ChvE binding is on the order of 0.1 to 1 μM (18, 25), whereas the 50% effective concentration (EC50) for the vir-inducing neutral sugars is significantly higher—on the order of 1 to 3 mM (18, 28). Given that ChvE is encoded on an operon homologous to an ABC transporter involved in neutral sugar utilization (18, 25, 29, 30), vir induction by neutral sugars was examined in strains carrying deletions in the transporters but still expressing wild-type ChvE. These studies found that deletion of the sugar transport capacity—as monitored by a decrease (or loss) of the capacity those strains have to use that particular sugar as a sole carbon source for growth—significantly decreased the EC50 for vir induction by that sugar (18, 31). This integration of the sugar transport activity with the sugar sensitivity for vir induction ensures that (i) at low sugar concentrations, sugar utilization is favored over vir induction and (ii) the vir system is induced only in environments with high sugar concentrations—likely found at a plant wound site—and not in soil, where free sugar is present but at lower concentrations (e.g., 10 to 100 μM [32]) than the EC50 for those sugars as vir inducers in wild-type Agrobacterium.
Given that vir-inducing phenols can be found in low to moderate concentrations in the soil (33–36), a similar mechanism(s) to ensure that high(er) levels of phenol are required for virulence gene expression might be expected. One piece of evidence supporting this concept is that, as described above, in the absence of high levels of inducing sugars, high levels of inducing phenols are required for vir induction. Are there other systems that affect sensitivity of the vir-inducing system for phenols? One expected phenotype of mutants in this case would be hypersensitivity to the vir-inducing activities of phenol derivatives such as 3,5-dimethoxy-4-hydroxyacetophenone (acetosyringone [AS]) (14). Campbell et al. (37) used a selectable marker (neomycin phosphotransferase) under the control of a vir promoter to select for spontaneous kanamycin-resistant mutants at low, normally noninducing levels of AS. The isolated mutants were indeed hypersensitive to AS for vir induction and (i) were not the result of Ti plasmid (and hence virA) genes and (ii) exhibited distinct phenol specificity that suggested some type of ligand/receptor binding interaction. However, the gene(s) responsible for these phenotypes was not identified.
Here, we used a similar selection method (vir promoter driving a selectable marker) but employed transposon mutagenesis so as to be able to identify the gene(s) responsible for the mutant phenotype. This work resulted in the identification of an RND-type efflux pump homologous to the MexE/MexF/OprN pump in Pseudomonas aeruginosa. Mutations in this operon confer on the mutant strains AS hypersensitivity for virulence gene induction, hypersensitivity to the toxic effects of chloramphenicol, and hypervirulence in the tobacco leaf virulence assay. In addition, we demonstrate phenol-specific differences in hypersensitivity for virulence gene expression and in competition with chloramphenicol for the putative efflux pump.
RESULTS
Isolation of mutants hypersensitive to vir-inducing activities of AS.
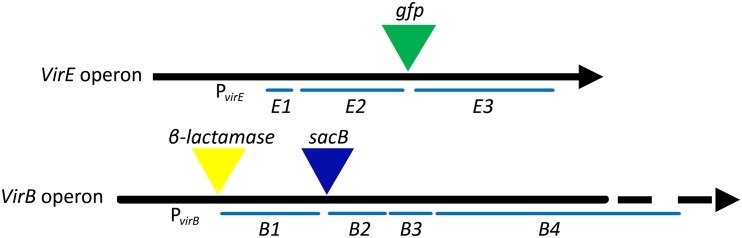
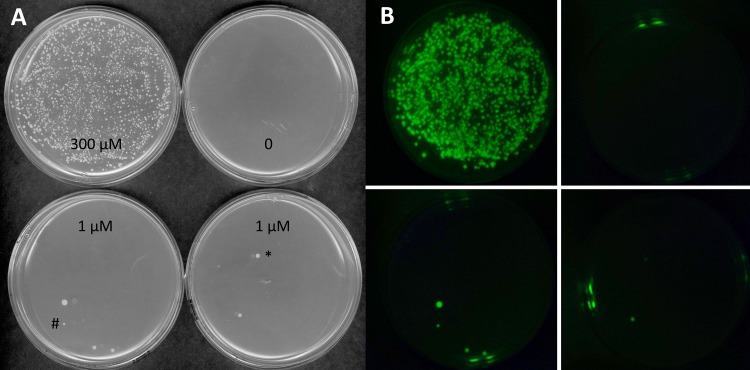
The objective of selecting mutants generated through transposon mutagenesis that are hypersensitive to AS for vir induction required us to construct selectable and screenable markers driven by vir promoters. This was achieved by constructing a version of pTiA6 so that it has β-lactamase (bla) fused to the start codon of virB1, levansucrase (sacB) fused to the start codon of virB2, and green fluorescent protein (GFP; encoded by gfp), with its own start codon, inserted between virE2 and virE3 (Fig. 1; see Materials and Methods for constructions). The β-lactamase provides positive selection (carbenicillin resistance [Carbr]) and levansucrase provides negative selection (sucrose toxicity), both driven by the virB promoter, and GFP expression provides a screenable marker driven by the virE promoter. Only the positive Carbr selection and GFP expression phenotypes were utilized in this study. Demonstration of the vir-controlled levansucrase as a negative selection agent is the subject of other studies in progress. Strain AB3012 carries this modified Ti plasmid in an otherwise wild-type C58 strain and was used as the target for transposon mutagenesis utilizing pRL27 (38). This plasmid cannot replicate in Agrobacterium but carries a transposon that moves a neomycin-phosphotransferase gene (nptII) and an oriR6K origin of replication into target DNA, resulting in kanamycin resistance and the capacity to recover target DNA adjacent to the insertion (38). Preliminary studies indicated that AB3012 cells can survive carbenicillin challenge in AB induction (ABI) medium at 10 μM AS but not at lower concentrations (data not shown). Electrocompetent AB3012 cells were prepared and electroporated with either pBBR1MCS-2 (control [39]) or pRL27 and, after recovery, plated directly onto ABI medium supplemented with carbenicillin, kanamycin, and 0, 1, or 300 μM AS. The cells electroporated with pBBR1MCS-2—which replicates in Agrobacterium and carries kanamycin resistance, but does not transpose—yielded carbenicillin-resistant colonies only at 300 μM AS (data not shown). In contrast, a small number of carbenicillin-resistant colonies grew at 1 μM AS from AB3012 cells electroporated with pRL27 (Fig. 2A). With one exception (indicated by * in Fig. 2A) these colonies also expressed GFP (Fig. 2B), indicating that constitutive expression from the virB promoter was not responsible for the carbenicillin resistance phenotype. A total of nine confirmed mutants that are AS hypersensitive for vir induction were isolated from approximately 100,000 kanamycin-resistant cells (see Materials and Methods).
FIG 1.
Map showing insertion sites of gfp, β-lactamase, and sacB genes in pTiA6 of A. tumefaciens strain AB3012.
FIG 2.
Acetosyringone (AS) sensitivity for induction of carbenicillin resistance of transposon mutants. AB3012 cells electroporated with pRL27 and plated onto AB induction (ABI) medium supplemented with 100 μg/ml carbenicillin and AS, as indicated. Approximately 2,500 cells were plated onto the 300 μM AS plate and 25,000 cells were plated on others. Plates were incubated at 25°C for 5 days (A) Visible light. (B) GFP.
Identification of the MexE/MexF/AmeC efflux pump.
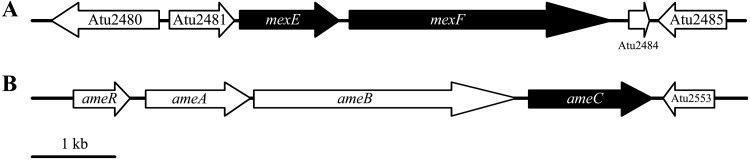
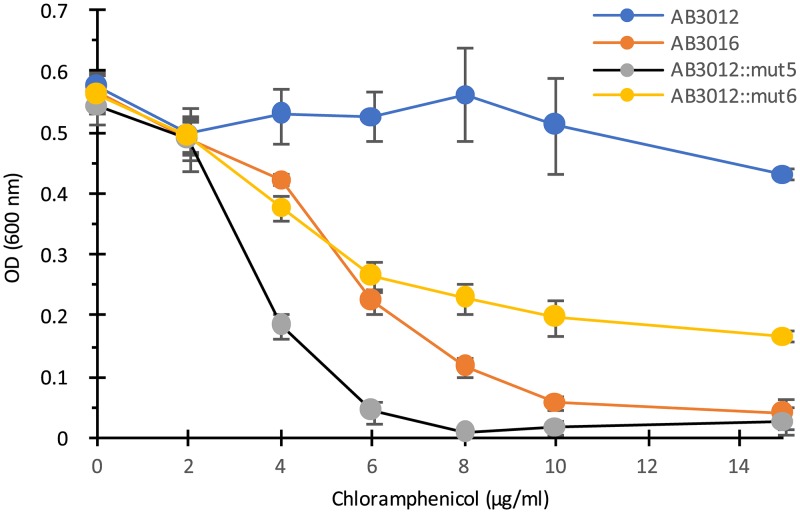
Genomic DNA samples were prepared from the transposon-generated mutants, digested with BfaI or BamHI (which do not cut within the transposon), ligated, and transformed into a PIR2 strain of Escherichia coli which allows for replication of any plasmid carrying the oriR6K origin of replication that is present in the transposed DNA (38). The recovered plasmids were sequenced using primers that read out of the transposon and into the host DNA at the insertion site. Eight out of the nine insertions mapped to an operon homologous to the mexE-mexF-oprN operon of Pseudomonas aeruginosa (40), which encodes an efflux pump of the RND family (Fig. 3; see also Table S1 in the supplemental material). Interestingly, the Agrobacterium operon carrying the homologs to mexE and mexF does not include an oprN homolog. The remaining mutant mapped to the ameC (also sometimes annotated as nodT) gene of C58 (Fig. 3), which is an oprN homolog of another apparent RND efflux operon (ameA, ameB, and ameC) (41). This mutant (indicated by # in Fig. 2A) contrasted with the other eight in that it grew very slowly on agar-solidified medium, although it grew at a similar rate to those of the other mutants and the wild type when tested in liquid medium (see, e.g., Fig. 4). Because the P. aeruginosa efflux pump encoded by mexE-mexF-oprN provides chloramphenicol resistance (42), we tested our mutants for chloramphenicol sensitivity. The results show that insertion mutations in ameC and mexE result in strains that are significantly more sensitive than the wild-type AB3012 strain to chloramphenicol (Fig. 4), consistent with a model in which these are components of an efflux pump. Earlier studies using a similar selection protocol identified spontaneous mutant strains derived from strain A348 that were AS hypersensitive for virulence gene induction but did not assign an underlying mechanism to them (37). These mutants were therefore tested for chloramphenicol sensitivity and were found to be hypersensitive to this antibiotic (see Fig. S2 in the supplemental material), suggesting they may be in the mexE-mexF-ameC-encoded efflux pump as well. Sequencing of the mexE, mexF, and ameC genes in the mutant strains would be required to confirm this.
FIG 3.
Map of mexE-mexF (A) and ameA-ameB-ameC (B) operons.
FIG 4.
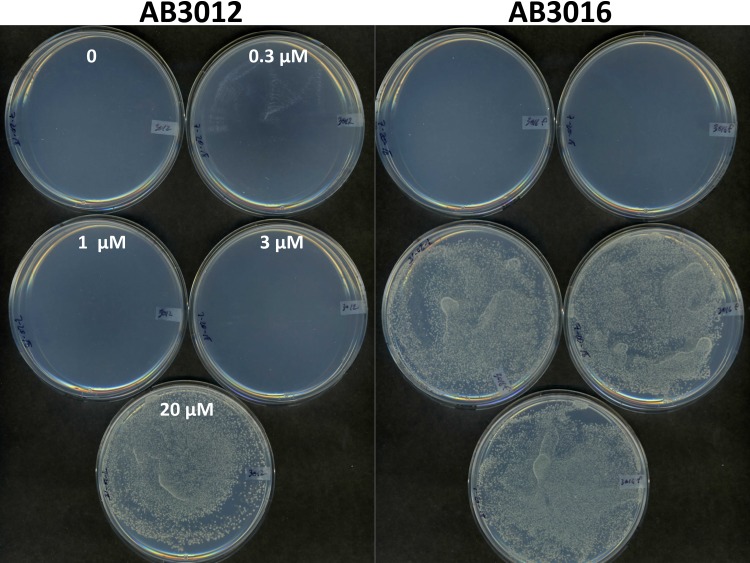
Growth of wild-type strain AB3012, AB3012 mutants 5 and 6 with transposon insertions in ameC and mexE, respectively, and the ΔmexE mutant AB3016 in various doses of chloramphenicol. Optical density at 600 nm (OD600) of cultures was measured after 20 h of incubation at 25°C. Error bars are standard deviations (SD); n = 3.
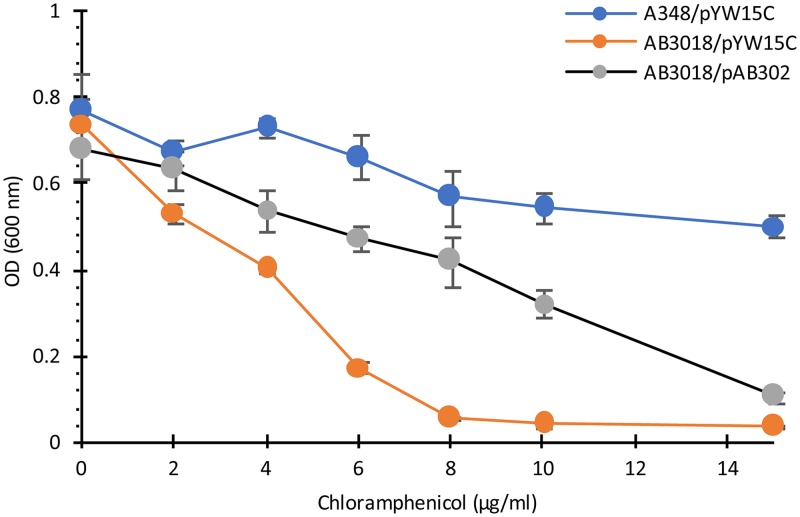
To further characterize this proposed pump, an in-frame deletion of mexE was made in AB3012 (see Materials and Methods). This strain, AB3016, is chloramphenicol hypersensitive (Fig. 4) and is also AS hypersensitive for vir induction, as determined by carbenicillin resistance assay and quantitative analysis of gfp expression (Fig. 5; see also Fig. S1 in the supplemental material). To confirm that deletion of mexE is responsible for the chloramphenicol hypersensitivity, this deletion was made in the wild-type strain A348, which carries wild-type pTiA6 and none of the markers present on the Ti plasmid of AB3012, yielding strain AB3018. This strain exhibited the same chloramphenicol sensitivity as that of AB3016 (Fig. 6). Additionally, pAB302, which carries the mexE open reading frame (ORF) driven by the PN25 promoter of pYW15C (43), was tested for its capacity to complement the mutant phenotype of AB3018. The results show that complementation is observed, namely, AB3018 with pAB302 is significantly more resistant than AB3018 carrying the vector pYW15C to chloramphenicol (Fig. 6). It does not, however, exhibit the same level of resistance as the wild-type strain A348 carrying pYW15C, suggesting that expression of mexE from a promoter different from the promoters driving expression of mexF and ameC may result in stoichiometry that does not yield wild-type levels of pump activity.
FIG 5.
Capacity of wild-type (AB3012) or ΔmexE (AB3016) mutant strains to grow in the presence of 100 μg/ml carbenicillin and various doses of AS, as shown. Approximately 1,000 cells were inoculated per plate and grown at 25°C for 3 days.
FIG 6.
Complementation of ΔmexE strain AB3018. Cells of wild-type strain A348 with pYW15c and strain AB3018 with pYW15c or pAB302 (expressing mexE from the PN25 promoter of pYW15c) were grown in ABI medium supplemented with chloramphenicol as indicated. OD600 of cultures was measured after 20 h of incubation. Error bars are SD; n = 3.
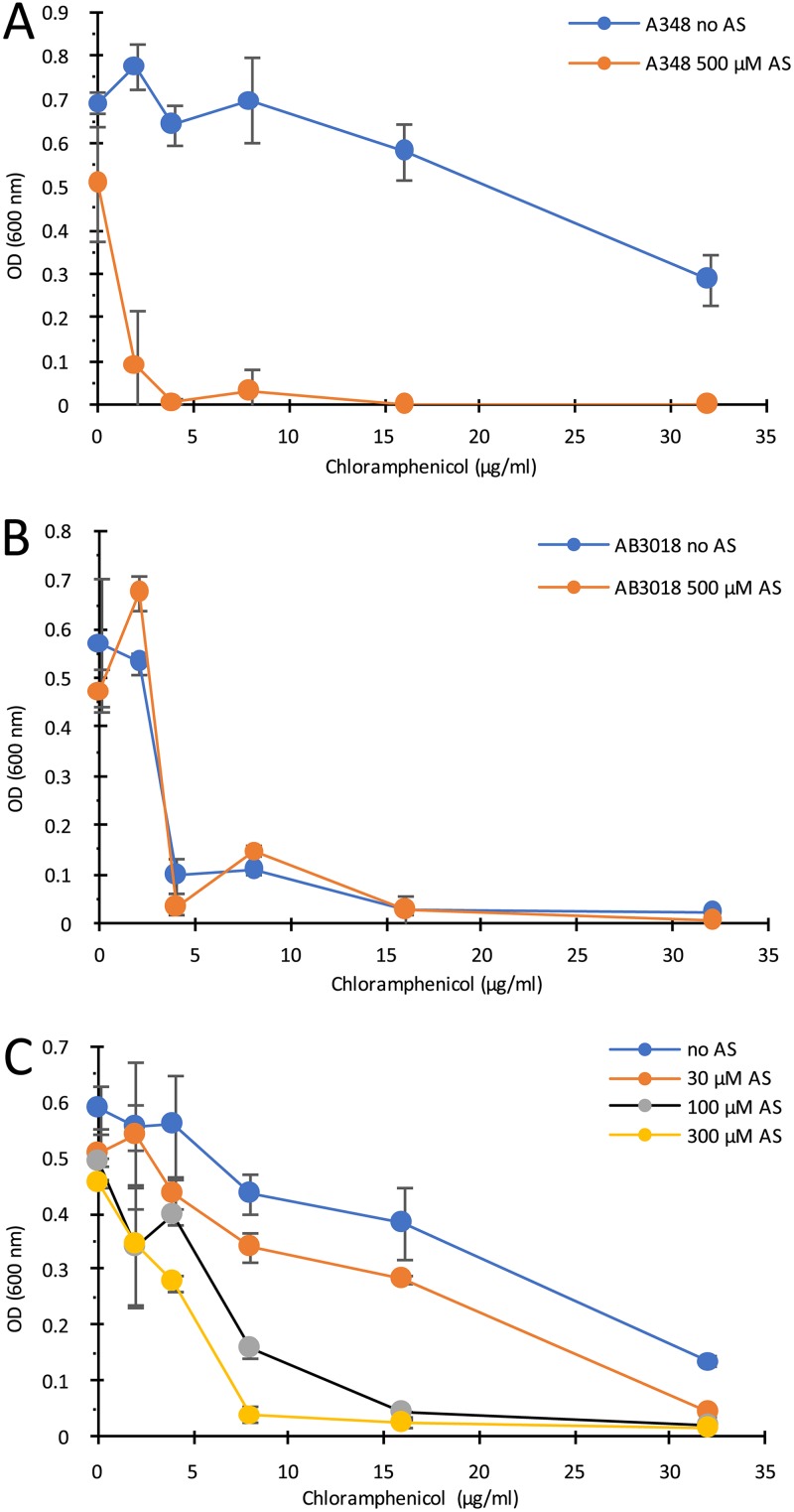
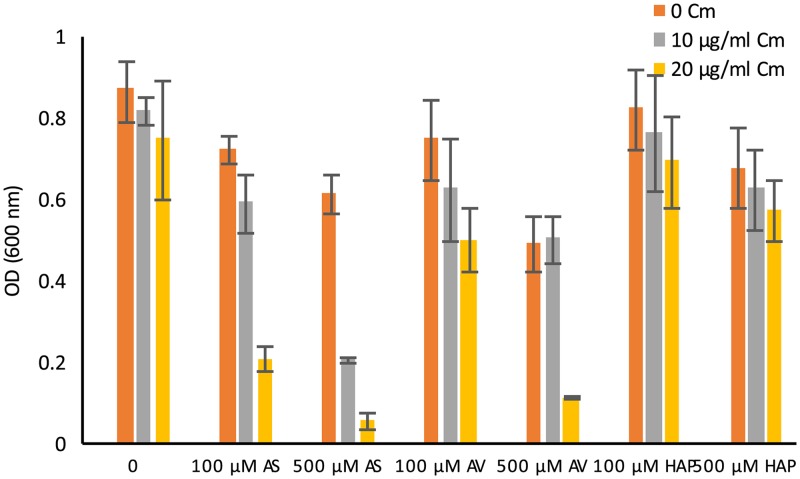
If the putative efflux pump in Agrobacterium can export both AS and chloramphenicol from the cells, then competition at the pump between these substrates might be expected. This was tested via growth assays examining a chloramphenicol dose response in the presence or absence of AS. Wild-type strain A348 was significantly more sensitive to the toxic effects of chloramphenicol in the presence of 500 μM AS (Fig. 7A), whereas the ΔmexE mutant strain AB3018 was equivalently sensitive to chloramphenicol in the presence or absence of 500 μM AS (Fig. 7B), indicating that the competition is at the efflux pump. The same results were observed when strains AB3016 and AB3012 were used in these competition assays (data not shown). Consistent with the proposed competition between these putative substrates is the dose-dependent relationship of AS to chloramphenicol toxicity; as the AS dose increased, the dose of chloramphenicol required for toxicity toward the wild-type strain A348 decreased (Fig. 7C). This is the case even though the levels of AS used in this experiment exhibited very modest growth inhibition (14%, 16%, and 23% for 30, 100, and 300 μM AS, respectively) when tested in the absence of chloramphenicol.
FIG 7.
Competition between AS and chloramphenicol for the MexE/MexF/AmeC efflux pump. (A and B) A348 (A) or AB3018 (B) cells grown in ABI medium plus or minus 500 μM AS at various chloramphenicol concentrations. OD600 measured after 20 h incubation. (C) A348 grown with 0, 30, 100, or 300 μM AS and various chloramphenicol concentrations. OD600 was measured after 20 h. Error bars are SD; n = 3.
Substrate specificity of pump activity.
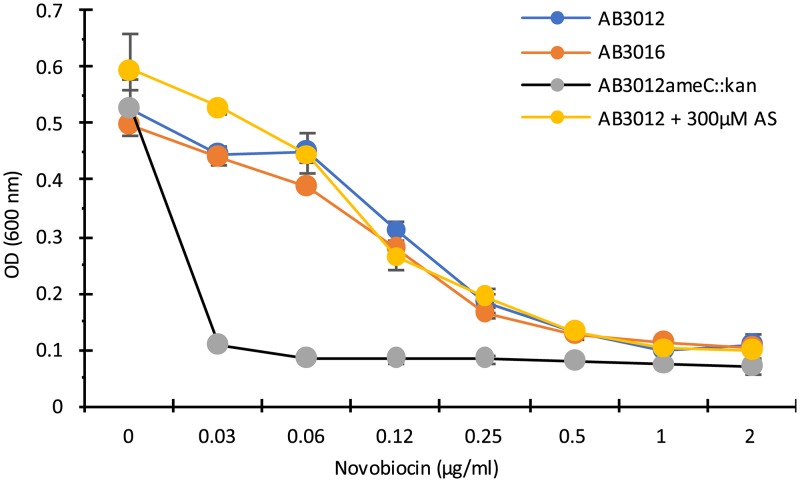
The apparent capacity of the MexE/MexF/AmeC pump to utilize diverse substrates, which include chloramphenicol and AS, is consistent with previous studies showing the diversity of substrates utilized by the MexE/MexF/OprN pump of P. aeruginosa (e.g., tetracycline [42]) and by the unidentified AmeC-containing pump of A. tumefaciens strain C58 (e.g., novobiocin [41]). To further characterize the range of MexE/MexF/AmeC substrates, the sensitivity of ΔmexE mutant strains to tetracycline and novobiocin was tested. Strains A348 and AB3018 (ΔmexE) showed the same sensitivity to tetracycline in either the presence or absence of AS (Fig. S3). Similarly, strains AB3012 (wild type) and AB3016 (ΔmexE) showed the same sensitivity to novobiocin (Fig. 8), as did strains A348 and AB3018 (data not shown). However, the strain carrying an insertion mutation in ameC (AB3012ameC::kan) was also tested. Consistent with the results of Peng and Nester (41), this strain was hypersensitive to novobiocin, and AS did not compete with novobiocin when both were provided to strain AB3012 (Fig. 8). These results suggest that AmeC may be involved in some pump, other than the MexE/MexF/AmeC pump identified here, that provides protection from novobiocin.
FIG 8.
Growth of wild-type strain AB3012, AB3012 with insertion in ameC, and ΔmexE strain AB3016 with various doses of novobiocin. OD600 of cultures was measured after 20 h of incubation at 25°C. Error bars are SD; n = 3.
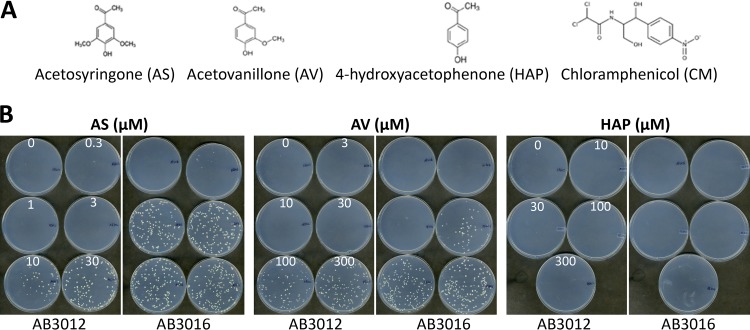
A key feature of the phenol-mediated induction of vir gene expression in Agrobacterium is that a wide variety of phenol derivatives can serve as vir inducers (44), and within this group there is a broad range of effectiveness. For example, 3,5-dimethoxy-4-hydroxyacetophenone (AS) (Fig. 9A) is a potent inducer, whereas 3-methoxy-4-hydroxyacetophenone (acetovanillone [AV]) is required in significantly higher concentrations than AS to achieve vir gene expression. 4-Hydroxy-acetophenone (HAP) is generally not an active vir inducer, although it can serve in this capacity when high levels of ChvE are present (45) or in the case of certain mutations in the VirA protein (46). To determine whether disruption of the putative MexE/MexF/AmeC efflux pump affects the sensitivity to AV and HAP for vir gene expression, these molecules were tested for their capacity to induce carbenicillin resistance in AB3012 or AB3016 (AB3012 ΔmexE mutant). The results show that, as expected, AB3016 is hypersensitive to the vir-inducing activity of AS by 10- to 20-fold compared to AB3012, whereas it is only slightly more sensitive to AV, and neither strain yields carbenicillin-resistant growth in response to HAP (Fig. 9B). This result demonstrates that disruption of the MexE/MexF/AmeC does not cause a general increase in sensitivity to potential vir inducers, a phenotype also observed in the AS-hypersensitive mutants isolated by Campbell et al. (37). The phenol specificity profile of the efflux pump was also tested using the competition assay between the phenol and chloramphenicol described above. The same pattern of phenol sensitivity was observed. In wild-type strain A348, a lower dose of AS was required to compete with chloramphenicol—and thereby to increase the toxic effects of this antibiotic—than that of AV, and HAP did not compete at any of the concentrations tested (Fig. 10). The fact that AS, AV, and HAP had similar effects on growth in the absence of chloramphenicol indicates that general phenol toxicity is not a critical component involved in the AS-chloramphenicol competition.
FIG 9.
(A) A Structures of acetosyringone (AS), acetovanillone (AV), 4-hydroxyacetophenone (HAP), and chloramphenicol. (B) AB3012 and AB3016 plated (∼200 cells per plate) on ABI medium plus 10 mM glucose, 100 μg/ml carbenicillin, and concentrations of AS, AV, and HAP as indicated. Incubated for 3 days at 25°C.
FIG 10.
Competition between various phenol derivatives and chloramphenicol. Wild-type strain A348 grown in ABI plus various concentrations of phenol derivatives and chloramphenicol, as indicated. OD600 measured after 20 h. Error bars are SD; n = 3.
Given the vir induction results described above, one possibility for the apparent lack of competition with chloramphenicol by AV or HAP is that the AS-mediated vir induction is indirectly affecting the sensitivity of the bacteria to chloramphenicol toxicity. This was tested by examining the putative AS-chloramphenicol competition in strain A136 (lacking the Ti plasmid and therefore incapable of virulence gene expression) and strain A348 (carrying the wild-type pTiA6). The results show that both strains exhibit the same response to the AS-chloramphenicol supplements in the medium, namely, as the AS dose increases, the concentration of chloramphenicol required for toxicity decreases (see Fig. S4 in the supplemental material). Similar results (data not shown) were obtained when these two strains were tested at pH 6.8 (nonpermissive for vir gene expression) rather than at pH 5.5 (permissive for vir induction), used in the assays presented here. These experiments demonstrate that the proposed competition between AS and chloramphenicol for the MexE/MexF/AmeC pump is independent of virulence gene expression. Taken together with results of assays on the phenol specificity for competition described above, these results suggest that the MexE/MexF/AmeC pump has a higher capacity to interact with AS than with the other two phenols tested, indicating that the methoxy groups may play a crucial role in phenol recognition by the pump. Tests of a broader range of phenolic derivatives is necessary to confirm this.
Virulence properties of the ΔmexE mutant.
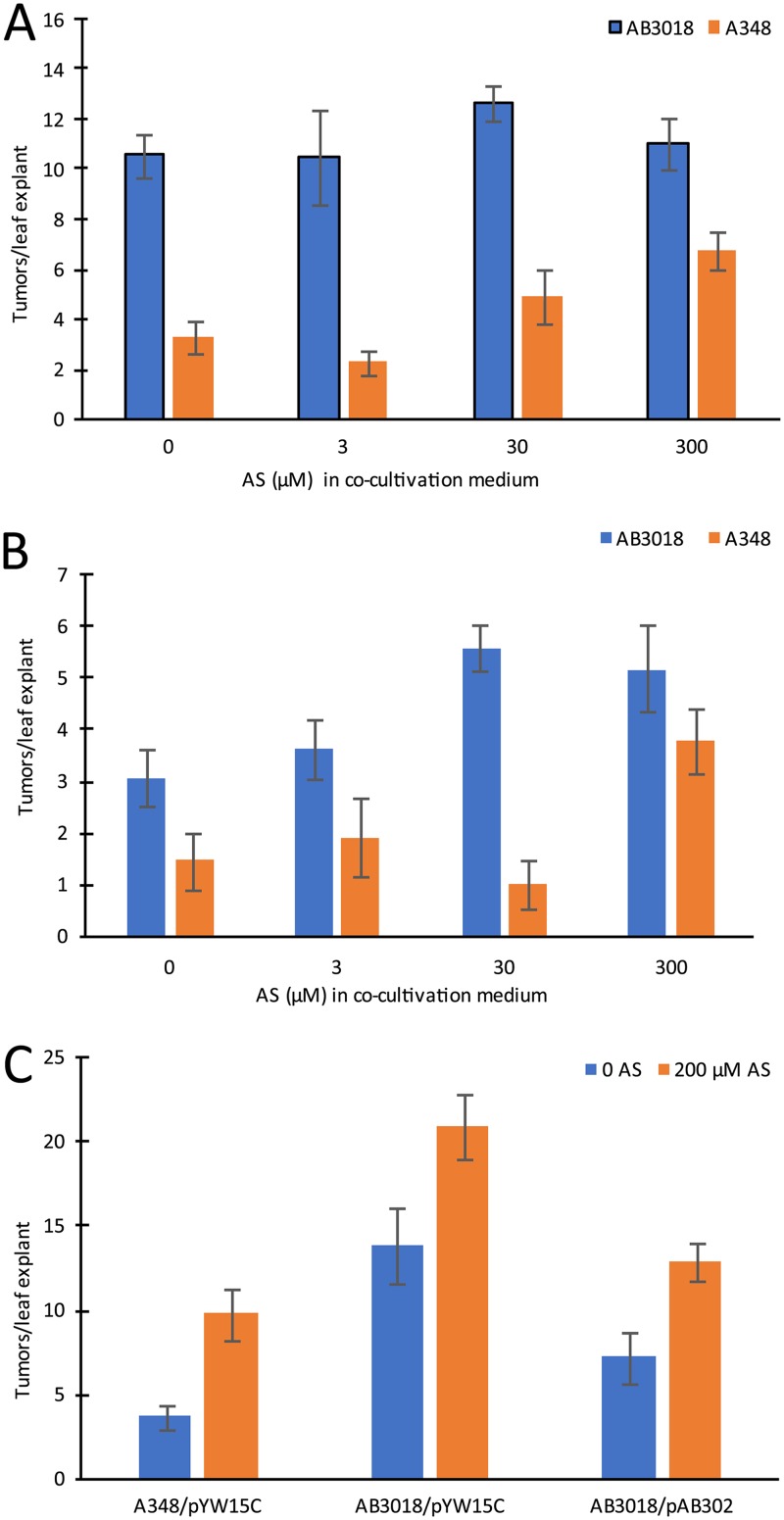
The tumor-inducing activity of strain A348 in at least some plant tissues is phenol inducer limited, and with these tissues, the prediction is that the AS-hypersensitive strains would induce greater numbers of tumors than the wild type strain. For example, exogenous AS provided in the cocultivation medium used for transformation of tobacco leaf tissues can dramatically stimulate tumor formation, and strains that are AS hypersensitive for vir induction are hypervirulent in such assays (26, 37). Therefore, the response of tobacco leaf explants to AB3018 (ΔmexE) and A348 when cocultivated in the presence of various AS concentrations was tested. The results demonstrate that the explants of young, expanding leaves (with approximately 50% of the area of fully expanded leaves) cocultivated with AB3018 in the absence of exogenous AS produce significantly more tumors then when such explants are cocultivated with A348 (Fig. 11A; see also Fig. S5 in the supplemental material). Further, supplemental AS in the cocultivation medium stimulates higher levels of tumor formation in explants cocultivated with A348, whereas this treatment did not further stimulate tumorigenesis by AB3018 (Fig. 11A). When the same experiment was carried out with explants from an older, fully expanded leaf on the same plant, AB3018 still induced more tumors than A348 when cocultivated in the absence of AS, but in this case, the leaf explants responded with more tumors when the cocultivation medium was supplemented with AS, although at a lower concentration of AS than that required to stimulate tumor formation in response to A348 (Fig. 11B). These results demonstrate that, indeed, the AB3018 mutant that is hypersensitive to AS for virulence gene expression is also hypervirulent in tobacco leaf explants. Moreover, the results suggest that there may be a difference in vir inducer concentration in expanding versus fully expanded leaves, with the former having significantly higher levels of endogenous inducer than the latter. Complementation studies using expanding leaves as the source of leaf explants demonstrated that expression of mexE from plasmid pAB302 significantly reduced the tumor-inducing activity of AB3018 (Fig. 11C), although, as was the case in the chloramphenicol sensitivity assays described above (Fig. 6) the complementation was not complete. In contrast to the results with tobacco, when Kalanchoe daigremontiana leaves were tested, AB3018 and A348 inoculations resulted in equivalent tumor formation (see Fig. S6 in the supplemental material), suggesting that the wound sites on these leaves have either high (nonlimiting) concentrations of the phenolic inducer or (possibly) a phenolic inducer that is not a substrate for the MexE/MexF/AmeC efflux pump.
FIG 11.
Virulence assays of wild-type strain A348 and AB3018 (ΔmexE). N. tabacum cv. H425 leaf explants cocultivated with A348 or AB3018 on Murashige and Skoog (MS) medium supplemented with various AS concentrations, as indicated, for 2 days at 25°C and then transferred to MS medium containing 200 μg/ml timentin and incubated for a further 12 days at 25°C. Mean number of tumors per explant and the standard error of the mean (n = 12) are shown. (A) Explants from an expanding leaf. (B) Explants from a fully expanded leaf. (C) Complementation of AB3018 by pAB302. A348/pYW15c, AB3018/pYW15C, and AB3018/pAB302 cocultivated with tobacco leaf explants for 48 h at 25°C with or without 200 μM AS, as shown, followed by transfer to MS medium plus timentin. Mean number of tumors per explant and the standard error of the mean (n = 12) are shown.
Competition between ΔmexE and wild-type strains.
Efflux pumps are often involved in protecting cells from toxic components of the environment (47). Given that the wound sites on plants—sites that are particularly susceptible to Agrobacterium-mediated plant transformation—can be a source of potentially toxic phenols (14, 16, 17), we tested the hypothesis that the putative MexE/MexF/AmeC efflux pump provides protection to Agrobacterium at the wound site by examining the relative fitness of ΔmexE and wild-type strains at such sites. To distinguish between the two strains, plasmid pMP7604, which is stable in Agrobacterium in the absence of selection and carries a constitutively expressed version of mCherry (48), was electroporated into A348 and AB3018. Nicotiana tabacum cv. H425 stem segments were placed onto water agar with the basal end up and inoculated with a 1:1 mix of either A348/pMP7604 and AB3018 or A348 and AB3018/pMP7604 at an optical density at 600 nm (OD600) of 0.1. After 5 days of incubation, the bacteria were recovered from the wound site and plated onto MG/L medium. The results (see Table S2 in the supplemental material) did not reveal any competitive advantage of the wild-type strain. This suggests that the phenolic concentration at the wound site is not sufficiently high to affect the survival of strains lacking the pump and is consistent with the observation that even at very high concentration, e.g., above saturating concentrations in terms of vir induction (500 μM), AS is only modestly inhibitory to agrobacterial growth (e.g., see Fig. 7C).
DISCUSSION
Expression of the Ti plasmid virulence genes in Agrobacterium tumefaciens is a significant commitment of energy resources (12), suggesting that selective pressures would yield a system (or systems) to ensure that expression occurs only at sites that would be productive in terms of plant transformation. Given that soils can contain significant levels of vir-inducing phenols (33–36), we hypothesized that there could be mechanisms that cause the bacteria to respond only to high(er) levels of the inducing phenols, such as those found at the wound sites on host plants. Using a β-lactamase gene driven by the virB promoter of the Ti plasmid as a selectable marker (Fig. 1), we carried out transposon mutagenesis and selected for mutants that were hypersensitive to AS vir-inducing activities (Fig. 2). Eight of the nine mutants isolated carried an insertion in an operon carrying genes that are homologous to mexE and mexF of the mexE-mexF-oprN operon of Pseudomonas aeruginosa. This operon encodes an efflux pump of the RND family (40) in which MexF is the inner membrane-localized transporter and MexE is an “adaptor protein” spanning the periplasmic space, interacting with MexF and OprN, an outer membrane-localized channel. Substrates are recognized and transported via the MexF protein via energy supplied by the proton motive force through the periplasmic space and outer membrane (for a review, see reference 49).
While the Agrobacterium mexE-mexF operon (see Fig. 3A) does not carry an oprN homolog encoding the outer membrane protein, one transposon mutant was in a gene (ameC) in a previously examined operon encoding a putative RND family efflux pump of unknown function (41) (Fig. 3B). Those studies did show, however, that deletion of ameC resulted in increased sensitivity to several antibiotics and detergents, whereas deletion of the other genes in that operon (ameA and ameB) did not yield these sensitive phenotypes. The authors suggested that AmeC may also be the outer membrane protein of some other unknown pump. Based on the data presented here, we propose that AmeC is the outer membrane protein of an efflux pump that has MexF as the inner membrane component and MexE as the periplasm-spanning protein that interacts with MexF and AmeC to construct a functional pump that can export AS and chloramphenicol. Biochemical assays will be required to confirm this. Additionally, because insertion mutations in ameC result in a novobiocin-hypersensitive phenotype, while deletion of mexE does not (41) (Fig. 9), and because mutations in AmeA and AmeB do not result in novobiocin resistance, AmeC may be part of three different efflux pumps. The loss of activity of one of these other pumps might be also responsible for the “slow growth on agar” phenotype observed in the strain carrying an insertion mutation in ameC (Fig. 2A). The utilization of an outer membrane component of an RND family efflux pump encoded on a different operon than the inner membrane and membrane-spanning components has been observed in other cases (50).
Strains with mutations that disrupt or delete parts or all of the MexE/MexF/AmeC pump are hypersensitive to the vir-inducing activity of the phenolic acetosyringone (AS) by approximately 10-fold compared with strains having the wild-type pump intact (Fig. 5 and 9B; see Fig. S1 in the supplemental material). The pump activity is predicted to reduce the level of AS that might accumulate in the cytoplasm—the proposed site of phenol recognition by VirA (51)—thereby setting up a situation in which higher levels of AS are required for induction than would be the case in the absence of the pump. The biological significance of the MexE/MexF/AmeC pump activity is demonstrated by the data showing that in the tobacco leaf explant virulence assay—where inducing phenols are limiting for tumor initiation (26, 37)—the ΔmexE strain is hypervirulent compared to the wild type (Fig. 11 and Fig. S5). Interestingly, these experiments revealed that younger, expanding tobacco leaves apparently have higher endogenous concentrations of phenolic vir inducers than the fully expanded leaf. An alternative hypothesis is that the fully expanded leaves produce an inhibitor of the virulence induction system. When tested on K. daigremontiana leaves, the ΔmexE strain was not hypervirulent compared to the wild type (Fig. S6), suggesting that the wound sites on leaves of this plant are not phenol limited for transformation. Another possibility is that the inducing phenol(s) of these leaves is not a substrate for the MexE/MexF/AmeC pump.
Given that chloramphenicol is a known substrate for the MexE/MexF/OprN pump of Pseudomonas aeruginosa (40, 42), we tested the mexE and ameC transposon mutants, as well as the ΔmexE strains, and found that they are hypersensitive to this antibiotic compared to strains with the wild-type pump (Fig. 4). If both AS and chloramphenicol are substrates for the MexE/MexF/AmeC pump, then competition between them for that pump might be expected. When provided AS at various concentrations, wild-type strains, but not the ΔmexE strains, significantly increase their chloramphenicol sensitivity (Fig. 7). This suggests that there is competition between these substrates and that the MexE/MexF/AmeC pump is the likely site of that competition. This hypothesis is supported by the observations that neither general phenol toxicity (Fig. 10) nor vir gene expression (Fig. S4) is responsible for the proposed competition. Finally, these results also provide an example of competition between nontoxic (to the bacteria) concentrations of a putative efflux pump substrate (AS) and other, more toxic substrates (chloramphenicol).
Earlier studies (52, 53) have demonstrated the presence of a chloramphenicol acetyltransferase gene (catB) in the C58 strain of A. tumefaciens, and this was proposed as a key element in chloramphenicol resistance of this strain. CatB activity is induced by subinhibitory levels of chloramphenicol via a mechanism proposed to involve translation attenuation. However, no strains carrying a catB deletion were examined in terms of sensitivity to chloramphenicol toxicity. The role of the MexE/MexF/AmeC pump in controlling the chloramphenicol sensitivity observed in our studies demonstrates an alternative means of providing resistance to the toxic effects of this antibiotic. As in the case in other systems (54), these results suggest that the efflux pump provides the first line of defense against antibiotics, and, as their concentrations rise, other metabolic activities provide yet another line of defense. Further characterization of the role of CAT activity in the chloramphenicol resistance of C58 derivatives will clarify how these two mechanisms of resistance are integrated with one another—for example, in the absence of the efflux pump, is a lower concentration of chloramphenicol required for induction of CAT activity?
Substrate specificity of RND efflux pumps is generally quite broad (e.g., see references 49 and 55), and the mechanisms underlying this revolve around the capacity of the substrates to interact with two different binding pockets in the pump. The observation here that both AS and chloramphenicol can be substrates for the MexE/MexF/AmeC pump is consistent with this. It was therefore surprising to find that modest alterations of the phenol structure—elimination of one (AV) or both (HAP) of the methoxy groups on the phenol ring—reduced or eliminated its capacity to be a pump substrate, as measured by either activation of the vir genes (Fig. 9B) or competition with chloramphenicol (Fig. 10). The observation that AV and HAP continue to be weak or noninducers of vir gene expression in the ΔmexE strains and are poor or noncompetitors with chloramphenicol for the MexE/MexF/AmeC pump raises the possibility that there is selection pressure on the pump to recognize and pump out active vir inducers. Examination of this hypothesis would entail characterization of the MexE/MexF/AmeC pump from a range of agrobacterial and related genera and species. Finally, the fact that the normally weak or noninducers appear not to be substrates of the MexE/MexF/AmeC pump suggests that they could accumulate to higher levels than that of AS in the cytoplasm of wild-type strains when provided at the same concentration and thus, on a molar basis, may be even weaker inducers of virulence gene expression than AS. However, while phenol derivatives such as the ones studied here are thought to enter the cell via partitioning across the inner membrane, their metabolic fate once they enter the cytoplasm (e.g., see reference 56) and their actual intracellular concentration have yet to be examined in cells lacking the MexE/MexF/AmeC efflux pump.
Efflux pump substrates have a variety of functions that range from antibiotic activity to bacterial virulence and intercellular signaling (47). Given that the MexE/MexF/AmeC pump substrates described here are involved in two of these functions (chloramphenicol toxicity and phenol-mediated virulence gene induction), an important question is whether both of these activities are crucial for bacterial success. That the pump provides chloramphenicol resistance to C58 suggests that it can play a protective role in soil environments, where chloramphenicol production by other bacteria (e.g., Streptomyces venezuelae [57]) could be a significant issue. However, the pump also appears to play a second key role, that of causing the bacterium to require “higher” levels of vir inducers in the environment before vir induction can occur. The observation that the pump mutants are sensitive to the vir-inducing activities of very low AS levels (0.3 to 1 μM AS), in comparison to the wild-type strain, indicates that the pump is clearing low, nontoxic levels of this phenol. This is consistent with the hypothesis that there is a selective advantage in suppressing the response to low levels of vir-inducing phenols. In the soil environment, this would ensure that low levels of phenol would not be able to induce vir expression that would be of no use to bacteria found in that setting. A question for future studies is the issue of when and how a pump such as MexE/MexF/AmeC evolved the capacity to pump alternative substrates that can then integrate with and help control critical cellular processes that are not apparently related to one another—in this case, chloramphenicol resistance and virulence gene expression.
The conclusion that Agrobacterium is utilizing an “unrelated” cellular activity—in this case. the MexE/MexF/AmeC pump—to suppress the response to low concentrations of a vir inducer is consistent with earlier studies on the role of ABC transporters in modulating the response of Agrobacterium to vir-inducing sugars (18, 31, 58). In all of these cases, the activity of the transporters appears to keep the inducing ligand concentration lower at the site of signal perception—the periplasm in the case of ChvE and sugars and the cytoplasm in the case of VirA and phenols. Taken together, these studies provide strong evidence that Agrobacterium has integrated cellular transport activities with the virulence regulating system so as to ensure that vir induction occurs only in environments that are expected to have high levels of vir inducers, such as at a wound site on a plant.
MATERIALS AND METHODS
Bacteria, media, and growth conditions.
Escherichia coli strains were grown in Luria-Bertani (LB) medium (Sigma) with appropriate antibiotics at 37°C. Agrobacterium tumefaciens strains were routinely maintained on MG/L medium at 25°C (59) (see Table 1 for strains and plasmids). Antibiotics were used at the following concentrations (in liquid or solid medium, respectively) for A. tumefaciens: 30 or 100 μg/ml (carbenicillin; Sigma) and 10 or 50 μg/ml (kanamycin; Sigma). For vir gene induction, AB induction (ABI) medium (59) was used, except 3.9 g/liter morpholineethanesulfonic acid (MES; pH 5.5) was used with Casamino Acids omitted. Growth assays of Agrobacterium strains in the presence of various antibiotics (chloramphenicol [Tokyo Chemical Industry], novobiocin, and tetracycline [Sigma]) or phenol derivatives (acetosyringone, acetovanillone, and 4-hydroxyacetophenone; Acros Organics) were carried out by first growing overnight cultures from single colonies in MG/L medium, followed by overnight growth in ABI medium supplemented with 10 mM glucose, all at 25°C. Cells from the overnight ABI cultures were resuspended, added to ABI medium plus various supplements at a starting OD600 of 0.08, and grown overnight at 25°C on a spinning drum, and their OD600s were measured. Fluorescence images of plates were acquired following the procedure of Siryaporn and Goulian (60) using a D50 camera (Nikon, Melville, NY) connected to the fluorescence illuminator from a Zeiss microscope (2FL fluorescence adapter) with a 100-W mercury lamp and using filter sets for GFP and mCherry.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tcr)] | StrateGene |
| PIR2 | F− Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1 uidA(ΔMluI)::pir | Invitrogen |
| A. tumefaciens | ||
| A136 | C58 background without Ti plasmid | 63 |
| A348 | C58 background carrying pTiA6 | 63 |
| AB140 | Spontaneous AS-hypersensitive mutant of A348 | 37 |
| AB144 | Spontaneous AS-hypersensitive mutant of A348 | 37 |
| AB147 | Spontaneous AS-hypersensitive mutant of A348 | 37 |
| AB300 | A348 with deletion of chvE | 25 |
| AB3003 | AB300 with gfp ORF inserted between virE2 and virE3 | This study |
| AB3003C | AB3003 with β-lactamase ORF from pBBR1MCS4 inserted after virB1 start codon | This study |
| AB3009 | AB3003C with sacB ORF from pK18mobsacB inserted after virB2 start codon | This study |
| AB3012 | AB3009 with chvE returned to its chromosomal location | This study |
| AB3016 | AB3012 with mexE deletion | This study |
| AB3018 | A348 with mexE deletion | This study |
| MDG165 | 358mx virE::[lacZ::gfp] | 61 |
| Plasmids | ||
| pBBR1MCS2 | Broad-host-range plasmid cloning vector; Kanr | 39 |
| pBBR1MCS4 | Broad-host-range plasmid cloning vector; Ampr | 39 |
| pK18mobsacB | Suicide plasmid vector; Kanr | 62 |
| pYW15C | IncW/ColE1 expression vector with PN25 promoter; Carbr | 43 |
| pRL27 | Tn5-RL27 (Kanr oriR6K) delivery vector | 38 |
| pAB302 | pYW15C with mexE expressed from PN25 promoter; Carbr | This study |
| pAB303 | pK18mobsacB carrying sequences flanking mexE orf; Kanr | This study |
| pJZ150 | chvE plus flanking sequences in pK18mobsacB; Kanr | This study |
| pJZ151 | β-Lactamase ORF fused to virB1 start codon plus flanking upstream and downstream virB1 sequences in pK18mobsacB; Kanr | This study |
| pJZ152 | sacB ORF fused to virB2 start codon plus flanking virB1, virB2, and virB3 sequences in pK18mobsacB; Kanr | This study |
| pJZ153 | gfp ORF (including start codon) plus virE2 and virE3 sequences in pK18mobsacB; Kanr | This study |
Carbr, carbenicillin resistance; Kanr, kanamycin resistance.
Construction of strains and plasmids.
We used marker exchange eviction mutagenesis (62) as previously described (30) to create strain AB3003 carrying GFP between virE2 and virE3 in the plasmid pTiA6. Primer E2GFPE3.P1 was used with E2GFPE3.P2 (see Table S3 in the supplemental material for primers) to amplify an 800-bp fragment containing the 3′ part of the virE2 sequence and part of the sequence between virE2 and virE3. Primers E2GFPE3.P3 and E2GFPE3.P4 were used to amplify the GFP gene with its own ribosomal binding site (RBS) sequence from MDG165 (61). The 5′ ends of primers E2GFPE3.P2 and E2GFPE3.P3 were designed to include complementary sequence so that the products from the first two PCRs could function as self-annealing templates in a third PCR with primers E2GFPE3.P1 and E2GFPE3.P4 to generate a 1.6-kb fragment carrying part of the virE2 sequence and the whole GFP gene. Primer E2GFPE3.P5 was used with E2GFPE3.P6 to amplify an 800-bp fragment containing the 5′ part of the virE3 sequence and part of the sequence between virE2 and virE3. This PCR product was mixed with the previous PCR product (the 1.6-kb fragment) to do the fifth PCR with primers E2GFPE3.P1 and E2GFPE3.P6 to generate a 2.4-kb fragment carrying the 3′ part of the virE2 sequence, the whole GFP gene, the virE2 and virE3 intergenic region, and the 5′ part of the virE3 sequence. This 2.4-kb PCR fragment was digested with BamHI and PstI and cloned into pK18mobsacB (59). The resulting construct (pJZ153) was electroporated into strain AB300 (25). The double-deletion strains were obtained and confirmed with primers conE2GFPE3.P1 (upstream of the amplified 3′ part of the virE2 sequence) and conE2GFPE3.P2 (downstream of the amplified 5′ part of the virE3 sequence). The resulting strain was named AB3003.
To construct AB3003C, the β-lactamase gene from pBBR1MCS-4 (39) was inserted behind the start codon and RBS of virB1. Therefore, virB1 is inactive, since it is disrupted by the β-lactamase insertion. PCR using primers PvirB.P2 (SalI) and 4pvirB.P1 and pTiA6 as the substrate amplified the upstream sequence of virB1, yielding product 1, ending with the start codon of virB1 (ATG). PCR using the primers 4carb.P2 and 4carb.P1 and pBBR1MCS-4 as the substrate yielded product 2, which includes the β-lactamase open reading frame (exclusive of its start codon) and flanking sequences upstream and downstream of virB1. PCR using the primers 4virB1.P2 and virB1.P1 (EcoRI) and pTiA6 as the substrate yielded product 3, which has sequences from the 3′ end of the β-lactamase gene fused to sequences downstream of the virB1 start codon. PCR using the primers PvirB.P2 (SalI) and 4carb.P1 and products 1 and 2 as the substrates to yield product 4 (upstream virB1 including the start codon, β-lactamase sequence, and some downstream virB1 sequence). PCR using the primers PvirB.P2 (SalI) and virB1.P1 (EcoRI) and products 3 and 4 yielded product 5 (upstream virB1 including the start codon and β-lactamase sequence after the virB1 start codon), which was then digested with EcoRI and SalI and cloned into pK18mobsacB. This plasmid (pJZ151) was electroporated into AB3003, and the insertion was selected for as above and confirmed via PCR using the primers UpvirB and DownvirB1, yielding AB3003C.
The sacB open reading frame from pK18mobsacB (62) was moved into AB3003C behind the virB2 start codon to yield AB3009, as follows. The upstream sequence of virB2 with the start codon of virB2, which is used as the start codon of sacB in the final construct, was amplified from pTiA6 using the primers 6virB1.P1 (EcoRI) and 5virB1.P11 (product 1). The sacB ORF from pK18mobsacB without its own start codon and with overlapping sequence from just before and after the start codon of virB2 was amplified using the primers 5sacB.P1 and 5sacB.P2 (product 2). The sequences downstream of the virB2 start codon plus the sacB overlapping sequence at its 3′ end were amplified using the primers 5virB2.p22 and 5virB2.P2 (SalI) (product 3). The primers 6virB1.P1 (EcoRI) and 5sacB.P2 were used with products 1 and 2 to yield the upstream virB1 sequences and the virB2 start codon (product 4). The final product that included virB1 sequences, the virB2 start codon, the sacB ORF, and downstream virB2 and virB3 sequences was amplified using products 3 and 4 as the substrate (product 5). This was then cut with EcoRI and SalI and cloned into pK18mobsacB, yielding pJZ152. This plasmid was electroporated into AB3003 and, after kanamycin and sucrose selection as above, yielded strain AB3009, which was confirmed using the primers 5UpvirB1 and 5DownvirB2.
Wild-type chvE, and ∼500 bp of upstream and downstream sequences were amplified using the primers chvE.PI and chvE.PIV and cloned into the XmaI site of pK18mobsacB, yielding pK18mobsacB::chvE (=pJZ150). This plasmid was electroporated into AB3009 and, after kanamycin and sucrose selection as above, yielded strain AB3012. The presence of the wild-type chvE at its original location was confirmed using the primers K262SconP1 and DOWNDC3.
ΔmexE strains were constructed by first amplifying AB3012 genomic DNA with the primers dMexEP1 and dMexEP2 to yield fragment A, which carried sequences just upstream of the mexE start codon plus overlapping sequence just downstream of the stop codon of mexE. Primers dMexEP3 and dMexEP42 and AB3012 genomic DNA as the template were used to generate fragment B, which carried sequences downstream of the mexE stop codon and overlapping sequence just upstream of the mexE start codon. Fragment C, which carried the deletion of the mexE ORF with approximately 500 bp of upstream and downstream sequences, was amplified from fragments A and B using the primers dMexEP1 and dMexEP42. Fragment C was digested with EcoRI and XbaI and cloned into pK18mobsacB (=pAB303). This plasmid was electroporated into strains AB3012 and A348, followed by kanamycin and sucrose selection as above, yielding ΔmexE strains AB3016 and AB3018, respectively. The deletion of mexE in sucrose-resistant (Sucr) kanamycin-sensitive (Kans) colonies was confirmed via PCR with the primers CondMexEP1 and CondMexEP2, which are outside fragment C.
The mexE open reading frame was amplified with Phusion polymerase (NEB) using the primers MexEP9 and MexEP10, each of which carries mexE and pYW15C (43) sequences. pYW15C was cut with SalI and SacI, treated with calf intestine phosphatase, and gel purified using the Qiagen gel extraction protocol. The mexE fragment and digested vector were incubated with HiFi DNA assembly master mix (NEB) per the vendor’s instructions, transformed into E. coli XL1, and plated on LB plates supplemented with ampicillin (100 μg/ml; Sigma). Plasmid minipreps were made (Qiagen), and those with a diagnostic 520-bp HindIII fragment were sequenced using the primers pYW15CseqF and pYW15CseqR. The confirmed plasmid, pAB302, contains the mexE ORF driven by the PN25 promoter of pYW15C.
Transposon mutagenesis of AB3012 and identification of insertion sites.
E. coli strain BW20767/pRL27 (38) was grown on 4 plates of LB medium plus 25 μg/ml kanamycin. Cells were resuspended in 2 ml of LB medium per plate and spun down, and plasmid DNA was prepared via the Qiagen miniprep protocol. This DNA was concentrated via sodium acetate-ethanol precipitation, washed with 70% ethanol, and resuspended in 50 μl H2O. The DNA concentration was measured at approximately 100 ng/μl using a Thermo Scientific NanoDrop spectrophotometer. Electrocompetent cells of AB3012 were prepared, electroporated with 2.5 μl of the pRL27 plasmid preparation or 0.5 μl of pBBR1MCS-2 (39), resuspended in 1 ml of MG/L medium (59), and allowed to recover on a spinning drum for 3 h at 25°C. These cells were spun down and resuspended in MES buffer (20 mM, pH 5.5) and plated on agar plates of ABI medium (59) containing 100 μg/ml carbenicillin (Acros Organics) and 50 μg/ml kanamycin (Sigma) plus 10 mM glucose and acetosyringone (AS; Acros Organics) at 0, 1, or 200 μM AS. Additionally, 5 μl of cells was plated on MG/L medium plus 50 μg/ml kanamycin to estimate the number of cells carrying a transposon. Plates were examined for growth and GFP expression (via fluorescence microscopy) after 5 days.
Colonies that were Kanr (kanamycin resistant), Carbr, and GFP positive on 1 μM AS were restreaked on ABI medium plus 10 mM glucose, 1 μM AS, 100 μg/ml carbenicillin, and 50 μg/ml kanamycin and confirmed as Kanr, Carbr, and GFP positive. Individual colonies from each transposon hit were grown overnight in 7 ml of MG/L medium plus 50 μg/ml kanamycin. A 500-ml aliquot of the overnight culture was used to make −80°C stocks, and the remainder was used for genomic DNA preparation, as follows. Aliquots (3 ml) were spun down and resuspended in 250 μl Qiagen miniprep buffer 1, followed by addition of 250 μl of Qiagen miniprep buffer 2 and incubation for 5 min at room temperature. Qiagen buffer 3 (350 μl) was then added to this, and the entire mixture was subjected to shearing by vortexing, then 1 pass of the mixture through a 21-gauge needle, followed by three passes of the mixture through a 25-gauge needle. After centrifugation for 10 min, the supernatant was applied to a Qiagen spin column, and DNA was washed and eluted per the vendor’s instructions. To recover the transposon and flanking Agrobacterium sequences, the genomic DNA preparations were digested with either BamHI or BfaI, neither of which cuts inside the transposon, purified via Qiagen spin column, and ligated with T4 ligase. This was then purified via a Qiagen spin column and eluted with H2O, and the eluate was electroporated into E. coli Pir2 electrocompetent cells that allow plasmid replication via the oriR6K sequences in the transposon (38). After a 1-h recovery period in 1 ml of LB medium at 37°C, the cells were plated onto LB plates plus 50 μg/ml kanamycin and grown overnight at 37°C. Colonies that grew up were restreaked on LB plates plus kanamycin and were used to make Qiagen DNA minipreps that were then sequenced using the primers oriKR6K and TnmodRKan2.
Virulence assays.
Analysis of tumor initiation by various Agrobacterium strains on Nicotiana tabacum cv. H425 leaf explants and Kalanchoe daigremontiana leaves was carried out as described previously (18).
Bacterial competition assays.
Stems from vegetative Nicotiana tabacum cv. H425 greenhouse-grown plants were surface sterilized via 3 consecutive cycles of 7% chlorox-H2O rinse-70% ethanol and then 3 rinses in sterile H2O. Stem segments (4 mm thick) were excised and moved onto H2O-0.9% agar plates (basal cut surface up) and inoculated with 10 μl of a 1:1 mix of Agrobacterium strains A348 and AB3018/pMP7604 or A348/pMP7604 and AB3018 that had been grown overnight in MG/L medium and resuspended to an OD600 of 0.1 in 20 mM MES (pH 5.5). The inoculated stem pieces were incubated in the dark for 5 days at 25°C. The bacteria on the wound sites were then resuspended in 20 mM MES (pH 5.5), diluted in the same buffer, and plated on MG/L medium; after 3 days, colonies were counted and pictures of red fluorescence taken.
Supplementary Material
ACKNOWLEDGMENTS
The NSF BIO Division of Integrative Organismal Systems (IOS) provided funding to Andrew N. Binns under grant number IOS11211019. Funds were also provided by the University of Pennsylvania.
The funders had no role in the study design, data collection and interpretation, or decision to submit the work for publication.
We thank the Goulian and Hsu labs at Penn for helpful discussions and Mark Goulian and Manuela Roggiani for strain MDG165 and for assistance with fluorescence microscopy. We thank Arlene Wise and David Lynn for reading earlier versions of the manuscript. We also thank the anonymous reviewers, who provided very useful comments.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Otten L, Burr T, Szegedi E. 2008. Agrobacterium: a disease-causing bacterium, p 1–46. In Tzfira T, Citovsky V (ed), Agrobacterium: from biology to biotechnology. Springer, New York, NY. [Google Scholar]
- 2.Dessaux Y, Faure D. 2018. Niche construction and exploitation by Agrobacterium: how to survive and face competition in soil and plant habitats. Curr Top Microbiol Immunol 418:55–86. doi: 10.1007/82_2018_83. [DOI] [PubMed] [Google Scholar]
- 3.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 4.Ellis JG, Kerr A, Petit A, Tempe J. 1982. Conjugal transfer of nopaline and agropine Ti plasmids—the role of agrocinopines. Mol Gen Genet 186:269–274. doi: 10.1007/BF00331861. [DOI] [Google Scholar]
- 5.DeCleene M, DeLey J. 1976. The host range of crown gall. Bot Rev 42:389–466. doi: 10.1007/BF02860827. [DOI] [Google Scholar]
- 6.McCullen CA, Binns AN. 2006. Agrobacterium tumefaciens and plant cell interactions required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 7.Subramoni S, Nathoo N, Limov E, Yuan ZC. 2014. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci 5:322–323. doi: 10.3389/fpls.2014.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YG, Christie PJ. 2018. The Agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr Top Microbiol Immunol 418:233–260. doi: 10.1007/82_2018_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelvin SB. 2017. Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Genet 51:195–217. doi: 10.1146/annurev-genet-120215-035320. [DOI] [PubMed] [Google Scholar]
- 10.Stachel SE, Zambryski PC. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325–333. doi: 10.1016/0092-8674(86)90653-7. [DOI] [PubMed] [Google Scholar]
- 11.Winans SC, Kerstetter RA, Nester EW. 1988. Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J Bacteriol 170:4047–4054. doi: 10.1128/jb.170.9.4047-4054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platt TG, Bever JD, Fuqua C. 2012. A cooperative virulence plasmid imposes a high fitness cost under conditions that induce pathogenesis. Proc Biol Sci 279:1691–1699. doi: 10.1098/rspb.2011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahl G. 1982. Molecular biology of wound healing: the conditioning phenomenon, p 211–267. In Kahl G, Schell J (ed), Molecular biology of plant tumors. Academic Press, New York, NY. [Google Scholar]
- 14.Stachel SE, Messens E, Van Montagu M, Zambryski P. 1985. Identification of signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624–629. doi: 10.1038/318624a0. [DOI] [Google Scholar]
- 15.Dixon RA, Paiva NL. 1995. Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya A, Sood P, Citovsky V. 2010. The role of plant phenolics in defense and communication during Agrobacterium and Rhizobium infection. Mol Plant Pathol 11:705–719. doi: 10.1111/j.1364-3703.2010.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimoda N, Toyoda-Yamamoto A, Nagamine J, Usami S, Katayama M, Sakagami Y, Machida Y. 1990. Control of expression of Agrobacterium vir genes by synergistic actions of phenolic signal molecules and monosaccharides. Proc Natl Acad Sci U S A 87:6684–6688. doi: 10.1073/pnas.87.17.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Zhao J, DeGrado WF, Binns AN. 2013. Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals. Proc Natl Acad Sci U S A 110:678–683. doi: 10.1073/pnas.1215033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venturi V, Fuqua C. 2013. Chemical signaling between plants and plant pathogenic bacteria. Annu Rev Phytopathol 51:17–37. doi: 10.1146/annurev-phyto-082712-102239. [DOI] [PubMed] [Google Scholar]
- 20.Lin YH, Pierce BD, Fang F, Wise A, Binns AN, Lynn DG. 2014. Role of the VirA histidine kinase of Agrobacterium in the initial steps of pathogenesis. Front Plant Sci 5:195. doi: 10.3389/fpls.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y-W, Jin S, Sim W-S, Nester EW. 1995. Genetic evidence for direct sensing of phenolic compounds by the VirA protein of Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 92:12245–12249. doi: 10.1073/pnas.92.26.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohrke SM, Yang H, Jin S. 2001. Reconstitution of acetosyringone-mediated Agrobacterium tumefaciens virulence gene expression in the heterologous host Escherichia coli. J Bacteriol 183:3704–3711. doi: 10.1128/JB.183.12.3704-3711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cangelosi GA, Ankenbauer RG, Nester EW. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc Natl Acad Sci U S A 87:6708–6712. doi: 10.1073/pnas.87.17.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao R, Lynn DG. 2007. Environmental pH sensing: resolving the VirA/VirG two component system inputs for Agrobacterium pathogenesis. J Bacteriol 189:6048–6056. doi: 10.1128/JB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He F, Nair GR, Soto CS, Chang Y, Hsu L, Ronzone E, DeGrado WF, Binns AN. 2009. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J Bacteriol 191:5802–5813. doi: 10.1128/JB.00451-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banta LM, Joerger RD, Howitz VR, Campbell AM, Binns AN. 1994. Glu 255 outside the predicted ChvE binding site in VirA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J Bacteriol 176:3242–3249. doi: 10.1128/jb.176.11.3242-3249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair GR, Lai X, Wise AA, Rhee BW, Jacobs M, Binns AN. 2011. The integrity of the periplasmic domain of the VirA sensor kinase is critical for optimal coordination of the virulence signal response in Agrobacterium tumefaciens. J Bacteriol 193:1436–1448. doi: 10.1128/JB.01227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ankenbauer RG, Nester EW. 1990. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol 172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemner JM, Liang X, Nester EW. 1997. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J Bacteriol 179:2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Binns AN. 2011. Characterization of the mmsAB-araD1 (gguABC) genes of Agrobacterium tumefaciens. J Bacteriol 193:6586–6596. doi: 10.1128/JB.05790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Binns AN. 2014. GxySBA ABC transporter of Agrobacterium tumefaciens and its role in sugar utilization and vir gene expression. J Bacteriol 196:3150–3159. doi: 10.1128/JB.01648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunina A, Kuzyakov Y. 2015. Sugars in soil and sweets for microorganisms: review of origin, content, composition, and fate. Soil Biol Biochem 90:87–100. doi: 10.1016/j.soilbio.2015.07.021. [DOI] [Google Scholar]
- 33.Kuiters AT, Sarink HM. 1986. Leaching of phenolic compounds from leaf and needle litter of several deciduous and coniferous trees. Soil Biol Biochem 18:475–480. doi: 10.1016/0038-0717(86)90003-9. [DOI] [Google Scholar]
- 34.Johnson RM, Pregitzer KS. 2007. Concentration of sugars, phenolic acids, and amino acids in forest soils exposed to elevated atmospheric CO2 and O3. Soil Biol Biochem 39:3159–3166. doi: 10.1016/j.soilbio.2007.07.010. [DOI] [Google Scholar]
- 35.De Fuedis M, Cardelli V, Massaccesi L, Hoffman D, Berns AE, Bol R, Cooco S, Corti G, Agnelli A. 2017. Altitude affects the quality of the water-extractable organic matter (WEOM) from rhizophere and bulk soil in European beech forests. Geoderma 302:6–13. doi: 10.1016/j.geoderma.2017.04.015. [DOI] [Google Scholar]
- 36.Chen Y, Chen W, Lan Y, Wang KT, Wu YC, Zhong XL, Ying KY, Li JY, Yang GD. 2019. Determination of 18 phenolic acids in tobacco and rhizosphere by ultra high performance liquid chromatography combined with triple quadrupole mass spectrometry. J Sep Sci 42:816–825. doi: 10.1002/jssc.201800819. [DOI] [PubMed] [Google Scholar]
- 37.Campbell AM, Tok JB, Zhang J, Wang Y, Stein M, Lynn DG, Binns AN. 2000. Xenognosin sensing in virulence: is there a phenol receptor in Agrobacterium tumefaciens? Chem Biol 7:65–76. doi: 10.1016/S1074-5521(00)00065-X. [DOI] [PubMed] [Google Scholar]
- 38.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 39.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 40.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechère JC. 1997. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol 23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 41.Peng WT, Nester EW. 2001. Characterization of a putative RND-type efflux system in Agrobacterium tumefaciens. Gene 270:245–252. doi: 10.1016/s0378-1119(01)00468-1. [DOI] [PubMed] [Google Scholar]
- 42.Maseda H, Yoneyama H, Nakae T. 2000. Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa. Antimicrob Agents Chemother 44:658–664. doi: 10.1128/aac.44.3.658-664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Mukhopadhyay A, Howitz VR, Binns AN, Lynn DG. 2000. Construction of an efficient expression system for Agrobacterium tumefaciens based on the coliphage T5 promoter. Gene 242:105–114. doi: 10.1016/s0378-1119(99)00541-7. [DOI] [PubMed] [Google Scholar]
- 44.Palmer AG, Gao R, Maresh J, Erbil WK, Lynn DG. 2004. Chemical biology of multi-host/plant pathogen interactions: chemical perception and metabolic complementation. Annu Rev Phytopathol 42:439–464. doi: 10.1146/annurev.phyto.41.052002.095701. [DOI] [PubMed] [Google Scholar]
- 45.Peng W-T, Lee Y-W, Nester EW. 1998. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar binding protein ChvE. J Bacteriol 180:5632–5638. doi: 10.1128/JB.180.21.5632-5638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang C-H, Zhu J, Winans SC. 1996. Pleiotropic phenotypes caused by genetic ablation of the receiver module of the Agrobacterium tumefaciens VirA protein. J Bacteriol 178:4710–4716. doi: 10.1128/jb.178.15.4710-4716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. 2009. Functional role of bacterial multidrug efflux pumps in microbial natural systems. FEMS Microbiol Rev 33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 48.Lagendijk EL, Validov S, Lamers GEM, de Weert S, Bloemberg GV. 2010. Genetic tools for tagging Gram-negative bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol Lett 305:81–90. doi: 10.1111/j.1574-6968.2010.01916.x. [DOI] [PubMed] [Google Scholar]
- 49.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim Biophys Acta 1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Q, Li XZ, Srikumar R, Poole K. 1998. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomanas aeruginosa independent of MexAB. Antimicrob Agents Chemother 42:1682–1688. doi: 10.1128/AAC.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang CH, Winans SC. 1992. Functional roles assigned to the periplasmic, linker, and receiver domains of the Agrobacterium tumefaciens VirA kinase. J Bacteriol 174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tennigkeit J, Matzura H. 1991. Nucleotide sequence analysis of a chloramphenicol-resistance determinant from Agrobacterium tumefaciens and identification of its gene product. Gene 98:113–116. doi: 10.1016/0378-1119(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 53.Rogers EJ, Rahman MS, Hill RT, Lovett PS. 2002. The chloramphenicol-inducible catB gene in Agrobacterium tumefaciens is regulated by translation attenuation. J Bacteriol 184:4296–4300. doi: 10.1128/jb.184.15.4296-4300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rahman T, Yarnall B, Doyle DA. 2017. Efflux transporters at the forefront of antimicrobial resistance. Eur Biophys J 46:647–653. doi: 10.1007/s00249-017-1238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramaswamy VK, Vargiu AV, Malloci G, Drier J, Ruggerone P. 2018. Molecular determinants of the promiscuity of MexB and MexY multidrug transporters of Pseudomonas aeruginosa. Front Microbiol 9:1144. doi: 10.3389/fmicb.2018.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brencic A, Eberhard A, Winans SC. 2004. Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol Microbiol 51:1103–1115. doi: 10.1046/j.1365-2958.2003.03887.x. [DOI] [PubMed] [Google Scholar]
- 57.Berendsen B, Pikkemaat M, Römkens P, Wegh R, van Sisseren M, Stolker L, Nielen M. 2013. Occurrence of chloramphenicol in crops through natural production by bacteria in soil. J Agric Food Chem 61:4004–4010. doi: 10.1021/jf400570c. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, Binns AN. 2016. Involvement of the Agrobacterium tumefaciens galacturonate tripartite ATP-independent (TRAP) transporter GaaPQM in virulence gene expression. Appl Environ Microbiol 82:1136–1146. doi: 10.1128/AEM.02891-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wise AA, Liu Z, Binns AN. 2006. Culture and maintenance of Agrobacterium strains, p 3–13. In Wang K. (ed), Agrobacterium protocols, 2nd ed, vol 1 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 60.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol 70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goulian M, van der Woude M. 2006. A simple system for converting lacZ to gfp reporter fusions in diverse bacteria. Gene 372:219–226. doi: 10.1016/j.gene.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 62.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 63.Garfinkel DJ, Nester EW. 1980. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol 144:732–743. doi: 10.1128/JB.144.2.732-743.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.