CF patients are chronically infected by polymicrobial communities. The two dominant bacterial pathogens that infect the lungs of CF patients are P. aeruginosa and S. aureus, with ∼30% of patients coinfected by both species. Such coinfected individuals have worse outcomes than monoinfected patients, and both species persist within the same physical space. A variety of host and environmental factors have been demonstrated to promote P. aeruginosa-S. aureus coexistence, despite evidence that P. aeruginosa kills S. aureus when these organisms are cocultured in vitro. Thus, a better understanding of P. aeruginosa-S. aureus interactions, particularly mechanisms by which these microorganisms are able to coexist in proximal physical space, will lead to better-informed treatments for chronic polymicrobial infections.
KEYWORDS: alginate, cystic fibrosis, Pseudomonas aeruginosa, Staphylococcus aureus, polymicrobial, mucoid, Nanostring, PQS, HQNO, pyoverdine, pyochelin, siderophores
ABSTRACT
Cystic fibrosis (CF) patients chronically infected with both Pseudomonas aeruginosa and Staphylococcus aureus have worse health outcomes than patients who are monoinfected with either P. aeruginosa or S. aureus. We showed previously that mucoid strains of P. aeruginosa can coexist with S. aureus in vitro due to the transcriptional downregulation of several toxic exoproducts typically produced by P. aeruginosa, including siderophores, rhamnolipids, and HQNO (2-heptyl-4-hydroxyquinoline N-oxide). Here, we demonstrate that exogenous alginate protects S. aureus from P. aeruginosa in both planktonic and biofilm coculture models under a variety of nutritional conditions. S. aureus protection in the presence of exogenous alginate is due to the transcriptional downregulation of pvdA, a gene required for the production of the iron-scavenging siderophore pyoverdine as well as the downregulation of the PQS (Pseudomonas quinolone signal) (2-heptyl-3,4-dihydroxyquinoline) quorum sensing system. The impact of exogenous alginate is independent of endogenous alginate production. We further demonstrate that coculture of mucoid P. aeruginosa with nonmucoid P. aeruginosa strains can mitigate the killing of S. aureus by the nonmucoid strain of P. aeruginosa, indicating that the mechanism that we describe here may function in vivo in the context of mixed infections. Finally, we investigated a panel of mucoid clinical isolates that retain the ability to kill S. aureus at late time points and show that each strain has a unique expression profile, indicating that mucoid isolates can overcome the S. aureus-protective effects of mucoidy in a strain-specific manner.
IMPORTANCE CF patients are chronically infected by polymicrobial communities. The two dominant bacterial pathogens that infect the lungs of CF patients are P. aeruginosa and S. aureus, with ∼30% of patients coinfected by both species. Such coinfected individuals have worse outcomes than monoinfected patients, and both species persist within the same physical space. A variety of host and environmental factors have been demonstrated to promote P. aeruginosa-S. aureus coexistence, despite evidence that P. aeruginosa kills S. aureus when these organisms are cocultured in vitro. Thus, a better understanding of P. aeruginosa-S. aureus interactions, particularly mechanisms by which these microorganisms are able to coexist in proximal physical space, will lead to better-informed treatments for chronic polymicrobial infections.
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive disorder with significant morbidity and mortality, affecting >70,000 people worldwide (1, 2). Decreased function of the cystic fibrosis transmembrane regulator (CFTR), an epithelial cell chloride ion transporter, causes increased viscosity of airway surface liquid and decreased mucociliary clearance of microorganisms in the CF airway (3, 4). Over time, these microorganisms form complex, polymicrobial communities, which are highly tolerant to antibiotic treatment; the airway damage due to these chronic infections eventually leads to respiratory failure (5–15). The development of novel CFTR-targeted therapeutics has markedly improved patient survival (2, 16–19), but despite these recent advances, the progression of CF pulmonary disease is one of chronic infection and inflammation punctuated by periods of clinical exacerbation, which cause irreversible damage to lung tissue (2, 7, 20–22).
Pseudomonas aeruginosa and Staphylococcus aureus are the two opportunistic pathogens most commonly isolated from CF patients’ lungs (2), and coinfection with both microbes is common (23, 24). Coinfection with P. aeruginosa and S. aureus alters antibiotic tolerance (25–28) and enhances virulence (29) in chronic infection. Furthermore, CF patients who are coinfected with both P. aeruginosa and S. aureus have poorer clinical outcomes than those who are monoinfected (23).
Studies of P. aeruginosa-S. aureus interactions have demonstrated that P. aeruginosa kills S. aureus in vitro, and this interaction is thought to contribute to the dominance of P. aeruginosa over S. aureus as CF patients age (30, 31). Considerable progress has been made in elucidating the mechanism of S. aureus killing mediated by P. aeruginosa (32–35). P. aeruginosa secretes quorum sensing-regulated antimicrobial exoproducts during acute infection, including 2-heptyl-4-hydroxyquinoline N-oxide (HQNO), siderophores, rhamnolipids, and phenazines. HQNO and the two P. aeruginosa siderophores pyoverdine and pyochelin have been shown to drive S. aureus toward a fermentative lifestyle (33), while rhamnolipids disrupt cell membrane integrity (36). Phenazines inhibit S. aureus metabolism and play roles in iron acquisition and biofilm development (37–39).
Chronic CF lung infection is marked by the emergence of mucoid P. aeruginosa isolates, which overproduce the exopolysaccharide alginate (40, 41). The overproduction of alginate by P. aeruginosa causes the transcriptional downregulation of HQNO, siderophores, and rhamnolipids, leading to decreased virulence toward S. aureus (32, 33, 42). The mechanism of alginate overproduction is typically due to a mutation in the mucA gene (43–45), a negative regulator of σ22 (AlgT/U) (43, 44, 46). The derepression of σ22 promotes the transcription of many genes, including the alginate biosynthetic operon. Disruption of algD, the first gene in the alginate biosynthetic operon, restores wild-type levels of antimicrobial exoproducts, demonstrating that alginate production is sufficient for antimicrobial exoproduct downregulation independent of other transcriptional effects of AlgT/U (32).
In this study, we asked how exogenous alginate influences the production of antistaphylococcal antimicrobial compounds by P. aeruginosa. We show that exogenous alginate is sufficient to protect S. aureus from P. aeruginosa in coculture, likely via the transcriptional downregulation of several genes whose products are required for P. aeruginosa to kill S. aureus. We also show that a three-way coculture between mucoid and nonmucoid P. aeruginosa strains and S. aureus results in attenuated killing of S. aureus, raising the possibility that the mechanism that we identified functions in vivo in patients with such mixed infections. Furthermore, the analysis of mucoid isolates of P. aeruginosa indicates that some isolates are able to kill S. aureus after prolonged coculture and that the mechanism of killing is due to diverse transcriptional changes, indicating a dynamic and evolving competition between these two microbes.
RESULTS
Exogenous alginate protects S. aureus in coculture with P. aeruginosa.
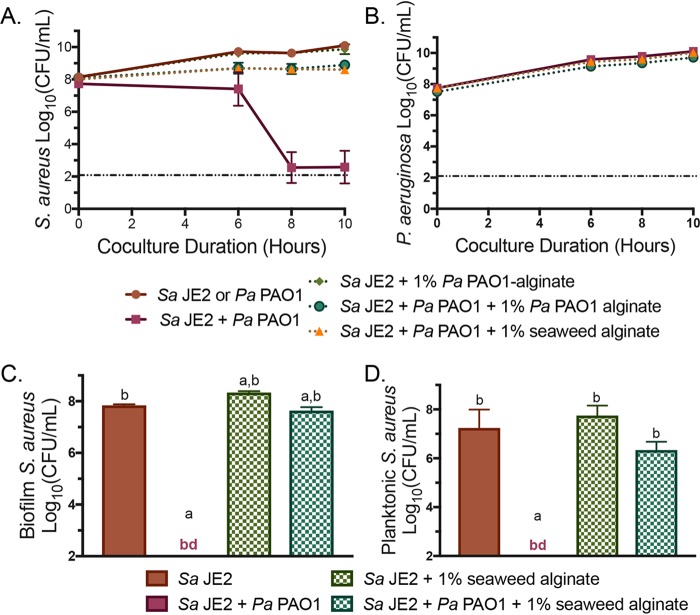
To determine whether the presence of exogenous alginate can promote coexistence between P. aeruginosa and S. aureus, we investigated a coculture with 1% seaweed-derived alginate (Fig. 1A and B; see also Fig. S1A and B in the supplemental material).
FIG 1.
Exogenous alginate protects S. aureus JE2 from P. aeruginosa PAO1 in coculture. (A and B) S. aureus (Sa) JE2 (A) and P. aeruginosa (Pa) PAO1 (B) growth curves over 10 h in liquid coculture in TSB with or without 1% seaweed-derived or 1% P. aeruginosa PAO1-derived alginate. CFU per milliliter were enumerated at the indicated time points and log10 transformed. The dashed line at 2 log10 CFU/ml indicates the limit of detection. (C and D) Biofilm (C) and planktonic (D) S. aureus JE2 growth after 16 h of static coculture in 1/2× MEM plus l-Gln and l-Arg with or without 1% seaweed-derived alginate. CFU per milliliter were enumerated and log10 transformed. Significance was determined by one-way ANOVA with Dunnett’s posttest. a, P < 0.05 with S. aureus JE2 as the reference; b, P < 0.0001 with P. aeruginosa PAO1 plus S. aureus JE2 as the reference; bd, below detection.
Because there are minor structural differences between seaweed-derived alginate and P. aeruginosa-produced alginate (46), with seaweed-derived alginate lacking acetylation, we also isolated alginate from the mucoid P. aeruginosa PAO1 mucA22 strain (see Materials and Methods) and added this P. aeruginosa PAO1-derived alginate to P. aeruginosa-S. aureus cocultures (Fig. 1A and B; Fig. S1A and B). Cultures were grown planktonically in tryptic soy broth (TSB) and plated onto Pseudomonas isolation agar (PIA) and mannitol salt agar (MSA) to quantify CFU of P. aeruginosa and S. aureus, respectively, over a 10-h coculture. Seaweed-derived and P. aeruginosa-derived alginate protected S. aureus equally well from killing by P. aeruginosa (Fig. 1A; Fig. S1A). One percent seaweed-derived alginate did not affect P. aeruginosa growth, while P. aeruginosa-derived alginate reduced the growth of P. aeruginosa by a small but statistically significant amount (Fig. 1B; Fig. S1B). We measured alginate production by two mucoid P. aeruginosa strains and determined that native levels of alginate production are ∼0.1% to 0.3% (wt/vol) under our culture conditions (Fig. S1C). We next expanded our coculture conditions to demonstrate that a range of exogenous alginate concentrations (0.25% to 2%) protected S. aureus from P. aeruginosa in this planktonic coculture model (Fig. S2 and Fig. S3A to D).
Bacterial interactions can change dramatically under different conditions, so we used a coculture model previously described by our laboratory to determine whether exogenous alginate also protects P. aeruginosa from S. aureus in a biofilm-based coculture model (25, 33). After an initial 1-h attachment phase followed by the removal of planktonic cells, cocultures of P. aeruginosa and S. aureus were incubated statically for 16 h, and CFU were then enumerated separately for the planktonic and biofilm fractions (see Materials and Methods). Biofilm coculture experiments were performed with P. aeruginosa PAO1 (Fig. 1C and D; Fig. S3E and F) and P. aeruginosa PA14 (Fig. S3G to J). S. aureus was killed by P. aeruginosa PAO1 in this biofilm model and protected from P. aeruginosa PAO1 with 1% exogenous alginate in both the biofilm (Fig. 1C) and planktonic (Fig. 1D) fractions. P. aeruginosa PAO1 growth was not affected by exogenous alginate in either fraction (Fig. S3E and F). Results were similar for P. aeruginosa PA14, except that exogenous alginate also modestly boosted P. aeruginosa PA14 growth (Fig. S3G to J). Experiments with similar results were also performed planktonically with the same minimal medium used for biofilm coculture experiments (Fig. S4). Taken together, these data indicate that exogenous alginate can modulate the killing of S. aureus by P. aeruginosa and that exogenous alginate alone is sufficient to protect S. aureus from P. aeruginosa under a variety of growth conditions.
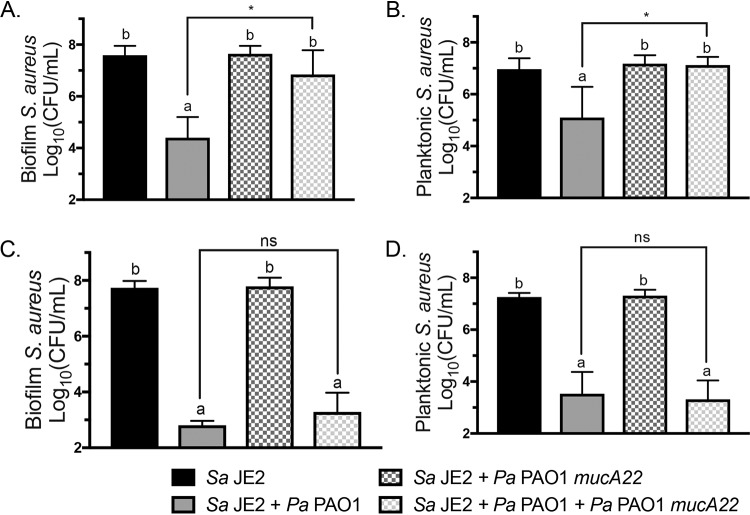
Triculture of S. aureus with wild-type and mucoid P. aeruginosa strains delays S. aureus killing.
Because exogenous alginate can modify interactions between P. aeruginosa and S. aureus, we hypothesized that mucoid P. aeruginosa might affect interactions between wild-type P. aeruginosa and S. aureus. Furthermore, we wanted to determine whether native levels of alginate production can impact nonmucoid P. aeruginosa interactions with S. aureus. We therefore designed a triculture experiment where S. aureus was either monocultured, cocultured with nonmucoid P. aeruginosa PAO1 or mucoid P. aeruginosa PAO1 mucA22, or cultured with both nonmucoid and mucoid P. aeruginosa strains (Fig. 2; Fig. S5). Each strain of P. aeruginosa was inoculated such that experiments with both mucoid and nonmucoid P. aeruginosa strains contained 2× the initial concentration of P. aeruginosa, to ensure that the total concentration of wild-type P. aeruginosa remained consistent across all conditions. S. aureus survival increased significantly in both the biofilm and planktonic fractions after 12 h (Fig. 2A and B) but not after 16 h (Fig. 2C and D) of triculture relative to coculture of S. aureus JE2 and nonmucoid P. aeruginosa PAO1. These results indicate that mucoid strains can impact interactions between nonmucoid P. aeruginosa and S. aureus by delaying S. aureus killing.
FIG 2.
Culture with mucoid P. aeruginosa PAO1 mucA22 delays S. aureus JE2 killing by wild-type P. aeruginosa PAO1. Shown are data for biofilm triculture on plastic with S. aureus JE2, P. aeruginosa PAO1, and P. aeruginosa PAO1 mucA22 in MEM plus l-Gln and l-Arg. CFU per milliliter were enumerated on mannitol salt agar (MSA) and log10 transformed. S. aureus JE2 viability in the biofilm (A) and planktonic (B) fractions after 12 h and S. aureus JE2 viability in the biofilm (C) and planktonic (D) fractions after 16 h of coculture were determined. Significance was determined by one-way ANOVA with Dunnett’s posttest. a, P < 0.05 with S. aureus JE2 as the reference; b, P < 0.05 with P. aeruginosa PAO1 plus S. aureus JE2 as the reference; *, P < 0.05; ns, not significant.
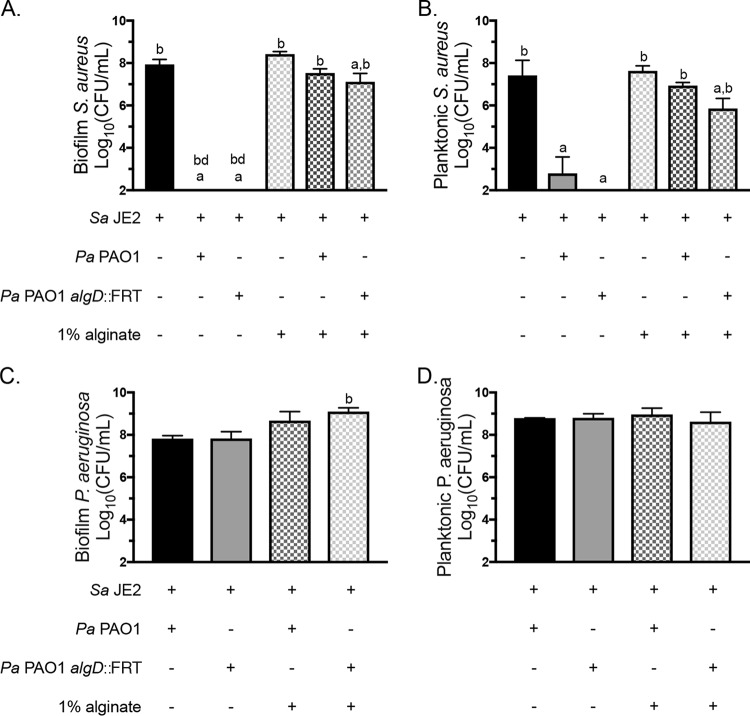
Alginate synthesis is not required for protection by exogenous alginate.
We hypothesized that exogenous alginate might protect S. aureus by stimulating P. aeruginosa to produce alginate and therefore decrease virulence toward S. aureus via known changes that accompany mucoidy (32). We used a P. aeruginosa strain that cannot synthesize alginate, P. aeruginosa PAO1 algD::FRT (flippase recognition target), to test whether the ability to self-produce alginate is required for S. aureus protection by exogenous alginate. Using the biofilm coculture model, we measured a significant increase in S. aureus survival in the presence of P. aeruginosa PAO1 algD::FRT with exogenous alginate in both the biofilm and planktonic fractions (Fig. 3A and B). The growth of P. aeruginosa PAO1 and P. aeruginosa PAO1 algD::FRT was nonsignificantly and significantly increased, respectively, by exogenous alginate, but in the biofilm fraction only (Fig. 3C and D). Therefore, the ability to self-produce alginate is not required to reduce virulence toward S. aureus in the presence of exogenous alginate.
FIG 3.
Alginate synthesis is not required for S. aureus protection. S. aureus JE2 was cocultured in 1/2× MEM plus l-Gln and l-Arg on plastic with P. aeruginosa PAO1 or P. aeruginosa PAO1 algD::FRT with or without 1% seaweed-derived alginate. (A and B) S. aureus JE2 viability in the biofilm (A) and planktonic (B) fractions after 16 h. (C and D) P. aeruginosa PAO1 viability in the biofilm (C) and planktonic (D) fractions after 16 h. Significance was determined by one-way ANOVA with Dunnett’s posttest. a, P < 0.05 with S. aureus JE2 as the reference; b, P < 0.05 with P. aeruginosa PAO1 plus S. aureus JE2 as the reference; bd, below detection.
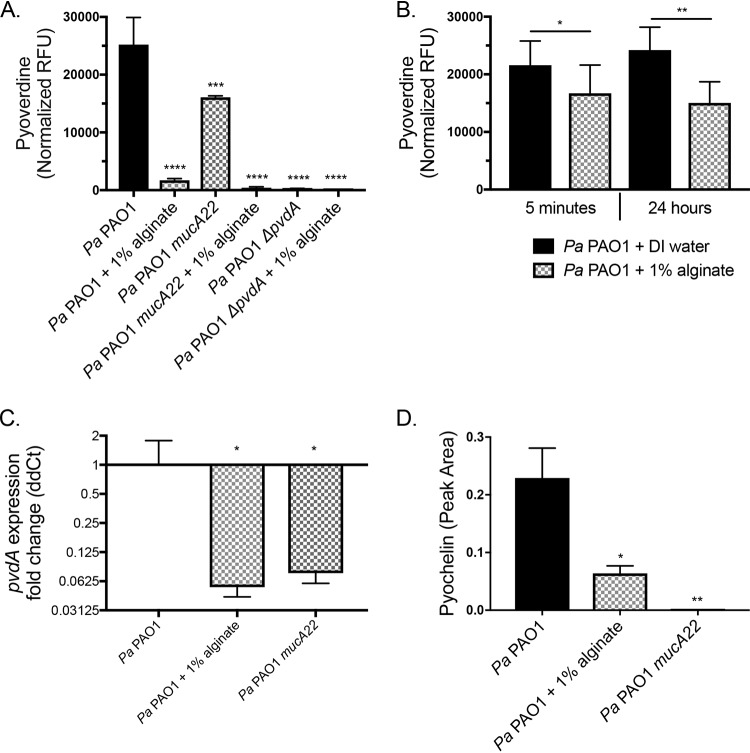
Exogenous alginate transcriptionally decreases siderophore production by P. aeruginosa.
S. aureus is protected from mucoid P. aeruginosa due to the transcriptional downregulation of genes in the mucoid strain of P. aeruginosa required to produce the iron chelator pyoverdine, rhamnolipid surfactant, and the redox-active molecule HQNO (32). Thus, we investigated whether exogenous alginate might function via a similar mechanism. Pyoverdine production was quantified by measuring relative fluorescence units (RFU) with excitation at 400 nm and absorbance at 460 nm of the supernatant from P. aeruginosa grown with or without 1% exogenous alginate. P. aeruginosa PAO1 mucA22 and P. aeruginosa PAO1 ΔpvdA were included as controls. Pyoverdine was decreased significantly when P. aeruginosa PAO1 was cultured with exogenous alginate (Fig. 4A), and this outcome was not altered by the presence of S. aureus (Fig. S6A) Interestingly, while mucoid P. aeruginosa PAO1 mucA22 produced significantly less pyoverdine than wild-type P. aeruginosa PAO1, the addition of exogenous alginate further decreased pyoverdine production by the mucoid strain.
FIG 4.
Exogenous alginate decreases P. aeruginosa PAO1 siderophore production. (A and B) P. aeruginosa PAO1 was grown on plastic in 1/2× MEM plus l-Gln and l-Arg with or without 1% alginate for 16 h at 37°C with 5% CO2. Supernatants were collected from the planktonic fraction. (A) Pyoverdine was quantified by measuring RFU of the supernatants at 400-nm excitation and 460-nm emission wavelengths and normalizing the values to CFU per milliliter of the planktonic fraction. (B) Supernatants were diluted 1/2-fold in DI water or 2% alginate (for a final concentration of 1% alginate) and incubated statically at 37°C with 5% CO2 for 5 min or 24 h. Pyoverdine was quantified by measuring RFU of the supernatants at 400-nm excitation and 460-nm emission wavelengths. Significance was determined by a paired t test. (C and D) P. aeruginosa PAO1 was grown in 25 ml TSB with shaking for 8 h. (C) pvdA expression was quantified by qRT-PCR, and the ΔΔCT value was calculated relative to P. aeruginosa PAO1 rpoD expression. (D) Pyochelin was quantified by LC-MS/MS as described in Materials and Methods. Significance determined by one-way ANOVA with Dunnett’s posttest comparison to P. aeruginosa PAO1 for each experiment unless otherwise indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (for all statistical tests).
We performed a quenching experiment to test whether exogenous alginate can absorb pyoverdine from the medium or mask its signal in our assay. P. aeruginosa PAO1 supernatants were diluted 1/2-fold with deionized (DI) water or 2% exogenous alginate, for a final concentration of 1% alginate. Pyoverdine was quantified after additional 5-min and 24-h incubations at 37°C. There was a small but significant and repeatable decrease in total pyoverdine measured when supernatants were diluted in alginate versus water (Fig. 4B), indicating that alginate may absorb a small portion of the pyoverdine present or partially mask its signal in our assay, but this effect does not explain the robust decrease in pyoverdine observed in Fig. 4A. Additionally, we measured the concentration of iron in alginate to determine whether alginate was introducing an exogenous iron source (Fig. S6B). The 1% seaweed-derived alginate increases iron concentrations ∼9 to 10 μM above the baseline level in the medium, which may contribute to the strong decrease of pyoverdine production in the presence of exogenous alginate.
To test whether pyoverdine production is reduced at the transcriptional level by exogenous alginate, we cultured P. aeruginosa PAO1 in the presence and absence of exogenous alginate and measured the transcription of pvdA, a gene essential for pyoverdine production, by reverse transcription-quantitative PCR (qRT-PCR). The mucoid strain P. aeruginosa PAO1 mucA22 was used as a control, as this strain transcriptionally downregulates pvdA (32). The expression of pvdA was significantly reduced when P. aeruginosa PAO1 was grown with 1% exogenous alginate (Fig. 4C), and this outcome was not altered by the presence of S. aureus (Fig. S6C). Consistent with previously reported data (32), pvdA expression was also decreased in the mucoid strain carrying the mucA22 mutation. When the level of pyoverdine production is low, P. aeruginosa will typically increase the production of pyochelin, the other major P. aeruginosa siderophore. We therefore used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to quantify pyochelin. P. aeruginosa PAO1 also reduced pyochelin production upon exposure to exogenous alginate (Fig. 4D). Taken together, these results indicate that exogenous alginate reduces the production of both P. aeruginosa siderophores pyoverdine and pyochelin, likely via mechanisms including transcriptional downregulation, alginate acting as a source of iron, and/or partial sequestration of siderophores that are produced.
Exogenous alginate posttranscriptionally decreases rhamnolipid production by P. aeruginosa.
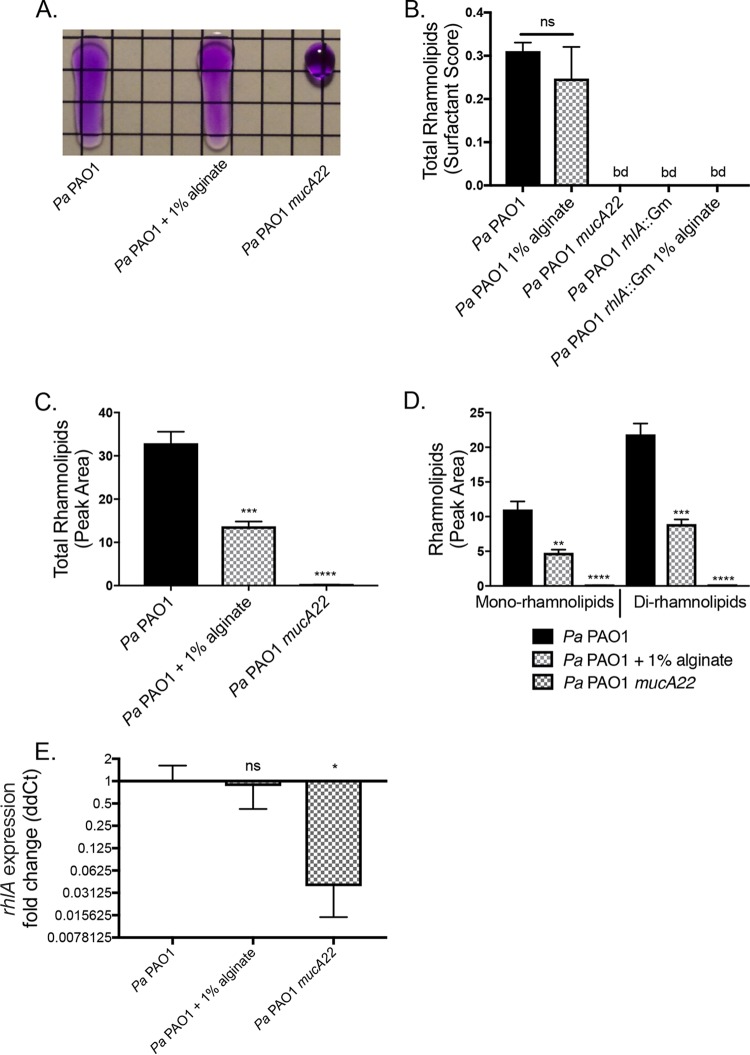
Rhamnolipids are surfactants produced by P. aeruginosa that intercalate into the cytoplasmic membrane of S. aureus (47). Surfactants disrupt the surface tension of liquids, a characteristic that we used to quantify rhamnolipid production by P. aeruginosa using the drop collapse assay (Fig. 5A). We observed a minor, nonsignificant decrease in total rhamnolipid production by wild-type P. aeruginosa PAO1 in the presence of 1% exogenous alginate. The rhamnolipid-deficient strain P. aeruginosa PAO1 rhlA::Gm was not affected by the presence of exogenous alginate (Fig. 5B), and the presence of S. aureus did not significantly alter the outcome of this assay (Fig. S6D).
FIG 5.
Rhamnolipid production is posttranscriptionally altered by exogenous alginate. P. aeruginosa PAO1 was grown in TSB liquid culture for 8 h with or without 1% alginate. Supernatants were collected by centrifugation to remove cell debris and sterile filtering. Cell pellets were snap-frozen for subsequent expression analyses. (A) Representative drop collapse assay image. P. aeruginosa PAO1 supernatants were prepared from mucoid and nonmucoid P. aeruginosa PAO1 strains and serially diluted 1:2 in PBS. Surfactant activity was assessed by placing a 20-μl droplet of each supernatant dilution on plastic, placing the droplet at a 90° angle for 10 s, and assessing migration (PBS was supplemented with 0.01% crystal violet to aid visualization). Surfactant activity was quantified as the reciprocal of the highest dilution at which the drop migrates. (B) Rhamnolipid production by P. aeruginosa PAO1 quantified by drop collapse and normalized to CFU per milliliter to determine the surfactant score. (C and D) Rhamnolipid quantification by LC-MS/MS for total rhamnolipids (C) and mono- and dirhamnolipids (D). (E) Expression of the rhlA gene was quantified by qRT-PCR, and the ΔΔCT value was calculated relative to P. aeruginosa PAO1 rpoD. Significance was determined for each experiment by one-way ANOVA with Dunnett’s posttest comparison to P. aeruginosa PAO1. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Because the drop collapse assay is an estimate of total rhamnolipids, we also used LC-MS/MS to quantify total and specific rhamnolipids in P. aeruginosa PAO1 supernatants. The overall rhamnolipid quantity significantly decreased by ∼50% with the addition of exogenous alginate when measured by this more sensitive method (Fig. 5C). The fold change decreases were similar for both mono- and dirhamnolipids (Fig. 5D). To determine whether the reduced rhamnolipid production was due to transcriptional regulation, we measured the relative expression levels of rhlA for P. aeruginosa PAO1 with or without 1% alginate and P. aeruginosa PAO1 mucA22. The levels of transcription of rhlA were not significantly different for P. aeruginosa PAO1 with and without 1% exogenous alginate (Fig. 5E), and the presence of S. aureus did not alter this outcome (Fig. S6E).
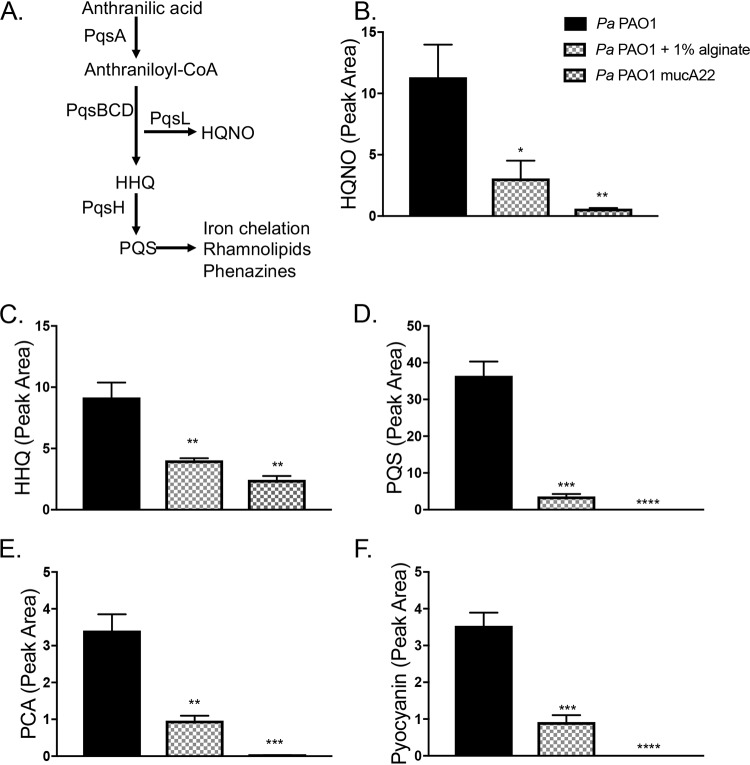
Exogenous alginate downregulates PQS quorum sensing.
The PQS (Pseudomonas quinolone signal) (2-heptyl-3,4-dihydroxyquinoline) quorum sensing system, which impacts a range of P. aeruginosa virulence factors (Fig. 6A), is known to be downregulated by mucoid P. aeruginosa strains (32, 48). We therefore measured the impact of exogenous alginate on several products regulated by the PQS quorum sensing system, with P. aeruginosa mucA22 as a control. Both self-produced and exogenous alginates decrease the production of several key PQS-regulated products, including HQNO, HHQ (4-hydroxy-2-heptylquinoline), and PQS (Fig. 6B to D), as well as downstream factors such as the phenazines PCA (phenazine-1-carboxylic acid) and pyocyanin (Fig. 6E and F).
FIG 6.
Exogenous alginate downregulates PQS quorum sensing. (A) The PQS quorum sensing regulon. PQS synthesis is dependent on the pqsABCD genes located in the pqs operon. Both HHQ and HQNO have direct antimicrobial properties, while PQS is the ligand for PqsR/MvfR. When the PQS level is high, this ligand interacts with PqsR/MvfR to positively regulate many downstream virulence factors. PQS has a direct iron-chelating function and promotes the expression of siderophore-encoding genes. The downstream effects listed here focus on effects relevant to this study and are not exhaustive. (B to F) LC-MS/MS was used to quantify HQNO (B), HHQ (C), PQS (D), PCA (E), and pyocyanin (F) produced by P. aeruginosa. Panel B includes the key for all graphs in this figure. Significance was determined for each experiment by one-way ANOVA with Dunnett’s posttest comparison to P. aeruginosa PAO1. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Expression profiles of P. aeruginosa in the presence of exogenous alginate.
To determine how exogenous alginate impacts P. aeruginosa gene expression more generally, we used the Nanostring PAV2 code set (49) to compare P. aeruginosa gene expression levels in the presence and absence of exogenous alginate and S. aureus. Nansotring is a digital multiplexed technology for direct quantification of RNA transcripts. The PAV2 code set used in this study contains probes for 75 transcripts associated with P. aeruginosa genes known or suspected to be expressed in the CF airway (49). P. aeruginosa PAO1 and P. aeruginosa PA14 were cultured to mid-log phase and then subcultured into fresh medium with or without 1% alginate. Samples were collected after an additional 45 min of growth, and expression was analyzed by using Nanostring according to the manufacturer’s protocols.
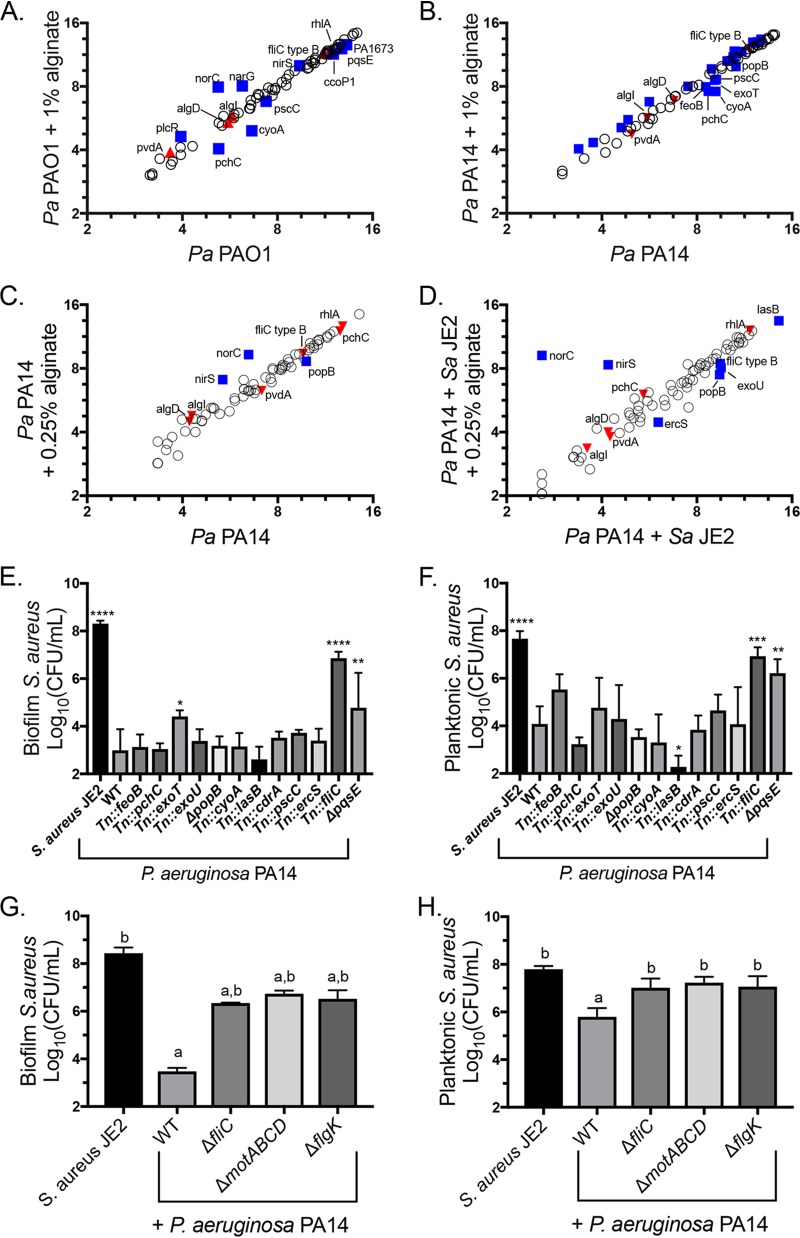
Ten genes were significantly differentially regulated by P. aeruginosa PAO1 in the presence of exogenous alginate (Fig. 7A). We previously used the same Nanostring code set to compare mucoid and nonmucoid P. aeruginosa PAO1 gene expression levels and measured a significant downregulation of pvdA, rhlA, pchC, fliC type B, and norC as well as a significant upregulation of algI and algD. Of these genes, only pchC is significantly differentially regulated in the same direction across both studies, indicating that transcriptional changes due to exogenous alginate are not broadly similar to the effects of self-produced alginate. We observed an overlap in significant transcriptional changes in response to exogenous alginate between P. aeruginosa PA14 and P. aeruginosa PAO1 for five genes: cyoA, narG, norC, pchC, and pscC (compare Fig. 7A and B). P. aeruginosa PA14 also had a large number of genes with small but significant transcriptional upregulation in the presence of exogenous alginate (Tables S1 and S2).
FIG 7.
Alginate alters the expression of P. aeruginosa genes essential for S. aureus killing. (A to D) Raw Nanostring counts were normalized to values for the positive controls and three housekeeping genes (rpoD, ppiD, and fbp) and log2 transformed. Three biological replicates were performed under each condition. Significantly differentially expressed genes were determined by an unpaired t test followed by the two-stage linear step-up procedure of Benjamini et al. (57) (with a q value of 1% for false discovery) and are marked by blue squares and labeled with the gene name. Genes significantly differentially expressed by mucoid P. aeruginosa in a previous study (32) but not by P. aeruginosa in the presence of exogenous alginate are marked by red triangles. (A and B) P. aeruginosa PAO1 (A) and P. aeruginosa PA14 (B) subcultured into TSB or TSB plus 1% alginate during the mid-log growth phase for 45 min. For clarity, only significantly downregulated genes are labeled in panel B. (C and D) P. aeruginosa PA14 monocultured with 0.25% alginate for 8 h (C) and cocultured with S. aureus JE2 in TSB with 0.25% alginate for 8 h (D). (E and F) S. aureus JE2 survival in the biofilm (E) and planktonic (F) fractions after 16 h of coculture with the indicated P. aeruginosa PA14 mutants corresponding to genes downregulated in one or more Nanostring experiments. Significance was determined by one-way ANOVA with Dunnett’s posttest comparison to S. aureus plus P. aeruginosa PA14. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (G and H) S. aureus JE2 survival in the biofilm (G) and planktonic (H) fractions after 16 h of coculture with P. aeruginosa PA14 motility mutants. Significance was determined by one-way ANOVA with Dunnett’s posttest. a, P < 0.05 with S. aureus JE2 as the reference; b, P < 0.05 with P. aeruginosa PAO1 plus S. aureus JE2 as the reference. WT, wild type.
To understand how long exposure to exogenous alginate and the presence of S. aureus affect P. aeruginosa transcription, we cultured P. aeruginosa PA14 for 8 h with or without 0.25% alginate and with or without S. aureus. Nanostring counts were normalized and analyzed as described above (Tables S3 and S4). Only three genes were significantly differentially expressed by P. aeruginosa PA14 relative to P. aeruginosa PA14 with 0.25% alginate (nirS, norC, and popB) (Fig. 7C). When P. aeruginosa PA14 was cocultured with S. aureus, the same three genes had altered expression levels, as did ercS, exoU, fliC type B, and lasB (Fig. 7D). Notably, fliC type B is also downregulated by mucoid P. aeruginosa relative to wild-type P. aeruginosa (32). The significantly differentially regulated genes that we identified had two distinct patterns of gene regulation. For fliC type B, lasB, exoU, and popB, the presence of alginate and S. aureus had additive effects (Fig. S7A and Table S4). For nirS, norC, cdrA, and ercS, the effects of exogenous alginate masked the effects of S. aureus. The data set was further analyzed by clustering and principal-component analysis, with results demonstrating that S. aureus has a larger effect on overall gene expression than exogenous alginate (Fig. S8A and B). This difference is driven by a strong impact of S. aureus on genes related to iron acquisition, including pvdA (Fig. S9A and B; Table S4).
We hypothesized that some of the genes transcriptionally downregulated by P. aeruginosa in the presence of exogenous alginate might contribute to S. aureus protection in coculture. We focused on downregulated genes because the Nanostring code set genes are related to virulence and pathogenesis, making it unlikely that the upregulation of any of these genes would positively impact S. aureus survival. We utilized a P. aeruginosa PA14 nonredundant transposon mutant library (50) to test whether any of these factors had a direct impact on S. aureus survival. We performed a biofilm coculture experiment for P. aeruginosa PA14 mutants of each gene identified as being significantly downregulated in any Nanostring experiment and found that disruption of exoT, fliC, or pqsE significantly enhanced S. aureus survival in the biofilm fraction in the presence of P. aeruginosa PA14 (Fig. 7E). The fliC and pqsE mutants also enhanced S. aureus survival in the planktonic fraction (Fig. 7F). Only the fliC mutation altered P. aeruginosa PA14 viable counts, which were slightly but significantly increased in the biofilm fraction (Fig. S10A and B).
Because fliC mutants do not produce flagella, we suspected that functional flagella might be required for S. aureus killing. We therefore tested flgK and motABCD mutants in the biofilm coculture model and saw that these mutations also prevented S. aureus killing by P. aeruginosa PA14 (Fig. 7G and H) without disrupting P. aeruginosa PA14 growth (Fig. S10C and D). Because flgK mutants are missing the flagellar cap and motABCD mutants produce a nonfunctional flagellum, these findings indicate that flagellar function is required for the full virulence of P. aeruginosa toward S. aureus. Thus, both self-produced and exogenous alginates likely reduce P. aeruginosa-mediated virulence toward S. aureus, at least in part, by downregulating flagellar genes.
Characterization of mucoid clinical isolates for the ability to kill S. aureus in coculture.
Our previous work (23, 32) demonstrated that most mucoid P. aeruginosa strains do not kill S. aureus when cocultured in vitro. Interestingly, other previous work reported by our laboratory indicated that at least one mucoid clinical isolate, P. aeruginosa FRD1, can kill S. aureus in coculture (33). To determine whether other mucoid clinical isolates could kill S. aureus during in vitro coculture, in part to understand the basis by which these “mucoid killers” could function, we selected a panel of mucoid clinical isolates identified in previous studies (32) to coculture with S. aureus.
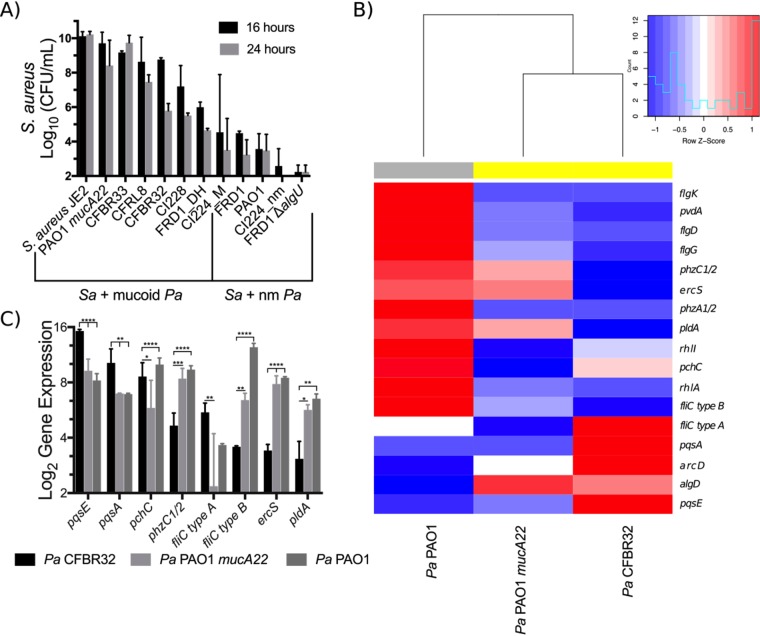
We cocultured P. aeruginosa clinical isolates with S. aureus JE2 and enumerated viable cells of S. aureus out to 24 h of coculture due to the slow growth of some of the clinical isolates relative to laboratory strains. The ability of mucoid clinical isolates to kill S. aureus in coculture varies by strain (Fig. 8A), with some strains such as FRD1 killing robustly and others such as CFBRPA33 showing no killing of S. aureus. Mucoidy still provides some protection from S. aureus even at late time points, as nonmucoid revertants for FRD1 and Cl2224 killed S. aureus more efficiently than the mucoid strains at 16 and 24 h. We quantified reversion rates for P. aeruginosa FRD1 and P. aeruginosa CFBRPA32 to determine whether killing by these strains is due to high rates of reversion to nonmucoid phenotypes and observed reversion rates of <0.1% after 24 h of culture (Table S5).
FIG 8.
Mucoid clinical isolates have various effects on S. aureus in coculture. (A) S. aureus JE2 viable counts after 16 and 24 h of coculture in flasks with shaking at 225 rpm in TSB with P. aeruginosa clinical isolates. nm, nonmucoid. (B and C) Log2 transformation of Nanostring counts normalized to values for positive controls and three housekeeping genes (rpoD, ppiD, and fbp) for the indicated transcripts for the clinical isolate P. aeruginosa CFBRPA32 (a “mucoid killer”), mucoid laboratory strain P. aeruginosa PAO1 mucA22, and wild-type laboratory strain P. aeruginosa PAO1 after 24 h of culture in flasks with shaking at 225 rpm in TSB. Data are from two biological replicates per strain. Gene expression was analyzed by two-way ANOVA followed by Tukey’s multiple-comparison test. (B) Heat map and dendrogram of all genes significantly differentially regulated between any two strains. Expression values are displayed as within-row z-scores. Yellow indicates mucoid strains, and gray indicates nonmucoid strains. (C) All genes significantly differentially regulated between P. aeruginosa CFBRPA32 and P. aeruginosa PAO1 mucA22. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We used the Nanostring code set described above to generate expression data for P. aeruginosa PAO1, CFBRPA32 (a mucoid killer), and the mucoid laboratory strain P. aeruginosa PAO1 mucA22 after 24 h of growth. Nanostring counts were normalized as described above and then analyzed by two-way analysis of variance (ANOVA) followed by Tukey’s posttest among all three strains (Tables S6 and S7). Of the 75 transcripts measured, 17 genes were significantly differentially regulated between at least one pair of strains (Fig. 8B).
Clustering by the expression of differentially regulated genes revealed that the expression profile of CFBRPA32 is more similar to that of the mucoid laboratory strain P. aeruginosa PAO1 mucA22 than to that of nonmucoid P. aeruginosa PAO1 (Fig. 8B). However, eight transcripts were significantly differentially regulated between P. aeruginosa CFBRPA32 and P. aeruginosa PAO1 mucA22: pqsE, pqsA, pchC, phzC1-phzC2, fliC type A, fliC type B, ercS, and pldA (Fig. 8C). The increased expression levels of pqsE, pqsA, and pchC indicate a potential role for both quorum sensing and iron acquisition in the ability of CFBRPA32 to kill S. aureus. The CFBRPA32 pqsE expression level is >50-fold higher than that of P. aeruginosa PAO1 mucA22 and >125-fold higher than that of P. aeruginosa PAO1. Because pqsE is essential for P. aeruginosa killing of S. aureus, high expression levels may contribute to the ability of P. aeruginosa CFBRPA32 to kill S. aureus at late time points.
We chose to perform our Nanostring experiment with CFBRPA32 because there are publicly available Nanostring data (49) for several other mucoid clinical isolates that we examined (CFRL8, CI2224, CI228, and FRD1). We reanalyzed these data to determine whether we could identify an expression pattern shared between mucoid killers (Fig. S11). However, there was no individual gene other than algD that correlated with S. aureus survival. Taken together, our data indicate that mucoid P. aeruginosa strains with the ability to kill S. aureus in coculture likely do so via strain-specific mechanisms, and furthermore, the functions of some of the genes identified in our analysis of mucoid killers overlap functions downregulated by exogenous alginate.
DISCUSSION
P. aeruginosa and S. aureus coexist within the CF lung, where a heterogeneous array of microorganisms, the host immune system, and nutrient availability all contribute to a complex and dynamic environment (7, 12, 31). Here, we demonstrate that alginate, which is known to impact both host-pathogen and pathogen-pathogen interactions, also indirectly impacts pathogen-pathogen interactions (51, 52). Protection of S. aureus from P. aeruginosa by exogenous alginate is consistent across a variety of nutritional and physiological contexts, given that protection occurs in liquid and biofilm modes of growth and in both rich and minimal media with two strains of P. aeruginosa (PAO1 and PA14) (Fig. 1; see also Fig. S1 to S4 in the supplemental material). Furthermore, alginate production by a mucoid strain of P. aeruginosa is sufficient to delay S. aureus killing by a nonmucoid strain of P. aeruginosa when all three strains are cultured together (Fig. 2; Fig. S5), demonstrating the potential for mucoid strains to alter interactions between nonmucoid P. aeruginosa and S. aureus strains in vivo.
While P. aeruginosa robustly kills S. aureus in vitro, it has been demonstrated that a reduction of any single virulence factor will greatly diminish the ability of P. aeruginosa to kill S. aureus. We demonstrate that the production of P. aeruginosa siderophores, pyoverdine and pyochelin, is decreased when P. aeruginosa is cultured in exogenous alginate and that this decrease is due at least in part to the transcriptional downregulation of pvdA (Fig. 4). Exogenous alginate, like self-produced alginate, causes the transcriptional downregulation of pvdA expression, indicating that self-produced alginate may also lead to the decreased expression of this gene via an outside-in signaling mechanism. There is precedence for exogenous exopolysaccharides regulating transcription, as the exopolysaccharide Psl is known to act as an extracellular signal to increase biofilm formation by stimulating c-di-GMP synthesis, although the mechanism is still unknown (53). Notably, the decrease in siderophore production may be due in part to the presence of iron in seaweed-isolated alginate (Fig. S6B). An additional factor confounding the role of siderophores in our studies is that S. aureus itself also has a large impact on the expression of iron acquisition genes, as seen by the strong induction of brfB and repression of pvdA, pchC, and phuR by S. aureus (Fig. S9B).
We also demonstrate a modest decrease in total rhamnolipids across multiple culture methods, which is not dependent on transcriptional regulation (Fig. 5). However, it is unclear whether this modest rhamnolipid decrease is enough to affect S. aureus survival. Exogenous alginate also interferes with P. aeruginosa quorum sensing, as demonstrated by the decreases in the levels of HQNO, alkylquinolones, and phenazines (Fig. 6). The combined interference with HQNO, siderophore, and phenazine production likely contributes to S. aureus survival in the presence of exogenous alginate. Finally, we demonstrate that a functional flagellum is required for S. aureus killing. While flagella are considered virulence factors in the host-pathogen context, their role in P. aeruginosa-S. aureus interactions has only recently been demonstrated (54).
Together, our findings demonstrate that exogenous alginate has the capacity, at least in part, to affect P. aeruginosa strains in a manner analogous to that of self-produced alginate, that is, by reducing the production of antistaphylococcal factors. An interesting question that remains is whether the response of P. aeruginosa to exogenous alginate is specific; it is possible that the mechanism by which P. aeruginosa senses and responds to alginate is due, for example, to increased viscosity. Alternatively, alginate could be enforcing spatial structure or influencing S. aureus gene expression.
Interestingly, the presence of alginate can be overcome in some mucoid clinical isolates through unique rewiring of gene expression (Fig. 8), with the apparent increased expression of genes coding for one or more key antistaphylococcal factors. Thus, endogenous and exogenous alginates seem to impact an overlapping set of functions that modulate the interaction between P. aeruginosa and S. aureus. Our observations are not entirely surprising in the context of the evolving nature of both P. aeruginosa and S. aureus within the CF lung. Finally, our study highlights the importance of the physical environment within the CF lung in the outcomes of polymicrobial interactions and the need for further studies examining how different specific aspects of the CF lung environment can impact both polymicrobial interactions and overall patient outcomes.
MATERIALS AND METHODS
Detailed protocols and strains used in this study are available in the supplemental material.
Media.
All experiments were performed in tryptic soy broth (TSB) or minimal essential medium (MEM). MEM was supplemented with 2 mM l-glutamine (l-Gln) with or without 0.4% l-arginine (l-Arg). For experiments with alginate, MEM was diluted 1/2-fold with sterile DI water or alginate. Seaweed alginate was prepared by dissolving alginic acid sodium salt (Sigma) in DI water and autoclaving to sterilize.
Cocultures.
Cultures grown overnight were diluted to a final optical density (OD) of 0.05 and inoculated at a 1:1 ratio unless otherwise indicated. For experiments in MEM, strains were centrifuged and resuspended in MEM plus l-Gln prior to dilution. Strains in flasks were grown with shaking at 225 rpm at 37°C, and tubes were placed on a culture wheel at 37°C. A 100-μl total volume was inoculated in a plastic 96-well plate for biofilm cultures. At the indicated time points, culture samples were serially diluted in phosphate-buffered saline (PBS) and plated onto Pseudomonas isolation agar (PIA) or mannitol salt agar (MSA) to quantify viable P. aeruginosa and S. aureus bacteria, respectively.
Alginate preparation and quantification.
P. aeruginosa PAO1 mucA22 was cultured in 25 ml TSB overnight with shaking at 225 rpm at 37°C, and alginate was isolated as previously described (32), with minor modifications (detailed in the supplemental material). The concentration of alginate in solution was determined by the carbazole method described previously by Knutson and Jeanes (55).
Siderophore quantification.
The relative fluorescence of supernatants was quantified at 400-nm excitation and 460-nm emission wavelengths. Measurements were normalized to CFU per milliliter.
Drop collapse assay.
Supernatants were serially diluted 1/2-fold in PBS and were scored as the reciprocal of the highest dilution at which the drop collapsed. Scores were normalized to CFU per milliliter.
qRT-PCR.
Cell cultures were pelleted (centrifugation for 2 min at 14,000 rpm at 4°C) and immediately frozen in ethanol cooled with dry ice. RNA was isolated with TRIzol (Zymo Research) as described by the manufacturer. DNA was removed by three sequential treatments with Turbo DNase (Invitrogen), cDNA was prepared (RevertAid first-strand cDNA synthesis kit), and qRT-PCR was performed with SsoFast EvaGreen supermix (Bio-Rad).
LC-MS/MS.
Detailed descriptions of LC-MS/MS supernatant extraction, analysis, feature finding, and molecular annotation are available in the supplemental material.
Nanostring.
Total RNA was prepared as described above for qRT-PCR. The PAV2 code set (49) was incubated with total RNA according to the manufacturer’s protocol.
Iron quantification.
Alginate isolated from P. aeruginosa PAO1 mucA22 was lyophilized, and iron was quantified by inductively coupled plasma mass spectrometry (ICP-MS) following nitric acid digestion of organic material according to the method described previously by Heck et al. (56).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R37 AI83256-06 to G.A.O. and T32AI007363 to C.E.P. Additional support was provided by the CF-Research Development Program (STANTO07R0), DartCF, and the BBC (P30-DK117469). The Phelan lab is supported by the ALSAM Foundation (L. S. Skaggs Professorship and Therapeutic Innovation Award) and the NIH (R35 GM128690-01).
We thank D. A. Hogan for providing access to clinical isolates, A. E. Baker for PA14 ΔfliC, and Tom Hampton for providing advice on expression data analysis. Clinical CF isolates CFBRPA32 and CFBRPA33 were provided by the CF Biospecimen Registry at the Children’s Healthcare of Atlanta and Emory University CF Discovery Core.
Footnotes
Supplemental material is available online only.
For a commentary on this article, see https://doi.org/10.1128/JB.00040-20.
REFERENCES
- 1.Cutting GR. 2015. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cystic Fibrosis Foundation. 2017. Patient registry annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 3.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Kurbatova P, Bessonov N, Volpert V, Tiddens HAWM, Cornu C, Nony P, Caudri D, CRESim Working Group. 2015. Model of mucociliary clearance in cystic fibrosis lungs. J Theor Biol 372:81–88. doi: 10.1016/j.jtbi.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 5.Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD. 2003. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 41:3548–3558. doi: 10.1128/jcm.41.8.3548-3558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bittar F, Richet H, Dubus J-C, Reynaud-Gaubert M, Stremler N, Sarles J, Raoult D, Rolain J-M. 2008. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS One 3:e2908. doi: 10.1371/journal.pone.0002908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filkins LM, O’Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther 21:595–599. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, Sogin ML, Czum J, Ashare A. 2016. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Koff EM, de Winter-de Groot KM, Bogaert D. 2016. Development of the respiratory tract microbiota in cystic fibrosis. Curr Opin Pulm Med 22:623–628. doi: 10.1097/MCP.0000000000000316. [DOI] [PubMed] [Google Scholar]
- 13.Leclair LW, Hogan DA. 2010. Mixed bacterial-fungal infections in the CF respiratory tract. Med Mycol 48:S125–S132. doi: 10.3109/13693786.2010.521522. [DOI] [PubMed] [Google Scholar]
- 14.Lynch SV, Bruce KD. 2013. The cystic fibrosis airway microbiome. Cold Spring Harb Perspect Med 3:a009738. doi: 10.1101/cshperspect.a009738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sibley CD, Rabin H, Surette MG. 2006. Cystic fibrosis: a polymicrobial infectious disease. Future Microbiol 1:53–61. doi: 10.2217/17460913.1.1.53. [DOI] [PubMed] [Google Scholar]
- 16.Mogayzel PJ, Naureckas ET, Robinson KA, Mueller G, Hadjiliadis D, Hoag JB, Lubsch L, Hazle L, Sabadosa K, Marshall B, Pulmonary Clinical Practice Guidelines Committee. 2013. Cystic fibrosis pulmonary guidelines. Chronic medications for maintenance of lung health. Am J Respir Crit Care Med 187:680–689. doi: 10.1164/rccm.201207-1160oe. [DOI] [PubMed] [Google Scholar]
- 17.Wainwright CE, Elborn JS, Ramsey BW. 2015. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med 373:1783–1784. doi: 10.1056/NEJMc1510466. [DOI] [PubMed] [Google Scholar]
- 18.Van Goor F, Hadida S, Grootenhuis PDJ, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A, Zhou J, McCartney J, Arumugam V, Decker C, Yang J, Young C, Olson ER, Wine JJ, Frizzell RA, Ashlock M, Negulescu P. 2009. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci U S A 106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiman L, Siegel JD, LiPuma JJ, Brown RF, Bryson EA, Chambers MJ, Downer VS, Fliege J, Hazle LA, Jain M, Marshall BC, O’Malley C, Pattee SR, Potter-Bynoe G, Reid S, Robinson KA, Sabadosa KA, Schmidt HJ, Tullis E, Webber J, Weber DJ, Cystic Fibrous Foundation, Society for Healthcare Epidemiology of America. 2014. Infection prevention and control guideline for cystic fibrosis: 2013 update. Infect Control Hosp Epidemiol 35:S1–S67. doi: 10.1086/676882. [DOI] [PubMed] [Google Scholar]
- 20.Sanders DB, Bittner RCL, Rosenfeld M, Redding GJ, Goss CH. 2011. Pulmonary exacerbations are associated with subsequent FEV1 decline in both adults and children with cystic fibrosis. Pediatr Pulmonol 46:393–400. doi: 10.1002/ppul.21374. [DOI] [PubMed] [Google Scholar]
- 21.Sanders DB, Bittner RCL, Rosenfeld M, Hoffman LR, Redding GJ, Goss CH. 2010. Failure to recover to baseline pulmonary function after cystic fibrosis pulmonary exacerbation. Am J Respir Crit Care Med 182:627–632. doi: 10.1164/rccm.200909-1421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Sullivan BP, Freedman SD. 2009. Cystic fibrosis. Lancet 373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 23.Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 35:947–953. doi: 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 24.Limoli DH, Hoffman LR. 2019. Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 74:684–692. doi: 10.1136/thoraxjnl-2018-212616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orazi G, O’Toole GA. 2017. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8:e00873-17. doi: 10.1128/mBio.00873-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. 2014. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaudoin T, Yau YCW, Stapleton PJ, Gong Y, Wang PW, Guttman DS, Waters V. 2017. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 3:25. doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP, Gode CJ, Lachiewicz AM, Wolfgang MC, Conlon BP. 2017. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol 15:e2003981. doi: 10.1371/journal.pbio.2003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alves PM, Al-Badi E, Withycombe C, Jones PM, Purdy KJ, Maddocks SE. 2018. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog Dis 76:fty003. doi: 10.1093/femspd/fty003. [DOI] [PubMed] [Google Scholar]
- 30.Pernet E, Guillemot L, Burgel P-R, Martin C, Lambeau G, Sermet-Gaudelus I, Sands D, Leduc D, Morand PC, Jeammet L, Chignard M, Wu Y, Touqui L. 2014. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat Commun 5:5105. doi: 10.1038/ncomms6105. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen AT, Oglesby-Sherrouse AG. 2016. Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl Microbiol Biotechnol 100:6141–6148. doi: 10.1007/s00253-016-7596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O’Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rüger M, Ackermann M, Reichl U. 2014. Species-specific viability analysis of Pseudomonas aeruginosa, Burkholderia cepacia and Staphylococcus aureus in mixed culture by flow cytometry. BMC Microbiol 14:56. doi: 10.1186/1471-2180-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. 2015. Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J Bacteriol 197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharali P, Saikia JP, Ray A, Konwar BK. 2013. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf B Biointerfaces 103:502–509. doi: 10.1016/j.colsurfb.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hotterbeekx A, Kumar-Singh S, Goossens H, Malhotra-Kumar S. 2017. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front Cell Infect Microbiol 7:106. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noto MJ, Burns WJ, Beavers WN, Skaar EP. 2017. Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J Bacteriol 199:e00221-17. doi: 10.1128/JB.00221-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parad RB. 1996. Heterogeneity of phenotype in two cystic fibrosis patients homozygous for the CFTR exon 11 mutation G551D. J Med Genet 33:711–713. doi: 10.1136/jmg.33.8.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schurr MJ, Yu H, Martinez-Salazar JM, Boucher JC, Deretic V. 1996. Control of AlgU, a member of the sigma E-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol 178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, Di Serio C, Bragonzi A, Cirillo DM. 2014. Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A 90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, Doring G, Tummler B. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 45.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846. doi: 10.1128/IAI.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey DM, Wozniak DJ. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol 56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 47.Sotirova AV, Spasova DI, Galabova DN, Karpenko E, Shulga A. 2008. Rhamnolipid-biosurfactant permeabilizing effects on gram-positive and gram-negative bacterial strains. Curr Microbiol 56:639–644. doi: 10.1007/s00284-008-9139-3. [DOI] [PubMed] [Google Scholar]
- 48.Ryall B, Carrara M, Zlosnik JEA, Behrends V, Lee X, Wong Z, Lougheed KE, Williams HD. 2014. The mucoid switch in Pseudomonas aeruginosa represses quorum sensing systems and leads to complex changes to stationary phase virulence factor regulation. PLoS One 9:e96166. doi: 10.1371/journal.pone.0096166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gifford AH, Willger SD, Dolben EL, Moulton LA, Dorman DB, Bean H, Hill JE, Hampton TH, Ashare A, Hogan DA. 2016. Use of a multiplex transcript method for analysis of Pseudomonas aeruginosa gene expression profiles in the cystic fibrosis lung. Infect Immun 84:2995–3006. doi: 10.1128/IAI.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.May TB, Shinabarger D, Maharaj R, Kato J, Chu L, DeVault JD, Roychoudhury S, Zielinski NA, Berry A, Rothmel RK. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin Microbiol Rev 4:191–206. doi: 10.1128/cmr.4.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedersen SS. 1992. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl 28:1–79. [PubMed] [Google Scholar]
- 53.Irie Y, Borlee BR, O’Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 109:20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Limoli DH, Donegan NP, Warren EA, Cheung AL, O’Toole GA. 2019. Interspecies signaling generates exploratory motility in Pseudomonas aeruginosa. Elife 8:e47365. doi: 10.7554/eLife.47365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem 24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 56.Heck JE, Andrew AS, Onega T, Rigas JR, Jackson BP, Karagas MR, Duell EJ. 2009. Lung cancer in a U.S. population with low to moderate arsenic exposure. Environ Health Perspect 117:1718–1723. doi: 10.1289/ehp.0900566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y, Krieger AM, Yekutieli D. 2006. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93:491–507. doi: 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.