Archaeosine is ubiquitous in archaeal tRNA, where it is located at position 15. Based on its molecular structure, it was proposed to stabilize tRNA, and we show that loss of archaeosine in Thermococcus kodakarensis results in a strong temperature-sensitive phenotype, while there is no detectable phenotype when it is lost in Methanosarcina mazei. Measurements of tRNA stability show that archaeosine stabilizes the tRNA structure but that this effect is much greater when it is present in otherwise unmodified tRNA transcripts than in the context of fully modified tRNA, suggesting that it may be especially important during the early stages of tRNA processing and maturation in thermophiles. Our results demonstrate how small changes in the stability of structural RNAs can be manifested in significant biological-fitness changes.
KEYWORDS: Methanosarcina, Thermococcus, archaeosine, tRNA modification
ABSTRACT
Archaeosine (G+) is a structurally complex modified nucleoside found quasi-universally in the tRNA of Archaea and located at position 15 in the dihydrouridine loop, a site not modified in any tRNA outside the Archaea. G+ is characterized by an unusual 7-deazaguanosine core structure with a formamidine group at the 7-position. The location of G+ at position 15, coupled with its novel molecular structure, led to a hypothesis that G+ stabilizes tRNA tertiary structure through several distinct mechanisms. To test whether G+ contributes to tRNA stability and define the biological role of G+, we investigated the consequences of introducing targeted mutations that disrupt the biosynthesis of G+ into the genome of the hyperthermophilic archaeon Thermococcus kodakarensis and the mesophilic archaeon Methanosarcina mazei, resulting in modification of the tRNA with the G+ precursor 7-cyano-7-deazaguansine (preQ0) (deletion of arcS) or no modification at position 15 (deletion of tgtA). Assays of tRNA stability from in vitro-prepared and enzymatically modified tRNA transcripts, as well as tRNA isolated from the T. kodakarensis mutant strains, demonstrate that G+ at position 15 imparts stability to tRNAs that varies depending on the overall modification state of the tRNA and the concentration of magnesium chloride and that when absent results in profound deficiencies in the thermophily of T. kodakarensis.
IMPORTANCE Archaeosine is ubiquitous in archaeal tRNA, where it is located at position 15. Based on its molecular structure, it was proposed to stabilize tRNA, and we show that loss of archaeosine in Thermococcus kodakarensis results in a strong temperature-sensitive phenotype, while there is no detectable phenotype when it is lost in Methanosarcina mazei. Measurements of tRNA stability show that archaeosine stabilizes the tRNA structure but that this effect is much greater when it is present in otherwise unmodified tRNA transcripts than in the context of fully modified tRNA, suggesting that it may be especially important during the early stages of tRNA processing and maturation in thermophiles. Our results demonstrate how small changes in the stability of structural RNAs can be manifested in significant biological-fitness changes.
INTRODUCTION
The tRNA molecule is notable for harboring a stunning diversity of posttranscriptional chemical modifications, typically representing ∼10% to 20% of the nucleosides in a particular tRNA (1). To date, over 130 modified nucleosides have been structurally characterized (2, 3), which vary from simple methylation of the base or ribose to extensive “hypermodification” of the canonical bases, the latter of which can result in radical structural changes and involve multiple enzymatic steps to complete. While we are still far from a comprehensive understanding of the roles of tRNA modification, it has become clear that modified nucleosides are integral to tRNA function at many levels, influencing translation (4–8), tRNA structure and stability (1, 9–13), and regulatory events (14–16).
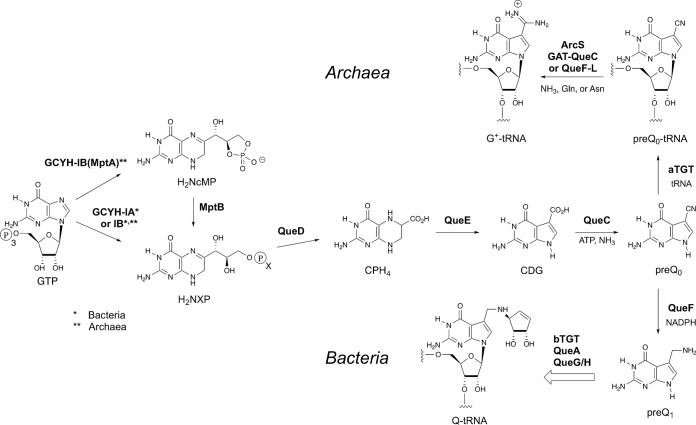
Among the most complex modifications known to occur in tRNA are the 7-deazaguanosine nucleosides archaeosine (G+) (17) and queuosine (Q) (18) (Fig. 1). Although both nucleosides share the core 7-deazaguanine structure, they are rigorously segregated with respect to phyla and location in the tRNA. Queuosine is ubiquitous throughout Bacteria and Eukarya (19), where it occurs specifically at the wobble position (20) in a subset of tRNAs (those coding for Tyr, His, Asp, and Asn). In contrast, archaeosine is present exclusively in the Archaea, where it is found in virtually all archaeal tRNAs at position 15 of the dihydrouridine loop (21), a site not modified in any tRNA outside the Archaea; in at least a few species, G+ is also present at position 13 (22).
FIG 1.
The biosynthetic pathways to archaeosine (G+) and queuosine (Q).
Despite the observed phylogenetic segregation, G+ and Q share a significant portion of their biosynthesis, and they remain the only modified nucleosides known for which a portion of the pathway occurs extrinsic to the tRNA, requiring the initial formation of a modified precursor base (23). All other modified nucleosides are formed exclusively via modification of a genetically encoded base in the RNA transcript. The pathway begins (Fig. 1) with the conversion of GTP to dihydroneopterin triphosphate (H2NTP) (Bacteria and Archaea) or the cyclic monophosphate (H2NcMP) (Archaea) by the enzyme GTP cyclohydrolase IA (GCYH-IA) in Bacteria (24) or GCYH-IB in Bacteria (25, 26) and Archaea (27), steps shared with the pterin pathways. After hydrolysis of H2NcMP (Archaea) by the enzyme MptB (28) the dihydroneopterin monophosphate (or triphosphate) is converted to carboxytetrahydropterin (CPH4) through the action of QueD (29), followed by the QueE-catalyzed ring contraction to 7-carboxy-7-deazaguanine (CDG) (30) and the formation of 7-cyano-7-deazaguanine (preQ0) by QueC (31). PreQ0 is the point of divergence in the bacterial and archaeal pathways, with preQ0 serving as the substrate for the enzyme tRNA-guanine transglycosylase (aTGT in Archaea, also known as 7-cyano-7-deazaguanine tRNA-ribosyltransferase), which catalyzes the exchange of the genetically encoded guanine-15 for preQ0 in archaeal tRNA. The preQ0-modified tRNA is converted to G+-modified tRNA by the action of either ArcS (32), QueF-L (33), or GAT-QueC (34), depending on the organism. In Bacteria, preQ0 is first reduced to preQ1 (35) before being inserted into specific bacterial tRNA at position 34 (the wobble position) by a bacterial tRNA-guanine transglycosylase (bTGT) (23) and further elaborated to Q-modified tRNA (36–38). Eukarya lack the de novo pathway and instead scavenge queuine, the free base of queuosine, from the environment, and a eukaryal TGT (eTGT) inserts queuine directly into the relevant tRNA (39), again at position 34.
The location of queuosine in the anticodons of specific bacterial and eukaryotic tRNAs suggests a role in modulating translational fidelity and efficiency, and studies are consistent with such a role (40–44). Archaeosine’s location at position 15, in the body of the tRNA, and its novel molecular structure led to a hypothesis that this modification functions (at least in part) to stabilize the structure of archaeal tRNA (17) via coulombic interactions of the positively charged formamidine group and the backbone phosphates in the vicinity. Notably, nucleotides (nt) 15 and 48 comprise the Levitt base pair, a conserved structural motif in the core of all tRNAs that is crucial for the overall structural integrity of tRNA. Computational studies revealed that the Levitt base pair H bonds are stronger in archaeosine-modified tRNA than in unmodified tRNA (45) due to the electron-withdrawing effect of the formamidine moiety (45), an effect that mimicked metal ion coordination to N-7 of guanine. Thus, two distinct mechanisms could be relevant to potential structural stabilization by G+.
To test the hypothesis that G+ serves to stabilize the structure of tRNA, we investigated the role of archaeosine both in vivo and in vitro. If, as proposed, G+ is important to tertiary structural stability of tRNA, this role would be especially critical in thermophilic organisms, where growth temperatures approach or exceed those needed to denature isolated tRNA, and G+-defective mutants should exhibit, at a minimum, a temperature-sensitive phenotype. Therefore, we carried out targeted gene knockouts of two genes in the G+ pathway in the hyperthermophile Thermococcus kodakarensis and investigated the consequences of these mutations for growth over a range of temperatures. As a complement, we also generated a knockout in the mesophile Methanosarcina mazei resulting in a strain lacking G+ and investigated its growth under a wide variety of growth conditions. To directly probe the structural impact of modification with preQ0 or G+, we investigated the thermal stability of tRNA possessing or lacking these modifications in the context of both fully modified tRNA isolated from T. kodakarensis strains and tRNA produced via in vitro transcription and modified with either preQ0 or G+ but lacking all other modifications.
We discovered that the genes of the G+ pathway are nonessential in both T. kodakarensis and M. mazei but that deletion strains of T. kodakarensis are temperature sensitive as predicted, consistent with the results of a recent genome-wide transposon mutagenesis screen (46) in which one of these genes (tgtA) was identified as important to thermophily. Additionally, we found that modification with G+ imparts a modest but measurable stabilizing effect on tRNA that is most apparent in tRNA transcripts that are otherwise unmodified.
RESULTS
T. kodakarensis and M. mazei mutant construction.
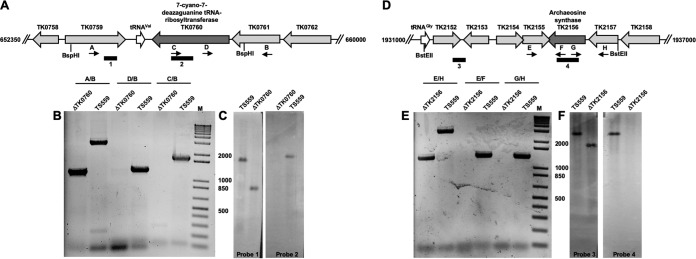
We targeted two genes encoding archaeosine biosynthetic proteins in the hyperthermophilic model archaeon Thermococcus kodakarensis for deletion from the genome. TK0760 (tgtA) encodes a homologue of the archaeal tRNA-guanine transglycosylase (aTGT) (UniProt accession no. Q5JHC0), while TK2156 (arcS) encodes a homologue of archaeosine synthase (ArcS) (UniProt accession no. Q5JHG7) (Fig. 2A and D, respectively); these enzymes catalyze the final, and only tRNA-dependent, steps in the biosynthesis of archaeosine (Fig. 1). Beginning with strain TS559, markerless deletion of the entire coding sequence of tgtA was possible, as was the deletion of most of arcS (to the exclusion of the 23 bp that overlap the divergent locus, TK2155). Deletion of each locus was confirmed by a series of diagnostic PCRs with purified genomic DNAs from each strain (Fig. 2B and E, respectively). Further confirmation of each deletion was provided by Southern blots of BspHI- and BstEII-digested preparations of genomic DNA from the T. kodakarensis ΔtgtA and ΔarcS strains, respectively (Fig. 2C and F). For each locus, probes complementary to the target gene (probes 2 and 4 [Fig. 2]) were unable to hybridize to any location on the genomes from the deletion strains, while probes complementary to adjacent sequences (probes 1 and 3 [Fig. 2]) did hybridize to genomic fragments that were shorter in deletion strains than those derived from strain TS559. In both instances, the difference in the size of the identified DNA fragment was consistent with the size of the target gene that was deleted.
FIG 2.
T. kodakarensis strains with TK0760 (7-cyano-7-deazaguanine tRNA-ribosyltransferase) and TK2156 (archaeosine synthase) markerlessly deleted. (A and D) Map of the T. kodakarensis genome surrounding TK0760 (A) and TK2156 (D) in the parental strain TS559, highlighting the binding positions of oligonucleotides that were used in diagnostic PCRs (panels B and E, respectively) and Southern blots (panels C and F, respectively). (B and E) PCRs with primer sets listed above each lane generate amplicons from genomic DNA purified from strains TS559, ΔTK0760, and ΔTK2156. The external primer pairs (A/B for TK0760; E/H for TK2156) generate smaller amplicons from ΔTK0760 and ΔTK2156 genomic DNAs, respectively, reflecting the loss of TK0760- or TK2156-coding sequences. Amplicons generated using one primer complementary to the target locus and one primer complementary to flanking sequences are generated only from TS559 genomic DNA, consistent with deletion of the TK0760- or TK2156-coding sequence, respectively. (C and F) Southern blots of digested total genomic DNA from strains TS559, ΔTK0760, and ΔTK2156 demonstrate deletion of TK0760 or TK2156, respectively. Blots developed with an amplicon complementary to the TK0760-coding sequences (probe 2) reveal a complementary target only from TS559 DNA, while an amplicon probe complementary to adjacent sequences (probe 1) within the same BspH1 fragment reveals a smaller target, consistent with deletion of TK0760-coding sequences. Blots developed with an amplicon complementary to the TK2156-coding sequences (probe 4) reveal a complementary target only from TS559 DNA, while an amplicon probe complementary to adjacent sequences (probe 3) within the same BstEII fragment reveals a smaller target, consistent with deletion of TK2156-coding sequences. Numbers between panels B and C and between panels E and F are DNA fragment sizes in base pairs. Lanes M, DNA standards.
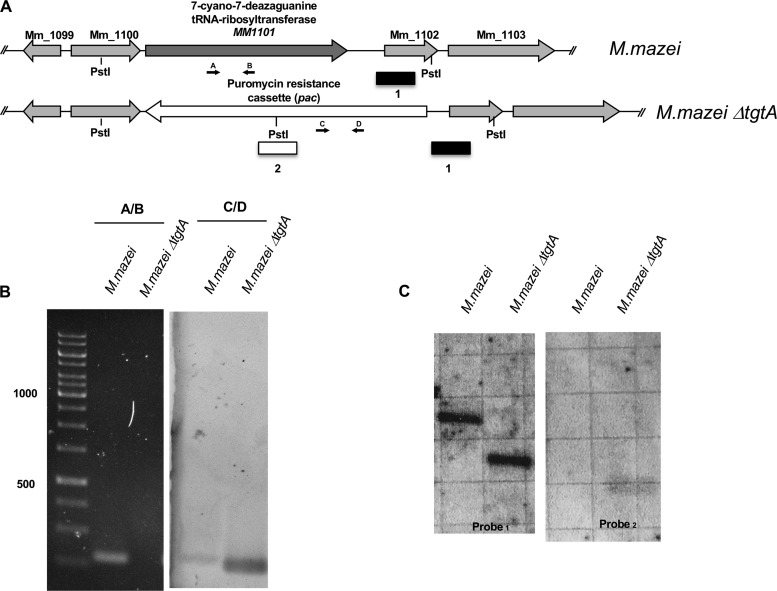
To investigate the consequences of archaeosine loss in a mesophile, the M. mazei gene MM1101 (tgtA), encoding aTGT (UniProt accession no. Q8PXW5) (47), was disrupted by the insertion of a puromycin resistance (pac) cassette by homologous recombination (Fig. 3A). Three independent puromycin-resistant transformants were isolated and grew at 37°C. The absence of the tgtA gene and presence of the puromycin resistance cassette were confirmed by both PCR and Southern hybridization (Fig. 3B and C, respectively).
FIG 3.
M. mazei strains with MM1101 (7-cyano-7-deazaguanidine tRNA ribosyltransferase) deleted. (A) Map of the M. mazei genome surrounding MM1101 (tgtA) in the parental strain M. mazei, highlighting the binding positions of oligonucleotides that were used in diagnostic PCRs (B) and Southern blots (C). (B) PCR with the primer sets listed above each lane generates amplicons from genomic DNA purified from wild-type and mutant (M. mazei ΔtgtA) strains. Amplicons generated by primers specific for the tgtA gene demonstrate the presence of tgtA in the wild type and loss of tgtA in M. mazei ΔtgtA. In contrast, amplicons generated from the puromycin (pac) cassette indicate that it is present in M. mazei ΔtgtA and absent in the wild-type strain. (C) Southern blots of PstI-digested total genomic DNA from M. mazei wild-type and ΔtgtA strains demonstrate loss of tgtA in M. mazei ΔtgtA. Blots developed with an amplicon complementary to sequences adjacent to tgtA (probe 1) reveal a smaller target, consistent with the deletion of tgtA and insertion of the pac cassette. Blots developed with an amplicon complementary to the pac cassette reveal a complementary target only in M. mazei ΔtgtA, consistent with a pac cassette insertion into the M. mazei ΔtgtA strain.
Nucleoside analysis of bulk tRNA from the T. kodakarensis and M. mazei cell lines.
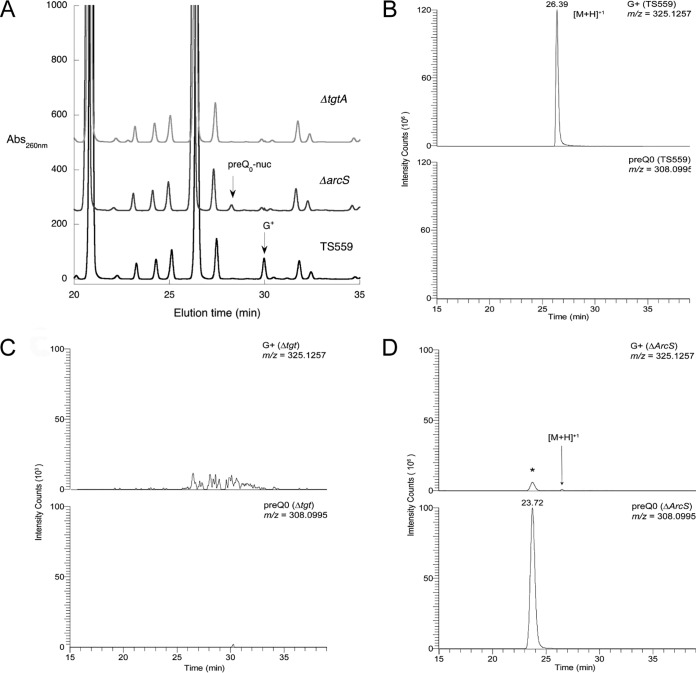
To confirm that tRNAs in the mutant strains were appropriately modified, purified tRNA from each of the T. kodakarensis and M. mazei strains was subjected to nuclease digestion and dephosphorylation, followed by high-pressure liquid chromatography (HPLC) analysis of the resulting nucleosides. The tRNAs from the three T. kodakarensis strains displayed the predicted pattern of modified nucleosides (Fig. 4A); preQ0 nucleoside and G+ were absent from the T. kodakarensis ΔtgtA strain, and preQ0 was present in the T. kodakarensis ΔarcS strain, with G+ being present only in the wild-type strain. Similarly, only the tRNA from the wild-type M. mazei strain contained G+ (see Fig. S1 in the supplemental material).
FIG 4.
Analysis of modification status of tRNA isolated from T. kodakarensis strains. (A) HPLC analysis of nucleoside digests of tRNA from T. kodakarensis TS559, the ΔarcS strain, and the Δtgt strain. (B) LC-MS analysis of nucleoside digests of tRNA from T. kodakarensis TS559, showing extracted ion chromatograms (XIC) of archaeosine (m/z 325.1257) (top) and preQ0 nucleoside (m/z 308.0994) (bottom). XIC relative abundances were scaled to the largest peak (archaeosine) at 106. Signal for preQ0 nucleoside was detected at background levels of 103. (C) LC-MS analysis of nucleoside digests of tRNA from the T. kodakarensis Δtgt strain, showing extracted ion chromatograms of archaeosine (m/z 325.1257) (top) and preQ0 nucleoside (m/z 308.0994) (bottom). Neither archaeosine nor preQ0 was detected at any appreciable levels. Chromatograms are scaled 103. (D) LC-MS analysis of nucleoside digests of tRNA from the T. kodakarensis ΔarcS strain, showing extracted ion chromatograms of archaeosine (m/z 325.1257) (top) and preQ0 nucleoside (m/z 308.0994) (bottom). For this run, G+ was detected at 1.6% the level of preQ0 nucleoside. The asterisk denotes the adduction of ammonium onto the preQ0 nucleoside during the electrospray process. Chromatograms are scaled 106. Analyses were carried out in triplicate for each of two independent preparations of tRNA.
To further address the modification status of the tRNA and confirm the peak assignments, we analyzed the tRNAs from the T. kodakarensis strains by liquid chromatography-mass spectrometry (LC-MS) (Fig. 4B to D). Analysis of the nucleoside digests from the isolated tRNAs from the T. kodakarensis cell lines confirmed the initial HPLC data with one exception; while no G+ was detected in the tRNA digests from either the T. kodakarensis ΔtgtA or ΔarcS strain by HPLC, LC-MS analysis was able to detect G+ in the T. kodakarensis ΔarcS samples, which varied from 1.6 to 6.6% of the intensity of that for preQ0 nucleoside (Fig. 4D).
Temperature-dependent growth of T. kodakarensis strains disrupted in archaeosine biosynthesis.
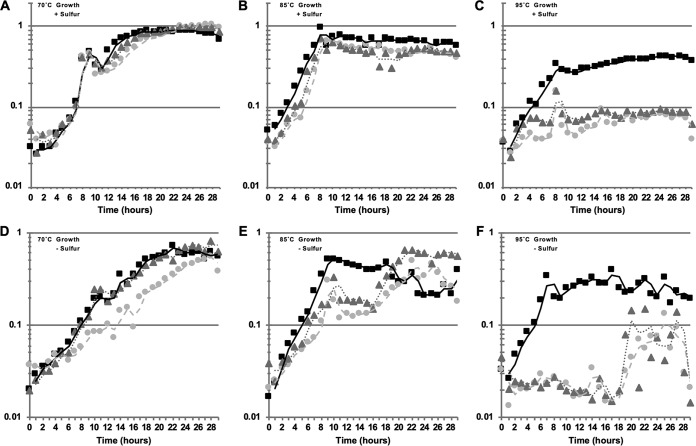
Deletion strains of T. kodakarensis were constructed at 85°C, and we noted that colonies from strains with tgtA or arcS deleted were slightly smaller than colonies produced by TS559. It was clear that loss of archaeosine biosynthesis was not lethal, but it appeared that loss of archaeosine biosynthesis did hinder growth. To more accurately measure the growth of each strain, we monitored the optical densities (ODs) of growing cultures while varying the incubation temperature to identify any potential role for archaeosine modification at reduced (70°C), optimal (85°C), or elevated (95°C) temperatures (Fig. 5). T. kodakarensis can support two radically different metabolic strategies based on the availability of elemental sulfur (S0) in the medium; thus, we monitored growth in the absence and presence of sulfur at three different temperatures.
FIG 5.
T. kodakarensis strains lacking tgtA or arcS are temperature sensitive. Culture growth was monitored by changes in optical density at 600 nm for cultures incubated at 70°C (A and D), 85°C (B and E), or 95°C (C and F). The results reported are the average values from minimally three independent experiments with triplicate biological replicates in each experiment. Cultures in panels A to C were provided 2 g/liter sulfur, while cultures in panels D to F received 5 g/liter pyruvate instead. Squares, TS559; triangles, ΔtgtA; circles, ΔarcS.
While deletion of tgtA or arcS had minimal or essentially no effect, respectively, on growth of T. kodakarensis cultures at 70°C (Fig. 5A and D), severe phenotypes were noted at elevated (95°C) temperatures, where neither deletion strain could support robust growth even after >30 h of incubation (Fig. 5C and F). Growth at the optimal temperature of 85°C was more modestly compromised for strains with arcS or tgtA deleted, with growth more severely affected in the absence of sulfur (Fig. 5B and E), an observation that extended to growth of the ΔarcS strain at 70°C.
Growth under diverse conditions of M. mazei strains disrupted in archaeosine biosynthesis.
We tested three independent M. mazei mutants with insertions in the tgtA gene for growth under various conditions relative to that of wild-type M. mazei. Growth was indistinguishable between wild-type and mutant strains at reduced (25°C), suboptimal (30°C), and optimal (37°C) growth temperatures (see Fig. S2 in the supplemental material). In order to test additional stress conditions, the M. mazei strains were grown under multiple conditions that have previously been determined to induce a stress response (48). These included the presence of metals (e.g., copper and nickel), high salt, the absence of sulfide, or the presence of antimicrobials. In each case, no difference in growth between the wild type and mutants was detected (see Fig. S3 in the supplemental material).
Thermal denaturation study of in vivo tRNAGln from T. kodakarensis and in vitro tRNAGln transcripts.
To directly probe for a potential structural role for G+ in tRNA, we investigated the thermal denaturation of tRNA extracted from the T. kodakarensis strains by measuring the hyperchromicity at 260 nm upon denaturation. In these experiments, the raw melt data were processed to obtain a differential melting profile (first-derivative plot of dA/dT versus temperature), which allowed the apparent melting temperature (Tm) to be easily determined over a range of magnesium chloride concentrations, from 0 to 10 mM in a buffer of 10 mM sodium cacodylate (pH 7.0) and 100 mM NaCl. Although it was recently reported that unfractionated tRNA from a T. kodakarensis strain lacking G+ exhibited a Tm 2°C lower than that of unfractionated tRNA from the wild-type strain (46), we were unable to observe discernible differences in the denaturation profiles of unfractionated tRNA from our three strains (data not shown), so we chose to investigate the behavior of a specific tRNA isolated from these strains and selected tRNAGln for further investigation.
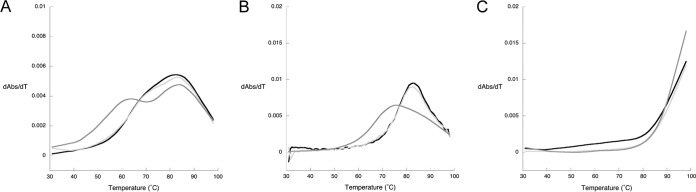
The tRNAGln isoacceptors were purified from the T. kodakarensis strains utilizing an affinity approach (49) as detailed in Materials and Methods. As with the unfractionated tRNA, the raw thermal denaturation data (Fig. 4) from the purified tRNAGln derived from the three strains were processed to obtain differential denaturation profiles (Fig. 6). Surprisingly, the tRNAGlns from the parental strain (TS559) containing G+ at position 15 and the ΔtgtA strain containing G behaved almost identically (Fig. 6). In the absence of Mg2+, both exhibited a slight shoulder at ∼70°C and a main transition (the TM) at ∼83°C (Fig. 6A). The TM is similar for tRNAGln from the T. kodakarensis ΔarcS strain (containing preQ0), but there is also a distinct shoulder in the latter at ∼64°C (Fig. 6A). At 100 μM Mg2+, the profiles for the tRNAGlns from the TS559 and T. kodakarensis ΔtgtA strains have lost the shoulder and exhibit a single well-defined TM at 83°C and 82°C, respectively (Fig. 6B). At the same Mg2+ concentration, the differential plot for the tRNAGln from the T. kodakarensis ΔarcS strain has coalesced into a very broad but asymmetric profile with the TM at ∼75°C. At 10 mM Mg2+, the tRNAGlns from all three strains denature at a temperature beyond the 98°C limit of the experiment (Fig. 6C).
FIG 6.
Thermal denaturation profiles (first derivative) of in vivo T. kodakarensis tRNAGln. The purified isoacceptor tRNAs from the Δtgt (light gray), ΔarcS (dark gray), and TS559 (black) strains were denatured in a background of 100 mM NaCl with no MgCl2 (A), 100 μM MgCl2 (B), or 10 mM MgCl2 (C).
To investigate the potential role of G+ in tRNA stability free from the effects of other modified nucleosides, we carried out thermal denaturation studies on tRNA produced through in vitro transcription and enzymatically modified to contain preQ0 or G+ at position 15. A tRNA transcript corresponding to T. kodakarensis tRNAGln(CUG) (with the 5′ adenosine substituted for guanosine) was prepared from a duplex DNA template as described in Materials and Methods. A portion of the tRNAGln transcript was then reacted in vitro with recombinant aTGT (Fig. 1) from Methanocaldococcus jannaschii (50) to replace the genetically encoded G at position 15 with preQ0. A portion of the preQ0-modified tRNA was then further reacted with recombinant M. jannaschii ArcS (32) to produce G+-modified tRNA (Fig. 1). Quantitation of preQ0 incorporation and subsequent conversion to G+ was carried out as described in Materials and Methods, and the modification state of the tRNA was confirmed by HPLC (see Fig. S5 in the supplemental material).
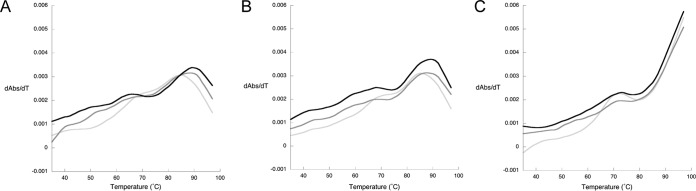
Similar to our observations with tRNAGln isolated from the T. kodakarensis ΔtgtA mutant, in the absence of magnesium the unmodified tRNAGln transcript exhibited a shoulder in the differential thermal denaturation plot at ∼70°C along with a TM of 84°C (Fig. 7A). In contrast, the effect of modification at position 15 on the tRNAGln transcript was markedly different than that observed for the in vivo-produced tRNA. The preQ0- and G+-modified tRNAGln transcripts both exhibit a TM significantly above that of the unmodified tRNAGln at 88 and 89°C, respectively. While both profiles also feature a shoulder (for the G+-modified tRNAGln transcript it is very distinct), these occur at a lower temperature (∼67°C) than for the unmodified transcript. Notably, the TMs for the modified transcripts are significantly higher than that observed in the fully modified tRNA isolated from T. kodakarensis. In the presence of 100 μM MgCl2 the TM increases to ∼86°C for the unmodified transcript and to 90°C for the G+-modified transcript, while the TM remains unchanged at 88°C for the preQ0-modified transcript (Fig. 7B). The shoulder persists in the profiles for all three tRNAs, with an increase of 1 to 2°C. In the presence of 10 mM MgCl2, the denaturation is not complete for any of the tRNAs at 98°C (Fig. 7C), the highest temperature reached in the experiment.
FIG 7.
Thermal denaturation profiles (first derivative) of in vitro-produced T. kodakarensis tRNAGln(CUG). The data correspond to the unmodified tRNA transcript possessing G at position 15 (light gray), the modified transcript possessing preQ0 (dark gray), and the modified transcript possessing G+ (black). The denaturing profiles were recorded in a background of 100 mM NaCl with no MgCl2 (A), 100 μM MgCl2 (B), or 10 mM MgC12 (C).
DISCUSSION
Archaeosine is a structurally complex modified nucleoside found in the tRNAs of Archaea, and it recently has been discovered in viral and bacterial DNAs (51). The proposals that G+ functions to stabilize tRNA tertiary structure (17, 45) prompted us to investigate this putative role in vivo through the construction and phenotypic characterization of T. kodakarensis and M. mazei strains that were disrupted in G+ biosynthesis and in vitro by directly measuring the thermal stability of tRNA in the presence and absence of G+.
Our observation of temperature sensitivity in T. kodakarensis lacking G+ is consistent with a role in structural stabilization of the tRNA by G+ and mirrors the results of a recent transposon mutagenesis study (46) in T. kodakarensis, which reported that disruption of the tgtA gene and loss of G+ modification was accompanied by loss of thermophily. Importantly, because we also observed this phenotype in the ΔarcS mutant, which possesses preQ0-modified tRNA, the loss of thermophily can be conclusively attributed to the unique physicochemical properties of G+. Interestingly, we observed no phenotypic differences between the wild-type and G+-deficient strains of the mesophile M. mazei under a range of growth conditions, including growth at suboptimal temperatures, while in Haloferax volcanii, also a mesophile and the only other organism in which loss of G+ has been investigated, loss of G+ was accompanied by cold sensitivity (52). Although both hot and cold tolerances can be rationalized by tRNA structural effects, the natures of these effects are typically in opposition to one another, with heat tolerance being associated with increasing structural rigidity and cold tolerance with relaxing structural rigidity, so the observation of both phenotypes accompanying loss of G+ is intriguing and may be due to the significant differences in the in vivo environments, most notably the very high salt concentrations in halophilic species.
While the presence or absence of G+ in tRNAGln isolated from T. kodakarensis had minimal impact on the overall stability of the otherwise fully modified tRNA, its presence had a significant effect on the stability of the tRNA transcripts, with the stabilizing effect manifested in a 4 to 5°C increase in the Tm depending on the concentration of MgCl2. The magnitude of the observed change in Tm is on the order of those for other modifications that have been characterized as structurally important (13) and approaches that for ribothymidine at position 54 of Escherichia coli tRNAMet (53), which contributes 6°C to the Tm of the tRNA. The fact that the effect is most pronounced for in vitro-transcribed tRNA, which is devoid of other modifications, suggests that this role may be most important in the early stages of folding and processing the nascent transcript. This interpretation is consistent with kinetic studies of aTGT, which revealed that the best substrates for the enzyme are unstructured RNAs (54, 55). While disruption of tRNA folding and/or processing due to the absence of G+ can easily account for the growth defects observed at higher temperatures for both T. kodakarensis mutants, we cannot rule out the possibility that otherwise fully modified tRNAs respond differentially to the presence or absence of G+, and some tRNA (other than tRNAGln) may exhibit more significant decreases in thermal stability in the absence of G+.
Surprisingly, deletion of arcS in T. kodakarensis did not completely abolish G+ biosynthesis, with the knockout strain displaying small amounts of G+ up to 6.6% of that of preQ0 nucleoside. While this low level of G+ was not significant in terms of the growth or thermal denaturation experiments, it does lead to the question of how G+ is formed in this mutant. The formation of G+ from preQ0-modified tRNA is the only step in the G+ pathway in which multiple nonhomologous enzymes that catalyze the same transformation have been discovered (Fig. 1); in addition to ArcS, the enzymes QueF-L (33, 34) and GAT-QueC (34) have also been shown to catalyze the formation of G+ from preQ0-modified tRNA. While a number of organisms possess more than one of these enzymes (34), neither QueF-L or GAT-QueC is present in T. kodakarensis (34). However, a number of organisms that possess genes encoding the rest of the G+ pathway lack genes encoding any of the three known enzymes that form G+ (34) (see Table S1 in the supplemental material), so it is likely that there exists at least one more enzyme responsible for G+ formation, and it may be present in T. kodakarensis.
Overall, both the in vivo results with T. kodakarensis and the in vitro biophysical studies (ours and those of Orita et al. [46]) support the original proposal that G+ is important for thermostability of archaeal tRNA (17) and demonstrate how small changes in the stability of structural RNAs can be manifested in significant biological-fitness changes. Nevertheless, the near ubiquity of G+ in the Archaea (it is absent only in Haloquadratum walsbyi), the majority of which are not thermophiles, argues for a more fundamental and universal role, but the absence of any distinct phenotypes in the M. mazei ΔtgtA mutant suggests that this role is a subtle one.
MATERIALS AND METHODS
General.
Buffers and salts of the highest grade available were purchased from Sigma-Aldrich unless otherwise noted. Diethyl pyrocarbonate (DEPC)-treated water was used for all solutions used for RNA-related assays (56). All buffers and solutions were otherwise prepared with Millipore MQ grade water. Dithiothreitol (DTT), isopropyl-β-d-thiogalactopyranoside (IPTG), kanamycin sulfate, DEPC, and ampicillin were purchased from RPI Corporation. [8-14C]guanine was purchased from PerkinElmer. Adenosine, guanosine, ATP, GTP, UTP, and CTP were all purchased from Sigma-Aldrich. Nickel-nitrile tetraacetic acid (Ni2+-NTA) was purchased from Qiagen and Sigma-Aldrich. Whatman GF-B filter disks were purchased from Fisher Scientific. Amicon centrifugal concentrators were from Millipore Sigma. Dialysis tubing was obtained from Thermo Fisher Scientific. Plasmid minikits were from Fermentas and Qiagen. Oligonucleotides were obtained from IDT or Operon. All reagents for SDS-PAGE were purchased from Bio-Rad. SDS-PAGE analysis was carried out using 12% (29:1 acrylamide-bisacrylamide) gels and visualized with Coomassie brilliant blue. DNA sequencing was carried out by the OHSU core facility in the Department of Molecular Microbiology and Immunology. The substrate preQ0 was synthesized as described previously (57), purified by reverse-phase HPLC, and stored at room temperature in dimethyl sulfoxide (DMSO). The recombinant aTGT (50) and ArcS (32) from M. jannaschii were overproduced and purified as previously described. An expression plasmid of a His6-tagged construct of the Δ172–173 mutant of T7 RNA polymerase (58) was provided by John Perona.
Instrumentation.
Analytical HPLC was performed on an Agilent 1100 series HPLC (G1312A binary pump and G1315A diode array detector). Preparative-scale separation was achieved using a Hitachi HPLC (L-6200 pump and L-4000 single-wavelength detector). UV-visible (UV-Vis) spectroscopy was carried out on a Varian Cary 100 Bio spectrophotometer fitted with a thermostat-controlled multicell holder.
T. kodakarensis strain construction.
T. kodakarensis strains with TGTa and ArcS markerlessly deleted were constructed essentially as described previously (59) using TS559 as the parental strain. Briefly, nonreplicative plasmids were temporarily integrated into the TS559 genome adjacent to the target locus and then excised through homologous recombination between direct repeats flanking the target gene. Markerless deletion of tgtA and the nonoverlapping sequences of arcS was confirmed by diagnostic PCRs using purified genomic DNAs as templates (Fig. 2B and E, respectively). The exact endpoints of the deletions were confirmed by sequencing amplicons from each locus generated with primers that bind to locations adjacent to each locus (primers A and B for tgtA and primers E and H for arcS). To confirm that neither tgtA nor arcS was relocated within the T. kodakarensis genome, total genomic DNA was purified, digested with either BstEII or BspHI, resolved, and transferred for Southern blotting as previously described (60). Two Southern blot probes (probes 1 and 2) were employed to confirm the deletion of tgtA, and two additional probes (probes 3 and 4) were used to confirm the deletion of arcS. Probe 1 was complementary to sequences within TK0759 that were located on the same BspHI fragment as tgtA, while probe 2 was complementary to tgtA sequences. Probe 3 was complementary to sequences within TK2152 and TK2153 that were located on the same BstEII fragment as arcS, while probe 4 was complementary to arcS sequences. Probe 1 was generated with the primer pair S.B. 760extF (5′-AGCAAGGGCGTGAACATCGAGTGGG-3′) and S.B. 760extR (5′-CCCTCTTCAAGGATTCTCTGCACG-3′), probe 2 was generated with the primer pair S.B. 760intF (5′-AAGGTAGCGAGGTGCTTGCCCTTGG-3′) and S.B. 760intR (5′-TGAAACCATCAGCCACCCGATCTTC-3′), probe 3 was generated with the primer pair 001-2153 (5′-CACCTTGAGGATATTAGTGATTGGC-3′) and 002-2151 (5′-CGTCTATTGAATACTGAGGTTTTCC-3′), and probe 4 was generated with the primer pair S.B. 2156intF (5′-TAGCGATAAGTCCTGTCCTCCTTTG-3′) and 002-2155 (5′-GGCCAAGTATGACATAGTAGTCACC-3′).
Growth of Thermococcus kodakarensis for tRNA isolation.
(i) Medium preparation. Growth medium contained (per liter) yeast extract (2.5 g), tryptone (2.5 g), NaCl (10.2 g), MgCl2·6H2O (2.4 g), MgSO4 (0.8 g), CaCl2·2H2O (0.4 g), KCl (0.3 g), sodium pyruvate (2.5 g), agmatine sulfate (0.6 g), 2 ml of a 500× vitamin stock solution (8 μM biotin, 5 μM folic acid, 50 μM pyridoxine, 15 μM thiamine, 15 μM riboflavin, 40 μM nicotinic acid, 20 μM Ca-pantothenate, 7 μM p-aminobenzoic acid, and 75 nM vitamin B12), and 2 ml of a 500× trace mineral stock solution [50 μM FeCl3, 5 μM MnCl2, 18.5 μM CoCl2, 7 μM CaCl2, 7.5 μM ZnCl2, 1.5 μM CuCl2, 1.6 μM H3BO3, 1 μM (NH4)2MoO4, 5 μM NiCl2, 850 nM NaSeO4, and 2 μM AlCl3]. The medium was prepared under N2 to remove all dissolved O2 (resazurin was added to 1 mg/liter) and autoclaved to sterilize. Before inoculation, the head gas was exchanged for 80:20 N2-CO2 to 10 lb/in2. To ensure fully anaerobic conditions, the growth medium was spiked with additional Na2S (from a 2.5% [wt/vol] stock) until resazurin remained colorless.
(ii) Cell growth. Starter cultures of T. kodakarensis (TS559 [wild type], ΔTK0760 [ΔtgtA], and ΔTK1256 [ΔarcS] strains) were grown at 60°C overnight in 10-ml cultures in Hungate tubes with a 1-ml inoculation from stock culture. The cells were then grown in 1-liter culture volumes. The medium and starter culture were brought to the target growth temperature before the entire starter culture was transferred to the larger flask and cells were allowed to grow for at least 16 h. The cells were then pelleted by centrifugation and frozen at −80°C until used.
Comparative growth profiles of T. kodakarensis strains.
The T. kodakarensis TS559, ΔTK0760, and ΔTK2156 strains were grown in sealed, 15-ml anaerobic tubes containing 10 ml ASW-YT medium (0.8× artificial seawater [ASW], 5 g/liter yeast extract, and 5 g/liter tryptone) with a headspace gas composition of 95% N2 and 5% H2 at one atmosphere of pressure. The medium was supplemented with vitamins and agmatine (as described above) and with either 5 g/liter pyruvate or 2 g/liter flowers of sulfur. Starter cultures were grown at 85°C, and the optical densities of cultures were monitored at 600 nm during subsequent growth at 70°, 85°, or 95°C. The results reported are the average values from minimally three independent experiments with triplicate biological replicates in each experiment.
Construction of M. mazei tgtA (MM1101) insertion mutants.
Methanosarcina mazei (DSM 3647) gene MM1101 (tgtA), encoding tRNA-guanine transglycosylase (aTGT), was disrupted by insertion of a puromycin resistance cassette in a manner similar to that for the disruption of the glnK gene (48). Briefly, ∼1,000 bp flanking the 5′and 3′regions (Fig. 3A) of tgtA were amplified from M. mazei genomic DNA. The primers for the 5′-flanking region, MM1101ko5primeF (AAAAAAGGTACCaaagcaatccataagtgaagc) (KpnI) and MM1101ko5primeRL (AAAAAGAATTCgccgcggttatagatgc) (EcoRI) (sequences in the M. mazei genome are in lowercase, and restriction sites are italicized) introduced KpnI and EcoRI restriction endonuclease cutting sites at the ends of the primers, while the primers for the 3′flanking region, Mm1101ko3primeF (AAAAAGAattcggaccttcccg) (EcoRI) and Mm1101ko3primeR (ttcaggatccctgccg) (BamHI) (sequences in the M. mazei genome are in lowercase, and restriction sites italicized) introduced an EcoRI site (a naturally occurring BamHI site was used for the reverse primer). Both PCR products were gel purified and introduced into pBluescript by cutting the plasmid and PCR products with EcoRI, KpnI, and BamHI, followed by ligation. The resulting plasmid, pKMSK1, was cut with EcoRI and ligated to an EcoRI-cut puromycin resistance cassette (pac cassette) (48), generating plasmid pKMSK2. Plasmid constructs were verified by DNA sequencing across ligation junctions. Plasmid pKMSK2 was cut with ScaI to generate a linear DNA with the pac cassette with ca. 1,000 bp of sequence flanking MM1101. This DNA was transformed into M. mazei with 1,2-dioleoyloxy-3-(trimethylammonium)propane (DOTAP) liposome-mediated transformation (48). Transformants were grown in the presence of puromycin, and three independent isolates, M. mazei ΔtgtA1, ΔtgtA2, and ΔtgtA3, were selected as single clones on plates containing puromycin. Insertion mutations were confirmed by PCR (Fig. 3B) with mutants containing the pac gene and lacking the tgtA gene and Southern blotting using flanking probes or pac probes. The flanking probe was made by PCR using primers Mma_attP_5′Flank (5′-GGCTTACTCCCGCTTTCTCT-3′) and Mma_attP_3′Flank (5′-TTGAGTTCCTCGCTTTCGAT-3′) and digoxigenin (DIG) nucleotide mix (Roche). The pac probe was made by PCR using primers KMSPacR (Mm1101_5′R_rc) (5′-GCATCTATAACCGCGGC-3′) and KMSPacF (Mm1101_3′F_rc) (5′-CGGGAAGGTCCCGAAT-3′) and DIG nucleotide mix (Roche).
Growth of M. mazei and mutants.
For growth at different temperatures, M. mazei cells were grown essentially as described previously (48). Cells were grown anaerobically in closed 5-ml culture tubes with 25 mM trimethylamine reduced with 2 mM cysteine and 1 mM sodium sulfide and an overpressure of N2-CO2. Cultures were supplemented with 100 μg/ml ampicillin or 100 μg/ml kanamycin to prevent bacterial growth. Mutants were selected with 2.5 μg/ml puromycin. Growth was monitored by measuring the optical density at 600 nm. For screening for growth changes of mutant strains under different conditions a microtiter plate assay modified for growth in anaerobic conditions was used (61). Reduction was performed only with cysteine and not with sodium sulfide. Growth was monitored until stationary phase was reached.
tRNA extraction from T. kodakarensis and M. mazei.
T. kodakarensis or M. mazei cells were suspended at 250 mg/ml in 100 mM ammonium acetate (pH 6.5) with 10 mM MgSO4 and 0.1 mM EDTA. An equal volume of saturated phenol mix (phenol-chloroform-isoamyl alcohol [25:24:1]) was added to lyse the cells, and after centrifugation to separate the phases, the bulk RNA was precipitated from the aqueous phase by adding 1/10 volume of 8.0 M ammonium acetate and two volumes of ethanol and cooling to –20°C for 2 h. The precipitated RNA was pelleted by centrifugation at 20,000 × g for 25 min at 4°C. The pellet was resuspended in 100 mM ammonium acetate (pH 6.5) with 10 mM MgSO4 and 0.1 mM EDTA, an equal volume of 8.0 M LiCl was added, and the mix was cooled at 4°C overnight. Precipitated rRNA species were removed by centrifugation (20,000 × g), followed by precipitation of the tRNA remaining in the supernatant with the addition of ammonium acetate-ethanol as described above.
To determine the modification state of the tRNA from each strain, the purified unfractionated tRNA samples were enzymatically digested and dephosphorylated as described preciously (62), followed by HPLC analysis on large (250- by 4.6-mm) or small (3- by 4.6-mm) Gemini columns (Phenomenex, 5 μm, C18). The mobile phase consisted of a variable gradient from 100% 25 mM ammonium acetate (pH 6.0) (solvent A) to a 60:40 mix of solvent A and solvent B (acetonitrile) over the course of 20 to 25 min.
Isolation of tRNA from T. kodakarensis for MS analysis.
Total RNA was extracted as described above; however, to prepare total tRNA for mass spectrometric (MS) analysis, solid-phase extraction was employed to reduce the counterion species present. Nucleobond RNA/DNA 400 columns (Macherey-Nagel) were employed to separate high-mass RNA molecules and total tRNA. Pelleted total tRNA was suspended in the appropriate buffer according to the manufacturer’s guidelines, and fractionation utilized a step gradient of salt concentrations with tRNA eluting in 0.65 M KCl and higher-mass molecules eluting in 1.15 M KCl. The RNA population in subfractions were confirmed by urea PAGE. The isolated tRNA was precipitated in 800 mM ammonium acetate-ethanol. This was repeated three times to replace the K+ with ammonium ions. The sample was then dried for subsequent LC-MS analysis.
The purified unfractionated tRNA samples were enzymatically digested and dephosphorylated as described preciously (62). Separation was accomplished by reversed-phase chromatography using an Acquity UPLC HSS T3 column (1.8 μm, 1 mm by 100 mm; Waters, Milford, MA) on a Vanquish Flex Quaternary UHPLC system (Thermo Fisher Scientific, San Jose, CA). The mobile phase A consisted of 5.3 mM ammonium acetate (pH 5.3) in LC-MS-grade water (Alfa Aesar, Haverhill, MA). Mobile phase B consisted of a 60:40 mixture of 5.3 mM ammonium acetate (pH 5.3) and acetonitrile (Honeywell Burdick & Jackson, Morris Plains, NJ) with a gradient of 0% B (from 0 to 1.8 min), 2% B at 3 to 3.5 min, 3% B at 4.1 min, 5% B at 7 min, 25% B at 9 min, 35% B at 15 min, 99% B at 15.5 min (hold for 4.5 min), 99% B at 20 min, and then returning to 0% B at 25.5 min at a flow rate of 100 μl min−1. The column temperature was set at 40°C.
High-resolution accurate mass analyses of nucleosides were performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) interfaced with an H-ESI electrospray source in positive-polarity mode. Full-scan data were acquired at a resolution of 120,000, mass range of 220 to 900 m/z, automatic gain control (AGC) of 7.5e4, and IT of 100 ms. Data-dependent top-speed tandem MS (MS/MS) spectra (1-s cycle, collision-induced dissociation [CID] of 42%) were acquired in the ion trap at a resolution of 15,000, AGC of 1.0e4, and IT of 150 ms. The other instrumental conditions were the following: quadrupole isolation of 1 m/z; retention factor (RF) of 35%; sheath gas, auxiliary gas, and sweep gas of 30, 10, and 0 arbitrary units, respectively; ion transfer tube temperature of 289°C; vaporizer temperature of 92°C; and spray voltage of 3,500 V. Data were analyzed using Xcalibur 4.0, Compound Discoverer 3.0, and MzVault 2.1 (Thermo Fisher Scientific).
Isolation of isoacceptor tRNA from T. kodakarensis.
To purify tRNAGln from the T. kodakarensis strains, we opted to employ an affinity approach based on hybridization with a DNA oligonucleotide complementary to a portion of the target tRNA (49). The area most distinct for Gln sequences among all T. kodakarensis tRNA sequences is from the anticodon stem-loop (ASL) leading to the 3′ end of the molecule. However, since both isoacceptors for the Gln-encoding tRNA are identical except for a single position in the anticodon, it was not possible to isolate the CUG or UUG isoacceptor free of the other. Nevertheless, we reasoned that a single nucleotide difference in the sequence of the anticodon loop (ACL) should be of no consequence for the overall stability of the tRNA, so the isolation of a mixture containing both isoacceptors would not compromise the experiment.
Potential DNA affinity oligonucleotides were designed by walking along the length of the tRNA in 3-nt steps beginning at position 26 (see Fig. S6A in the supplemental material). By first investigating the ability of each oligonucleotide to hybridize with an in vitro-synthesized tRNAGln transcript corresponding to T. kodakarensis tRNAGln(CUG) via native PAGE, we identified Aff3 as the best candidate for forming a stable hybrid with the in vivo tRNAGln from T. kodakarensis (Fig. S6B).
The streptavidin-agarose (Thermo Scientific) resin was activated by binding the Aff3 biotinylated oligonucleotide to the streptavidin (oligonucleotide at 15 μM in 10 mM Tris-HCl [pH 7.5] and 100 mM NaCl). For annealing of the tRNA to the immobilized DNA, the total tRNA was dissolved in annealing buffer (10 mM Tris-HCl [pH 7.5], 900 mM NaCl, 1 mM EDTA) and heated to 95°C for 5 min. After cooling to 85°C, the resin (preequilibrated in annealing buffer) was added and the slurry allowed to fully cool to room temperature with occasional mixing. The resin was pelleted by centrifugation (5,000 × g), and the unbound RNA was removed with the supernatant. Annealing buffer was added to wash the resin, followed by heating to 45°C for 5 min to remove nonspecifically bound tRNA, centrifugation, and removal of the supernatant. This process was repeated until the OD260 of the supernatant was below 0.01 absorbance unit (AU)/ml. Elution of the tRNAGln was achieved by resuspending the resin in 0.5 ml of elution buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl), heating the solution to 75°C for 5 min, and centrifuging to collect the unbound tRNAGln (see Fig. S7 in the supplemental material). The isolated tRNA was shown to be homogenous in both denaturing (urea-Tris-borate-EDTA [TBE]) and native (TB, 100 mM NaCl) PAGE (see Fig. S8 in the supplemental material).
Production of tRNA transcripts in vitro.
Double-stranded template DNA was designed based on the sequence of tRNAGln(CUG) from T. kodakarensis, with the exception that the native gene sequence was modified by changing the 5′ adenosine nucleotide to a guanosine (double underline) for enhanced transcription yield (63): 5′-GGCCCCGUGGUGUAGCGGCCAAGCAUGCGGGACUCUGGAUCCCGCGACCGGGGUUCGAAUCCCCGCGGGGCUACCA-3′. The template DNA was prepared from two DNA oligonucleotides that were designed with a 10-bp overlap at the center of the target sequence (underlined) and which contained 2′-O-methyl modifications on the two terminal 5-residues of the template strand (63) and the standard T7 promoter at the 5′ end of the nontemplate strand (bold): 5′-TAATACGACTCACTATAGGCCCCGTGGTGTAGCGGCCAAGCATGCGGGA-3′and 5′-mUmGGTAGCCCCGCGGGGATTCGAACCCCGGTCGCGGGATCCAGAGTCCCGCATGC-3′.
The complete template was generated by primer extension using the Klenow fragment (Fermentas) to create two fully complementary strands. The two oligonucleotides were mixed to a final concentration of 4 μM each in the presence of deoxynucleoside triphosphates (dNTPs) (600 μM each) and using the manufacturer’s reaction conditions. The primers were extended by cycling 25 times between 37°C and 10°C in 30-s pulses (Applied Biosystems 2720 thermal cycler). The DNA was then isolated by organic extraction (equal volume of 25:24:1 phenol-chloroform-isoamyl alcohol vortexed and then centrifuged at 20,000 × g for 5 min) and ethanol precipitation of the aqueous phase. The template was then resuspended in water at 10 μM.
RNA was transcribed from 1 μM DNA template in 30 mM Tris-HCl (pH 8.0), 40 mM MgCl2, 10 mM DTT, 0.1% Triton X-100, 100 μM spermidine, 2.5 mM NTP (individual nucleotides obtained from Sigma, stock made up in DEPC-treated water and stored at −80°C), 50 μg/ml of the Δ172–173 mutant of T7 RNA polymerase (58), and 1 U/ml of inorganic pyrophosphatase (Sigma). The reactions were run for 4 h at 37°C and quenched by ethanol precipitation. The recovered pellet was solubilized in DEPC-treated water and then mixed with an equal volume of formamide–5 mM EDTA. The reaction products were denatured at 95°C and then separated by denaturing urea PAGE (7 M urea, 10% acrylamide, 1× TBE; gel run at 18 W). The full-length product band was excised from the gel and the RNA extracted by overnight crush and soak in 800 mM ammonium acetate. The purified RNA was then precipitated with the addition of ethanol and the pellet resuspended in 1.0 mM sodium citrate (pH 6.3) and stored at −80°C.
Preparation of preQ0- and G+-modified tRNA.
The tRNAGln(CUG) transcript was modified by incorporation of preQ0 base at position 15 by the action of M. jannaschii aTGT. The activity of the enzyme was determined by substituting [8-14C]guanine in place of preQ0 in a standard reaction assay (50), which established the conditions for quantitative incorporation of preQ0. Reaction conditions were 50 mM succinate (pH 5.5), 20 mM MgCl2, 100 mM KCl, 2 mM DTT, 100 μM tRNA, and 1 mM preQ0. The reaction solution containing tRNA was heated at 80°C for 3 min before the addition of aTGT to a final concentration of 10 μM and incubation at 80°C for 1 h. The reaction was repeated for two more rounds of incorporation to ensure complete substitution with preQ0 base. The reaction was terminated by the addition of 1/10 volume of 8 M ammonium acetate. Reaction components were removed by phenol-chloroform extraction, and the tRNA was isolated by ethanol precipitation of the aqueous phase. The tRNA pellet was resuspended in 1.0 mM sodium citrate (pH 6.3) and stored at −80°C.
To produce G+-modified tRNA, a sample of preQ0-modified tRNA was suspended (50 μM) in 100 mM HEPES (pH 7.0), 0.5 M NaCl, 20 mM MgCl2, 5.0 mM glutamine, 1.0 mM DTT, and 10 μM M. jannaschii ArcS. The sample was reacted for 1 h at 40°C. The modified RNA was isolated as described above. Samples of both preQ0- and G+-modified tRNA were digested, dephosphorylated, and analyzed by HPLC as described above to confirm the modification status (see Fig. S5 in the supplemental material).
UV thermal denaturation studies.
All thermal denaturation studies were performed on a Cary 100 Bio UV-Vis spectrophotometer. Single-wavelength absorbance at 260 nm was used to record the unfolding of the tRNA species being studied. The temperature was maintained by a thermostat-controlled cell block holder. The thermal melt cycle was controlled by the Thermal program in the Cary Win UV software suite. Samples were prepared in 10 mM sodium cacodylate (pH 7.0) and 100 mM NaCl. This was supplemented with either EDTA or MgCl2 for experiments lacking or containing MgCl2, respectively. RNA was heated in buffer to 98°C and slowly cooled to 55°C, at which point EDTA or MgCl2 was added and the sample allowed to cool to room temperature. During analysis, the sample volume (120 μl) was covered with mineral oil to prevent evaporation. The raw absorbance-versus-temperature data were converted to a differential profile (dA260/dT versus temperature) and the Tm determined from these plots.
Supplementary Material
ACKNOWLEDGMENTS
The Iwata-Reuyl lab acknowledges David Draper for engaging discussions about Mg2+-RNA binding.
This work was supported with funding from NASA (NNX07AJ26G to K.M.S. and D.I.-R.), the Alexander von Humboldt Foundation (to R.S.A. and K.M.S.), and the Department of Energy, Basic Energy Sciences Division (DE-SC0014597 to T.J.S.).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bjork GR. 1995. Biosynthesis and function of modified nucleosides, p 165–206. In Soll D, RajBhandary UL (ed), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC. [Google Scholar]
- 2.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. 2011. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res 39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama S, Nishimura S. 1995. Modified nucleosides and codon recognition, p 207–223. In Soll D, RajBhandary UL (ed), tRNA: structure, biosynthesis, and function. ASM Press, Washington, DC. [Google Scholar]
- 5.Bjork GR. 1992. The role of modified nucleosides in tRNA interactions, p 23–85. In Hatfield DL, Lee BL, Pirtle RM (ed), Transfer RNA in protein synthesis. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 6.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. 1988. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T. 1988. A novel lysine-substituted nucleoside in the first position of the anti-codon of minor isoleucine tRNA from Escherichia coli. J Biol Chem 263:9261–9267. [DOI] [PubMed] [Google Scholar]
- 8.Thiaville PC, Legendre R, Rojas-Benítez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crécy-Lagard V. 2016. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb Cell 3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalak JA, Dalluge JJ, McCloskey JA, Stetter KO. 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33:7869–7876. doi: 10.1021/bi00191a014. [DOI] [PubMed] [Google Scholar]
- 10.Derrick WB, Horowitz J. 1993. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res 21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perret V, Garcia A, Puglisi J, Grosjean H, Ebel JP, Florentz C, Giege R. 1990. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie 72:735–743. doi: 10.1016/0300-9084(90)90158-D. [DOI] [PubMed] [Google Scholar]
- 12.Horie N, Hara-Yokoyama M, Yokoyama S, Watanabe K, Kuchino Y, Nishimura S, Miyazawa T. 1985. Two tRNAIle1 species from an extreme thermophile, Thermus thermophilus HB8: effect of 2-thiolation of ribothymidine on the thermostability of tRNA. Biochemistry 24:5711–5715. doi: 10.1021/bi00342a004. [DOI] [PubMed] [Google Scholar]
- 13.Lorenz C, Lunse CE, Morl M. 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7:35. doi: 10.3390/biom7020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persson BC. 1993. Modification of tRNA as a regulatory device. Mol Microbiol 8:1011–1016. doi: 10.1111/j.1365-2958.1993.tb01645.x. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Xu Z, Sheng J. 2018. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes 9:246. doi: 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geslain R, Pan T. 2011. tRNA: vast reservoir of RNA molecules with unexpected regulatory function. Proc Natl Acad Sci U S A 108:16489–16490. doi: 10.1073/pnas.1113715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. 1993. Structure of archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-β-d-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J Biol Chem 268:10076–10086. [PubMed] [Google Scholar]
- 18.Kasai H, Oashi Z, Harada F, Nishimura S, Oppenheimer NJ, Crain PF, Liehr JG, von Minden DL, McCloskey JA. 1975. Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid. 7–(4,5-cis-Dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry 14:4198–4208. doi: 10.1021/bi00690a008. [DOI] [PubMed] [Google Scholar]
- 19.Katz JR, Basile B, McCloskey JA. 1982. Queuine, a modified base incorporated posttranscriptionally into eukaryotic transfer RNA: wide distribution in nature. Science 216:55–56. doi: 10.1126/science.7063869. [DOI] [PubMed] [Google Scholar]
- 20.Kersten H. 1988. The nutrient factor queuine: biosynthesis, occurence in transfer RNA and function. Biofactors 1:27–29. [PubMed] [Google Scholar]
- 21.Sprinzl M, Hartmann T, Weber J, Blank J, Zeidler R. 1989. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 17:r1–r67. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamura T, Hirata A, Ohno S, Nomura Y, Nagano T, Nameki N, Yokogawa T, Hori H. 2016. Multisite-specific archaeosine tRNA-guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo-acidophilic archaeon. Nucleic Acids Res 44:1894–1908. doi: 10.1093/nar/gkv1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata-Reuyl D, de Crécy Lagard V. 2009. Enzymatic formation of the 7-deazaguanosine hypermodified nucleosides of tRNA, p 379–394. In Grosjean H. (ed), DNA and RNA modification enzymes: structure, mechanism, function and evolution. Landes Bioscience, New York, NY. [Google Scholar]
- 24.Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. 2008. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP cyclohydrolase I. J Bacteriol 190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaran B, Bonnett SA, Shah K, Gabriel S, Reddy R, Schimmel P, Rodionov DA, de Crecy-Lagard V, Helmann JD, Iwata-Reuyl D, Swairjo MA. 2009. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J Bacteriol 191:6936–6949. doi: 10.1128/JB.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. 2006. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J Biol Chem 281:37586–37593. doi: 10.1074/jbc.M607114200. [DOI] [PubMed] [Google Scholar]
- 27.Grochowski LL, Xu H, Leung K, White RH. 2007. Characterization of an Fe(2+)-dependent archaeal-specific GTP cyclohydrolase, MptA, from Methanocaldococcus jannaschii. Biochemistry 46:6658–6667. doi: 10.1021/bi700052a. [DOI] [PubMed] [Google Scholar]
- 28.Mashhadi Z, Xu H, White RH. 2009. An Fe2+-dependent cyclic phosphodiesterase catalyzes the hydrolysis of 7,8-dihydro-d-neopterin 2′,3′-cyclic phosphate in methanopterin biosynthesis. Biochemistry 48:9384–9392. doi: 10.1021/bi9010336. [DOI] [PubMed] [Google Scholar]
- 29.McCarty RM, Somogyi A, Bandarian V. 2009. Escherichia coli QueD is a 6-carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 48:2301–2303. doi: 10.1021/bi9001437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowling DP, Bruender NA, Young AP, McCarty RM, Bandarian V, Drennan CL. 2014. Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat Chem Biol 10:106–112. doi: 10.1038/nchembio.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelp MT, Bandarian V. 2015. A single enzyme transforms a carboxylic acid into a nitrile through an amide intermediate. Angew Chem Int Ed Engl 54:10627–10629. doi: 10.1002/anie.201504505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips G, Chikwana VM, Maxwell A, El-Yacoubi B, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. 2010. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J Biol Chem 285:12706–12713. doi: 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bon Ramos A, Bao L, Turner B, de Crécy-Lagard V, Iwata-Reuyl D. 2017. QueF-like, a non-homologous archaeosine synthase from the Crenarchaeota. Biomolecules 7:36. doi: 10.3390/biom7020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips G, Swairjo MA, Gaston KW, Bailly M, Limbach PA, Iwata-Reuyl D, de Crécy-Lagard V. 2012. Diversity of archaeosine synthesis in crenarchaeota. ACS Chem Biol 7:300–305. doi: 10.1021/cb200361w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Lanen SG, Reader JS, Swairjo MA, de Crecy-Lagard V, Lee B, Iwata-Reuyl D. 2005. From cyclohydrolase to oxidoreductase: discovery of nitrile reductase activity in a common fold. Proc Natl Acad Sci U S A 102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinzie SD, Thern B, Iwata-Reuyl D. 2000. Mechanistic studies of the tRNA-modifying enzyme QueA: a chemical imperative for the use of AdoMet as a “ribosyl” donor. Org Lett 2:1307–1310. doi: 10.1021/ol005756h. [DOI] [PubMed] [Google Scholar]
- 37.Miles ZD, McCarty RM, Molnar G, Bandarian V. 2011. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A 108:7368–7372. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zallot R, Ross R, Chen W-H, Bruner SD, Limbach PA, de Crécy-Lagard V. 2017. Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem Biol 12:844–851. doi: 10.1021/acschembio.6b01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boland C, Hayes P, Santa-Maria I, Nishimura S, Kelly VP. 2009. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem 284:18218–18227. doi: 10.1074/jbc.M109.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersten H, Kersten W. 1990. Biosynthesis and function of queuine and queuosine tRNAs, p B69–B108. In Gehrke CW, Kuo KCT (ed), Chromatography and modification of nucleosides, part B. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 41.Marks T, Farkas WR. 1997. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem Biophys Res Commun 230:233–237. doi: 10.1006/bbrc.1996.5768. [DOI] [PubMed] [Google Scholar]
- 42.Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ. 1999. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology 255:2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 43.Durand J, Okada N, Tobe T, Watarai M, Fukuda I, Suzuki T, Nakata N, Komatsu K, Yoshikawa M, Sasakawa C. 1994. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of E. coli K-12. J Bacteriol 176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakovich T, Boland C, Bernstein I, Chikwana VM, Iwata-Reuyl D, Kelly VP. 2011. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J Biol Chem 286:19354–19363. doi: 10.1074/jbc.M111.219576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliva R, Tramontano A, Cavallo L. 2007. Mg2+ binding and archaeosine modification stabilize the G15 C48 Levitt base pair in tRNAs. RNA 13:1427–1436. doi: 10.1261/rna.574407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orita I, Futatsuishi R, Adachi K, Ohira T, Kaneko A, Minowa K, Suzuki M, Tamura T, Nakamura S, Imanaka T, Suzuki T, Fukui T. 2019. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res 47:1964–1976. doi: 10.1093/nar/gky1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Bäumer S, Jacobi C, Brüggemann H, Lienard T, Christmann A, Bömeke M, Steckel S, Bhattacharyya A, Lykidis A, Overbeek R, Klenk H-P, Gunsalus RP, Fritz H-J, Gottschalk G. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4:453–461. [PubMed] [Google Scholar]
- 48.Ehlers C, Weidenbach K, Veit K, Deppenmeier U, Metcalf WW, Schmitz RA. 2005. Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Goe1. Mol Genet Genomics 273:290–298. doi: 10.1007/s00438-005-1128-7. [DOI] [PubMed] [Google Scholar]
- 49.Kazayama A, Yamagami R, Yokogawa T, Hori H. 2015. Improved solid-phase DNA probe method for tRNA purification: large-scale preparation and alteration of DNA fixation. J Biochem 157:411–418. doi: 10.1093/jb/mvu089. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y, Fox DT, Lacy JA, Van Lanen SG, Iwata-Reuyl D. 2000. Hypermodification of tRNA in thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J Biol Chem 275:28731–28738. doi: 10.1074/jbc.M002174200. [DOI] [PubMed] [Google Scholar]
- 51.Thiaville JJ, Kellner SM, Yuan Y, Hutinet G, Thiaville PC, Jumpathong W, Mohapatra S, Brochier-Armanet C, Letarov AV, Hillebrand R, Malik CK, Rizzo CJ, Dedon PC, de Crécy-Lagard V. 2016. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci U S A 113:E1452–1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blaby IK, Phillips G, Blaby-Haas CE, Gulig KS, El Yacoubi B, de Crécy-Lagard V. 2010. Towards a systems approach in the genetic analysis of archaea: accelerating mutant construction and phenotypic analysis in Haloferax volcanii. Archaea 2010:426239. doi: 10.1155/2010/426239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davanloo P, Sprinzl M, Watanabe K, Albani M, Kersten H. 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res 6:1571–1581. doi: 10.1093/nar/6.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomura Y, Ohno S, Nishikawa K, Yokogawa T. 2016. Correlation between the stability of tRNA tertiary structure and the catalytic efficiency of a tRNA-modifying enzyme, archaeal tRNA-guanine transglycosylase. Genes Cells 21:41–52. doi: 10.1111/gtc.12317. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe M, Nameki N, Matsuo-Takasaki M, Nishimura S, Okada N. 2001. tRNA recognition of tRNA-guanine transglycosylase from a hyperthermophilic archaeon, Pyrococcus horikoshii. J Biol Chem 276:2387–2394. doi: 10.1074/jbc.M005043200. [DOI] [PubMed] [Google Scholar]
- 56.Wolf B, Lesnaw JA, Reichmann ME. 1970. A mechanism of the irreversible inactivation of bovine pancreatic ribonuclease by diethylpyrocarbonate. A general reaction of diethylpyrocarbonate. A general reaction of diethylpyrocarbonate with proteins. Eur J Biochem 13:519–525. doi: 10.1111/j.1432-1033.1970.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 57.Migawa MT, Hinkley JM, Hoops GC, Townsend LB. 1996. A two step synthesis of the nucleoside Q precursor 2-amino-5-cyanopyrrolo[2,3-d]pyrimidin-4-one (PreQ0). Synth Commun 26:3317–3322. doi: 10.1080/00397919608004641. [DOI] [Google Scholar]
- 58.Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. 1997. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J Mol Biol 269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- 59.Hileman TH, Santangelo TJ. 2012. Genetics techniques for Thermococcus kodakarensis. Front Microbiol 3:195. doi: 10.3389/fmicb.2012.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santangelo TJ, Cubonova L, James CL, Reeve JN. 2007. TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J Mol Biol 367:344–357. doi: 10.1016/j.jmb.2006.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bang C, Schilhabel A, Weidenbach K, Kopp A, Goldmann T, Gutsmann T, Schmitz RA. 2012. Effects of antimicrobial peptides on methanogenic archaea. Antimicrob Agents Chemother 56:4123–4130. doi: 10.1128/AAC.00661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pomerantz SC, McCloskey JA. 1990. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol 193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- 63.Sherlin LD, Bullock TL, Nissan TA, Perona JJ, Lariviere FJ, Uhlenbeck OC, Scaringe SA. 2001. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA 7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.