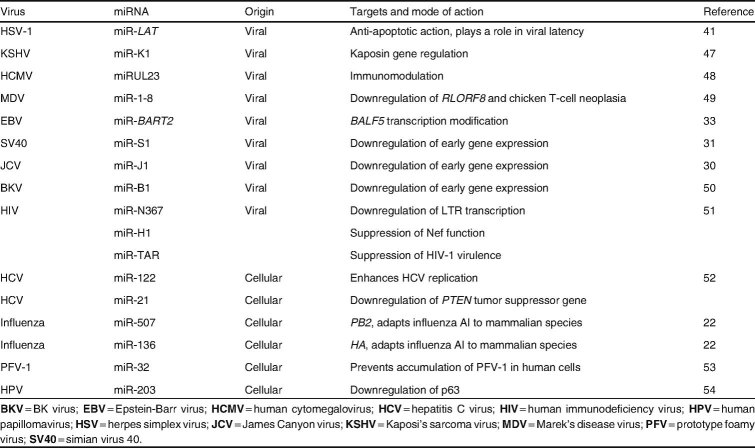
Abstract
New therapeutic approaches are urgently needed for serious diseases, including cancer, cardiovascular diseases, viral infections, and others. A recent direction in drug development is the utilization of nucleic acidbased therapeutic molecules, such as antisense oligonucleotides, ribozymes, short interfering RNA (siRNA), and microRNA (miRNA). miRNAs are endogenous, short, non-coding RNA molecules. Some viruses encode their own miRNAs, which play pivotal roles in viral replication and immune evasion strategies. Conversely, viruses that do not encode miRNAs may manipulate host cell miRNAs for the benefits of their replication. miRNAs have therefore become attractive tools for the study of viral pathogenesis. Lately, novel therapeutic strategies based on miRNA technology for the treatment of viral diseases have been progressing rapidly. Although this new generation of molecular therapy is promising, there are still several challenges to face, such as targeting delivery to specific tissues, avoiding off-target effects of miRNAs, reducing the toxicity of the drugs, and overcoming mutations and drug resistance. In this article, we review the current knowledge of the role and therapeutic potential of miRNAs in viral diseases, and discuss the limitations of these therapies, as well as strategies to overcome them to provide safe and effective clinical applications of these new therapeutics.
Keywords: Human Immunodeficiency Virus, Epstein Barr Virus, miRNA Expression Profile, HCMV Infection, Severe Acute Respiratory Syndrome
1. Introduction
RNA interference (RNAi) is a system in living cells that regulates the activation and silencing of gene expression. RNAi governs the regulation of host cell genes, mainly through two types of RNA molecules: small interfering RNA (siRNA) and microRNA (miRNA).[1] siRNAs are a class of double-stranded RNA molecules, 20–25 nucleotides in length, which play different roles in cellular biology. Experimental application or targeting of siRNAs in various cell types and animal models has shown promise for the potential treatment of diseases induced by viruses such as hepatitis C, influenza, and HIV.[2–4] The suppression of viral gene expression by siRNAs makes them very attractive for antiviral therapy, and some siRNAs are already being used in clinical trials.[5] However, siRNAs still have a long way to go before being brought to market, because of their potential side effects. One of the major concerns in using siRNAs as molecular therapeutics is their induction of a strong immune response.
miRNAs, which were discovered in 1993,[6] are a group of non-coding, single-stranded RNA molecules, ranging in size from 19 to 25 nucleotides.[7] It is now believed that miRNAs compose one percent of the total human genome. miRNAs are widely expressed in various species, including viruses. The first virally encoded miRNA was discovered in the Epstein Barr virus (EBV) genome.[8] Now, there are more than 141 identified miRNAs encoded by 15 viruses, with more expected to be identified in the near future as a result of the improvement in online prediction and validation tools.[9] Cellular miRNAs in animals seem to be conserved, while virally encoded miRNAs are highly variable even within the same group of viruses.[10] This may be due to the high frequency of viral mutations relative to eukaryotes. miRNAs play an important role in regulation of almost one-third of all known human mRNAs.[11] Most miRNAs have a specific tissue expression profile. Their unique expression pattern may explain their roles in different biologic activities, such as cellular differentiation, environment adaptation, oncogenesis, and host-pathogen interaction.[12]
The therapeutic potential of miRNAs was first realized with the discovery that downregulation of miR-15 and miR-16 is associated with development of B-cell leukemia.[13] Shortly after that, the potential for treatment of several other cancers was realized.[14] Scientists have aimed to control the expression level of key genes via manipulation of cellular or viral miRNAs to treat disease. These approaches include anti-miRNA oligonucleotides (AMOs), peptide nucleic acids, and miRNA sponges.[15] Initial experiments have obtained promising results in controlling various viral infections.[16–18] In addition, it has been found that restoring or over-expressing certain miRNAs may also be beneficial for reverse pathologic conditions, especially in cancer treatments.[19]
The goal of this article is to review the recent progress in the understanding of the roles of miRNAs in viral diseases, and to discuss the potential of these molecules in serving as a therapeutic target or as a useful therapeutic tool. We also specifically highlight the major obstacles faced by miRNA technology in both therapeutics and vaccine strategies.
2. Roles and Therapeutic Potential of MicroRNAs (miRNAs) in Viral Infections
Viruses are obligate intracellular parasites. They lack the essential machinery required for their replication. Thus, viruses adopt several clever strategies to ensure the success of their replication in a suitable host, one of which is manipulation of the host miRNAs to modify the cellular environment for their own benefit.[20] Some DNA viruses are capable of encoding their own miRNAs to modulate both the viral and cellular protein expression in order to provide a favorable environment for viral replication.[12] Answering back, certain host miRNAs alter the cell gene expression to defend the cells against the viral infection by interfering with viral proteins or other cellular factors as a type of immune response against these particular viruses.[21] Therefore, the relationship between viruses and miRNAs is complicated, to say the least. Since miRNAs play essential roles in viral infections, they are considered to be promising therapeutic targets in infectious diseases. Their endogenous nature, small size, and flexible function make miRNAs very good candidates, as they may trigger lower immunogenic responses and have fewer side effects than siRNAs.[12,14] In view of the current data regarding the roles of different viral and cellular miRNAs in various viral replication cycles, we believe that manipulation of these miRNAs will have a promising therapeutic role in infectious diseases.[12,14,22–25] Currently, there are more than 700 miRNAs encoded by the human genome alone.[12] We will discuss the roles and therapeutic potential of cellular as well as viral miRNAs (if any) in the pathogenesis and treatment of different viral diseases.
2.1 Human Polyomaviruses (HPyVs)
Human polyomaviruses (HPyVs) are a group of oncogenic, circular, non-enveloped, double-stranded DNA (dsDNA) viruses.[26] Five polyomaviruses have been found to infect humans. Of particular interest are two strains of these viruses, named (according to the initials of the first affected patients) BK virus (BKV) and James Canyon virus (JCV).[27] The reservoir species for human infection is the rhesus macaque. Humans have also acquired simian virus 40 (SV40) infection from contaminated poliovirus vaccines, and recent studies have reported horizontal transmission between people.[28] HPyVs induce a wide range of tumors affecting almost all body organs (including the brain, bones, colon, pancreas, stomach, and urogenital tract), as well as lymphomas and leukemia.[28,29] The viral genome of SV40 encodes five proteins; two large T antigen (LT), one small T antigen (st-Ag), and three that encode the capsid proteins (Vp1, Vp2 and Vp3). HPyVs are able to encode viral miRNAs for their own benefit. Both BKV and JCV encode the same miRNA, named miR-J1. It is upregulated in the brain in progressive multifocal leukoencephalopathy syndrome, suggesting a major role in this particular disease.[30]
SV40 encodes miRNAs called v-miRNAs during the late stages of infection.[31] They are complementary to the viral mRNAs produced at the early stage of viral infection.[31] These v-miRNAs slow down the expression of the viral T-antigen genes and lower the level of interferon (IFN)-γ produced by cytotoxic T lymphocytes, thus reducing the influx of inflammatory cells and facilitating evasion of the immune response.[31]
2.2 Human Papillomaviruses (HPVs)
Human papillomaviruses (HPVs) are also oncogenic viruses.[32] They are usually associated with different forms of both benign and malignant tumors, especially those affecting the skin and the genital tract.[32] These viruses are usually classified, on the basis of their virulence, into either low- or high-pathogenic variants.[26] Only a few HPV strains can produce miRNAs during their replication.[33] For example, HPV-31 encodes viral miRNAs at a very early stage of the infection, but these miRNAs are usually degraded once latent infection takes place.[34] In another independent study, both HPV-31 and HPV-18 were found to encode viral miRNAs, but they are not involved in cell transformation or cancer development.[33]
It is well known that host miR-34a is involved in inhibition of abnormal cell growth in tumors.[35] miR-34a inhibits cell cycle progression at the G2 phase and subsequently induces apoptosis.[36] Studies have reported success using the tumor suppressor complex (mRNA-cellular miRNA-34a), which targets downstream genes of tumor protein p53 (TP53). miR-34a is usually downregulated during HPV infection in primary keratinocytes.[36] Thus, restoration of the normal expression level of this miRNA is a potential strategy for therapeutic intervention.[36]
2.3 Adenoviruses
Adenoviruses are a group of non-enveloped dsDNA viruses. Over 50 serotypes have been identified in different clinical diseases, such as respiratory, gastrointestinal, urogenital, and eye diseases.[36]Adenovirus usually encodes several small non-coding RNA molecules called virus-associated RNAs, such as VA1 and VA2.[37] They facilitate immune evasion by inhibiting dsRNA-induced protein kinase R (PKR), which blocks IFN-α activity.[38] One study reported that adenovirus VA1-RNA interferes with the biogenesis of host miRNAs and the function of siRNA/shRNA (short hairpin RNA), through inhibition of the nuclear transport of the pre-miRNAs and the shRNAs and direct inhibitory action of Dicer.[37] Usually, a small part of the VA1 RNA is subjected to processing by Dicer, and this results in generation of miRNA. Use of anti-miRNA antisense inhibitors (2′-O-methyl AMO) to downregulate this miRNA resulted in inhibition of virus production.[39]
2.4 Herpesviruses
Herpesviruses are a group of enveloped dsDNA viruses, classified into three subfamilies (α, β, and γ). They are characterized by induction of latent infections in their target hosts.[40] These virus-encoded miRNAs play important roles in the establishment of latent infection, as well as the pathogenesis of virally induced diseases. According to the most recent studies, herpesviruses utilize their encoded miRNAs in a wide range of biologic functions, such as inhibition of apoptosis, immune evasion, control of cellular proliferation, and regulation of viral replication.[41–43] In the following section, we will discuss herpesvirus-encoded miRNAs.
2.4.1 Herpes Simplex Virus (HSV)-1
One of the most important genes encoded by herpes simplex virus (HSV) is called the latency-associated transcript (LAT).[44] This gene does not encode proteins but may be involved in the production of miRNAs or in cell survival after viral infection.[44] There has been debate around the origin of miR-LAT as to whether it is a virus-encoded or cell-encoded miRNA.[41] This miRNA is believed to act by downregulating transforming growth factor (TGF)-β and SMAD3. TGF-β plays an important role in cell proliferation and induction of apoptosis. SMAD3 is a signaling pathway mediator, which is triggered by the action of TGF-β.[41] HSV-1 miR-LAT-ICP34.5 has been recently discovered during HSV-1 infection.[45] According to bioinformatic analysis, the HSV-1 genome encodes 24 miRNAs, eight of which were found to be conserved between both HSV1 and HSV-2, and thus are believed to be functional.[46] Six of these miRNAs are upregulated in the trigeminal ganglia of mice infected with HSV-1. These miRNAs are encoded by LAT (table I).[46] In addition, quantitative reverse transcription PCR showed that both miR-1 and miR-6 were highly expressed in Vero cells infected with HSV-1 — as high as 1200 and 300 copies, respectively — whereas the other four miRNAs showed downregulation, with only 40 copies per infected cell.[55] The miRNA expression profile during HSV-1 infection revealed that several miRNAs among the eight candidates mentioned earlier were upregulated. Those miRNAs were believed to play major roles in induction of the latent phase of viral infection. This assumption was based on comparison between the miRNA expression profiles of the latent and active infections. For example, miR-H2 was found to be upregulated in latent infection to a level of 63 000 copies/cell, compared with less than 40 copies/cell during active infection.[56] Furthermore, miRNA-2 inhibits the production of the infected cell polypeptide (ICP)-0 protein, which is responsible for triggering the active phase of HSV infection. Another upregulated miRNA, miR-6, is responsible for downregulation of ICP4, which is responsible for the increased expression level of many HSV genes during the active phase of HSV-1 infection.[55,56]
Table I.
Overview of selected microRNAs (miRNAs) involved in the pathogenesis of viral diseases
2.4.2 Human Cytomegalovirus (HCMV)
Human cytomegalovirus (HCMV) is a herpesviruses affecting humans, and can result in acute or latent infections.[57] The form of infection largely depends on the immune status of the affected host.[57] It can be fatal in immune-compromised patients, such as those with AIDS or recent organ transplants.[57] It may also be responsible for birth defects and congenital abnormalities in pregnant women.[21] As with other herpesviruses, there is evidence supporting the presence of miRNAs that modulate viral pathogenesis in different tissues.[21] Specifically, HCMV has been recently reported to encode 15 miRNAs.[21] The most commonly expressed three miRNAs during the active phase of HCMV infection are miR-UL23-5p, miR-UL23-3p, and miR-US24,[48] which target different cellular proteins, such as transcription factors (HFN3 and TGIF2), receptors (CD206 receptors for interleukin-18), and other proteins involved in signal transduction pathways, such as RAB2L.[48] HCMV is able to induce the latent phase of infection by a cunning immune-evasion strategy through the action of miR-UL112, which targets several cellular proteins such as major histocompatibility complex (MHC) class I associated proteins, especially the MHC class I-related chain B (MICB).[58,59] miR-UL112-1 also regulates early viral protein expression, such as immediate early protein (IE)-72. IE72 is a key mediator in the shift from the latent to the active phase of viral infection, because it suppresses IE1.[58,59] Thus, if the HCMV infection is synchronized with overexpression of the miR-UL112, the IE1 protein expression level is greatly reduced, which mediates the latent phase of infection.[43] It is also thought that miR-UL112-1 targets another viral protein called UL114.[58] Downregulation of UL114 protein, using miR-UL112-1, results in inhibition of viral DNA replication and subsequently triggers the latent phase of infection, making the virus able to evade the host immune system.[34] Exploitation of this mechanism is being considered as a potential therapeutic strategy.[21]
2.4.3 Epstein-Barr Virus (EBV)
EBV is another oncogenic virus affecting humans. It is usually associated with induction of latent infection in more than 95% of affected patients.[60] In most cases, benign tumors develop; in some cases, however, malignant tumors may also develop, such as Hodgkin’s lymphoma, T-cell lymphoma, nasopharyngeal carcinoma, and gastric tumor.[61] During the active phase of EBV infection, more than 100 genes are usually expressed, whereas only 11 of them are expressed during latent infection.[21] It has been recently reported that EBV encodes more than 20 miRNAs.[33] These miRNAs are divided into two groups: one group is encoded from the intronic regions of a gene called BART, which is expressed at high levels in epithelial cells, but at lower levels in B cells, in the latent phase of infection; the other group is encoded from the untranslated region of a gene called BHRF1, a viral BCL2 homolog that prevents apoptosis.[8] Although the functions of most EBV-encoded miRNAs have not been completely identified, it has been found that miRNA-BART2 targets BALF5 mRNA, a viral DNA polymerase, and miRNA-BART2 induces cleavage in the 3′ untranslated region (UTR) of BALF5, resulting in inhibition of the lytic viral infection (figure 1 and table I).[63] According to bioinformatic prediction data, EBV miRNA-BART5 is believed to target PUMA, a modulator of apoptosis in the BCL2 protein group regulated by TP53.[64] In some cases, PUMA can also induce apoptosis through the TP53-independent pathway. Therefore, targeting PUMA with EBV miR-BART5 results in suppression of its action in apoptosis.[64]
Fig. 1.
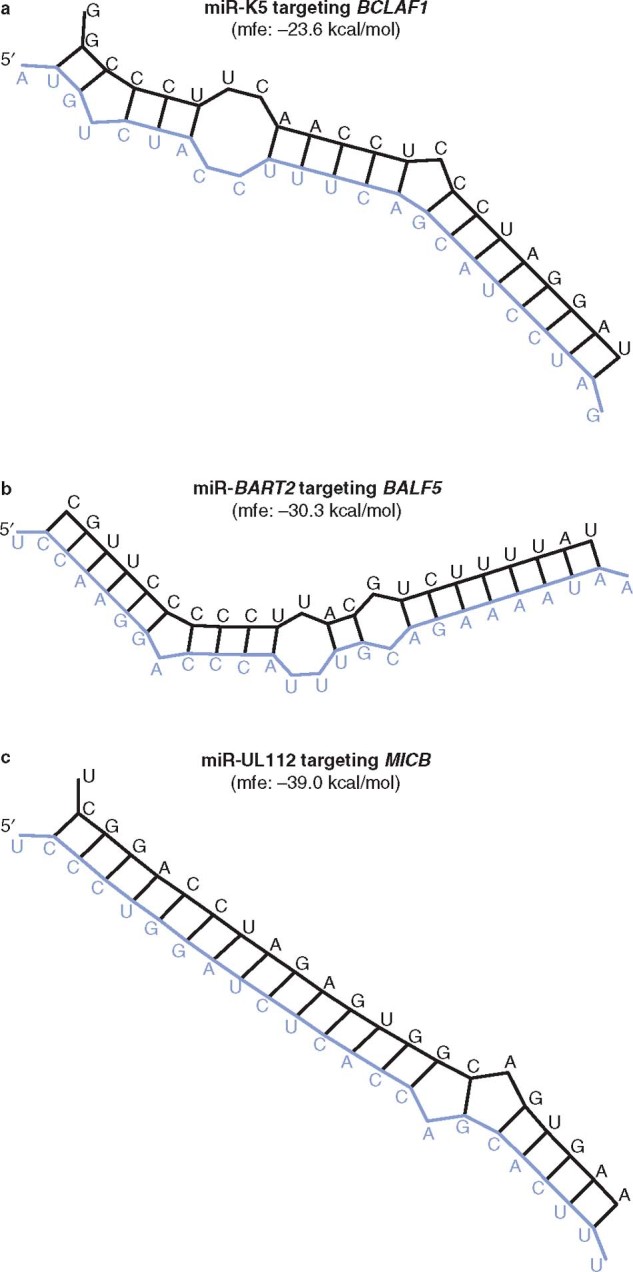
Secondary structures of viral microRNAs (miRNAs [black sequences]) hybridizing with corresponding targets of host genes (blue sequences). The viral miRNA sequence and the 3′ untranslated region (UTR) sequence of the targeted host and viral genes were obtained from the referenced articles or from the National Center for Biotechnology Information (NCBI) website and then submitted to the RNAhybrid software program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). The predicted secondary structure for the hybridization between viral miRNAs and their targets are generated by the RNAhybrid program. (a) miR-K5 encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV), targeting host gene BCLAF1 (BCL2-associated transcription factor 1).[62] (b) miR-BART2 encoded by EBV, targeting the viral DNA polymerase gene BALF5.[63] (c) miR-UL112 encoded by herpesviruses, targeting host gene MICB (MHC class I-related chain B).[59] mfe = minimum free energy.
Other EBV-encoded miRNAs target the IFNγ-inducible chemokine CXCL11.[65] Without CXCL11, EBV is able to evade the host immune response and subsequently enhance EBV replication.[65] The same group performed bioinformatic analysis and showed that CXCL11 contains a target sequence for miR-BHRF1-3 at its 3′UTR sequence. Targeting CXCL11 using this viral encoded miRNA will have an immunomodulatory effect on the viral-induced tumor.[65] Therefore, it is believed that targeting BHRF1–3 could be a good therapeutic approach for viral EBV-induced tumors.[65]
2.4.4 Kaposi’s Sarcoma Virus (KSHV)
KSHV is one of the gamma-herpes viruses groups and is usually associated with Kaposi’s sarcoma infection, from which it acquired its name.[47] Like other herpesviruses, KSHV usually induces latent infections.[47] KSHV encodes several miRNA candidates from the genomic region spanning 4 kilobases between ORF12 and ORF71.[66] KSHV-encoded miRNAs target important genes involved in cell proliferation, modulation of the host immune system, apoptosis, and angiogenesis. For example, miR-K5 targets the mRNA of the BCL2 (prosurvival gene) interacting protein called BCLAF1.[62] This leads to reactivation of the KSHV lytic infection.[62] Thus KSHV-encoded miRNAs play an important role in the virus/host interactions, and silencing of those miRNAs using different approaches, particularly the AMO, is therefore a promising therapeutic strategy against such virus infection (figure 1).[62]
2.4.5 Marek’s Disease Virus (MDV)
Marek’s disease virus (MDV) is one of the alpha herpesviruses, characterized by rapid production of T-cell lymphomas in chickens.[67] The viral genome encodes several miRNAs, which are mainly encoded from the oncogenes, especially Meq and LAT.[68] The miRNAs of this virus are highly expressed in MDV-transformed cells, suggesting an essential role in MDV oncogenesis. However, their target genes have not been well identified.[68,69] It is known that several host cell miRNAs — such as miR-17, miR-92, miR-21, and let-7i — are involved in the cancer development induced by MDV in chickens.[68] They are most often upregulated in an MSB-1 (MDV-transformed CD4+ T-cell line derived from a spleen lymphoma induced by the BC-1 strain of MDV-1) library after MDV infection.[69] This makes for an ideal model for the study of miRNAs. Comparative analysis of miRNA expression profiles during MDV infection, using microarray analysis, may enable identification of specific miRNAs involved in neoplastic transformation when compared with non-infected cells. This may become a useful diagnostic approach through examination of miRNA signatures profiles.[70] This is supported by a recent report showing an association between downregulation of the host cellular miR-155 expression and upregulation of MDV-encoded miRNAs (M2-5P, M12-3P, M5-3P, M4-5P, M3-5P, M11-5P, M2-3P, M4-3P, and M3-3P), a potential marker for MDV-induced tumor formation.[69] Subsequent identification of the putative target of these miRNAs will lead to a better understanding of the molecular mechanism of MDV-induced tumorigeneses.[69]
2.5 Human Immunodeficiency Virus (HIV)
The human immunodeficiency virus (HIV) genome encodes several important genes, such as Nef, Vef, Tat, and Vpu.[71] Bioinformatic analysis suggests that the HIV genome encodes five pre-miRNAs, which are processed into ten mature miRNAs, but their definite functions are still not well identified.[72]
The HIV Nef gene is located at the 3′ end of the HIV genome and is highly expressed in early viral infection.[73] It has been shown to downregulate the expression levels of CD4, CD28, and MHC class I molecules.[73] HIV Nef encodes miR-N367, which has a unique function at both the transcriptional and translational levels, rather than at the post-transcriptional level.[51,74] The mechanism of action of miR-N367 is believed to be suppression of HIV promoter activity; however, further studies are required to explain the exact mechanism of such an action.[51] Several studies reported that downregulation of the Nef gene results in inhibition of HIV replication.[51,74] This may lead to production of low-pathogenic strains of HIV or may favor latent HIV infection.[74] Targeting of these viral miRNAs using different approaches, especially AMO treatment, could potentially have a therapeutic effect on HIV.[2]
2.6 Hepatitis C Virus (HCV)
Currently, there are more than 170 million people affected by hepatitis C virus (HCV) infection worldwide.[75] Drug resistance is one of the major hindrances in treating such viral infection.[76] HCV induces different forms of tumors in humans and is one of the major causes of liver diseases all over the world, resulting in hepatocellular carcinoma and, finally, complete liver failure.[77] It has been recently reported that host cellular miRNA-122 has two recognition sites in the 5′UTR of the HCV genome, resulting in upregulation of HCV infection.[78] Further investigation showed that interaction between miRNA-122 and the viral genome causes accumulation of viral RNA in the liver tissues (table I). Furthermore, the level of viral RNA in the liver tissues is usually controlled by miR-122 binding sites.[78] Interestingly, HCV infection also modulates cellular miRNA expression profiles. Following HCV infection, three miRNAs (miR-122, miR-100, and miR-10a) are upregulated, while other two miRNAs (miR-198 and miR-145) are downregulated.[78] Ura et al.[52] found that cyclin G1 acts as a putative target for miR-122. Use of a primate model targeting miR-122 with specific antagonists resulted in a reduction in the level of HCV replication in the affected livers, demonstrating the promise of this strategy.[79] In addition, a recent study demonstrated the therapeutic potential of silencing miR-122 in chronic HCV viral infection in primate models, whereby chimpanzees that were positive for HCV infection were treated with a specific LNA-modified oligonucleotide (SPC3649).[79] These LNA-oligomers targeted the complement sequence of miR-122 and resulted in a decrease in the duration of the viremia following acute HCV infection. There were no reports of any side effects or any viral resistance observed after the treatment.[79] This approach provided long-lasting effects in the HCV-infected animals, as well as great improvement in the liver pathology.[79] More clinical trials are needed to further confirm the promising results of this new molecular therapeutic approach.[78]
2.7 Hepatitis B Virus (HBV)
Hepatitis B virus (HBV) belongs to the genus Orthohepadnavirus in the family of Hepadnaviridae. HBV infection progresses into cirrhosis and hepatocellular carcinoma in most cases.[80] It is believed that HBV encodes miRNAs that regulate their own gene expression.[81] According to bioinformatic predictions, HBV encodes only one miRNA. However, several studies have failed to identify any cellular genes regulated by this virus-encoded miRNA, implying alternative gene expression mechanisms.[81,82] According to the miRNA expression profiles of several patients suffering from cirrhosis due to HBV infection, the host Hsa-miRNA-615-3p is usually upregulated. In recent clinical studies, Ura et al.[52] studied the role of different miRNAs in the pathogenesis of both HBV and HCV in the context of development of hepatocellular carcinoma in infected liver tissues.[52] In this study, the differential expression levels of 188 miRNAs from 12 HBV and 18 HCV patients were tested using qRT-PCR. According to this study, 19 miRNA candidates were highly expressed in both HBV and HCV infections, and 31 miRNAs served as markers for the severity of liver damage. It is known that HBV infection triggers pathways associated with DNA damage, recombination, and signal transduction pathways — whereas HCV infection usually triggers an immune response, antigen presentation, cell cycle progression, proteosome activation, and lipid metabolism. Therefore, the overall conclusion was that certain miRNAs may act as important mediators in the pathogenesis of both HBV and HCV infections.[52] These studies have paved the way to a new era in molecular antiviral therapy through modulation of the expression levels of those key miRNAs. There is now a new antiviral therapy for controlling HBV infection, using artificial miRNAs. This approach has revealed a dramatic reduction in HBV protein expression levels and a remarkable reduction in viral DNA replication in vitro.[83–85]
2.8 Severe Acute Respiratory Syndrome Virus (SARS)
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a single-stranded RNA virus, which belongs to the family Coronaviridae. Although several trials have been performed to treat this pathogen using conventional drugs such as ribavirin, antibiotics, anti-inflammatory steroids, and different kinds of immune stimulators, these approaches still lack viral specificity. SARS-CoV infection in bronchioalveolar stem cells (BASCs) is a prime example of how miRNA modulates the virus-host interaction. SARS-CoV is unable to replicate in well differentiated cells, so it has to control BASC cellular differentiation in order to establish a successful viral infection.[86] This virus usually hijacks cellular miRNAs such as miR-17*, miR-574-5p, and miR-214 for the benefits of its replication and immune evasion. The nucleocapsid and spike glycoproteins downregulate the expression levels of miR-223 and miR-98, respectively. This action enables the virus to hinder BASC cellular differentiation and the production of inflammatory chemokines, creating an environment that is optimal for virus replication. Restoration of the levels of miR-223 and miR-98 poses a potential novel approach in treating SARS-CoV infection.[86]
2.9 Influenza Virus
The influenza A outbreak of 2009 provided a warning about the urgent need for new alternative molecular therapeutic approaches for both the treatment and prophylaxis of such viral infections. Molecular therapy using miRNA technology may offer a new therapeutic approach to cope with the continuous changes in virus strains every year. Recent bioinformatics tools have paved the way for the discovery of new miRNAs and their target sequences for the design of nucleic acid-based therapeutics. For example, there are two human-encoded miRNAs that have potential binding sites within both the viral polymerase (PB2)and hemagglutinin (HA) genes (miR-507 and miR-136, respectively) [table I].[22] The target sequences of these two miRNAs are highly conserved among different influenza virus strains. The HA protein is involved in the attachment of the virus to its receptors, and the PB2 protein is an essential component in the ribonucleoprotein complex, needed for RNA transcription and replication.
The presence of human miRNAs that target conserved regions of influenza RNA suggests that the human genome has evolved to use this as a defense mechanism against infection. This supports the argument that targeting viral genes with miRNAs may be an effective strategy. This may also suggest that it is a futile attempt, since we have the miRNAs and yet still succumb to influenza infections. A recent study has been conducted to determine miRNA expression profiles after avian influenza virus (AIV) infection in chickens. This study showed changes in the cellular miRNA profile in response to the AIV infection, suggesting that the miRNAs play a role in the host-pathogen interaction during AIV infection.[87] Specifically, there were alterations in the miRNA profiles of miR-146, which had been previously reported to play a role in immune-related signal pathways in mammals.[87]
One recent study utilized miRNA technology in the development of influenza virus vaccines, whereby a new influenza A virus vaccine was developed using miRNA-based gene silencing. The method involves introducing an miRNA sequence of non-avian origin, known as a miRNA-responsive element (MRE), into the viral nucleoprotein gene, resulting in construction of new reassortant H1N1 and H5N1 viruses. With this strategy, the degree of the viral attenuation is controlled by the expression level of mir-124, which targets the introduced MRE sequence. This novel strategy offered a very good vaccine that was species specific, offering a high level of protection.[4] The nascent viruses were attenuated for mice, while they still propagated well in embryonated chicken eggs and were able to generate high levels of neutralizing antibodies in animals. This novel approach for influenza vaccine development may be used in combination with the currently available vaccine in order to increase both the safety and the efficacy of influenza virus vaccines in the near future.[4]
2.10 Coxsackievirus
Coxsackievirus, especially coxsackievirus B3 (CVB3), is the most common pathogen of human myocarditis. Anti-CVB3 drug development has been recently focused on a nucleic acid-based strategy. Our laboratory first reported the successful inhibition (>92%) of CVB3 replication in HeLa cells by transfection of siRNAs targeting viral protease 2A RNA. We also found that the antiviral effect was disrupted by mutations in the central strand region, and mismatch was tolerated near the 3′ end but not near the 5′ end of the siRNA;[88] furthermore, the siRNA effect was mediated by the antisense strand to the viral genome, rather than the sense strand complimentary to the viral negative-strand of the replicating intermediate. This finding was further conformed by another report.[89] When applied systemically to mice, siRNA targeting 2A had a significant protective effect if applied 6 and 14 hours after infection, including reduced viral replication and tissue injury, as well as an increased survival rate.[90]
Recently, we tested a packaging RNA (pRNA) vector (a component of the bacterial phage nano-motor) for targeted delivery of siRNAs. Through conjugation of a folate ligand to the pRNA vector, we specifically delivered the siRNAs targeting CVB3 2A to HeLa cells and HL-1 cardiomyocytes that expressed folate receptors.[91] In addition to the transfection of mature siRNAs, overexpression of shRNAs was also effective against CVB3 3D RNA polymerase and structural protein VP1, both in cells and in mice, where viral pancreatitis was significantly reduced.[92] Schubert et al.[89] used the SiDEx double expression vector to simultaneously transfect two siRNA sequences targeting the CVB3 3D RNA polymerase sequence in a green fluorescent protein (GFP) reporter construct. This double expression of both siRNAs successfully suppressed reporter expression despite the intentional introduction of an artificial point mutation (simulating an escape mutation or a miRNA target) that caused a mismatch with one of the two siRNAs.
3. Limitations of the Therapeutic Potential of miRNAs
As we have discussed above, there have been some promising results supporting the development of miRNAs for the treatment of several viral infections, and some of these miRNA-based drugs have reached the clinical trial stage. Despite this great progress, their clinical applications are still hampered by several challenges. In the following section, we briefly discuss the current obstacles or limitations facing miRNAs-based antiviral therapy.
3.1 Immune Responses to Viral Vectors
One of the major limitations for the use of miRNA-based antiviral therapy is the production of transgene-specific immunity.[93] Delivery of miRNAs using viral vectors usually results in the development of immune response against the viral vector. Basically, the delivery vector will stimulate an innate immune response in the forms of cytotoxic T-lymphocytes, humoral neutralizing antibody against their viral capsid proteins, and cytokine-mediated inflammatory responses in vivo.[93,94] Direct correlation between the immune response to the adenovirus capsid protein and the concentration of the viral vector has been reported; this interaction is usually associated with undesirable side effects in the host, especially if the construct moves from the target tissue into the blood circulation.[95]
3.2 Lack of Targeted miRNA Delivery Systems
The targeted delivery of siRNA, miRNA, and other nucleic acid-based therapies is another major concern of using these molecular therapeutic approaches. In contrast to the great progress in local administration of both siRNA- and miRNA-based therapies in the eyes, lung, and vagina, systemic delivery to target organs such as the liver, heart, and intestine is still undergoing optimization.[96,97] It is interesting to note that some studies have shown success in administering siRNAs via the intracerebral route; however, the risk is that foreign nucleic acid may be delivered to the central nervous system.[93]
3.3 Lack of Established miRNA Standard Analysis Techniques
Several laboratory techniques — such as real-time PCR, micro-array analysis, Luminex bead arrays, Northern blotting, in situ hybridization, formalin fixation, and paraffin embedding — are currently in use in miRNA detection and quantification. However, all of these techniques still require further optimization.[98–100] Once they are optimized, a clear choice for sensitivity and specificity will emerge, and this approach will allow early and sensitive detection of miRNA expression in different disease syndromes. This will have a great impact on the early tracking of serious viral diseases.[101]
3.4 Off-Target Effects and Unidentified Targets of miRNAs
The off-target effects of siRNAs were one of the major concerns in earlier studies using both siRNA and shRNA technologies in gene therapy. As a new generation of molecular gene therapy, miRNAs would be expected to have a high degree of specificity for their targets. However, since miRNA action is based on imperfect base pairing with the target sequence in most circumstances, the specificity will be lower than that of siRNA. This prediction has been confirmed by recent clinical trials, followed up by microarray analysis, which revealed possible off-target effects of miRNAs.[102] Another follow-up study by Birmingham et al.,[103] using the combination of bioinformatics and microarray analysis, found that using either the siRNA or the miRNA could result in off-target silencing. In addition, in vivo studies have revealed that one miRNA may target several genes at the same time, and the targets are not clearly identified. This suggests diverse modes of action of a given single miRNA. On the other hand, one gene may be regulated by several miRNAs,[104] indicating that the mode of action is more complicated than expected. Since drug therapies must precisely target the virus in question and nothing else, a large undertaking is needed to gather all possible information regarding all targets of each miRNA that is being considered for drug development.[104,105]
3.5 Mutations and Resistance
Although the currently used viral vectors in miRNA delivery are non-pathogenic, there is always the possibility of mutations within those viral vectors. These mutations may not only result in abnormal gene expression of the viral miRNA construct but may also cause possible insertion of vectors into the human genome, increasing the risk of cancer.[23] Moreover, the targeted viruses (especially the RNA viruses) are prone to mutation, which may drive drug resistance. There are currently two possible approaches to conquer these issues: one is the targeting of cellular factors that are essential for virus replication or use of more than one miRNA for the same target gene; the other possible solution is the targeting of several conserved regions of the viral genome by different siRNAs or miRNAs.
4. Concluding Remarks and Future Prospects
Viruses are among the most common causes of human diseases. Because of the unique biologic properties of viruses, there is no effective and specific antiviral therapy available so far. Several vaccines and antiviral drugs have shown a limited degree of efficacy for prophylaxis and treatment of some viral infections. However, high mutation rates enable viral diseases to emerge and re-emerge frequently. Thus, new strategies for drug and vaccine development must be devised to fight the threat of viral diseases to human health. Recent advances in the understanding of miRNA structure, function, and particularly their association with the molecular pathogenesis of a variety of complex diseases, have served as a theoretical basis for drug development. On the one hand, as key factors for viral replication and latency, miRNAs are ideal targets for inhibition. In this regard, construction of mRNAs that contain multiple tandem binding sites of a given miRNA may be useful to produce decoys or ‘miRNA sponges’ to inhibit the function of a specific miRNA. In addition, chemically synthesized antisense RNA oligomers (‘antagomirs’) targeting a miRNA of interest could be also be a promising approach to inhibit miRNA activity. On the other hand, miRNA expression vectors can be used to overexpress specific miRNAs to achieve a long-term effect of reversing the imbalance of miRNA expression caused by infection. Further, introduction of pre-miRNA mimetics for transient replacement is another option for investigation.
In summary, although there are many limitations at present, we believe that with the rapid progress in miRNA research, these small molecules will become an invaluable target and a useful tool for basic research and drug development. It is expected that an miRNA-based antiviral therapy will become available for clinical application in the near future.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of BC and Yukon. Dr. Maged Gomaa Hemida is a recipient of the CIHR-IMPACT postdoctoral training fellowship. Xin Ye is supported by a UGF Award from the University of British Columbia.
The authors have no conflicts of interest that are directly relevant to the content of this review.
References
- 1.Elbashir S, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Couturier JP, Root-Bernstein RS. HIV may produce inhibitory microRNA (miRNAs) that block production of CD28, CD4 and some interleukins. J Theor Biol. 2005;235(2):169–84. doi: 10.1016/j.jtbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 4.Perez JT, Pham AM, Lorini MH, et al. MicroRNA-mediated species-specific attenuation of influenza A virus. Nature Biotechnol. 2009;27(16):572–6. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 5.Ashihara E, Kawata E, Maekawa T. Future prospect of RNA interference for cancer therapies. Curr Drug Targets. 2010;11(3):345–60. doi: 10.2174/138945010790711897. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Griffith-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeffer S. Identification of virus-encoded microRNAs. Science. 2004;304:734–6. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNA in hostvirus interaction. Nucleic Acids Res. 2008;37(4):1035–48. doi: 10.1093/nar/gkn1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2005;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38:S25–30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 13.Callin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell JC, Kirn D. MicroRNAs fine-tune oncolytic viruses. Nature Biotechnol. 2008;26(12):1346–8. doi: 10.1038/nbt1208-1346. [DOI] [PubMed] [Google Scholar]
- 15.Elbert MS, Nelson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elmen J, Lindow M, Schutz S, et al. LNA-mediated microRNAs silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 17.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44(1):55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Mattes J, Collision A, Foster PS. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr Opin Mol Ther. 2008;10(2):150–7. [PubMed] [Google Scholar]
- 19.Saumet A, Vetter G, Cougot N, et al. MicroRNA-associated therapies. In: Ying S-Y, et al., editors. Current perspectives in microRNAs (miRNA) New York: Springer-Verlag; 2008. pp. 395–429. [Google Scholar]
- 20.Scaria V, Hariharan M, Maiti S, et al. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68–76. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moens U. Silencing viral microRNA as a novel antiviral therapy? J Biomed Biotechnol. 2009;2009:419539. doi: 10.1155/2009/419539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3(68):1–9. doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castano D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–33. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3(6):375–87. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henke JI, Goergen D, Zheng J, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–10. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical applications. Nat Rev Cancer. 2008;2(5):342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 27.Shah KV. Human polyomavirus BKV and renal disease. Nephrol Dial Transplant. 2000;15:754–5. doi: 10.1093/ndt/15.6.754. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M, Abend JR, Johnson SF, et al. The role of polyomaviruses in human disease. Virology. 2009;384(2):266–73. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moens U, Van Ghelue M, Johnannessen M. Human polyomaviruses: molecular mechanisms for transformation and their association with cancer. In: Tuneley EI, editor. New research on oncogenic viruses. New York: Nova Science; 2008. pp. 1–63. [Google Scholar]
- 30.Seo GJ, Fink HL, O’Hara B, et al. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82(20):9823–8. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan CS, Grundhoff S, Tevethia J, et al. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cell. Nature. 2005;435(7042):682–6. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 32.Madkan VK, Cook-Norris RH, Steadman MC, et al. The oncogenic potential of human papillomaviruses: a review on the role of host genetics and environmental cofactors. Br J Dermatol. 2007;157(2):228–41. doi: 10.1111/j.1365-2133.2007.07961.x. [DOI] [PubMed] [Google Scholar]
- 33.Cai X, Li G, Laimonis AL, et al. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006;80(21):10890–3. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeffer SA, Swer M, Lagos-Quintana R, et al. Identification of microRNAs of the Herpesvirus family. Nat Methods. 2005;2:269–76. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 35.Ronald TKP, Carmen O, Leung N, et al. MicroRNA-34a suppresses invasion through downregulation of Notch 1 and Jagged 1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31(6):1037–44. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Brahmakshatriya V, Zhu H, et al. Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genomics. 2009;10:512. doi: 10.1186/1471-2164-10-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol. 2004;78:12868–76. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori K, Jüttermann R, Wienhues U, et al. Anti-interferon activity of adenovirus-2-encoded VAI and VAII RNAs in translation in cultured human cells. Virus Res. 1996;42(1–2):53–63. doi: 10.1016/0168-1702(95)01309-1. [DOI] [PubMed] [Google Scholar]
- 39.Anderson MG, Haasnoot PCJ, Xu N, et al. Suppression of RNA interference by adenovirus-associated RNA is processed to functional interfering RNAs involved in virus production. J Virol. 2005;79:9556–65. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman B, Pellet PE. The family Herpesviridae: a brief introduction. In: Knipe DM, Howley PM, editors. Fields virology. 4th. Philadelphia (PA): Lippincott Williams & Wilkins; 2001. pp. 2381–97. [Google Scholar]
- 41.Gupta A, Gartner JJ, Sethupathy P, et al. Anti-apoptic function of a microRNA encoded by HSV-1 latency-associated transcripts. Nature. 2006;442(7098):82–5. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 42.Stern-Ginossar V, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grey F, Meyers H, White EA, et al. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugden B. Virology: micro mystery. Nature. 2006;442(7098):33–4. doi: 10.1038/442033a. [DOI] [PubMed] [Google Scholar]
- 45.Roizman B, Knipe DM, Whitely RJ, et al. The replication of Herpes simplex viruses. In: Knipe DM, Howley MP, Griffin DE, et al., editors. Fields’ virology. 5th. New York: Lippincott Williams & Wilkins; 2007. pp. 2501–601. [Google Scholar]
- 46.Cui C, Griffiths A, Li G, et al. Prediction and identification of herpes simplex virus-1 encoded microRNAs. J Virol. 2006;80(11):5499–508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai X, Cullen BR. Transcriptional origin of Kaposi’s sarcoma associated herpesvirus microRNAs. J Virol. 2006;80:2234–42. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn W, Trang P, Zhong Q, et al. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 2005;41(3):186–91. doi: 10.1111/j.1462-5822.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 49.Burnside J, Bernberg E, Anderson A, et al. Marek’s disease virus encodes microRNAs that map to meq and the latency associated transcripts. J Virol. 2006;80:8778–86. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to auto-regulate viral gene expression. Virology. 2009;383(2):183–7. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Omoto S, Fujii Y. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86(3):751–5. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- 52.Ura S, Honda M, Yasmashita T, et al. Differential expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49(4):1098–112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 53.Lecellier CH, Dunoyer P, Arar K, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–60. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 54.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84(10):5212–21. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umbach JL, Kramer MF, Jurak I, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–3. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grondin B, DeLuca N. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J Virol. 2000;74(24):11504–10. doi: 10.1128/JVI.74.24.11504-11510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rafailidis PI, Mourtzoukou EG, Varbobitis LC, et al. Severe cytomegalovirus infection in apparently immunocompetent patients: a systematic review. Virology. 2008;5(47):1–7. doi: 10.1186/1743-422X-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grey F, Nelson J. Identification and function of human cytomegalovirus microRNAs. J Clin Virol. 2008;41(3):186–91. doi: 10.1016/j.jcv.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy E, Vanicek J, Robins H, et al. Suppression of immediate early viral gene expression by Herpesvirus encoded microRNAs: implications for latency. Proc Natl Acad Sci U S A. 2008;105(14):5453–8. doi: 10.1073/pnas.0711910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehraein Y, Lennerz C, Ehlhardt S, et al. Latent Epstein-Barr virus (EBV) infection and cytomegalovirus (CMV) infection in synovial tissue of autoimmune chronic arthritis determined by RNA-and DNA-in situ hybridization. Mod Pathol. 2004;17:781–9. doi: 10.1038/modpathol.3800119. [DOI] [PubMed] [Google Scholar]
- 61.Klein E, Kis LL, Klein G. Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions. Oncogene. 2007;26:1297–305. doi: 10.1038/sj.onc.1210240. [DOI] [PubMed] [Google Scholar]
- 62.Ziegelbauer JM, Sullivan CS, Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat Genet. 2009;41(1):130–4. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barth S, Pfuhl T, Mamiani A, et al. Epstein Barr virus encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acid Res. 2008;36:666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P, Yu J, Zhang L. The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci U S A. 2007;104:4054–9. doi: 10.1073/pnas.0700020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia T, O’Hara A, Araujo I, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68(5):1436–42. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ganem D, Ziegelbauer Y. MicroRNAs of Kaposi’s sarcoma-associated herpes virus. Semin Cancer Biol. 2008;18(6):437–40. doi: 10.1016/j.semcancer.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Calnek BW. Marek’s disease: a model for herpesvirus oncology. Crit Rev Microbiol. 1986;12(4):293–320. doi: 10.3109/10408418509104432. [DOI] [PubMed] [Google Scholar]
- 68.Xu H, Yao Y, Zhao Y, et al. Analysis of the expression profiles of Marek’s disease virus-encoded microRNAs by real-time quantitative PCR. J Virol Methods. 2008;149:201–8. doi: 10.1016/j.jviromet.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 69.Yao Y, Zhao Y, Smith L, et al. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J Gen Virol. 2008;90:1551–9. doi: 10.1099/vir.0.009902-0. [DOI] [PubMed] [Google Scholar]
- 70.Rosenfeld N, Aharonov R, Meri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–9. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 71.Weinberg MS, Morris KV. Are viral-encoded microRNAs mediating latent HIV-1 infection? DNA Cell Biol. 2006;25:223–31. doi: 10.1089/dna.2006.25.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bennasser Y, Le SY, Yeung ML, et al. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retro virology. 2004;1(43):1–5. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foster JL, Garcia JV. HIV-Nef: at the crossroads. Retrovirology. 2008;5:84. doi: 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Omoto S, Ito M, Tsutsumi Y, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Njoroge FG, Chen KX, Shihh NY, et al. Challenges in modern drug discovery: a case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection. Acc Chem Res. 2008;41(1):50–9. doi: 10.1021/ar700109k. [DOI] [PubMed] [Google Scholar]
- 76.Pawlotsky J. Mechanisms of drug resistance to current and future antiviral therapies for hepatitis C virus infection. Curr Hepat Rep. 2004;3(1):38–43. doi: 10.1007/s11901-004-0007-8. [DOI] [Google Scholar]
- 77.Persico M, Palmentieri B, Coppola LG, et al. Occurrence of HCC in asymptomatic HCV-related chronic hepatitis. Dig Dis Sci. 2002;47(11):2407–10. doi: 10.1023/A:1020527118782. [DOI] [PubMed] [Google Scholar]
- 78.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esau C, Davis S, Murray SF, et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 80.Hassan MM, Li D, El-Deeb AS, et al. Association between hepatitis B virus and pancreatic cancer. J Clin Oncol. 2008;26(28):4557–62. doi: 10.1200/JCO.2008.17.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jin WB, Wu FL, Kong D, et al. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124–6. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Lewis B. Targeted deletion of dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2003;105:2111–6. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ely A, Naidoo T, Mufaamadi S, et al. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16(6):1105–12. doi: 10.1038/mt.2008.82. [DOI] [PubMed] [Google Scholar]
- 84.Gao YF, Yu L, Wei W, et al. Inhibition of hepatitis B virus gene expression and replication by artificial microRNA. World J Gastroentrol. 2008;14(29):4684–9. doi: 10.3748/wjg.14.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li ZC, Zhang S, Huang C, et al. MicroRNAome of splenic macrophages in hypersplenism due to portal hypertension in hepatitis B virus-related cirrhosis. Exp Biol Med. 2008;233(11):1454–61. doi: 10.3181/0711-RM-321. [DOI] [PubMed] [Google Scholar]
- 86.Qin C. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J Pathol. 2005;206:251–9. doi: 10.1002/path.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, Wang HK, McCoy JP, et al. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;4:637–47. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuan J, Cheung PKM, Zhang HM, et al. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J Virol. 2005;79(4):2151–9. doi: 10.1128/JVI.79.4.2151-2159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schubert SG, Grunert HP, Zeichhardt H, et al. Maintaining inhibition: siRNA double expression vectors against coxsackieviral RNAs. J Mol Biol. 2005;346(2):457–65. doi: 10.1016/j.jmb.2004.11.074. [DOI] [PubMed] [Google Scholar]
- 90.Merl S, Michaelis C, Jaschke B, et al. Targeting 2A protease by RNA interference attenuates coxsackieviral cytopathogenicity and promotes survival in highly susceptible mice. Circulation. 2005;111(13):1583–92. doi: 10.1161/01.CIR.0000160360.02040.AB. [DOI] [PubMed] [Google Scholar]
- 91.Zhang M, Su Y, Guo S, et al. Targeted delivery of anti-coxsackievirus siRNAs using ligand-conjugated packaging RNAs. Antiviral Res. 2009;83:307–16. doi: 10.1016/j.antiviral.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim JY, Chung SK, Hwang HY, et al. Expression of short hairpin RNAs against the coxsackievirus B3 exerts potential antiviral effects in Cos-7 cells and in mice. Virus Res. 2007;125(1):9–13. doi: 10.1016/j.virusres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 93.Dyxhoorn M, Lieberman J. Prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu Rev Biomed Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- 94.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Therapy. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morral N, O’Neal WK, Rice K, et al. Lethal toxicity, severe endothelial injury, and a threshold effect with high doses of an adenoviral vector in baboons. Hum Gene Ther. 2002;13:143–54. doi: 10.1089/10430340152712692. [DOI] [PubMed] [Google Scholar]
- 96.Bitko V, Musiyenko A, Shulyayeva O, et al. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–5. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 97.Palliser D, Chowdhury D, Wang QY, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 98.Hafner M, Landgraf P, Ludwig J, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lim LP, Linsley PS. Mustering the micromanagers [comment] Nat Biotechnol. 2007;25:996–7. doi: 10.1038/nbt0907-996. [DOI] [PubMed] [Google Scholar]
- 100.Thompson RC, Deo M, Turner DL. Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods. 2007;43:153–61. doi: 10.1016/j.ymeth.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 102.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 103.Birmingham A, Anderson E, Sullivan K, et al. A protocol for designing siRNA with high functionality and specificity. Nat Protoc. 2007;2(9):2068–78. doi: 10.1038/nprot.2007.278. [DOI] [PubMed] [Google Scholar]
- 104.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 105.Zhao Y. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cadiogenesis. Nature. 2006;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]