Abstract
Picornavirus replication is critically dependent on the correct processing of a polyprotein precursor by 3C protease(s) (3Cpro) at multiple specific sites with related but non-identical sequences. To investigate the structural basis of its cleavage specificity, we performed the first crystallographic structural analysis of non-covalent complexes of a picornavirus 3Cpro with peptide substrates. The X-ray crystal structure of the foot-and-mouth disease virus 3Cpro, mutated to replace the catalytic Cys by Ala and bound to a peptide (APAKQ|LLNFD) corresponding to the P5–P5′ region of the VP1-2A cleavage junction in the viral polyprotein, was determined up to 2.5 Å resolution. Comparison with free enzyme reveals significant conformational changes in 3Cpro on substrate binding that lead to the formation of an extended interface of contact primarily involving the P4–P2′ positions of the peptide. Strikingly, the deep S1′ specificity pocket needed to accommodate P1′-Leu only forms when the peptide binds. Substrate specificity was investigated using peptide cleavage assays to show the impact of amino acid substitutions within the P5–P4′ region of synthetic substrates. The structure of the enzyme–peptide complex explains the marked substrate preferences for particular P4, P2 and P1 residue types, as well as the relative promiscuity at P3 and on the P′ side of the scissile bond. Furthermore, crystallographic analysis of the complex with a modified VP1-2A peptide (APAKE|LLNFD) containing a Gln-to-Glu substitution reveals an identical mode of peptide binding and explains the ability of foot-and-mouth disease virus 3Cpro to cleave sequences containing either P1-Gln or P1-Glu. Structure-based mutagenesis was used to probe interactions within the S1′ specificity pocket and to provide direct evidence of the important contribution made by Asp84 of the Cys-His-Asp catalytic triad to proteolytic activity. Our results provide a new level of detail in our understanding of the structural basis of polyprotein cleavage by 3Cpro.
Abbreviations: 3Cpro, 3C protease(s); FMDV, foot-and-mouth disease virus; PV, poliovirus; HRV, human rhinovirus; HAV, hepatitis A virus; TEVpro, tobacco etch virus NIa protease; PDB, Protein Data Bank; ESRF, European Synchrotron Radiation Facility
Keywords: X-ray crystallography, picornavirus, foot-and-mouth disease virus, 3C protease, peptide recognition
Introduction
Foot-and-mouth disease is a serious widespread viral disease of cloven-hoofed animals, including important agricultural species such as cattle, sheep, pigs and goats.1, 2 The virus spreads rapidly, and although endemic and epidemic situations can be controlled using vaccines that are based on inactivated virus particles, political and technical difficulties with the maintenance and use of vaccine stocks have stimulated the search for alternative means of tackling the disease, such as anti-viral drugs.3 The development of such treatments will demand a detailed knowledge of the molecular basis of viral replication. In this article, we focus on the structural basis of the cleavage activity of foot-and-mouth disease virus (FMDV) 3C protease(s) (3Cpro). As a highly conserved viral enzyme,4 FMDV 3Cpro is a potential drug target.
FMDV is a single-stranded positive-sense RNA virus belonging to the picornavirus family that also includes human pathogens such as poliovirus (PV), human rhinovirus (HRV) and hepatitis A virus (HAV). Picornaviruses share a common replication strategy in which the viral RNA genome is translated in the cytoplasm of infected cells as a long polyprotein precursor that is cleaved by virally encoded proteases to release functional proteins needed for the synthesis of new virions. In all cases, 3Cpro performs the majority of these cleavages (10 out of 13 for FMDV) by targeting specific sequences within the polyprotein.2, 5, 6
Structural studies on picornaviral 3Cpro revealed an overall fold that closely resembles the architecture of trypsin-like serine proteases.7, 8, 9, 10, 11 The peptide-binding cleft, which contains the active site at its centre, is located at the interface between two β-barrels. Unusually, picornaviral 3Cpro possess a Cys-His-Asp/Glu catalytic triad at the centre of this cleft, instead of the Ser-His-Asp arrangement of active-site residues that is commonly found in serine proteases.12, 13, 14
Most investigations of the cleavage specificity of picornaviral 3Cpro have relied on extensive sequence analyses of cleavage junctions within the polyprotein3, 15 or on in vitro cleavage assays using protein or peptide substrates.8, 16, 17, 18, 19, 20 There is a surprising degree of variability in the sequences of polyprotein junctions that are cleaved by picornaviral 3Cpro, but these studies have nevertheless been valuable in identifying residues in the substrate that are important determinants of cleavage. Most picornavirus 3Cpro cleave the peptide bond between a highly conserved P1-Gln/P1′-Gly pair15, 21 (where P1 and P1′ denote the first amino acids on the N-terminal and C-terminal sides of the scissile bond, respectively; in this notation,22 the corresponding subsites on the enzyme are labelled S1 and S1′). FMDV 3Cpro is an interesting exception to this general pattern, since it exhibits a preference either for substrates in which P1-Gln is followed by a relatively large apolar amino acid in the P1′ position or for P1′Glu/P1′Gly junctions.3 HAV 3Cpro can also tolerate larger P1′ amino acids in substrates with P1-Gln.21 Other positions in the peptide also contribute to substrate recognition; in general, there is a preference for a hydrophobic residue at P4,16, 18, 19 and the nature of the P2 and P2′ amino acids, which vary between different picornavirus families, can also be important.8
Extensive structural analysis of trypsin-like serine proteases has revealed a common mode of substrate binding in which the polypeptide sequence recognised by the enzyme lies in an extended conformation (similar to a β-strand) within the peptide-binding cleft.23 These enzymes typically recognise the alternating positions of the side chains along the sequence that are characteristic of an extended backbone conformation, and this serves to place the scissile bond in the correct orientation at the active site. The primary specificity determinants in trypsin-like serine proteases are commonly located within the P4–P1 sequence,24, 25, 26 but can extend well beyond this core region for enzymes that are highly specific.27 Crystallographic analysis of enzyme–substrate complexes reveals that viral 3C-like proteases, such as tobacco etch virus NIa protease (TEVpro)28 or severe acute respiratory syndrome coronavirus main protease,29 fall into the latter group, since contacts with the enzyme can involve up to five or six amino acids on either side of the cleavage site. However, although both of these proteases are ‘3C-like’ in structure and have the common trypsin-like fold, they exhibit substantial differences from one another and from genuine picornaviral 3Cpro, especially in the loops that define the peptide-binding surface.
Previous structural analyses of picornavirus 3Cpro–peptide complexes have used relatively short covalently attached peptides or peptide-mimicking inhibitors. In most cases, inhibitors were designed to bind to the S4–S1 subsites,24, 25, 26 although there is one report of the structure of an inhibitor bound to the S1′–S2′ subsites.30 These studies are a powerful complement to cleavage assays, since they have helped to elucidate the structural basis of amino acid recognition by these enzymes, particularly on the N-terminal side of the scissile bond. However, they cannot give a complete picture of protease–substrate interactions since, in each case, the peptide is covalently attached to the protease and represents, at most, one-half of the peptide substrate recognised by the enzyme.
Here we report the first crystallographic analysis of a picornaviral 3Cpro complexed with an intact peptide substrate that spans P5–P5′. Our structural analysis of FMDV 3Cpro is combined with peptide cleavage assays that have examined the effects on the cleavage rate of amino acid variations within P5–P4′ and of mutations in the peptide-binding cleft of the enzyme. This concerted investigation provides a detailed and insightful description of the interactions between enzyme and substrate. Most strikingly, we find that significant conformational adaptation by the enzyme is important for substrate recognition. This helps to explain several important aspects of specificity differences between 3Cpro from different picornaviruses.
Results
Peptide cleavage assays
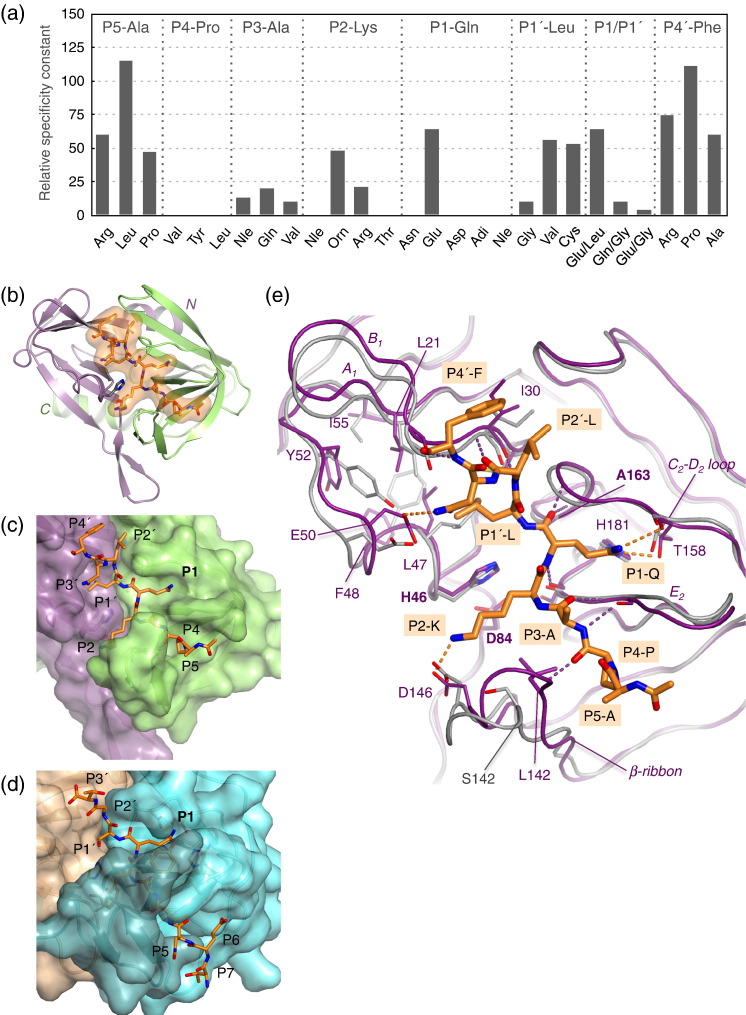
In conjunction with our structural analysis, we performed an extensive investigation of the impact of sequence variation in the peptide on the rate of cleavage by FMDV 3Cpro. For these assays, we synthesised a series of variants of FRET4, a modified peptide based on the P5–P8′ region of the VP1-2A cleavage junction31—one of the 10 sites cleaved by FMDV 3Cpro in the viral polyprotein. Cleavage of FRET4 produces a readily detectable increase in fluorescence31 (Materials and Methods). The FRET4 variants, which contained single amino acid substitutions at positions P5–P1′ and P4′ or a double substitution at P1 and P1′, were used in cleavage assays to determine the relative value of the specificity constant (k cat/K m) in each case (Fig. 1a).
Fig. 1.
Overview of peptide recognition by FMDV 3Cpro. (a) Relative cleavage activity of FMDV 3Cpro against variants of the FRET4 substrate with amino acid substitutions in positions P5–P4′. The activity is normalised with respect to the cleavage rate observed with wild-type substrate (Materials and Methods). The amino acids in the wild-type sequence are given at the top of each section; the identity of the substituted amino acid is given along the bottom. Note that, in the P1–P1′ panel, the Glu-Leu and Gln-Gly data bars are for single P1 and P1′ mutants, respectively, but are duplicated here for ease of comparison with the Glu-Gly double mutant. (b) Schematic view of the co-crystal structure of 3Cpro (with N-terminal and C-terminal β-barrels shown in lilac and green, respectively) complexed with the VP1-2A peptide. The peptide is depicted in sticks colour coded by atom type (carbon, orange; oxygen, red; nitrogen, blue), with its van der Waals surface shown as a semitransparent surface. (c) Surface representation of FMDV 3Cpro complexed with the VP1-2A peptide (sticks), coloured as in (b). (d) Surface representation of tobacco etch virus NIa 3C-like protease complexed with a P7–P3′peptide.28 The N-terminal and C-terminal β-barrels are shown in cyan and tan, respectively. (e) Superposition of the free (gray) and peptide-bound (purple) forms of FDMV 3Cpro. The viewpoint is similar to that of (b). Backbone–backbone hydrogen bonds are shown as purple broken lines; other hydrogen bonds are shown in orange. Selected side chains are shown as sticks; secondary structural features that show greatest movement on peptide binding are labelled. Note that the unbound structure of FMDV 3Cpro has a Ser at position 142.12
A detailed account of the results of these experiments will be presented alongside a description of the structural features of peptide–protease interactions at each position in the substrate sequence (see the text below). Overall, it is notable that, within the context of the VP1-2A sequence, variation at all positions from P4 to P1′ (inclusive) can reduce the rate of peptide cleavage by FMDV 3Cpro to between 0% and 50% of the rate observed with the control peptide (Fig. 1a). In contrast, variation at the P5 and P4′ positions at the extremities of the peptide had little impact on peptide cleavage. With one exception, these results are consistent with a previous alanine-scanning analysis of the peptide.8 They confirm the importance of positions P4, P2, P1 and P1′ for substrate recognition, but reveal, for example, that residues at the P3 position can also contribute to cleavage specificity. The only inconsistency noted is that substitution of P4′-Phe (by Arg, Pro or Ala) had little effect on peptide cleavage in the fluorescence assay, whereas it had been found previously in an HPLC-based assay that substitution by Ala abrogated cleavage.8 The origin of this discrepancy is unknown, but the structural results are consistent with more recent findings indicating that the P4′ residue is unlikely to contribute significantly to substrate specificity (see the text below).
Structure determination
Recombinant-type A1061 FMDV 3Cpro, inactivated by mutation of the active site Cys to Ala (C163A) and containing the C95K and C142L substitutions necessary for enhanced solubility,12 was purified from Escherichia coli and co-crystallised with a 5-fold molar excess of the decameric peptide VP1-2A (Materials and Methods). This peptide spans the P5–P5′ positions22 and contains the sequence APAKQ|LLNFD from the VP1-2A junction in the polyprotein that was cut most rapidly in peptide cleavage assays.8 The protease–peptide complex yielded crystals in space group P212121 that diffracted X-rays to 2.5 Å. The structure was determined by molecular replacement phasing using the structure of the free enzyme [Protein Data Bank (PDB) ID 2j92 12] as search model. There are two molecules in the asymmetric unit, and the initial-difference electron density map revealed clear density for the peptide bound to each molecule. The density was of high quality, consistent with full occupancy of the binding site, and readily allowed determination of the side-chain conformations for 9 of the 10 amino acids (P5–P4′) in the peptide. The electron density at the C-terminal end of the peptide was too weak to permit incorporation of the P5′-Asp residue into the refined model (Supplementary Fig. 1a). Atomic structures of the two 3Cpro–peptide complexes in the crystal were built using O32 and refined with CNS33 to yield a model with an R free of 26.7% and excellent stereochemistry (Table 1 ).
Table 1.
Data collection and refinement statistics
| Diffraction data | ||
| Peptide complex | VP1-2A | VP1-2Am |
| Synchrotron | ESRF ID23-2 | ESRF ID14-2 |
| Space group | P212121 | P21 |
| a, b, c (Å) | 64.8, 75,3, 86.0 | 64.0, 75.1, 86.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 99.3, 90 |
| Resolution range (Å)a | 30.3–2.5 (2.66–2.5) | 63.2–2.7 (2.87–2.7) |
| Independent reflections | 14,044 | 16,981 |
| Multiplicityb | 4.4 (2.5) | 2.4 (2.6) |
| Completeness (%) | 93.4 (78.5) | 76.1 (60.8) |
| I/σI | 9.1 (2.5) | 8.6 (3.2) |
| Rmerge (%) | 13.9 (34.9) | 8.6 (38.6) |
| Model refinement | ||
| Non-hydrogen atoms: protein/water | 2958/33 | 6044/52 |
| Rcryst (%) | 21.2 | 23.6 |
| Rfree (%) | 26.7 | 29.1 |
| r.m.s. bond lengths (Å) | 0.008 | 0.008 |
| r.m.s. bond angles (°) | 1.29 | 1.33 |
| Average B-factor (Å2) | 46.7 | 32.1 |
| Ramachandran plot (% favoured/allowed) | 87.2/10.5 | 85.1/14.3 |
| PBD ID | 2wv4 | 2wv4 |
Resolution ranges for the highest-resolution shells are given in parentheses.
Values for the outermost-resolution shell are given in parentheses.
An identical approach was applied to the preparation and structure determination of the complex of 3Cpro with the modified VP1-2Am peptide, which contains a Gln-to-Glu substitution at the P1 position. Although this peptide differs by only one amino acid from VP1-2A, the 3Cpro–VP1-2Am complex yielded crystals of a different space group (P21) that diffracted to a slightly lower resolution (2.7 Å). The data set is only 76% complete, but averaging across the four molecules in the asymmetric unit yielded a good-quality electron density map (Supplementary Fig. 1b) that provided a clear indication of the bound conformation of residues P5–P4′ of the VP1-2Am peptide. The refined model has an R free of 29.1% (Table 1).
Overview of the structure
The overall structures of the 3Cpro–peptide complexes obtained with VP2-2A and VP1-2Am are very similar to one another (Supplementary Fig. 2). There are differences in the backbone conformations of residues 105–109 at the C-terminal end of the polypeptide linking the two β-barrels of the enzyme, indicative of flexibility in this portion of the backbone. However, this is unlikely to impact peptide recognition, since this part of the linker runs along the underside of the C-terminal β-barrel, opposite to the surface containing the peptide-binding cleft.
When viewed with the interdomain cleft aligned vertically (as in Fig. 1b and c), the peptide is observed to bind largely within a deep surface groove that is oriented diagonally and intersects the cleft at the active site. Consequently, residues P5–P1 on the amino-terminal side of the scissile bond mainly contact the C-terminal β-barrel, while residues P1′–P5′ interact primarily with the N-terminal β-barrel. The length of the recessed groove is sufficient to accommodate residues P4–P2′ of the peptide, although residues outside this core are also in contact with the surface of the protease.
Conformational changes associated with binding
Comparison of the bound and free12 forms of FMDV 3Cpro reveals several conformational changes that can be ascribed to peptide binding because they are observed for the six independent structures found in the asymmetric units of the crystals of both 3Cpro–peptide complexes, all of which have different packing environments in the crystal. These differences extend across both β-barrels (Fig. 1e; Supplementary Movie 1). On the P5–P1 side, the β-ribbon (residues 138–150) moves in to contact the peptide, and there are adjustments in the backbone of the polypeptide that form the flanks of the S1 subsite (β-strand E2 and the C-terminal end of the C2–D2 loop) (Fig. 1e); on the P1′–P5′ side of the peptide, the most significant changes in backbone conformation occur in residues 47–53 and in the nearby A1–B1 pair of β-strands (residues 18–31) that are in closest contact with the peptide. These two segments of polypeptide are altered due to a propagated series of side-chain movements initiated by the rotation of the side chain of Leu47 that is necessary to accommodate the P1′-Leu residues of the peptide (see the text below).
Structural basis of peptide binding specificity
The extensive malleability of the peptide-binding surface allows the protease to form intimate contacts with the substrate along most of its length. In common with other trypsin-like proteases, there is a network of hydrogen bonds between the peptide and the protease backbones that mimics the interactions between protein β-strands. In the case of the complex between FMDV 3Cpro and the VP1-2A peptide, there are nine such main-chain hydrogen bonds in total, extending from P4-Pro to P5′-Asp (Fig. 1e). These main-chain contacts are bolstered by hydrogen bonds and apolar contacts between the peptide side chains and the protease. We will consider each of these in detail.
P5-Ala at the N-terminus of the peptide is largely solvent exposed (Fig. 2a). The side chain points towards the hydrophobic side chain of Met143, but is too far away (4.7 Å) to make contact. This lack of contact accounts for the finding that substitution by Arg, Leu or Pro at this position has only a modest effect on substrate cleavage, reducing the rate by 2-fold at most (Fig. 1a).
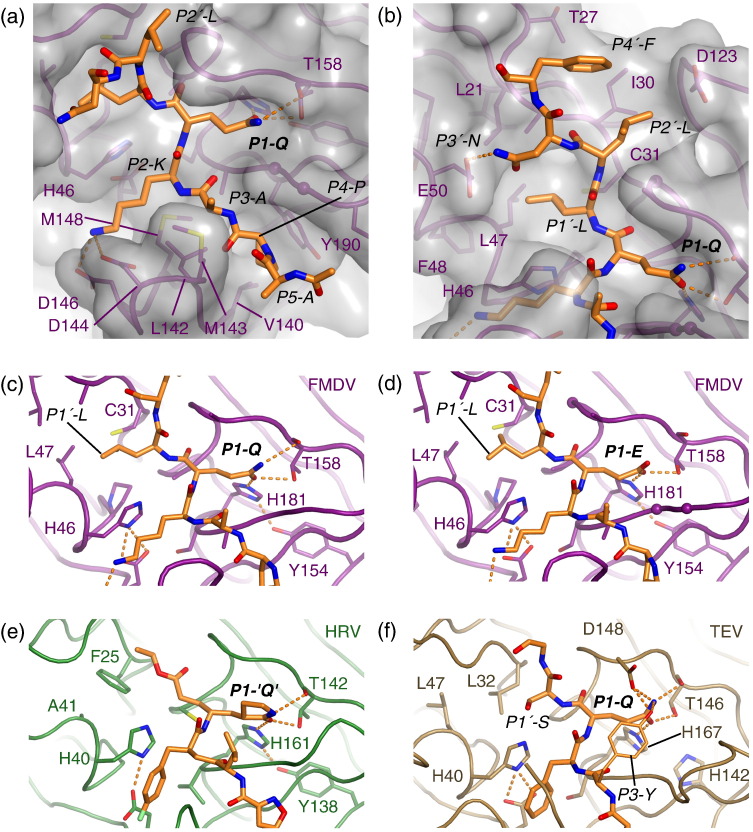
Fig. 2.
Structural details of peptide recognition by FMDV 3Cpro. (a) Close-up view of the binding of the P5–P1 residues from the VP1-2A peptide. The protease backbone is depicted as a smooth ribbon covered by a semitransparent gray van der Waals surface; selected side chains are shown. (b) Close-up view of the binding of the P1′–P4′ residues from the VP1-2A peptide. Detailed views of the P1–P1′ interactions for (c) FMDV 3Cpro complexed with VP1-2A and for (d) FMDV 3Cpro complexed with VP1-2Am. The two hydrogen bonds made by OE1 from the P1-Glu side chain to Thr158 and His181 are shown as dotted lines; atom OE2 from the P1-Glu side chain is too distant from Thr158 to form a hydrogen bond. (e) HRV 3Cpro complexed irreversibly with a peptide-like inhibitor.25 (f) Tobacco etch virus NIa 3C-like protease complexed with a P7–P3′ peptide.28
P4-Pro, by contrast, is accommodated in a shallow apolar depression that marks the beginning of the peptide-binding groove and is formed by the side chains of Leu142 (Cys in the wild-type protein12), Val140 and Tyr190 (Fig. 2a). The P4-Pro side chain is not completely buried, since one flank containing the Cδ atom faces outward and is solvent exposed. This part of the side chain is quite close to the carbonyl oxygen of Val141 and to the side-chain hydroxyl of Tyr190 (4.2 Å in each case). Intriguingly, mutation of the P4 residue to Val, Leu or Ile completely abrogated cleavage of the fluorescent substrate (Fig. 1a), consistent with a previous observation that an Ala substitution at this position also prevented cleavage.8 These results suggest that, at least within the context of the VP1-2A peptide, there is a strict requirement for P4-Pro. Although the structure suggests that Ala and Val side chains, which are similar in size to Pro, should be accommodated in the S4 pocket, they may nevertheless affect the binding conformation sufficiently to prevent proper presentation of the scissile bond to the active site. This is a rather surprising result given the evident malleability of the protease. It is also unexpected, since Ala, Val and Ile are observed at P4 in other FMDV polyprotein cleavage junctions, albeit within the context of different sequences (see Table 2 in Birtley et al. 8).
The side chain of P3-Ala points towards the solvent, but packs against Met143 at the apical tip of the β-ribbon that alters its conformation on peptide binding to contact the P side of the substrate (Fig. 2a; Supplementary Movie 1). This explains why, in common with other similar proteases, there is no strong preference for particular residues at this position in natural cleavage junctions.3 In sequences cleaved by FMDV 3Cpro, Val, Gln, Glu, His, Phe, Ser, Arg and Ala—all of which could make some hydrophobic contact with the side chain of Met143—are found at P3. Nevertheless, variation of this residue to Gln, Glu or Val within the context of the VP1-2A peptide reduces cleavage rates 5-fold to 10-fold, demonstrating that variation at the P3 position can affect cleavage (Fig. 1a). It may be that these larger side chains affect the positioning of the β-ribbon, which then has a knock-on effect on peptide interactions at other subsites.
The P2-Lys side chain inserts into the cleft between the β-ribbon and the body of the N-terminal β-barrel of the protease (Fig. 2a). On one side, the Lys side chain primarily contacts the side chain of His46 (the central residue of the catalytic triad), whereas on the other side, the aliphatic portion of the Lys contacts the apolar side chains of Leu142 and Met148. At the distal end of the pocket, which is open to solvent, there are salt bridges from the amine group at the tip of the Lys side chain to two acidic residues from the β-ribbon, Asp144 (3.6 Å) and Asp146 (2.8 Å) (Fig. 2a). Formation of the S2 pocket appears to be dependent on peptide binding, since Leu142, Asp144 and Asp146 all adjust their positions in the presence of substrate (Fig. 1e). The salt bridges to the pair of Asp residues in the β-ribbon explain the strong preference for P2-Lys in natural substrates (although it is not yet clear why this preference is strongest in substrates with P1-Gln3). Consistent with the acidic nature of the distal end of the S2 pocket, incorporation at P2 of norleucine (which is structurally similar to Lys but lacks the amine group) abrogates cleavage, whereas substitution by Arg or ornithine (both of which have a positive charge at the tip of the side chain) only modestly reduces the rate of peptide cleavage (Fig. 1a). Curiously, substitution by Thr also abrogates cleavage, even though this residue is found in other natural FMDV 3Cpro substrates;8 it may be that Thr is tolerated at P2 only in the context of the variation of the amino acids at other positions in the peptide substrate.
The side chain of P1–Gln is accommodated by small (0.7 Å) movements of the two backbone segments that form the flanks of the S1 pocket (residues 158–161 and 183–186) (Fig. 1e). The P1 side chain makes three hydrogen bonds to the pocket: two between the carbonyl group and the side chains of His181 and Thr158 (2.6–2.9 Å) and one from the amide group to the backbone carbonyl of Thr158 (3.2–3.5 Å) (Fig. 2a and c). The pocket therefore displays good chemical complementarity to the P1-Gln side chain. However, peptide cleavage assays previously showed that incorporation of Glu at this position only reduces the cleavage rate by a factor of 2,8 consistent with the common occurrence of P1-Glu in FMDV 3Cpro substrates.3 To investigate this finding in more detail, we determined the crystal structure of 3Cpro bound to a modified VP1-2A peptide, in which P1-Gln is substituted by P1-Glu (VP1-2Am). The structure reveals that the P1-Glu side chain binds in a manner that is almost identical with that observed for P1-Gln in the VP1-2A peptide (Fig. 2c and d). One oxygen atom from the P1-Glu side-chain carboxylate (OE1) makes two hydrogen bonds (ranging in length from 2.4 to 2.8 Å over the four independent structures) to His181 and Thr158, but the other carboxylate oxygen (OE2) is more distant from the main-chain carbonyl oxygen of Thr158 (3.2–3.7 Å) and—assuming that the side-chain carboxylate is deprotonated—cannot form a hydrogen bond. The fact that the OE2 oxygen is also exposed to solvent (Fig. 2d) may also attenuate any negative impact on binding by allowing access to positive counterions in the cytoplasm of infected cells.
It therefore appears that the primary interactions with the P1 side chain of the substrate are the two shorter hydrogen bonds made with the side chains of His181 and Thr158. The longer separation between the carbonyl oxygen of Thr158 and the P1-Gln amide or the P1-Glu carboxylate oxygen (OE2) is likely to reduce the impact of these groups on peptide binding, thus allowing FMDV 3Cpro to cleave substrates containing either type of residue in the P1 position. The importance of the hydrogen bonds to the side chains of His181 and Thr158 is underscored by the finding that incorporation of uncharged (norleucine), longer (adipic acid) or shorter (Asp) carboxylated side chains at P1 prevents cleavage of the substrate by FMDV 3Cpro (Fig. 1e).
The binding of the P1′-Leu side chain is unusual because it is accommodated in a pocket that forms only when the protease interacts with the peptide substrate (Fig. 2b). In the free enzyme, Leu47 packs against Cys31 to form the floor of a shallow S1′ pocket, but peptide binding was found to induce the rotation of the side chain of Leu47 towards the core of the N-terminal β-barrel, thereby creating a much deeper compartment to accommodate the peptide side chain (Fig. 1e). The largely apolar sides of the S1′ pocket are formed on one side by Pro44, His46, Leu47 and the aliphatic flank of Glu50, and by Ala29 and Cys31 on the other side. The displacement of Leu47 has significant knock-on effects, leading to displacement of Phe48, Tyr52, Leu21 and Ile55; there is also alteration of the main-chain conformation of residues 48–52 (Fig. 1e; Supplementary Movie 1).
As noted previously,3 large hydrophobic side chains at the P1′ position are preferred in natural FMDV 3Cpro substrates containing P1-Gln. This correlation of P1 and P1′ amino acids seems to be necessary for efficient cleavage, since substitution of P1′-Leu by Gly yielded a 10-fold reduction in cleavage rate, whereas introduction of Val or Cys at P1′ reduced the rate of cleavage by only 2-fold (Fig. 1a). Examination of the structure suggests that a P1′-Gly side chain would be too small even to make stabilising contacts with the shallow form of the S1′ pocket.
Since the bound conformations of VP1-2A (P1-Gln) and VP1-2Am (P1-Glu) peptides are very similar, it was no surprise to find that incorporation of P1′-Gly within the context of the VP1-2Am sequence also gave a 10-fold reduction in the rate of cleavage (Fig. 1a). However, these results cannot explain the observation that natural FMDV substrates with P1-Glu are most frequently observed to have Gly at P1′.3 We suggest that sequences beyond the residues on either side of the scissile bond are important in ensuring that it is presented optimally to the active site.
P2′-Leu binds at one end of the recessed peptide-binding groove (Fig. 1, Fig. 2). The side chain packs against Ala160, Gly161, Ile30 and the Cβ group of Asp123, and against the side chain of P4′-Phe from the peptide, but is still only partially shielded from solvent (Fig. 2b). The interaction between the P2′ and the P4′ side chains may not be replicated in other peptides, since neither position is particularly well conserved in polyprotein junctions cleaved by 3C.34 The apolar nature of the S2′ binding pocket explains the general preference for P2′ residues with some hydrophobic functionality (Ser, Lys, Thr and Leu in P1-Gln substrates; Pro, Gly, Leu and Ile in P1-Glu substrates).
The P3′-Asn side chain is almost completely solvent exposed, but is positioned to form a hydrogen bond with Glu50 on the enzyme surface (Fig. 2b). However, there is little sequence conservation at this position in FMDV 3Cpro substrates, and the most common alternative P3′ residues (Tyr, Ala and Ile) would not maintain this interaction. The exposed position of P3′-Asn explains why a 5-((2-aminoethyl)amino)naphthalene-1-sulfonic acid group can be added to an Asp side chain at this position in the FRET4 peptide substrate without compromising its ability to be cleaved by FMDV 3Cpro.31
P4′-Phe is also largely solvent exposed and primarily contacts a small apolar patch on the upper surface of the C-terminal β-barrel formed by the side chain of Ile30 and the methyl group of Thr27 (Fig. 2b). The hydrophobic nature of this patch explains the general preference for residues with apolar side chains at P4′.3 Consistent with this notion, substitution of P4′-Phe by Arg, Pro or Ala—all of which have significant apolar features—only modestly reduced the rate of peptide cleavage (Fig. 1e).
The position of P5′-Asp is unknown, since there is no electron density for this residue. It appears to make no stabilising contacts with the protein, in agreement with the observation that mutation to Ala at P5′ had no effect of the rate of peptide cleavage.8
Mutagenic analysis of the S1′ specificity pocket
As described above, the co-crystal structure of 3Cpro with the VP1-2A peptide revealed that binding of the peptide necessitated movement of Leu47 to create a pocket deep enough to accommodate the P1′-Leu side chain (Fig. 1e). This S1′ pocket is the site of an interesting difference in specificity between FMDV 3Cpro, on one hand, and 3Cpro from HRV and PV, on the other hand. Whereas FMDV 3Cpro preferentially cleaves Gln-Xaa junctions (where Xaa is predominantly Leu, Ile or Thr),3 HRV and PV 3Cpro have a marked preference for cleavage at Gln-Gly junctions.15 Comparison of the protease structures readily accounts for this difference, since the Cys31-Leu47 residues that form the floor of the shallow S1′ pocket in the free form of FMDV 3Cpro are replaced by Phe25-Ala41 in 3Cpro from HRV (PDB ID 1cqq 25) and PV (PDB ID 1l1n 10) (Fig. 2c and e; Supplementary Fig. 4). Examination of these structures suggests that steric hindrance would prevent the movement of Phe25 that would be necessary to accommodate a large P1′ side chain.
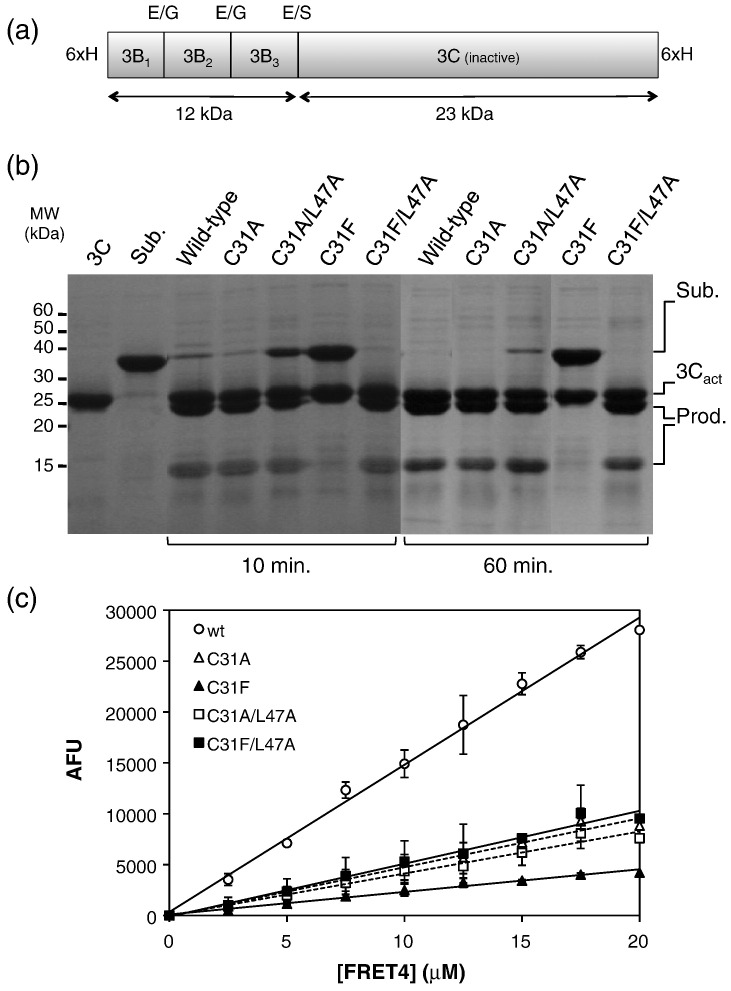
We explored this idea by mutating the S1′ pocket on FMDV 3Cpro to make it resemble HRV 3Cpro and by testing the effect on the cleavage activity of a 3BC polyprotein substrate that contains Glu-Ser and Glu-Gly cleavage junctions (i.e., they have a small P1′ side chains) (Fig. 3a; Materials and Methods). In this assay, the primary cleavage observed is located at the Glu-Ser junction; further cleavage at the Glu-Gly junctions within 3B1–3B2–3B3 may be impeded by the low solubility of this fragment (Fig. 3a and b). Under the assay conditions used, wild-type FMDV 3Cpro cleaves over 95% of the substrate within 10 min. The single mutation C31F drastically reduced the ability of the protease to cleave this protein substrate; no cleavage was evident after a 1-h incubation (Fig. 3b), and very partial cleavage was observed after 4 h (Supplementary Fig. 3). We estimate that the reduction in activity is approximately 200-fold. In contrast, mutation to the much smaller Ala side chain (C31A) had essentially no effect on the proteolytic activity of 3Cpro in this assay (Fig. 3b). The considerable reduction in protease activity due to the C31F substitution is probably due to the steric strain that arises from the close packing of the large side chains of Phe31 and Leu47 in the S1′ binding pocket of the mutant, which may have a knock-on effect on the position of His46, a key component of the catalytic triad (compare Fig. 2c and e; Supplementary Fig. 4). This interpretation is strongly supported by the finding that the C31F/L47A double mutation, which reduces the size of the side chain opposite Phe31 and generates an S1′ pocket that resembles that found in HRV and PV 3Cpro, restores near-wild-type cleavage activity (Fig. 3b). The C31A/L47A double mutant is slightly impaired compared to the wild-type protease, perhaps because the P1′ residue is bound less tightly in the enlarged S1′ pocket.
Fig. 3.
Mutagenic analysis of S1′ specificity. (a) Schematic diagram of the inactive polyprotein precursor, 3BC, (containing three cleavage junctions) used as substrate in these cleavage assays (see Materials and Methods).35 The P5–P5′ sequences of the three cleavage junctions in this polyprotein are: 3B1/3B2, LPQQE|GPYAG; 3B2/3B3, PVVKE|GPYEG; 3B3/3C, LIVTE|SGAPP. The sizes of the primary cleavage products are indicated. (b) SDS-PAGE analysis of polyprotein precursor digestion by FMDV 3Cpro S1′ mutants at 10 and 60 min. Labels on the right-hand side indicate the substrate (Sub.), the active 3Cpro (3Cact) and the primary cleavage products (Prod.). (c) Comparative activity of FMDV 3Cpro S1′ mutants measured in the fluorescent peptide cleavage assay using the synthetic FRET4 substrate,31 which contains the VP1-2A cleavage junction (Materials and Methods). Data shown are the average fluorescence units from three experiments ± SEM.
We also investigated the impact of these S1′ mutations on the cleavage of the fluorescent FRET4 substrate, which is based on the sequence of the VP1-2A peptide and contains a Gln-Leu cleavage junction; this substrate therefore places a large hydrophobic side chain (P1′-Leu) in the S1′ pocket. As found previously, the C31F mutant showed the greatest impairment in cleavage activity, although the 7-fold reduction in cleavage rate compared to that in wild type was much less than the ∼ 200-fold reduction observed in the polyprotein cleavage assay (Fig. 3b). The precise reason for the difference in the magnitudes of the effects is not clear, but may relate to the different sequences that are being cleaved in the two experiments.
Strikingly, and in contrast to the result obtained with a substrate containing a small P1′ amino acid (Fig. 3b), the double mutation C31F/L47A did not rescue wild-type activity with the FRET4 substrate (although, as expected, the enzyme was slightly more active than the C31F single mutant) (Fig. 3c). Overall, these results are consistent with structural data: The C31F/L47A double mutation would be predicted to make the S1′ pocket on FMDV 3Cpro resemble the inflexible shallow depression found in the HRV enzyme (Supplementary Fig. 4), which has a preference for substrates with a small P1′ residue.
Curiously, the single C31A mutation reduced the rate of cleavage of the fluorescent VP1-2A peptide by about 3-fold (Fig. 3c). This loss of activity possibly occurred because the P1′-Leu of the substrate was bound less snugly by the enzyme. As before, the C31A/L47A double mutation caused only a very slight further reduction in cleavage activity.
Mutagenic analysis of the catalytic Asp
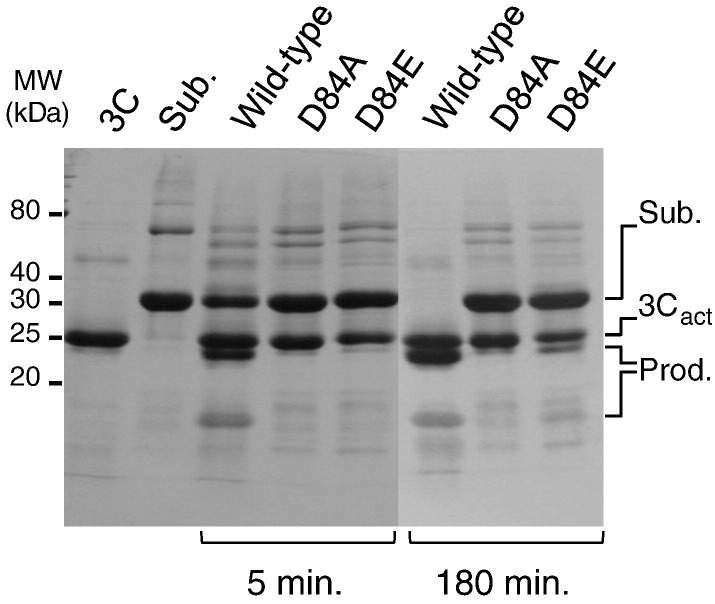
The crystal structure of the 3Cpro–peptide complex reveals that the conformation of the catalytic triad is unchanged by peptide binding (Fig. 1e). In particular, it shows that Asp84, the acidic member of the catalytic triad, maintains a hydrogen-bond interaction with His46, supporting the idea that this unusual class of trypsin-like cysteine proteases requires a full Cys-His-Asp triad for catalytic activity.8, 14 We tested this hypothesis by mutating Asp84 (Fig. 4 ). The mutation D84E, which maintains a carboxylate side chain, reduced cleavage activity by about 2 orders of magnitude under our experimental conditions: whereas wild-type 3Cpro cleaved ∼ 50% of the polyprotein in 5 min at 37 °C, the D84E mutant cleaved < 10% of the substrate after 180 min (Fig. 4). The mutation D84A was even more deleterious: There was no discernable cleavage of the substrate after 3 h, consistent with a reduction in activity of at least 3 orders of magnitude.
Fig. 4.
Mutagenic analysis of D84 from the catalytic triad. SDS-PAGE analysis of polyprotein precursor digestion by wild-type FMDV 3Cpro and the D84A and D84E mutants at 5 and 180 min. The substrate is the same as depicted in Fig. 3a.
These findings were confirmed in more quantitative assays using the fluorescent FRET4 peptide substrate (Supplementary Fig. 5). Cleavage by the D84E mutant was about 200 times slower than cleavage by wild-type FMDV 3Cpro. Cleavage by the D84A mutant was at least 1000 times slower and was beneath the detection level of the assay.
Our results indicate that mutation of Asp84 is even more deleterious to 3Cpro activity than was reported previously.36 This apparent discrepancy is likely due to the greater sensitivity of our assay, which examined cleavage over a range of time points (Fig. 4). Our findings confirm the importance of Asp84 for cleavage activity by FMDV 3Cpro and explain the strict conservation of this residue in all serotypes of FMDV.4
Role of β-ribbon in catalysis
The contacts that the side chain of Leu142 in the β-ribbon makes with apolar features on P4-Pro and P2-Lys in the co-crystal structure explain the importance of this residue to catalytic activity (Fig. 1, Fig. 2).12 In FMDV 3Cpro, this residue is strictly conserved as Cys,4 which is also apolar. Previously, we mutated C142 to prevent aggregation of the protein due to formation of intermolecular disulphide bonds, but found that substitution by a large hydrophobic side chain was necessary to preserve the catalytic activity of the protease.12, 13 Although slightly larger, Leu is a reasonable structural mimic of Cys, and the structure of the 3Cpro–peptide complex confirms our previous conclusion that the apolarity of C142 in the wild-type protein is necessary for its interaction with the P4 and P2 residues to ensure a correct presentation of the polypeptide substrate to the active site.12 Consistent with this idea, introduction of the C142L mutation into a type O1K FMDV had no significant effect on the infectivity of the virus in tissue culture (Fig. 5 ). This observation and the fact that the equivalent residue in HRV and PV 3Cpro is a leucine (Leu127) nevertheless beg the question, “Why is Cys142 strictly conserved in FMDV 3Cpro?” We speculate that this may confer a selective advantage during infection of natural hosts by permitting inactivation of the protease on cell lysis.
Fig. 5.
Infectivity of the C142L mutants of O1K FMDV. Single-step growth curves of wild-type O1K FMDV and two mutant viruses, both containing the C142L mutation in the β‑ribbon of 3Cpro. Virus yield was determined by measuring the number of FMDV RNA genomes in a real-time RT-PCR assay (Materials and Methods).
Discussion
An extensive flexible interface for peptide recognition
We have determined the crystal structure of FMDV 3Cpro in complex with a specific peptide substrate, providing the first detailed view of enzyme–substrate interactions for this important group of picornavirus proteases. The mode of peptide binding is similar to that observed in HRV and HAV 3Cpro complexed with covalently attached peptide-like inhibitors24, 25, 26 and with peptide complexes of other trypsin-like proteases37 in that the peptide adopts a linear extended conformation that places the P1 side chain into the S1 specificity pocket within the C-terminal β-barrel, immediately adjacent to the catalytic triad. The mode of binding in FMDV 3Cpro involves the five main-chain–main-chain hydrogen bonds between the enzyme and the P3–P1′ positions on the peptide that are conserved in trypsin-like proteases and appear to be necessary for proper positioning of the scissile bond in the active site.27
However, the structure reveals that the enzyme–substrate interaction extends well outside this core region; all side chains from P4–P4′ are involved to a greater or lesser extent in contacts with the surface of 3Cpro. Overall, the structure of the complex is highly consistent with the results of experiments performed to probe the effect of variation in side chains at different positions within the peptide substrate on cleavage rates, which suggest that the region P4–P2′ contributes most to peptide specificity. This is probably because these six residues occupy the deepest part of the surface groove that forms the peptide-binding cleft (Fig. 1c).
There are, nevertheless, some unexpected results from the cleavage assays that are not readily explained by the structure. For example, it is notable that, within the context of the VP1-2A cleavage junction sequence, conservative substitution of P4-Pro with residues such as Val or Ala that occur at this position in other junctions leads to complete abrogation of peptide cleavage (Fig. 1a). The effect of these substitutions is all the more surprising given the evident flexibility displayed by the protease (Fig. 1e; Supplementary Movie 1), but it is perhaps a reflection of the important role played by sites remote from the scissile bond in peptide cleavage efficiency.27, 37
The extensive interface of contact between the peptide and the protease in FMDV 3Cpro exhibits similarities to that observed in the 3C-like TEVpro, which was solved in complex with a peptide that spanned positions P7–P3′28 (Fig. 1c and d). Superposition of the two structures shows that the backbone geometry is similar over the common P4–P3′ region. Structural similarity is closest for the core segment centred on the scissile bond (P2–P2′; root-mean-squared deviation over C a = 0.14 Å), but there are significant structural deviations outside this region, dictated by the differing topologies of the peptide-binding cleft. In particular, TEVpro provides a much more enclosed binding site for the P4–P3 residues due to the close apposition of the C-terminal β-ribbon with the E2–F2 loop in the C-terminal β-barrel (Fig. 1d).
The FMDV 3Cpro–peptide interaction is also reminiscent of the extended interface observed in the structures of specific protein inhibitors of trypsin and trypsin-like proteases such as chymotrypsin and collagenase.38, 39, 40, 41, 42 These inhibitors are generally composed of a small stable domain that interacts with the target enzyme by inserting an extended internal loop, constrained at both ends, into the peptide-binding cleft of the target enzyme. The extent of the interaction varies from P3–P2′ in the complex of trypsin with pancreatic trypsin inhibitor41 to P7–P4′ for ecotin bound to collagenase.39 These inhibitors strongly mimic highly specific substrates, but are usually cleaved extremely slowly because the rigidity of the enzyme–peptide interface prevents the conformational changes necessary to attain the tetrahedral intermediate at the scissile bond and because the tightly packed interface prevents access of water molecules that would be necessary for the deacylation step of catalysis.43 In contrast, in 3Cpro, extensive sequence recognition must also be allied with sufficient flexibility to permit catalysis.
Although the residual flexibility in the 3Cpro–peptide complex necessary for catalysis is not detectable by crystallography, our results do reveal that structural adjustments are made across the peptide-binding surface on substrate binding. Particularly notable are the movements of the β-ribbon to form the S2–S4 subsites, the strands flanking the S1 pocket and the large rotation of Leu47 within the S1′ pocket to accommodate the large P1′ side chain (Fig. 1e). These observations are broadly consistent with previous NMR analyses of HRV 3Cpro, which revealed flexibility throughout the enzyme that was attenuated by interactions between the protein and a peptide-like inhibitor spanning P6–P1.24 These marked conformational changes associated with peptide binding to FMDV 3Cpro are in contrast to the relatively rigid lock-and-key binding of some protein inhibitors to trypsin-like proteases38, 42 and underscore the importance of the structural analysis of the peptide-bound form in obtaining a more complete picture of the substrate basis of cleavage specificity. This information is of particular relevance to inhibitor design, although further structural analyses of FMDV 3Cpro bound to different peptide sequences will be needed to map out the full extent of structural changes that accompany substrate binding.
The conformational changes propagated by the rotation of Leu47 to accommodate P1′-Leu in the FMDV 3Cpro substrate were unanticipated. Our structural and mutagenic analyses suggest that such side-chain movements are very unlikely to occur in HRV or PV 3Cpro, since the F25/A41 pair of residues that lie on either side of the pocket entrance (equivalent to C31/L47 in FMDV 3Cpro) does not have the flexibility to accommodate large P1′ side chains (Fig. 2c–e). This explains the strict requirement for small P1′ side chains (most often Gly) in substrates for 3Cpro from HRV and PV.15, 21 In contrast, HAV 3Cpro, like FMDV 3Cpro, is tolerant of larger P1′ residues (e.g., R, V and M).21 Since it has an M29/A45 pair of residues flanking the S1′ pocket, we suggest that the Met29 side chain may have the ability to move to accommodate larger P1′ side chains.
S1 specificity
The crystal structures of FMDV 3Cpro, in complex with the VP1-2A and VP1-2Am peptides, show that P1-Gln or P1-Glu binds similarly within the S1 specificity pocket, with both making hydrogen bonds from a side-chain oxygen atom to the side chains of Thr158 and His181; Tyr154 also plays an important role in determining P1 specificity, since it stabilises the orientation of His181 (Fig. 2c and d). This structural arrangement accounts well for the observation that a Gln-to-Glu substitution at P1 within the VP1-2A peptide only modestly reduces peptide cleavage (Fig. 1a and e).
However, the bound conformation of P1-Gln in FMDV 3Cpro is also very similar to the conformation of the modified glutamine observed in the S1 pocket when the peptide-like inhibitor AG7088 is bound to HRV 3Cpro.25 Although the glutamine side chain was altered to incorporate a lactam ring, glutamine-specific hydrogen bonds were observed in S1. It is therefore difficult to understand why FMDV 3Cpro readily cleaves substrates with P1-Gln or P1-Glu, while HRV 3Cpro exhibits a strong preference for P1-Gln substrates.15, 17, 20 The discrepancy is all the more remarkable, since the residues primarily responsible for P1-Gln or Glu recognition in FMDV 3Cpro (Thr158, Tyr154 and His181) are conserved in the P1-Gln-specific HRV 3Cpro and in the P1-Glu-specific V8 protease from Staphylococcus aureus.44
Given the close structural similarity between FMDV and HRV 3Cpro in the S1 pocket, why does HRV 3Cpro discriminate so strongly against P1-Glu? In the coronavirus main protease from tobacco etch virus, the selectivity for P1-Gln is enforced by an interaction between the Gln side-chain amide and the carboxylate of Asp148 (Fig. 2f);28 clearly, the presence of this negatively charged side chain strongly disfavours binding of substrates with P1-Glu. However, HRV 3Cpro lacks an equivalent repulsive group (Fig. 2e), and it remains unclear how discrimination against P1-Glu substrates is achieved.
It may be worth considering the impact of remote sites on P1 selectivity in these enzymes. As noted previously,3 FMDV 3Cpro substrates with P1-Glu most commonly have a small residue at P1′ and are less likely to conserve Lys at P2. Perhaps the correlated amino acid variations at these and other positions have a positive impact on the cleavage of P1-Glu substrates. Further exploration of this notion will benefit from a structural analysis of a complex of FMDV 3Cpro with a natural P1-Glu substrate.
Materials and Methods
Expression and purification of FMDV 3Cpro
The original construct that expresses a C-terminally truncated, catalytically inactive form of the type A10 FMDV 3Cpro containing C95K, C142S and C163A substitutions (see Table 2 in Birtley and Curry13) was modified for co-crystallisation trials using QuikChange (Stratagene) to engineer an S142L mutation. This generates a form of 3Cpro that has wild-type binding activity but remains soluble at purified protein concentrations in excess of 10 mg/ml.12 This modified construct was used to generate additional 3Cpro variants by the same technique to add mutations to the active site and the S1′ subsite. FMDV 3C proteins were produced in E. coli and purified essentially as reported previously,12, 13 although the gel-filtration step was omitted for samples that were used in cleavage assays. The purified proteins were concentrated to 5–6 mg/ml in 50 mM Hepes (pH 7.1), 400 mM NaCl and 1 mM β-mercaptoethanol, and stored in small aliquots at − 80 °C before use in protease cleavage assays.
A modified pET-FMDV3CXS construct,13, 35 inactivated by the introduction of the C163A mutation in 3C, was used to express the 35-kDa fragment (3BC) of the FMDV A10 polyprotein that corresponds to 3B1–3B2–3B3–3BC (Fig. 3a). The protein, which has a non-cleavable C-terminal His-tag, was produced in BL21(DE3) E. coli and purified to about 95% homogeneity on TALON beads (BD Biosciences) using the same protocol as for the active 3Cpro mutants.
Peptide synthesis
The peptides for co-crystallisation (VP1/2A, APAKQLLNFD; VP1/2Am, APAKELLNFD)—synthesised by standard Fmoc solid-phase synthesis as described previously,8 with acetylation of the N-terminus and amidation of the C-terminus to block charges—were purified by reverse-phase HPLC. Sequences were confirmed by mass spectroscopy. The FRET4 substrate (and sequence variants) used in peptide cleavage assays was prepared as reported.12, 31
Complex formation and crystallisation
3Cpro–peptide complexes were prepared at a 1:5.5 molar ratio of enzyme to peptide, since this has worked well for related proteases.28 Typically, 50 μl of 3Cpro at 17 mg/ml protein in 100 mM Hepes (pH 7), 400 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 2 mM β-mercaptoethanol and 0.01% (wt/vol) sodium azide was mixed with 7 μl of 30 mM peptide dissolved in the same buffer, yielding a final protein concentration of 14.9 mg/ml. The complex was incubated with rotation at room temperature for 1 h and used immediately in sitting-drop vapour diffusion crystallisation trials. Optimised crystals of 3Cpro bound to APAKQLLNFD were obtained by mixing 1 μl of complex with 3 μl taken from a 1-ml reservoir composed of 40–42.5% polyethylene glycol 400, 0.2 M LiSO4 and 0.1 M Tris (pH 8.0). Similar conditions were used to crystallise the complex with APAKELLNFD peptide: 41–43% polyethylene glycol 400, 0.2 M LiSO4 and 0.1 M Tris (pH 8.0).
Crystals were soaked for a few seconds in mother liquor incorporating 20% (vol/vol) glycerol and flash cooled on beamlines ID23-2 and ID14-2 at the European Synchrotron Radiation Facility (ESRF; Grenoble, France) in a stream of N2 at 100 K. Diffraction data were processed with MOSFLM and scaled using SCALA from the CCP4 suite of programs.45 The data for the 3Cpro–VP1-2A complex crystals were phased with Phaser (version 1.3.2)46 using the unliganded structure of 3Cpro (PDB ID 2j92 12) as search model after removal of the most mobile surface loops (residues 74–80 and 138–150). This procedure located two molecules in the asymmetric unit, with an overall log likelihood gain of 1319. The protease structure from this model was then used to solve the structure of the 3Cpro–VP1-2Am complex, again using Phaser.46 In that case, four molecules were identified in the asymmetric unit; the overall log likelihood gain was 3981. Manual model adjustment and refinement of both models were performed with O32 and CNS,47 respectively.
Polyprotein cleavage assays
The cleavage activity of TALON-purified 3Cpro mutants was assessed using a protolytically inactive form of FMDV strain A1061 3BC precursor as substrate.13, 35 Working stocks of 3Cpro and the 3BC substrate (Fig. 3a) were prepared at 2 mg/ml in 50 mM Hepes (pH 7.1), 400 mM NaCl and 20 mM DTT. In each assay, equal volumes of substrate and 3Cpro were mixed to give a final concentration of each of 1 mg/ml and incubated at 37 °C for varying times up to 17 h. Digestion reactions were stopped by direct addition of 5 μl of 2× SDS sample buffer to 5 μl of the reaction mixture, immediately followed by heating at 95 °C for 2 min.
Fluorescent peptide cleavage assays
Assays were performed in 96-well plates essentially as described previously,12 with minor modifications. Fifty to 100 μl of varying concentrations of the FRET4 fluorogenic substrate31 were added to an equal volume of enzyme. The final enzyme concentrations were typically 1–2 μM (wild type), 5 μM (C31A, C31F, C31A/L47A and C31F/L47A mutants) and 10 μM (D84A and D84E mutants). Reactions were performed in triplicate at 37 °C in 100 mM NaH2PO4/Na2HPO4 (pH 7.4), 1 mM ethylenediaminetetraacetic acid, 1 mM Tris(2-carboxyethyl)-phosphine HCl and 5% glycerol. Relative hydrolysis rates were determined by monitoring fluorescence at 2-min intervals for 20–60 min in a Cytofluor microplate reader or a Molecular Devices SpectraMax M2e. The excitation wavelength used was 335 nm, and emission was measured at 460 nm. Initial hydrolysis rates were recorded (typically from 0 to 20 min) for each substrate concentration, and specificity constants were calculated by fitting to the Michaelis–Menten equation.
Infectivity assays
The generation of the C142L sequence change within the whole virus (type O1K) was achieved by overlap PCR using the pT7S3 infectious copy cDNA plasmid48 as template, using the flanking primers 3AEcofor (AGGCAGCAATTGAATTCTTTGA) and 3CPstrev (CGTTGCCTCCTGCAGAGTGA) plus the degenerate mutagenic oligonucleotides 3CmutC142Lfor (ACTTACAAGGACATTGTGGTTCTNATGGACGGAGACACCATG) and 3CmutC142Lrev (CCGTCCATNAGAACCACAATGTCCTTGTAAGT). Up to four different codons (CTN) for leucine (L) could have been generated to replace the Cys (C) residue codon (TGC) in this procedure, but only two were used. The mutant PCR product, including the EcoRI (nt 5058)-PstI (nt 6209) region (numbered according to accession no. X00871), was inserted into the pCR-XL-TOPO vector (Invitrogen), and individual plasmids were sequenced. The EcoRI-PstI fragments containing the CTA (C142L1) and CTG (C142L2) codons were excised and reconstructed into pT7S3 via an intermediate plasmid containing the BamHI fragment (nt 2909–7117). The full-length wild-type and mutant plasmids were linearised with HpaI, and RNA transcripts were prepared using T7 RNA polymerase (T7 Megascript kit; Ambion). The transcripts were introduced into BHK cells by electroporation, essentially as described previously.49 Complete cytopathic effect was apparent following overnight incubation. Rescued virus harvests were passaged once in BHK cells, and then RNA was extracted using a viral RNA extraction kit (Qiagen). RT-PCRs, with or without reverse transcriptase, were performed using the primers (3AEcofor and 3CPstrev, as described above) for 25 cycles. The expected products were only observed in the presence of the reverse transcriptase (this ensured that products were derived from the viral RNA and not from residual cDNA template), then the amplicons were sequenced using the same primers and maintained the respective wild-type or mutant sequences as anticipated (data not shown).
A single-step growth curve of the rescued viruses was performed within BHK cells, which were infected with wild-type or mutant viruses in parallel, and the cells were harvested at t = 0, 2, 4 and 6 h postinfection directly into RLT buffer (Qiagen), the first stage of the RNA extraction procedure. FMDV RNA was quantified by a standard diagnostic real-time RT-PCR assay, as described previously,50, 51 using amounts of RNA and cDNA that ensured that both cDNA synthesis and real-time PCR were within the linear range of the assays. The numbers of viral genomes in equal aliquots of RNA were determined by reference to a dilution series of RNA transcripts.
Molecular Graphics
All structural images were prepared with PyMol52. The morph animation shown in Supplementary Movie 1 was generated using eMovie53.
Accession numbers
Coordinates and structure factors have been deposited in the PDB with accession numbers 2wv4 and 2wv5.
Acknowledgements
We would like to thank the staff on beamlines ID23-2 and ID14-2 at the ESRF for assistance with X-ray data collection. S.C. and R.J.L. are grateful for grant support provided by the Biotechnology and Biological Sciences Research Council. We also thank Preben Normann, Inge Nielsen and Tina Frederiksen (National Veterinary Institute, Lindholm), and Imperial College students Siew Lian Low and George Wong for excellent technical assistance. S.R.K. was supported by an Engineering and Physical Sciences Research Council PhD studentship. N.R.-R. was funded by a Marie Curie Host Fellowship for Early Stage Research Training. T.R.S. was funded by a PhD studentship awarded by Imperial College.
Edited by G. Schulz
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.10.048
Appendix A.
Morph animation showing conformational changes in FMDV 3Cpro on peptide binding. Viewpoint is the same as in Fig. 1e. Further description of key features can be found in the audio commentary accompanying the video.
References
- 1.Sutmoller P., Barteling S.S., Olascoaga R.C., Sumption K.J. Control and eradication of foot-and-mouth disease. Virus Res. 2003;91:101–144. doi: 10.1016/s0168-1702(02)00262-9. [DOI] [PubMed] [Google Scholar]
- 2.Grubman M.J., Baxt B. Foot-and-mouth disease. Clin. Microbiol. Rev. 2004;17:465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curry S., Roqué-Rosell N., Zunszain P.A., Leatherbarrow R.J. Foot-and-mouth disease virus 3C protease: recent structural and functional insights into an antiviral target. Int. J. Biochem. Cell Biol. 2007;39:1–6. doi: 10.1016/j.biocel.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrillo C., Tulman E.R., Delhon G., Lu Z., Carreno A., Vagnozzi A., et al. Comparative genomics of foot-and-mouth disease virus. J. Virol. 2005;79:6487–6504. doi: 10.1128/JVI.79.10.6487-6504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skern T., Hampoelz B., Guarné A., Fita I., Bergmann E., Petersen J., James M.N.G. In: Molecular Biology of Picornaviruses. Semler B.L., Wimmer E., editors. ASM Press; Washington, DC: 2002. Structure and function of picornavirus proteases; pp. 199–212. [Google Scholar]
- 6.Belsham G.J. Translation and replication of FMDV RNA. Curr. Top. Microbiol. Immunol. 2005;288:43–70. doi: 10.1007/3-540-27109-0_3. [DOI] [PubMed] [Google Scholar]
- 7.Allaire M., Chernaia M.M., Malcolm B.A., James M.N. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 8.Birtley J.R., Knox S.R., Jaulent A.M., Brick P., Leatherbarrow R.J., Curry S. Crystal structure of foot-and-mouth disease virus 3C protease: new insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 2005;280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 9.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., et al. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 10.Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 11.Lee C.C., Kuo C.J., Ko T.P., Hsu M.F., Tsui Y.C., Chang S.C., et al. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J. Biol. Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweeney T.R., Roqué-Rosell N., Birtley J.R., Leatherbarrow R.J., Curry S. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the beta-ribbon in proteolysis. J. Virol. 2007;81:115–124. doi: 10.1128/JVI.01587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birtley J.R., Curry S. Crystallization of foot-and-mouth disease virus 3C protease: surface mutagenesis and a novel crystal-optimization strategy. Acta Crystallogr. Sect. D. 2005;61:646–650. doi: 10.1107/S0907444905007924. [DOI] [PubMed] [Google Scholar]
- 14.Yin J., Bergmann E.M., Cherney M.M., Lall M.S., Jain R.P., Vederas J.C., James M.N. Dual modes of modification of hepatitis A virus 3C protease by a serine-derived beta-lactone: selective crystallization and formation of a functional catalytic triad in the active site. J. Mol. Biol. 2005;354:854–871. doi: 10.1016/j.jmb.2005.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blom N., Hansen J., Blaas D., Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordingley M., Callahan P., Sardana V., Garsky V., Colonno R. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J. Biol. Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 17.Cordingley M.G., Register R.B., Callahan P.L., Garsky V.M., Colonno R.J. Cleavage of small peptides in vitro by human rhinovirus 14 3C protease expressed in Escherichia coli. J. Virol. 1989;63:5037–5045. doi: 10.1128/jvi.63.12.5037-5045.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jewell D.A., Swietnicki W., Dunn B.M., Malcolm B.A. Hepatitis A virus 3C proteinase substrate specificity. Biochemistry. 1992;31:7862–7869. doi: 10.1021/bi00149a017. [DOI] [PubMed] [Google Scholar]
- 19.Pallai P.V., Burkhardt F., Skoog M., Schreiner K., Bax P., Cohen K.A., et al. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J. Biol. Chem. 1989;264:9738–9741. [PubMed] [Google Scholar]
- 20.Long A.C., Orr D.C., Cameron J.M., Dunn B.M., Kay J. A consensus sequence for substrate hydrolysis by rhinovirus 3C proteinase. FEBS Lett. 1989;258:75–78. doi: 10.1016/0014-5793(89)81619-9. [DOI] [PubMed] [Google Scholar]
- 21.Seipelt J., Guarne A., Bergmann E., James M., Sommergruber W., Fita I., Skern T. The structures of picornaviral proteinases. Virus Res. 1999;62:159–168. doi: 10.1016/s0168-1702(99)00043-x. [DOI] [PubMed] [Google Scholar]
- 22.Schechter I., Berger A. On the size of the active site in proteases: I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 23.Perona J.J., Craik C.S. Structural basis of substrate specificity in the serine proteases. Protein Sci. 1995;4:337–360. doi: 10.1002/pro.5560040301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjorndahl T.C., Andrew L.C., Semenchenko V., Wishart D.S. NMR solution structures of the apo and peptide-inhibited human rhinovirus 3C protease (serotype 14): structural and dynamic comparison. Biochemistry. 2007;46:12945–12958. doi: 10.1021/bi7010866. [DOI] [PubMed] [Google Scholar]
- 25.Matthews D.A., Dragovich P.S., Webber S.E., Fuhrman S.A., Patick A.K., Zalman L.S., et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl Acad. Sci. USA. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin J., Cherney M.M., Bergmann E.M., Zhang J., Huitema C., Pettersson H., et al. An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors. J. Mol. Biol. 2006;361:673–686. doi: 10.1016/j.jmb.2006.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perona J.J., Craik C.S. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem. 1997;272:29987–29990. doi: 10.1074/jbc.272.48.29987. [DOI] [PubMed] [Google Scholar]
- 28.Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., III, Kapust R.B., et al. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 29.Xue X., Yu H., Yang H., Xue F., Wu Z., Shen W., et al. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergmann E.M., Cherney M.M., McKendrick J., Frormann S., Luo C., Malcolm B.A., et al. Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV. Virology. 1999;265:153–163. doi: 10.1006/viro.1999.9968. [DOI] [PubMed] [Google Scholar]
- 31.Jaulent A.M., Fahy A.S., Knox S.R., Birtley J.R., Roque-Rosell N., Curry S., Leatherbarrow R.J. A continuous assay for foot-and-mouth disease virus 3C protease activity. Anal. Biochem. 2007;368:130–137. doi: 10.1016/j.ab.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 32.Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these maps. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brunger A.T. Version 1.2 of the Crystallography and NMR System. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 34.Curry S., Roqué-Rosell N., Sweeney T.R., Zunszain P.A., Leatherbarrow R.J. Structural analysis of foot-and-mouth disease virus 3C protease: a viable target for antiviral drugs? Biochem. Soc. Trans. 2007;35:594–598. doi: 10.1042/BST0350594. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Ross-Smith N., Proud C.G., Belsham G.J. Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: identification of the eIF4AI cleavage site. FEBS Lett. 2001;507:1–5. doi: 10.1016/s0014-5793(01)02885-x. [DOI] [PubMed] [Google Scholar]
- 36.Grubman M.J., Zellner M., Bablanian G., Mason P.W., Piccone M.E. Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology. 1995;213:581–589. doi: 10.1006/viro.1995.0030. [DOI] [PubMed] [Google Scholar]
- 37.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 38.Frigerio F., Coda A., Pugliese L., Lionetti C., Menegatti E., Amiconi G., et al. Crystal and molecular structure of the bovine alpha-chymotrypsin–eglin c complex at 2.0 Å resolution. J. Mol. Biol. 1992;225:107–123. doi: 10.1016/0022-2836(92)91029-o. [DOI] [PubMed] [Google Scholar]
- 39.Perona J.J., Tsu C.A., Craik C.S., Fletterick R.J. Crystal structure of an ecotin–collagenase complex suggests a model for recognition and cleavage of the collagen triple helix. Biochemistry. 1997;36:5381–5392. doi: 10.1021/bi9617522. [DOI] [PubMed] [Google Scholar]
- 40.Tsunogae Y., Tanaka I., Yamane T., Kikkawa J., Ashida T., Ishikawa C., et al. Structure of the trypsin-binding domain of Bowman–Birk type protease inhibitor and its interaction with trypsin. J. Biochem. 1986;100:1637–1646. doi: 10.1093/oxfordjournals.jbchem.a121872. [DOI] [PubMed] [Google Scholar]
- 41.Huber R., Kukla D., Bode W., Schwager P., Bartels K., Deisenhofer J., Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor: II. Crystallographic refinement at 1.9 Å resolution. J. Mol. Biol. 1974;89:73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- 42.Fujinaga M., Sielecki A.R., Read R.J., Ardelt W., Laskowski M., James M.N. Crystal and molecular structures of the complex of alpha-chymotrypsin with its inhibitor turkey ovomucoid third domain at 1.8 Å resolution. J. Mol. Biol. 1987;195:397–418. doi: 10.1016/0022-2836(87)90659-0. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski M., Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 44.Prasad L., Leduc Y., Hayakawa K., Delbaere L.T. The structure of a universally employed enzyme: V8 protease from Staphylococcus aureus. Acta Crystallogr. Sect. D. 2004;60:256–259. doi: 10.1107/S090744490302599X. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computer Project No. 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., et al. Crystallography and NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Ellard F.M., Drew J., Blakemore W.E., Stuart D.I., King A.M. Evidence for the role of His-142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J. Gen. Virol. 1999;80:1911–1918. doi: 10.1099/0022-1317-80-8-1911. [DOI] [PubMed] [Google Scholar]
- 49.Nayak A., Goodfellow I.G., Woolaway K.E., Birtley J., Curry S., Belsham G.J. Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J. Virol. 2006;80:9865–9875. doi: 10.1128/JVI.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belsham G.J., Normann P. Dynamics of picornavirus RNA replication within infected cells. J. Gen. Virol. 2008;89:485–493. doi: 10.1099/vir.0.83385-0. [DOI] [PubMed] [Google Scholar]
- 51.Reid S.M., Grierson S.S., Ferris N.P., Hutchings G.H., Alexandersen S. Evaluation of automated RT-PCR to accelerate the laboratory diagnosis of foot-and-mouth disease virus. J. Virol. Methods. 2003;107:129–139. doi: 10.1016/s0166-0934(02)00210-0. [DOI] [PubMed] [Google Scholar]
- 52.Delano W.L. DeLano Scientific; San Carlos, CA, USA: 2002. The PyMOL molecular graphics system. [Google Scholar]
- 53.Hodis E., Schreiber G., Rother K., Sussman J.L. eMovie: a storyboard-based tool for making molecular movies. Trends Biochem. Sci. 2007;32:199–204. doi: 10.1016/j.tibs.2007.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morph animation showing conformational changes in FMDV 3Cpro on peptide binding. Viewpoint is the same as in Fig. 1e. Further description of key features can be found in the audio commentary accompanying the video.