Summary
Background
Characterising sexual networks with transmission of sexually transmitted infections might allow identification of individuals at increased risk of infection. We aimed to investigate sexual mixing in Neisseria gonorrhoeae transmission networks between women, heterosexual men, and men who report sex with men (MSM), and between people with and without HIV.
Methods
In this cross-sectional observational study, we whole-genome sequenced N gonorrhoeae isolates from the archive of the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP).w Isolates that varied by five single nucleotide polymorphisms or fewer were grouped into clusters that represented sexual networks with N gonorrhoeae transmission. Clusters were described by gender, sexual risk group, and HIV status.
Findings
We sequenced 1277 N gonorrhoeae isolates with linked clinical and sociodemographic data that were collected in five clinics in England during 2013–16 (July 1 to Sept 30 in 2013–15; July 1 to Sept 9 in 2016). The isolates grouped into 213 clusters. 30 (14%) clusters contained isolates from heterosexual men and MSM but no women and three (1%) clusters contained isolates from only women and MSM. 146 (69%) clusters comprised solely people with negative or unknown HIV status and seven (3%) comprised only HIV-positive people. 60 (28%) clusters comprised MSM with positive and negative or unknown HIV status.
Interpretation
N gonorrhoeae molecular data can provide information indicating risk of HIV or other sexually transmitted infections for some individuals for whom such risk might not be known from clinical history. These findings have implications for sexual health care, including offering testing, prevention advice, and preventive treatment, such as HIV pre-exposure prophylaxis.
Funding
National Institute for Health Research Health Protection Research Unit; Wellcome; Public Health England.
Introduction
Gonorrhoea, caused by the bacterium Neisseria gonorrhoeae, is an urgent threat to public health because of the development of antimicrobial resistance and impending threat of untreatable infections. Although rare in the general population,1 diagnoses of N gonorrhoeae infection have risen substantially in England in the past decade, primarily among men who report sex with men (MSM).2 N gonorrhoeae is concentrated in sexual networks wherein network members report high numbers of previous partners and concurrent sexual partnerships.3 In the UK, some young adults, MSM, and people of black Caribbean ethnicity are at particular risk.2 Assortative sexual mixing might also concentrate infection in these groups, whereas disassortative sexual mixing fosters wider dissemination of infection across the population.3
Compared with women and men who have sex exclusively with women, MSM are more likely to engage in sexual behaviour that puts them at risk of sexually transmitted infections (STIs), including condomless sex with multiple sexual partners.4 MSM living with HIV have the highest rates of N gonorrhoeae diagnoses,5 possibly linked to the adoption of HIV seroadaptive behaviours that accentuate STI risk, especially condomless anal sex.6, 7 Among MSM without HIV, N gonorrhoeae infection is a strong predictor of subsequent HIV infection.8
Characterising sexual networks and understanding sexual mixing patterns that facilitate N gonorrhoeae transmission can help predict and explain the distribution of infection in the population and might allow the identification of individuals at risk of other STIs, such as HIV and hepatitis A and B.9 However, sexual network analysis is complicated by uncontactable cases and incomplete epidemiological data, even after intensive contract tracing.10 Collecting information about socially sensitive behaviours, such as sexual behaviour, can be subject to reporting bias, and people with shared epidemiological characteristics or risk behaviours might not necessarily share a sexual network.
Research in context.
Evidence before this study
Our previous systematic review of studies published up to March 31, 2017, which investigated the use of molecular epidemiology for understanding the transmission of Neisseria gonorrhoeae in sexual networks found that few whole-genome sequencing (WGS) studies used broad sampling strategies and had sufficient patient information to investigate and describe sexual networks with N gonorrhoeae transmission. To update that review, we searched PubMed using the terms “Neisseria gonorrhoeae” OR “gonorrh*” AND “molecular epidemiology” for papers published up to March 31, 2019. Our search identified 26 studies. Nine studies used WGS, of which seven focused on the genetic determinants and spread of antimicrobial resistance in N gonorrhoeae. Six studies (three using WGS) only included isolates with a particular antimicrobial resistance phenotype or molecular strain type and provided sparse epidemiological information. A study from New Zealand that included 398 N gonorrhoeae isolates found a high frequency of sexual mixing between heterosexual or bisexual populations and men who report sex with men (MSM). A study from Brighton, UK, that included 100 isolates, predominantly from MSM, found that most sexual networks comprised people with differing HIV statuses. To our knowledge, no large WGS study including N gonorrhoeae isolates from multiple geographically dispersed sexual health clinics, people from various demographic and sexual behaviour groups, and across multiple years has been done in England.
Added value of this study
We did one of the largest molecular epidemiological studies of N gonorrhoeae to date, analysing WGS and epidemiological data from 1277 cases of N gonorrhoeae reported from five clinics in England between 2013 and 2016. To our knowledge, this study is the first to use WGS data to describe the sexual mixing patterns of women, heterosexual men, and MSM, and to identify the extent of sexual mixing between MSM with identical or different HIV statuses. We found that WGS data identified a considerable number of sexual networks containing men who report only heterosexual behaviours and MSM and extensive sexual mixing between MSM with HIV and those without or with unknown status. We showed that phylogenomic analyses provided additional information about sexual mixing patterns in N gonorrhoeae transmission networks that might not have been evident from epidemiological data alone.
Implications of all the available evidence
Reconstructing sexual networks from epidemiological data alone can be complicated because ascertaining whether individuals with similar reported characteristics share a sexual network can be difficult, and epidemiological data is often incomplete because of uncontactable individuals. By providing information about the sexual networks a patient belongs to and probable sexual health risks, real-time WGS data on N gonorrhoeae could influence clinical decision making and support tailored and relevant interventions. Sexual networks containing only isolates from heterosexual men and MSM suggest groups of heterosexual-identifying men might exist who could benefit from enhanced sexual health testing and prevention messages that would normally be targeted to MSM. Evidence of sexual mixing between MSM with HIV and those without HIV or with unknown status suggests that WGS could help identify men who might be at considerable and unrecognised risk of HIV infection. Such insights might improve prevention of HIV and other sexually transmitted infections by guiding decisions on preventive interventions, including HIV pre-exposure prophylaxis and hepatitis A and B testing and vaccination.
Phylogenomic analysis can resolve some of the uncertainties in epidemiological investigations by differentiating genetically related clusters of infections, which probably belong to a shared sexual network. Incorporating epidemiological and sexual behaviour data into phylogenetic analyses can thus provide insight into sexual mixing patterns and related public health implications. Studies of HIV and hepatitis B have identified sexual networks of MSM that include men who report exclusively female partners.11, 12
Phylogenomic analysis of N gonorrhoeae with linked epidemiological data has been used to map the spread of antimicrobial resistance across sexual networks and between countries and investigate the characteristics of N gonorrhoeae transmission chains.13, 14, 15, 16, 17, 18, 19, 20, 21 However, few studies have included sufficient clinical and sexual behaviour data to explore sexual mixing patterns between different sociodemographic groups and how mixing might influence STI and HIV transmission risk.22
To inform public health infection control efforts, we whole-genome sequenced more than 1200 N gonorrhoeae isolates with linked clinical and sociodemographic data collected in five sexual health clinics in England over a 4-year period. We investigated sexual mixing in N gonorrhoeae transmission networks between MSM, men reporting heterosexual behaviours, and women, and between MSM with and without HIV.
Methods
Isolate sampling
For this cross-sectional observational study, N gonorrhoeae isolates were obtained from the archive of the Gonococcal Resistance to Antimicrobials Surveillance Programme (GRASP).23 GRASP is a sentinel programme that tests antimicrobial susceptibility of consecutive isolates received from all culture-positive N gonorrhoeae diagnoses made between July 1, and Sept 30, 2013–15, and between July 1, and Sept 9, 2016, in 26 sexual health clinics in England and Wales. Since 2000, GRASP has collected 1200–2500 isolates annually. Necessary for susceptibility testing, GRASP only includes N gonorrhoeae cultures, which are available for just under half of all gonorrhoea diagnoses in England.24 For this study, five GRASP clinics (two in London and one each in Birmingham, Bristol, and Liverpool) were selected to provide isolates that were broadly representative of people with gonorrhoea in England in terms of sociodemographic and behavioural characteristics, including gender, sexual risk group, age, ethnicity, and HIV status. Eligibility for sequencing was defined as all archived isolates from these clinics between 2013 and 2016, which were the most recent years with available GRASP data.
Public Health England (PHE) has permission to process patient confidential data obtained by GRASP under Regulation 3 of the Health Service (Control of Patient Information) Regulations 2002. Information governance advice and ethics approval for this study were granted by the PHE Research Ethics and Governance Group.
Epidemiological data collection
The GRASP archive includes clinical, sociodemographic, and behavioural data collected through PHE's STI surveillance system (GUMCAD)25 and directly from participating GRASP clinics. GUMCAD contains longitudinal patient records that enabled us to investigate whether people with a negative or unknown HIV status at the time of N gonorrhoeae diagnosis were subsequently diagnosed with HIV.
Isolation and whole-genome sequencing of isolates
Single colonies were isolated on non-selective GCVIT agar (Difco BBL GC II Agar Base plus 1% Vitox; Oxoid, Altrincham, UK). DNA was extracted from a subculture of a single colony of each isolate using the automated QIAsymphony DNA Mini Kit (Qiagen, Hilden, Germany). Whole-genome sequencing (WGS) was done at the Wellcome Sanger Institute (Hinxton, UK) with the Illumina HiSeq-X-Ten system and processed in the Sanger WGS data management pipeline. Further details are described in appendix 1 (p 1) Sequence data are available on the European Nucleotide Archive with use of study accession identifier ERP022090. Metadata for sequences are provided in appendix 2.
Data analysis
To assess sample representativeness, we compared the epidemiological characteristics of the sequenced isolates with those of non-sequenced isolates (those that met study collection criteria but were not viable for sequencing) among all eligible study isolates and all gonorrhoea diagnoses in England from Jan 1, 2013, to Dec 31, 2016. Significant differences were assessed using a two-sample proportions Z test with a threshold for significance of 0·05.
A phylogenetic tree with genetic recombination events removed was created using Gubbins (version 2.4.0)26 with the RAxML tree building option (appendix 1 p 1) and the default GTR+gamma substitution model. The number of single nucleotide polymorphisms (SNPs) between each pair of isolates was estimated from the branch length distance, which was extracted from the phylogenetic tree using SeaView (version 4.7).27 Branch lengths are an estimate of the total number of substitutions that have occurred between a pair of isolates. To translate this estimate into an estimated SNP difference, the branch length was multiplied by the number of nucleotides in the sequence alignment that were used to create the phylogenetic tree.
The number of SNPs between each pair of isolates was used to define clusters of genetically similar isolates representing sexual networks with N gonorrhoeae transmission. No standardised method exists to define a genetic cluster representing a sexual network. De Silva and colleagues14 predicted that isolates collected on the same day and up to nine SNPs apart and isolates collected within a year of each other and up to 14 SNPs apart might be part of the same transmission network. Studies by Fifer and colleagues17 and Kwong and colleagues19 found that the SNP difference between known contacts ranged from zero to five. We chose to group isolates into clusters using a SNP threshold of five or fewer SNPs, meaning that within a cluster each isolate must be no more than five SNPs different to at least one other isolate in the cluster. This SNP threshold was chosen as a conservative estimate for people who are closely linked within a sexual network. We did a sensitivity analysis to assess the effect of different thresholds on cluster size and potential implications for interpretation using threshold cutoffs of three or fewer, ten or fewer, and 14 or fewer SNPs (appendix 1 p 1).
Clusters were described by gender, HIV status, and sexual risk group, including all women, heterosexual men, and MSM (women who have sex exclusively with women were not considered as a separate group in this analysis because of small numbers). Univariate and multivariable analysis were used to assess differences in the epidemiological characteristics of people who were clustered with others from the same or different sexual risk groups. The epidemiological characteristics explored were diagnosis year, clinic location, age, ethnicity, country of birth, symptomatic infection, diagnosed with an STI (excluding HIV) in the previous year, number of sexual partners in the 3 months before diagnosis, and sex while travelling outside of the UK in the 3 months before diagnosis (herewith described as having travel-associated sexual partnerships). Significance was assessed by whether the CI of the odds ratio (OR) included 1·0 and whether the χ2 p value was less than 0·05. A multivariable logistic regression model using a forward stepwise approach was developed including variables identified as being significantly associated with the outcome in the univariate model. The likelihood ratio test was used to assess whether explanatory variables should remain in the multivariable model, using a significance threshold of 0·05. The same methods were used to assess differences between the epidemiological characteristics of clusters comprising only MSM with a negative or unknown HIV status and those comprising MSM with negative or unknown and positive HIV status. Missing data are reported in appendix 1 (p 2); records with missing data were not included in the analyses.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
1277 (91%) of 1407 eligible isolates were successfully sequenced. The remaining 130 (9%) isolates were not sequenced because the bacteria were no longer viable. Sequencing success was lower for isolates collected in 2016 than for those from other years (251 [79%] of 319) in 2016, 367 [92%] of 398 in 2015, 333 [97%] of 342 in 2014, and 326 [94%] of 348 in 2013). Sequenced isolates represented 21% of all 6184 GRASP isolates and 8–10% of all gonorrhoea diagnoses in each of the selected clinics during the study period: for the two London clinics, 186 (9%) of 1981 and 386 (8%) of 4865 isolates were sequenced, respectively; for the three clinics outside of London, sequencing was done for 163 (10%) of 1682, 138 (8%) of 1812, and 404 (9%) of 4335 isolates, respectively. The sequenced isolates represented 1% (1277 of 146 369) of all gonorrhoea diagnoses in England during the study period (appendix 1 p 2). The 1277 isolates came from 1256 people, of whom 1235 (98%) had one and 21 (2%) had two gonorrhoea diagnoses.
MSM were overrepresented in this study compared with N gonorrhoeae diagnoses nationally (766 [60%] of 1277 vs 72 660 [50%] of 146 369; p<0·0001). 226 (18%) of 1277 participants were known to be living with HIV, compared with 16 171 (11%) of 146 369 people infected with gonorrhoea nationally (p<0·0001). Few epidemiological data were missing (<10% for most variables; appendix 1 p 2), although data on number of partners and travel-associated partnerships were not available from one clinic in London.
We identified 213 clusters comprising 630 (49%) of 1277 sequenced isolates, including 414 (54%) of 766 isolates from MSM, 91 (44%) of 206 isolates from women, and 125 (41%) of 304 isolates from heterosexual men. Most clusters contained two isolates (135 [63%] of 213; range of 2–21 isolates; appendix 1 p 7). Results from the sensitivity analysis showed minimal effect of various SNP thresholds on cluster size and description (appendix 1 pp 3, 4).
Among the 213 clusters, 108 (51%) only included isolates from MSM and 49 (23%) only included isolates from women and heterosexual men (table 1). A small percentage of clusters included only women, only heterosexual men, or isolates from all three sexual risk groups (table 1). 30 (14%) clusters comprised isolates from heterosexual men and MSM but no women and three (1%) comprised only women and MSM (table 1). The 30 clusters of heterosexual men and MSM included the two largest clusters (comprising 21 and 11 isolates), which contained isolates from multiple years (appendix 1 p 5).
Table 1.
Description of 213 clusters according to sexual risk group of isolates
| Number of clusters (n=213) |
Number of isolates in cluster |
|||
|---|---|---|---|---|
| Women | Heterosexual men | MSM | ||
| Only women | 9 (4%) | 20 | 0 | 0 |
| Only heterosexual men | 9 (4%) | 0 | 18 | 0 |
| Only MSM | 108 (51%) | 0 | 0 | 323 |
| Only women and heterosexual men | 49 (23%) | 60 | 63 | 0 |
| Only women and MSM | 3 (1%) | 3 | 0 | 3 |
| Only heterosexual men and MSM | 30 (14%) | 0 | 36 | 80 |
| Women, heterosexual men, and MSM | 5 (2%) | 8 | 8 | 8 |
Data are n or n (%). Heterosexual men were men who reported sexual activity exclusively with women. MSM=men who reported sex with men.
Of the 125 heterosexual men who clustered, 63 (50%) clustered with only women, 36 (29%) with only MSM, 18 (14%) with only heterosexual men, and eight (6%) with MSM and women (table 1). There were no significant differences between the heterosexual men who clustered with only women and those who clustered with only MSM (appendix 1 p 6).
Of the 414 MSM who clustered, 323 (78%) clustered with only MSM, 80 (19%) with only heterosexual men, eight (2%) with women and heterosexual men, and three (1%) with only women (table 1). MSM who clustered with heterosexual men were less likely to be from a clinic in London and less likely to be born outside the UK than were those from MSM-only clusters (table 2). The association with country of birth did not remain after adjusting for geographical location (outside of London vs London).
Table 2.
Epidemiological characteristics of MSM who clustered with heterosexual men and other MSM versus those who clustered with MSM only
| Total isolates (n=403) | Clustered with isolates from heterosexual men and MSM (n=80) | Clustered only with isolates from MSM (n=323) | OR (95% CI)* | p value | |
|---|---|---|---|---|---|
| Year† | |||||
| 2013 | 113 | 24 (30%) | 89 (28%) | 1·00 | .. |
| 2014 | 116 | 27 (34%) | 89 (28%) | 1·12 (0·60–2·10) | 0·71 |
| 2015 | 121 | 22 (28%) | 99 (31%) | 0·82 (0·43–1·57) | 0·56 |
| 2016 | 53 | 7 (9%) | 46 (14%) | 0·56 (0·22–1·42) | 0·22 |
| Clinic site | |||||
| Outside of London | 155 | 53 (66%) | 102 (32%) | 1·00 | .. |
| London | 248 | 27 (34%) | 221 (68%) | 0·23 (0·14–0·40)† | <0·0001 |
| Age (years)‡ | |||||
| ≤24 | 80 | 13 (16%) | 67 (21%) | 1·00 | .. |
| 25–34 | 176 | 36 (45%) | 140 (43%) | 1·32 (0·66–2·67) | 0·43 |
| ≥35 | 146 | 31 (39%) | 116 (36%) | 1·38 (0·67–2·82) | 0·38 |
| Ethnicity | |||||
| White | 279 | 69 (87%) | 240 (76%) | 1·00 | .. |
| Black Caribbean | 18 | 5 (6%) | 13 (4%) | 1·34 (0·46–3·89) | 0·60 |
| Black African | 8 | 0 | 8 (3%) | .. | .. |
| Black other | 2 | 0 | 2 (<1%) | .. | .. |
| Asian | 15 | 1 (1%) | 14 (4%) | 0·25 (0·03–1·94) | 0·15 |
| Other | 12 | 0 | 12 (4%) | .. | .. |
| Mixed | 32 | 4 (5%) | 28 (9%) | 0·50 (0·17–1·47) | 0·20 |
| Country of birth | |||||
| UK | 242 | 62 (78%) | 180 (56%) | 1·00 | .. |
| Not UK | 133 | 17 (21%) | 116 (36%) | 0·42 (0·23–0·77)§ | 0·0035 |
| Symptomatic infection¶ | |||||
| No | 115 | 18 (26%) | 97 (35%) | 1·00 | .. |
| Yes | 235 | 52 (74%) | 183 (65%) | 1·53 (0·85–2·77) | 0·16 |
| Diagnosed with an STI (excluding HIV) in the previous year | |||||
| No or unknown | 268 | 60 (75%) | 208 (65%) | 1·00 | .. |
| Yes | 134 | 20 (25%) | 114 (35%) | 0·61 (0·35–1·06) | 0·08 |
| HIV status | |||||
| Negative or unknown | 281 | 59 (74%) | 222 (69%) | 1·00 | .. |
| Positive | 122 | 21 (26%) | 101 (31%) | 0·78 (0·45–1·36) | 0·38 |
| Number of sexual partners in the 3 months before diagnosis‡ | |||||
| None | 6 | 3 (5%) | 3 (2%) | 1·00 | .. |
| One | 63 | 21 (34%) | 42 (27%) | 0·50 (0·09–2·75) | 0·42 |
| Two or more | 146 | 37 (61%) | 109 (71%) | 0·34 (0·06–1·78) | 0·18 |
| Travel-associated sexual partnership in the 3 months before diagnosis | |||||
| No | 195 | 54 (89%) | 141 (92%) | 1·00 | .. |
| Yes | 20 | 7 (12%) | 13 (8%) | 1·41 (0·53–3·72) | 0·49 |
Data are n or n (%), unless otherwise indicated. Heterosexual men were men who reported sexual activity exclusively with women. MSM=men who reported sex with men. OR=odds ratio. STI=sexually transmitted infection.
Unadjusted ORs from univariate analyses, indicating the epidemiological characteristics of isolates from MSM who are more likely to cluster with isolates from heterosexual men and MSM than only isolates from MSM.
Adjusted OR 0·29 (95% CI 0·17–0·52), p<0.0001 (adjusted for country of birth).
Results for tests for trend were p=0·19 for year, p=0·42 for age, and p=0·10 for sexual partners.
The association that was found for country of birth did not remain when adjusted for geographical location in the multivariable model (outside of London vs in London; adjusted OR 0·73 [95% CI 0·38–1·41], p=0·36).
Symptomatic infection was defined in the Gonococcal Resistance to Antimicrobials Surveillance Programme as discharge at any site or dysuria.
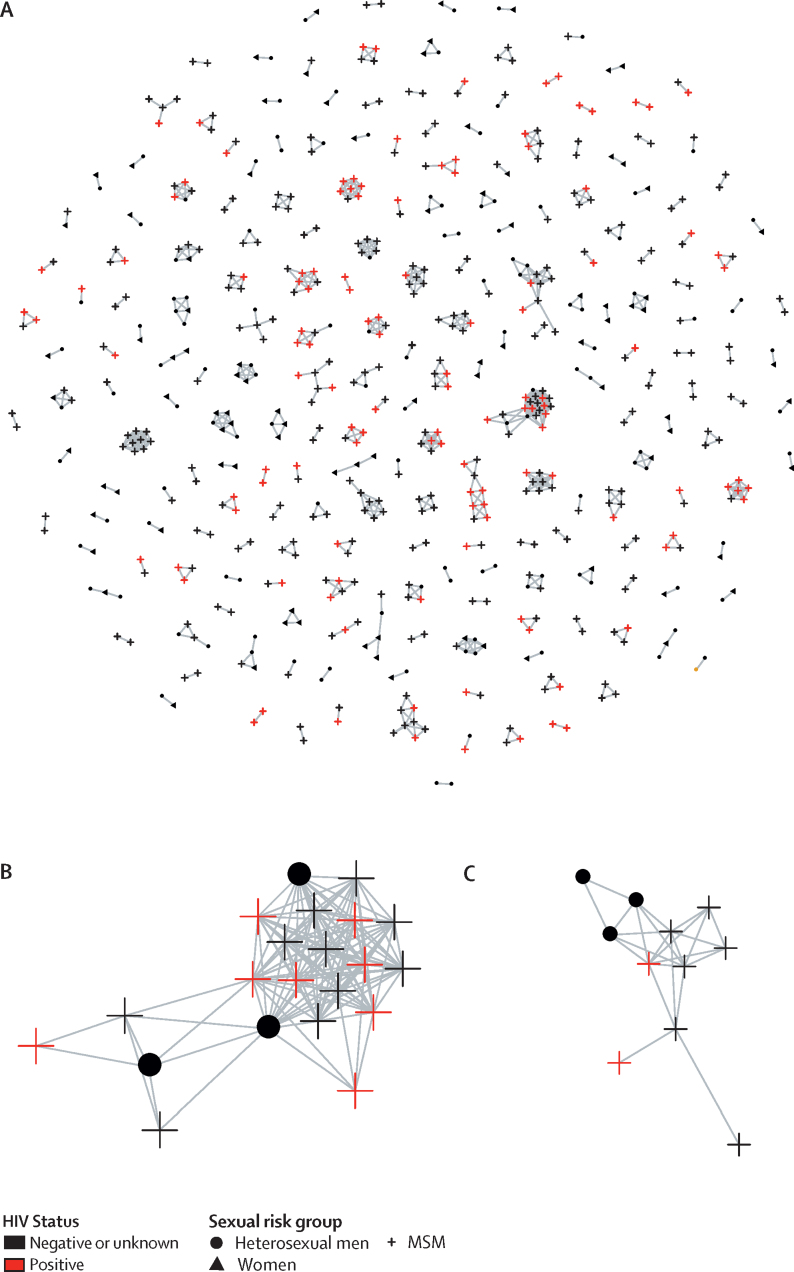
146 (69%) of 213 clusters comprised only isolates from people with HIV-negative or HIV-unknown status and seven (3%) comprised only isolates from people with HIV (appendix 1 p 4). Each of the seven HIV-positive clusters contained only two isolates, all from MSM. 60 (28%) clusters comprised both people with HIV and people without HIV or with unknown HIV status. Of these 60 clusters, 51 (85%) only contained isolates from MSM, eight (13%) included isolates from heterosexual men and MSM, and one (2%) contained only two isolates, both from heterosexual men (figure; appendix 1 p 4). Within the eight clusters containing MSM and heterosexual men, which included the two largest clusters (appendix 1 p 5), 21 (26%) of 80 MSM were HIV-positive and all 36 heterosexual men were HIV-negative or had unknown HIV status.
Figure.
Clusters of genetically similar Neisseria gonorrhoeae isolates, by sexual risk group and HIV status
Genetic similarity between isolates was defined as five or fewer single nucleotide polymorphisms. Image was created with the visualisation tool MicrobeTrace. (A) All clusters (213 in total, including 630 isolates). (B) Largest cluster, comprising 21 isolates. (C) Second largest cluster, comprising 11 isolates. MSM=men who reported sex with men.
Of the 292 MSM with HIV-negative or HIV-unknown status who clustered with other MSM, 124 (42%) clustered with at least one HIV-positive MSM. MSM with negative or unknown HIV status who were diagnosed in a London clinic were more likely to be clustered with HIV-positive MSM than were those who were diagnosed outside of London (table 3). Clustering of MSM with negative or unknown HIV status with HIV-positive MSM was less likely to occur in 2016 than in 2013 (table 3).
Table 3.
Epidemiological characteristics of MSM with negative or unknown HIV status who clustered with MSM with negative or unknown HIV status versus those who clustered with HIV-positive MSM
| Total isolates (n=222) | Clustered with isolates from MSM with negative or unknown HIV status only (n=120) | Clustered with isolates from HIV-positive MSM (n=102) | Adjusted odds ratio (95% CI)* | p value† | |
|---|---|---|---|---|---|
| Year‡ | |||||
| 2013 | 56 | 29 (52%) | 27 (48%) | 1 | .. |
| 2014 | 60 | 26 (43%) | 34 (57%) | 1·37 (0·65–2·91) | 0·41 |
| 2015 | 68 | 36 (53%) | 32 (47%) | 0·94 (0·46–1·95) | 0·87 |
| 2016 | 38 | 29 (77%) | 9 (24%) | 0·33 (0·13–0·85) | 0·021 |
| Clinic location | |||||
| Outside of London | 86 | 59 (69%) | 27 (31%) | 1 | .. |
| London | 136 | 61 (45%) | 75 (55%) | 2·65 (1·47–4·74) | 0·0007 |
| Age, years‡ | |||||
| ≤24 | 55 | 37 (67%) | 18 (33%) | .. | .. |
| 25–34 | 104 | 52 (50%) | 52 (50%) | .. | .. |
| ≥35 | 63 | 31 (49%) | 32 (51%) | .. | .. |
| Ethnicity | |||||
| White | 168 | 92 (55%) | 76 (45%) | .. | .. |
| Black Caribbean | 4 | 1 (25%) | 3 (75%) | .. | .. |
| Black African | 5 | 1 (20%) | 4 (80%) | .. | .. |
| Black other | 1 | 1 (100%) | 0 | .. | .. |
| Asian | 10 | 3 (30%) | 7 (70%) | .. | .. |
| Other | 8 | 5 (63%) | 3 (38%) | .. | .. |
| Mixed | 21 | 14 (67%) | 7 (33%) | .. | .. |
| Country of birth | |||||
| UK | 136 | 77 (57%) | 59 (43%) | .. | .. |
| Not UK | 76 | 38 (50%) | 38 (50%) | .. | .. |
| Symptomatic infection§ | |||||
| No | 67 | 37 (55%) | 30 (45%) | .. | .. |
| Yes | 126 | 65 (52%) | 61 (48%) | .. | .. |
| Diagnosed with an STI (excluding HIV) in the previous year | |||||
| No or unknown | 161 | 90 (56%) | 71 (44%) | .. | .. |
| Yes | 61 | 30 (49%) | 31 (51%) | .. | .. |
| Number of sexual partners in the 3 months before diagnosis‡ | |||||
| None | 3 | 1 (33%) | 2 (67%) | .. | .. |
| One | 32 | 19 (59%) | 13 (41%) | .. | .. |
| Two or more | 89 | 51 (57%) | 38 (43%) | .. | .. |
| Travel-associated sexual partnership in the 3 months before diagnosis | |||||
| No | 113 | 63 (56%) | 50 (44%) | .. | .. |
| Yes | 11 | 8 (73%) | 3 (27%) | .. | .. |
Data are n or n (%), unless otherwise indicated. MSM=men who reported sex with men. STI=sexually transmitted infection.
Adjusted odds ratios from multivariable analyses are shown, indicating the epidemiological chracteristics of MSM with negative or unknown HIV status that cluster with HIV-positive MSM; results are adjusted for the association with clinic location and year of diagnosis.
The p-value for the likelihood ratio test for the overall association between the exposure (all categories) and the outcome was 0·0008.
Results for tests for trend were p=0·025 for year, p=0·057 for age, and p=0·79 for sexual partners.
Symptomatic infection was defined in the Gonococcal Resistance to Antimicrobials Surveillance Programme as discharge at any site or dysuria.
In the longitudinal patient records held in GUMCAD, we observed that 33 (3%) of 1051 patients who were HIV-negative or had unknown HIV status at the time of gonorrhoea diagnosis were subsequently diagnosed with HIV. All were MSM and 25 (75%) were diagnosed within 1 year of their gonorrhoea diagnosis. Patient follow-up varied between 3 months and 4 years because data were censored at the end of 2016. Among those diagnosed with HIV after gonorrhoea diagnosis, 17 (52%) clustered with others in the study and eight (24%) were in clusters containing isolates from both HIV-positive people and people with negative or unknown HIV status.
Discussion
This large molecular epidemiology study showed how sequencing data can be used to gain insights into sexual transmission networks transmitting STIs. Within a sample from England, we observed men who reported exclusively heterosexual behaviours who were part of sexual networks with MSM and many sexual networks comprising both HIV-positive MSM and MSM with negative or unknown HIV status. In both cases, the molecular data provided additional risk-profiling information indicating high risk of HIV and other blood-borne viruses, which might otherwise not be indicated from the patient's clinical history. This information has implications for sexual health-care, including offering testing and prevention advice and management to people perceived to be at increased risk. Our findings complement existing evidence on sexual mixing and show the diversity and complexity of N gonorrhoeae sexual networks. Few WGS studies of N gonorrhoeae have benefited from clinical and behavioural data.21 To our knowledge, our study of N gonorrhoeae is the largest and most representative in England to date and one of the largest internationally. We sampled isolates from a geographically dispersed subset of clinics in England and although we found some significant differences between the characteristics of the study isolates and those of all gonorrhoea diagnoses in England, the absolute differences were small. Therefore, our study probably provides reliable insights into the molecular epidemiology of N gonorrhoeae in England.
The observed differences in patient characteristics between the study and national cases and the small sizes of clusters probably reflect a bias in the patient groups and isolates that were successfully cultured and the GRASP sampling frame.28 Just under 50% of isolates from gonorrhoea diagnoses in England are successfully cultured, and culture is more likely to be successful from genital and symptomatic infections than from extragenital and asymptomatic infections and less likely to be successful from women and MSM than from heterosexual men.24 The poor availability of cultures, short period of data collection in GRASP (3 months per year), and inclusion of a small number of clinics also explain why most clusters identified in this study were small and 647 cultures did not cluster at all. As with all WGS studies of N gonorrhoeae, an entirely representative sample of the gonococcal population cannot be achieved given many infections are never diagnosed. Therefore, we were unable to fully explore the patterns of sexual mixing for underrepresented groups, especially women. Additionally, data on sexual behaviour, other than sexual risk group, were sparse; more detailed information collected in larger studies might improve the power to detect significant differences between people clustered in distinct groups.
The low proportion of re-infection in our study—only 21 (2%) of 1256 people included in our study had more than one gonorrhoea diagnosis—was probably due to the restricted time period of GRASP, which means that reinfections occurring 4–9 months after the initial infection would not be captured in our study. Anyone attending a different sexual health clinic would also not be identified as a reinfection in our study because it is not possible to trace patients between clinics in GUMCAD
Although we could not distinguish between direct and indirect sexual transmission or establish the direction of transmission, the tight SNP threshold that was used to define clusters meant that people within the same cluster were likely to belong to the same recent transmission network. However, we could not confirm that our chosen SNP threshold truly identified closely linked cases within a sexual network as partner notification data were not available. As with other N gonorrhoeae studies, the high frequency of DNA recombination requires computational strategies to identify SNP differences arising through mutation rather than recombination. We identified and removed recombinant DNA, but some probably remained, which might have led to incorrect inferences about relatedness for some isolates.
We found that mixed clusters of men reporting heterosexual behaviour and MSM were considerably more common than those of MSM and women, which might provide evidence of sexual mixing between gay-identifying and heterosexual-identifying MSM. We also identified clusters containing only heterosexual men, which might represent a group of heterosexual-identifying men having sex with other heterosexual-identifying men. However, it is possible that heterosexual men and MSM were not directly linked but shared female partners who were underrepresented in this study because of difficulty in collecting cultures from women,24 which might also explain the small size of most clusters. We found no differences in epidemiological characteristics between heterosexual men clustering only with MSM and those clustering only with women. Another study has found that men in sexual networks with MSM who report only heterosexual behaviours are more likely to be of black African ethnicity.11 A British population-based survey found that of 190 men reporting a same-sex partner, 54 (28%) identified as heterosexual.29 Where differences exist between identity and same-sex behaviour in men, public health and clinical practice interventions and messaging that target only MSM who identify as gay or bisexual might miss and even disenfranchise heterosexual-identifying MSM.
We found some evidence of serosorting among MSM, as some N gonorrhoeae clusters only comprised people of the same HIV status. As seen in a previous study,21 a larger number of clusters comprised both MSM with HIV and those with negative or unknown HIV status, which is perhaps unsurprising given the inherent limitations of serosorting as an HIV risk-reduction strategy (ie, undiagnosed infections and misreporting of status).6, 7 Even in small clusters of people with mixed HIV status, we were able to show subsequent HIV diagnoses among some MSM with HIV-negative or HIV-unknown status at gonorrhoea diagnosis.
Most epidemiological characteristics of MSM who were HIV-negative or HIV-unknown who clustered with HIV-positive MSM could not be distinguished from those who clustered only with other HIV-negative or HIV-unknown MSM. Increased availability and use of antiretroviral therapies, including treatment as prevention in HIV-positive and PrEP in HIV-negative MSM, might explain the substantial number of clusters of people with mixed HIV status. Sexual behaviours, specifically condom use, have probably been affected by the increase in HIV-positive MSM with an undetectable viral load and in PrEP use in HIV-negative MSM, which might lead to N gonorrhoeae but not HIV transmission. With reduced HIV transmission risk, HIV sero-adaptive behaviours might have declined or become more complex.
National guidelines for sexual health services vary for different populations according to their sexual and other risk behaviours. For example, MSM are routinely offered extra-genital testing for chlamydia and gonorrhoea and testing and vaccination for hepatitis A and B, whereas most heterosexual men are not offered such services.30 To choose the appropriate management strategy, clinicians often use self-reported behavioural data from patients to assess their risk. However, patients might be reluctant to report some sexual behaviours because of concerns about confidentiality and stigma, or they might not know that they belong to a sexual network that increases their risk of HIV and other blood-borne viruses.
Our findings suggest that sequencing data on N gonorrhoeae could, when combined with epidemiological data, be used to improve understanding of a person's sexual network and HIV and blood-borne virus risk. Although the absolute risk of infection transmission could not be quantified in this study, if available in a clinically relevant timeframe, WGS data could provide insight into sexual risk profiles and support patient management, including extra-genital testing for chlamydia and gonorrhoea, HIV PrEP, testing and vaccination for hepatitis A and B, and behavioural risk-reduction advice.
Implementation of WGS to support patient and partner risk management raises ethical issues about communication and interpretation of genomic data in an appropriately sensitive manner that takes account of patient confidentiality and safeguards to avoid deductive disclosure about transmission. Information derived from WGS data should be communicated to the patient without revealing specific details about the sexual network they are predicted to be a part of. Further research is needed to explore the ethical and patient communication issues around the use of WGS to inform clinical management.
Several practical and cost implications should be considered before WGS is implemented in this manner. Expansion of facilities and expertise is required to conduct WGS, analyse WGS data, and share WGS data between local and national health-care professionals. Results would need to be sufficiently prompt (<2 weeks) to enable clinical and public health decision making. Real-time WGS would require direct sequencing from urine or swabs rather than from culture to facilitate faster sample processing and better coverage.
Acknowledgments
Acknowledgments
This study was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HRPU) in Blood Borne and Sexually Transmitted Infections at University College London (Steering Committee: Caroline Sabin [Director], Anthony Nardone [PHE Lead], Catherine Mercer, Gwenda Hughes, Jackie Cassell, Greta Rait, Samreen Ijaz, Tim Rhodes, Kholoud Porter, Sema Mandal and William Rosenberg) in partnership with Public Health England (PHE), in collaboration with London School of Hygiene & Tropical Medicine. The study was also funded by the Wellcome Trust (grant number 098051). We thank the clinical and laboratory staff who contributed to the Gonococcal Resistance to Antimicrobials Surveillance Programme during the study. The views expressed are those of the authors and not necessarily those of the NIHR, the Department of Health and Social Care, or PHE.
Contributors
KT, NF, SH, and GH designed, initiated, and coordinated the study. KT, GH, MC, RP, HM, and HF contributed to data collection and management. KT analysed the data with support from LS-B and SH. All authors contributed to interpretation of the data. KT wrote the first draft of the paper and all authors read, commented on, and approved the final manuscript.
Declaration of interests
We declare no competing interests.
Supplementary Materials
References
- 1.Sonnenberg P, Clifton S, Beddows S. Prevalence, risk factors, and uptake of interventions for sexually transmitted infections in Britain: findings from the National Surveys of Sexual Attitudes and Lifestyles (Natsal) Lancet. 2013;382:1795–1806. doi: 10.1016/S0140-6736(13)61947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Public Health England Sexually transmitted infections and screening for chlamydia in England: 2018. June 7, 2019. https://www.gov.uk/government/statistics/sexually-transmitted-infections-stis-annual-data-tables (accessed Jan 7, 2020).
- 3.Ghani AC, Swinton J, Garnett GP. The role of sexual partnership networks in the epidemiology of gonorrhea. Sex Transm Dis. 1997;24:45–56. doi: 10.1097/00007435-199701000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Mercer CH, Prah P, Field N. The health and well-being of men who have sex with men (MSM) in Britain: evidence from the third National Survey of Sexual Attitudes and Lifestyles (Natsal-3) BMC Public Health. 2016;16:525. doi: 10.1186/s12889-016-3149-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malek R, Mitchell H, Furegato M. Contribution of transmission in HIV-positive men who have sex with men to evolving epidemics of sexually transmitted infections in England: an analysis using multiple data sources, 2009–2013. Euro Surveill. 2015;20:21093. doi: 10.2807/1560-7917.es2015.20.15.21093. [DOI] [PubMed] [Google Scholar]
- 6.Aghaizu A, Wayal S, Nardone A. Sexual behaviours, HIV testing, and the proportion of men at risk of transmitting and acquiring HIV in London, UK, 2000–13: a serial cross-sectional study. Lancet HIV. 2016;3:e431–e440. doi: 10.1016/S2352-3018(16)30037-6. [DOI] [PubMed] [Google Scholar]
- 7.Daskalopoulou M, Rodger AJ, Phillips AN. Condomless sex in HIV-diagnosed men who have sex with men in the UK: prevalence, correlates, and implications for HIV transmission. Sex Transm Infect. 2017;0:1–9. doi: 10.1136/sextrans-2016-053029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward H, Ronn M. Contribution of sexually transmitted infections to the sexual transmission of HIV. Curr Opin HIV AIDS. 2010;5:305–310. doi: 10.1097/COH.0b013e32833a8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fichtenberg CM, Muth SQ, Brown B, Padian NS, Glass TA, Ellen JM. Sexual network position and risk of sexually transmitted infections. Sex Transm Infect. 2009;85:493–498. doi: 10.1136/sti.2009.036681. [DOI] [PubMed] [Google Scholar]
- 10.Smolarchuck C, Wensley A, Padfield S, Fifer H, Lee A, Hughes G. Persistence of an outbreak of gonorrhoea with high-level resistance to azithromycin in England, November 2014—May 2018. Euro Surveill. 2018;23:1800287. doi: 10.2807/1560-7917.ES.2018.23.23.1800287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragonnet-Cronin M, Hue S, Hodcroft EB. Non-disclosed men who have sex with men in UK HIV transmission networks: phylogenetic analysis of surveillance data. Lancet HIV. 2018;5:e309–e316. doi: 10.1016/S2352-3018(18)30062-6. [DOI] [PubMed] [Google Scholar]
- 12.Shankar AG, Mandal S, Ijaz S. An outbreak of hepatitis B in men who have sex with men but identify as heterosexual. Sex Transm Infect. 2016;92:227. doi: 10.1136/sextrans-2015-052490. [DOI] [PubMed] [Google Scholar]
- 13.Grad YH, Kirkcaldy RD, Trees D. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis. 2014;14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Silva D, Peters J, Cole K. Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis. 2016;16:1295–1303. doi: 10.1016/S1473-3099(16)30157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papp JR, Abrams AJ, Nash E. Azithromycin resistance and decreased ceftriaxone susceptibility in Neisseria gonorrhoeae, Hawaii, USA. Emerg Infect Dis. 2017;23:830–832. doi: 10.3201/eid2305.170088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckley C, Forde BM, Trembizki E, Lahra MM, Beatson SA, Whiley DM. Use of whole genome sequencing to investigate an increase in Neisseria gonorrhoeae infection among women in urban areas of Australia. Sci Rep. 2018;8:1503. doi: 10.1038/s41598-018-20015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fifer H, Cole M, Hughes G. Sustained transmission of high-level azithromycin-resistant Neisseria gonorrhoeae in England: an observational study. Lancet Infect Dis. 2018;18:573–581. doi: 10.1016/S1473-3099(18)30122-1. [DOI] [PubMed] [Google Scholar]
- 18.Harris SR, Cole MJ, Spiteri G. Public health surveillance of multidrug-resistant clones of Neisseria gonorrhoeae in Europe: a genomic survey. Lancet Infect Dis. 2018;18:758–768. doi: 10.1016/S1473-3099(18)30225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong JC, Chow EPF, Stevens K. Whole-genome sequencing reveals transmission of gonococcal antibiotic resistance among men who have sex with men: an observational study. Sex Transm Infect. 2018;94:151–157. doi: 10.1136/sextrans-2017-053287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RS, Seemann T, Heffernan H. Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J Antimicrob Chemother. 2018;73:353–364. doi: 10.1093/jac/dkx405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters J, Cresswell F, Amor L. Whole genome sequencing of Neisseria gonorrhoeae reveals transmission clusters involving patients of mixed HIV serostatus. Sex Transm Infect. 2018;94:138–143. doi: 10.1136/sextrans-2017-053198. [DOI] [PubMed] [Google Scholar]
- 22.Town K, Bolt H, Croxford S. Neisseria gonorrhoeae molecular typing for understanding sexual networks and antimicrobial resistance transmission: a systematic review. J Infect. 2018;76:507–514. doi: 10.1016/j.jinf.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Public Health England Gonococcal resistance to antimicrobials surveillance programme (GRASP): protocol. 2014. https://www.gov.uk/government/publications/gonococcal-resistance-to-antimicrobials-surveillance-programme-grasp-protocol/gonococcal-resistance-to-antimicrobials-surveillance-programme-grasp-protocol (accessed Nov 20, 2018).
- 24.Mohammed H, Ison CA, Obi C. Frequency and correlates of culture-positive infection with Neisseria gonorrhoeae in England: a review of sentinel surveillance data. Sex Transm Infect. 2015;91:287–293. doi: 10.1136/sextrans-2014-051756. [DOI] [PubMed] [Google Scholar]
- 25.Savage EJ, Mohammed H, Leong G, Duffell S, Hughes G. Improving surveillance of sexually transmitted infections using mandatory electronic clinical reporting: the genitourinary medicine clinic activity dataset, England, 2009–2013. Euro Surveill. 2014;19:20981. doi: 10.2807/1560-7917.es2014.19.48.20981. [DOI] [PubMed] [Google Scholar]
- 26.Croucher NJ, Page AJ, Connor TR. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 28.Hughes G, Nichols T, Ison CA. Estimating the prevalence of gonococcal resistance to antimicrobials in England and Wales. Sex Transm Infect. 2011;87:526–531. doi: 10.1136/sextrans-2011-050071. [DOI] [PubMed] [Google Scholar]
- 29.Geary RS, Tanton C, Erens B. Sexual identity, attraction and behaviour in Britain: the implications of using different dimensions of sexual orientation to estimate the size of sexual minority populations and inform public health interventions. PLoS One. 2018;13:e0189607. doi: 10.1371/journal.pone.0189607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clutterbuck D, Asboe D, Barber T. 2016 United Kingdom national guideline on the sexual health care of men who have sex with men. 2018. https://www.bashhguidelines.org/media/1162/msm-2016.pdf (accessed Nov 20, 2018). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.