Abstract
In the past 20 years, there have been several approaches to achieve cardioprotection or cardiac regeneration using a vast variety of cell therapies and remote ischemic pre-conditioning (RIPC). To date, substantial proof that either cell therapy or RIPC has the potential for clinically relevant cardiac repair or regeneration of cardiac tissue is still pending. Preclinical trials indicate that the secretome of cells in situ (during RIPC) as well as of transplanted cells may exhibit cardioprotective properties in the acute setting of cardiac injury. The secretome generally consists of cell-specific cytokines and extracellular vesicles (EVs) containing microRNAs (miRNAs). It is currently hypothesized that a subset of known miRNAs play a crucial part in the facilitation of cardioprotective effects. miRNAs are small non-coding RNA molecules that inhibit post-transcriptional translation of messenger RNAs (mRNAs) and play an important role in gene translation regulation. It is also known that one miRNAs usually targets multiple mRNAs. This makes predictability of pharmacokinetics and mechanism of action very difficult and could in part explain the inferior performance of various progenitor cells in clinical studies. Identification of miRNAs involved in cardioprotection and remodeling, the composition of miRNA profiles, and the exact mechanism of action are important to the design of future cell-based but also cell-free cardioprotective therapeutics. This review will give a description of miRNA with cardioprotective properties and a current overview on known mechanism of action and potential missing links. Additionally, we will give an outlook on the potential for clinical translation of miRNAs in the setting of myocardial infarction and heart failure.
Keywords: microRNA, extracellular vesicles, second generation cell therapies, translation, cardioprotection, secretome
Introduction
Cardiovascular disease remains one of the most important challenges clinicians face today. Despite huge efforts in prevention and undeniable progress in acute and short-term survival, disease progression over long-term still burdens our health care system without a real curative approach. The reason: regenerative capacity of an adult human heart is quite limited due to the low turnover rate of cardiomyocytes and lack of a sufficient pool of tissue resident progenitors (Tzahor and Poss, 2017). This renders the human heart very susceptible to any form of acute or chronic injury with a critical loss of cardiomyocytes or their function, ultimately leading to the clinical manifestation of heart failure. For these facts, cardiac medicine has been identified as a promising field for application of regeneration technologies. In the past 20 years, cell-based therapies have aimed to either induce ‘de novo’ generation or stimulate tissue dormant progenitors to differentiate into mature cardiomyocytes (Behfar et al., 2014). The promising results from in vitro studies and pre-clinical trials have led to a large number of clinical trials that for the most part investigated the therapeutic effect of different bone marrow, adipose- or neonatal tissue derived progenitor cells (first generation cell therapy). With limited pre-clinical data available in hindsight, a rush into first clinical trials in the early and mid 2000s has largely failed to demonstrate any meaningful results for patients with cardiac disease. More recently, myocardium resident cardiac progenitor cells, iPSCs, and conditioned progenitor cells have also been investigated in that context (second generation cell therapy) (Cambria et al., 2017). However, they as well failed to reach respective primary endpoints in most clinical trials; that is to say reduction in scar size and stabilization or improvement of cardiac function (Moyé, 2014; Gyöngyösi et al., 2015). As a result, the idea of a regenerating heart has been abandoned by many scientists. Luckily, the failure of such clinical studies has also led to post hoc analyses of the mechanism of action by which various cell types exhibit cardioprotection in the injured heart. Today, there is consensus that paracrine mediators, released by transplanted cells upon injury signals, mediate protection and limit adverse myocardial remodeling (Madonna et al., 2019). There is evidence emerging, that mediators can limit the extend of cardiomyocyte loss during acute injury and positively impact adverse myocardial remodeling in the chronic setting. Within the past 10 years, microRNAs (miRNAs) have come into focus as the next generation “cell” therapy studies have demonstrated that the paracrine secretion of nano- and macrovesicles containing miRNAs are mainly responsible for the cardioprotective effect of cellular therapies. Preclinical trials were able to demonstrate that miRNAs or extracellular vesicles (EVs) containing miRNAs were capable to reproduce the cellular effects of cardioprotection (Behfar and Terzic, 2019; Madonna et al., 2019; Maring et al., 2019). Within the scope of this review, we intend to provide a general overview of the potential role of miRNAs in cardioprotection and elaborate on potential use of miRNAs as a therapeutic agent.
Micro RNAs – One Shoe Fits All?
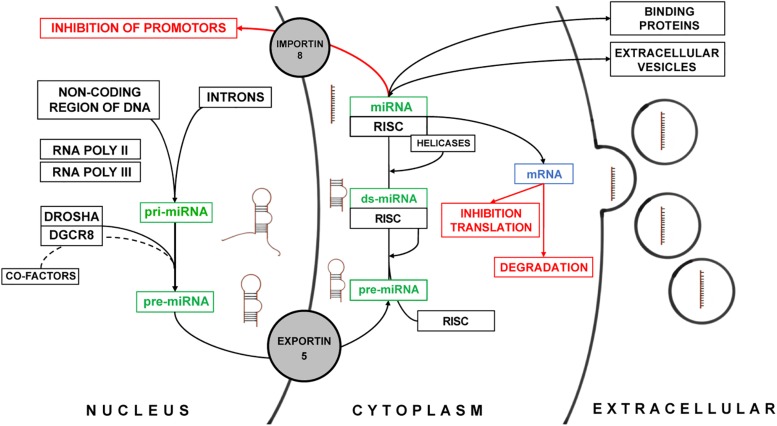
MicroRNAs play an important role in the inhibition of messenger RNA (mRNA) translation in the cytoplasm but have been also identified to regulate transcription in the nuclear compartment of mammalian cells (Bartel, 2004). The biosynthesis was long believed to be linear and universal for all miRNAs. However, recent functional studies demonstrated, that a multitude of alternate miRNA-specific biosynthesis pathways exist and that they require a plethora of regulatory mechanisms – many of which still need to be identified (Lee et al., 1993; Bartel, 2004) (Figure 1). miRNA are encoded within the entire genome and are usually arranged in clusters (Rodriguez et al., 2004). Most miRNA genes are located in the non-coding areas of the genome. In some cases, miRNAs are located within the introns of protein coding genes such as miR-103 which is located within in the intron of pantothenate kinase 1, 2, and 3 together with miR-107 (Rodriguez et al., 2004; Lin et al., 2006). miRNAs are transcribed by RNA Polymerases II and III into a primary miRNA (pri-miRNA). The pri-miRNA consists of a terminal loop region, a stem and two single-stranded flanking RNA regions up- and downstream of the hairpin (Cai et al., 2004; Lee et al., 2004). For the canonical pathway of miRNA maturation, the terminal single stranded RNA region is then spliced by the complex of Drosha and DGCR8 protein (DiGeorge critical region 8) (Lee et al., 2003). The DGCR8 protein has a binding and proof-reading domain that ensures the correct binding and identification of the cleavage site for the Drosha protein, a RNase III enzyme. For some miRNAs additional factors may be required for the correct splicing of the hairpin precursor (Guil and Cáceres, 2007; Davis et al., 2008; Michlewski et al., 2008) before the precursor miRNA (pre-miRNA) is transported from the nucleus into the cytoplasm by Exportin-5 (Yi et al., 2003). Exportin-5 not only acts as a transporter but also as an additional “proof-reader” only transporting correctly processed pre-miRNA (Zeng and Cullen, 2004). In the cytoplasm, the pre-miRNA – still consisting of the terminal loop and the double stranded stem – binds to the RNA-induced silencing complex (RISC) (Gregory et al., 2005). This protein complex contains RNase Dicer, double stranded RNA binding domain proteins (Tar RNA binding protein), PACT (protein activator of PKR), and Argonaute-2 (MacRae et al., 2008). The latter is the key component that mediates the miRNA inhibition of transcription of mRNAs (Diederichs and Haber, 2007). The terminal loop of pre-miRNAs is spliced off, whereas for some of the miRNAs both the guide and passenger strand can serve as individual mature miRNAs (Matranga et al., 2005). In general, the passenger strand is degraded after unwinding of the double stranded pre-miRNA by helicases (Leuschner et al., 2006). Similar to the processing of miRNAs in the nucleus some miRNAs are dependent on additional co-factors for maturation (Hutvágner et al., 2001; Diederichs et al., 2008). This highlights again, that miRNA biosynthesis is subject to an integrate regulatory machinery which we only just begin to understand. Upon maturation, miRNAs as part of the RISC complex either inhibit the translation of their target mRNA or they are packaged into EVs contained in multivesicular bodies to be released in the extracellular space (Turchinovich et al., 2012). The encapsulation of miRNAs does not happen at random and is also a highly regulated process (Turchinovich et al., 2012; Villarroya-Beltri et al., 2013). This allows for the dynamic response of cells in regard to which miRNAs are released upon microenvironmental cues. A study by Villarroya-Beltri et al. (2013) has shown that specific motifs within the sequence of miRNA determine their localization in either the cytoplasm or EVs. They have also identified heterogeneous nuclear ribonucleoproteins such as hnRNPA2B1 and hnRNPA1 that specifically bind to these motifs and are also present in EVs. Independent of their origin, all miRNAs have in common to alter gene expression via inhibitory mechanisms of posttranscriptional modification of mRNAs. The mechanism of inhibition depends on the complementarity of the miRNA sequence to its target mRNA. miRNAs with high complementarity direct the RISC to the mRNA and initiate the degradation. Lower complementarity can lead to inhibition of ribosomal translation of the target mRNA. It is overall hypothesized that the specificity of miRNAs is determined by the quality and stability of base pairing to their respective targets. Furthermore, miRNAs have also been identified to perform transcriptional silencing by targeting promotor regions within the heterochromatin. The aforementioned regulation of biosynthesis, maturation, and biological effects of numerous miRNA can be influenced by changes in the cellular microenvironment in a non-linear manner – posing a major challenge for scientists to predict possible effects of miRNA in vivo (Lee et al., 1993; Liu B. et al., 2014).
FIGURE 1.
Summary of the biosynthesis and the biological effect of miRNAs. The different stages of miRNA are schematically depicted next to the green description of miRNA. The biological effect of miRNAs is marked in red.
Cardioprotective Mirnas – a Two-Sided Sword
For some time now, it has been hypothesized that factors released by cells under stress can induce a protective effect in neighboring or remote tissues (Kharbanda et al., 2002; Chen et al., 2008; Hausenloy et al., 2015). We now know that the key player in transmitting these signals are miRNAs. In the extracellular compartment, miRNAs are usually transported via EVs or binding proteins to protect them from degradation by nucleases (Li et al., 2012; Boon and Dimmeler, 2015). Similarly, the effect of therapeutic cell preparations exhibits their protective effect via the transmission of EVs loaded with miRNAs. Over the course of the past 10 years the sera of patients undergoing an ischemic event as well as the secretory profile of most cells utilized for cardioprotective purposes have been characterized (Baglio et al., 2015; Barile et al., 2016; Barile and Vassalli, 2017; Shao et al., 2017; Bellin et al., 2019). Each year, an increasing number of pathways are identified that either hint toward protection or damage of the myocardium (Barile et al., 2014, 2016; Gallet et al., 2016; Ciullo et al., 2019). Many functional studies have preceded these in-depth analyses of miRNA as cardioprotective agents, remote ischemic pre-conditioning (RIPC) being one of the most prominent examples (Kharbanda et al., 2002; Hausenloy et al., 2015). Here, a different organ or the heart itself is exposed to brief, non-fatal ischemia/reperfusion. Pre-clinical models show that RIPC can increase the survival of cardiomyocytes upon injury and positively impacts the myocardial remodeling (Konstantinov et al., 2005; Wei et al., 2011). In RIPC, circulating miRNA and EVs play an important role (Frey et al., 2018; Spannbauer et al., 2019). Even though circulating miRNAs and EVs have been identified as key mediators of that cardioprotective effect, it is a rather crude and a non-targeted therapeutic approach. Most when RIPC was investigated in clinical trials, none of the primary endpoints predicted by preclinical studies were met (Brevoord et al., 2012). Similar to experience from clinical trials investigating cell therapies, the exact mechanisms of action of RIPC were not fully understood and may explain the failed translation. However, the data collected on miRNAs from these studies laid the groundwork for many functional studies investigating the cardioprotective effects of miRNAs.
In recent years, a plethora of mechanistical studies for the downstream effect of various miRNA were conducted. Here, miRNAs were either investigated as diagnostic markers or as potential targets for therapies (Barile et al., 2016). The in-depth analysis of regulation of the identified miRNAs and their downstream targets revealed that some miRNAs had contradictory effects in the myocardium (Table 1). Furthermore, to the best of our knowledge no study performed any concurrent analysis to identify potential targets that are not related to a cardioprotective effect. In this next section, we highlight a selection of miRNAs that have been associated with a cardioprotective potential but also bear the risk of adverse or off-target effects. Most of these miRNAs were identified either in EVs of therapeutic cell products or as biomarkers during acute and chronic myocardial injury. The knowledge of their exact mechanism of action is therefore highly recommended. The following miRNAs are only a small example of miRNAs that are commonly identified but not limited to their cardioprotective potential. The selection of miRNA should not be seen as a comprehensive summary of all known cardioprotective miRNAs which would be beyond the scope of this review. Others have provided more detailed lists of cardioprotective miRNAs (Varga et al., 2015; Wendt et al., 2018). The pre-clinical experience with the following miRNAs should highlight the potential and pitfalls we as scientist may face when designing therapeutic strategies with miRNAs.
TABLE 1.
Summarizes a selection of miRNAs that have been identified with either a harmful or cardioprotective property in cardiovascular disease.
| miRNA | Disease model | Releasing cell type | Experimental approach | Experimental model | Effect | Identified targets | Recipient cell | Off-target effects | PMID |
| miR-665 | I/R | CM | Suppression of miR-665 via dexmedetomidine | Rat heart Langendorff preparation | Improved LVDP during reperfusion | AK1, Cbr2 | Cardiac cells | Not investigated | 31026731 |
| HF | – | I.m. injection with anti-sense miRNA plasmids | Rat model of HF | Improved LVEF, reduced CM apoptosis, improved Mc ultrastructure | GLP1R | Cardiac cells | Not investigated | 30666648 | |
| HF | Global | I.v. injection of rAAV miR-665 inhb. | Murine model of LV pressure overload | Improved LVEF, reduced fibrosis, improved vascularization | CD34 | Global | Not investigated | 30243022 | |
| – | Human CM | In vitro gain and loss of function in human CM | Mechanistic model | – | Cbr1 and Cbr2 | Human CM | – | 25111814 | |
| miR-132 | I/R | – | Loss of function in vivo, gain of function in vitro | Murine hind limb ischemia | Slower perfusion recovery, less collateralization, modulation of RAS-MAPK signaling | Rasa1 and Spred1 | – | Not investigated | 25016614 |
| Afib | – | In vitro loss and gain of function in CF | Mechanistic model | In human and dog with Afib decreased expr. miR-132 in atrium | CTGF | CF | – | 28731126 | |
| DCM | – | In vitro analysis of cardiac cell isolates from DCM rats, overexpression of miR-132 | DCM rat model | Activation of PI3K/Akt pathway, CM apoptosis down | PTEN | – | – | 30271437 | |
| AMI | BM-MSCex electroporated with miR-132 | I.m. injections with MSCex | Murine model of AMI | Increased LVEF, enhanced neovascularization in BZ | Rasa1 | HUVECS | Not investigated | 30216493 | |
| miR-132 + miR-126 | DMap | – | Transfection of aortic rings with miR-132, miR-126 | Endothelial sprouting in aortic rings under high glucose | Decreased EC apoptosis, improved endothelial sprouting | Spred1 | HUVECS, Ecs | – | 31179325 |
| miR-210 + miR-132 + miR-146a-3p | AMI | CPCs | I.v. injection with CPCex rich in miR-210, miR-132, miR-146a-3p vs. Fibex | Murine model of AMI | Less CM apoptosis, enhanced angiogenesis in BZ, improved LVEF | EFNA3, PTP1b | – | Not investigated | 28731126 |
| miR-126 | AMI | AT-MSCs overexpressing miR-126 | I.m. injection of AT-MSCex | Murine model of AMI | Increased neoangiogenesis | Not investigated | – | Not investigated | 29241208 |
| miR-126-5p | Endothelial injury | – | KO of EC Dicer and rescue experiment with miR-126-5p transfection | CA injury | Endothelial Dicer processes pre-mir-126 into mir-126-3p (guide strand) and the passenger strand-5p. 5p is involved in dendothelial repair and proliferation by targeting the Notch1 inhibitor Delta-like homolog 1 (Dlk1) | Dlk1 | ECs | Not investigated | 30213595 |
| mir-210 | AMI | – | Observational study | AMI in rats | Increased levels of miR-210 | – | – | – | 31596148 |
| AMI | BM-MSCs | BM-MSCs rich in miR-210 vs. BM-MSCs with miR-210 silencing | Murine model of myocardial infarction | Increase LVEF, increased neoangiogenesis | EFNA3 | – | Not investigated | 28249798 | |
| I/R | BM-EPCs | BM-EPCs gain and loss of miR-210 | Murine hind limb ischemia | With miR-210 improved perfusion recovery and collateralization | EFNA3 | ECs | Not investigated | 29908843 | |
| I/R | – | Loss and gain of function | In vitro in H9c2 cells, mechanistic model | CXCR4 | H9c2 | – | 29710553 | ||
| mir-206 | Afib | Lentiviral overexpression of miR-206 in PVFP | Canine model of Afib | Overexpression of miR-206 increased incidence of Afib | GCH1 | – | Not investigated | 29436714 | |
| Afib | – | Overexpression of miR-206 in murine hearts | Transgenic mouse model | Overexpression led to decreased lifespan and arrhythmias | Cx43 | – | Not investigated | 30322759 | |
| AMI | – | Cardiac specific expression of miR-206 | Murine model of AMI | CM hypertrophy and increases survival under AMI | FBPP1 | – | Not investigated | 26333362 | |
| HF | – | Increased expression of miR-206 via HMGB1 | Murine model of AMI | Increased collagenolytic activity, decreases myocardial fibrosis | TIMP3 | CF | Not investigated | 21731608 | |
| AMI | – | In vitro loss and gain of function, with in vivo confirmation | Murine model of AMI | Reduced CM apoptosis, improved LVEF | ATG3 | H9c2 | Not investigated | 30551524 | |
| miR-206 + miR-216b | AMI | – | Via HDC gain and loss influence expression of miR-206, miR-216b | Murine model of AMI | Targets Atg13 and reduces autophagy upon hypoxia. miR-206 is induced via histamine | ATG3 | – | Not investigated | 29880830 |
| miR-206 + miR-1 | DMap | – | In vitro loss and gain of function | In vitro in H9c2 cells, mechanistic model | Increased CM apoptosis | Hsp60 | H9c2 | – | 20655308 |
| miR-146a | DCM | CF, CM | AAV9 mediated overexpression of miR146a in vivo and in vitro | Murine model of LV pressure overload | Decreased myocardial contractility | SUMO1 | CM | Not investigated | 30355233 |
| AMI | EPCs | EPC injection in BZ | AMI in rats | Downregulation of miR-146a and reduced CM apoptosis and increased VEGF expression | – | – | Not investigated | 30344699 | |
| – | – | Lentiviral overexpression of miR-146a in H92c | In vitro in H9c2 cells, mechanistic model | Increases MMP9, may reduce fibrosis in injured heart | FOS, AP1 | CM | – | 26112171 | |
| Sepsis induced cardiac dysfunction | – | Transfection of mice with miR-146a, in vitro H9c2 and macrophages | Murine sepsis model | Attenuation of sepsis induced myocardial dysfunction | IRAK, TRAF6 | H9c2, J774 macrophages | 26048146 | ||
| DoxDCM | CM | In vitro overexpression and suppression of miR-146a | In vitro in H9c2 cells, mechanistic model | Induction of cell death upon Dox treatment | ErbB4 | H9c2 | – | 20495188 | |
| AMI | AT-MSCs overexpressing miR-146a | I.m. injections of AT-MSCex native and overexpressing miR-146a | AMI in rats | Decreased CM apoptosis, decreased inflammation, decreased fibrosis | EGR1 | H9c2 | Not investigated | 30362610 | |
| miR-146a-5p | DoxDCM | CPCs | CPCex rich in miR-146a-5p vs. Fibex | DoxDCM model in rats | Decreased CM apoptosis | Traf6, Smad4, Nox4, Mpo | – | Not investigated | 31098627 |
| miR-146a + miR-155 | – | DC | Injection of endotoxin exposed mice with Dcex rich in miR-155 and miR-146a | Murine model of endotoxin inflammation | miR-146a attenuates inflammation, miR-155 increases inflammation | – | – | Not investigated | 26084661 |
| miR-22 | AMI | – | AAV9 overexpression of miR-22 | AMI in rats | Decreased CM apoptosis, decreases infarct size | CBP | – | Not investigated | 24338162 |
| – | – | Overexpression of miR-22 in murine lungs, zebrafish and ECs in vitro | – | VEC | EC | Not investigated | 28112401 | ||
| AMI, HF | – | Gain and loss study on miR-22 | In vitro in CF, mechanistic model | Overexpression limits expression of Col1α1, Col3α1 | TGFβR | CF | – | 27997889 | |
| AMI | – | Gain and loss study on miR-22 | In vitro in rat CMs | Overexpression prevented autophagy and apoptosis in CMs | p38α | CM | – | ||
| AMI | – | I.m. injection of miR-22 inhib., loss and gain function in vitro | AMI in rats | Inhibition decreases infarct size, reduces CM apoptosis | Sirt1, PGC1α | H9c2 | Not investigated | 27174562 | |
| DCM | – | miR-22 deficient mice and gain and loss function in H9c2 | Murine model of left ventricular pressure over load | miR-22 suppression led to left ventricular dilation | PReBPb | H9c2 | Not investigated | 22570371 | |
| DCM | – | Gain and loss of miR-22 in mice | Murine model of left ventricular pressure overload | Overexpression of miR-22 protected from DCM | Sirt1, Hdac4 | – | Not investigated | 23524588 | |
| – | – | Gain and loss study in H9c2 | In vitro in H9c2 cells, mechanistic model | Prevents the activation of NFkB/Caspase3 mediated apoptosis upon stress | p65 | H9c2 | – | 30504734 | |
| AMI | – | In vitro gain and loss study, in vivo miR-22 KO mice | Murine model of AMI | In miR-21 KO mice decreased survival, decreased LVEF, increased scar size | KBTBD7 | Macrophages | 29991775 | ||
| miR-21 | AMI | – | AAV9 overexpression of miR-21 | AMI in rats | Promotes cardiac fibroblast activation and CF to myofibroblast transformation (CMT) | Jagged1 | – | Not investigated | 29808534 |
| – | Targets PDCD4 to reduce apoptosis after HIF-1alpha activated expression of mir21 | 29170412 | |||||||
| AMI | – | In vitro in CF, in vivo induction of miR-21 via TGF-β1 | Murine model of AMI | Increased fibrosis in the heart upon AMI | Smad7 | CF | Not investigated | 28817807 | |
| – | – | Gain and loss study in H9c2 cells | In vitro in H9c2 cells, mechanistic model | Inhibits autophagy and apoptosis upon I/R partially via the Akt/mTOR pathway | – | H9c2 | – | 27680680 | |
| – | – | Exposure of H9c2 cells with CPC derived EVs rich in miR-21 | In vitro in H9c2 cells, mechanistic model | Targets PDCD4 when CDC derived exosomes are added to CMs | – | H9c2 | – | 27336721 | |
| – | – | Gain and loss study in PBMCs | In vitro in human PBMCs, mechanistic model | Via targeting SMAD7, mir-21 can reduce the number of circulating Tregs | Smad7 | Human Tregs | – | 26383248 | |
| – | – | Gain and loss study in H9c2 cells | In vitro in H9c2 cells, mechanistic model | Proof for a positive feedback loop between mir-21 and HIF-1alpha which reduces apoptosis upon hypoxia and stress | PTEN | H9c2 | – | 24983504 | |
| miR-21-5p | – | – | Gain and loss study in H9c2 | In vitro in H9c2 cells, mechanistic model | Modulation of reliance on glycolytic or fatty acid oxidation in mitochondria | – | H9c2 | – | 30657727 |
| – | BM-MSCs | Exposure of H9c2 cells with BM-MSCex rich in miR-21a-5p | In vitro in H9c2 cells, mechanistic model | Reduction of CM apoptosis upon stress. This was identified in EVs from MSCs (miR-21a-5p). | PDCD4, PTEN, Peli1 an dFasL | H9c2 | – | 29698635 |
AAV, adeno-associated-virus; Afib, atrial fibrillation; AMI, acute myocardial infarction; AT-MSCex, AT-MSC exosomes; AT-MSCs, adipose tissue derived mesenchymal stem cells; BM-EPCs, bone marrow derived endothelial progenitor cells; BM-MSCex, BM-MSC exosomes; BM-MSCs, bone marrow derived mesenchymal stem cells; CF, cardiac fibroblast; CPCs, cardiac progenitor cells; DCex, dendritic cell exosomes; DCM, dilated cardiomyopathy; DCs, dendritic cells; DMap, diabetic myocardial microangiopathy; DoxDCM, doxorubicin induced dilated cardiomyopathy; ECs, endothelial cells; EPCs, endothelial progenitor cells; HF, heart failure; HUVECs, human umbilical vein endothelial cells; I/R, ischemia reperfusion.
MicroRNAs – The Good, the Bad, and the Ugly
Off-Target Effects of miRNAs
Especially studies investigating RIPC have identified clusters of cardioprotective miRNAs (Varga et al., 2015). In these studies as well as in those that used EVs, an undefined cocktail of miRNAs was systemically applied. In both cases, defining the mode of action is virtually impossible. Usually, hundreds of miRNAs can be identified in RIPC and even in EV preparations numerous miRNAs can be found, some of which taken by themselves have been identified as damaging to the myocardium. As an example, miR-665 has been identified in sera and serum exosomes of patients with heart failure (Li et al., 2016; Fan et al., 2019). MiR-665 directly targets the cannabinoid receptor 2 (CbR2) and adenylate kinase 1 (AK1) (Möhnle et al., 2014; Lin et al., 2019). In a rat heart Langendorff preparation, inhibition of CbR2 and AK1 leads to the upregulation of pro-apoptotic genes such as B cell lymphoma 2 (Bcl-2) and Bcl-2-associated X protein (Bax) and caspase-3 in H9c2 cells, a commonly used rat cardiomyocyte cell line (Yu et al., 2019). In a murine model of myocardial infarction, AAV9 transfection with miR-665 antisense miRNA led to improvement of cardiac function (Fan et al., 2018). Here, the group has identified glucagon-like peptide-1 receptor (GLP1R) as the target for miR-665. Inhibition of miR-665 expression led to increased cAMP signaling in the heart via the promotor GLP1R and reduced apoptotic events upon ischemia/reperfusion injury in the heart. Identification of harmful miRNAs like miRNA-665 in autologous exosome preparations from patients with heart failure could serve as quality markers and help prevent potential off-target effects. In some cases, off target effects or cardioprotective miRNAs have already been identified. miRNA-206, for example, has been associated with both cardioprotective, but also damaging effects upon overexpression (Table 1). Transcription of miR-206 can be induced via histamine release upon myocardial stress in mice (Ding et al., 2018). In vitro, miR-206 can prevent apoptosis in hypoxic conditions by targeting autophagy related protein 2 (ATG3) (Kong et al., 2019). In a murine model of acute myocardial infarction, this prevented ubiquitination of cytosomal proteins and apoptosis (Ding et al., 2018). By cardiac-specific overexpression of miR-206 is has been shown that forkhead box protein P1 induces hypertrophy of cardiomyocytes which during stress can prevent cardiomyocyte apoptosis (Yang et al., 2015). By inhibiting metalloproteinase inhibitor 3 (TIMP3), miR-206 has also been shown to attenuate cardiac fibrosis in the setting of chronic heart failure (Limana et al., 2011). However, two independent groups have also uncovered two mechanisms by which miR-206 increases the risk of cardiac arrhythmias. miR-206 targets connexin 43 (Cx43), an important component of gap junctions in the myocardium in a transgenic mouse model (Roell et al., 2007). Low expression of Cx43 has been associated with cardiac arrhythmias such as atrial fibrillation. Additionally, Wei et al. (2018) uncovered that miR-206 binds to GTP cyclohydrolase I (GCH1) in a canine model of atrial fibrillation (Afib). GCH1 is the rate limiting enzyme in de novo synthesis of tetrahydrobiopterin (BH4). Decreased expression of BH4 was associated with shortened refractory times in atrial cardiomyocytes in humans, which led to Afib (Wei et al., 2018). In this example, different groups were able to demonstrate a cardioprotective effect of miR-206. At the same time potential off target effects were uncovered that could impede the translation of important signal transduction proteins in the myocardium. These off-target effects were never investigated in the murine models that investigated the cardioprotective effects for this miRNA. It has also been shown that miRNA have species specific effects. A recently published large animal study demonstrated that the delivery of miR-199 via adenoviral transfer to the myocardium of pigs resulted in improved contractility and myocardial mass in the short term (Gabisonia et al., 2019). After 1 month, however, the pigs died of arrhythmias. Histological analysis revealed that the myocardium was infiltrated with proliferating cells displaying a poorly differentiated myoblastic phenotype. This off-target effect of miR-199 was not identified in small animal studies. Results like these raise the question whether results from murine models or even porcine models are really translatable into clinical applications without additional safeguards that can predict these off-target and adverse effects.
Neoangiogenesis – Good for Cardioprotection, Bad for Cancer Progression
Neoangiogenesis plays an important role in protecting the myocardium in the border zones from infarcts from myocardial remodeling. Therefore, many miRNAs such as miR-132, miR-126, and miR-210 have been investigated for their angiogenic potential (Table 1). The transcription of most of these miRNAs is dependent on the expression VEGF (Lei et al., 2015; Chodari et al., 2019). For instance, miR-132 expression is dependent in the promotor cAMP response element-binding protein as the transcription factor, which is induced by VEGF stimulation (Chodari et al., 2019). EVs from cardiac progenitor cells (CPCs) rich in miR-132 inhibit the translation of Ras GTPase activating protein (p120RasGAP) which promotes proliferation and sprouting of endothelial cells thus improving neovascularization in vivo and in vitro (Barile et al., 2014). In a murine model of acute myocardial infarction this led to improved left ventricular ejection fraction on a functional level and improved vascularization in the infarct border zone on a histological level (Barile et al., 2014). Similarly, EVs from adipose-derived mesenchymal stem cells (AT-MSCs) that overexpressed miR-126, also improved cardiac function and resulted in a denser microvasculature in the infarct border zone in rats (Luo et al., 2017). Functional studies have shown that miR-126 is highly depended on the activation of endothelial dicer RNA Polymerase III. Only the passenger strand miR-126-5p can bind to the Notch1 inhibitor Delta-like homolog 1 (Dlk1) (Zhou Z. et al., 2018). The notch signaling pathway is crucial to endothelial cell differentiation and endothelial sprouting (Mack and Iruela-Arispe, 2018). While there is no direct evidence linking the cardioprotective effect of miR-132 to cardiomyocytes, there is evidence that miR-132 can also reduce fibrosis in by targeting connective tissue growth factor (Zhang C.-J. et al., 2018). Furthermore, miR-132 has also been identified in a rat model of DCM to target phosphatase and tensin homolog (PTEN) (Ma et al., 2018). Suppression of PTEN activates the phosphoinositide 3-kinases/protein kinase B pathway (PI3K/Akt-pathway) which facilitates cardiomyocyte and endothelial proliferation alike (Zhang C.-J. et al., 2018). miR-210 has also been shown to promote angiogenesis in the myocardium (Mutharasan et al., 2011). In contrast to miR-126 and miR132, the expression of miR-210 depends on HIF-1alpha, which is also released under hypoxic stress (Barile et al., 2014). In both murine models of acute myocardial infarction and hind limb ischemia it has been shown that miR-210 encapsulated by EVs promotes angiogenesis in endothelial cells as well as suppresses apoptosis in cardiomyocytes by targeting Ephrin A3 (Barile et al., 2014; Wang et al., 2017; Besnier et al., 2018). While the upregulation or substitution of all aforementioned miRNAs are associated with a pro-angiogenic profile in the setting of myocardial infarction, they have also been identified in cancer biogenesis and metastasis formation. Also, in the field of cancer biology the data for these miRNAs is heterogeneous and in depending on cancer type they are both associated as a positive and negative prognostic marker. The heterogeneity of these results and their role in tumor progression could, however, pose an obstacle for their use as a cardioprotective agent. As a solution, patients susceptible to certain cancers that depend on overexpression of these miRNA need to be identified to prevent adverse effects from a hypothetical therapeutic miRNA.
How to Deal With Contradictory Results
miR-146a is part of a negative feedback loop in the canonical pathway of NFkB activation. miR-146a binds to the mRNA encoding for interleukin-1 receptor associated kinase 1 (IRAK1) and tumor necrosis factor receptor-associated receptor 6 (TRAF6). Both of this receptor bound factors are essential for the IL-1 and TNFalpha activation of NFkB (Fish and Cybulsky, 2015; Gao et al., 2015; Milano et al., 2019). As part of that negative feedback loop miR-146a is highly expressed in atherosclerotic plaques. However, the overexpression of this miRNA can also attenuate the inflammatory response as shown in gain and loss studies in ApoE deficient mice, where miRNA-146a plays an important part in the attenuation of atherosclerotic plaques (Fish and Cybulsky, 2015). In a rat model of AMI, EVs from AT-MSCs overexpressing miR-146a targeted the early growth response protein 1 in cardiomyocytes decreasing cardiomyocyte apoptosis, cardiac fibrosis and ultimately improving the heart function (Pan et al., 2019). Milano et al. (2019) demonstrated that the passenger strand miR-146a-5p reduced the inflammatory signaling pathways by directly targeting TRAF6, SMAD4, IRAK1, NADPH oxidase 4 (NOX4), and myeloperoxidase (MPO) in a model of doxorubicin/trastuzumab induced cardiomyopathy. Interestingly, another group showed that with treatment of doxorubicin alone an upregulation of miR146a occurs, targeting the receptor tyrosine-protein kinase erbB-4 (ErbB4). Here, a negative correlation between miR-146a and cardiac function was confirmed and explained by the suppression of the ErbB4 dependent neuregulin1/ErbB pathway, which is essential for adult cardiac function (Horie et al., 2010). It is, however, important to note that these results were only obtained from in vitro experiments. Both miR-21 and -22 have been associated with cardioprotective properties by reducing cardiomyocyte apoptosis and have been found in most EVs from therapeutic cell product isolates to date (Barile et al., 2016; Gallet et al., 2016). While there is evidence, that miR-22 also exhibits an anti-fibrotic effect during myocardial remodeling, overexpression of miR-21 is clearly associated with promoting cardiac fibrosis (Table 1). miR-21 directly targets jagged1 and SMAD7 in rat hearts when overexpressed via AAV9 (Zhou X.-L. et al., 2018). In the aforementioned experiment, jagged1 suppression activates cardiac fibroblast proliferation and facilitates cardiac fibroblast to myofibroblast transformation. In a murine model of AMI, suppression of SMAD7 via miR-21 led to an increased expression of Collagen 1 alpha, alpha-smooth muscle actin (alpha-SMA) and F-actin. In a similar model of AMI in rats, AVV9 mediated overexpression of miR-22 let to the inhibition of CBP-associated factor AP1 (Yang et al., 2014). Downregulation of this promotor leads to the activation of MMP9, which in turn can reduce cardiac fibrosis. Additionally, miR-22 binds to the mRNA coding for the TGFbeta receptor 1 (Hong et al., 2016). In mice, silencing of miR-22 led to increased expression of Collagen 1 alpha 1 and 3alpha1 and an overall increased amount of cardiac fibrosis after myocardial infarction. Regarding the anti-apoptotic effect of miR-22, there are contradictory results published to date. Gurha et al. (2012) and Du et al. (2016) both demonstrated that SIRT1 and PGC1alpha are both targeted by miR-22 in cardiomyocytes. In both experiments, overexpression of miR-22 lead to increased cell death and reduction in myocardial mass whereas in the studies published by Huang et al. (2013) and Hong et al. (2016) cardiac mass increased upon miR-22 overexpression and ischemia/reperfusion injury. Huang et al. (2013) also demonstrate the targeting of SIRT1 but in their hands, cardiomyocyte hypertrophy was the predominant finding. Hong et al. (2016) demonstrated decreased apoptosis levels in the myocardium and linked that effect to the inhibition of CREB. For miR-21 the anti-apoptotic effects is shown more robustly between different research teams (Table 1). miR-21 expression is under direct control of HIF1alpha but can also influence the expression of HIF1alpha itself in a positive feedback loop (Liu Y. et al., 2014). The miRNA targets programmed cell death protein 4 (PCDP4) and reduces cell apoptosis upon hypoxic stress. This effect has been demonstrated with EVs containing high levels of miR-21 from different cell types such as CPCs and MSCs (Xiao et al., 2016; Luther et al., 2018). Both miR-21 and miR-22 exemplify the various outcomes that can be seen when working with miRNAs in pre-clinical models. Even within one species there are variations and sometime contradictory results that can be elaborated by other groups on both ends of the aisle. Especially miRNAs that can promote fibrosis can have severe and unwanted effects in the injured heart and may lead to increase in scar mass. Here, patient screening and good patient selection may help to prevent these contradictory results. This can only be achieved by understanding all pathways that can be altered by each respective miRNA and tools to identify patients that may be susceptible to treatments with certain miRNAs.
Summary
The data that has been collected on miRNAs targeting cardioprotection so far exemplifies the importance of knowing the relevant targets of miRNAs, since introduction of foreign miRNAs via exosomal transfer or other clinical relevant approaches may have some severe side effects. In this context, it is also worth noting that all of the aforementioned miRNAs are also involved in tumor biology. Especially miRNAs that impact neoangiogenesis in myocardial repair are important factors in tumor angiogenesis as well. In addition, most of the mechanistical studies have focused on the interaction of one miRNA on multiple targets. Upon ischemia/reperfusion or transfer of exogenous EVs the interaction of numerous miRNAs on multiple potential targets needs to be taken into account. With current methods, experimental data usually depicts a single linear arm in a complex matrix of interactions between miRNAs, mRNAs and transcription factors. Our current understanding of these complex matrices is only rudimental at best. And it may also be one of the main reasons why none of the clinical trials with cell-based therapies or RIPC have delivered the expected results we anticipated from pre-clinical experience. As repeatedly highlighted by numerous experts in the field, understanding the pathways by which a single- or a collection of miRNAs in an exosome, facilitate cardioprotection will be crucial for successful clinical translation (Madonna et al., 2016, 2019; Behfar and Terzic, 2019). Combined effort of computational models and artificial intelligence in merging and interpreting the acquired data, might help us in the future to achieve this goal. Identifying such ‘pathway’ matrices will be detrimental in defining quality standards for therapeutic exosome or single miRNA-based products. The past experience from cell-based therapies have taught us that preclinical data in the field of cardiac regeneration or cardioprotection does not necessarily translate into therapeutic success in the clinical setting. Going forward, we will have to deepen our understanding of miRNA interactions by strengthen our efforts to collaborate with bioinformaticians for more sophisticated predictive algorithms.
Author Contributions
TN-S and ME contributed to the conception and design of this review. VE, HR, and HM helped to organize the article database and supported the literature review. TN-S wrote the first draft of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. TN-S is a participant in the BIH Charité Clinician Scientist Program funded by the Charité -Universitätsmedizin Berlin and the Berlin Institute of Health.
References
- Baglio S. R., Rooijers K., Koppers-Lalic D., Verweij F. J., Pérez Lanzón M., Zini N., et al. (2015). Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res. Ther. 6:127. 10.1186/s13287-015-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile L., Lionetti V., Cervio E., Matteucci M., Gherghiceanu M., Popescu L. M., et al. (2014). Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res. 103 530–541. 10.1093/cvr/cvu167 [DOI] [PubMed] [Google Scholar]
- Barile L., Moccetti T., Marbán E., Vassalli G. (2016). Roles of exosomes in cardioprotection. Eur. Heart J. 38 1372–1379. 10.1093/eurheartj/ehw304 [DOI] [PubMed] [Google Scholar]
- Barile L., Vassalli G. (2017). Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 174 63–78. 10.1016/j.pharmthera.2017.02.020 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Behfar A., Crespo-Diaz R., Terzic A., Gersh B. J. (2014). Cell therapy for cardiac repair–lessons from clinical trials. Nat. Rev. Cardiol. 11 232–246. 10.1038/nrcardio.2014.9 [DOI] [PubMed] [Google Scholar]
- Behfar A., Terzic A. (2019). Regeneration for all: an Odyssey in biotherapy. Eur. Heart J. 40 1033–1035. 10.1093/eurheartj/ehz095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin G., Gardin C., Ferroni L., Chachques J., Rogante M., Mitrečić D., et al. (2019). Exosome in cardiovascular diseases: a complex world full of hope. Cells 8:166. 10.3390/cells8020166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnier M., Gasparino S., Vono R., Sangalli E., Facoetti A., Bollati V., et al. (2018). miR-210 enhances the therapeutic potential of bone-marrow-derived circulating proangiogenic cells in the setting of limb ischemia. Mol. Ther. 26 1694–1705. 10.1016/j.ymthe.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon R. A., Dimmeler S. (2015). MicroRNAs in myocardial infarction. Nat. Rev. Cardiol. 12 135–142. 10.1038/nrcardio.2014.207 [DOI] [PubMed] [Google Scholar]
- Brevoord D., Kranke P., Kuijpers M., Weber N., Hollmann M., Preckel B. (2012). Remote ischemic conditioning to protect against ischemia-reperfusion injury: a systematic review and meta-analysis. PLoS One 7:e42179. 10.1371/journal.pone.0042179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C. H., Cullen B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10 1957–1966. 10.1261/rna.7135204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambria E., Pasqualini F. S., Wolint P., Günter J., Steiger J., Bopp A., et al. (2017). Translational cardiac stem cell therapy: advancing from first-generation to next-generation cell types. npj Regen. Med. 2:17. 10.1038/s41536-017-0024-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Tredget E. E., Wu P. Y. G., Wu Y., Wu Y. (2008). Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One 3:e1886. 10.1371/journal.pone.0001886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodari L., Dariushnejad H., Ghorbanzadeh V. (2019). Voluntary wheel running and testosterone replacement increases heart angiogenesis through miR-132 in castrated diabetic rats. Physiol. Int. 106 48–58. 10.1556/2060.106.2019.06 [DOI] [PubMed] [Google Scholar]
- Ciullo A., Biemmi V., Milano G., Bolis S., Cervio E., Fertig E. T., et al. (2019). Exosomal expression of CXCR4 targets cardioprotective vesicles to myocardial infarction and improves outcome after systemic administration. Int. J. Mol. Sci. 20:468. 10.3390/ijms20030468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008). SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454 56–61. 10.1038/nature07086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S., Haber D. A. (2007). Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131 1097–1108. 10.1016/j.cell.2007.10.032 [DOI] [PubMed] [Google Scholar]
- Diederichs S., Jung S., Rothenberg S. M., Smolen G. A., Mlody B. G., Haber D. A. (2008). Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc. Natl. Acad. Sci. U.S.A. 105 9284–9289. 10.1073/pnas.0800803105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Abudupataer M., Zhou Z., Chen J., Li H., Xu L., et al. (2018). Histamine deficiency aggravates cardiac injury through miR-206/216b-Atg13 axis-mediated autophagic-dependant apoptosis. Cell Death Dis. 9:694. 10.1038/s41419-018-0723-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J.-K., Cong B.-H., Yu Q., Wang H., Wang L., Wang C.-N., et al. (2016). Upregulation of microRNA-22 contributes to myocardial ischemia-reperfusion injury by interfering with the mitochondrial function. Free Radic. Biol. Med. 96 406–417. 10.1016/j.freeradbiomed.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Fan J., Li H., Nie X., Yin Z., Zhao Y., Zhang X., et al. (2018). MiR-665 aggravates heart failure via suppressing CD34-mediated coronary microvessel angiogenesis. Aging (Albany NY) 10 2459–2479. 10.18632/aging.101562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhang X., Nie X., Li H., Yuan S., Dai B., et al. (2019). Nuclear miR-665 aggravates heart failure via suppressing phosphatase and tensin homolog transcription. Sci. China Life Sci. 1–13. 10.1007/s11427-018-9515-1 [DOI] [PubMed] [Google Scholar]
- Fish J. E., Cybulsky M. I. (2015). ApoE attenuates atherosclerosis via miR-146a. Circ. Res. 117 3–6. 10.1161/CIRCRESAHA.115.306733 [DOI] [PubMed] [Google Scholar]
- Frey U. H., Klaassen M., Ochsenfarth C., Murke F., Thielmann M., Kottenberg E., et al. (2018). Remote ischaemic preconditioning increases serum extracellular vesicle concentrations with altered micro−RNA signature in CABG patients. Acta Anaesthesiol. Scand. 63 483–492. 10.1111/aas.13296 [DOI] [PubMed] [Google Scholar]
- Gabisonia K., Prosdocimo G., Aquaro G. D., Carlucci L., Zentilin L., Secco I., et al. (2019). MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 569 418–422. 10.1038/s41586-019-1191-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet R., Dawkins J., Valle J., Simsolo E., de Couto G., Middleton R., et al. (2016). Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur. Heart J. 38 201–211. 10.1093/eurheartj/ehw240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Wang X., Zhang X., Ha T., Ma H., Liu L., et al. (2015). Attenuation of cardiac dysfunction in polymicrobial sepsis by MicroRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J. Immunol. 195 672–682. 10.4049/jimmunol.1403155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory R. I., Chendrimada T. P., Cooch N., Shiekhattar R. (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123 631–640. 10.1016/j.cell.2005.10.022 [DOI] [PubMed] [Google Scholar]
- Guil S., Cáceres J. F. (2007). The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 14 591–596. 10.1038/nsmb1250 [DOI] [PubMed] [Google Scholar]
- Gurha P., Abreu-Goodger C., Wang T., Ramirez M. O., Drumond A. L., Van Dongen S., et al. (2012). Targeted deletion of MicroRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation 125 2751–2761. 10.1161/CIRCULATIONAHA.111.044354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyöngyösi M., Wojakowski W., Lemarchand P., Lunde K., Tendera M., Bartunek J., et al. (2015). Meta-analysis of cell-based CaRdiac stUdiEs (ACCRUE) in patients with acute myocardial infarction based on individual patient data. Circ. Res. 116 1346–1360. 10.1161/CIRCRESAHA.116.304346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausenloy D. J., Candilio L., Evans R., Ariti C., Jenkins D. P., Kolvekar S., et al. (2015). Remote ischemic preconditioning and outcomes of cardiac surgery. N. Engl. J. Med. 373 1408–1417. 10.1056/NEJMoa1413534 [DOI] [PubMed] [Google Scholar]
- Hong Y., Cao H., Wang Q., Ye J., Sui L., Feng J., et al. (2016). MiR-22 may suppress fibrogenesis by targeting TGFβR I in cardiac fibroblasts. Cell. Physiol. Biochem. 40 1345–1353. 10.1159/000453187 [DOI] [PubMed] [Google Scholar]
- Horie T., Ono K., Nishi H., Nagao K., Kinoshita M., Watanabe S., et al. (2010). Acute doxorubicin cardiotoxicity is associated with miR-146a-induced inhibition of the neuregulin-ErbB pathway. Cardiovasc. Res. 87 656–664. 10.1093/cvr/cvq148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.-P., Chen J., Seok H. Y., Zhang Z., Kataoka M., Hu X., et al. (2013). MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 112 1234–1243. 10.1161/CIRCRESAHA.112.300682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A. E., Bálint É, Tuschl T., Zamore P. D. (2001). A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293 834–838. 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Kharbanda R. K., Mortensen U. M., White P. A., Kristiansen S. B., Schmidt M. R., Hoschtitzky J. A., et al. (2002). Transient limb ischemia induces remote ischemic preconditioning in vivo. Circulation 106 2881–2883. 10.1161/01.CIR.0000043806.51912.9B [DOI] [PubMed] [Google Scholar]
- Kong F., Jin J., Lv X., Han Y., Liang X., Gao Y., et al. (2019). Long noncoding RNA RMRP upregulation aggravates myocardial ischemia-reperfusion injury by sponging miR-206 to target ATG3 expression. Biomed. Pharmacother. 109 716–725. 10.1016/j.biopha.2018.10.079 [DOI] [PubMed] [Google Scholar]
- Konstantinov I. E., Arab S., Li J., Coles J. G., Boscarino C., Mori A., et al. (2005). The remote ischemic preconditioning stimulus modifies gene expression in mouse myocardium. J. Thorac. Cardiovasc. Surg. 130 1326–1332. 10.1016/j.jtcvs.2005.03.050 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 75 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425 415–419. 10.1038/nature01957 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K. H., Lee S., Baek S. H., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23 4051–4060. 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., van Mil A., Brandt M. M., Grundmann S., Hoefer I., Smits M., et al. (2015). MicroRNA-132/212 family enhances arteriogenesis after hindlimb ischaemia through modulation of the Ras-MAPK pathway. J. Cell. Mol. Med. 19 1994–2005. 10.1111/jcmm.12586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner P. J. F., Ameres S. L., Kueng S., Martinez J. (2006). Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7 314–320. 10.1038/sj.embor.7400637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Fan J., Yin Z., Wang F., Chen C., Wang D. W. (2016). Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget 7 33–45. 10.18632/ONCOTARGET.6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhu D., Huang L., Zhang J., Bian Z., Chen X., et al. (2012). Argonaute 2 complexes selectively protect the circulating MicroRNAs in cell-secreted microvesicles. PLoS One 7:e46957. 10.1371/journal.pone.0046957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limana F., Esposito G., D’Arcangelo D., Di Carlo A., Romani S., Melillo G., et al. (2011). HMGB1 attenuates cardiac remodelling in the failing heart via enhanced cardiac regeneration and miR-206-mediated inhibition of TIMP-3. PLoS One 6:e19845. 10.1371/journal.pone.0019845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Feng D. G., Xu J. (2019). microRNA-665 silencing improves cardiac function in rats with heart failure through activation of the cAMP signaling pathway. J. Cell. Physiol. 234 13169–13181. 10.1002/jcp.27987 [DOI] [PubMed] [Google Scholar]
- Lin S. L., Miller J. D., Ying S. Y. (2006). Intronic microRNA (miRNA). J. Biomed. Biotechnol 2006:26818. 10.1155/JBB/2006/26818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li J., Cairns M. J. (2014). Identifying miRNAs, targets and functions. Brief. Bioinform. 15 1–19. 10.1093/bib/bbs075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Nie H., Zhang K., Ma D., Yang G., Zheng Z., et al. (2014). A feedback regulatory loop between HIF-1α and miR-21 in response to hypoxia in cardiomyocytes. FEBS Lett. 588 3137–3146. 10.1016/j.febslet.2014.05.067 [DOI] [PubMed] [Google Scholar]
- Luo Q., Guo D., Liu G., Chen G., Hang M., Jin M. (2017). Exosomes from MiR-126-overexpressing Adscs Are therapeutic in relieving acute myocardial ischaemic injury. Cell. Physiol. Biochem. 44 2105–2116. 10.1159/000485949 [DOI] [PubMed] [Google Scholar]
- Luther K. M., Haar L., McGuinness M., Wang Y., Lynch T. L., IV, Phan A., et al. (2018). Exosomal miR-21a-5p mediates cardioprotection by mesenchymal stem cells. J. Mol. Cell. Cardiol. 119 125–137. 10.1016/j.yjmcc.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Ma T., Chen Y., Chen Y., Meng Q., Sun J., Shao L., et al. (2018). MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018:3290372. 10.1155/2018/3290372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack J. J., Iruela-Arispe M. L. (2018). NOTCH regulation of the endothelial cell phenotype. Curr. Opin. Hematol. 25 212–218. 10.1097/MOH.0000000000000425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae I. J., Ma E., Zhou M., Robinson C. V., Doudna J. A. (2008). In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. U.S.A. 105 512–517. 10.1073/pnas.0710869105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madonna R., Van Laake L. W., Botker H. E., Davidson S. M., De Caterina R., Engel F. B., et al. (2019). ESC working group on cellular biology of the heart: position paper for cardiovascular research: tissue engineering strategies combined with cell therapies for cardiac repair in ischaemic heart disease and heart failure. Cardiovasc. Res. 115 488–500. 10.1093/cvr/cvz010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madonna R., Van Laake L. W., Davidson S. M., Engel F. B., Hausenloy D. J., Lecour S., et al. (2016). Position paper of the European society of cardiology working group cellular biology of the heart: cell-based therapies for myocardial repair and regeneration in ischemic heart disease and heart failure. Eur. Heart J. 37 1789–1798. 10.1093/eurheartj/ehw113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maring J. A., Lodder K., Mol E., Verhage V., Wiesmeijer K. C., Dingenouts C. K. E., et al. (2019). Cardiac progenitor cell–derived extracellular vesicles reduce infarct size and associate with increased cardiovascular cell proliferation. J. Cardiovasc. Transl. Res. 12 5–17. 10.1007/s12265-018-9842-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D. P., Zamore P. D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123 607–620. 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- Michlewski G., Guil S., Semple C. A., Cáceres J. F. (2008). Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell 32 383–393. 10.1016/j.molcel.2008.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano G., Biemmi V., Lazzarini E., Balbi C., Ciullo A., Bolis S., et al. (2019). Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc. Res. 116 383–392. 10.1093/cvr/cvz108 [DOI] [PubMed] [Google Scholar]
- Möhnle P., Schütz S. V., Schmidt M., Hinske C., Hübner M., Heyn J., et al. (2014). MicroRNA-665 is involved in the regulation of the expression of the cardioprotective cannabinoid receptor CB2 in patients with severe heart failure. Biochem. Biophys. Res. Commun. 451 516–521. 10.1016/j.bbrc.2014.08.008 [DOI] [PubMed] [Google Scholar]
- Moyé L. (2014). DAMASCENE and meta-ecological research: a bridge too far. Circ. Res. 115 484–487. 10.1161/CIRCRESAHA.114.304767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutharasan R. K., Nagpal V., Ichikawa Y., Ardehali H. (2011). microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am. J. Physiol. Circ. Physiol. 301 H1519–H1530. 10.1152/ajpheart.01080.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Alimujiang M., Chen Q., Shi H., Luo X. (2019). Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction-induced myocardial damage via downregulation of early growth response factor 1. J. Cell. Biochem. 120 4433–4443. 10.1002/jcb.27731 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffiths-Jones S., Ashurst J. L., Bradley A. (2004). Identification of mammalian microRNA host genes and transcription units. Genome Res. 14 1902–1910. 10.1101/gr.2722704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roell W., Lewalter T., Sasse P., Tallini Y. N., Choi B.-R., Breitbach M., et al. (2007). Engraftment of connexin 43-expressing cells prevents post-infarct arrhythmia. Nature 450 819–824. 10.1038/nature06321 [DOI] [PubMed] [Google Scholar]
- Shao L., Zhang Y., Lan B., Wang J., Zhang Z., Zhang L., et al. (2017). MiRNA-sequence indicates that mesenchymal stem cells and exosomes have similar mechanism to enhance cardiac repair. Biomed Res. Int. 2017:4150705. 10.1155/2017/4150705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannbauer A., Traxler D., Lukovic D., Zlabinger K., Winkler J., Gugerell A., et al. (2019). Effect of ischemic preconditioning and postconditioning on exosome-rich fraction microRNA levels, in relation with electrophysiological parameters and ventricular arrhythmia in experimental closed-chest reperfused myocardial infarction. Int. J. Mol. Sci. 20:2140. 10.3390/ijms20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Weiz L., Burwinkel B. (2012). Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 37 460–465. 10.1016/j.tibs.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Tzahor E., Poss K. D. (2017). Cardiac regeneration strategies: staying young at heart. Science 356 1035–1039. 10.1126/science.aam5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z. V., Giricz Z., Bencsik P., Madonna R., Gyongyosi M., Schulz R., et al. (2015). Functional genomics of cardioprotection by ischemic conditioning and the influence of comorbid conditions: implications in target identification. Curr. Drug Targets 16 904–911. 10.2174/1389450116666150427154203 [DOI] [PubMed] [Google Scholar]
- Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., et al. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 4:2980. 10.1038/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Chen C., Yang D., Liao Q., Luo H., Wang X., et al. (2017). Mesenchymal stem cells-derived extracellular vesicles, via miR-210, improve infarcted cardiac function by promotion of angiogenesis. Biochim. Biophys. Acta Mol. Basis Dis. 1863 2085–2092. 10.1016/j.bbadis.2017.02.023 [DOI] [PubMed] [Google Scholar]
- Wei J., Zhang Y., Li Z., Wang X., Chen L., Du J., et al. (2018). GCH1 attenuates cardiac autonomic nervous remodeling in canines with atrial-tachypacing via tetrahydrobiopterin pathway regulated by microRNA-206. Pacing Clin. Electrophysiol. 41 459–471. 10.1111/pace.13289 [DOI] [PubMed] [Google Scholar]
- Wei M., Xin P., Li S., Tao J., Li Y., Li J., et al. (2011). Repeated remote ischemic postconditioning protects against adverse left ventricular remodeling and improves survival in a rat model of myocardial infarction. Circ. Res. 108 1220–1225. 10.1161/CIRCRESAHA.110.236190 [DOI] [PubMed] [Google Scholar]
- Wendt S., Goetzenich A., Goettsch C., Stoppe C., Bleilevens C., Kraemer S., et al. (2018). Evaluation of the cardioprotective potential of extracellular vesicles – a systematic review and meta-analysis. Sci. Rep. 8:15702. 10.1038/s41598-018-33862-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Pan Y., Li X. H., Yang X. Y., Feng Y. L., Tan H. H., et al. (2016). Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. 7:e2277. 10.1038/cddis.2016.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yang J., Chen L., Ding J., Li S., Wu H., et al. (2014). MicroRNA-22 targeting CBP protects against myocardial ischemia-reperfusion injury through anti-apoptosis in rats. Mol. Biol. Rep. 41 555–561. 10.1007/s11033-013-2891-x [DOI] [PubMed] [Google Scholar]
- Yang Y., Del Re D. P., Nakano N., Sciarretta S., Zhai P., Park J., et al. (2015). miR-206 mediates YAP-induced cardiac hypertrophy and survival. Circ. Res. 117 891–904. 10.1161/CIRCRESAHA.115.306624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin Y., Macara I. G., Cullen B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17 3011–3016. 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Yang W., Wang W., Wang Z., Pu Y., Chen H., et al. (2019). Involvement of miR-665 in protection effect of dexmedetomidine against oxidative stress injury in myocardial cells via CB2 and CK1. Biomed. Pharmacother. 115:108894. 10.1016/j.biopha.2019.108894 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Cullen B. R. (2004). Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32 4776–4785. 10.1093/nar/gkh824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-J., Huang Y., Lu J.-D., Lin J., Ge Z.-R., Huang H. (2018). Upregulated microRNA-132 rescues cardiac fibrosis and restores cardiocyte proliferation in dilated cardiomyopathy through the phosphatase and tensin homolog-mediated PI3K/Akt signal transduction pathway. J. Cell. Biochem. 120 1232–1244. 10.1002/jcb.27081 [DOI] [PubMed] [Google Scholar]
- Zhou X.-L., Xu H., Liu Z.-B., Wu Q.-C., Zhu R.-R., Liu J.-C. (2018). miR-21 promotes cardiac fibroblast-to-myofibroblast transformation and myocardial fibrosis by targeting Jagged1. J. Cell. Mol. Med. 22 3816–3824. 10.1111/jcmm.13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Schober A., Nazari-Jahantigh M. (2018). Dicer promotes endothelial recovery and limits lesion formation after vascular injury through miR-126-5p. Int. J. Cardiol. 273 199–202. 10.1016/j.ijcard.2018.09.006 [DOI] [PubMed] [Google Scholar]