Abstract
Olfactory receptor-78 (Olfr-78) is a recently identified G protein-coupled receptor activated by short-chain fatty acids acetate and propionate. A suggested role for this receptor exists in the prostate where it may influence chronic inflammatory response leading to intraepithelial neoplasia. Olfr-78 has also been shown to be expressed in mouse colon. Short-chain fatty acids and their receptors are well known to modulate inflammation in the gut. Considering this possibility, we first explored if colitis regulated Olfr-78 expression in the gut, where we observed a significant reduction in the expression of Olfr-78 transcript in mouse models of dextran sodium sulfate (DSS)- and 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis. To more directly test this, mice deficient in Olfr-78 were administered with DSS in water for 7 days and were found to have increased expression of IL-1β and inflammatory signs in colon compared with control mice. Next, we explored the expression of its human counterpart olfactory receptor family 51, subfamily E, member 2 (OR51E2) in human intestinal samples and observed that it was in fact also expressed in human colon samples. RNA sequence analysis revealed significant changes in the genes involved in infection, immunity, inflammation, and colorectal cancer between wild-type and Olfr-78 knockout mice. Collectively, our findings show that Olfr-78 is highly expressed in colon and downregulated in DSS- and TNBS-induced colitis, and DSS-treated Olfr-78 null mice had increased colonic expression of cytokine RNA levels, suggesting a potential role for this receptor in intestinal inflammation. Future investigations are needed to understand how Olfr-78/OR51E2 in both mouse and human intestine modulates gastrointestinal pathophysiology.
Keywords: inflammation, intestine, Olfr-78
INTRODUCTION
Short-chain fatty acids (SCFA) are produced by microbial fermentation of dietary fiber and have long been appreciated to play an essential role in colonic health. The most commonly studied SCFAs are acetate, propionate, and butyrate (28). A growing body of evidence indicates that SCFAs collectively mediate their important cellular processes, such as proliferation, differentiation, apoptosis, and immune response, at least in part via activation of G protein-coupled receptors (GPCRs; see Refs. 12 and 50). Thus far, SCFAs have been described to activate three GPCRs: free fatty acid receptors 2 and 3 [FFA2 and -3, also referred to as G protein-coupled receptor (GPR)-43 and -41], and niacin/butyrate receptor GPR109A, where acetate and propionate activate GPR-43 and -41 while butyrate is the ligand for GPR109A (37). GPR-43 and -41 are expressed in colonic epithelial cells and peripheral blood mononuclear cells (18). Additional studies have demonstrated the expression of GPR-43/41 mainly in L-type enteroendocrine cells and suggested a role in regulation of glucagon-like peptide-1 (GLP-1) and peptide YY-1 (PYY-1) production (13, 31, 39, 48). Although knockout (KO) mouse models of GPR-43 and -41 suggested the importance of these receptors in chronic inflammatory conditions such as colitis, asthma, obesity, and arthritis, there is a lack of consensus as to whether GPR-43, in particular, is pro- or anti-inflammatory with contradictory findings from different studies. These data raise the possibility that compensatory mechanisms involving other SCFA-activated GPCRs might play a role (3).
Importantly, one of the olfactory receptors, a subclass of GPCRs, olfactory receptor 78 (Olfr-78; in humans referred to as OR51E2, olfactory receptor family 51, subfamily E, member 2), is expressed in multiple tissues, including intestinal tissue, including enteroendocrine cells, and activated by acetate and propionate (38, 42, 52). Olfactory receptors, the largest in the genome (7, 22), are more commonly recognized for sensing odorant molecules in the nose to initiate a neuronal response that triggers the perception of a smell. In more recent years, the functional role of olfactory receptors in nonolfactory organs has been reported by others. Interestingly, this receptor, Olfr-78, has been implicated in blood pressure regulation (38) and inhibition of prostate cancer cell proliferation (30). Ligand-induced activation of OR51E2 (the human isoform) inhibits melanocyte proliferation, differentiation, and melanogenesis (15). Although the expression of Olfr-78 is reported in enteroendocrine cells and epithelial cells of the colon by immunofluorescence and RT-PCR, respectively (13), its functional significance in gastrointestinal pathophysiology has yet to be investigated.
In the present study, we observed that the expression of Olfr-78 mRNA was significantly reduced in the colon of mouse models of dextran sodium sulfate (DSS)- and 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis and TNF-α-treated HT-29 cells. Therefore, we investigated the effect of DSS colitis (3% DSS in water for 7 days) in mice deficient of Olfr-78. These studies showed that, in response to DSS colitis, Olfr-78 KO mice exhibited higher levels of IL-1β expression, epithelial cell destruction, and inflammatory infiltration compared with their wild-type (WT) counterparts, implying a role in immunomodulatory function of this receptor in intestinal inflammation. Because this receptor has not been reported to be expressed in the human intestine, we also examined the expression of its human counterpart OR51E2 along the length of the human intestine. Our data showed that there was marginal increase in the expression of Olfr-78/OR51E2 mRNA levels in the colon compared with ileum and jejunum of both the mouse and the human intestine.
MATERIALS AND METHODS
Olfr78−/− Mice
Heterozygous (Olfr78+/−) mice purchased from Jackson Laboratory were bred at the Biological Resources Laboratory, University of Illinois at Chicago. All animal experiments performed were approved by the animal care committees of the University of Illinois at Chicago and Jesse Brown Veterans Affairs Medical Center (Chicago, IL). WT (Olfr78+/+) and Olfr-78 KO (Olfr−/−) mice were generated by breeding Olfr-78 heterozygous mice. Eight- to ten-week-old mice were used in the studies. Genotyping was carried out on tail clippings.
Mouse and Human Tissues/Cells
Colonic cell lines.
All of the cell lines used in the study were purchased from ATCC and free of mycoplasma contamination. Mouse intestinal tissues were obtained from 8- to 10-wk-old C57BL/6J black mice. Human intestinal samples were obtained from human organ donors. Specifically, RNA was extracted from the mucosal scrapings of five healthy organ donor intestines. The intestinal tissue was obtained through the Gift of Hope organ and tissue donor network of Illinois after the harvest of transplantable organs. These investigations were approved by an exempt protocol from the Institutional Review Board of the University of Illinois at Chicago and Jesse Brown Veterans Affairs Medical Center. Briefly, the intestine was divided into nine sections separating the different segments as duodenum, jejunum, ileum, cecum, ascending colon, transverse colon, descending colon, sigmoid colon, and rectum. The intestinal segments were cleaned with an ice-cold 0.9% NaCl solution, and scraped mucosa was frozen at 80°C in RNAlater solution. RNA was extracted from these tissues with the use of an RNAeasy kit (Qiagen, Valencia, CA), and purity of RNA was determined with the Agilent Bioanalyzer 2100. Purified RNA was reverse transcribed and amplified with the gene-specific primers using Syber green master mix (Qiagen).
PCR Primers
All of the primers used in the study are presented in Table 1.
Table 1.
Primers used in the study
| Sequence | |
|---|---|
| Human primers | |
| OR51E2 | (F) 5′-GTCTTACTGCCATTCTGCTGGT-3′ |
| (R) 5′-TGACACACAGGTTCCAAAGG-3′ | |
| GAPDH | (F) 5′-GAAATCCCATCACCATCTTCC-3′ |
| (R) 5′-AAATGAGCCCCAGCCTTCT-3′ | |
| Mouse primers | |
| Olfr78 | (F) 5′-GGCATTTGGGACTTGTGTGT-3′ |
| (R) 5′-AGCACCGTAGATGATGGGATT-3′ | |
| Cyp2d12 | (F) 5′-TGCTGTTGACCTGTGGAGAG-3′ |
| (R) 5′-GCCCAGCCTGAGTAGTGAAG-3′ | |
| St6galnac1 | (F) 5′-CCTTTCCTGGTTCAGGCATA-3′ |
| (R) 5′-GGGGCGGTATATTCTCCAAT-3′ | |
| TRIM12a | (F) 5′-CCTGTGAGTGCAGATTGTGG-3′ |
| (R) 5′-TGTTGGCCACATTTCGATTA-3′ | |
| TRIM30d | (F) 5′-AGAGCTGACTGGGAGAACCA-3′ |
| (R) 5′-GCTCCAACTGATGCTGAACA-3′ | |
| Abca5 | (F) 5′-TGGTAGCCGCAAGTCTTTCT-3′ |
| (R) 5′-CGATGCCTGCAGAACTGTAA-3′ | |
| St8sia5 | (F) 5′-CTTGTCCAGGTGCTGCAATG-3′ |
| (R) 5′-AGGGCATTTCCTTGGGAAACA-3′ | |
| CXCL-12 | (F) 5′-GCTCTGCATCAGTGACGGTA-3′ |
| (R) 5′-TAATTTCGGGTCAATGCACA-3′ | |
| IL-1β | (F) 5′-GCAACTGTTCCTGAACTCAACT-3′ |
| (R) 5′-ATCTTTTGGGGTCCGTCAACT-3′ | |
| CXCL-1 | (F) 5′-GCAACTGTTCCTGAACTCAACT-3′ |
| (R) 5′-AATTGTATAGTGTTGTCAGAAGCCA-3′ | |
| CCL-3 | (F) 5′-TTCTCTGTACCATGACACTCTGC-3′ |
| (R) 5′-CGTGGAATCTTCCGGCTGTAG-3′ | |
| GAPDH | (F) 5′-TGTGTCCGTCGTGGATCTGA-3′ |
| (R) 5′-CCTGCTTCACCACCTCTTGAT-3′ |
OR51E2, olfactory receptor family 51, subfamily E, member 2; Olfr78, olfactory receptor-78; cyp2d12, cytochrome P-450, family 2, subfamily d, polypeptide 12; st6galnac1, ST6 N-acetylgalactosaminide α2,6-sialyltransferase 1; TRIM12a, tripartite motif containing protein 12a; TRIM30d, tripartite motif-containing protein 30d; abca5, ATP-binding cassette transporter A5; st8sia5, ST8 α-N-acetyl-neuraminide α2,8-sialytransferase 5; CXCL-12, C-X-C motif chemokine 12; CXCL-1, C-X-C motif chemokine 1; F, forward; R, reverse.
TNF Treatment of HT-29 Cells
HT-29 cells were cultured in 12-well Transwell inserts in McCoy’s media with 10% fetal bovine serum and 20 mM HEPES, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 2 μg/mL gentamicin for 12 to 14 days. Monolayers were treated with and without TNF (100 ng/mL) for 6–24 h in culture medium supplemented with 0.2% BSA.
Cecal Contents and Microbiome Analysis
Whole cecal contents collected from six WT and five KO mice were stored at −80°C until further use. Microbiome analysis was performed using the shotgun whole genome sequencing as described earlier (40). Sequencing files were filtered for low-quality reads (<25 on Phred quality score), short reads (<100 bp), or any mouse reads. The remaining high-quality microbial reads were assembled into longer contigs using MetaVelvet (29). Each sample taxonomic profile was annotated using the ensemble classifier WEVOTE (27), with basic classifiers Kraken (51), Clark (34), and BLASTN (1). Statistical analysis was performed in R (version 3.6.0). The phyloseq package (26) was used to agglomerate microbiome abundance data, taxonomic data, and metadata. Normalization was performed by edgeR (41) for differential species analysis.
DSS and TNBS Mouse Models
For the studies related to evaluating Olfr-78 mRNA expression, 8- to 10-wk-old C57BL/6J black mice received either 3.5% DSS (MP Biomedicals, Solon, OH) in drinking water for 7 days or no DSS in water (control group). For the TNBS-induced colitis model, mice were first presensitized with 3.75 mg of TNBS in 100 μL of 50% ethanol by applying on the shaved dorsal skin, and the following day 100 μL of 3.5 mg TNBS in 40% ethanol was slowly administered intrarectally under anesthesia using a lubricated 3.5-Fr silicon catheter (Harvard Apparatus, Holliston, MA; see Ref. 46).
DSS-induced colitis in WT and Olfr-78 KO mice.
Eight- to ten-week-old wild-type (WT) and OLFR-78−/− KO mice (n = 11 WT and 15 KO/both sexes) were administered with DSS (3.0%) in drinking water for 7 days. Next, mice were euthanized, colon tissues were collected for histopathological and myeloperoxidase (MPO) assay, and RNA isolated from mucosal scrapings of distal colon were used for RT-PCR to evaluate cytokine and chemokine levels.
Hematoxylin and Eosin Staining and Histopathological Score
To study the histology of the distal colon and to understand the extent of inflammation, hematoxylin and eosin (H&E) staining was employed. Staining was conducted on formalin-fixed paraffin-embedded distal colonic tissue sections of 5 µm thickness. The tissues were deparaffinized as follows. First slides were heated to 60°C and immersed in xylene (Fisher) for 20 min. These slides were then gradually immersed in solutions of ethanol of decreasing concentrations (100%, 90%, 70%, and 50% vol/vol ethanol) to rehydrate the tissues. Finally, slides were immersed in distilled water. Following the rehydration process, slides were stained with the H&E staining kit (Scytek Laboratories) per the manufacturer’s protocol. Histopathological score was determined in a blinded manner for parameters listed, inflammatory infiltrate, epithelial damage, edema, crypt destruction, erosion and ulceration, and goblet cell damage. A score of 1−3 was assigned where 1 was mild, 2 was moderate, and 3 was severe, based on previously published scoring systems (14).
Periodic Acid-Schiff Staining
Periodic acid-Schiff (PAS) staining was used to determine the levels of mucus and goblet cells in mice distal colonic tissues. Paraffin-embedded distal colonic tissue sections were stained with PAS staining (Sigma) per the manufacturer’s protocol after deparaffinization as described in the section above. PAS reagent binds to glycoproteins (neutral and acidic mucins) and lipids in the secreted mucus and stains goblet cells in magenta.
Cytokine and Chemokine Analysis
RNA was extracted from mouse colonic mucosal samples using a Qiagen RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The quality and quantity of total RNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Extracted RNA was amplified by a Brilliant SYBR Green qRT-PCR Master Mix kit (Agilent, Santa Clara, CA) using mouse gene-specific primers for IL-1β, C-X-C motif chemokine (CXCL)-1, and CCL-3. The relative mRNA levels were expressed as a percentage of control normalized to GAPDH used as an internal control gene.
Myeloperoxidase Activity
Myeloperoxidase (MPO) activity in distal colons of WT and Olfr-78 KO mice was measured as described earlier (16). Briefly, distal colonic tissue from DSS-treated WT and Olfr-78 KO mice was homogenized in hexadecyltrimethylammonium bromide (HTAB) buffer containing 0.5% wt/vol HTAB in potassium phosphate buffer, pH 6.0. The tissue homogenate was centrifuged at 13,000 rpm for 5 min, and clear supernatant was collected. MPO activity was measured by a colorimetric assay involving hydrogen peroxide-dependent oxidation of o-dianisidine dihydrochloride (Sigma) catalyzed by MPO (6). Absorbance was read at 450 nm using a microplate reader (FLUOstar; BMG Labtech, Cary, NC). MPO activity was expressed as units per mg of tissue.
RNA Seq Analysis
Seq analysis was performed at the Northwestern University core facility.
RNA seq library construction.
To generate RNA-sequencing libraries, RNA quality was determined with the Agilent Bioanalyzer 2100, accepting RNA integrity numbers (RIN) of >7 and quantified using Qubit. Directional mRNA libraries were prepared using the Illumina TruSeq mRNA Sample Preparation Kits per the manufacturer’s instructions. Briefly, polyadenylated mRNAs were captured from total RNA using oligo(dT) selection. Next, samples were converted to cDNA by reverse transcription, and each sample was ligated to Illumina-sequencing adapters containing unique barcode sequences. Barcoded samples were then amplified by PCR and the resulting cDNA libraries were quantified using qPCR. Finally, equimolar concentrations of each cDNA library were pooled and sequenced on the Illumina HiSeq 4000.
RNA seq analysis.
The quality of DNA reads, in FASTQ format, was evaluated using FastQC. Reads were trimmed to remove Illumina adapters from the 3′-ends using cutadapt. Trimmed reads were aligned to the Mus musculus genome (mm10) using STAR (11). Read counts for each gene were calculated using htseq-count (2) in conjunction with a gene annotation file for mm10 obtained from Ensembl (http://useast.ensembl.org/index.html). Normalization and differential expression were calculated using DESeq2 (20). The cutoff for determining significantly differentially expressed genes was an false discovery rate-adjusted P value <0.05.
Statistical Analysis
Data are presented as means ± SE. Differences between groups were analyzed by Student’s t test or one-way analysis of variance (ANOVA) followed by Tukey’s test. P value ≤ 0.05 was considered statistically significant.
RESULTS
Olfr-78/OR51E2 RNA is Highly Expressed in Colon of Mouse and Human Intestine
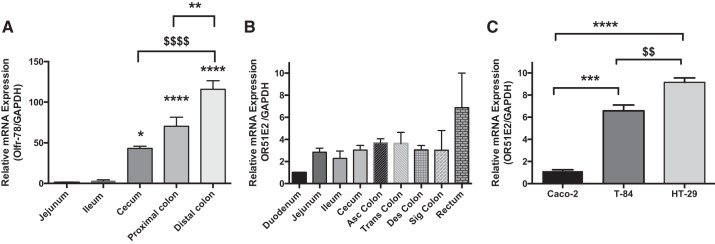
Expression of Olfr-78 RNA in enteroendocrine L cells and mouse intestine has previously been reported by immunostaining and RT-PCR, respectively (13). However, the distribution of Olfr-78 counterpart OR51E2 mRNA along the length of the human intestine has not been investigated. Therefore, we first examined the expression of OR51E2 transcript levels along the length of the human intestine and compared with the Olfr78 mRNA levels in the mouse intestine. We observed that Olfr78 mRNA levels along the length of the mouse intestine were greater in cecum and colon compared with small intestine (duodenum, jejunum, and ileum; see Fig. 1A). In human intestine, almost uniform OR51E2 mRNA levels were observed throughout the intestinal segments (Fig. 1B). Overall, the results of PCR indicated that Olfr-78/OR51E2 is present in both the murine and the human intestine, but in mouse intestine the expression was predominantly higher in colon. Considering these findings, we analyzed OR51E2 levels in Caco-2, T-84, and HT-29 human colonic cancer cell lines to explore the feasibility of using these cells in Olfr-78/OR51E2 signaling studies. Compared with Caco-2 cells, HT-29 and T-84 cells displayed significantly greater levels of OR51E2 transcript (Fig. 1C).
Fig. 1.
Olfactory receptor-78 (Olfr-78)/olfactory receptor family 51, subfamily E, member 2 (OR51E2) expression in mouse and human intestinal samples and colon cancer cell lines. A: total RNA isolated from mucosal scrapings of different regions of mouse intestine was analyzed for Olfr-78 expression by real-time RT-PCR and normalized with GAPDH mRNA (n = 3; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, $$P < 0.01, and $$$$P = 0.0001). B: RNA from different regions of human intestine was extracted and analyzed for OR51E2 expression by real-time RT-PCR and normalized with GAPDH RNA. C: RNA from Caco-2, T-84, and HT-29 colonic epithelial cells was isolated and subjected to real-time RT-PCR using OR51E2-specific primers and normalized with GAPDH mRNA.
Olfr-78 Transcripts Were Significantly Downregulated in Mouse Models of DSS- and TNBS-Induced Colitis
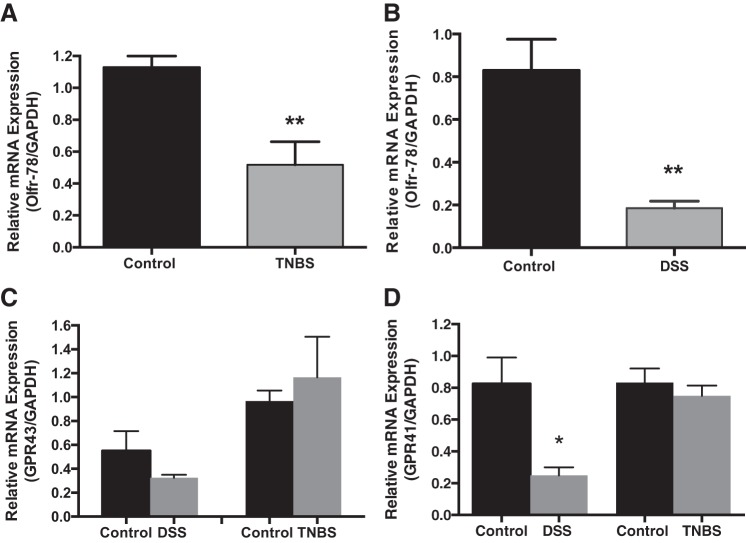
Because of the presence of higher Olfr-78 levels in the colon (Fig. 1), we reasoned that its expression may be regulated in intestinal inflammatory conditions, especially colitis, considering the role of SCFAs and their receptors in colitis. Therefore, we first evaluated Olfr-78 mRNA levels in mouse models of DSS- and TNBS-induced colitis by RT-PCR. As shown in Fig. 2, A and B, Olfr-78 mRNA levels were significantly reduced in distal colons of TNBS- and DSS-treated mice, respectively, compared with control mice. As Olfr-78-related SCFA receptors GPR-41/43 have also been shown to play a role in immune modulation in colitis (24, 45), we also examined their mRNA levels in the colonic tissue of these mice. Whereas no significant change in GPR-43 mRNA was noticed (Fig. 2C), GPR-41 levels were significantly reduced in DSS-induced colitis but not TNBS-induced colitis (Fig. 2D). These results suggest a possible role for Olfr-78 in both DSS- and TNBS-induced intestinal inflammatory conditions.
Fig. 2.
Olfactory receptor-78 (Olfr-78) expression in colitis models. A: colonic Olfr-78 expression is reduced in mouse models of 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis, where real-time RT-PCR was performed on total RNA extracted from mucosal scrapings of distal colons from control and TNBS-treated mice using Olfr-78-specific primers and normalized with GAPDH RNA (n = 4, **P < 0.01). B: colonic Olfr-78 expression is reduced in mouse model of dextran sodium sulfate (DSS)-induced colitis, where real-time RT-PCR was performed on total RNA extracted from mucosal scrapings of distal colons from control and DSS-treated mice using Olfr-78 gene-specific primers and normalized with GAPDH mRNA (n = 4, **P < 0.01). C and D: G protein-coupled receptor (GPR) 43/41 expression is not altered in TNBS- and DSS-induced mouse colitis models. Real-time RT-PCR was performed on RNA isolated from proximal and distal colons of control and DSS- and TNBS-treated mice using GPR-43/41 gene-specific and GAPDH primers (n = 4, *P < 0.05).
Mice Deficient in Olfr-78 Displayed Higher Level of Intestinal Inflammation
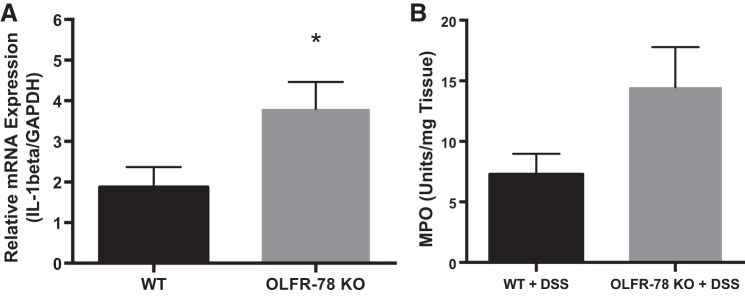
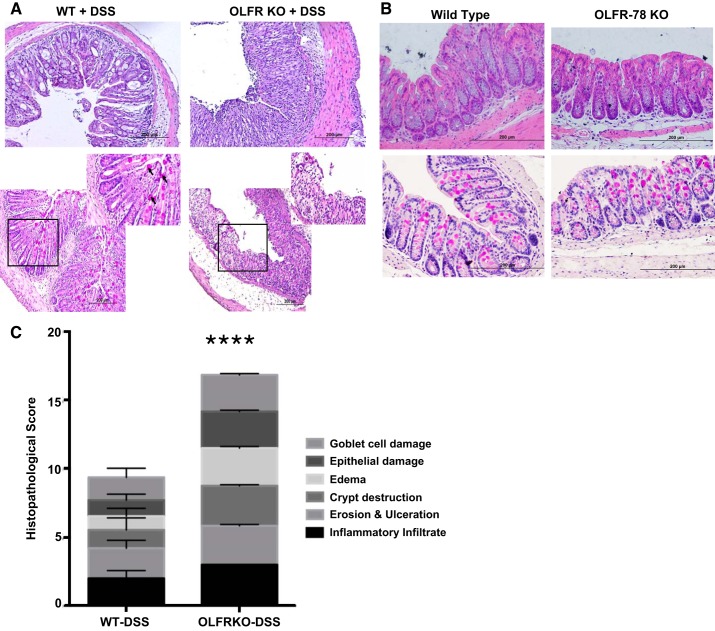
Decreased expression of colonic Olfr-78 transcripts in DSS- and TNBS-treated mice indicated that Olfr78 KO mice may be more susceptible to inflammation. Hence, we induced colitis by administering DSS in drinking water to KO and WT animals. Colonic mRNA expression of proinflammatory cytokine IL-1β (one of the key cytokines known to be induced in DSS colitis) was higher in male Olfr-78 KO mice compared with male WT mice (Fig. 3A). However, colonic mRNA expression of chemokines CXCL-3 and CCL-1 was not altered (data not shown). As a surrogate marker of inflammation, myeloperoxidase (MPO) assay was also performed on these samples to indicate neutrophil infiltration. Although there was a marked increase in the MPO activity in Olfr-78 KO mice compared with WT mice, a statistical significance was not observed (Fig. 3B). The development of colitis was also investigated by histological examination of colonic sections. Colons from KO mice treated with DSS displayed intense inflammatory infiltration and crypt distortion and decreased mucin content (Fig. 4A). Histological examination and mucin content as determined by PAS staining of colons from WT and Olfr-78 KO mice not treated with DSS did not show any differences (Fig. 4B). H&E-stained sections were scored by a blinded observer to assess the inflammatory cell infiltration, crypt distortion, edema, goblet cell damage, and epithelial damage. Colons from KO mice treated with DSS had higher histological damage scores compared with WT mice treated with DSS (Fig. 4C). Because there was a significant reduction of Olfr-78 transcript in mouse models of colitis, we next evaluated the effect of inflammatory agents such as TNF-α on Olfr-78 expression. HT-29 cells (human colonic epithelial cells that express higher levels of OR51E2 compared with other cells tested; Fig. 1C) were treated with and without TNF-α for 24 h, and RNA from these cells was analyzed for OR51E2 expression by RT-PCR. Significantly lower levels of OR51E2 mRNA were observed in TNFα-treated cells but not in control cells (Fig. 5).
Fig. 3.
A: increased expression of IL-1β in colons of olfactory receptor-78 (Olfr-78) knockout (KO) mice treated with dextran sodium sulfate (DSS). Wild-type (WT) and Olfr-78 KO male mice were administered 3% DSS in water for 7 days and RNA from distal colon was isolated for the analysis of proinflammatory cytokine IL-1β expression by real-time RT-PCR (n = 5 for WT and 7 for Olfr-78 KO; *P < 0.05). B: increased myeloperoxidase (MPO) activity in Olfr-78 KO mice treated with DSS compared with WT mice treated with DSS.
Fig. 4.
Structural changes in colons of wild-type (WT) and olfactory receptor-78 (Olfr-78) KO mice treated with dextran sodium sulfate (DSS). A: formalin-fixed distal colon tissues from WT and Olfr-78 knockout (KO) mice treated with DSS were subjected to H&E staining (top) assessing structural changes and Periodic acid-Schiff staining (PAS; bottom) for mucin production by goblet cells. Boxed area in black borders is zoomed in on the top right corner to show mucin staining (n = 3 for WT and 6 for KO). B: same as in A except formalin-fixed distal colon tissues from WT and Olfr-78 KO mice not treated with DSS were used. C: histological scores from H&E- and PAS-stained sections are presented. ****P < 0.0001.
Fig. 5.
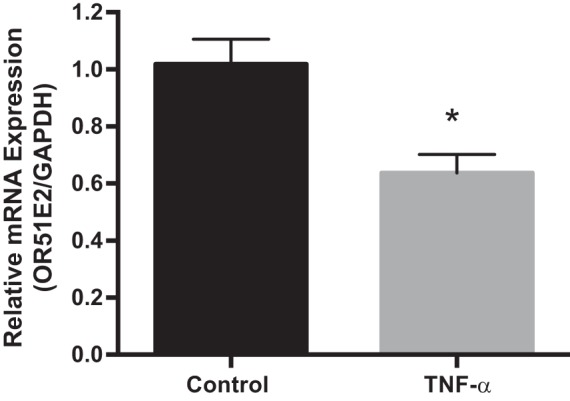
TNF-α treatment reduced olfactory receptor family 51, subfamily E, member 2 (OR51E2) RNA expression in the human colonic cell line. RNA was extracted from control and TNF-α-treated (100 ng/mL) HT-29 colonic cells and analyzed for the expression of OR51E2 by real-time RT-PCR and normalized with GAPDH (n = 3, *P < 0.05).
Microbiome Analysis of Cecal Contents of WT and Olfr-78 KO Mice
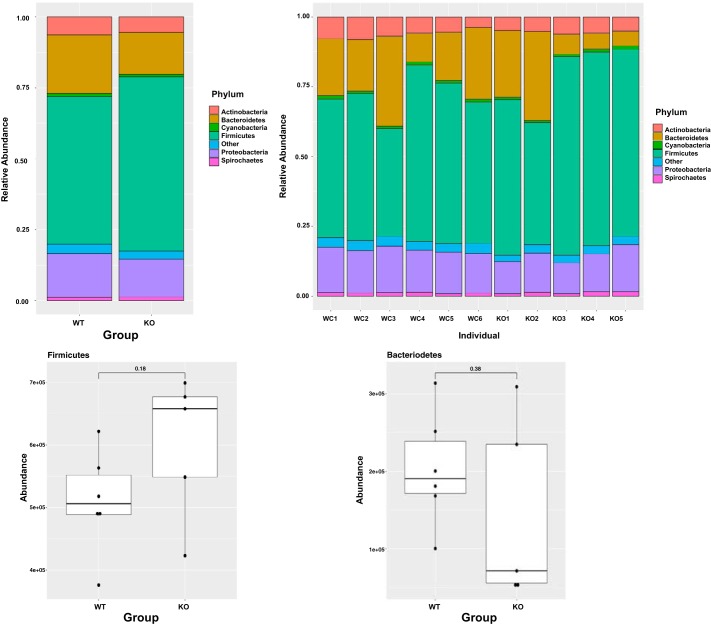
Because intestinal commensal microbes play an essential role in colonic health, we next investigated the microbial population of cecal contents of Olfr-78 WT and KO mice. Although α- and β-diversities of microbial populations were not significantly different between WT and KO groups (data not shown), there were changes in the abundance of bacteria from phyla such as Bacteroidetes, Firmicutes, and proteobacteria. Increased proportion of Firmicutes to Bacteroidetes and decreased abundance of microbes belonging to proteobacteria were noted in the cecal contents of Olfr-78 KO mice compared with WT mice (Fig. 6). We also observed significant changes in few bacterial species belonging to phyla Bacteroidetes, Firmicutes, Actinobacteria, and proteobacteria between WT and KO mice (as reported in Table 2).
Fig. 6.
Top: bar plots representing changes in the microbial abundance at phyla level in cecal contents of wild-type (WT) and olfactory receptor-78 (Olfr-78) knockout (KO) mice. Bottom: box plots showing the differences in the abundance of Firmicutes and Bacteriodetes between WT and Olfr-78 KO mice. Differential testing was conducted by two-sample t test. P < 0.05 was considered significant.
Table 2.
Significant changes in bacterial species from WT and Olfr-78 KO
| No. | Phylum | Species | WT Mean | KO Mean | P Value |
|---|---|---|---|---|---|
| 1 | Bacteroidetes | Bacteroidetes helcogenes | 4,671.98 | 2,161.13 | 0.044 |
| 2 | Bacteroidetes | Prevotella ruminicola | 4,474.86 | 1,762.32 | 0.043 |
| 3 | Actinobacteria | Gordonibacter pamelaeae | 4,053.04 | 222.62 | 0.034 |
| 4 | Actinobacteria | Olsenella uli | 1,605.21 | 702.95 | 0.003 |
| 5 | Chlorobi | Pelodictyon luteolum | 1,016.37 | 270.01 | 0.015 |
| 6 | Actinobacteria | Streptosporangium roseum | 907.35 | 331.49 | 0.014 |
| 7 | Proteobacteria | Alicycliphilus denitrificans | 841.12 | 373.1 | 0.013 |
| 8 | Proteobacteria | Thiocystis violascens | 822.39 | 340.17 | 0.016 |
| 9 | Proteobacteria | [Enterobacter] aerogenes | 822.37 | 423.34 | 0.008 |
| 10 | Proteobacteria | Burkholderia cenocepacia | 725 | 243.49 | 0.018 |
| 11 | Proteobacteria | Burkholderia sp. CCGE1002 | 687.57 | 204.01 | 0.019 |
| 12 | Proteobacteria | Komagataeibacter medellinensis | 681.4 | 195.31 | 0.044 |
| 13 | Firmicutes | Butyrate-producing bacterium SM4/1 | 3,089.02 | 4,667.15 | 0.022 |
| 14 | Proteobacteria | Ralstonia pickettii | 497.22 | 960.68 | 0.017 |
| 15 | Firmicutes | Desulfosporosinus orientis | 314.98 | 836.22 | 0.030 |
| 16 | Firmicutes | Paenibacillus sp. FSL R5-0912 | 213.1 | 667.2 | 0.032 |
| 17 | Actinobacteria | Corynebacterium aurimucosum | 244.52 | 611.99 | 0.030 |
WT, wild type; Olfr78, olfactory receptor-78; KO, knockout.
RNA seq Analysis Revealed Gene Expression Changes in WT and Olfr-78 KO Mice
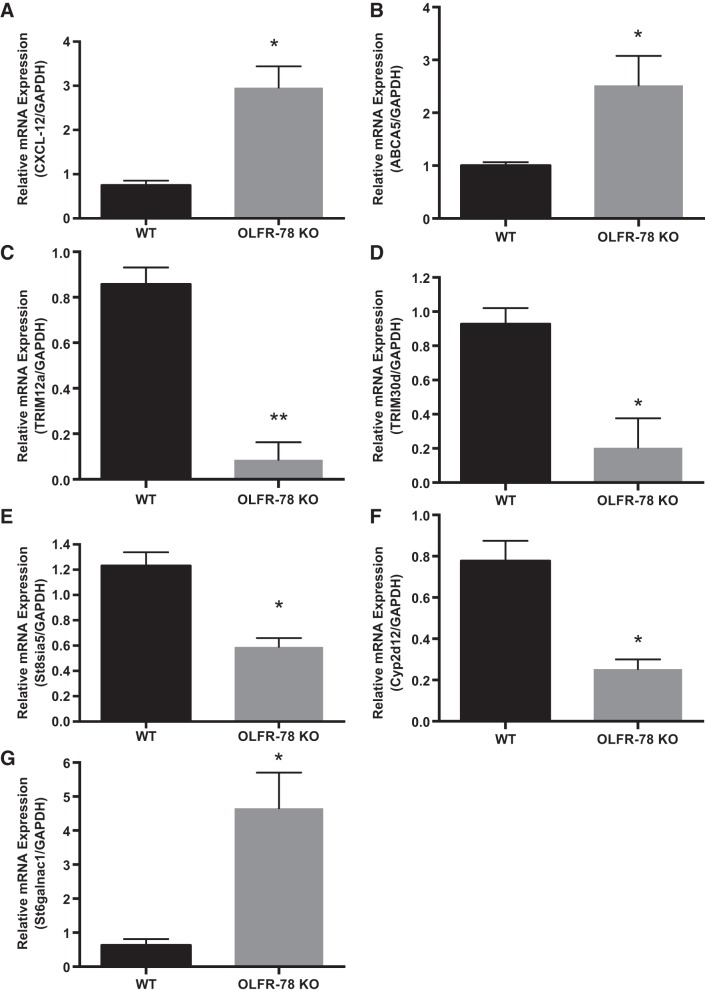
Finally, we explored for genes differentially expressed between WT and Olfr-78 KO mice by RNA-sequencing analysis of distal colon isolated from these mice. The expression of >400 genes was found to be significantly different between the two groups. The key genes that were significantly (log2) changed and are known to have a potential role in intestinal inflammation, immunity, infection, and colorectal cancer (CRC) included: tripartite motif containing protein 12a (Trim12a), tripartite motif containing protein 30d (Trim30d), ST6 N-acetylgalactosaminide α2,6-sialyltransferase 1 (st6galnac1), ATP-binding cassette transporter A5 (abca5), cytochrome P-450, family 2, subfamily d, polypeptide 12 (cyp2d12), chemokine ligand Cxcl12, and ST8 α-N-acetyl-neuraminide α2,8-sialytransferase 5 (st8sia5). Confirmation of these gene expression patterns by real-time PCR is presented in Fig. 7, A–G.
Fig. 7.
Validation of few up- and downregulated genes identified by RNA seq analysis of distal colon tissue of wild-type (WT) and olfactory receptor-78 (Olfr-78) knockout (KO) mice. Total RNA was extracted from mucosal scrapings of distal colons from WT and Olfr-78 KO mice and real-time RT-PCR was performed using the gene-specific primers indicated on y-axis and normalized with GAPDH mRNA (n = 3, *P < 0.05 and **P < 0.01).
DISCUSSION
Despite the fact that Olfr-78 is primarily detected in olfactory tissues, recent reports documented its expression in diverse tissues, such as kidney, lungs, prostate, melanocytes, endothelium, and enteroendocrine L cells of colon (13, 15, 38, 53). However, its functional role in most of these tissues is largely unknown. In this study, we studied differential expression of Olfr-78/OR51E2 mRNA along the length of the mouse and human intestines, respectively. While we observed that Olfr-78/OR51E2 RNA is expressed along the entire length of the mouse and human intestines, Olfr-78 expression in mouse was significantly higher in the colon rather than in small intestine. In addition, OR51E2 also appears to be expressed in colonic epithelial cells as is evident by their expression in Caco-2, HT-29, and T-84 cells (Fig. 1C). However, empirical data on protein expression could not be generated because of lack of reliable antibodies against this receptor. We tested several commercial antibodies using protein extracts from human cell lines and mouse intestinal segments with proper controls and our KO mice, and none were confirmed to be specific. There is a consensus opinion in the field of GPCRs that lack of reliable antibodies against these receptors is a major impediment in the detection of endogenous proteins.
The identification and localization of Olfr-78 receptor in mouse colon is intriguing, since colonic epithelial cells are challenged by a variety of external stimuli that are capable of triggering immune inflammatory responses that could alter the epithelial barrier function. We detected significant reduction in Olfr-78 but not GPR-43 mRNA levels in both DSS- and TNBS-induced mouse models of colitis and significantly lower GPR-41 mRNA levels in DSS-induced but not in TNBS-induced colitis (Fig. 2, A–D). This could be interpreted as Olfr-78 playing a more important role in both chemical (DSS)-induced and immune-based (TNBS) inflammatory conditions in contrast to involvement of GPR-41 in chemical-induced colitis only. Moreover, Olfr-78 KO mice administered with DSS showed increased intestinal inflammatory response, since histological scores revealed marked intestinal epithelial destruction, edema, goblet cell damage, and intense inflammatory infiltration compared with WT mice (Fig. 4, A and C). We also observed significantly higher levels of IL-1β and elevated but not statistically significant MPO activity in colons of these KO mice (Fig. 3, A and B). In this regard, it has been shown that proinflammatory cytokines are elevated in DSS- and TNBS-induced colitis (17), a feature commonly noticed in the inflamed gut of ulcerative colitis patients. Moreover, RNA seq data showed significant reduction in TRIM proteins (which play a role in innate immunity) in Olfr-78 KO mice compared with WT (Fig. 7, C and D). Collectively, these findings suggest that Olfr-78 might play a role in the regulation of immune responses in inflammatory conditions.
The majority of previous studies used GPR-41/43 KO mice to investigate the significance of GPR-41/43 in inflammatory response. For example, Maslowski et al. reported that, in DSS- and TNBS-induced mouse colitis models, GPR-43−/− but not WT mice showed increased morbidity and inflammation as evaluated by histological score of colonic tissue, greater MPO activity, increased CD44+IL17+ T cells in the mesenteric lymph nodes, and elevated levels of proinflammatory cytokines in colon tissue (24). Smith et al. used DSS-induced and T cell transfer models of colitis, respectively, to show that colonic tissue of GPR-43 KO mice harbored elevated proinflammatory and reduced anti-inflammatory markers and decreased T-reg cells (45). Taken together, these studies established that GPR-43 reduces the severity of acute and chronic colitis-induced inflammatory response while GPR-43 KO mice show exacerbated inflammation under these conditions. However, two studies reported contradictory findings with lower immune response and colonic inflammation but increased mortality in knockout mice compared with the control group (25, 44).
Although these studies implicated GPR-43 in inflammatory diseases, their exact role as to whether protective or causative is not clear, and findings are inconsistent between the studies (3). In this context, our identification of significant reduction in Olfr-78 but not GPR-43 mRNA levels in mouse models of colitis bears considerable importance and suggests a number of interesting possibilities. Notably, inflammatory stimuli such as TNF-α, LPS, and granulocyte macrophage-colony-stimulating factor activate GPR-43 promoter leading to its increased expression (4). However, we did not notice any change in GPR-43 RNA levels in colitis, and, in contrast, Olfr-78 levels were downregulated. Our findings imply that there is a negative correlation between Olfr-78 expression and inflammation associated with colitis, and it is likely because of the downregulation of Olfr-78 by inflammatory stimuli. To investigate further whether reduced Olfr-78 expression is a cause or effect of colitis, we analyzed its expression in vitro in the HT-29 colonic cell line treated with inflammatory stimuli such as TNF-α, and, consistent with colitis models, we observed a reduction in the levels of OR51E2 (Fig. 5). In addition, our studies with Olfr78 KO with DSS colitis show enhanced gut inflammation. Therefore, overall our results bear considerable significance and highlight the importance of Olfr-78/OR51E2 in gut inflammation.
Recent evidence suggests that changes in the composition of the gut microbiota are linked to multiple systemic disorders (8, 23). In the current study, sequencing of cecal microbiomes of WT and Olfr-78 KO mice revealed alterations in gut microbial composition (Fig. 6). Olfr-78 KO mice had greater abundance of Firmicutes compared with their WT counterparts. Elevated levels of Firmicutes have been shown to be associated with obesity, diabetes, and inflammation (5, 49). Thus, our data showing increased Firmicutes levels support the increased DSS-induced intestinal inflammatory changes in Olfr-78KO mice. Recently, metagenomic analysis using different cohorts involving normal and colon cancer human subjects indicated that abundance of Gordinobacter bacteria is elevated in normal human subjects, whereas levels of Desulfovibrio species are higher in colon cancer patients. Desulfovibrio bacteria produce choline converting enzyme (Cutc), which converts dietary tryptophan and choline to trimethylamine N-oxide, which is linked to metabolic disorders (47). In this regard, we observed significant decrease in Gordinobacter and significant increase in Desulfosporosinus species of bacteria in KO mice compared to WT mice (Table 2). This probably might explain the increased susceptibility of KO mice to DSS-induced colitis. We have not performed microbiome analysis of cecal samples isolated from DSS-treated WT and Olfr-78 KO mice. However, an interesting study by Lee et al. (19) demonstrated that DSS-treated mice had more abundance of bacteria belonging to phylum Firmicutes over phylum Bacterioidetes. These data correlate with our finding that Olfr-78 KO (not treated with DSS) mice had more Firmicutes than Bacetrioidetes compared with WT mice not treated with DSS. In our future studies, we will investigate the microbiome analysis of cecal contents isolated form WT and Olfr-78 KO mice treated with DSS.
Olfr-78 is similar to GPR-41 and -43, which are also expressed in colonic epithelial and immune cells (18), and GPR-41/43 are also in enteroendocrine L cells (48). GPR-41 and -43 are more well-characterized SCFA receptors than Olfr-78 and have been shown to play essential roles in regulating various cellular physiological processes. For example, GPR-41 and -43 regulate the production of GLP-1 in L cells. Acetate and propionate, agonists of GPR-41/43, reduced GLP-1 secretion in mixed primary cultures from colon of GPR-43 KO mice but not in WT mice. However, with GPR-41 KO mice, the decrease in GLP-1 was not as pronounced as that of GPR-43 KO mice (48). Additionally, GPR-43 KO mice had significantly reduced GLP-1 protein and reduced glucagon and PYY mRNA expression in colonic tissue compared with WT. However, knockout of GPR-41 did not have a significant effect on colonic glucagon or GLP-1 levels. Because GPR-41/43 and Olfr-78 are activated by acetate and propionate, Olfr-78 signaling might also play an important role in regulating GLP-1 and PYY-1 secretion. It is worth noting that GPR-41/43 and Olfr-78 signal through different G protein α-subunits. In contrast to activation of the Giα pathway by GPR-41/43, Olfr-78 stimulates the Gsα pathway, leading to increased cAMP production (43). In this regard, notably, Pluznick et al. showed that Olfr-78 activation increases, whereas GPR-41 activation reduces, blood pressure (38). Chang et al. showed that Olfr-78 activation by lactate in carotid body during hypoxic conditions elevated blood pressure to restore normal blood pressure and prevent hypotension (9). This interplay of GPR-41 and Olfr-78 signaling might switch physiological effects to maintain homeostasis. Because GPR-41/43 and Olfr-78 couple to different Gα subunits and have opposing effects on blood pressure, it is plausible that Olfr-78 and GPR-41 may have disparate physiological effects in colon.
RNA seq analysis of distal colon revealed significant changes in gene expression in KO mice compared with WT mice. Particularly genes involved in infection and immunity, inflammation, and colorectal carcinogenesis (Trim12a, TRIM30d, st6galnac1, st8sia5, cyp2d12, abca5, and cxcl12) were significantly altered between WT and KO mice as shown in Fig. 7. A significant increase in chemokine ligand CXCL-12 is noticed in Olfr-78 KO mice than in WT mice (Fig. 7A). In this regard it is worth noting that CXCL-12 chemokine and its receptor CXCR-4 had been shown to increase proliferation and metastasis of CRC via MAPK/PI3K/activator protein-1 signaling (21). We observed significant increase in ABCA5 RNA in KO mice, which has been shown to play a role in colon cancer development (Fig. 7B and Ref. 33). Our findings show a significant decrease in TRIM12a and -30d (Fig. 7, C and D). Tripartite motif containing proteins (TRIM proteins) belong to the E3 ubiquitin ligase family, are induced by type I and II interferons, and exhibit antiviral and antibacterial properties. They also play a crucial role in innate immunity (35). As shown in Fig. 7E, we observed a significant decrease in st8sia5 levels in KO mice compared with WT. Interestingly, significant reduction of st8sia5 in FoxO3 KO mice, which develop colon tumors, and in mice treated with azoxymethane and dextran sulfate sodium (AOM/DSS), which induce inflammation-mediated colon cancer, was reported (36). Also in this study authors reported that, in inflammatory bowel disease patients, a significant reduction in st8sia5 levels was noticed (36). In the same model of inflammation-induced CRC using AOM/DSS, it has been demonstrated that cyp2d12 levels are significantly elevated, and, in our study, we noticed an increase in cyp2d12 RNA in KO mice (Fig. 7F and Ref. 10). Increased expression of st6galnac1 leads to CRC progression by activating the Akt pathway in conjunction with galectin3 (32). Importantly, we noticed significantly elevated levels of st6galnac1 in Olfr-78 KO mice (Fig. 7G). Thus, cecal microbial changes observed in KO mice correlated with gene expression changes related to intestinal inflammation and colorectal cancer.
Collectively, our data suggests that Olfr-78 plays a role in intestinal inflammation. Moreover, its deficiency leads to altered gut microbial composition and gene expression changes that enhance the susceptibility to intestinal inflammation and possibly CRC progression. Next steps will be to explore how Olfr-78 mediates inflammation and if modulating the receptor can improve inflammatory conditions.
GRANTS
These studies were supported by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development: merit Review Awards: BX002011 (P. K. Dudeja) and BX003382 (B. T. Layden); VA Senior Research Career Scientist Award 1K6BX005242 (P. K. Dudeja); and National Institutes of Health (NIDDK) National Institute of Diabetes and Digestive and Kidney Diseases Grants DK54016, DK81858, DK92441 (P. K. Dudeja), and DK104927 (B. T. Layden).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.K., P.K.D., and B.T.L. conceived and designed research; K.K., A.N.A., S.P., D.J., A.K., Y.C., and Y.X. performed experiments; K.K., A.N.A., S.P., D.J., A.K., Y.C., Y.X., P.K.D., and B.T.L. analyzed data; K.K., P.K.D., and B.T.L. interpreted results of experiments; A.N.A., S.P., D.J., and A.K. prepared figures; K.K. drafted manuscript; P.W.F., D.L.P., P.K.D., and B.T.L. edited and revised manuscript; K.K., A.N.A., S.P., D.J., A.K., Y.C., Y.X., P.W.F., D.L.P., P.K.D., and B.T.L. approved final version of manuscript.
REFERENCES
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169, 2015. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang Z, Ding JL. GPR41 and GPR43 in obesity and inflammation–protective or causative? Front Immunol 7: 28, 2016. doi: 10.3389/fimmu.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ang Z, Er JZ, Ding JL. The short-chain fatty acid receptor GPR43 is transcriptionally regulated by XBP1 in human monocytes. Sci Rep 5: 8134, 2015. doi: 10.1038/srep08134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 8: 42, 2016. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley PP, Priebat DA, Christensen RD, Rothstein G. Measurement of cutaneous inflammation: estimation of neutrophil content with an enzyme marker. J Invest Dermatol 78: 206–209, 1982. doi: 10.1111/1523-1747.ep12506462. [DOI] [PubMed] [Google Scholar]
- 7.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175–187, 1991. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 8.Casén C, Vebø HC, Sekelja M, Hegge FT, Karlsson MK, Ciemniejewska E, Dzankovic S, Frøyland C, Nestestog R, Engstrand L, Munkholm P, Nielsen OH, Rogler G, Simrén M, Öhman L, Vatn MH, Rudi K. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther 42: 71–83, 2015. doi: 10.1111/apt.13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang AJ, Ortega FE, Riegler J, Madison DV, Krasnow MA. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 527: 240–244, 2015. doi: 10.1038/nature15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Robertis M, Arigoni M, Loiacono L, Riccardo F, Calogero RA, Feodorova Y, Tashkova D, Belovejdov V, Sarafian V, Cavallo F, Signori E. Novel insights into Notum and glypicans regulation in colorectal cancer. Oncotarget 6: 41237–41257, 2015. doi: 10.18632/oncotarget.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell 48: 612–626, 2012. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischer J, Bumbalo R, Bautze V, Strotmann J, Breer H. Expression of odorant receptor Olfr78 in enteroendocrine cells of the colon. Cell Tissue Res 361: 697–710, 2015. doi: 10.1007/s00441-015-2165-0. [DOI] [PubMed] [Google Scholar]
- 14.Geboes K, Riddell R, Ost A, Jensfelt B, Persson T, Löfberg R. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut 47: 404–409, 2000. doi: 10.1136/gut.47.3.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelis L, Jovancevic N, Veitinger S, Mandal B, Arndt HD, Neuhaus EM, Hatt H. Functional characterization of the odorant receptor 51E2 in human melanocytes. J Biol Chem 291: 17772–17786, 2016. doi: 10.1074/jbc.M116.734517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JJ, Shajib MS, Manocha MM, Khan WI. Investigating intestinal inflammation in DSS-induced model of IBD. J Vis Exp (60): 3678, 2012. doi: 10.3791/3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145: 396–406.e10, 2013. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 18.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 278: 25481–25489, 2003. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 19.Lee KW, Kim M, Lee CH. Treatment of dextran sulfate sodium-induced colitis with mucosa-associated lymphoid tissue lymphoma translocation 1 inhibitor MI-2 is associated with restoration of gut immune function and the microbiota. Infect Immun 86: e00091-18, 2018. doi: 10.1128/IAI.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J, Su H, Yu B, Guo T, Gong Z, Qi J, Zhao X, Du J. CXCL12 gene silencing down-regulates metastatic potential via blockage of MAPK/PI3K/AP-1 signaling pathway in colon cancer. Clin Transl Oncol 20: 1035–1045, 2018. doi: 10.1007/s12094-017-1821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci USA 101: 2584–2589, 2004. doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22: 589–599, 2017. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461: 1282–1286, 2009. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A, Izawa S, Kondo Y, Ito Y, Tamura Y, Yanamoto K, Noda H, Tanabe A, Okaniwa N, Yamaguchi Y, Iwamoto T, Kasugai K. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm Bowel Dis 19: 2848–2856, 2013. doi: 10.1097/01.MIB.0000435444.14860.ea. [DOI] [PubMed] [Google Scholar]
- 26.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217, 2013. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metwally AA, Dai Y, Finn PW, Perkins DL. WEVOTE: weighted voting taxonomic identification method of microbial sequences. PLoS One 11: e0163527, 2016. doi: 10.1371/journal.pone.0163527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7: 189–200, 2016. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namiki T, Hachiya T, Tanaka H, Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40: e155, 2012. doi: 10.1093/nar/gks678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem 284: 16218–16225, 2009. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nøhr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S, Jones RM, Offermanns S, Schwartz TW. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 154: 3552–3564, 2013. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 32.Ogawa T, Hirohashi Y, Murai A, Nishidate T, Okita K, Wang L, Ikehara Y, Satoyoshi T, Usui A, Kubo T, Nakastugawa M, Kanaseki T, Tsukahara T, Kutomi G, Furuhata T, Hirata K, Sato N, Mizuguchi T, Takemasa I, Torigoe T. ST6GALNAC1 plays important roles in enhancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget 8: 112550–112564, 2017. doi: 10.18632/oncotarget.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtsuki S, Kamoi M, Watanabe Y, Suzuki H, Hori S, Terasaki T. Correlation of induction of ATP binding cassette transporter A5 (ABCA5) and ABCB1 mRNAs with differentiation state of human colon tumor. Biol Pharm Bull 30: 1144–1146, 2007. doi: 10.1248/bpb.30.1144. [DOI] [PubMed] [Google Scholar]
- 34.Ounit R, Wanamaker S, Close TJ, Lonardi S. CLARK: fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics 16: 236, 2015. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozato K, Shin DM, Chang TH, Morse HC III. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol 8: 849–860, 2008. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penrose HM, Cable C, Heller S, Ungerleider N, Nakhoul H, Baddoo M, Hartono AB, Lee SB, Burow ME, Flemington EF, Crawford SE, Savkovic SD. Loss of forkhead box O3 facilitates inflammatory colon cancer: transcriptome profiling of the immune landscape and novel targets. Cell Mol Gastroenterol Hepatol 7: 391–408, 2019. doi: 10.1016/j.jcmgh.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 5: 202–207, 2014. doi: 10.4161/gmic.27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 110: 4410–4415, 2013. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Psichas A, Sleeth ML, Murphy KG, Brooks L, Bewick GA, Hanyaloglu AC, Ghatei MA, Bloom SR, Frost G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int J Obes 39: 424–429, 2015. doi: 10.1038/ijo.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 469: 967–977, 2016. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez M, Siwko S, Zeng L, Li J, Yi Z, Liu M. Prostate-specific G-protein-coupled receptor collaborates with loss of PTEN to promote prostate cancer progression. Oncogene 35: 1153–1162, 2016. doi: 10.1038/onc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal 2: ra9, 2009. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A, Franke A, Ott S, Häsler R, Nikolaus S, Fölsch UR, Rose-John S, Jiang HP, Li J, Schreiber S, Rosenstiel P. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 183: 7514–7522, 2009. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- 45.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573, 2013. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.te Velde AA, Verstege MI, Hommes DW. Critical appraisal of the current practice in murine TNBS-induced colitis. Inflamm Bowel Dis 12: 995–999, 2006. doi: 10.1097/01.mib.0000227817.54969.5e. [DOI] [PubMed] [Google Scholar]
- 47.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, Beghini F, Manara S, Karcher N, Pozzi C, Gandini S, Serrano D, Tarallo S, Francavilla A, Gallo G, Trompetto M, Ferrero G, Mizutani S, Shiroma H, Shiba S, Shibata T, Yachida S, Yamada T, Wirbel J, Schrotz-King P, Ulrich CM, Brenner H, Arumugam M, Bork P, Zeller G, Cordero F, Dias-Neto E, Setubal JC, Tett A, Pardini B, Rescigno M, Waldron L, Naccarati A, Segata N. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 25: 667–678, 2019. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61: 364–371, 2012. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 50.Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients 3: 858–876, 2011. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46, 2014. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia C, Ma W, Wang F, Hua Sb, Liu M. Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene 20: 5903–5907, 2001. doi: 10.1038/sj.onc.1204803. [DOI] [PubMed] [Google Scholar]
- 53.Xu LL, Stackhouse BG, Florence K, Zhang W, Shanmugam N, Sesterhenn IA, Zou Z, Srikantan V, Augustus M, Roschke V, Carter K, McLeod DG, Moul JW, Soppett D, Srivastava S. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res 60: 6568–6572, 2000. [PubMed] [Google Scholar]