Abstract
Aims:
The solute carrier family 2 (SLC2) genes are comprised of 14 members which are essential for the maintenance of glucose uptake and survival of tumour cells. This study was performed to investigate the associations of SLC2 family gene expression with mortality in acute myeloid leukemia (AML).
Methods:
Clinical features and SLC2 family gene expression data were obtained from The Cancer Genome Atlas and Gene Expression Omnibus database. The associations between SLC2 family gene expression and clinicopathologic features were analyzed using linear regression model. Kaplan-Meier survival, univariate, multivariate survival analyses and validation analysis were performed to analyze the associations between SLC2 family gene expression and patients’ overall survival.
Results:
Patient mortality was positively associated with age and cytogenetic risk in AML patients. Kaplan-Meier survival analysis suggested that patients with high SLC2A5 and SLC2A10 expression showed poorer survival than those with low SLC2A5 and SLC2A10 expression. In contrast, patients with high SLC2A13 expression exhibited better prognosis than those with low SLC2A13 expression (P < 0.05 for all cases, log rank test). Multivariate survival analysis and validation analysis confirmed that high expression of SLC2A5 and SLC2A10 and low expression of SLC2A13 were associated with increased mortality (P = 0.00, Odd ratio [OR]:4.05, 95% Confidence Interval [CI]: 1.73-10.22; P = 0.00, OR: 3.66, 95% CI: 1.54-9.25; and P = 0.01, OR: 0.26, 95% CI: 0.09-0.68, respectively).
Conclusion:
SLC family gene expression, such as SLC2A5, SLC2A10 and SLC2A13, was significantly associated with prognosis of AML patients, their expression levels might become useful prognostic biomarkers in AML.
Keywords: Acute myeloid leukemia, SLC2A5, SLC2A10, SLC2A13, Overall survival
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous hematological diseases, characterized by abnormal accumulation of blast cells in the bone marrows and peripheral blood.1 Acute myeloid leukemia is the most common myeloid leukemia; the incidence rate is largely age-related, with a prevalence of 3.8 cases per 100 000 rising to 17.9 cases per 100 000 adults aged 65 years and older. Risk factors of the disease include exposure to ionizing radiation, benzene, and cytotoxic chemotherapy.1 Cytogenetic abnormalities have been widely used as cytogenetic markers for evaluating the prognosis of patients with AML, such as positive prognostic factors T(8;21) and inv(16)/t(16;16) and negative prognostic factor inv(3)/t(3;3).2 Although cytogenetic alterations show clinical utility for risk stratification, they might not be applicable for a large fraction of AML samples without structural abnormalities.3,4 Therefore, identifying new prognostic biomarkers is critical to the survival of patients with AML.
The solute carrier family 2 (SLC2) genes encode glucose transporter (GLUT) proteins, which are members of the major facilitator superfamily of membrane transporters. The GLUT proteins comprise 14 family members that could be stratified into 3 classes based on sequence similarity: class 1 (GLUTs 1-4, 14), class 2 (GLUTs 5, 7, 9, and 11), and class 3 (GLUTs 6, 8, 10, 12, and 13).5 Of the 14 GLUT proteins, the roles of GLUT1 have been well established in a wide range of cancer types. Expression of GLUT1 is associated with aggressive tumor grade and poor survival outcomes in breast cancer6 and thyroid cancer.7 The GLUT1 expression was significantly related to tumor differentiation, lymph node metastasis, tumor size, advanced tumor stages, and overall survival (OS) of patients with lung cancer. The increased expression of GLUT1 was associated with shorter disease-free survival in lung cancer.8 Expression of GLUT1 protein was significantly correlated with advanced tumor stages and poor differentiation in patients with hepatocellular carcinoma. Inhibition of GLUT1 expression impaired the proliferative and migratory capabilities of hepatocellular carcinoma cells.9
Up to date, it remains unknown with respect to the associations among OS, clinical characteristics, and expression of SLC2 family genes in AML. Therefore, this study was conducted to address these issues by a variety of statistical methods using a large set of AML patient data from The Cancer Genome Atlas (TCGA)10 and Gene Expression Omnibus (GEO) database.
Materials and Methods
Acquisition of SLC2 Family Gene Expression and Clinical Feature Data
Since French–American–British classification M3 has distinct biologic features and favorable outcome compared to other subtypes, only non-M3 patients were included in the study. Expression of SLC2 family gene, IDH1, IDH2, DNMT3A, NPM1, FLT3, and CEBPA mutation and clinical feature data of 157 patients with non-M3 AML were obtained from the TCGA database10 (TCGA data set). Clinicopathologic characteristics analyzed in the study included patients’ age, gender, percentage of bone marrow blast cells (PBMBC), cytogenetic risk, CD33, CD34, IDH1, IDH2, DNMT3A, NPM1, FLT3, CEBPA mutation statuses, neoadjuvant treatment, survival status, and follow-up data. The SLC2A2 gene which has no expression values in 90% AML samples was eliminated from the study. To validate the associations of SLC2 family gene expression with OS, SLC2 family gene expression and clinical data of 397 non-M3 AML patients were downloaded from the Tyner study11 (Validation dataset) in the GEO database. Clinicopathologic characteristics in the validation data set included patients’ age, gender, PBMBC, cytogenetic risk, IDH1, DNMT3A, NPM1, FLT3-IDT, and CEBPA mutation statuses, chemotherapy, bone marrow transplant, targeted therapy, survival status, and follow-up data. All patients provided written inform consent. As the data sets included in the study were downloaded from public databases, the study did not need the approval of an ethics committee.
Associations Among Clinical Characteristics, Mortality, and Expression of SLC2 Family Genes
Associations between clinical characteristics, mortality, and expression of SLC2 family genes were analyzed by various statistical methods. Student t test was used to compare the age and PBMBC between patients with AML who died and were alive. Fisher exact test was used to analyze the associations between mortality and patients’ gender, cytogenetic risk, CD33, CD34, IDH1, IDH2, DNMT3A, NPM1, FLT3, CEBPA mutation statuses, chemotherapy, bone marrow transplant, and targeted therapy. Linear regression model was applied to evaluate the associations between clinical features and SLC2 family gene expression. Pearson correlation was used to analyze the correlation between different SLC2 messenger RNA (mRNA) expression. All statistical analyses were performed in R (version 3.2.2), and P < .05 indicates statistical significance. Protein–protein interaction (PPI) network was built by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to characterize the interactions of SLC2 family genes at the protein level.12
Associations Between OS and SLC2 Family Gene Expression
To characterize the associations of SLC2 family gene expression and patients’ OS in the TCGA and validation data sets, receiver operating characteristic (ROC) curves were built with the function of ROC of the R package of pROC.13 The optimal cutoff values were determined by the pROC package, and patients with AML were divided into high and low expression groups based on the cutoff values. “High-expression” group referred to the patients who exhibited higher SLC2 family gene expression levels than the cutoff values, whereas the “low-expression” group was those patients who showed lower expression levels than the cutoff values. Kaplan-Meier survival analysis was performed, and the log-rank test was utilized to compare the difference in survival rates between the high- and low-expression groups using the R package of survival.14 To evaluate the odds ratios (OR) of high SLC2 family gene expression on patients’ OS, univariate and multivariate survival analyses were performed using logistic regression model. Both OR and 95% confidence interval (CI) of OR were extracted from the univariate and multivariate survival analyses; P < .05 indicates statistical significance.
Results
General Characteristics of 157 Patients With AML in the TCGA Data Set
The mean age of 157 patients with AML was 56.04, and the minimum and maximum age were 18 and 88 years, respectively. The average PBMBC was 41.5 at diagnosis. Cytogenetic risk evaluation showed 17, 102, and 36 patients had favorable, intermediate, and poor outcomes, respectively, and the cytogenetic risk for 2 patients was missing. In all, 118 and 107 cancer samples showed CD33- and CD34-positive results, while 13 and 24 AML samples were CD33- and CD34 negative, respectively. In all, 16, 17, 43, 48, 44, and 13 AML samples had IDH1, IDH2, DNMT3A, NPM1, FLT3, and CEBPA mutations, respectively. Forty-three patients with AML underwent neoadjuvant treatment, and 114 patients didn’t have neoadjuvant treatment. On the last day of follow-up, 109 patients with AML died, and 48 were alive. The mean follow-up time was 517.33 days (interquartile range, 153-699 days).
The Associations Between Mortality and Clinicopathologic Characteristics in AML
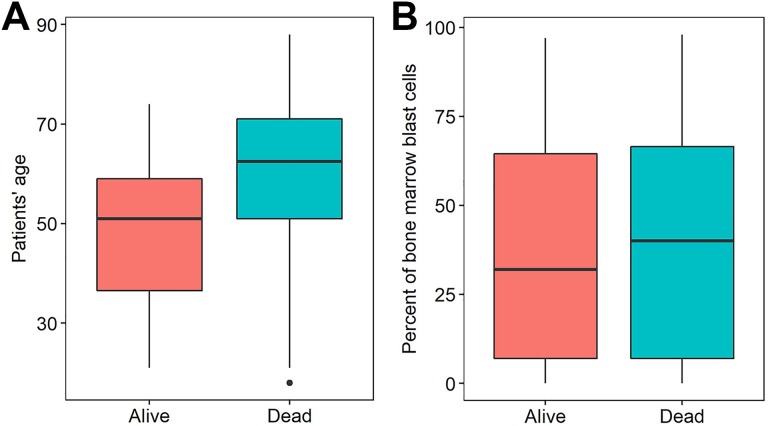
Among the clinicopathologic characteristics, patients with older age exhibited significantly worse mortality than those with younger age (P < .0001, Student t test, Figure 1A and Table 1). Patients’ mortality was significantly associated with cytogenetic risk (P = .001, Fisher exact test; Table 1). However, the remaining factors, gender, CD33, CD34 status, PBMBC, IDH1, IDH2, DNMT3A, NPM1, FLT3, CEBPA mutations, and neoadjuvant treatment did not exhibit a significant association with OS in patients with AML (P > .05 for all cases, Fisher exact test or Student t test; Figure 1B and Table 1).
Figure 1.
Patients’ age (A) and percentage of bone marrow blast cells (B) were significantly different between patients with AML who were died or alive. AML indicates acute myeloid leukemia.
Table 1.
Association Between the Clinicopathologic Characteristics and Patients’ Mortality in Patients With AML.
| Variables | Group | Alive | Dead | P Value | Statistical Method |
|---|---|---|---|---|---|
| Age | 49.63 | 58.86 | .00 | Student t test | |
| PBMBC | 44.25 | 40.28 | .48 | Student t test | |
| Gender | Female | 21 | 50 | .86 | Fisher exact test |
| Male | 27 | 59 | |||
| CD33 | Negative | 5 | 8 | .53 | Fisher exact test |
| Positive | 34 | 84 | |||
| CD34 | Negative | 9 | 15 | .63 | Fisher exact test |
| Positive | 34 | 73 | |||
| Cytogenetic risk | Favorable | 10 | 7 | .03 | Fisher exact test |
| Intermediate | 30 | 72 | |||
| Poor | 8 | 28 | |||
| IDH1 mutation | Mutant | 7 | 9 | .26 | Fisher exact test |
| Wild-type | 41 | 100 | |||
| IDH2 mutation | Mutant | 5 | 12 | 1 | Fisher exact test |
| Wild-type | 43 | 97 | |||
| DNMT3A mutation | Mutant | 10 | 33 | .25 | Fisher exact test |
| Wild-type | 38 | 76 | |||
| NPM1 mutation | Mutant | 15 | 33 | 1 | Fisher exact test |
| Wild-type | 33 | 76 | |||
| CEBPA mutation | Mutant | 4 | 9 | 1 | Fisher exact test |
| Wild-type | 44 | 100 | |||
| FLT3 mutation | Mutant | 12 | 32 | .7 | Fisher exact test |
| Wild-type | 36 | 77 | |||
| Neoadjuvant treatment | Yes | 12 | 31 | .7 | Fisher exact test |
| No | 36 | 78 |
Abbreviations: AML, acute myeloid leukemia; PBMBC, percentage of bone marrow blast cells.
The Associations Between Clinicopathologic Characteristics and SLC2 Family Gene Expression in AML
A linear regression model was applied to study the associations between clinicopathologic characteristics and mRNA expression values of SLC2 family genes. Patients’ age was positively correlated with SLC2A5, SLC2A8, and SLC2A10 mRNA expression. The PBMBC was negatively correlated with SLC2A1, SLC2A3, SLC2A4, SLC2A6, SLC2A9, SLC2A10, and SLC2A14 expression (Table 2). CD33 status was negatively associated with SLC2A4, SLC2A7, and SLC2A14. CD34 status was positively associated with SLC2A1 and negatively associated with SLC2A9 and SLC2A13 expression (Table 2). Cytogenetic risk was positively associated with SLC2A1, SLC2A4 and SLC2A5 and negatively associated with SLC2A7 and SLC2A13 expression (Table 2). IDH1 mutation was positively associated with SLC2A13 expression. NPM1 mutation was positively associated with SLC2A13 and negatively associated with SLC2A4, SLC2A7, SLC2A12, and SLC2A14 expression (Table 2). CEBPA mutation was positively associated with SLC2A1 and negatively associated with SLC2A8 expression (Table 2). FLT3 mutation was negatively correlated with SLC2A1, SLC2A4, and SLC2A7 expression (Table 2). Neoadjuvant treatment was negatively correlated with SLC2A1 and SLC2A4 expression (Table 2).
Table 2.
Linear Regression Analysis Between Clinicopathologic Characteristics and SLC2 Family Gene Expression in Patients With AML.a
| Gene | Age | Gender (male) | PBMBC | CD33 | CD34 | Cytogenetic Risk | IDH1 Mutation | IDH2 Mutation | DNMT3A Mutation | NPM1 Mutation | CEBPA Mutation | FLT3 Mutation | Neoadjuvant Treatment |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC2A1 | - | + | + | + | - | - | |||||||
| SLC2A3 | - | ||||||||||||
| SLC2A4 | - | - | + | — | -- | - | |||||||
| SLC2A5 | + | ++ | |||||||||||
| SLC2A6 | - | ||||||||||||
| SLC2A7 | - | - | - | - | |||||||||
| SLC2A8 | + | - | |||||||||||
| SLC2A9 | — | - | |||||||||||
| SLC2A10 | ++ | -- | |||||||||||
| SLC2A11 | |||||||||||||
| SLC2A12 | - | ||||||||||||
| SLC2A13 | - | — | - | + | +++ | ||||||||
| SLC2A14 | — | - | - |
Abbreviations: AML, acute myeloid leukemia; SLC2, solute carrier family 2; PBMBC, percentage of bone marrow blast cells.
a +, ++, +++ denote positive correlation with P value < .05, P value < .01, and P value < .001, respectively. -, --, --- stand for negative correlation with P value < .05; P value < .01, and P value < .001, respectively.
The Gene–Gene and Protein–Protein Interactions of SLC2 Family Genes
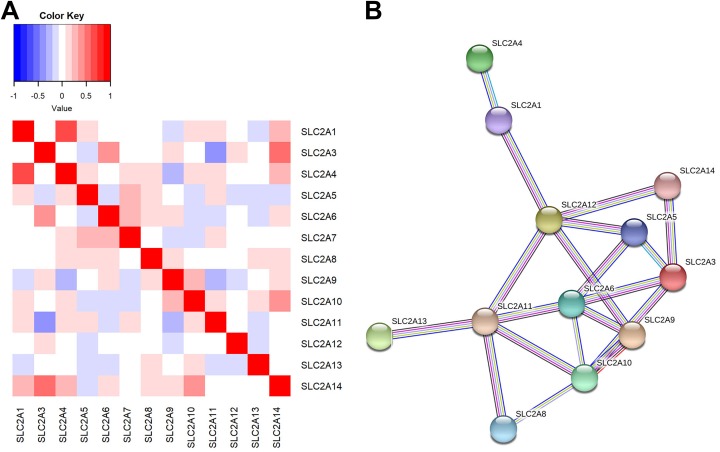
In order to characterize the coexpression between different SLC2 family gene expression in AML, the correlations of SLC2 family gene expression were analyzed. The expression of SLC2 family genes was highly correlated; of the 13 SLC2 family members, SLC2A7 was significantly coexpressed with SLC2A4, SLC2A5, SLC2A6, SLC2A9, and SLC2A11. SLC2A4, SLC2A6, SLC2A9, SLC2A11, and SLC2A14 showed significant correlation with the expression of 4 other family members (P < .05 for all cases, Pearson correlation; Figure 2A). Whereas, no significant correlation was observed between SLC2A12 and other SLC2 family members (P > .05 for all cases, Pearson correlation; Figure 2A). We also applied STRING to develop the PPI network for SLC2 family genes. The PPI network comprised 12 nodes and 19 edges, with a median node degree of 3.17 (Figure 2B). As expected, the PPI network exhibited more interactions than the randomly imputed interactions using similar size of proteins (PPI enrichment P value <.0001). The gene–gene and PPI networks indicated that the SLC2 family genes were coexpressed and exhibited extensive homology at the protein level.
Figure 2.
The solute carrier family 2 gene–gene and protein–protein interaction networks. A, Gene–gene interaction network of SLC2 family genes. B, Protein–protein interaction network of SLC2 family genes. Network nodes and edges represent proteins and protein–protein associations, respectively. Light blue lines represent known interactions from the curated Kyoto Encyclopedia of Genes and Genomes database, purple lines represent experimentally determined protein–protein interactions, green lines denote genes that are frequently observed in each other’s genomic neighborhood, black lines stand for genes where expression are correlated across a large number of experiments, dark blue lines indicate gene families with similar occurrence patterns across genomes, and light gray lines indicate proteins with amino acid sequence similarity. SLC2 indicates the solute carrier family 2.
Survival Analyses Between Patient Mortality and SLC2 Family Gene Expression in AML
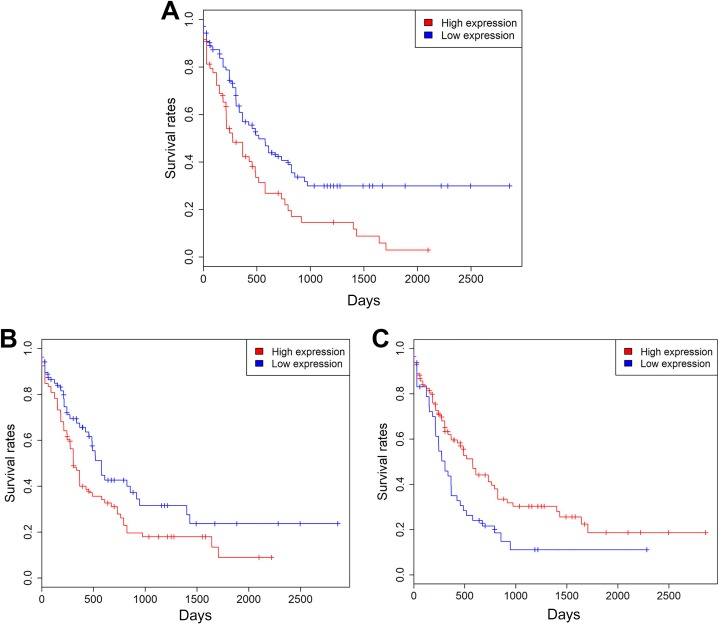
To evaluate the predictive capability of SLC2 family gene expression for patients’ survival, ROC curves were built with the R package of pROC. Then, the 157 patients with AML in the TCGA data set were divided into low- and high-expression groups based on cutoff values. Kaplan-Meier survival analysis suggested that patients with high expression levels of SLC2A4, SLC2A12, and SLC2A13 exhibited a favorable prognosis. In contrast, patients with high expression levels of SLC2A5, SLC2A8, and SLC2A10 were associated with a poor prognosis (P < .05 for all cases, log rank test; Figure 3 and Supplementary Table 1). Univariate analysis using logistic regression model showed that decreased SLC2A4 and SLC2A13 expression was significantly associated with increased mortality (P = .04, OR:0.47, 95% CI: 0.23-0.95; P = .01, OR: 0.34, 95% CI: 0.14-0.75, respectively). Increased SLC2A5 and SLC2A10 expression was associated with increased mortality (P = .00, OR: 3.42, 95% CI: 1.63-7.69; P = .01, OR: 2.59, 95% CI: 1.3-5.33, respectively). Then, multivariate analyses were applied between patients’ survival and the mortality-associated features, including patients’ age, cytogenetic risk, and SLC2A4, SLC2A5, SLC2A10, and SLC2A13 expression levels. Multivariate survival analyses confirmed that high SLC2A5 and SLC2A10 expression and low SLC2A13 expression were associated with increased mortality (P = .00, OR: 4.05, 95% CI: 1.73-10.22; P = .00, OR: 3.66, 95% CI: 1.54-9.25; and P = .01, OR: 0.26, 95% CI: 0.09-0.68, respectively; Table 3).
Figure 3.
Kaplan-Meier survival analysis shows SLC2A5 (A), SLC2A10 (B), and SLC2A13 (C) expression levels were significantly associated with clinical outcomes in the TCGA data set. TCGA indicates The Cancer Genome Altas.
Table 3.
Multivariate Analyses Between Patients’ Overall Survival and the Mortality-Associated Features and SLC2A4, SLC2A5, SLC2A10, and SLC2A13 Expression Levels.
| Clinical Features | OR (95% CI) | P Value |
|---|---|---|
| Age | 1.04 (1.01-1.06) | .01 |
| Cytogenetic risk | 1.17 (0.56-2.51) | .67 |
| SLC2A4 | 0.52 (0.23-1.15) | .11 |
| SLC2A5 | 4.05 (1.73-10.22) | .00 |
| SLC2A10 | 3.66 (1.54-9.25) | .00 |
| SLC2A13 | 0.26 (0.09-0.68) | .01 |
Abbreviations: OR, odd ratio; CI, confidence interval.
Validation Analyses
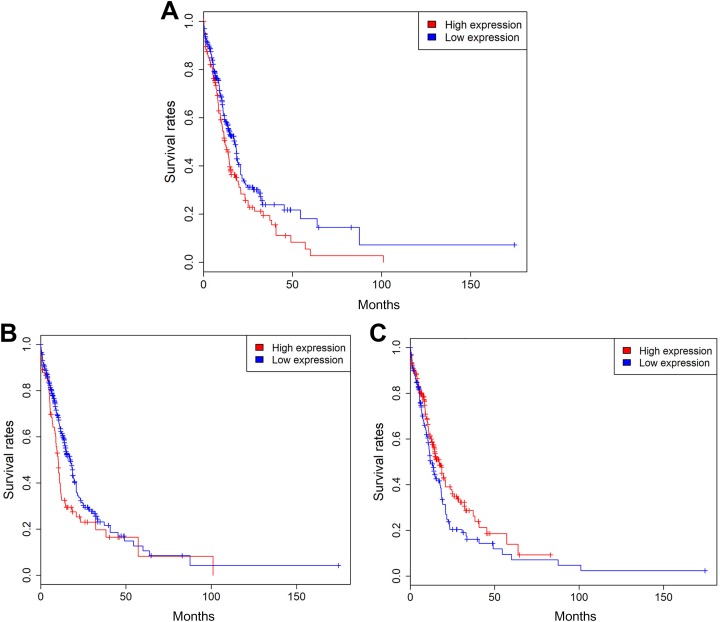
In order to further validate the findings mentioned earlier, the associations among SLC2 family gene expression and mortality, clinical factors were analyzed in 397 non-M3 AML samples of the validation data set. The detailed clinical characteristics of patients with AML in the validation set are shown in Supplementary Table 2. As expected, patient’s age, cytogenetic risk, chemotherapy, bone marrow transplant, and targeted therapy exhibited a significant association with OS in patients with AML (P < .05 for all cases, Fisher exact test or Student t test, Supplementary Table 2). The other factors PBMBC, gender, IDH1, DNMT3A, NPM1, FLT3-IDT, and CEBPA mutation statuses did not show significant association with OS in patients with AML (P > .05 for all cases, Fisher exact test or Student t test; Supplementary Table 2). The associations between SLC2 gene expression and clinical factors are presented in Supplementary Table 3. Many SLC2 genes were significantly correlated with clinical characteristics in patients with AML (P < .05 for all cases, linear regression model; Supplementary Table 3). Kaplan-Meier survival analysis suggested that patients with high expression levels of SLC2A3, SLC2A4, SLC2A13, and SLC2A14 exhibited a favorable prognosis. In contrast, patients with high expression of SLC2A5 and SLC2A10 were associated with a poor prognosis (P < .05 for all cases, log rank test; Figure 4). Univariate analysis using logistic regression model showed that decreased SLC2A13 expression was significantly associated with increased mortality (P = .00, OR: 0.54, 95% CI: 0.35-0.81). Increased SLC2A5 and SLC2A10 expression was associated with increased mortality (P = .00, OR:2.04, 95% CI: 1.31-3.21; P = .02, OR:1.88, 95% CI: 1.12-3.22). Then, multivariate analyses were applied between patients’ survival and the mortality-associated features, including patients’ age, cytogenetic risk, chemotherapy, bone marrow transplant, targeted therapy, and SLC2A5, SLC2A10, and SLC2A13 expression levels. Multivariate survival analyses confirmed that high SLC2A10 expression was associated with increased mortality (P = .03, OR: 1.97, 95% CI: 1.07-3.73; Supplementary Table 4).
Figure 4.
Kaplan-Meier survival analysis shows SLC2A5 (A), SLC2A10 (B), and SLC2A13 (C) expression levels were significantly associated with clinical outcomes in the validation data set.
Discussion
In the present study, we investigated the associations between mortality, clinicopathologic characteristics, and SLC2 family gene expression in AML. As expected, we found patients’ age and cytogenetic risk were significantly associated with mortality in patients with AML. Recent study has revealed IDH1, IDH2, DNMT3A, NPM1, FLT3-IDT, and CEBPA mutations and mixed lineage leukaemia translocations improve risk stratification for patients with AML.15-17 However, IDH1, IDH2, DNMT3A, NPM1, FLT3-IDT, and CEBPA mutations were not associated with mortality in this study. This might be because the number of patients with AML is limited, and the follow-up time is relatively short; therefore, a large number of AML samples with long-term follow-up might be needed to validate the associations between IDH1, IDH2, DNMT3A, NPM1, FLT3-IDT, CEBPA mutation and OS in AML.
Many SLC2 family members are significantly correlated with clinical features of patients with AML. We found 3 genes, SLC2A5, SLC2A10, and SLC2A13, showed significant associations with mortality, which might have clinical values. SLC2A10 encodes a member of the GLUT family. The encoded protein, GLUT10, plays an important role in the regulation of glucose homeostasis. Mutations in this gene have been associated with arterial tortuosity syndrome18. To date, the role of SLC2A10 in AML remains unclear, and this is the first study that reported the association of SLC2A10 with OS in AML. The increased SLC2A10 expression enhances uptake and utilization of glucose in AML cells, leading to increased viability of AML cells and inferior OS of patients with AML.
SLC2A5 encodes the protein of GLUT5, which plays a major role in the fructose transportation in mammalian cells.19 Overexpression of SLC2A5 has been observed in lung adenocarcinoma,20 Philadelphia chromosome-positive acute lymphoblastic leukemia,21 and AML.22 In line with the finding of this study, patients with overexpression of SLC2A5 have poor prognosis in lung cancer20 and AML.22 The increased expression in the sugar transporter GLUT5 encoded by SLC2A5 enhanced uptake and utilization of fructose in the primary leukemic cells of patients with AML, which caused enhanced proliferation of AML cells and inferior OS. Expression of SLC2A5 was significantly correlated with grade of differentiation, pelvis invasion, and breaking capsule in clear cell carcinoma.23 Silencing SLC2A5 expression inhibits cell proliferation, invasion, and induces cellular apoptosis. Enhanced expression of SLC2A5 increases cell proliferation, invasion, and tumorigenic capability in lung cancer.20
SLC2A13, as a proton (H+) myo-inositol transporter24, has an important role in the metabolism of phosphatidyl inositol and could accomplish tumorigenic effect by influencing the function of Akt.25 SLC2A13 is more frequently present in lung cancer samples with good differentiation and without lymph node invasion.26 SLC2A13 is consistently increased in the cancer stem cells in oral squamous cell carcinoma, it might function as a potential marker for cancer stem cells.27 So far, there is no study regarding the involvement of SLC2A13 in AML, and for the first time, our study reported increased expression of SLC2A13 was associated with decreased mortality in patients with AML. As SLC2A13 accomplishes tumorigenic effect by influencing the function of Akt, the impact of SLC2A13 expression on the OS of patients with AML is possibly mediated by regulating the function of Akt; however, the detailed mechanism still needs further studies. SLC2A5, SLC2A10, and SLC2A13 expression profiling may have certain advantage and clinical applicability in patients with AML when compared to prognosis assessment by either cytogenetics or mutation profiling which are limited to a fraction of patients with AML. Patients with AML with low expression of SLC2A13 and high expression of SLC2A5 and SLC2A10 are expected to have an inferior prognosis. Therefore, more aggressive treatment or frequent follow-up may be needed for these patients. SLC2A5, SLC2A10, and SLC2A13 may also provide promising cancer targets for patients with AML. For instance, drugs inhibiting SLC2A5 gene significantly suppressed fructose-induced proliferation, colony growth, and migration of AML cells with enhanced fructose utilization.22
Conclusion
In conclusion, high SLC2A13 expression is associated with decreased mortality, whereas, high SLC2A5 and SLC2A10 expression is associated with increased mortality in patients with AML. The solute carrier family 2 gene expression represents a potentially prognostic biomarker to predict survival in AML.
Supplemental Material
S_Tables for The Solute Carrier Family 2 Genes Are Potential Prognostic Biomarkers in Acute Myeloid Leukemia by Binbin Lai, Yanli Lai, Yanli Zhang, Miao Zhou, Lixia Sheng and Guifang OuYang in Technology in Cancer Research & Treatment
Abbreviations
- AML
acute myeloid leukemia
- CI
confidence interval
- GEO
Gene Expression Omnibus
- GLUT
glucose transporter
- mRNA
messenger RNA
- OR
odd ratio
- OS
overall survival
- PBMBC
percentage of bone marrow blast cells
- PPI
protein–protein interaction
- ROC
receiver operating characteristic
- SLC2
the solute carrier family 2
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TCGA
The Cancer Genome Altas.
Footnotes
Authors’ Note: The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was funded by the traditional Chinese medicine administration of Zhejiang Province (Grant No. 2015ZZ018), the National Science Foundation of Zhejiang Province (Grant No. LY17H160005).
Supplemental Material: Supplemental material for this article is available online.
ORCID iDs: Miao Zhou  https://orcid.org/0000-0002-5463-4784
https://orcid.org/0000-0002-5463-4784
Guifang OuYang  https://orcid.org/0000-0002-0915-2372
https://orcid.org/0000-0002-0915-2372
References
- 1. Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. [DOI] [PubMed] [Google Scholar]
- 2. Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18(2):115–136. doi:10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 3. Walter MJ, Payton JE, Ries RE, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci U S A. 2009;106(31):12950–12955. doi:10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bullinger L, Krönke J, Schön C, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010;24(2):438–449. doi:10.1038/leu.2009.263. [DOI] [PubMed] [Google Scholar]
- 5. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34(2):121–138. doi:10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang SS, Chun YK, Hur MH, et al. [Expression of glucose transporter 1 in human breast carcinoma and its clinical significance]. Sichuan da xue xue bao Yi xue ban J Sichuan Univ Med Sci Ed. 2002;93(10):1123–1128. http://www.ncbi.nlm.nih.gov/pubmed/12417042. [Google Scholar]
- 7. Chai YJ, Yi JW, Oh SW, et al. Upregulation of SLC2 (GLUT) family genes is related to poor survival outcomes in papillary thyroid carcinoma: analysis of data from the Cancer Genome Atlas. Surgery. 2017;161(1):188–194. doi:10.1016/j.surg.2016.04.050. [DOI] [PubMed] [Google Scholar]
- 8. Zhang B, Xie Z, Li B. The clinicopathologic impacts and prognostic significance of GLUT1 expression in patients with lung cancer: a meta-analysis. Gene. 2019;689:76–83. doi:10.1016/j.gene.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 9. Amann T, Maegdefrau U, Hartmann A, et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol. 2009;174(4):1544–1552. doi:10.2353/ajpath.2009.080596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi:10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. doi:10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016;45(D1):D362–D368. doi:10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77 doi:10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fox J. Cox proportional-hazards regression for survival data—the Cox proportional-hazards model. Most. 2002;2008(June):1–18. doi:10.1016/j.carbon.2010.02.029. [Google Scholar]
- 15. Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi:10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin LI, Chen CY, Lin DT, et al. Characterization of CEBPA mutations in acute myeloid leukemia: Most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res. 2005;11(4):1372–1379. doi:10.1158/1078-0432.CCR-04-1816. [DOI] [PubMed] [Google Scholar]
- 17. Fathi AT, Chen Y-B. Treatment of FLT3-ITD acute myeloid leukemia. Am J Blood Res. 2011;1(2):175–189. http://www.ncbi.nlm.nih.gov/pubmed/22432079%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC3301423. [PMC free article] [PubMed] [Google Scholar]
- 18. Ritelli M, Chiarelli N, Dordoni C, et al. Arterial Tortuosity Syndrome: Homozygosity for two novel and one recurrent SLC2A10 missense mutations in three families with severe cardiopulmonary complications in infancy and a literature review. BMC Med Genet. 2014;15(1). doi:10.1186/s12881-014-0122-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Douard V, Ferraris RP. The role of fructose transporters in diseases linked to excessive fructose intake. J Physiol. 2013;591(2):401–414. doi:10.1113/jphysiol.2011.215731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weng Y, Fan X, Bai Y, et al. SLC2A5 promotes lung adenocarcinoma cell growth and metastasis by enhancing fructose utilization. Cell Death Discov. 2018;4(1):38 doi:10.1038/s41420-018-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao P, Huang J, Zhang D, et al. SLC2A5 overexpression in childhood philadelphia chromosome-positive acute lymphoblastic leukaemia. Br J Haematol. 2018;183(2):242–250. doi:10.1111/bjh.15580. [DOI] [PubMed] [Google Scholar]
- 22. Chen WL, Wang YY, Zhao A, et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30(5):779–791. doi:10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Medina Villaamil V, Aparicio Gallego G, Valbuena Rubira L, et al. Fructose transporter GLUT5 expression in clear renal cell carcinoma. Oncol Rep. 2011;25(2):315–323. doi:10.3892/or.2010.1096. [DOI] [PubMed] [Google Scholar]
- 24. Uldry M, Ibberson M, Horisberger JD, Chatton JY, Riederer BM, Thorens B. Identification of a mammalian H+-myo-inositol symporter expressed predominantly in the brain. EMBO J. 2001;20(16):4467–4477. doi:10.1093/emboj/20.16.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brognard J, Clark AS, Ni Y, Dennis PA. Akt/pbotein kinace B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61(10):3986–3997. [PubMed] [Google Scholar]
- 26. Bankovic J, Stojsic J, Jovanovic D, et al. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer. 2010;67(2):151–159. doi:10.1016/j.lungcan.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 27. Lee DG, Lee JH, Choi BK, et al. H+-myo-inositol transporter SLC2A13 as a potential marker for cancer stem cells in an oral squamous cell carcinoma. Curr Cancer Drug Targets. 2011;11(8):966–975. doi:10.2174/156800911797264752 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S_Tables for The Solute Carrier Family 2 Genes Are Potential Prognostic Biomarkers in Acute Myeloid Leukemia by Binbin Lai, Yanli Lai, Yanli Zhang, Miao Zhou, Lixia Sheng and Guifang OuYang in Technology in Cancer Research & Treatment