Abstract
Background
Understanding factors driving virological failure, including the contribution of HIV drug resistance mutations (DRM), is critical to ensuring HIV treatment remains effective. We examine the contribution of drug resistance mutations for low viral suppression in HIV-positive participants in a population-based sero-prevalence survey in rural South Africa.
Methods
We conducted HIV drug resistance genotyping and ART analyte testing on dried blood spots (DBS) from HIV-positive adults participating in a 2014 survey in North West Province. Among those with virologic failure (> 5000 copies/mL), we describe frequency of DRM to protease inhibitors (PI), nucleoside reverse transcriptase inhibitors (NRTI), and non-nucleoside reverse transcriptase inhibitors (NNRTI), report association of resistance with antiretroviral therapy (ART) status, and assess resistance to first and second line therapy. Analyses are weighted to account for sampling design.
Results
Overall 170 DBS samples were assayed for viral load and ART analytes; 78.4% of men and 50.0% of women had evidence of virologic failure and were assessed for drug resistance, with successful sequencing of 76/107 samples. We found ≥1 DRM in 22% of participants; 47% were from samples with detectable analyte (efavirenz, nevirapine or lopinavir). Of those with DRM and detectable analyte, 60% showed high–level resistance and reduced predicted virologic response to ≥1 NRTI/NNRTI typically used in first and second-line regimens.
Conclusions
DRM and predicted reduced susceptibility to first and second-line regimens were common among adults with ART exposure in a rural South African population-based sample. Results underscore the importance of ongoing virologic monitoring, regimen optimization and adherence counseling to optimize durable virologic suppression.
Keywords: South Africa; HIV drug resistance; Surveillance, viral suppression; Virological failure; Antiretroviral therapy (ART); ART adherence
Background
The UNAIDS HIV epidemic targets for detection, sustained antiretroviral therapy (ART), and viral suppression propose that overall community viral suppression should reach 73% by 2020 [1]. With testing and treatment increasingly available and universal in sub-Saharan Africa, home to the majority of HIV cases and greatest need for treatment scale up [2], meeting this goal should be attainable, particularly as ART has increasingly reduced morbidity and mortality and has been demonstrated to substantially reduce further transmission [3–6]. To reap the benefits of ART and achieve widespread viral suppression, however, both access to and consistent adherence to medication is critical for achieving durable viral suppression and preventing drug resistance [3, 7–10].
Among patients in clinical research cohorts in sub-Saharan Africa with access to virologic monitoring, viral suppression at 12 months has been estimated between 84 and 90% among those on ART [11–13]. In South Africa, findings of the National HIV Prevalence, Incidence, Behaviour and Communication Survey 2017 similarly estimated that 89.9 and 82.1% of females and males on ART were virally suppressed [14]. Less is known about prevalence of viral suppression in the general population, particularly in rural areas, where monitoring is less consistent [15]. One population-based study covering 32 rural Kenyan and Ugandan communities noted that 45% of HIV-positive individuals had evidence of viral suppression prior to intensive interventions to improve ART initiation [16]. The recent Universal Test and Treat trial in Kwa Zulu Natal similarly noted high viral suppression rates for those on ART (85%), but an overall suppression rate of 49% among all PLHIV in 2016 [17], lower than the 2016–2017 PopART trial estimates of 63–72% virally suppressed in community cohorts [18].
While known factors contribute to virological failure (intermittent adherence to medication resulting in non-suppressive drug levels in a setting of ongoing viral replication, transmitted resistance), it remains unknown what proportion of those undergoing virological failure can be attributed to each factor. Studies in sub-Saharan Africa have found drug resistance mutations in 6–14% of ART-naïve patients [11, 19, 20], and 84–89% of those with virological failure who initiated ART ≥12 months prior [19, 21]. Results from a systematic review and meta-analysis estimated a prevalence of pretreatment non-nucleotide reverse-transcriptase inhibitor (NNRTI) resistance of 11% in southern Africa [22]. Results from one of few population-based studies in South Africa indicate that transmitted resistance is increasing; from 0% in 2010, to 5 and 7% in 2011 and 2012, respectively [23]. Understanding factors driving virological failure, including the contribution of both pre-treatment drug resistance and acquired resistance, is critical to ensuring treatment remains effective and that existing first-line regimens can be preserved. Data are scarce on the prevalence of genotypic resistance in rural areas of Sub-Saharan Africa [15] and rarely is genotypic resistance data available from population-based sampling, coincident with both reported adherence and known ART exposure.
We conducted a population-based bio-behavioral survey in 2014 to characterize the HIV care continuum in a rural district of North West Province, an area of the country with substantial burden of disease and little available data [24]. We noted that while > 90% of men and women in care reported taking ART, only an estimated 29% of men and 60% of women in care achieved virologic suppression (< 5000 copies/mL) measured from dried blood spots (DBS). To understand the discrepancy between reported ART intake and viral suppression and assess contributing factors to the low suppression rates, we assessed all HIV-positive participants for antiretroviral drug exposure and performed drug resistance testing among those not suppressed. In this manuscript we examine the potential contribution of drug resistance mutations for low viral suppression in this HIV-positive, rural South African, population-based sample, discussing implications for future programming.
Methods
Study setting
Data were collected from January–March 2014 in Lekwa-Teemane and Greater Taung sub-districts, within Dr. Ruth Segomotsi Mompati (RSM) District of North West Province, South Africa. RSM is comprised of both rural and peri-urban areas, with an economy centered on beef production and agriculture. The study area includes approximately 230,000 people, the majority of whom speak Setswana. North West Province has the fourth highest HIV prevalence in South Africa, with an estimated prevalence of 20.3% in the adult population 15–49 years [25] and 29.2% in the antenatal population [26]. One quarter of households in North West are food insecure, reporting lack of money to buy food in the past year [27].
Data collection
The 2014 bio-behavioral survey employed multi-stage cluster sampling, with twenty-three enumeration areas (EAs) selected proportionate to size in each sub-district by Statistics South Africa (StatsSA) using 2011 census data. In each EA up to 36 inhabited dwelling units (DUs) were randomly selected from the StatsSA sampling frame for inclusion in the sample and one adult (18–49 years) was randomly selected per DU for participation. Local, trained fieldworkers assessed eligibility criteria, including age (18–49), residence in the home, and ability to consent; obtained written informed consent; and conducted a survey by computer-assisted personal interviewing in a private location at the participant’s home. Survey questions included HIV testing history, known status, history of HIV care and treatment, ART initiation, and medication adherence. The fieldworkers then referred consenting participants for HIV rapid testing and DBS sample collection. Full data collection procedures have been described elsewhere [24].
For individuals consenting to HIV testing and sample collection, serostatus was determined using the Alere Determine HIV-1/2 rapid antibody test with finger-stick capillary blood (Alere Medical Co.,Ltd., Chiba, Japan) and, if reactive, confirmed using the First Response HIV 1–2.0 Rapid Whole Blood Test (Premier Medical Corporation Ltd., Daman, India). Participants with HIV-positive or discrepant results were asked to provide finger-prick blood for DBS using a Munktell filter card (Ahlstrom Munktell, Helsinki, Finland). Participants who declined HIV rapid testing in their home could provide blood for DBS for laboratory HIV diagnosis (serology: ELISA confirmed with Western blot).
Laboratory methods
Viral Load: DBS cards were dried, stored under desiccant at ambient temperature, transported to the testing laboratory (Clinical Laboratory Services, Johannesburg) within six days of collection, and stored at − 70 °C. Viral load testing was performed using the COBAS AmpliPrep for sample preparation and COBAS TaqMan HIV-1 2.0 test (Roche Applied Science, Pleasanton CA, USA; lower limit of quantification 400 copies/mL) a previously validated method [28]. We opted to use a viral suppression cut-off of < 5000 copies/mL, a higher threshold than that recommended for plasma, as there is no definitive cut-point using DBS [29].
HIV Drug Resistance testing was performed using archived DBS with a VL > 5000 copies/mL at the National Institute for Communicable Diseases, Johannesburg, RSA. Spots were lysed in 2 ml of NucliSENS lysis buffer (Biomerieux, Germany) for 2 h at room temperature. Total nucleic acid was extracted using the NucliSENS EasyMAG® automated system according to the manufacturer’s instructions. Amplification of a 1084 bp polymerase chain reaction (PCR) fragment consisting of codons 1–99 of protease (Pro) and codons 1–250 of reverse transcriptase (RT) was performed as previously described [30]. Editing of sequences was performed using Recall software v2.10. Drug Resistance Mutations (DRM) and inferred susceptibility to individual ARV for RT and Pro regions were determined using the Stanford Drug Resistance Database V8.4 (http://hivdb.stanford.edu) [31].
ART Drug Exposure was determined by a validated qualitative liquid chromatography-tandem mass spectrometry (LC MS/MS) method for the determination of the presence or absence of various antiretroviral analytes against cutoff samples analysed at a known concentration (0.02 μg/ml). A method was validated for the qualitative determination of 9 antiretroviral drugs from DBS, and consisted of a protein precipitation, followed by high performance liquid chromatography with MS/MS detection using a gradient elution. Deuterated internal standards were used for each analyte. An AB Sciex API 4000 mass spectrometer at unit resolution in the multiple reaction monitoring (MRM) mode was used to monitor the transition of each of the protonated precursor ions. Analytes were assessed for the following three drugs: efavirenz (EFV), lopinavir (LPV), nevirapine (NVP), accounting for the majority of first and second line regimens, as tenofovir and emtricitabine are typically given in combination with efavirenz [32].
Analysis
Analyses aimed to characterize the scope of HIV drug resistance and types of DRM by treatment history among viremic HIV-positive participants. All analyses were weighted to account for sample design, with the exception of counts displayed in Fig. 1. Weights were created using the inverse probability of selection at each stage (EA, DU and person) and adjusted for non-response to reflect the municipality, age group and sex distributions within the target population [33]. Using the full survey population, we calculated weighted sample sizes, weighted proportions and 95% confidence intervals (CIs) to describe participant demographic characteristics and HIV-care indicators. We used Stata’s ‘subpop’ command to calculate estimates specific to HIV-positive participants, including full participant data in the calculation of standard errors. We used chi-square statistics to assess demographic differences between the HIV suppressed and non-HIV suppressed participants. When self-report and biomarkers were discrepant (i.e. participant stated not initiating ART but had positive analytes), we utilized the biomarker to indicate ART status.
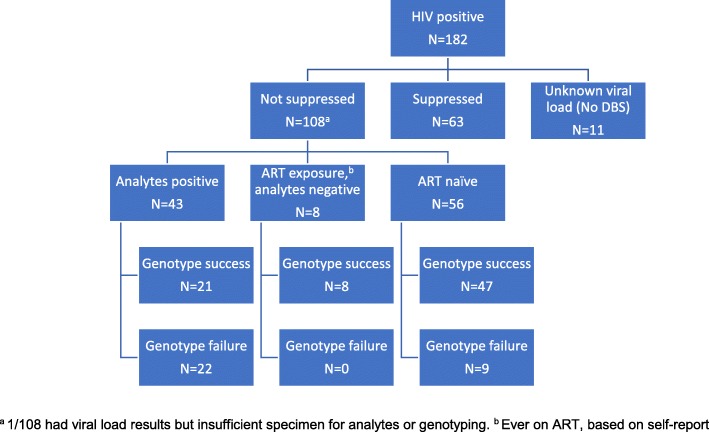
Fig. 1.
Participant viral suppression, medication history, and resistance status, North West Province, South Africa
Virological failure was defined as ≥5000 copies/mL measured from DBS, a conservative cut-off because of the potential for contributions of pro-viral DNA in the quantification using DBS instead of plasma RNA [29, 34]. We assessed presence of genotypic resistance to protease inhibitors (PI), NRTIs, and NNRTIs, and the association with ART status. ART status was categorized in three groups: those who had evidence of ART analytes, those who reported ART initiation but did not have evidence of analytes (likely inconsistent or interrupted treatment), and those who were ART-naïve, having reported never initiating ART and having no evidence of analytes.
Overall 71.7% of participants consented to HIV-testing (749/1044). Of participants with confirmed HIV-positive status, 94% provided DBS (171/182), however one DBS card had insufficient specimen for genotyping. To assess whether the population with missing DBS data differed from the population with complete data, we used multiple imputation to create 50 copies of the dataset with simulated values based on observed data; our analyses with imputed datasets produced adjusted estimates for uncertainty. Missing HIV and viral suppression status were imputed based on age, sex, education, food insecurity, and lifetime number of sexual partners. Imputation of missing viral load also incorporated ART initiation, adherence, and months since diagnosis. All analyses were performed using Stata 14 (StataCorp, College Station, TX, USA).
Results
Among 182 seropositive participants confirmed by serial rapid testing or HIV DNA PCR, DBS were available for viral load and analyte testing from 170 participants (11 did not provide DBS and one had insufficient specimen for genotyping; Fig. 1). A total of 107 dB samples from individuals with viral load ≥5000 copies/ml were assayed for sequence-based drug resistance testing. While younger people were less likely to provide DBS, no differences were found in DBS provision by sex or education. Using imputed values for viral load and HIV status, viral suppression was not significantly different across DBS provision status.
Participant characteristics
The final weighted sample reflects the sex and age distribution of the HIV-positive population of the sub-districts (Table 1). Overall, only 38.4% of the HIV-positive population had ≤5000 copies/mL detected (referred to in this manuscript as virally suppressed). Those who were suppressed were far more likely to be female than male; however, viral suppression status did not differ by age or education level. Not surprisingly, the majority of those who were virally suppressed were aware of their infection (86.0%); 14.0% of those suppressed reported being HIV-negative or unknown during the survey. Those who were suppressed were also more likely to have initiated ART (95.3%) than those who were not suppressed (80.3%). Only 33.3% of those not suppressed had evidence of ART analytes, compared to 75.5% of those suppressed.
Table 1.
Participant characteristics by viral load status, North West Province, South Africa, 2014a
| Not Suppressed (≥ 5000 copies/ml) wgt n = 7981 |
Suppressed (< 5000 copies/ml) wgt n = 4976 |
||||
|---|---|---|---|---|---|
| wgt % | 95% CI | wgt % | 95% CI | P-value | |
| Overall | 61.6 | (51.4–70.9) | 38.4 | (29.2–48.6) | |
| Sex | |||||
| Male | 51.1 | (41.0–61.1) | 21.1 | (10.4–38.2) | 0.003 |
| Female | 48.9 | (38.9–59.1) | 78.9 | (61.8–89.6) | |
| Age group | |||||
| 18–29 Years | 22.2 | (11.4–38.7) | 19.0 | (9.2–35.1) | 0.868 |
| 30–39 Years | 45.0 | (31.0–59.9) | 42.7 | (28.1–58.7) | |
| 40–49 Years | 32.8 | (22.1–45.6) | 38.3 | (24.5–54.4) | |
| Education | |||||
| Primary or Less | 30.2 | (20.3–42.4) | 35.3 | (21.6–52.0) | 0.759 |
| Some Secondary | 39.3 | (30.0–49.5) | 39.8 | (28.0–53.0) | |
| Completed Secondary | 30.5 | (22.2–40.2) | 24.8 | (13.2–41.8) | |
| Prior knowledge of HIV serostatusb | |||||
| Known Positive | 54.2 | (41.7–66.1) | 86.0 | (68.6–94.6) | 0.005 |
| Unknown | 45.8 | (33.9–58.3) | 14.0 | (5.4–31.4)e | |
| ART initiationb | |||||
| No | 19.7 | (10.4–34.3) | 4.8 | (1.4–15.1) | 0.040 |
| Yes | 80.3 | (65.7–89.6) | 95.3 | (85.0–98.6) | |
| Time on ARTb | |||||
| < 1 year | 23.4 | (14.4–35.9) | 13.8 | (7.4–24.3) | 0.134 |
| 1–3 years | 42.9 | (29.4–57.5) | 33.7 | (18.7–52.8) | |
| > 3 years | 33.7 | (21.9–48.0) | 52.5 | (30.8–58.5) | |
| Analytes foundc | |||||
| Yes | 33.3 | (25.3–42.4) | 75.5 | (57.9–87.4) | < 0.001 |
| No | 66.7 | (57.6–74.7) | 24.5 | (12.6–42.2) | |
| CD4 category (Pima)d | |||||
| ≤ 350 cells/μL | 57.8 | (42.8–71.5) | 28.2 | (15.7–45.2) | 0.018 |
| > 350 cells/μL | 42.2 | (28.6–57.2) | 71.9 | (54.8–84.3) | |
aWeights account for sampling, non-response, and age/sex of target population
bBased on self-report
cAssessed for EFV, LPV, NVP
dAmong those for whom Pima CD4 results were available
eRespondents may be mis-reporting status, be elite controllers, or on ART without understanding of their condition [35]
Resistance patterns
Among the 107 HIV-positive participants with a DBS viral load > 5000 copies/mL and specimens for genotyping, sequences were successfully generated for 71% (n = 76), with greater sequencing failures among those who were positive for ART analytes compared to those who were ART-naïve or were negative for ART analytes but with reported prior ART exposure (Fig. 1). Seventy-five (99%) viruses sequenced were pure consensus subtype C using two independent typing tools. One was pure consensus D; none were found to be unique recombinant forms (subtype data not shown).
In weighted analyses, 21.7% of those sequenced had evidence of DRM (Table 2). Resistance was most prominent in those who were initiated on ART with negative analyte results, who likely took ART inconsistently or had interrupted treatment. In this group, 51.1% (95% CI 11.6–89.3%) had any resistance mutation, as compared to 46.6% (95% CI 24.6–70.0%) of those sequenced with positive analyte results and 8.0% (95% CI 3.1–18.9%) among those sequenced who were ART-naïve.
Table 2.
Resistance by ART Status and drug class, North West Province, South Africa, 2014a
| Analyte Positivec | ART experienced,b analyte negative | ART naïveb | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Percent | 95% CI | Percent | 95% CI | Percent | 95% CI | Percent | 95% CI | |
| Genotype result | wgt na = 2643 | wgt na = 677 | wgt na = 4623 | wgt na = 7943 | ||||
| No resistance | 24.4 | 14.4–38.3 | 49.0 | 10.7–88.4 | 72.6 | 55.2–85.1 | 54.6 | 43.4–65.4 |
| Any resistance | 21.3 | 9.1–42.4 | 51.1 | 11.6–89.3 | 6.3 | 2.4–15.5 | 15.1 | 8.2–26.3 |
| Genotyping failure | 54.3 | 35.4–72.0 | 0.0 | – | 21.1 | 9.4–40.7 | 30.3 | 20.3–42.6 |
| Resistance - among those genotyped | wgt na = 1209 | wgt na = 677 | wgt na = 3650 | wgt na = 5536 | ||||
| No resistance | 53.4 | 30.0–75.7 | 49.0 | 10.7–88.4 | 92.0 | 81.0–96.9 | 78.3 | 64.3–87.9 |
| Any resistance | 46.6 | 24.6–70.0 | 51.1 | 11.6–89.3 | 8.0 | 3.1–18.9 | 21.7 | 12.1–35.7 |
| Drug class resistance | wgt na = 564 | wgt na = 346 | wgt na = 291 | wgt na = 1201 | ||||
| Single class | ||||||||
| NRTI only | 0.0 | – | 0.0 | – | 0.0 | – | 0.0 | – |
| NNRTI only | 0.0 | – | 73.1 | 27.9–95.0 | 63.0 | 18.1–92.9 | 36.3 | 17.7–60.2 |
| PI only | 0.0 | – | 12.2 | 1.4–58.6 | 11.5 | 1.2–58.0 | 6.3 | 1.3–25.7 |
| Dual class | ||||||||
| NRTI/NNRTI | 100.0 | – | 14.7 | 1.6–64.1 | 0.0 | – | 51.2 | 28.3–73.6 |
| PI/NRTI | 0.0 | – | 0.0 | – | 25.5 | 3.3–77.6 | 6.2 | 0.7–37.3 |
| PI/NNRTI | 0.0 | – | 0.0 | – | 0.0 | – | 0.0 | – |
aWeights account for sampling, non-response, and age/sex of target population, bBy self report, c Assessed EFV, LPV, NVP
Sequencing results showed a predominance of NNRTI resistance (Table 2). The most common resistance pattern was dual class NRTI/NNRTI resistance at 51.2% (95% CI 28.3–73.6%), followed by single class resistance to NNRTI at 36.3% (95% CI 17.7–60.2%). However, specific mutations differed by presence of analytes. Of the 21 participants successfully sequenced with positive NVP or EFV results, 10 showed two or more DRM, all of whom had both NRTI and NNRTI resistance. The predominant NRTI mutations were M184V (8/10) and K65R (5/10) with three participants harboring both mutations. (Table 3). Among those with NNRTI resistance, the predominant mutations were at codons 100, 101, 103, 106 and 181. In contrast, among participants without evidence of being on ART (analyte negative – regardless of reported ART initiation), 10 harbored at least one major NRTI, NNRTI or PI DRM. Two showed NRTI resistance (M184V) and six harbored NNRTI DRM at codons 101 and 103. The major PI mutations M46I, I50I/V and V82I, which confer low to intermediate resistance to LPV/r used in second line therapy, were detected in three participants. (Table 3).
Table 3.
Resistance mutations, North West Province, South Africa, 2014
| Sex | Age Group | Months on ART | ARV analyte exposure | Viral load | Drug resistance mutation by drug class | ||||
|---|---|---|---|---|---|---|---|---|---|
| EFV | NVP | LPV | NRTI | NNRTI | PI | ||||
| Male | 18–29 | 47 | – | – | – | 7327 | Wild-Type | Wild-Type | V82L |
| Female | 30–39 | 25 | + | + | – | 8580 | M184V | Y188L | Wild-Type |
| Male | 30–39 | N/A | – | – | – | 18,800 | Wild-Type | Wild-Type | I50V |
| Female | 30–39 | 21 | + | – | – | 27,474 | K65R, Y115F | V106M, Y181C | Wild-Type |
| Male | 40–49 | 92 | – | – | – | 35,300 | M184V | K103N, Y188L | Wild-Type |
| Female | 40–49 | 117 | + | – | – | 36,873 | M41L, M184V, T215F | K101E, V106M, G190A | Wild-Type |
| Male | 30–39 | N/A | – | – | – | 49,500 | M184I | Wild-Type | M46I |
| Female | 30–39 | 90 | + | – | – | 53,600 | K65R, M184V | K101P, K103S | Wild-Type |
| Female | 30–39 | 55 | + | – | – | 56,600 | M184V | K103N | Wild-Type |
| Male | 30–39 | 49 | + | – | – | 73,200 | D67G, T69D, K70R, M184V, K219Q | L100I, K103N | Wild-Type |
| Female | 30–39 | N/A | – | – | – | 95,059 | Wild-Type | K103N | Wild-Type |
| Female | 30–39 | 47 | + | – | – | 95,100 | K65R, M184V | Y181C, G190S | Wild-Type |
| Female | 30–39 | 61 | + | – | – | 120,000 | K65R, T69del | K101E, Y181C, G190S | Wild-Type |
| Male | 18–29 | N/A | – | – | – | 122,000 | Wild-Type | K101E | Wild-Type |
| Male | 30–39 | 22 | – | – | – | 135,000 | Wild-Type | K103N | Wild-Type |
| Female | 30–39 | 17 | – | – | – | 145,000 | Wild-Type | V106M | Wild-Type |
| Male | 40–49 | N/A | – | – | – | 390,000 | Wild-Type | K103N | Wild-Type |
| Male | 18–29 | N/A | + | – | – | 415,000 | K65R, Y115F, M184V | V106M, G190A | Wild-Type |
| Male | 18–29 | 15 | – | – | – | 490,000 | Wild-Type | K103N, G190A | Wild-Type |
| Male | 30–39 | 67 | + | – | – | 799,000 | M41L, D67N, K70E, Y115F, M184V | K103N, V106M | Wild-Type |
Treatment-specific resistance
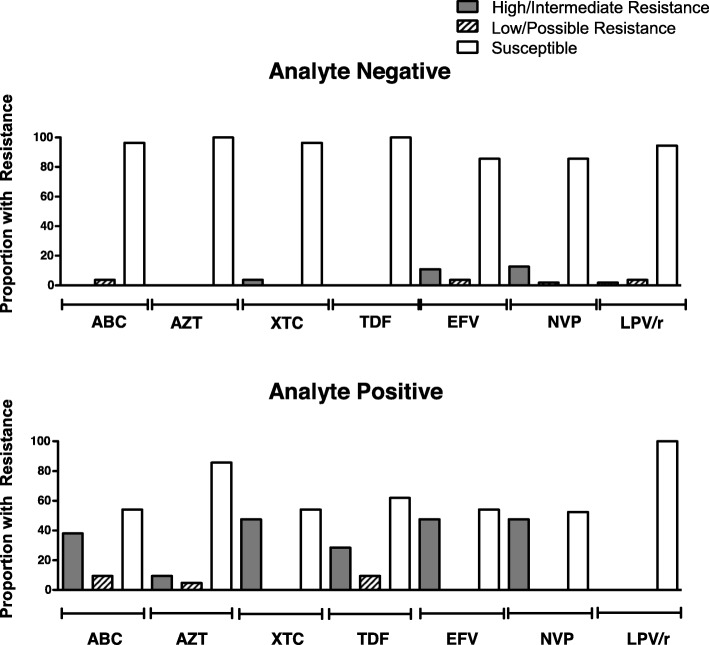
There were significant differences in treatment-specific resistance among the analyte positive and negative participants. Among those who were analyte negative, fewer than 5 % demonstrated resistance to azidothymidine (AZT), abacavir (ABC), lamivudine or emtricitabine (XTC), tenofovir disoproxil fumarate (TDF), LPV/r and thus few would fail to suppress using the first-line regimens in South Africa. Among those who were analyte positive, over 10% and as high as 47% showed any resistance to XTC, TDF, EFV, NVP, and ABC and thus are predicted to show reduced susceptibility to first- or second-line regimes containing these antiretrovirals (Fig. 2). Among the participants sequenced, three carried M184V/I with K65R, mutations conferring resistance to the regimen used for oral HIV pre-exposure prophylaxis (PrEP) (FTC/TDF).
Fig. 2.
Treatment-specific resistance among analyte negative and analyte positive participants, North West Province, South Africa
Discussion
We set out to examine the contribution of HIV drug resistance mutations to rates of virologic failure after noting that almost half of HIV-positive participants reporting ART initiation in a population-based sero-prevalence survey had evidence of virologic failure, despite 93% of those in care reporting near perfect adherence [24]. In exploring the factors behind poor viral suppression rates in this rural South African population, drug resistance mutations appear to be making a substantial contribution to sub-optimal outcomes, particularly for those in treatment. Among those sequenced, evidence of DRM was present in 22% of the population and in 47% of those with evidence of ART analytes, with implications for efficacy of treatment regimens.
Little is known about the prevalence of DRM both prior to and following treatment initiation in the general population. One population-based study in South Africa found evidence of DRM in 7% of ART-naïve patients in rural KwaZulu-Natal in 2012, similar to our findings of DRMs in 8% of our 2014 North West population sequenced pre-treatment. Clinical cohort studies in sub-Saharan Africa have found a similar prevalence of drug resistance mutations in 6–11% of ART-naïve patients [11, 20, 22]. If our study participants reported ART initiation accurately, 8% would represent the baseline estimate of transmitted resistance in the population, most of which was NNRTI only (63.0%), with fewer mutations representing PI/NRTI resistance (25.5%) and even fewer being single PI resistance (11.5%). For these patients, DRM could lead to first-line failure. Recent studies have demonstrated that the prevalence of transmitted and acquired drug resistance is increasing in RSA [20]. The cost in a program as large as South Africa’s makes baseline DRM testing prior to ART initiation to inform regimen selection largely infeasible; however NHLS has instituted alert reports for adherence counseling and DRM testing should be made available when treatment failure is evident.
Resistance was particularly high in our sample among those who had initiated ART, and most prevalent among those who had reported initiation but had no evidence of ART analytes, e.g. probable non-adherence. Non-adherence is likely a big contributor to lack of viral suppression and acquired resistance in this sample. It is not possible to distinguish between acquired and transmitted resistance in our data, however, patterns of resistance by ART status provide insights about contributions of acquired and transmitted resistance in this population-based sample. The large majority of DRMs in this population occurred in the ART-initiated population, implying a high likelihood of acquired resistance playing a central role, particularly with all mutations demonstrating NRTI/NNRTI (first-line) dual resistance among the analyte positive group. In fact, research has noted that a high proportion of adults and children who received first line ART will develop virological failure during the first 5 years, with 70–90% of patients in virologic failure acquiring drug-resistant HIV [36]. While this estimate of DRM is higher than that observed in our sample, we were unable to sequence 30% of samples and the majority (22/31) of those were ART positive with viral load measures close to the cut-off (i.e. 20,000 copies or less) for virologic failure. If all ART positive participants with genotyping failure had resistance mutations, this would represent 75.6% of analyte positive participants, coinciding with current estimates [36]. Indeed, if participants were partially suppressed on ART and therefore had positive analytes and lower but not suppressed viral loads, they would be more prone to generate resistance and may lead to an underestimate of DRMs in this population.
The extremely high rates of dual NRTI/NNRTI resistance among the analyte positive group has important implications for the future of the current first-line treatment. Currently the South African national program recommends that providers consider switching patients on the first-line drug regimen if the patient has experienced virological failure (VL > 1000 copies/mL) on at least two occasions two months apart despite good adherence. Our data indicate that approximately one-third of those who report ART intake are negative for analytes, making a premature decision to switch regimens potentially costly and could encourage further DRM. With South Africa currently starting up introduction of a new triple combination regimen of tenofovir disoproxil fumarate, lamivudine (3TC), and dolutegravir (TLD) [37, 38], some progress in stemming resistance to NNRTI could be imminent. However, even with availability of the new regimen, routine monitoring of resistance remains critical [39]. Finally, circulating drug-resistant viruses may have implications for persons initiating PrEP with TDF/FTC. The few seroconversions among individuals enrolled in PrEP trials occurred among those who started PrEP during unrecognized acute HIV infection, and the emergence of resistance primarily involved M184V/I, which can be selected for under the selective pressure of FTC [40–42].
A strength of this study is the population-based sample in an understudied area with data on viral load, ART analytes, and DRMs. Our data also has several limitations. Only 70% of the samples were sequenced, with failure being highest for those not suppressed on ART and with comparatively lower viral load measures, which could lead to an underestimation of DRMs if that population is intermittently adherent. Studies restricted to ART-naïve populations using the same medium for genotyping have sequencing success of closer to 92% [43]. Additionally, ART initiation and adherence are based on self-report; however, the use of analyte data as a biological marker of initiation and adherence improves the study’s accuracy. We cannot distinguish between acquired and transmitted resistance in our data, and, finally, we used DBS instead of plasma for the assays. Though DBS offer a feasible, low-resource alternative for viral load and DRM surveillance, they have well documented limitations – notably cellular HIV DNA contributes to copy number when using whole blood instead of blood plasma samples [29]. We therefore used a conservative threshold of 5000 copies/mL for defining viral suppression, and, as a result, run the risk of over-estimating viral suppression. Even so, viral suppression in this study aligns well with data from the NHLS at the time of the study, which suggested that viral suppression among PLHIV in care (defined as < 400 copies/mL plasma) ranged from 52 to 75% for North West Province, depending on the district [44], similar to our estimates among those on treatment.
Conclusion
We found that both non-adherence and drug resistance mutations likely play a key role in virologic failure in this rural community in North West Province, South Africa. Increasing ART coverage can significantly lower the risk of new HIV infections in South Africa [45]; however, gains in treatment expansion will be lost if inconsistent adherence is not addressed and resistance continues to increase. There is growing evidence that pre-treatment drug resistance is increasing in sub-Saharan Africa [22, 23], making it essential that policies around treatment regimens are reviewed and updated frequently, and that surveillance be instituted in both HIV-positive, ART-naïve populations and among those failing treatment, including monitoring of drug exposure for non-adherence and treatment failure despite adherence. Countries should consider instituting sentinel sites for baseline drug resistance testing in settings where feasible.
Acknowledgements
We thank the data collection teams and site supervisors, including Jessica Morris, Elsie Raphela, and Charles Koenaite. We thank Lebogang Ntswane and Jay Gilvidys for community entry and training support. We thank the North West Provincial Department of Health (DoH), Dr. Ruth Segomotsi Mompati District DoH, Lekwe Teemane and Greater Taung Sub-district DoH, and the Provincial Research Committee for ongoing support of this project. We thank Clinical Laboratory Services for ART analyte testing. We thank the participants for their generosity and willingness to be a part of this study.
Abbreviations
- ABC
Abacavir
- ART
Antiretroviral therapy
- AZT
Azidothymidine
- DBS
Dried blood spots
- DRM
HIV drug resistance mutations
- DU
Dwelling unit
- EA
Enumeration area
- EFV
Efavirenz
- LC MS/MS
Liquid chromatography-tandem mass spectrometry method
- LPV
Lopinavir
- MRM
Multiple reaction monitoring
- NNRTI
Non-nucleoside reverse transcriptase inhibitors
- NRTI
Nucleoside reverse transcriptase inhibitors
- NVP
Nevirapine
- PCR
Polymerase chain reaction
- PI
Protease inhibitors
- PrEP
HIV pre-exposure prophylaxis
- Pro
Protease
- RSM
Dr. Ruth Segomotsi Mompati District
- RT
Reverse transcriptase
- StatsSA
Statistics South Africa
- TDF
Tenofovir disoproxil fumarate
- TLD
Dolutegravir
- UNAIDS
Joint United Nations Programme on HIV/AIDS
- XTC
Lamivudine or emtricitabine
Authors’ contributions
SAL, TL, and SB designed the study; SL wrote the manuscript; ACM analyzed data and assisting with writing; TL, AP, and GH designed and implemented laboratory protocols and contributed to writing and interpretation of findings; JSG and LMP managed study implementation and data collection in the field; HG provided overall study coordination; HMT assisted with writing and interpretation of findings. All authors read and approved the final manuscript.
Funding
This project has been supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the US Centers for Disease Control and Prevention under the terms of Cooperative Agreement 5U2GGH000324–02. Additional support for TL was provided through the University of California San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research (CFAR), an NIH-funded program (P30 AI027763). The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the views of CDC or the NIH. Funding bodies played no role in the design of the study; collection, analysis, and interpretation of the data; or writing the manuscript.
Availability of data and materials
The datasets generated and analysed during the current study are not publicly available to ensure confidentiality of participants’ private health information. De-identified, aggregate data will be made available from the corresponding author upon reasonable request and with IRB approval.
Ethics approval and consent to participate
The protocol was approved by the Committee for Human Research at the University of California, San Francisco; the Human Subjects Division at University of Washington; the Human Sciences Research Council Research Ethics Committee in South Africa; the Policy, Planning, Research, Monitoring and Evaluation Committee for the North West Provincial Department of Health; and the CDC’s Center for Global Health, Human Research Protection.
Local, trained fieldworkers assessed ability to consent and obtained written informed consent for the survey, HIV rapid testing, and DBS sample collection. Participants who declined HIV rapid testing in their home could provide blood for DBS for laboratory HIV diagnosis (serology: ELISA confirmed with Western blot).
Consent for publication
N/A
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.UNAIDS: 90–90-90: An ambitious treatment target to help end the AIDS epidemic . Joint United Nations Programme on HIV/AIDS (UNAIDS) 2014. [Google Scholar]
- 2.UNAIDS . Global AIDS Update 2016. Geneva: Joint United Nations Programme on HIV/AIDS; 2016. [Google Scholar]
- 3.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, Hakim JG, Kumwenda J, Grinsztejn B, Pilotto JH, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granich RM, Gilks CF, Dye C, De Cock KM, Williams BG. Universal voluntary HIV testing with immediate antiretroviral therapy as a strategy for elimination of HIV transmission: a mathematical model. Lancet. 2009;373(9657):48–57. doi: 10.1016/S0140-6736(08)61697-9. [DOI] [PubMed] [Google Scholar]
- 5.Bartlett JA, Fath MJ, Demasi R, Hermes A, Quinn J, Mondou E, Rousseau F. An updated systematic overview of triple combination therapy in antiretroviral-naive HIV-infected adults. AIDS. 2006;20(16):2051–2064. doi: 10.1097/01.aids.0000247578.08449.ff. [DOI] [PubMed] [Google Scholar]
- 6.Bor J, Herbst AJ, Newell ML, Barnighausen T. Increases in adult life expectancy in rural South Africa: valuing the scale-up of HIV treatment. Science. 2013;339(6122):961–965. doi: 10.1126/science.1230413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bangsberg DR, Hecht FM, Charlebois ED, Zolopa AR, Holodniy M, Sheiner L, Bamberger JD, Chesney MA, Moss A. Adherence to protease inhibitors, HIV-1 viral load, and development of drug resistance in an indigent population. AIDS. 2000;14(4):357–366. doi: 10.1097/00002030-200003100-00008. [DOI] [PubMed] [Google Scholar]
- 8.Hogg RS, Heath K, Bangsberg D, Yip B, Press N, O'Shaughnessy MV, Montaner JS. Intermittent use of triple-combination therapy is predictive of mortality at baseline and after 1 year of follow-up. AIDS. 2002;16(7):1051–1058. doi: 10.1097/00002030-200205030-00012. [DOI] [PubMed] [Google Scholar]
- 9.Paterson DL, Swindells S, Mohr J, Brester M, Vergis EN, Squier C, Wagener MM, Singh N. Adherence to protease inhibitor therapy and outcomes in patients with HIV infection. Ann Intern Med. 2000;133(1):21–30. doi: 10.7326/0003-4819-133-1-200007040-00004. [DOI] [PubMed] [Google Scholar]
- 10.Wood E, Montaner JS, Yip B, Tyndall MW, Schechter MT, O'Shaughnessy MV, Hogg RS. Adherence and plasma HIV RNA responses to highly active antiretroviral therapy among HIV-1 infected injection drug users. CMAJ. 2003;169(7):656–661. [PMC free article] [PubMed] [Google Scholar]
- 11.Boender TS, Hoenderboom BM, Sigaloff KC, Hamers RL, Wellington M, Shamu T, Siwale M, Labib Maksimos EE, Nankya I, Kityo CM, et al. Pretreatment HIV drug resistance increases regimen switches in sub-Saharan Africa. Clin Infect Dis. 2015;61(11):1749–1758. doi: 10.1093/cid/civ656. [DOI] [PubMed] [Google Scholar]
- 12.Boender TS, Sigaloff KC, McMahon JH, Kiertiburanakul S, Jordan MR, Barcarolo J, Ford N, Rinke de Wit TF, Bertagnolio S. Long-term Virological outcomes of first-line antiretroviral therapy for HIV-1 in low- and middle-income countries: a systematic review and meta-analysis. Clin Infect Dis. 2015;61(9):1453–1461. doi: 10.1093/cid/civ556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kharsany ABM, Cawood C, Khanyile D, Lewis L, Grobler A, Puren A, Govender K, George G, Beckett S, Samsunder N, et al. Community-based HIV prevalence in KwaZulu-Natal, South Africa: results of a cross-sectional household survey. Lancet HIV. 2018;5(8):e427–e437. doi: 10.1016/S2352-3018(18)30104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Human Sciences Research Council (HSRC) The Fifth South African National HIV Prevalence, Incidence, Behaviour and Communication Survey, 2017: HIV Impact Assessment Summary Report. Cape Town: HSRC Press; 2018. [Google Scholar]
- 15.Rossouw TM, Nieuwoudt M, Manasa J, Malherbe G, Lessells RJ, Pillay S, Danaviah S, Mahasha P, van Dyk G, de Oliveira T. HIV drug resistance levels in adults failing first-line antiretroviral therapy in an urban and a rural setting in South Africa. HIV Med. 2017;18(2):104–114. doi: 10.1111/hiv.12400. [DOI] [PubMed] [Google Scholar]
- 16.Petersen M, Balzer L, Kwarsiima D, Sang N, Chamie G, Ayieko J, Kabami J, Owaraganise A, Liegler T, Mwangwa F, et al. Association of Implementation of a universal testing and treatment intervention with HIV diagnosis, receipt of antiretroviral therapy, and viral suppression in East Africa. Jama. 2017;317(21):2196–2206. doi: 10.1001/jama.2017.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwuji CC, Orne-Gliemann J, Larmarange J, Balestre E, Thiebaut R, Tanser F, Okesola N, Makowa T, Dreyer J, Herbst K, et al. Universal test and treat and the HIV epidemic in rural South Africa: a phase 4, open-label, community cluster randomised trial. Lancet HIV. 2018;5(3):e116–e125. doi: 10.1016/S2352-3018(17)30205-9. [DOI] [PubMed] [Google Scholar]
- 18.Hayes RJ, Donnell DJ, Floyd S, Mandla N, Bwalya J, Shanaube K, Moore A, Eshleman SH, Fraser C, El-Sadr WM, et al. Impact of universal testing and treatment in Zambia and South Africa: HPTN071(POPART). In: Conference on Retroviruses and Opportunistic Infections (CROI). vol. Abstract #92. Seattle; 2019. http://www.croiconference.org/sessions/impact-universal-testing-and-treatment-zambia-and-south-africa-hptn071popart.
- 19.Boender TS, Kityo CM, Boerma RS, Hamers RL, Ondoa P, Wellington M, Siwale M, Nankya I, Kaudha E, Akanmu AS, et al. Accumulation of HIV-1 drug resistance after continued virological failure on first-line ART in adults and children in sub-Saharan Africa. J Antimicrob Chemother. 2016;71(10):2918–2927. doi: 10.1093/jac/dkw218. [DOI] [PubMed] [Google Scholar]
- 20.Steegen K, Carmona S, Bronze M, Papathanasopoulos MA, van Zyl G, Goedhals D, MacLeod W, Sanne I, Stevens WS. Moderate levels of pre-treatment HIV-1 antiretroviral drug resistance detected in the first South African National Survey. PLoS One. 2016;11(12):e0166305. doi: 10.1371/journal.pone.0166305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunt GM, Dokubo EK, Takuva S, de Oliveira T, Ledwaba J, Dube N, Moodley P, Sabatier J, Deyde V, Morris L, et al. Rates of virological suppression and drug resistance in adult HIV-1-positive patients attending primary healthcare facilities in KwaZulu-Natal, South Africa. J Antimicrob Chemother. 2017;72(11):3141–3148. doi: 10.1093/jac/dkx252. [DOI] [PubMed] [Google Scholar]
- 22.Gupta RK, Gregson J, Parkin N, Haile-Selassie H, Tanuri A, Andrade Forero L, Kaleebu P, Watera C, Aghokeng A, Mutenda N, et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis. 2018;18(3):346–355. doi: 10.1016/S1473-3099(17)30702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manasa J, Danaviah S, Lessells R, Elshareef M, Tanser F, Wilkinson E, Pillay S, Mthiyane H, Mwambi H, Pillay D, et al. Increasing HIV-1 drug resistance between 2010 and 2012 in adults participating in population-based HIV surveillance in rural KwaZulu-Natal, South Africa. AIDS Res Hum Retroviruses. 2016;32(8):763–769. doi: 10.1089/aid.2015.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippman SA, Shade SB, El Ayadi AM, Gilvydis JM, Grignon JS, Liegler T, Morris J, Naidoo E, Prach LM, Puren A, et al. Attrition and opportunities along the HIV care continuum: findings from a population-based sample, north West Province, South Africa. J Acquir Immune Defic Syndr. 2016;73(1):91–99. doi: 10.1097/QAI.0000000000001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shisana O, Rehle T, Simbayi LC, Zuma K, Jooste S, Zungu N, Labadarios D, Onoya D, et al. South African national HIV prevalence, incidence and behaviour survey, 2012. Cape Town: HSRC Press; 2014. [DOI] [PubMed] [Google Scholar]
- 26.National Department of Health. The National Antenatal Sentinel HIV and Syphilis Prevalence Survey, South Africa, 2015. Pretoria; 2017.
- 27.Statistics South Africa. Community Survey 2016. Available at: http://cs2016.statssa.gov.za. Accessed June 2019.
- 28.Prach LM, Puren A, Lippman SA, Carmona S, Stephenson S, Cutler E, Barnhart S, Liegler T. Design and implementation of an external quality assessment program for HIV viral load measurements using dried blood spots. J Clin Microbiol. 2015;53(3):964–966. doi: 10.1128/JCM.02698-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkin NT. Measurement of HIV-1 viral load for drug resistance surveillance using dried blood spots: literature review and modeling of contribution of DNA and RNA. AIDS Rev. 2014;16(3):160–171. [PubMed] [Google Scholar]
- 30.Zhou Z, Wagar N, DeVos JR, Rottinghaus E, Diallo K, Nguyen DB, Bassey O, Ugbena R, Wadonda-Kabondo N, McConnell MS, et al. Optimization of a low cost and broadly sensitive genotyping assay for HIV-1 drug resistance surveillance and monitoring in resource-limited settings. PLoS One. 2011;6(11):e28184. doi: 10.1371/journal.pone.0028184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee SY, Fessel WJ, Zolopa AR, Hurley L, Liu T, Taylor J, Nguyen DP, Slome S, Klein D, Horberg M, et al. HIV-1 protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J Infect Dis. 2005;192(3):456–465. doi: 10.1086/431601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Department of Health, Republic of South Africa . National Consolidated Guidelines for the Prevention of Mother-to-Child Transmission of HIV (PMTCT) and the Management of HIV in Children, Adolescents and Adults. Pretoria: Department of Health, Republic of South Africa; 2014. [Google Scholar]
- 33.Levy PS, Lemeshow SA. Sampling of populations: methods and applications., 4th edn. Hoboken: Wiley; 2008. [Google Scholar]
- 34.Bertagnolio S, Parkin NT, Jordan M, Brooks J, Garcia-Lerma JG. Dried blood spots for HIV-1 drug resistance and viral load testing: a review of current knowledge and WHO efforts for global HIV drug resistance surveillance. AIDS Rev. 2010;12(4):195–208. [PubMed] [Google Scholar]
- 35.Mooney AC, Campbell CK, Ratlhagana MJ, Grignon JS, Mazibuko S, Agnew E, Gilmore H, Barnhart S, Puren A, Shade SB, et al. Beyond social desirability Bias: investigating inconsistencies in self-reported HIV testing and treatment behaviors among HIV-positive adults in north West Province, South Africa. AIDS Behav. 2018;22(7):2368–2379. doi: 10.1007/s10461-018-2155-9. [DOI] [PubMed] [Google Scholar]
- 36.Inzaule SC, Ondoa P, Peter T, Mugyenyi PN, Stevens WS, de Wit TFR, Hamers RL. Affordable HIV drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan Africa. Lancet Infect Dis. 2016;16(11):e267–e275. doi: 10.1016/S1473-3099(16)30118-9. [DOI] [PubMed] [Google Scholar]
- 37.Venter WDF, Moorhouse M, Sokhela S, Fairlie L, Mashabane N, Masenya M, Serenata C, Akpomiemie G, Qavi A, Chandiwana N, et al. Dolutegravir plus two different Prodrugs of Tenofovir to treat HIV. N Engl J Med. 2019;381(9):803–15. [DOI] [PubMed]
- 38.Department of Health, Republic of South Africa . 2019 ART Clinical Guidelines for the Management of HIV in Adults, Pregnancy, Adolescents, Children, Infants and Neonates. Pretoria: Department of Health, Republic of South Africa; 2019. [Google Scholar]
- 39.Inzaule SC, Hamers RL, Doherty M, Shafer RW, Bertagnolio S, Rinke de Wit TF. Curbing the rise of HIV drug resistance in low-income and middle-income countries: the role of dolutegravir-containing regimens. Lancet Infect Dis. 2019;19(7):e246–e252. doi: 10.1016/S1473-3099(18)30710-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehman DA, Baeten JM, McCoy CO, Weis JF, Peterson D, Mbara G, Donnell D, Thomas KK, Hendrix CW, Marzinke MA, et al. Risk of drug resistance among persons acquiring HIV within a randomized clinical trial of single- or dual-agent preexposure prophylaxis. J Infect Dis. 2015;211(8):1211–1218. doi: 10.1093/infdis/jiu677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liegler T, Abdel-Mohsen M, Bentley LG, Atchison R, Schmidt T, Javier J, Mehrotra M, Eden C, Glidden DV, McMahan V, et al. HIV-1 drug resistance in the iPrEx preexposure prophylaxis trial. J Infect Dis. 2014;210(8):1217–1227. doi: 10.1093/infdis/jiu233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margot NA, Waters JM, Miller MD. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob Agents Chemother. 2006;50(12):4087–4095. doi: 10.1128/AAC.00816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, DeVos J, Medina-Moreno S, Wagar N, Diallo K, Beard RS, Zheng DP, Mwachari C, Riwa C, Jullu B, et al. Utilization of dried blood spot specimens can expedite nationwide surveillance of HIV drug resistance in resource-limited settings. PLoS One. 2018;13(9):e0203296. doi: 10.1371/journal.pone.0203296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Department of Health, Republic of South Africa, Directorate: Monitoring and Evaluation: Health Indicators Update: Antiretroviral Indicators. Pretoria; 2013.https://www.uio.no/studier/emner/matnat/ifi/INF5761/v14/timeplan/artprogramme-overview-from-2004-to-2013_ndoh-2013.pdf.
- 45.Tanser F, Barnighausen T, Grapsa E, Zaidi J, Newell ML. High coverage of ART associated with decline in risk of HIV acquisition in rural KwaZulu-Natal, South Africa. Science. 2013;339(6122):966–971. doi: 10.1126/science.1228160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are not publicly available to ensure confidentiality of participants’ private health information. De-identified, aggregate data will be made available from the corresponding author upon reasonable request and with IRB approval.