Abstract
Background:
Echocardiographic quantification of left ventricular (LV) ejection fraction (EF) relies on either manual or automated identification of endocardial boundaries followed by model-based calculation of end-systolic and end-diastolic LV volumes. Recent developments in artificial intelligence resulted in computer algorithms that allow near automated detection of endocardial boundaries and measurement of LV volumes and function. However, boundary identification is still prone to errors limiting accuracy in certain patients. We hypothesized that a fully automated machine learning algorithm could circumvent border detection and instead would estimate the degree of ventricular contraction, similar to a human expert trained on tens of thousands of images.
Methods:
Machine learning algorithm was developed and trained to automatically estimate LVEF on a database of >50 000 echocardiographic studies, including multiple apical 2- and 4-chamber views (AutoEF, BayLabs). Testing was performed on an independent group of 99 patients, whose automated EF values were compared with reference values obtained by averaging measurements by 3 experts using conventional volume-based technique. Inter-technique agreement was assessed using linear regression and Bland-Altman analysis. Consistency was assessed by mean absolute deviation among automated estimates from different combinations of apical views. Finally, sensitivity and specificity of detecting of EF ≤35% were calculated. These metrics were compared side-by-side against the same reference standard to those obtained from conventional EF measurements by clinical readers.
Results:
Automated estimation of LVEF was feasible in all 99 patients. AutoEF values showed high consistency (mean absolute deviation =2.9%) and excellent agreement with the reference values: r=0.95, bias=1.0%, limits of agreement =±11.8%, with sensitivity 0.90 and specificity 0.92 for detection of EF ≤35%. This was similar to clinicians’ measurements: r=0.94, bias=1.4%, limits of agreement =±13.4%, sensitivity 0.93, specificity 0.87.
Conclusions:
Machine learning algorithm for volume-independent LVEF estimation is highly feasible and similar in accuracy to conventional volume-based measurements, when compared with reference values provided by an expert panel.
Keywords: echocardiography, endocardium, left ventricular function, machine learning, observer variation
Clinical Perspective.
Echocardiographic quantification of left ventricular ejection fraction relies on either manual or automated identification of endocardial boundaries followed by model-based calculation of end-systolic and end-diastolic left ventricular volumes. Recent developments in artificial intelligence resulted in computer algorithms that allow near automated detection of endocardial boundaries and measurement of left ventricular volumes and function. However, boundary identification is still prone to errors limiting accuracy in certain patients. This initial feasibility study demonstrated that machine learning algorithm for estimation of left ventricular ejection fraction that avoids image segmentation and volume measurements and instead mimics a human expert’s eye, is highly feasible and can yield results that are in close agreement with what highly experienced readers measure using conventional methodology. This approach may prove to have important clinical implications because of its fast and fully automated nature. It is conceivable that in the future, the starting point of interpretation of an echocardiography exam would include automated estimates of ventricular function the reader would be presented with along with the images.
Introduction
See Editorial by Leeson and Fletcher
Despite the well-known limitations of ejection fraction (EF), it remains the most commonly clinically used echocardiographic measure of left ventricular (LV) performance. Current echocardiography guidelines1 emphasize the importance of accurate quantification of LV EF, since multiple indications for therapeutic interventions rely on cutoff values of this parameter. Quantitative evaluation of LV EF requires measurement of end-systolic and end-diastolic volumes, which traditionally rely on tracing endocardial boundaries at these 2 phases of the cardiac cycle, followed by model-based calculations. This methodology is associated with a considerable inter-observer variability that stems from individual differences in the perception of the blood tissue interface in the presence of endocardial trabeculae, especially at end-systole.2
Although LV EF calculation is recommended by the guidelines, many clinical laboratories find this practice too tedious and time-consuming for routine use, and echocardiographers commonly rely on their ability to visually estimate LV EF on the basis of years of experience acquired through the interpretation of thousands of exams. While usually this results in a qualitative grading of LV function as normal, mildly, moderately or severely reduced, some readers report their findings as a narrow range of values, for example, 40% to 45%. Although this methodology is highly subjective, studies have shown that when performed by expert readers, it may be relatively accurate when compared with actual measurements, while other studies reported significant inter-reader variability underscoring the need for systematic quantification.3–7
It is widely accepted that the solution to this conundrum lies in the development of automated techniques, which have surged in the recent years with the rapid developments in computer hardware and software technology. Most recently, machine learning techniques, commonly known as artificial intelligence, have been employed to automatically identify LV endocardial boundaries.8–12 These algorithms use for training large databases of echocardiographic images of variable quality depicting a wide range of pathologies, in which endocardial boundaries have been either traced or confirmed by experts. The training consists of identifying image features and patterns and associating them with endocardial boundary position. This knowledge is then used to automatically identify boundaries in images that are not part of the training set. This approach has indeed resulted in successful near automated quantification of LV volumes and EF in a majority of unselected patients undergoing echocardiographic examinations.13 Nevertheless, boundary identification is prone to errors due to suboptimal image quality, artifacts, and extremely unusual image features secondary to different pathologies. As a result, there is a certain percentage of patients in whom these algorithms may not be sufficiently robust.
Accordingly, we hypothesized that a different approach could be implemented to develop a fully automated machine learning algorithm that would mimic what an experienced human eye and brain do, when they estimate LV EF without tracing the endocardial borders and calculating ventricular volumes. We postulated that instead, similar to a human eye, given a sufficiently large and heterogeneous learning data set, the computer could be trained to directly estimate the dimensionless degree of ventricular contraction and expansion, independently of ventricular size. The aim of this study was to develop such an algorithm and test its feasibility and accuracy for automated quantification of LV EF by a well-trained expert computer against reference values provided by a panel of human experts using conventional volume-based measurements.
Methods
Data and materials used in this study will not be made publicly available.
Machine Learning Algorithm
Our unconventional machine learning algorithm was developed to estimate LV EF without measuring LV end-systolic and end-diastolic volumes. This alternative approach assumes that the ventricle contracts throughout systole simultaneously along its long axis and in the radial direction, so that its corresponding dimensions L and R change over time according to 2 time-dependent dimensionless contraction coefficients CL(t) and CR(t), (Figure 1). Using these coefficients, LV volume throughout the cardiac cycle can be described by the following function of time:
Figure 1.
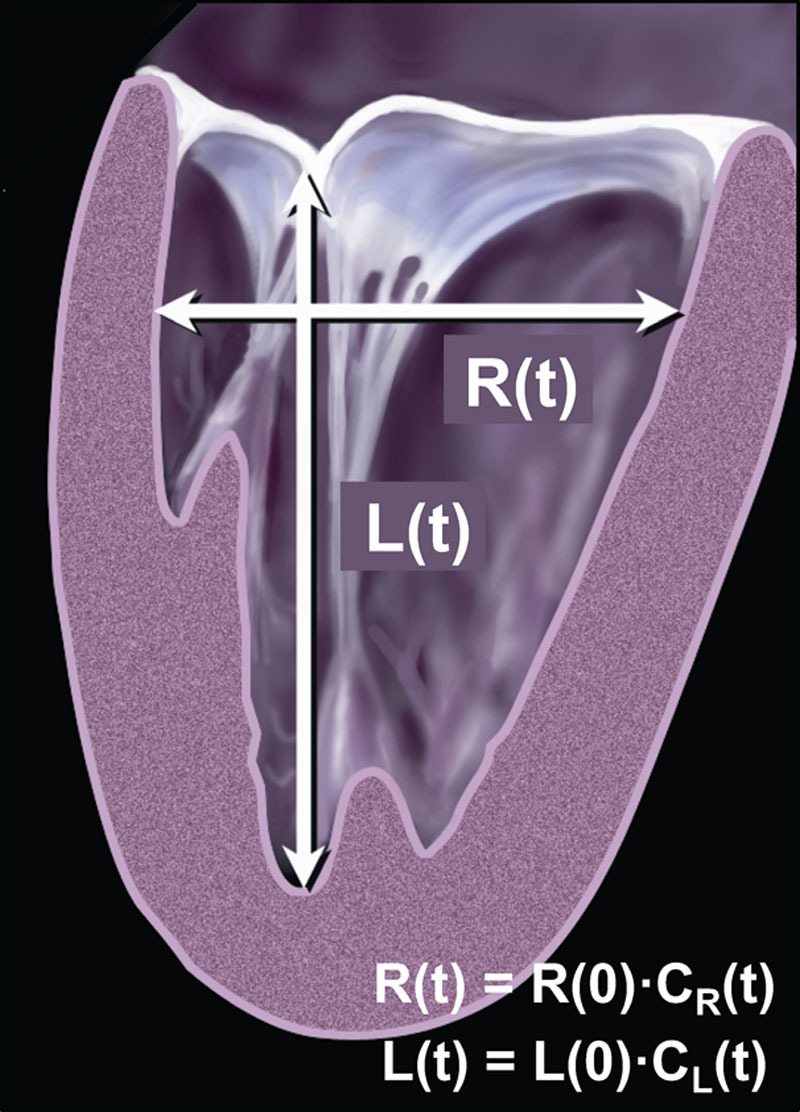
Schematic representation of the left ventricle with its longitudinal and radial dimensions L and R, which change over time from its initial values at time 0, namely L(0) and R(0) according to dimensionless time-dependent contraction coefficients CL(t) and CR(t), which can be used to calculate ejection fraction (see text for details).
where V(0) is the volume at time 0, and CL(t) and CR(t) reach their minimum values at the end of systole and their maximum value of 1.0 at the end of diastole. By definition of LV EF as the difference between the maximum volume at end-diastole (ED) and minimum volume at end-systole (ES) normalized by the former, it can be expressed in terms of the above 2 contraction coefficients as follows:
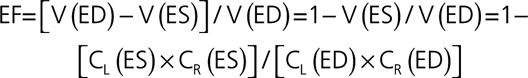 |
Assuming that ED is time 0, both CL(ED) and CR(ED) would equal 1, while CL(ES) and CR(ES) are their minimum values, this expression is reduced to:
which allows calculation of EF from the estimated minimum contraction coefficients in the longitudinal and radial direction without measuring the volumes. For example, if during systole, the ventricle shortens by 14%, CL would reach minimum value of 0.86, and if at the same time its radial dimension shortens by 30%, corresponding to the minimum CR value of 0.70, this would result in EF of 40%:
Our machine learning algorithm was designed to train the computer to estimate the minimum values of the above 2 contraction coefficients, CL-min and CR-min, at the end of contraction. Briefly, we used a deep learning technique, which does not use any sort of explicit tracking methodology, but instead lets the neural network decide from the data itself what the best approach to handle the data would be. In other words, the algorithm was not guided by the developers as to what should be detected or tracked throughout the cardiac cycle. Instead, the algorithm was allowed to derive from the thousands of images the features and visual patterns necessary to estimate EF in agreement with the reference values obtained by human readers using conventional methodology. The neural network was constrained to report the amplitude of change in ventricular dimensions, roughly the equivalent of the above contraction coefficients. Importantly, the neural network had the total freedom in choosing to track relative sizes/dimensions of physiological features and speckle patterns. It is likely that it uses a combination of these.
Algorithm Training
The algorithm developed on the basis of the above described principle (AutoEF, BayLabs inc, San Francisco, CA) was implemented in Python and trained using Keras (https://keras.io/) with a Tensorflow (https://www.tensorflow.org/) backend to train and deploy the Neural Networks. The training was performed on a database of >50 000 echocardiographic studies from the Minneapolis Heart Institute over a period of 10 years, using the following equipment: ACUSON/Siemens SEQUOIA (N=17,359), SC2000 (N=6308), CX50 (N=4472); Philips iE33 (N=7279), EPIQ 7C (N=706); General Electric Vivid-I (N=14,957), Vivid 7 (N=839). Contrast-enhanced images were not used. Training included the use of multiple apical 2- and 4-chamber views available as part of each individual exam and LV EF values measured over the years by clinicians interpreting these studies using conventional methodology (biplane Simpson technique), recommended by the American Society of Echocardiography guidelines.1 After this training, the algorithm was designed to provide fully automated estimates of LV EF on any pair of apical 2- and 4-chamber views.
Performance Testing
This algorithm was tested on an independent group of 99 patients undergoing clinically indicated echocardiographic examinations at the Minneapolis Heart Institute (age 66±16, 62 males, 37 females). The patients were retrospectively selected from the database into 3 equally sized subgroups of LV EF: 0% to 35%, >35 and ≤55%, and >55%, to have a wide range of uniformly represented LV function. In addition, these patients were selected into 3 equally sized subgroups of body mass index: 0 to 25, 26 to 30, and >30 kg/m2, to fairly represent the inter-patient differences in body habitus. No additional criteria, such as image quality, were used to exclude patients. The study was approved by the Institutional Review Board with a waiver of consent.
Images were acquired using a random mix of available equipment similar to the training data set. Each patient underwent imaging in apical 2- and 4-chamber views, which were saved in multiple loops. Three sonographers independently selected the pair of best two loops for analysis (one for each view), resulting in 3 distinct automated EF estimates. These values were used to assess measurement reproducibility when applied to different image loops, with the understanding that the fully automated, deterministic nature of the algorithm inherently has zero inter- and intraobserver variability when applied to the same image pair. Reproducibility (or consistency) was assessed by calculating the mean absolute difference between the 3 EF estimates obtained in each patient.
To validate the automated EF estimates, and on the understanding that the role of such automated system would be to assist cardiologists in the interpretation of echocardiograms in their usual clinical environment, we have decided to test them against an accepted reference standard most relevant to the field. Therefore, reference values were obtained using conventional volume-based technique (biplane Simpson technique) by 3 experts, board-certified echocardiographers (Drs Lang, Asch, and Abraham). To ensure that these reference values reliably represented the LV function of each patient, each reader analyzed all 3 pairs of apical views in a random order, resulting in a total of 9 independent conventional EF measurements per patient. Averaging these 9 values resulted in a single reference EF value per patient.
Once these 99 reference values were obtained, validation consisted of comparing for each of the 99 patients all 3 automated EF estimates (total N=297) to the corresponding reference value. Inter-technique agreement was assessed using linear regression, intraclass correlation (ICC), and Bland-Altman analysis of biases and limits of agreement (LOA). In addition, as an alternative to Bland-Altman bias, which can be zero in the presence of wide LOA, inter-technique agreement was also assessed using MAD, which was calculated for the 297 automated estimates obtained in the 99 patients and the corresponding 99 reference values. Both MAD and Bland-Atman metrics were expressed in absolute EF units, namely percent of ED volume.
Finally, we calculated the sensitivity, specificity, negative and positive predictive values and overall accuracy of the detection of severely reduced LV function, reflected by EF ≤35%. In addition, κ-statistics were used to test the inter-technique agreement in the ability to detect severely reduced LV function, according to the above definition. The calculated κ-coefficients were judged as follows: 0 to 0.2 low, 0.2 to 0.4 moderate, 0.4 to 0.6 substantial, 0.6 to 0.8 good, and >0.8 excellent.
To put in perspective all these performance metrics of the automated EF estimates, they were compared against the same reference standard side-by-side with those obtained from conventional EF measurements performed by clinical readers who interpreted these studies at the Minneapolis Heart Institute that provided them to us. This included comparisons of the results of the linear regression, Bland-Altman analysis, MAD, ICC, and the accuracy of detection of EF ≤35%. In contrast, consistency of the automated EF estimates was not compared with that of the clinical readers, which included only one EF value per patient.
Finally, the latter EF measurements performed by clinical readers were used to create a mathematical de-trending correction. Specifically, we assumed that these measurements when compared against reference values provided by the expert panel, would yield a regression equation that is not an identity function y=x, but y=Ax-B (where the slope A≠1 and the intercept B≠0), which would indicate a trend of error. These coefficients A and B were then used to devise a linear de-trending procedure, which would essentially force the above data set of conventional clinical EF measurements to best fit the identity function. Then, this de-trending procedure with the same coefficients A and B was applied to the automated EF estimates. The corrected automated EF estimates were tested for reproducibility and accuracy, using the same metrics initially used for the uncorrected estimates, namely consistency MAD, linear regression, Bland-Altman biases and LOA (defined as ±1.96 SD around the mean), MAD and the accuracy of detection of EF ≤35%.
Statistical Analysis
Data were expressed as mean values±standard deviations. Significance of biases between the automated EF estimates and the reference measurements was assessed using 2-tailed paired Student t tests. Similarly, the biases between the clinical reads and the expert-provided reference standard were tested using the same methodology. To rule out the effects of data clustering due to dependency in observations (namely 3 EF estimates in the same patient), the 3 estimates were compared against the reference values separately, one at a time. This included linear regression, ICC, and Bland-Altman analyses. All analyses, including basic statistics, were performed using Microsoft Excel. Additional statistical analyses were performed using Prism software (GraphPad Software, San Diego, CA).
Results
The only user input required by the automated algorithm is the selection of the 2 apical views. Once this is done, the time to obtain an EF value was in the order of magnitude of 1 to 5 seconds on a standard personal computer. The algorithm was able to analyze all 297 pairs of apical views obtained in the 99 patients that comprised the testing set.
Repeated automated EF measurements from different pairs of apical images resulted in MAD =2.9±2.0%, reflecting high level of consistency. These automated measurements were in excellent agreement with the reference standard, reflected by a high correlation with r=0.95 (P<0.001; CI, 0.938–0.960), ICC =0.92 (CI, 0.90–0.936), a minimal bias of 1.0% with LOA of ±12.1% (Figure 2, left), MAD =5.1±3.5%. This was comparable to or slightly better than the accuracy of the clinical readers’ measurements: r=0.94 (P<0.001; CI, 0.925–0.952), ICC =0.90 (CI, 0.876–0.920), a bias of 1.4% (P=0.65) and LOA of ±13.8% (Figure 2, right), and MAD =5.8±3.9% (P=0.23). Separate comparisons for the individual estimates (N=99 each) showed similar correlations, biases and LOA to those obtained for the 3 estimates combined (Table 1), indicating that data clustering due to dependency in observations did not have major effect.
Figure 2.
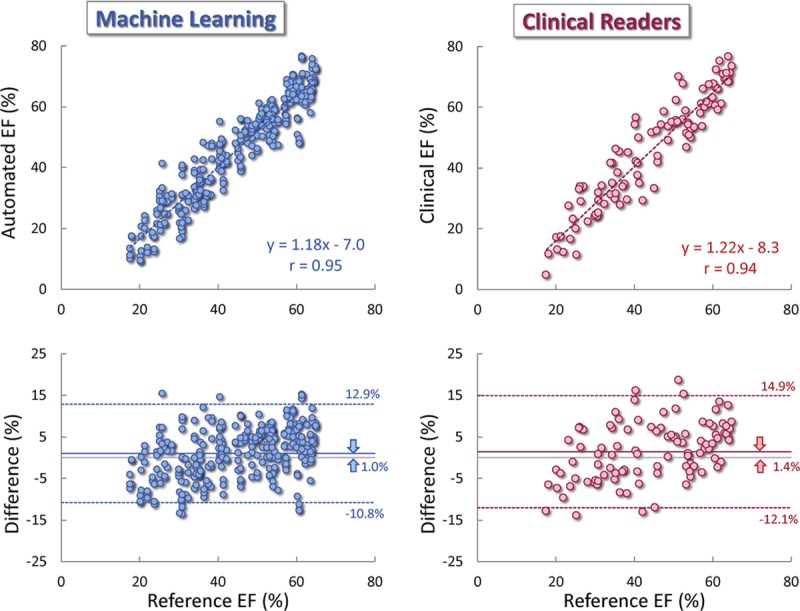
Agreement between the machine learning-based automated ejection fraction (EF) measurements (left), side-by-side with the clinical measurements (right) against reference values obtained by averaging measurements by a panel of 3 experts: linear regression (top) and Bland-Altman analysis (bottom).
Table 1.
Effects of Data Clustering Due to Dependency in Observations, Namely 3 EF Estimates in the Same Patient
| All 3 Estimates | Estimate 1 | Estimate 2 | Estimate 3 | ||
|---|---|---|---|---|---|
| Linear regression | r value | 0.95 | 0.94 | 0.94 | 0.95 |
| P value | 0.0000 | 0.0000 | 0.0000 | 0.0000 | |
| 95% CI | (0.938–0.96) | (0.912–0.959) | (0.912–0.959) | (0.926–0.966) | |
| Intraclass correlation | ICC | 0.92 | 0.92 | 0.92 | 0.93 |
| 95% CI | (0.900–0.936) | (0.883–0.946) | (0.883–0.946) | (0.897–0.953) | |
| Bland-Altman analysis | Bias | 1.0 | 1.1 | 0.8 | 1.1 |
| LOA (2SD) | 12.1 | 12.2 | 12.4 | 11.7 |
Results of linear regression and Bland-Altman analysis (both against the reference values), performed for all 3 estimates combined vs each of the 3 estimates separately. See text for details. EF indicates ejection fraction; ICC, indicates intraclass correlation; LOA, limits of agreement.
In terms of the ability to correctly identify patients with severely reduced LV function (EF ≤35%), the automated analysis resulted in high sensitivity, specificity, negative and positive predictive values, and accuracy, which reflected a comparable or slightly better diagnostic performance than the clinical reads, when both were compared against the same reference standard (Table 2). Similarly, κ value was higher for the automated analysis than for the clinical reads: 0.806 (judged as excellent), compared with 0.745 (judged as good), respectively (Table 2).
Table 2.
Diagnostic Performance of the Machine-Learning Algorithm for Automated Evaluation of LVEF in Terms of its Ability to Identify Patients With Severely Reduced LV Function (EF ≤35%)
| Sensitivity | Specificity | NPV | PPV | Accuracy | κ | |
|---|---|---|---|---|---|---|
| Auto EF | 0.90 | 0.92 | 0.96 | 0.83 | 0.92 | 0.806 |
| (95% CI) | (0.82–0.95) | (0.88–0.95) | (0.92–0.98) | (0.74–0.89) | (0.88–0.95) | (0.733–0.880) |
| Clinical reads | 0.93 | 0.87 | 0.97 | 0.74 | 0.89 | 0.745 |
| (95% CI) | (0.76–0.99) | (0.77–0.93) | (0.89–0.99) | (0.58–0.86) | (0.81–0.94) | (0.606–0.885) |
Data in parentheses represent 95% CI. EF indicates ejection fraction; LV, left ventricular; NPV, indicates negative predictive value; and PPV, positive predictive value.
The de-trending correction resulted in a slight improvement in the agreement with the reference standard (Figure 3), reflected by a correlation unchanged from the original automated EF data (r=0.95; P<0.001; CI, 0.938–0.960), but higher ICC =0.94 (CI, 0.925–0.952), smaller bias (−0.4% versus 1.0%; P<0.001) with narrower LOA (±8.9% versus ±12.1%), and smaller MAD (3.5±2.8% versus 5.1±3.5%; P<0.001). The de-trending correction also improved the reproducibility of the automated measurements from different image pairs, as reflected by a lower consistency MAD value (2.1±1.5% versus 2.9±2.0%; P<0.001).
Figure 3.
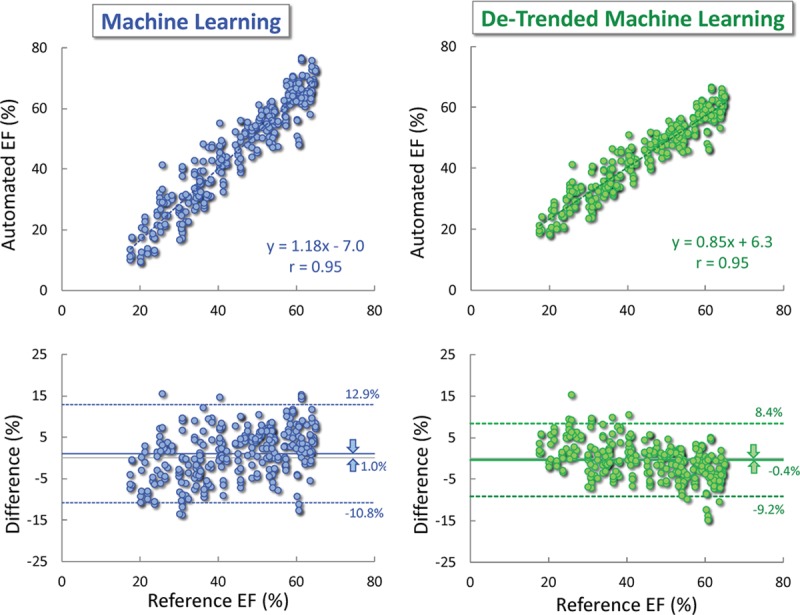
Agreement between the machine learning-based automated ejection fraction (EF) measurements without (left) and with de-trending correction (right) against reference values obtained by averaging measurements by a panel of 3 experts: linear regression (top) and Bland-Altman analysis (bottom).
Discussion
In this study, we tested the feasibility and accuracy of a novel fully automated, machine learning algorithm for the quantification of LV EF without first identifying endocardial borders and measuring LV volumes at end-systole and end-diastole. Testing included 99 patients representing a wide range of EF and image quality as it relates to body habitus. We found that the novel algorithm was feasible in every patient included in the test group, and when compared with the reference standard of conventional measurements performed by a panel of experts, the automated estimates were highly accurate. Importantly, the accuracy was similar to that of the conventional analysis by independent clinical readers. In addition, the automated analysis yielded highly consistent results when applied to different pairs of apical 2- and 4-chamber views. Finally, we demonstrated that a simple mathematical de-trending correction based on parameters derived from conventional measurements, further improved the accuracy and consistency of the automated analysis by almost eliminating the inter-technique bias and minimizing the LOA.
The rapid technological evolution of the past decades that brought about an exponential increase in computing power has spurred the development of artificial intelligence in virtually every area of our lives. It is probably not unreasonable to think that we are only at the beginning of this technological revolution that already surpasses the imagination of most of us. In the era of ubiquitous devices that allow us to control almost anything around us with a few words, it is only natural that the sweeping change in perception of what can and what cannot be done without direct human involvement has not spared the field of medical imaging.14,15 Specifically, machine learning is rapidly proving its usefulness in cardiac imaging, where it allows automated identification of cut-planes and cardiac structures and lends itself to an increasing variety of automated measurements that up to now have relied on sometimes extensive user input.16,17 While the details of how machine learning works are difficult to understand for most who are not professionals in this area, we all know that it requires for training purposes big data that includes every possible anatomic and functional variation, to reach accurate performance. Fortunately, echocardiography has been in existence long enough to offer the developers data sets of hundreds of thousands of exams containing millions of images traced, measured, and characterized by human experts over the years.
The novel machine-learning algorithm tested in this study is an example of such collaboration that resulted in a new tool that was designed to automatically provide an answer to probably the most commonly encountered question in echocardiography, namely the quantification of LV function. The novel approach at the basis of this tool was based on the idea that if a human eye and brain can learn with experience how to estimate EF without measuring LV volumes and making calculations, then machine learning could be harnessed to train a computer to perform this task. One of the advantages of this approach is that it does not rely on accurate image segmentation throughout the entirety of the blood-tissue interface, which is known to be difficult when image quality is suboptimal, and as a result is known as the major source of inaccuracy. A well-trained human eye knows how to extrapolate endocardial border position in areas where it is poorly defined, and estimate the magnitude of ventricular contraction despite such gaps. This concept was translated into the simple mathematical formalism described above and implemented into a machine-learning algorithm to mimic what an expert’s eye does, after being trained on a sufficiently large number of cases representing most ventricular geometries and functional abnormalities.
Nonetheless, one might legitimately question our approach since LV volumes are meaningful too, and are often part of the clinical decision making. However, LV EF is the critical parameter expected to be included in every echo report, and by no means do we advocate ignoring LV volumes. This study aimed at testing the feasibility of a fully automated quantitative evaluation of EF without any human involvement by taking advantage of a novel machine learning approach to strengthen the baseline data available by default before any user supervised analysis is performed. We envision this information being included with the images the moment the reader opens the study. This does not preclude the reader from using any additional means to measure and report LV volumes, when deemed necessary, with the understanding that in most cases, a statement that LV size is normal may suffice.
Our study provided initial validation in a group of patients, who were not part of the large training set. One might question the fact that cardiac magnetic resonance, which is frequently referred to as the gold standard for cardiac chamber quantification18 was not used as a reference for comparisons. However, this was neither an oversight nor an attempt to cut corners in the design of our study, but rather intentional and directly related to our goal, which was testing the novel algorithm as a potential substitute for the conventional echocardiographic methodology. We could not have learned how close these 2 techniques are by comparing either one or both of them to a cardiac magnetic resonance reference. It is well established by previous studies that echocardiography underestimates ventricular volumes resulting in rather inconsistent inter-modality differences in EF,2 and such comparisons would likely confirm this knowledge, while leaving the above key question unanswered. We felt that the only way to know whether the automated technique is potentially good enough to replace the conventional measurement methodology is to compare the former head-to-head against a strong reference obtained using the latter. To achieve this goal, we averaged EF measurements obtained from multiple images (9 pairs of apical views obtained by 3 different sonographers and analyzed by 3 different expert readers, who are leaders in the field and were instrumental in developing the ASE guidelines). Comparisons against this reference showed excellent agreement, indicating the potential value of the newly developed automated approach. Moreover, by comparing the accuracy of the automated approach side-by-side with that of the conventional measurements by independent clinical readers proved once again in a different way that the automated approach is at least as good as human readers, with the clear advantage of its fast and fully automated nature.
Limitations
In this era of big data, one might see the relatively small size of our test group (99 patients) as a limitation. Indeed, 99 is a disproportionately small number compared with 50 000 studies used for training the software. However, it is important to remember that this was the first study to test the feasibility of this novel algorithm and evaluate its ability to compete with conventional measurements by human experts. Also, the actual number of automated estimates that were tested was 297, since each of the 3 pairs of apical views obtained in each patient were tested independently. This sample provided high levels of statistical confidence (evidenced by very low P values in every comparison where differences were noted), indicating that further increase in the sample would be highly unlikely to affect the findings.
One inherent limitation of the approach tested in this study is that in the absence of endocardial boundaries, the potential user would not be able to confirm the accuracy of the automated EF estimates by verifying the accuracy of the endocardial boundary position and tracking. However, when he/she feels that the estimate may not be accurate, there would still be the option of using the conventional methodology to compare and verify.
One might see as a limitation the fact that the training set included 50 000 studies acquired over an extended period of time using imaging equipment from a variety of vendors, and was thus nonuniform in image quality and other characteristics. However, this may also be seen as a strength of our study design, as the applicability of the tested approach is more likely to be widely generalizable.
Another limitation is related to the de-trending correction, which was based on the trend noted in the measurements made by the clinical readers of the 99 patients in the test group. This is somewhat problematic because the software was trained on a data set generated by the same laboratory that provided the test set, suggesting that if the 99 studies were analyzed by readers from another institution who may be using a slightly different measurement methodology, the de-trending parameters might be different. In fact, a close examination of the results of the de-trending shows that, despite the fact that it improved both the accuracy and consistency of the automated estimates, it might have actually resulted in an over-correction (Figure 2, right). This is evidenced by the regression slope A <1 and a positive intercept B, as well as a negative (albeit minimal) bias, with mostly negative inter-technique differences in the high EF range, all being the opposite of the trends seen in the noncorrected data (Figure 2, left). Accordingly, the ultimate value of the de-trending correction remains to be proven.
Conclusions
This initial feasibility study demonstrated that machine learning algorithm for estimation of LV EF that avoids image segmentation and volume measurements and instead mimics a human expert’s eye, is highly feasible and can yield results that are in close agreement with what highly experienced readers measure using conventional methodology. This approach may prove to have important clinical implications because of its fast and fully automated nature. It is conceivable that in the future, the starting point of interpretation of an echocardiography exam would include automated estimates of ventricular function the reader would be presented with along with the images.
Footnotes
The Data Supplement is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCIMAGING.119.009303.
Acknowledgment
We wish to thank Dr Richard Bae and the Echocardiographic laboratory at Minneapolis Heart Institute for providing the data sets used in this study.
Sources of Funding
The study was supported by Bay Labs Inc.
Disclosures
In addition to employment relationships between several authors (Dr Poilvert, M. Adams, N. Romano, Dr Hong, and Dr Martin) and Bay Labs Inc. Drs Abraham, Mor-Avi, and Lang served as paid consultants to this company for work related to this article. The other authors report no conflicts.
References
- 1.Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, Lancellotti P, Muraru D, Picard MH, Rietzschel ER, Rudski L, Spencer KT, Tsang W, Voigt JU. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American society of echocardiography and the European Association of cardiovascular imaging. J Am Soc Echocardiogr. 2015;28:1.e14–39.e14. doi: 10.1016/j.echo.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 2.Mor-Avi V, Jenkins C, Kühl HP, Nesser HJ, Marwick T, Franke A, Ebner C, Freed BH, Steringer-Mascherbauer R, Pollard H, Weinert L, Niel J, Sugeng L, Lang RM. Real-time 3-dimensional echocardiographic quantification of left ventricular volumes: multicenter study for validation with magnetic resonance imaging and investigation of sources of error. JACC Cardiovasc Imaging. 2008;1:413–423. doi: 10.1016/j.jcmg.2008.02.009 [DOI] [PubMed] [Google Scholar]
- 3.Rich S, Sheikh A, Gallastegui J, Kondos GT, Mason T, Lam W. Determination of left ventricular ejection fraction by visual estimation during real-time two-dimensional echocardiography. Am Heart J. 1982;104:603–606. doi: 10.1016/0002-8703(82)90233-2 [DOI] [PubMed] [Google Scholar]
- 4.Hope MD, de la Pena E, Yang PC, Liang DH, McConnell MV, Rosenthal DN. A visual approach for the accurate determination of echocardiographic left ventricular ejection fraction by medical students. J Am Soc Echocardiogr. 2003;16:824–831. doi: 10.1067/S0894-7317(03)00400-0 [DOI] [PubMed] [Google Scholar]
- 5.Mele D, Campana M, Sclavo M, Seveso G, Aschieri D, Nesta F, D’Aiello I, Ferrari R, Levine RA. Impact of tissue harmonic imaging in patients with distorted left ventricles: improvement in accuracy and reproducibility of visual, manual and automated echocardiographic assessment of left ventricular ejection fraction. Eur J Echocardiogr. 2003;4:59–67. doi: 10.1053/euje.2002.0619 [DOI] [PubMed] [Google Scholar]
- 6.Thavendiranathan P, Popović ZB, Flamm SD, Dahiya A, Grimm RA, Marwick TH. Improved interobserver variability and accuracy of echocardiographic visual left ventricular ejection fraction assessment through a self-directed learning program using cardiac magnetic resonance images. J Am Soc Echocardiogr. 2013;26:1267–1273. doi: 10.1016/j.echo.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 7.Kusunose K, Shibayama K, Iwano H, Izumo M, Kagiyama N, Kurosawa K, Mihara H, Oe H, Onishi T, Onishi T, Ota M, Sasaki S, Shiina Y, Tsuruta H, Tanaka H; JAYEF Investigators. Reduced variability of visual left ventricular ejection fraction assessment with reference images: the Japanese Association of Young Echocardiography Fellows Multicenter Study. J Cardiol. 2018;72:74–80. doi: 10.1016/j.jjcc.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 8.Tsang W, Salgo IS, Medvedofsky D, Takeuchi M, Prater D, Weinert L, Yamat M, Mor-Avi V, Patel AR, Lang RM. Transthoracic 3D echocardiographic left heart chamber quantification using an automated adaptive analytics algorithm. JACC Cardiovasc Imaging. 2016;9:769–782. doi: 10.1016/j.jcmg.2015.12.020 [DOI] [PubMed] [Google Scholar]
- 9.Medvedofsky D, Mor-Avi V, Amzulescu M, Fernández-Golfín C, Hinojar R, Monaghan MJ, Otani K, Reiken J, Takeuchi M, Tsang W, Vanoverschelde JL, Indrajith M, Weinert L, Zamorano JL, Lang RM. Three-dimensional echocardiographic quantification of the left-heart chambers using an automated adaptive analytics algorithm: multicentre validation study. Eur Heart J Cardiovasc Imaging. 2018;19:47–58. doi: 10.1093/ehjci/jew328 [DOI] [PubMed] [Google Scholar]
- 10.Tamborini G, Piazzese C, Lang RM, Muratori M, Chiorino E, Mapelli M, Fusini L, Ali SG, Gripari P, Pontone G, Andreini D, Pepi M. Feasibility and accuracy of automated software for transthoracic three-dimensional left ventricular volume and function analysis: comparisons with two-dimensional echocardiography, three-dimensional transthoracic manual method, and cardiac magnetic resonance imaging. J Am Soc Echocardiogr. 2017;30:1049–1058. doi: 10.1016/j.echo.2017.06.026 [DOI] [PubMed] [Google Scholar]
- 11.Levy F, Dan Schouver E, Iacuzio L, Civaia F, Rusek S, Dommerc C, Marechaux S, Dor V, Tribouilloy C, Dreyfus G. Performance of new automated transthoracic three-dimensional echocardiographic software for left ventricular volumes and function assessment in routine clinical practice: comparison with 3 tesla cardiac magnetic resonance. Arch Cardiovasc Dis. 2017;110:580–589. doi: 10.1016/j.acvd.2016.12.015 [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Feng H, Ni L, Wang H, Gao D. Realization of fully automated quantification of left ventricular volumes and systolic function using transthoracic 3D echocardiography. Cardiovasc Ultrasound. 2018;16:2 doi: 10.1186/s12947-017-0121-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medvedofsky D, Mor-Avi V, Byku I, Singh A, Weinert L, Yamat M, Kruse E, Ciszek B, Nelson A, Otani K, Takeuchi M, Lang RM. Three-dimensional echocardiographic automated quantification of left heart chamber volumes using an adaptive analytics algorithm: feasibility and impact of image quality in nonselected patients. J Am Soc Echocardiogr. 2017;30:879–885. doi: 10.1016/j.echo.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Summers RM. Machine learning and radiology. Med Image Anal. 2012;16:933–951. doi: 10.1016/j.media.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Ginneken B. Fifty years of computer analysis in chest imaging: rule-based, machine learning, deep learning. Radiol Phys Technol. 2017;10:23–32. doi: 10.1007/s12194-017-0394-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Gopalakrishnan V. An overview and evaluation of recent machine learning imputation methods using cardiac imaging data. Data (Basel). 2017;2:8. doi: 10.3390/data2010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, Lassen MH, Fan E, Aras MA, Jordan C, Fleischmann KE, Melisko M, Qasim A, Shah SJ, Bajcsy R, Deo RC. Fully automated echocardiogram interpretation in clinical practice. Circulation. 2018;138:1623–1635. doi: 10.1161/CIRCULATIONAHA.118.034338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer CM, Barkhausen J, Flamm SD, Kim RJ, Nagel E; Society for Cardiovascular Magnetic Resonance Board of Trustees Task Force on Standardized Protocols. Standardized cardiovascular magnetic resonance imaging (CMR) protocols, society for cardiovascular magnetic resonance: board of trustees task force on standardized protocols. J Cardiovasc Magn Reson. 2008;10:35 doi: 10.1186/1532-429X-10-35 [DOI] [PMC free article] [PubMed] [Google Scholar]


