Abstract
One of the most statistically significant loci to result from large-scale GWAS of schizophrenia is 10q24.32. However, it is still unclear how this locus is involved in the pathoaetiology of schizophrenia. The hypothesis that presynaptic dopamine dysfunction underlies schizophrenia is one of the leading theories of the pathophysiology of the disorder. Supporting this, molecular imaging studies show evidence for elevated dopamine synthesis and release capacity. Thus, altered dopamine function could be a potential mechanism by which this genetic variant acts to increase the risk of schizophrenia. We therefore tested the hypothesis that the 10q24.32 region confers genetic risk for schizophrenia through an effect on striatal dopamine function. To this aim we investigated the in vivo relationship between a GWAS schizophrenia-associated SNP within this locus and dopamine synthesis capacity measured using [18F]-DOPA PET in healthy controls. 92 healthy volunteers underwent [18F]-DOPA PET scans to measure striatal dopamine synthesis capacity (indexed as Kicer) and were genotyped for the SNP rs7085104. We found a significant association between rs7085104 genotype and striatal Kicer. Our findings indicate that the mechanism mediating the 10q24.32 risk locus for schizophrenia could involve altered dopaminergic function. Future studies are needed to clarify the neurobiological pathway implicated in this association.
Keywords: schizophrenia, 10q24.32, psychosis, dopamine synthesis capacity, PET, striatum, imaging
1. Introduction
Large-scale Genome-Wide Association Studies (GWAS) have identified a number of common genetic variations conferring risk for schizophrenia in human populations (Pardiñas et al., 2018; Ripke et al., 2014, 2013, 2011). Nevertheless, it remains challenging to assign the association signal at risk loci to specific genes or mechanisms, which is a requirement if genetic findings are to help elucidate the pathophysiology of the disorder. In this sense it is important to identify methods to investigate the biological pathways linked to risk loci and thus help identify potential pharmacological targets (Birnbaum and Weinberger, 2017; Schubert et al., 2014).
One of the best supported genome-wide significant loci from large-scale GWAS of schizophrenia is the 10q24.32 region (Duarte et al., 2016; Pardiñas et al., 2018; Ripke et al., 2014). Potential specific molecular mechanisms of genetic risk related to this locus have been recently identified. In particular, increased expression of BLOC-1 related complex subunit 7 (BORCS7) and of a novel arsenite methyltransferase isoform, namely AS3MTd2d3, have been found in the brains of patients with schizophrenia relative to controls (Li et al., 2016b). In the general population, the full isoform of arsenite methyltransferase is involved in arsenic metabolism (Sumi and Himeno, 2012), and arsenic toxicity has been implicated in central nervous system (CNS) dysfunction (Tyler and Allan, 2014) and psychosis (Ratnaike, 2003). However, the AS3MTd2d3 isoform lacks arsenite methyltransferase activity (Li et al., 2016b); therefore a different, and still unknown, mechanism is likely to be implicated in the biological process leading to risk for schizophrenia.
The hypothesis that dopamine dysfunction underlies schizophrenia is one of the leading theories of the pathophysiology of the disorder (Abi-Dargham et al., 2000; Davis et al., 1991; Heinz et al., 2003; Heinz and Schlagenhauf, 2010; Howes et al., 2015, 2012; Howes and Kapur, 2009; Laruelle and Abi-Dargham, 1999) and is supported by findings from genetic studies of the disorder through genome-wide significant association at a locus implicating the DRD2 dopamine receptor (Pardiñas et al., 2018; Ripke et al., 2014). Molecular imaging studies show evidence for presynaptic striatal dopamine dysfunction, in particular increased striatal dopamine synthesis and release capacity, in patients with schizophrenia (Abi-Dargham et al., 2009; Hietala et al., 1999; Howes et al., 2013, 2009; Jauhar et al., 2017a; Kumakura et al., 2007; Meyer-Lindenberg et al., 2002; Mizrahi et al., 2012; Reith et al., 1994) with a large effect size on meta-analysis (Cohen d = 0.79) (Howes et al., 2012). Moreover, increased dopamine synthesis capacity is also reported in individuals at increased clinical risk of schizophrenia (Egerton et al., 2013; Howes et al., 2011a), as well as in first-degree relatives of patients with schizophrenia (Huttunen et al., 2008), and linked to transition to the disorder (Howes et al., 2011b). Thus, altered dopamine synthesis capacity could be a potential mechanism by which genetic variation acts to increase risk of schizophrenia.
Our study aimed to test the hypothesis that the association signal at the 10q24.32 locus confers genetic risk for schizophrenia through an effect on striatal dopamine function. We therefore investigated the in vivo relationship between the GWAS schizophrenia-associated SNP rs7085104 (the top expression quantitative trait locus (eQTL) for AS3MT and BORCS7 (Li et al., 2016b)) and dopamine synthesis capacity, measured using [18F]-DOPA positron emission tomography (PET) in healthy controls. We hypothesised that carriers of the rs7085104 risk allele (A) would show increased striatal dopamine synthesis capacity relative to controls who did not carry the risk allele.
2. Methods
2.1. Participants
We studied 92 healthy volunteers (demographics in Table 1). Inclusion criteria were: minimum age 18 years, good physical health with no history of major medical condition and capacity to give written informed consent. Exclusion criteria were: history of significant head trauma, history of neurological disorder, presence of any significant medical disorder or treatment, pregnancy or breastfeeding, a diagnosis of past or current psychiatric disorders using the Structured Clinical Interview for DSM-IV (SR et al., 1996) including alcohol or any other substance dependence or abuse, a family history of any psychotic disorder in first- or second- degree relatives. PET data from some participants have been included in previous publications (Bloomfield et al., 2014a, 2014b; Dahoun et al., 2018; Froudist-Walsh et al., 2017; Jauhar et al., 2017b).
Table 1. Demographic characteristics of the sample.
| AA | GA | GG | Total | |
|---|---|---|---|---|
| N | 43 | 41 | 8 | 92 |
| Age (yr ± SD) | 28.28 ± 8.34 | 31.17 ± 8.46 | 32.50 ± 12.47 | 29.93 ± 8.84 |
| Gender (male/female) | 26/17 | 21/20 | 5/3 | 52/40 |
| Ancestry cluster (EUR/AFR/ASI/other) | 31/9/0/3 | 27/7/2/5 | 6/0/1/1 | 64/16/3/9 |
| PET scanner (scanner 1/scanner 2/scanner 3) | 18/20/5 | 16/13/12 | 3/2/3 | 37/35/20 |
| Tobacco smoking status (non-smoker/smoker) | 36/7 | 31/10 | 6/2 | 73/19 |
| Kicer (1/min) whole striatum (± SD) | 0.013 ± 0.001 | 0.013 ± 0.001 | 0.012 ± 0.001 | 0.013 ± 0.001 |
| Kicer (1/min) associative striatum (± SD) | 0.013 ± 0.001 | 0.013 ± 0.001 | 0.012 ± 0.001 | 0.013 ± 0.001 |
| Kicer (1/min) limbic striatum (± SD) | 0.013 ± 0.001 | 0.013 ± 0.001 | 0.012 ± 0.001 | 0.013 ± 0.001 |
| Kicer (1/min) sensorimotor striatum (± SD) | 0.013 ± 0.001 | 0.014 ± 0.002 | 0.012 ± 0.001 | 0.013 ± 0.001 |
2.2. SNP Selection and Genotype determination
We chose rs7085104 as the sole polymorphism of interest for this study given cumulative evidence for its importance in the 10q24 associated region: (i) a recent finding implicating it as the top SNP eQTL for AS3MT and BORCS7 (Li et al., 2016b), (ii) it was the index SNP at this locus in previous schizophrenia GWAS association (Ripke et al., 2013) and (iii) in a recent genome-wide methylation study rs7085104 was found to be a methylation QTL in human fetal brain for AS3MT (Hannon et al., 2015).
DNA was extracted from whole blood samples or cheek swabs using standard procedures (Freeman et al., 2003). Genotyping was performed at Cardiff University, using HumanCore Exome 1.1 arrays (“Psych-chip”, Illumina, San Diego, California, USA). Genotype quality control (QC) was performed according to standard parameters (Anderson et al., 2010). The variant of interest for our analysis, rs7085104, was directly genotyped and passed QC in all the subjects.
2.3. Population structure
The top 10 principal components of the sample were generated using PC-AiR (Conomos et al., 2015) on the full set of genotypes, and included as covariates of no interest in all the analyses, in order to correct for population stratification.
Moreover, to confirm the robustness of the findings, the analyses were repeated in a sub-group of individuals of European ancestry. To this aim, each individual was assigned to a cluster (European, African, Asian) on the basis of their ancestry score (threshold for the attribution to a specific cluster: 0.7). Ancestry scores were calculated using ADMIXTURE (Alexander et al., 2009) (Version 1.3.0) with the Human Genome Diversity Panel (HGPD-CEPH) (Li et al., 2008) as a reference.
2.4. PET scanning
[18F]-DOPA PET scans were used to measure striatal dopamine synthesis capacity (indexed as the influx rate constant Kicer).
2.4.1. Image acquisition
Dynamic scans were acquired in three-dimensional mode (transaxial resolution of ~5 mm full width at half maximum ((NEMA), 2007) using three different PET scanners: one was a ECAT HR+ 962 PET scanner (CTI/Siemens, Knoxville, Tennessee) while the other two were Siemens Biograph HiRez XVI PET-CT scanners (Siemens Healthcare, Erlangen, Germany). Subjects were asked to refrain from eating and drinking (except water) for at least 12 hours before the scans. One hour before the scan, all participants received 400 mg entacapone, a peripheral catechol-o-methyl-transferase inhibitor which decreases the formation of radiolabelled metabolites that may cross the blood–brain barrier (Cumming et al., 1993; Guttman et al., 1993), and 150 mg carbidopa, a peripheral aromatic acid decarboxylase inhibitor which reduces the peripheral metabolism of the tracer (Garnett et al., 1983). Approximately 150 MBq of radioactive [18F]-DOPA was administered by intravenous injection followed by 95 minutes of dynamic PET scan. PET data were reconstructed using filterback projection and corrected for tissue attenuation and scatter (full details are reported in (Bloomfield et al., 2014a, 2014b; Froudist-Walsh et al., 2017; Jauhar et al., 2017b)).
2.4.2. Analysis of PET data
PET image analysis was performed as previously described (Dahoun et al., 2018). In summary, frames were realigned to a single reference frame, employing a mutual information algorithm (Studholme et al., 1996; Turkheimer et al., 1999). The transformation parameters were then applied to the corresponding attenuated-corrected dynamic images, creating a movement-corrected dynamic image, which was used in the analysis. Realigned frames were then summated to create an individual motion-corrected reference map for the brain tissue segmentation. The striatum was sub-divided into sub-regions as previously described (Howes et al., 2009; Martinez et al., 2003) to create a Region of Interest (ROI) map (Egerton et al., 2010). SPM8 (http://www.fil.ion.ucl.ac.uk/spm) was used to normalize a tracer-specific ([18F]-DOPA) template together with the ROI map to each individual PET summation image (Howes et al., 2009). The striatal influx constant (Kicer) was calculated relative to uptake in the reference region using a graphical approach adapted for a reference tissue input function (Howes et al., 2009). To control for effects of scanner model, PET scanners were included as covariates of no interest in all analyses.
2.5. Statistical analysis
ANOVA and χ2 tests were used to compare demographics age and gender as functions of rs7085104 genotype. ANCOVA analyses were performed to explore the correlation between genotype and dopamine synthesis capacity (fixed factor: rs7085104 genotype [three groups: GG, GA, AA, dependent variable: Kicer, covariates of no interest: age, gender, scanner, top 10 genetic principal components). Exploratory analyses were conducted using associative striatum, limbic striatum and sensorimotor striatum Kicer as dependent variables. All these were performed in SPSS (IBM Corp. Released 2016. IBM SPSS Statistics for Mac, Version 24.0.0.1 Armonk, NY: IBM Corp.). GraphPad Prism 7.02 (http://www.graphpad.com/) was used to plot the main results.
The R package RobustSNP (So and Sham, 2011) was used to confirm the genetic model suggested by the LSD post hoc analysis. The robust tests were performed including the same covariates of no interest used for the ANCOVAs.
3. Results
Demographic (± SD) and Kicer values included are reported in Table 1.
Genotype groups were in Hardy-Weinberg Equilibrium (X2= 0.16, p= 0.69) and did not differ in terms of age (p= 0.22), sex (X2= 0.86, p= 0.65), PET scanner (X2= 5.66, p= 0.23) or tobacco smoking status (X2= 0.94, p= 0.62).
The frequency of the schizophrenia risk allele in our sample approximated that reported in the public NCBI database dbSNP (Sherry, 2001). Specifically, the risk allele (A) of rs7085104 had a reported frequency of 0.62 in the latter, while the frequency for our sample was 0.69.
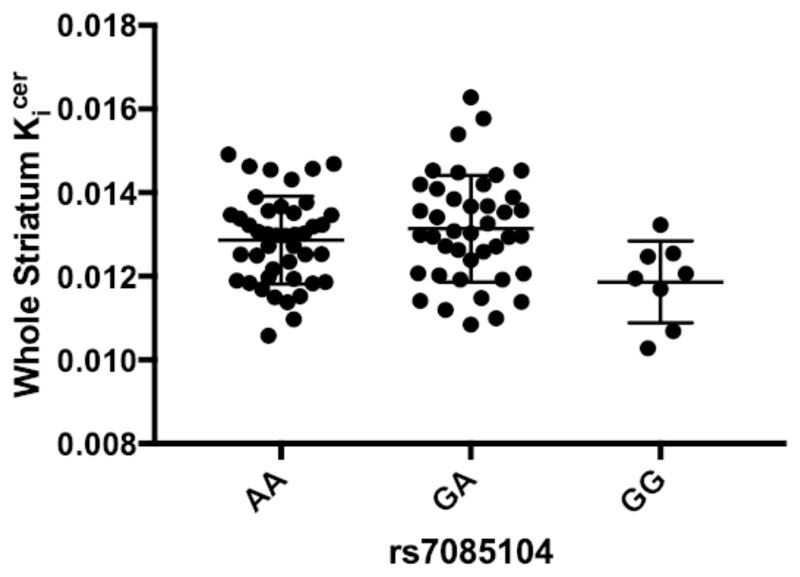
We found a significant association between rs7085104 genotype and whole striatal Kicer (F(2,76)= 4.660, p= 0.012) [Fig. 1]. LSD post-hoc analyses showed that risk allele carriers had significantly elevated Kicer compared to subjects with the genotype GG (GA vs GG, p= 0.003; GA vs AA, p= 0.275; AA vs GG, p= 0.020, Hedges’ g= 0.957), suggesting a dominant effect of A alleles. The RobustSNP association test confirmed the dominant model for this association (ZDominant Model= 2.337, pDominant Model= 0.019).
Figure 1. Effect of AS3MT rs7085104 on whole striatum Kicer.
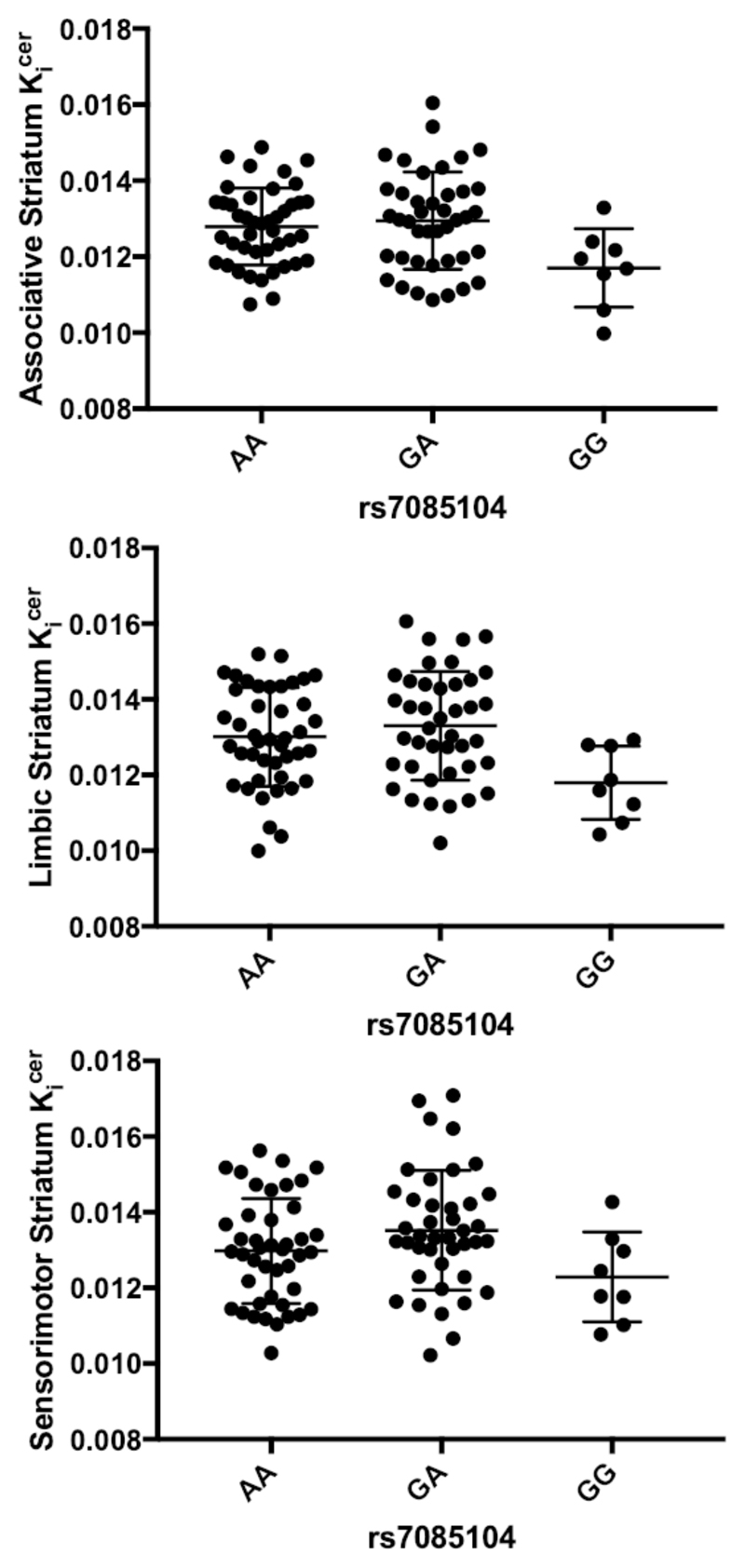
The exploratory analyses in the striatal subdivisions revealed an effect of genotype on dopamine synthesis capacity for all the regions. Specifically, we found an effect of rs7085104 genotype on associative Kicer (F(2,76)= 4.075, p= 0.021; LSD post-hoc contrasts: GA vs GG, p= 0.006; GA vs AA, p= 0.424; AA vs GG, p= 0.020) [Fig. 2a], limbic Kicer (F(2,76)= 5.453, p= 0.006; LSD post-hoc contrasts: GA vs GG, p= 0.001; GA vs AA, p= 0.406; AA vs GG, p= 0.007) [Fig. 2b], and sensorimotor Kicer (F(2,76)= 4.100, p= 0.020; LSD post-hoc contrasts: GA vs GG, p= 0.008; GA vs AA, p= 0.126; AA vs GG, p= 0.071) [Fig. 2c]. RobustSNP association tests confirmed the dominant effect of A alleles on dopamine synthesis capacity for all the striatal subdivisions (associative Kicer : ZDominant Model= 2.217, pDominant Model= 0.0266; limbic Kicer : ZDominant Model= 2.480, pDominant Model= 0.013; sensorimotor Kicer : ZDominant Model= 2.345, pDominant Model= 0.019).
Figure 2. Exploratory analyses: effect of AS3MT rs7085104 on Kicer in the different striatal subdivisions.
The sensitivity analyses performed in the sub-group of individuals of European Ancestry confirmed the association of rs7085104 genotype with whole striatal Kicer (F(2,48)= 6.102, p= 0.004) and all the striatal subdivisions: associative Kicer (F(2,48)= 4.835, p= 0.012); limbic Kicer (F(2,48)= 6.935, p= 0.002); and sensorimotor Kicer (F(2,48)= 6.091, p= 0.004).
4. Discussion
We report for the first time an in vivo association between the schizophrenia-associated SNP rs7085104 and striatal dopamine synthesis capacity. Specifically, we found that the rs7085104 risk allele (A) carriers show higher striatal Kicer than the subjects homozygous for the G allele. It is not clear at present whether the possible dominant effect we have detected on dopamine synthesis capacity matches the observed effect of this SNP as a schizophrenia risk allele, since the latter has at the moment only been assessed by GWAS based on simple additive models (Pardiñas et al., 2018; Ripke et al., 2014, 2013). In order to resolve this, explicit modelling of dominance effects in large-scale schizophrenia samples would have to be performed, as has been explored in other psychiatric conditions (Leblond et al., 2019; Van der Auwera et al., 2018). At the moment, dominant effects are not currently considered to explain much of the variance of human complex traits in general, though they have been shown to exist at some particular loci (Zhu et al., 2015).
These results offer new insights into the biological mechanisms that could underlie the association of the locus 10q24.32 with schizophrenia. A genetic risk variant on chromosome 10q24.32 has recently been demonstrated to correlate with the expression of the gene AS3MT (Duarte et al., 2016). AS3MT codes for an arsenic methyltransferase (Lin et al., 2002; Sumi and Himeno, 2012). Since arsenic can result in CNS toxicity and neurological sequelae, including the development of psychosis (Ratnaike, 2003; Tyler and Allan, 2014), it has been speculated that dysregulation of arsenic metabolism mediates the relationship between this genetic locus and schizophrenia. However, Li and colleagues (Li et al., 2014) have demonstrated that the top GWAS SNP for this locus is not associated with the full-length isoform of the AS3MT transcript (AS3MTfull) but with the isoform AS3MTd2d3, which lacks arsenite methyltransferase activity. Therefore, alterations in arsenic metabolism may not explain the association between rs7085104 and schizophrenia. Our data suggest that the dopaminergic pathway is involved in this association.
The rs7085104 SNP might impact dopamine synthesis capacity through its association with the expression of BORCS7 (Li et al., 2016b). Specifically, the risk genotype (A) is associated with upregulation of this protein, which forms one of the subunits of BLOC-one-related complex (BORC). BORC in turn makes up subunits of lysosome-related organelles complex 1 (BLOC-1), which has been shown to be involved in modulation of dopaminergic neurotransmission (Iizuka et al., 2007; Nagai et al., 2010).
There is accumulating evidence to support the longstanding neurodevelopmental hypothesis of adult-onset psychiatric disorders and in particular schizophrenia (Birnbaum et al., 2015; Howes and Murray, 2014; Jaffe et al., 2014; Weinberger, 1987). Therefore, it is noteworthy that the expression of AS3MTd2d3 and BORCS7 is up-regulated in early neuronal differentiation (Li et al., 2016b). In consideration of the fact that human dopaminergic innervation is also present in the early phases of neurodevelopment (Money and Stanwood, 2013; Zecevic and Verney, 1995), it can be speculated that the genetic variant examined in our study exerts its effect on the dopaminergic system early in the neurodevelopment, increasing risk for schizophrenia (Hannon et al., 2015).
Our study was conducted in healthy subjects; therefore, future work should focus on patients with schizophrenia and other major psychiatric disorders. Supporting this assertion of broader implications of 10q24.32 in serious mental illness, rs7085104 and two other SNPs in linkage disequilibrium (rs7914558 (D’= 0.956; r2 = 0.689 in Europeans) and rs11191580 (D’= 1; r2= 0.178 in Europeans)) within this chromosomal region are also associated with bipolar disorder, major depressive disorder, schizophrenia, autism spectrum disorder, and attention deficit-hyperactivity disorder (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Li et al., 2016a; Psychiatric GWAS Consortium Bipolar Disorder Working Group et al., 2011; Ripke et al., 2011). In this context, it is noteworthy that expression of AS3MTd2d3 is elevated in patients not only with schizophrenia but also major depression when compared with healthy subjects. Dopamine dysregulation, namely elevated dopamine synthesis capacity, has been demonstrated in patients with bipolar disorder with psychosis as well as in schizophrenia (Jauhar et al., 2017a). It could be speculated that the mediator of the association of this locus with major psychiatric disorders is the dopaminergic system.
Strengths and Limitations
Since there is evidence of increased striatal dopamine synthesis capacity in schizophrenia (Howes et al., 2012), the increased striatal dopamine synthesis capacity of the risk allele carriers could explain the association of this genetic variant with schizophrenia (Ripke et al., 2014).
However, it should be considered that association does not necessarily imply causality. Pre-clinical studies are needed to clarify the mechanism that links the genetic risk given by polymorphisms within 10q24.32-33 with dysregulation in the dopaminergic system. Moreover, in consideration of the small proportion of subjects with the genotype GG for the SNP rs7085104 and of the fact that some imaging studies based on a candidate gene approach have shown contradictory findings (Bogdan et al., 2017), replications with a higher number of subjects are needed. Nevertheless, to our knowledge, this study is one of the largest PET studies of association of a genetic variant with dopamine function (Dahoun et al., 2018; Gluskin and Mickey, 2016; Laakso et al., 2005; Shumay et al., 2017; Wiers et al., 2017; Wu et al., 2012).
We note that, for the SNP described herein, the allele associated with increased schizophrenia risk is the most common allele in the general population. Whilst risk alleles for disorders are generally minor alleles (Kido et al., 2018), this is not always the case and several risk alleles identified by GWAS have frequencies higher than 50% in the general population (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Mulle, 2012). This is in line with the evidence that schizophrenia is a polygenic disorder; and many common variants contribute to risk and protection, each with small effects (Weinberger, 2019). These small effects have likely enabled a proportion of these risk variants to become highly represented in the general population as a consequence of genetic drift or balancing selection (Owen et al., 2016; Pardiñas et al., 2018).
A relevant limitation was the use of data from three different PET scanners. However, to control for this, PET scanners were included as covariates of no interest in all the analyses. Anyway, there was no difference in the genotype distribution across the three scanners. Furthermore, some participants were smokers; however, groups did not differ for smoking status and dopamine synthesis capacity is not altered in moderate smokers (Bloomfield et al., 2014a); and it is not clear if even heavy smoking has a significant effect on dopamine synthesis capacity (Ashok et al., 2019).
Our sample was not homogeneous for ethnicity, thus genetic principal components were included as covariates in all the analyses, in order to correct for population stratification. However, a recent GWAS has shown an association of common genetic variants within the locus 10q24.32 with schizophrenia in the Han Chinese population (Yu et al., 2016). Moreover, a variable number tandem repeat (VNTR) in the first exon of AS3MT is associated with AS3MTd2d3 mRNA expression not only in Caucasian subjects but also in African Americans (Li et al., 2014). This would indicate that the association of this locus with schizophrenia is ethnicity-independent.
Implications
Studies such as this can help move from a genomic region of association to highlighting potential mechanisms through which candidate SNPs within the region are acting to increase the risk for schizophrenia. Our study can be considered a step towards translation of one of the most statistically significant findings in the genetics of schizophrenia, the association of the locus 10q24.32, towards the identification of novel treatment targets in line with recent proposals (Birnbaum & Weinberger, 2017; Schubert et al., 2014).
The modest effect size of the association between rs7085104 and dopamine synthesis capacity is in line with the fact that the risk for schizophrenia conferred by each of the schizophrenia GWAS-significant loci is relatively small (Ripke et al., 2014). In this regard, the contribution of this SNP to the variation in the striatal Kicer - as can be deduced from the difference between the subjects homozygous for the risk variant and the individuals homozygous for the G allele (Hedges’ g= 0.957) - is in line with the effect size of the association of AS3MTd2d3 expression with diagnosis of schizophrenia (effect size= 0.813 (Li et al., 2016b)). However, given the modest sample size of G homozygotes in our study, our effect size estimate should be considered as preliminary and warrants replication. Moreover, in view of the fact the risk allele is common in the general population, it is important to note that other genetic and environmental factors, such as psychosocial stressors, cannabis use, and obstetric complications are likely to combine to lead to schizophrenia (Birnbaum and Weinberger, 2017; Howes et al., 2017).
4.1. Conclusions
The results from the present work indicate that the mechanism mediating the 10q24.32 risk locus for schizophrenia could involve altered dopaminergic function. Future studies are needed to clarify the neurobiological pathway implicated in this association.
Highlights.
Subjects genotyped for the GWAS schizophrenia-associated SNP rs7085104 underwent 18F-DOPA PET.
rs7085104 is associated with striatal dopamine synthesis capacity.
We found an effect of genotype on dopamine function for all the striatal subdivisions.
The association of the 10q24.32 locus with schizophrenia could be mediated by dopaminergic function.
Acknowledgements
This research was supported by Medical Research Council (grant MC-A656-5QD30), Maudsley Charity (grant 666), the US Brain & Behavior Research Foundation, Wellcome Trust (grant 094849/Z/10/Z to Professor Howes), the National Institute for Health Research Biomedical Research Centre at South London and Maudsley National Health Service Foundation Trust, King’s College London. Dr Jauhar is supported by a Sim Fellowship, from the Royal College of Physicians, Edinburgh. Dr Veronese is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. Dr Dahoun was supported by a EU-FP7 MC-ITN IN-SENS grant (grant number 607616) and by the National Institute for Health Research (NIHR) at Oxford Health NHS Foundation Trust. Dr Bloomfield is supported by a UCL Excellence Fellowship and by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
We acknowledge the Cardiff University MRC CNGG Core Team, especially Lucinda Hopkins and Lesley Bates, for laboratory sample management and genotyping of the healthy volunteer sample.
We would like to thank Prof. D.R. Weinberger for his helpful comments and suggestions.
Footnotes
Financial Disclosures and conflict of interests
Professor Howes has received investigator-initiated research funding from and/or participated in advisory/speaker meetings organised by Astra-Zeneca, Autifony, BMS, Eli Lilly, Heptares, Janssen, Lundbeck, Lyden-Delta, Otsuka, Servier, Sunovion, Rand and Roche. Neither Professor Howes nor his family have been employed by or have holdings/a financial stake in any biomedical company.
Dr D'Ambrosio reported no biomedical financial interests or potential conflicts of interest.
Dr Dahoun reported no biomedical financial interests or potential conflicts of interest.
Dr Pardiñas reported no biomedical financial interests or potential conflicts of interest.
Dr Veronese reported no biomedical financial interests or potential conflicts of interest.
Dr Bloomfield reported no biomedical financial interests or potential conflicts of interest.
Dr Jauhar reported no biomedical financial interests or potential conflicts of interest.
Dr Bonoldi reported no biomedical financial interests or potential conflicts of interest.
Dr Rogdaki reported no biomedical financial interests or potential conflicts of interest.
Dr Froudist-Walsh reported no biomedical financial interests or potential conflicts of interest.
Professor Walters reported no biomedical financial interests or potential conflicts of interest.
References
- (NEMA), N.E.M.A. Performance Measurements of Positron Emission Tomographs. Rosslyn, VA: NEMA; 2007. Stand. Publ. NU 2-2007. [Google Scholar]
- Abi-Dargham A, Rodenhiser J, Printz D, Zea-Ponce Y, Gil R, Kegeles LS, Weiss R, Cooper TB, Mann JJ, Van Heertum RL, Gorman JM, et al. Increased baseline occupancy of D2 receptors by dopamine in schizophrenia. Proc Natl Acad Sci U S A. 2000;97:8104–8109. doi: 10.1073/pnas.97.14.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Dargham A, van de Giessen E, Slifstein M, Kegeles LS, Laruelle M. Baseline and Amphetamine-Stimulated Dopamine Activity Are Related in Drug-Naïve Schizophrenic Subjects. Biol Psychiatry. 2009;65:1091–1093. doi: 10.1016/j.biopsych.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok AH, Mizuno Y, Howes OD. Tobacco smoking and dopaminergic function in humans: a meta-analysis of molecular imaging studies. Psychopharmacology (Berl) 2019 doi: 10.1007/s00213-019-05196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum R, Jaffe AE, Chen Q, Hyde TM, Kleinman JE, Weinberger DR. Investigation of the prenatal expression patterns of 108 schizophrenia-associated genetic loci. Biol Psychiatry. 2015;77:e43–e51. doi: 10.1016/j.biopsych.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Birnbaum R, Weinberger DR. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017 doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- Bloomfield MA, Pepper F, Egerton A, Demjaha A, Tomasi G, Mouchlianitis E, Maximen L, Veronese M, Turkheimer F, Selvaraj S, Howes OD. Dopamine function in cigarette smokers: an [(1)(8)F]-DOPA PET study. Neuropsychopharmacology. 2014a;39:2397–2404. doi: 10.1038/npp.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield MAP, Morgan CJA, Egerton A, Kapur S, Curran HV, Howes OD. Dopaminergic function in cannabis users and its relationship to cannabis-induced psychotic symptoms. Biol Psychiatry. 2014b;75:470–478. doi: 10.1016/j.biopsych.2013.05.027. [DOI] [PubMed] [Google Scholar]
- Bogdan R, Salmeron BJ, Carey CE, Agrawal A, Calhoun VD, Garavan H, Hariri AR, Heinz A, Hill MN, Holmes A, Kalin NH, et al. Imaging Genetics and Genomics in Psychiatry: A Critical Review of Progress and Potential. Biol Psychiatry. 2017;82:165–175. doi: 10.1016/j.biopsych.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Miller MB, Thornton TA. Robust Inference of Population Structure for Ancestry Prediction and Correction of Stratification in the Presence of Relatedness. Genet Epidemiol. 2015;39:276–293. doi: 10.1002/gepi.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming P, Leger GC, Kuwabara H, Gjedde A. Pharmacokinetics of plasma 6-[18F]fluoro-L-3,4-dihydroxyphenylalanine ([18F]Fdopa) in humans. J Cereb Blood Flow Metab. 1993;13:668–675. doi: 10.1038/jcbfm.1993.85. [DOI] [PubMed] [Google Scholar]
- Dahoun T, Pardiñas AF, Veronese M, Bloomfield MAP, Jauhar S, Bonoldi I, Froudist-Walsh S, Nosarti C, Korth C, Hennah W, Walters J, et al. The effect of the DISC1 Ser704Cys polymorphism on striatal dopamine synthesis capacity: an [18F]-DOPA PET study. Hum Mol Genet. 2018;27:3498–3506. doi: 10.1093/hmg/ddy242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Kahn RS, Ko G, Davidson M. Dopamine in schizophrenia: A review and reconceptualization. Am J Psychiatry. 1991;148:1474–1486. doi: 10.1176/ajp.148.11.1474. [DOI] [PubMed] [Google Scholar]
- Duarte RRR, Troakes C, Nolan M, Srivastava DP, Murray RM, Bray NJ. Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. Am J Med Genet B Neuropsychiatr Genet. 2016:806–814. doi: 10.1002/ajmg.b.32445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerton A, Chaddock CA, Winton-Brown TT, Bloomfield MA, Bhattacharyya S, Allen P, McGuire PK, Howes OD. Presynaptic striatal dopamine dysfunction in people at ultra-high risk for psychosis: findings in a second cohort. Biol Psychiatry. 2013;74:106–112. doi: 10.1016/j.biopsych.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Egerton A, Demjaha A, McGuire P, Mehta MA, Howes OD. The test-retest reliability of 18F-DOPA PET in assessing striatal and extrastriatal presynaptic dopaminergic function. Neuroimage. 2010;50:524–531. doi: 10.1016/j.neuroimage.2009.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B, Smith N, Curtis C, Huckett L, Mill J, Craig IW. DNA from buccal swabs recruited by mail: Evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav Genet. 2003;33:67–72. doi: 10.1023/A:1021055617738. [DOI] [PubMed] [Google Scholar]
- Froudist-Walsh S, Bloomfield MAPP, Veronese M, Kroll J, Karolis VR, Jauhar S, Bonoldi I, McGuire PK, Kapur S, Murray RM, Nosarti C, et al. The Effect Of Perinatal Brain Injury On Dopaminergic Function And Hippocampal Volume In Adult Life. bioRxiv. 2017;6:1–20. doi: 10.7554/eLife.29088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett ES, Firnau G, Nahmias C. Dopamine visualized in the basal ganglia of living man. Nature. 1983 doi: 10.1097/00004728-198402000-00054. [DOI] [PubMed] [Google Scholar]
- Gluskin BS, Mickey BJ. Genetic variation and dopamine D2 receptor availability: a systematic review and meta-analysis of human in vivo molecular imaging studies. Transl Psychiatry. 2016;6:e747. doi: 10.1038/tp.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Leger G, Reches A, Evans A, Kuwabara H, Cedarbaum JM, Gjedde A. Administration of the new COMT inhibitor OR-611 increases striatal uptake of fluorodopa. Mov Disord. 1993;8:298–304. doi: 10.1002/mds.870080308. [DOI] [PubMed] [Google Scholar]
- Hannon E, Spiers H, Viana J, Pidsley R, Burrage J, Murphy TM, Troakes C, Turecki G, O’Donovan MC, Schalkwyk LC, Bray NJ, et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2015;19:48–54. doi: 10.1038/nn.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz A, Romero B, Gallinat J, Juckel G, Weinberger DR. Molecular brain imaging and the neurobiology and genetics of schizophrenia. Pharmacopsychiatry. 2003;36:152–157. doi: 10.1055/s-2003-45123. [DOI] [PubMed] [Google Scholar]
- Heinz A, Schlagenhauf F. Dopaminergic dysfunction in schizophrenia: Salience attribution revisited. Schizophr Bull. 2010;36:472–485. doi: 10.1093/schbul/sbq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietala J, Syvälahti E, Vilkman H, Vuorio K, Räkköläinen V, Bergman J, Haaparanta M, Solin O, Kuoppamäki M, Eronen E, Ruotsalainen U, et al. Depressive symptoms and presynaptic dopamine function in neuroleptic-naive schizophrenia. Schizophr Res. 1999;35:41–50. doi: 10.1016/S0920-9964(98)00113-3. [DOI] [PubMed] [Google Scholar]
- Howes O, Bose S, Turkheimer F, Valli I, Egerton A, Stahl D, Valmaggia L, Allen P, Murray R, McGuire P. Progressive increase in striatal dopamine synthesis capacity as patients develop psychosis: a PET study. Mol Psychiatry. 2011a;16:885–886. doi: 10.1038/mp.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes O, McCutcheon R, Stone J. Glutamate and dopamine in schizophrenia: an update for the 21st century. J Psychopharmacol. 2015;29:97–115. doi: 10.1177/0269881114563634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Bose SK, Turkheimer F, Valli I, Egerton A, Valmaggia LR, Murray RM, McGuire P. Dopamine synthesis capacity before onset of psychosis: a prospective [18F]-DOPA PET imaging study. Am J Psychiatry. 2011b;168:1311–1317. doi: 10.1176/appi.ajp.2011.11010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Kambeitz J, Kim E, Stahl D, Slifstein M, Abi-Dargham A, Kapur S. The nature of dopamine dysfunction in schizophrenia and what this means for treatment. Arch Gen Psychiatry. 2012;69:776–786. doi: 10.1001/archgenpsychiatry.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III--the final common pathway. Schizophr Bull. 2009;35:549–562. doi: 10.1093/schbul/sbp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, McCutcheon R, Owen MJ, Murray RM. The Role of Genes, Stress, and Dopamine in the Development of Schizophrenia. Biol Psychiatry. 2017;81:9–20. doi: 10.1016/j.biopsych.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Montgomery AJ, Asselin MC, Murray RM, Valli I, Tabraham P, Bramon-Bosch E, Valmaggia L, Johns L, Broome M, McGuire PK, et al. Elevated striatal dopamine function linked to prodromal signs of schizophrenia. Arch Gen Psychiatry. 2009;66:13–20. doi: 10.1001/archgenpsychiatry.2008.514. [DOI] [PubMed] [Google Scholar]
- Howes OD, Murray RM. Schizophrenia: An integrated sociodevelopmental-cognitive model. Lancet. 2014;383:1677–1687. doi: 10.1016/S0140-6736(13)62036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, Williams M, Ibrahim K, Leung G, Egerton A, McGuire PK, Turkheimer F. Midbrain dopamine function in schizophrenia and depression: a post-mortem and positron emission tomographic imaging study. Brain. 2013;136:3242–3251. doi: 10.1093/brain/awt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen J, Heinimaa M, Svirskis T, Nyman M, Kajander J, Forsback S, Solin O, Ilonen T, Korkeila J, Ristkari T, McGlashan T, et al. Striatal dopamine synthesis in first-degree relatives of patients with schizophrenia. Biol Psychiatry. 2008;63:114–117. doi: 10.1016/j.biopsych.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence That the BLOC-1 Protein Dysbindin Modulates Dopamine D2 Receptor Internalization and Signaling But Not D1 Internalization. J Neurosci. 2007;27:12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, Gao Y, Jia Y, Maher BJ, Hyde TM, Kleinman JE, et al. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2014;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhar S, Nour MM, Veronese M, Rogdaki M, Bonoldi I, Azis M, Turkheimer F, McGuire P, Young AH, Howes OD. A Test of the Transdiagnostic Dopamine Hypothesis of Psychosis Using Positron Emission Tomographic Imaging in Bipolar Affective Disorder and Schizophrenia. JAMA Psychiatry. 2017a doi: 10.1001/jamapsychiatry.2017.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauhar S, Veronese M, Rogdaki M, Bloomfield M, Natesan S, Turkheimer F, Kapur S, Howes OD. Regulation of dopaminergic function: an |[lsqb]|18F|[rsqb]|-DOPA PET apomorphine challenge study in humans. Transl Psychiatry. 2017b:7. doi: 10.1038/tp.2016.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido T, Sikora-Wohlfeld W, Kawashima M, Kikuchi S, Kamatani N, Patwardhan A, Chen R, Sirota M, Kodama K, Hadley D, Butte AJ. Are minor alleles more likely to be risk alleles? BMC Med Genomics. 2018;11:1–11. doi: 10.1186/s12920-018-0322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura Y, Cumming P, Vernaleken I, Buchholz H-G, Siessmeier T, Heinz A, Kienast T, Bartenstein P, Grunder G. Elevated [18F]Fluorodopamine Turnover in Brain of Patients with Schizophrenia: An [18F]Fluorodopa/Positron Emission Tomography Study. J Neurosci. 2007;27:8080–8087. doi: 10.1523/JNEUROSCI.0805-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakso A, Pohjalainen T, Bergman J, Kajander J, Haaparanta M, Solin O, Syvalahti E, Hietala J. The A1 allele of the human D2 dopamine receptor gene is associated with increased activity of striatal L-amino acid decarboxylase in healthy subjects. Pharmacogenet Genomics. 2005;15:387–391. doi: 10.1097/01213011-200506000-00003. [DOI] [PubMed] [Google Scholar]
- Laruelle M, Abi-Dargham A. Dopamine as the wind of the psychotic fire: new evidence from brain imaging studies. J Psychopharmacol. 1999;13:358–371. doi: 10.1177/026988119901300405. [DOI] [PubMed] [Google Scholar]
- Leblond CS, Cliquet F, Carton C, Huguet G, Mathieu A, Kergrohen T, Buratti J, Lemière N, Cuisset L, Bienvenu T, Boland A, et al. Both rare and common genetic variants contribute to autism in the Faroe Islands. npj Genomic Med. 2019;4:1. doi: 10.1038/s41525-018-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation. Science (80-.) 2008;319:1100 LP–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Li L, Chang H, Peng T, Li M, Xiao X. Evidence of AS3MTd2d3-Associated Variants within 10q24.32-33 in the Genetic Risk of Major Affective Disorders. Mol Neuropsychiatry. 2016a;2:213–218. doi: 10.1159/000452998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y, Chen Q, Li C, Jia Y, Ohi K, Maher BJ, et al. A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med. 2016b;22:649–656. doi: 10.1038/nm.4096. [DOI] [PubMed] [Google Scholar]
- Li M, Tao R, Jaffe AE, Zhang F, Chen D, Kleinman JE, Hyde TM, Shin JH, Weinberger DR. A human-specific isoform of AS3MT regulated by a human-unique variation explains susceptibility to psychiatric illness. Neuropsychopharmacology. 2014;39:S397–S398. [Google Scholar]
- Lin S, Shi Q, Brent Nix F, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Martinez D, Slifstein M, Broft A, Mawlawi O, Hwang D-RR, Huang Y, Cooper T, Kegeles L, Zarahn E, Abi-Dargham A, Haber SN, et al. Imaging human mesolimbic dopamine transmission with positron emission tomography. Part II: amphetamine-induced dopamine release in the functional subdivisions of the striatum. J Cereb Blood Flow Metab. 2003;23:285–300. doi: 10.1097/01.WCB.0000048520.34839.1A. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Miletich RS, Kohn PD, Esposito G, Carson RE, Quarantelli M, Weinberger DR, Berman KF. Reduced prefrontal activity predicts exaggerated striatal dopaminergic function in schizophrenia. Nat Neurosci. 2002;5:267–271. doi: 10.1038/nn804. [DOI] [PubMed] [Google Scholar]
- Mizrahi R, Addington J, Rusjan PM, Suridjan I, Ng A, Boileau I, Pruessner JC, Remington G, Houle S, Wilson AA. Increased stress-induced dopamine release in psychosis. Biol Psychiatry. 2012;71:561–567. doi: 10.1016/j.biopsych.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Money KM, Stanwood GD. Developmental origins of brain disorders: roles for dopamine. Front Cell Neurosci. 2013;7:1–14. doi: 10.3389/fncel.2013.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG. Schizophrenia genetics: Progress, at last. Curr Opin Genet Dev. 2012;22:238–244. doi: 10.1016/j.gde.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Nagai T, Kitahara Y, Shiraki A, Hikita T, Taya S, Kaibuchi K, Yamada K. Dysfunction of dopamine release in the prefrontal cortex of dysbindin deficient sandy mice: An in vivo microdialysis study. Neurosci Lett. 2010 doi: 10.1016/j.neulet.2009.12.071. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. doi: 10.1016/S0140-6736(15)01121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018 doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group, P. Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Rietschel M, Blackwood D, Corvin A, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnaike RN. Acute and chronic arsenic toxicity. Postgrad Med J. 2003;79:391LP–396. doi: 10.1136/pmj.79.933.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith J, Benkelfat C, Sherwin A, Yasuhara Y, Kuwabara H, Andermann F, Bachneff S, Cumming P, Diksic M, Dyve SE, Etienne P, et al. Elevated dopa decarboxylase activity in living brain of patients with psychosis. Proc Natl Acad Sci U S A. 1994;91:11651–4. doi: 10.1073/pnas.91.24.11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Dushlaine CO, Chambert K, Moran JL, Anna K, Akterin S, Bergen S, Collins AL, Crowley JJ, Kim Y, Lee SH, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1–26. doi: 10.1038/ng.2742.Genome-wide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans Pa, Lee P, Bulik-Sullivan B, Collier Da, Huang H, Pers TH, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders A, Kendler K. Genome-wide association study identifies five new schizophrenia loci. Nat. 2011;43:969–976. doi: 10.1038/ng.940.Genome-wide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CR, Xi HS, Wendland JR, O’Donnell P. Translating human genetics into novel treatment targets for schizophrenia. Neuron. 2014;84:537–541. doi: 10.1016/j.neuron.2014.10.037. [DOI] [PubMed] [Google Scholar]
- Sherry ST. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Wiers CE, Shokri-Kojori E, Kim SW, Hodgkinson CA, Sun H, Tomasi D, Wong CT, Weinberger DR, Wang G-J, Fowler JS, et al. New repeat polymorphism in the AKT1 gene predicts striatal dopamine D2/D3 receptor availability and stimulant-induced dopamine release in the healthy human brain. J Neurosci. 2017;37:4982–4991. doi: 10.1523/JNEUROSCI.3155-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Sham PC. Robust association tests under different genetic models, allowing for binary or quantitative traits and covariates. Behav Genet. 2011;41:768–775. doi: 10.1007/s10519-011-9450-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SR FMB, Gibbon M, JBW W. Am Psychiatr. Press Washington, DC; 1996. Structured Clinical Interview for DSM-IV Axis I Disorders, Clinician Version (SCID-CV) [Google Scholar]
- Studholme C, Hill DL, Hawkes DJ. Automated 3-D registration of MR and CT images of the head. Med Image Anal. 1996;1:163–175. doi: 10.1016/s1361-8415(96)80011-9. [DOI] [PubMed] [Google Scholar]
- Sumi D, Himeno S. Role of Arsenic (+3 Oxidation State) Methyltransferase in Arsenic Metabolism and Toxicity. Biol Pharm Bull. 2012;35:1870–1875. doi: 10.1248/bpb.b212015. [DOI] [PubMed] [Google Scholar]
- Turkheimer FE, Brett M, Visvikis D, Cunningham VJ. Multiresolution analysis of emission tomography images in the wavelet domain. J Cereb Blood Flow Metab. 1999;19:1189–1208. doi: 10.1097/00004647-199911000-00003. [DOI] [PubMed] [Google Scholar]
- Tyler CR, Allan AM. The Effects of Arsenic Exposure on Neurological and Cognitive Dysfunction in Human and Rodent Studies: A Review. Curr Environ Heal reports. 2014;1:132–147. doi: 10.1007/s40572-014-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera S, Peyrot WJ, Milaneschi Y, Hertel J, Baune B, Breen G, Byrne E, Dunn EC, Fisher H, Homuth G, Levinson D, et al. Genome-wide gene-environment interaction in depression: A systematic evaluation of candidate genes. Am J Med Genet Part B Neuropsychiatr Genet. 2018;177:40–49. doi: 10.1002/ajmg.b.32593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR. Thinking about schizophrenia in an era of genomic medicine. Am J Psychiatry. 2019 doi: 10.1176/appi.ajp.2018.18111275. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Wiers CE, Towb PC, Hodgkinson CA, Shen P-H, Freeman C, Miller G, Lindgren E, Shokri-Kojori E, Demiral ŞB, Kim SW, Tomasi D, et al. Association of genetic ancestry with striatal dopamine D2/D3 receptor availability. Mol Psychiatry. 2017:1–6. doi: 10.1038/mp.2017.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, O’Keeffe D, Politis M, O’Keeffe GC, Robbins TW, Bose SK, Brooks DJ, Piccini P, Barker RA. The catechol-O-methyltransferase Val158Met polymorphism modulates fronto-cortical dopamine turnover in early Parkinson’s disease: A PET study. Brain. 2012;135:2449–2457. doi: 10.1093/brain/aws157. [DOI] [PubMed] [Google Scholar]
- Yu H, Yan H, Li J, Li Z, Zhang X, Ma Y, Mei L, Liu C, Cai L, Wang Q, Shi Y, et al. Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol Psychiatry. 2016;1:7. doi: 10.1038/mp.2016.212. [DOI] [PubMed] [Google Scholar]
- Zecevic N, Verney C. Development of the catecholamine neurons in human embryos and fetuses, with special emphasis on the innervaton of the cerebral cortex. J Comp Neurol. 1995;351:509–535. doi: 10.1002/cne.903510404. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Bakshi A, Vinkhuyzen AAE, Hemani G, Lee SH, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, Esko T, Milani L, Mägi R, et al. Dominance Genetic Variation Contributes Little to the Missing Heritability for Human Complex Traits. Am J Hum Genet. 2015;96:377–385. doi: 10.1016/j.ajhg.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]