Abstract
Infectious diseases are a major global public health problem. Multiple agents are now recognized to cause indistinguishable illnesses. The term ‘syndrome’ applies to such situations, for which early and rapid diagnosis of the infecting agent would enable prompt and appropriate therapy. Public health physicians would also get timely information on the specific etiology of the infectious syndrome, facilitating intervention at the community level in the face of outbreaks or epidemics. A variety of molecular techniques have been evaluated for rapid diagnosis of infectious syndromes. These techniques include real-time multiplex PCR, DNA microarray, loop-mediated isothermal amplification, and other similar assays. This review surveys such state-of-the-art technologies.
Keywords: Respiratory Syncytial Virus, West Nile Virus, Fluorescence Resonance Energy Transfer, Dengue Virus, Japanese Encephalitis Virus
1. Introduction
A number of diverse infectious agents cause diseases that present with similar signs and symptoms characteristic of an ‘infectious syndrome.’ This classically sets them apart from other clinical infectious conditions that are caused by an easily identifiable organism or a small number of related organisms (e.g. typhoid fever). Some such syndromes may have a bacterial, viral, or other etiology. Diseases of infectious etiology, including common infectious diseases such as dengue fever, are increasingly presenting with protean manifestations or with atypical manifestations. Presently, a paradigm shift is occurring in the clinician’s approach to the patient as having an ‘acute febrile illness’ (the syndromic approach), in which the clinician recognizes that there are multiple different infectious agents that could potentially cause the disease. Successful treatment of such patients requires identification of the specific causative agent and therefore relies on laboratory diagnostic packages provided for specific syndromes.
Many of the descriptions of such syndromes have been based on isolation of the pathogen and/or serologic diagnosis. This information on the etiology has enhanced understanding of the pathogenesis and epidemiology of these syndromes, but the methods that have been employed have been cumbersome and slow. Presently, the approach is to develop a single technologic ‘platform’-based diagnosis for distinct infectious syndromes, with a rapid turn-around time (<8 hours). Developments in molecular detection have facilitated such an approach using PCR and its modifications, DNA microarrays, and nanotechnology. The use of multiplex PCR (mPCR) and DNA microarrays has already begun to move from the research arena into clinical service laboratories. However, nanotechnology is still an emerging field with the exciting potential for diagnosis of infectious diseases. Nanomaterials are versatile and can be engineered into biofunctionalized particles. The field of pathogen detection has begun to exploit the unique optical and magnetic properties of nanoscale materials, such as fluorescent nanoparticles, metallic nanostructures, and super-paramagnetic nanoparticles, for bioimaging and detection of infectious micro-organisms.[1]
This article lists important and widely recognized entities of infectious syndromes and reviews the recent literature regarding molecular and nanotechnology-based methods used in their diagnosis. A literature search on this topic reveals a paucity of specific information that would give insights into syndromes of infectious origin. There are few research laboratories and biotechnology companies trying to develop useful devices for infectious syndromes. The available literature has certain limitations from a geographic point of view because the researchers have developed platforms for comprehensive diagnosis of important syndromes in their regions. The review shows the lacunae that researchers in tropical countries must address when developing appropriate platforms that are relevant to their regions for use in hospitals and public health laboratories.
To date, research has focused on the generation of genomic information directly from samples such as tissue or blood. In certain instances, generation of information on the genotype of the infectious agent could help to formulate treatment. An illustrative example is the treatment of hepatitis C virus (HCV) infection, where the duration of pegylated interferon (PEG-interferon) therapy is determined by the genotype of the viral strain infecting the patient. Certain post-amplification methods — which include Sanger sequencing and pyro sequencing of target genes, reverse hybridization, and Luminex analysis — are coming into use in Western countries.[2]
A survey of the published literature indexed in PubMed revealed the use of five different molecular assay technologies that have recently been developed for detection of various microbial/viral infections. The principles and intended uses of the assays are discussed in this review.
2. Important Infectious Disease Syndromes and Molecular Approaches to Syndromic Diagnosis
2.1 Exanthematous Fever
The term ‘exanthem’ refers to a rash on the skin, which occurs generally in children. The rash can spread all over the body. It may be caused by micro-organisms and viruses (infectious causes). However, it should be noted that the development of this clinical condition may also be due to exposure to toxins or drugs, or it can result from autoimmune disease (noninfectious causes). Some of the infectious etiologies include measles virus, rubella virus, varicella zoster virus (VZV), parvovirus B19, human herpesvirus (HHV)-6, Epstein-Barr virus (EBV), enteroviruses, and dengue virus. Bacterial infections such as scarlet fever, Staphylococcus aureus infections (toxic shock syndrome), meningococcemia, typhoid fever, and rickettsial infection may also present as fevers with exanthema.
Research in veterinary diagnostics has made some progress, such as establishing a definitive diagnosis of vesicular or vesicular-like lesions in livestock animals. It is difficult to achieve a differential etiologic diagnosis of foot-and-mouth disease virus (FMDV), vesicular stomatitis virus (VSV), and swine vesicular disease virus (SVDV). A rapid method has been described, using padlock probes and microarrays to detect and simultaneously differentiate the three viruses in a single reaction (the padlock probe/microarray assay). The assay had a lower limit of detection of 144 median tissue culture infective doses (TCID50) per assay for FMDV, 720 TCID50 per assay for VSV, and 29 TCID50 per assay for SVDV. Although this method was not evaluated on clinical samples, the authors concluded that it is comparable to real-time PCR.[3]
Likewise, a high-throughput multiplexed assay has been developed for laboratory use to detect and differentiate FMDV from other similar disease-causing viruses. The assay simultaneously screens for five RNA and two DNA viruses by using multiplexed reverse-transcription PCR (mRT-PCR) amplification coupled with a microsphere hybridization array and flow-cytometric detection. The sensitivity of mRT-PCR for FMDV was 93.5% and the specificity was 91.9%, but this evaluation was performed only on a panel of clinically confirmed positive and negative cases.[4]
An interesting study described the application of a long oligonucleotide microarray assay for identification of viruses that are known to cause vesicular or vesicular-like lesions in livestock animals. The results suggested that the microarray assay could be a valuable tool for diagnosis of vesicular and vesicular-like diseases.[5] The assay was used on epithelium samples from vesicular lesions, but the authors reported difficulty with template preparation. Furthermore, the assay had varied sensitivity for different viruses. Further validation of the probe sets and optimization of sample preparation procedures are needed.
In our opinion, it would be useful to have a diagnostic device that could correctly identify the etiology of important fever with rash, such as measles, rubella, parvovirus, HHV-6, rickettsial rash, and scarletinal rash in humans.
2.2 Neurologic Infections
Central nervous system (CNS) infections are acutely life threatening and warrant immediate diagnosis to enable specific antimicrobial/antiviral therapy. The etiology of bacterial meningitis consists of Streptococcus pneumoniae, Haemophilus influenzae type b, Neisseria meningitidis, and Listeria monocytogenes. Several viral agents that cause aseptic meningitis include members of the genus enterovirus, herpes simplex virus (HSV), and mumps virus.
Encephalitis is an acute inflammation of the white matter of the brain. Encephalitis with meningitis (inflammation of the meninges) is known as meningoencephalitis. Symptoms include headache, fever, confusion, drowsiness, and fatigue. More severe symptoms include seizures/convulsions, tremors, hallucinations, and memory problems. Viral encephalitis may be sporadic or epidemic in its occurrence. A common cause of encephalitis in humans is infection with HSV; this condition occurs sporadically in all parts of the globe. In contrast, some viral encephalitis can occur as epidemics in geographically restricted areas (dependent on vector distribution) — e.g. Japanese encephalitis, which occurs in South and Southeast Asia.
Encephalitis can also be caused by a bacterial infection, such as bacterial meningitis, spreading directly to the brain (primary encephalitis) or it may be a complication of a current infectious disease, such as syphilis (secondary encephalitis). Certain parasitic or protozoal infestations, such as toxoplasmosis, malaria, or primary amoebic meningoencephalitis, can also cause encephalitis in people with compromised immune systems. Lyme disease and/or Bartonella henselae may also cause encephalitis.
Boriskin et al.[6] developed a microarray-based virus detection assay for 13 viral agents that cause meningitis or encephalitis, by targeting 38 target genes. The clinical sensitivity and specificity of the assay were established as being 93% and 100%, respectively. The results were comparable to those of uniplex PCR, which is considered a gold standard. The authors investigated a variety of DNA and RNA neurotropic viruses, including members of the Herpesviridae and Picornaviridae families. The utility of this assay is limited because it is virus specific and strain-specific variations cannot be detected, which could be a limitation for RNA viruses because of genetic variation (nucleotide polymorphism) among RNA virus strains.
In the antemortem diagnosis of infectious meningoencephalitis affecting humans, molecular testing has gained significant currency. A modification of PCR, which uses consensus degenerate hybrid primers (CODEHOP), has been used to identify and characterize novel infectious agents. Molecular testing is also gaining acceptance in veterinary infectious disease diagnosis. Tests such as conventional and real-time PCR have been shown to be useful in diagnosis of CNS infections caused by agents such as canine distemper virus, Toxoplasma gondii, Neospora caninum, rickettsial species, and others. The CODEHOP approach has been used for canine meningoencephalitis, including assays for rickettsial organisms, Borrelia spp., Bartonella spp., and some viral families.[7] It must be emphasized, though, that such reported innovations need thorough evaluation in a clinical setting against tests that could be accepted as a gold standard.
Zheng et al.[8] developed an mPCR-based DNA microarray for simultaneous detection, as well as species identification, of seven human herpes viruses — namely HSV-1, HSV-2, VZV, EBV, cytomegalovirus (CMV), HHV-6A, and HHV-6B. Primers and oligonucleotide probes were targeted at the highly conserved regions of the DNA polymerase gene of human herpes viruses. DNA microarrays were made by printing the oligonucleotide probes onto special glass slides. After amplification and labeling with Cy5, the PCR products were hybridized on the microarrays chips. The technique was compared with TaqMan probe-based real-time PCR. The microarray had a lower limit of detection of 10 copies/µL and showed no cross-reaction. Evaluation was performed on clinical cerebrospinal fluid (CSF) samples from children. The assay had sensitivity of 96.2% and specificity of 99.3%. Figure 1 shows the principle of the PCR-based DNA microarray.
Fig. 1.
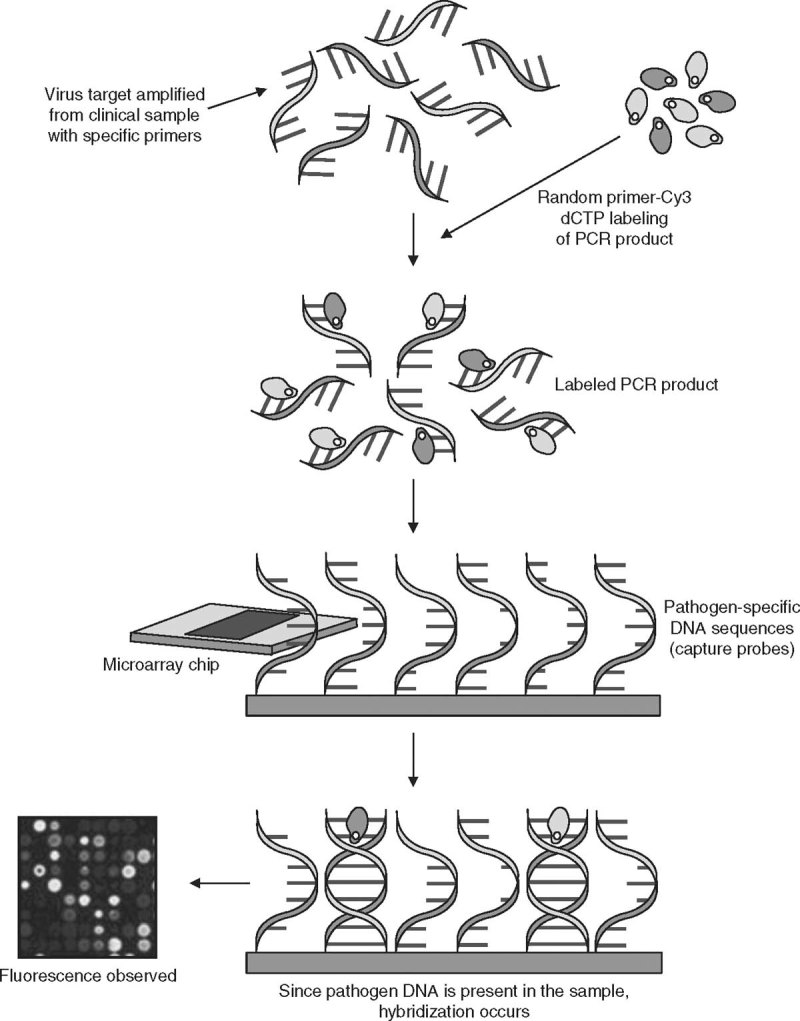
Principle of PCR-based DNA microarray detection of specific viral genes. A specific viral target gene is amplified from a clinical sample, using species-specific primers. The amplified PCR products are labeled with a random primer incorporated with Cy3 deoxycytidine triphosphate (dCTP). The labeled PCR products bind to the pathogen-specific oligonucleotide capture probe immobilized on the microarray chip. If pathogen DNA is present and amplified from the sample, hybridization with the capture probe is detected by analysis of fluorescence, using an Affymetrix 428 array scanner (Affymetrix, Santa Clara, CA, USA) with the aid of ImaGene software (version 4.2; BioDiscovery, Marina del Rey, CA, USA).
In a landmark development for diagnosis of neurologic infections, Shi et al.[9] reported an evaluation of a PCR micro-array that could detect seven viruses, namely HSV-1, HSV-2, VZV, EBV, CMV, HHV-6A, and HHV-6B. The mPCR product was genotyped by DNA microarray technology. The multiplex primers and oligonucleotide probes were specific for the highly conserved regions of the DNA polymerase gene. The assay did not show any non-specificity. Like the assay developed by Zheng et al.,[8] the assay developed by Shi et al.[9] had a lower limit of detection of 10 copies of viral load. The authors tested CSF samples from children and reported sensitivity of 91.7% and specificity of 100%, similar to the results reported by Zheng et al.[8]
A study by Gaeta et al.[10] reported a quantitative real-time PCR assay for detection of six viruses causing meningitis and meningoencephalitis in CSF samples. The assay was able to detect HSV-1, HSV-2, EBV, CMV, HHV-6, and VZV, correlating with clinical conditions. Herpes viral co-infections were documented in several patients. The lower limit of detection was 250 genome copies/mL or 10 copies per reaction. The diagnostic sensitivity ranged from 89.8% to 94.1% for the different viruses that were tested, which would be acceptable for use in clinical laboratories.
A rapid, quantitative, real-time, reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) assay was developed for detection of Japanese encephalitis virus (JEV), targeting the envelope gene. The lower limit of detection was less than 1 plaque-forming unit (PFU). The assay was specific to JEV; it did not falsely detect other flaviviruses such as dengue virus and West Nile virus (WNV).[11] Clinical evaluation of such tests is essential before a decision can be made on their usefulness. Figure 2 shows the principle of the LAMP assay.
Fig. 2.
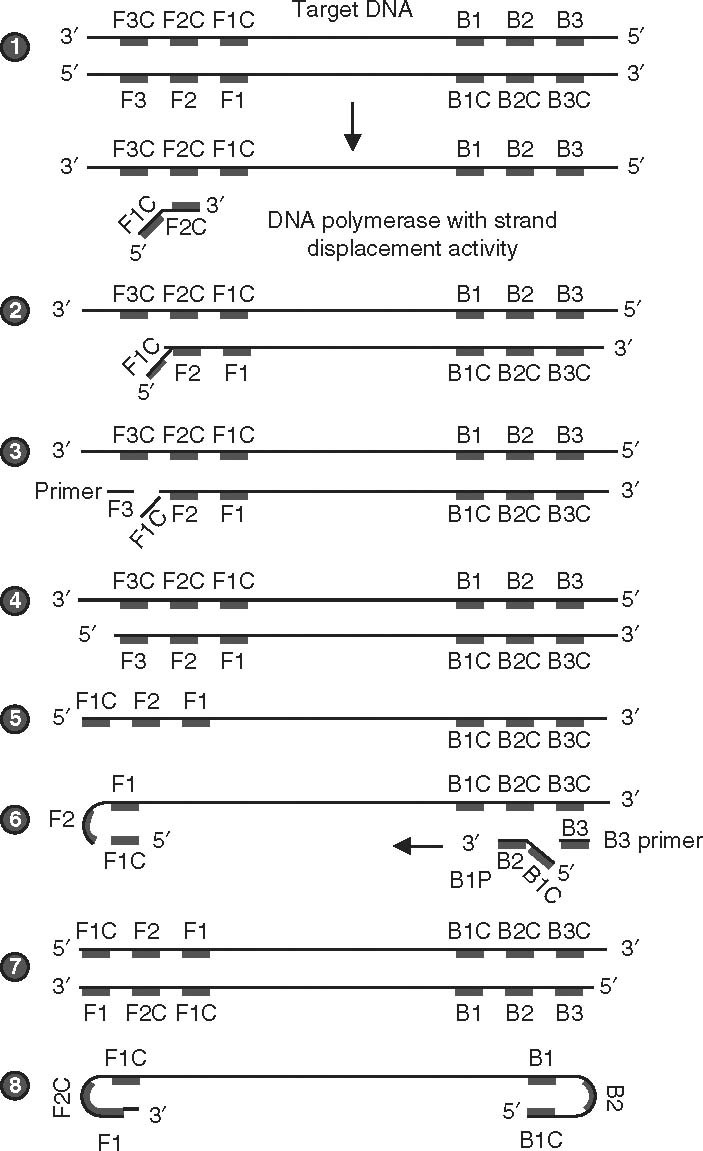
Sketch depicting the steps involved in the loop-mediated isothermal amplification (LAMP) assay. (1) The LAMP assay is entirely carried out at 65°C, when the double-stranded DNA (dsDNA) is in the state of dynamic equilibrium. In these conditions, one of the LAMP primers can anneal to the complimentary sequence of dsDNA (the target). Initiation of DNA synthesis is facilitated by DNA polymerase with strand displacement activity. The enzyme displaces and releases the single-stranded DNA (ssDNA). There is no denaturation step involved. The forward primers F1, F2, and F3 and the backward primers B1, B2, and B3 anneal to the complementary sequence (F1C, F2C, and F3C; B1C, B2C, and B3C) on one strand, extending the complementary strand sequence. Usually, six primers bind to about eight sites in the entire target sequence. (2) The DNA polymerase acts on the complementary DNA (cDNA) strand of the template DNA, starting from the 3′ end of the F2 region of the forward inner primer (FIP), and proceeding with strand synthesis. (3) The F3 primer anneals to the F3C region, releasing the FIP-linked complementary strand. (4) DNA synthesis proceeds from the F3 primer. (5) F1C (the FIP-linked complementary strand) is then released as a single strand because of displacement by the DNA strand synthesized from F3P, and the released single strand forms a stem-loop structure at the 5′ end. The loop formation occurs because F1C is complimentary to the F1 regions. (6) The ssDNA from step 5 serves as a template for backward inner primer (BIP)-initiated DNA synthesis. Subsequently, B3-primed strand displacement DNA synthesis occurs. The BIP anneals to the DNA strand produced in step 5. Starting from the 3′ end of the BIP, synthesis of the complementary strand takes place. This process allows the DNA to revert from a loop structure to a linear structure. The B3 primer anneals to the outside of the BIP and then, through the activity of DNA polymerase and starting at the 3′ end, the DNA synthesized from the BIP is displaced and released as a single strand before DNA synthesis from the B3 primer. (7) This process allows dsDNA synthesis. (8) After each amplification cycle, i.e. LAMP cycling, the BIP-linked complementary strand that is displaced forms a structure with a stem loop at each end (a dumbbell structure). The assays generally run for about 60 minutes, generating clear turbidity measured by a photometer. Usually, turbidity measured as absorbance of 0.1 is considered positive.
Since CNS infections are life threatening, an attempt should be made to develop assay systems suitable for comprehensive etiologic diagnosis, which would facilitate the prompt initiation of appropriate therapy. Such measures would be life saving and, to a large extent, would prevent neurologic sequelae.
2.3 Sexually Transmitted Infections
Sexually transmitted infections (STIs) are infections of the genitourinary tract that are spread by venereal transmission. They affect a wide age group from older teenagers to older adults, and their prevalence is increasing. STIs constitute an infectious syndrome caused by multiple agents, including bacteria, viruses, and parasites. Several of these infectious are clinically identifiable in males. In females, the infections may be asymptomatic. Several STIs affect fertility, especially among females (e.g. pelvic inflammatory disease, a consequence of chlamydial infection). This group of infections should be considered high priority for developing accurate and easy pan-diagnostic devices. Detailed laboratory investigations aid specific diagnosis in both sexes.
A commercial ‘closed system,’ the Cobas 4800 CT/NG test, has been successfully used for high-throughput identification of Chlamydia trachomatis and Neisseria gonorrhoeae infections.[12] The assay was used on urine samples and showed sensitivity of 94.5%, specificity of 99.5%, a negative predictive value of 98.8%, and a positive predictive value of 97.7% for C. trachomatis, and corresponding values of 92.9%, 100%, 99.7%, and 100%, respectively, for N. gonorrhoeae. For urethral/cervical swab specimens, the sensitivity, specificity, negative predictive value, and positive predictive value were 92.0%, 100%, 99.5%, and 100%, respectively, for C. trachomatis, and 100%, 99.4%, 100%, and 90.0%, respectively, for N. gonorrhoeae.
It is now very clear that important blood-borne viruses, such as human immunodeficiency virus (HIV), hepatitis B virus (HBV) and HCV, are efficiently transmitted through sexual routes. Thus, assays detecting these viruses could be used in screening blood and blood products and for patients attending STI clinics. A method has been developed, based on real-time mRT-PCR performed within the microarray hydrogel pads. In this assay, double-stranded amplification products are simultaneously detected using nonspecific SYBR Green I dye by testing in separate pads bearing 5′-immobilized specific primers. It has been evaluated on reference strains and some stored plasma samples. The lower limit of detection ranged from 10 to 15 genome equivalents per 25 µL reaction assay for the three viruses that were tested. The sensitivity and specificity were established to be 100%.[13]
Globally, human papillomavirus (HPV) is a very important STI agent. Multiple genotypes of this virus can cause genital warts, and certain types of HPV-16 and -18 are now known to be involved in the etiology of cervical cancers. Early diagnosis of infections of this virus is therefore important. A DNA microarray chip assay based on light-scattering of aggregated silica nanoparticle probes has been developed.[14] Here, target HPV DNA is sandwiched between the capture DNA immobilized on the chip and the probe DNA immobilized on the plain silica nanoparticle. The spots where the sandwich reaction occurs appear bright white and are readily distinguishable with the naked eye. Visual detection was most efficient using 286 nm silica nanoparticles and 200 pM of target DNA. The authors demonstrated the development of such a portable nanodevice for detection and genotyping of HPV.[14] The lower limit of detection was 100 pM with a simple microscope illumination system, and 200 pM to 1 nM with the naked eye. The advantages include the low cost of the light-scattering detector system and the ability to use the system without the device by naked-eye examination; also, plain silica particles are readily available.
de Méndez et al.[15] developed a line probe assay for detection and genotyping of HPV DNA in abnormal Papanicolaoustained cervicovaginal smears. This method was reported to be efficient, sensitive, fast, and useful in routine investigation for better clinical management of the patient. It should be pointed out that the potential application of the line probe assay has not been sufficiently exploited. Figure 3 illustrates the principle of this assay.
Fig. 3.
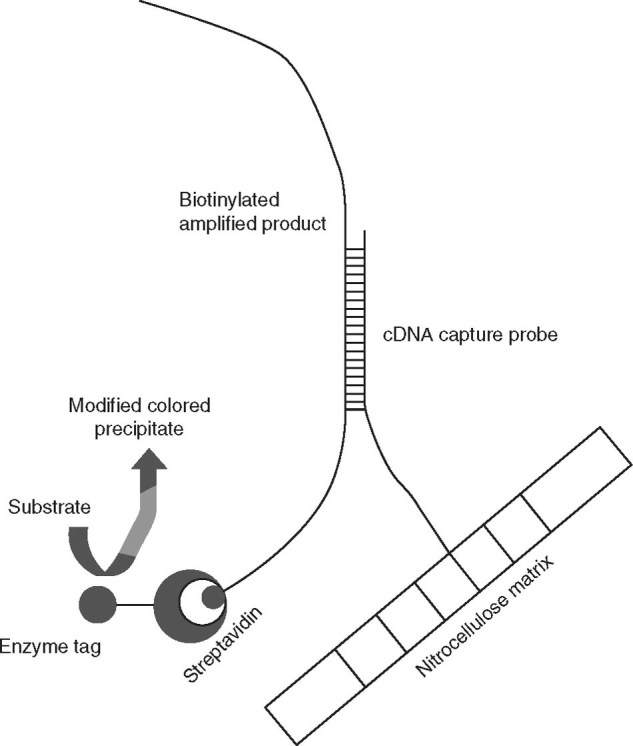
Schematic representation of the principle of the line probe assay. The nitrocellulose matrix (strip) carries pre-synthesized complementary DNA (cDNA) probes that are specific for genotypes of hepatitis C virus (HCV) immobilized at different locations; as many as 23 such probes, including amplification controls, are fixed on the matrix. The multiplex PCR product of HCV (produced with biotinylated primers) is applied in the reaction vessel containing the matrix. After hybridization, the captured amplicons are detected by a biotin-streptavidin enzyme conjugate with a substrate color reaction. The hybridization profile that is obtained is specific for each genotype, and this is interpreted with a standard chart.
Previously, Tang et al.[16] developed a visual gene detection technique with high sensitivity and specificity for clinical applications. The visual DNA microarray was capable of simultaneous, sensitive, and specific detection of HIV-1 and Treponema pallidum. This assay uses a gold label silver stain (GLSS) coupled with asymmetric mPCR. With this method, 5′-end amino-modified oligonucleotides are immobilized on a glass surface and used as the capturing probes to bind the complementary biotinylated target DNA. Gold-conjugated streptavidins are used in the microarray for specific binding to biotin. The microarray spots develop (black) as a result of the precipitation of silver onto nanogold particles that are bound to streptavidin. PCR products of HIV-1 and T. pallidum were used as positive controls. The results were comparable to real-time PCR findings. The lower limit of detection of the assay was 106 copies/mL of target DNA amplicons prepared by conventional PCR and mPCR, but 104 copies/mL when prepared by asymmetric PCR and asymmetric mPCR. Subsequently, Tang et al.[17] developed a GLSS-dependent visual protein micro-array technique for simultaneous, sensitive, and specific detection of Ureaplasma parvum and Chlamydia trachomatis. The authors used the N-terminus multiple-banded antigen of U. parvum and the major outer membrane protein of C. trachomatis. In this assay, the specific antigens are labeled with nanogold-staphylococcal protein A (SPA) and immobilized on a glass surface treated with 3-glycidoxypropyltrimethoxysilane. The bound antigens are recognized by the complementary target antibodies applied to the prepared microarray surface. The assay utilizes a ‘sandwich’ format, wherein the nanogold-SPA probe is used as an indicator and GLSS is applied to amplify the detection signals and produce black images on array spots, which are visible with the naked eye. The lower limit of detection of the protein microarray assay was 2 ng/mL. The sensitivity was comparable to that of the fluorescent detection method. The results were found to be consistent with ELISA and quantitative real-time PCR assay in clinical samples.
The current literature documents several successful attempts at developing diagnostic tools to aid diagnosis of the STI syndrome. The assays require evaluation at various laboratories for field testing before wide use.
2.4 Hemorrhagic Fever
The majority of hemorrhagic fevers are caused by viruses and so are often termed viral hemorrhagic fevers (VHFs). This refers to a group of illnesses that are caused by several distinct families of viruses. In general, the term VHF is used to describe a severe multisystem syndrome (i.e. multiple organ systems in the body are affected). Characteristically, the overall vascular system is damaged, and the body’s ability to regulate itself is impaired. These symptoms are often accompanied by hemorrhage (bleeding); however, the bleeding itself is rarely life threatening.[18]
The agents of VHF are all enveloped RNA viruses. Their survival is dependent on an animal or insect host, called the natural reservoir. The viruses are geographically restricted to the areas where their host species live. Humans are not the natural reservoir for any of these viruses. Humans are infected when they come into contact with infected hosts. However, with some viruses, after accidental transmission from the host, humans can transmit the virus to one another. Human cases or outbreaks of hemorrhagic fevers caused by these viruses occur sporadically and irregularly. The occurrence of outbreaks cannot be easily predicted. With a few noteworthy exceptions, there is no cure or established drug treatment for VHFs. Because other diseases have similar clinical symptoms, specific laboratory diagnostic tests are necessary to provide differential diagnosis, especially during outbreaks. Specific diagnosis would help public health officials to institute appropriate measures to control the outbreak.
VHFs are caused by four distinct families of RNA viruses: Arenaviridae (Lassa, Junin, Machupo, Sabia, and Guanarito), Filoviridae (Ebola and Marburg), Bunyaviridae (hantavirus), and Flaviviridae (dengue virus and Kyasanur forest disease virus). All types of VHF are characterized by fever along with bleeding disorders and can progress to high fever, shock, and death in extreme cases. Some of the VHF agents cause relatively mild illnesses, such as Scandinavian nephropathia epidemica, while others, such as African Ebola virus, can cause severe, life-threatening disease.
Dengue virus causes illnesses ranging from classical dengue fever (febrile illness) to dengue hemorrhagic fever and dengue shock syndrome. Dengue virus has four distinct serotypes, 1 through 4, each with the potential to cause clinical disease. Maneekan et al.[19] developed a single-tube mRT-PCR with a primer targeting the nonstructural protein region 5 (NS5) of the dengue virus genome, a highly conserved region for dengue virus serotyping. The authors compared the new assay with a well established conventional PCR. The mRT-PCR assay had sensitivity of 96.7% and specificity of 96.7%. The lower limit of detection ranged from 0.1 to 0.001 PFU per assay reaction for the different serotypes tested.
Trombley et al.[20] designed TaqMan-based real-time PCR assays for specific and absolute quantitative detection of multiple hemorrhagic fever viruses. The authors evaluated 48 formats and found 46 to be virus specific. The limit of detection for the assays ranged from 10 to 0.001 PFU per assay reaction. These assays provided qualitative and quantitative data. The clinical evaluation of these assays is awaited. Figure 4 depicts the TaqMan principle-based real-time PCR.
Fig. 4.
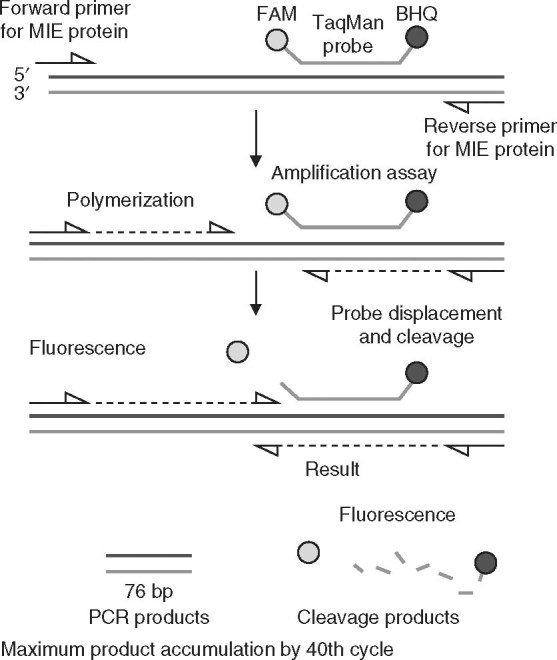
Diagrammatic outline of the TaqMan-based real-time PCR assay for detection of cytomegalovirus. The fluorophore 6-carboxyfluorescein (FAM) is used as the probe label and Black Hole Quencher® (BHQ) is used as the quencher dye. Forward and reverse primers are targeted toward major immediate early (MIE) protein. Amplification is measured dynamically as fluorescence intensity. Usually, a cut-off fluorescence intensity of 0.05 or more with a typical sigmoid amplification curve within the 40th cycle is considered positive.
It is now recognized that these infections are an emerging disease threat, and public health professionals will benefit from laboratories that are able to provide syndromic diagnosis. Initiatives toward this goal are in progress.
2.5 Hepatitis
Hepatitis is characterized by the presence of inflammatory cells in liver tissue. The condition can be self-limiting (healing on its own) or can progress to fibrosis (scarring) and cirrhosis. Hepatitis may occur with limited or no symptoms, but often leads to jaundice, anorexia (poor appetite), and malaise. Hepatitis is considered acute when it lasts less than 6 months and chronic when it persists for longer. A group of viruses known as the hepatitis viruses cause most cases of hepatitis worldwide, but hepatitis can also be due to toxins (notably alcohol, certain medications, and plants), other infections, and autoimmune diseases.
Acute hepatitis can be caused by hepatitis A virus (HAV), HBV, HCV, hepatitis D virus (HDV), hepatitis E virus (HEV), HSV, CMV, EBV, yellow fever virus, adenoviruses, non-viral infections with T. gondii, Leptospira species, and Coxiella burnetii. HBV (with or without superinfection with HDV) and HCV are important agents in blood-borne transmissions.
Nucleic acid testing platforms to detect HIV, HBV, and HCV include branched DNA assay and real-time mPCR assays for use in blood banks and blood product testing.[21] Because these viruses are either difficult or dangerous to culture, such molecular methods have become the gold standard.[22]
Tang et al.[23] developed a novel assay based on an electrochemical immunosensor array for simultaneous detection of 5-type hepatitis virus antigens (i.e. HAV, HBV, HCV, HDV, and HEV). The 5-type hepatitis virus antibodies were immobilized on an electrochemical sensor array, using nanogold particles and protein A as individual matrices. The immunosensor array was used to capture their corresponding antigens from the sample solution with a one-step capture format. The analyte detection depends on the electrical potential change from the native state to the analyte-bound state by using a two-electrode system. All five types of hepatitis virus antigens were identified by the immunosensor assay within 5 minutes, with a detection limit of ≤1.0 ng/mL. Moreover, the assay did not show any cross-reaction or nonspecific binding. This chip-based immunosensor array opens up new opportunities for high-throughput multi-analyte immunoassays for simple nanodevices that physicians could use at the point of care.
Jothikumar et al.[24] developed a real-time RT-PCR assay for quantitative detection of HAV and coxsackievirus B3 from environmental samples. The extracted RNA was compatible with quantitative RT-PCR with detection sensitivity of 0.8 PFU per reaction. On the basis of these results, the authors concluded that the developed assay could be applicable to detection of other pathogens in water and food.
Wang et al.[25] reported a quantum dot (QD)-DNA nanosensor based on fluorescence resonance energy transfer (FRET) for detection of DNA and a single mismatch in the HBV genome by examination of a synthetic 30-mer oligonucleotide containing the mutations rtm204m and rtm204v in the reverse transcriptase (RT) gene, indicating resistance to lamuvidine used in the treatment of HBV infection. The study used water-soluble cadmium selenide/zinc sulfide QDs with 3-mercaptopropionic acid. Subsequently, oligonucleotides were attached to the QD surface to form functional QD-DNA conjugates. Sandwiched hybrids were formed with the addition of DNA targets and Cy5-modified signal DNAs into the QD-DNA conjugates. The resulting assembly allows the Cy5 fluorophore (the acceptor) and the QD (the donor) to be brought into proximity, resulting in fluorescence emission from the acceptor by means of FRET, following illumination of the donor. The authors used an oligonucleotide ligation assay to detect single-base mutants in the HBV genome. They showed that if there was a single-base mismatch, this was recognized by the ligase, hence the detection probe was not ligated and no Cy5 emission was produced, because of the lack of FRET. The QD-DNA FRET-based nanosensor assay had a lower limit of detection of 4 nM. The advantages of this technique include the simplicity and efficiency of using a multi-label plate assay to detect single-base mutants in target DNA, using multicolor dyes, rather than using a fluorimeter. Fluorometric methods are more complicated and time consuming for detecting and characterizing signals from such a QD-DNA FRET system. This method is simple and efficient and could be potentially used for high-throughput multiplex detection of target DNA and the mutants. Figure 5 shows the principle of QDs.
Fig. 5.
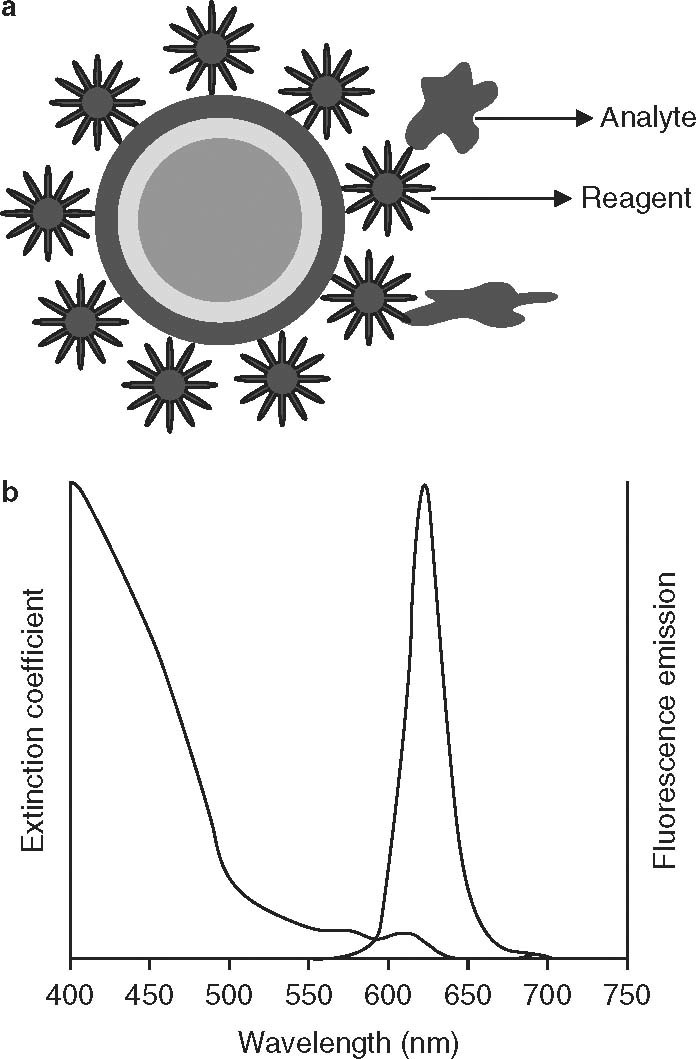
Graphic representation of the quantum dot (QD) principle. (a) QD nanocrystal used in analyte (microbial/viral antigen) detection. The QD (detection reagent) contains a core and shell component with the reagent, e.g. a monoclonal antibody specific for the analyte. (b) Optical principle underlying the detection of the analyte-reagent interaction on the surface of the QD, which results in a measurable variation that is detectable as a change in the photon emission pattern. QDs emit a narrow-band visible wavelength of light.
Hepatitis of different viral etiologies occurs in different parts of the globe in situations such as water-borne epidemics (HAV, HEV) and iatrogenic situations (HBV, HCV). The practicing physician will benefit from information on specific etiology for appropriate management with specific antiviral therapy. Quantitative assays such as those based on real-time PCR will give valuable information that will help in the monitoring of treatment.
2.6 Diarrheal Diseases
Diarrheal diseases affect all age groups and occur in many parts of the world, especially in developing countries, where children are particularly affected and can die from these diseases. These illnesses can occur as epidemics via either water-borne or food-borne routes. The etiology can be bacterial, viral, or protozoan. Early etiologic diagnosis could allow preventive public health measures and appropriate therapy.
Enterotoxigenic Escherichia coli (ETEC) is a common pathogen worldwide and causes infectious diarrhea, especially traveler’s diarrhea. Conventionally, the organisms were characterized by physiologic assays, immunoassays, and PCR-based methods for detection of ETEC (the heat-labile enterotoxin and/or the heat-stable enterotoxin). Separate serotyping methods using antisera were required to determine the ETEC serogroup. Wang et al.[26] developed a DNA microarray that could simultaneously detect enterotoxin genes and the 19 most common O-serogroup genes in ETEC strains. The random PCR strategy used by the authors documented an effective alternative to mPCR for detection of pathogens using DNA microarrays.
You et al.[27] developed a microarray technique for detection and identification of enteropathogenic bacteria at the species and subspecies levels, from stool specimens. The pathogens that were targeted were E. coli, Vibrio cholerae, Vibrio parahaemolyticus, Salmonella enterica, Campylobacter jejuni, Shigella, and Yersinia enterocolitica. The target was labeled with a fluorescence dye by mPCR and hybridized to the specific virulence gene probes immobilized on a microchip. The DNA microarray detected pathogens at concentrations as low as 58 genome copies/µL. The findings were compared and confirmed with species-specific conventional PCR, nested PCR, and sequencing. This technique appears to be an attractive diagnostic tool for rapid and simultaneous identification of multiple enteropathogenic pathogens in clinical practice, especially in patients with infectious diarrhea.
A combined mPCR and DNA microarray with tyramide signal amplification was developed by Jin et al.[28] for simultaneous detection of six human diarrheal pathogens, namely Yersinia enterocolitica, Shigella spp., Salmonella typhi, V. cholerae, E. coli O157:H7, and Brucella spp. The method also distinguished V. cholerae serotype O1 from serotype O139, and E. coli O1 57:H7 from O1 57:non-H7. The assay was found to be 100% specific for diagnostic detection and surveillance of multiple human pathogens, with a detection limit of 103 colony-forming units (CFU)/mL. The sensitivity of DNA microarray was based on the cut-off values and hybridization signals.
An mPCR assay was developed by Morin et al.[29] for simultaneous detection of E. coli O157:H7, V. cholerae O1, and S. typhi, targeting specific unique regions. The assay was found to detect as few as 30 cells of E. coli O157:H7 and S. typhi in clinical isolates. Assays such as these will need testing on clinical samples further to testing on isolates.
Li et al.[30] developed a DNA microarray, targeting O-serotype-specific genes to detect ten serotypes of Shigella (Shigella sonnei; Shigella flexneri type 2a; Shigella boydii types 7, 9, 13, 16, and 18; and Shigella dysenteriae types 4, 8, and 10) and five serotypes of E. coli (O55, O111, O114, O128, and O157). The detection sensitivity was 50 ng genomic DNA or 104 CFU/mL in mock stool specimens. The results suggest that the assay could be useful in clinical diagnosis, food safety, and epidemiologic surveillance.
Diarrheal diseases — especially those caused by diarrheagenic E. coli and certain viruses such as rotavirus, norovirus, and other similar viruses — are today recognized as a global problem. The problem is more severe in poor, developing countries in tropical regions. Specific etiologic information is important for public health professionals to implement preventive measures. Here, too, assays using the DNA-microarray or real-time mPCR platforms would be advantageous.
2.7 Ophthalmic Infections
Ophthalmic infections include infections of the conjunctiva (conjunctivitis), cornea (keratitis), both cornea and conjunctiva (keratoconjunctivitis), and the interior of the eye (endopthalmitis). These conditions are caused by different agents, which include bacteria, viruses, and fungi. Viral infections tend to occur as epidemics, and bacterial infections occur as small outbreaks.
Hlinomazová et al.[31] evaluated quantitative real-time PCR for detection of HSV1 in corneal swabs, an important causative agent in ophthalmic infections such as keratitis and keratouveitis. The EliGene® HSV1 RT kit is an in vitro medical diagnostic device, intended for use with the Applied Biosystems Real Time 7300 instrument (ABI7300; Applied Biosystems, Foster City, CA, USA). This kit allows absolute quantitative analysis of the PCR product, using EliDNA HSV1 QRT Standard (Elisabeth Pharmacon; Prague, Czech Republic). The detection limit described by the kit was 10 genome copies per reaction. The results correlated with the clinical picture of the disease and offered exact identification of the viral DNA in patients with herpes stromal keratitis.
A study was carried out using quantitative real-time PCR for detection of CMV in patients with epithelial, stromal, or endothelial keratitis of unknown origin. The authors investigated different types of sample, including tears, corneal scrapings, and aqueous humor specimens. The study documented the utility of the quantitative PCR assay for diagnosis and monitoring of the clinical course of the disease.[32]
Since infections of the eye have the potential to cause vision loss, early diagnosis and treatment are important. Because of the varied etiologies, a DNA microarray approach may hold the key.
2.8 Congenital Infections
Congenital infections are infections that are likely to affect the fetus in utero if the mother has an active infection. These are classically known as the ‘TORCH agents,’ which refer to infections with T. gondii, Rubella virus, CMV, and HSV. In developing countries, some medical experts extend this acronym to ‘TORCHS,’ including syphilis caused by T. pallidum. The list may also be extended to include less common agents in this condition, such as VZV, HBV, and HIV.
In HIV infection, maternal transmission from the infected mother to the newborn occurs mainly during the child’s birth. For molecular testing of HIV, either pro-viral DNA or RNA is targeted in blood or in plasma, respectively. Alternatively, HIV p24 antigen is tested to determine infection in infants, using heated plasma or serum samples for early diagnosis. Levels of this antigen are reported to be significantly higher in transmitting mothers than in non-transmitting mothers.[33]
Early diagnosis of maternal infections that have a bearing on fetal health and development of the newborn is of great importance to obstetricians and perinatalogists. Several infections are recognized in this category, and there have been some important developments in successful diagnosis of the multiple etiologies. For example, recent work toward a common molecular method for diagnosis of the TORCH group of infections has led to development of a QD-based protein microarray to simultaneously detect TORCH-related antibodies in sera. The technique combines the principles of immunofiltration and QD-labeled probes for detection.[34] The newly developed assay was compared with standard ELISA methods and was found to give statistically similar results, but with a quicker turn-around time, thus offering a rapid diagnostic tool. The development and commercialization of such assays by investigators represent a great stride forward in infectious disease diagnostics.
2.9 Febrile Illnesses
In this review, we use the term ‘febrile illness’ to refer to communicable (exogenous) infections that present with pyrexia of short duration (≤2 weeks, acute febrile illness). This would include certain viral and bacterial infections. Some of these, such as dengue fever and typhoid fever, may have some distinguishing features to enable the physician to make a provisional diagnosis (e.g. dengue fever is characterized by severe myalgia and retro-orbital pain, whereas typhoid fever usually presents with prodromal gastrointestinal symptoms). The other category is pyrexia of unknown origin (PUO). Very often, patients are treated empirically, and a specific diagnosis would obviously help in determining the appropriate use of antimicrobial or supportive therapy. The classical definition of PUO is fever of more than 102°F for at least 3 weeks’ duration with a cause that remains undiagnosed even after detailed investigations. Generally, PUO is caused by bacterial or viral agents. These include improperly investigated typhoid fever, melioidosis, tuberculosis, Q fever, early HIV infection, HBV infections, and EBV infections.
Salmonella spp. are very important causative agents of food-borne disease. A low-density DNA-microarray containing 281 oligonucleotide probes has been developed to detect a wide range of specific marker genes associated with antibiotic resistance, cell envelope structures, mobile genetic elements, and pathogenicity of this group of organisms. A recently published study indicated that this DNA microarray assay may be suitable for use in monitoring of food safety.[35] Such measures are being implemented in developed countries, including the US.
A rapid and sensitive microarray was designed for detection of Chikungunya virus, JEV, yellow fever virus, dengue virus, hantavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), and H5N1 avian influenza virus in patients with PUO. The panel included nine genera and 16 virus species. Consensus genus-specific primers were used to reverse transcribe the viral target genes so as to minimize the interference of other viruses. 70-mer oligonucleotides were used at the genus level and 50-mer oligonucleotides were used at the species level. The second-strand synthesis was carried out with random primers. The amplified products were labeled and processed for microarray analyses. We agree with the authors, who stated that this assay could potentially be used for clinical diagnosis.[36]
A one-step, single tube, real-time RT-LAMP assay was developed and evaluated with standard strains and clinical specimens spiked with different concentrations of virus strains for detecting the envelope gene of WNV.[37] The specificity of the RT-LAMP assay was validated by the absence of any cross-reaction with other, closely related members of the Flavivirus group. These results indicate that the RT-LAMP assay is extremely rapid, cost effective, and highly sensitive (detection limit = 0.1 PFU of virus), and has potential usefulness for rapid, comprehensive WNV surveillance along with virus isolation and/or serology.[37] The same group has developed a dengue virus serotype-specific RT-LAMP assay in clinical isolates and serum samples from confirmed cases, which did not cross react with WNV or JEV. The RT-LAMP assay showed sensitivity of 0.1–1 PFU of virus (10- to 100-fold more sensitive than RT-PCR). In patient serum samples with reference to the results of virus isolation, the assay had sensitivity of 100% and specificity of 93%.[38]
2.10 Respiratory Infections
Respiratory infections are caused by a variety of microbial and viral agents. Some are important as opportunistic infections. Communicable agents causing infections that are community acquired may cause lower respiratory tract infections (LRTIs) or upper respiratory tract infections (URTIs). LRTIs are more serious and may be caused by S. pneumoniae or viruses. Respiratory syncytial virus (RSV) causes serious LRTIs in young children in several parts of the globe. In recent years, two important global concerns have been avian influenza and pandemic swine influenza.
A study by Agrawal et al.[39] demonstrated the use of antibody-conjugated nanoparticles for rapid, sensitive, and quantitative detection of RSV. In this study, dual-color QDs or FRET nanobeads were used for virus detection. The nano-particles were characterized for sensitivity and specificity on cell culture lysates of different viruses. The assay needs to be evaluated on clinical samples.
Wang et al.[40] developed and evaluated a novel mPCR-based reverse line blot assay for simultaneous detection of 12 potential respiratory pathogens in children with community-acquired pneumonia. The pathogens included S. aureus, S. pneumoniae, Streptococcus pyogenes, Moraxella catarrhalis, H. influenzae, H. influenzae type b, Bordetella pertussis, Klebsiella pneumoniae, Legionella pneumophila, Mycobacterium tuberculosis, Chlamydia pneumoniae, and Mycoplasma pneumonia. S. pneumoniae and H. influenza (including type b) were the most important pathogens detected. Multiple pathogens (two or more) were detected in 35% of specimens. This study showed the assay to be a sensitive tool for identification of respiratory pathogens.
A recent development has been the use of DNA-microarray platform technology (the Virochip) for detection of viruses causing pediatric respiratory tract infections.[41] The Virochip was compared with conventional direct fluorescent antibody (DFA) and PCR-based testing for detection of respiratory viruses on nasopharyngeal aspirate samples. The Virochip showed sensitivity of 85–90% and specificity of ≥99% for RSV, influenza A, and the rhinoviruses/enteroviruses that were tested; it showed 19% greater detection than DFA. Interestingly, the Virochip was able to detect mixed infections as well, unlike DFA and PCR.[41]
In an important study on acute LRTIs, a cost-effective mPCR assay for use on nasopharyngeal aspirates was reported for detection of RSV, influenza viruses, parainfluenza viruses, and human metapneumovirus. The mPCR assay detected respiratory viruses in 35.2% of 301 samples. This study in young children showed predominance of RSV; mixed viral infections were detected in 18.8% of the children.[42]
Use of the more recently introduced RT-LAMP assay has been reported for highly pathogenic avian H5N1 influenza virus infection.[43] The authors reported 100-fold higher sensitivity of this assay compared with one-step RT-PCR. The assay was highly specific for the H5 subtype. The development of platforms for agents causing LRTIs is an exciting improvement in the field of molecular diagnosis of respiratory infections.
2.11 Opportunistic Infections
The term ‘opportunistic infection’ refers to a group of infections causing diseases such as encephalitis or lymphadenitis in primarily or secondarily immunocompromised patients (e.g. HIV-infected individuals or transplant recipients). Opportunistic infections can be caused by viruses, bacteria, protozoa, or fungi; the number of agents is very large, and pathogen-specific diagnosis is available for the vast majority of these. There are assays available that are designed to identify the pathogen causing encephalitis in immunocompromised patients; for example, real-time PCR assays for DNA neurotropic viruses.[44]
Use of a PCR-based, low-density DNA microarray has been reported for pathogens known to cause granulomatous lymphadenitis.[45] The assay was used on DNA extracted from paraffin-embedded tissue. The mPCR amplicons were hybridized to glass slides containing probes from Mycobacterium spp., Yersinia spp., Bartonella henselea, and T. gondii. The study found that the technique was useful in making a specific diagnosis, though it would benefit from inclusion of a broader range of probe sequences for additional species-specific assays.
The number of immunosuppressed patients is growing worldwide as a result of the widespread increase in transplantation programs in different parts of the globe, as well as the still-smoldering HIV pandemic; consequently, the frequency of opportunistic infections is also increasing. Presently, there are several platforms based on real-time mPCR or DNA microarrays for important viral infections, especially of the herpes group, as discussed in section 2.2.[6,46]
3. Geographic Region-Specific Etiology of the Syndrome
Data available from different areas of the globe indicate similarity in the etiology of certain infectious conditions, where the agents are latent in individuals and cause ‘endogenous’ infections, such as opportunistic infections, among immunosuppressed individuals. Many of these infections are not communicable, i.e. they are not transmissible to members of the community except in special settings. However, the vast majority of the infections are communicable (exogenous) and pose a public health threat because of their ability to cause outbreaks or epidemic infections. Such communicable diseases may spread globally (e.g. avian flu and pandemic influenza [swine flu]), whereas other infectious syndromes, such as VHFs and some acute febrile illnesses, occur in geographically restricted areas. The etiology of several such syndromes has to be studied from region to region to identify the individual pathogenic agents in a given syndrome. The individual agents causing geographically prevalent syndromes vary because of host and environmental factors (e.g. poor sanitation and unsafe water) and vectors. The vectors (arthropods) or animal carriers (zoonotic hosts) are different in different geographic areas. Thus, the etiology of infectious syndromes differs between North and South America, between Africa and Asia, and distinctly between temperate and tropical climates. For example, an important cause of febrile illness in India is S. typhi, which causes typhoid fever, whereas in the Far East, the most frequent cause of febrile illness is Burkholderia pseudomallei, which causes melioidosis.
In the context of geographic variation of pathogen and disease manifestation, one could cite the example of hantaviruses, which are widely distributed rodent-borne viruses.[47] This group of viruses can cause two important syndromes: hemorrhagic fever with renal syndrome (HFRS) in Eurasia, and hantavirus pulmonary syndrome in the Americas.[48] HFRS is a clinically distinct febrile illness, characterized by a hemorrhagic rash with renal dysfunction.
Seoul virus is a hantavirus that is transmitted by domestic rats (Rattus rattus) and has a global distribution because of the movement of rats to new areas through international shipping.[49] The disease caused by Seoul virus infection, HFRS, therefore occurs around the globe. Sin Nombre virus and other New World hantaviruses are transmitted by rodents of the subfamily Sigmodontinae (New World rats and deer mice)[50,51] and cause hantavirus cardiac pulmonary syndrome (HCPS) in the Americas. Andes virus is transmitted by long-tailed pygmy rice rats and causes a severe form of HCPS in South America.[52,53] Hence, geographic region-specific data are vital before designing laboratory diagnostic tools. Another outstanding example of geographic variation is the absence of yellow fever virus, which is transmitted by the mosquito Aedes aegyptii, in India. This vector is involved in transmission of both dengue virus and yellow fever virus in South America, whereas in India, the same species of mosquito is involved in transmission of dengue virus and certain other flaviviruses that are not common in South America. Interestingly, certain infectious agents are seen in geographically circumscribed areas — for example, Kyasunur forest disease is transmitted by hemophagous ticks and is seen only in the Shimoga district of Karnataka, India.[54] Similarly, Bartonella bacilliformis is transmitted by sandflies of genus Lutzomyia, causing Oroya fever, which is seen only in South America.[55] Rocky Mountain spotted fever is often considered endemic only in the US.[56]
It is our premise that there is a need for epidemiologic surveillance data to determine the identity of infectious agents causing an infectious syndrome that is predominant in each region, so that assays for appropriate syndromic diagnosis can be developed.
4. Conclusion
Infectious disease diagnosis has historically been based on clinical acumen and laboratory procedures such as media-based culture (for bacteria), cell culture (for viruses), microscopy, and serology. Often, laboratory investigations have focused on established pathogens that are linked to the clinical conditions, in essentially a ‘hit or miss’ approach. This approach is now considered by experts to be highly inefficient. It is imperative that comprehensive epidemiologic data are obtained to allow development of assays that could be used in a given region of the world or globally. Current trends in the development of molecular and nanotechnology-based diagnostic tools are very encouraging. The molecular techniques that hold special promise in this area are PCR-based DNA microarrays, realtime mPCR, and nanotechnology-based devices for nanoscale reactions, wherein the analyte is detected by a biosensor.
Acknowledgments
No sources of funding were used to prepare this review. The authors have no conflicts of interest that are directly relevant to the content of the review.
References
- 1.Tallury P, Malhotra A, Byrne LM, et al. Nanobioimaging and sensing of infectious diseases. Adv Drug Deliv Rev. 2010;62(4–5):424–37. doi: 10.1016/j.addr.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curr Opin Pediatr. 2009.
- 3.Banér J, Gyarmati P, Yacoub A, et al. Microarray-based molecular detection of foot-and-mouth disease, vesicular stomatitis and swine vesicular disease viruses, using padlock probes. J Virol Methods. 2007;143(2):200–6. doi: 10.1016/j.jviromet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Hindson BJ, Reid SM, Baker BR, et al. Diagnostic evaluation of multiplexed reverse transcription-PCR microsphere array assay for detection of foot-and-mouth and look-alike disease viruses. J Clin Microbiol. 2008;46(3):1081–9. doi: 10.1128/JCM.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack PJ, Amos-Ritchie RN, Reverter A, et al. Microarray-based detection of viruses causing vesicular or vesicular-like lesions in livestock animals. Vet Microbiol. 2009;133(1–2):145–53. doi: 10.1016/j.vetmic.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boriskin YS, Rice PS, Stabler RA, et al. DNA microarrays for virus detection in cases of central nervous system infection. J Clin Microbiol. 2004;42(12):5811–8. doi: 10.1128/JCM.42.12.5811-5818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nghiem PP, Schatzberg SJ. Conventional and molecular diagnostic testing for the acute neurologic patient. J Vet Emerg Crit Care (San Antonio) 2010;20(1):46–61. doi: 10.1111/j.1476-4431.2009.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng ZB, Wu YD, Yu XL, et al. DNA microarray technology for simultaneous detection and species identification of seven human herpes viruses. J Med Virol. 2008;80(6):1042–50. doi: 10.1002/jmv.21131. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Wu Y, Cai M, et al. Rapid diagnosis of herpetic encephalitis in children by PCR-microarray technology for simultaneous detection of seven human herpes viruses. Eur J Pediatr. 2010;169(4):421–5. doi: 10.1007/s00431-009-1038-5. [DOI] [PubMed] [Google Scholar]
- 10.Gaeta A, Verzaro S, Cristina LM, et al. Diagnosis of neurological herpesvirus infections: real time PCR in cerebral spinal fluid analysis. New Micro-biologica. 2009;32:333–40. [PubMed] [Google Scholar]
- 11.Hiroko T, Tomoyoshi K. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription loop-mediated isothermal amplification. Microbiol Immunol. 2006;50(5):379–87. doi: 10.1111/j.1348-0421.2006.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 12.Rockett R, Goire N, Limnios A, et al. Evaluation of the Cobas 4800 CT/NG test for detecting Chlamydia trachomatis and Neisseria gonorrhoeae. Sex Transm Infect. 2010;86(6):470–3. doi: 10.1136/sti.2010.042812. [DOI] [PubMed] [Google Scholar]
- 13.Khodakov DA, Zakharova NV, Gryadunov DA, et al. An oligonucleotide microarray for multiplex real-time PCR identification of HIV-1, HBV, and HCV. Biotechniques. 2008;44(2):241–6. doi: 10.2144/000112628. [DOI] [PubMed] [Google Scholar]
- 14.Piao JY, Park EH, Choi K, et al. Direct visual detection of DNA based on the light scattering of silica nanoparticles on a human papillomavirus DNA chip. Talanta. 2009;80(2):967–73. doi: 10.1016/j.talanta.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 15.de Méndez MT, Bosch AL. Detection and genotyping of human papillomavirus DNA using polymerase chain reaction short PCR fragment 10-line probe assay in abnormal Papanicolaou-stained cervicovaginal smears. Acta Cytol. 2009;53(5):540–7. doi: 10.1159/000325382. [DOI] [PubMed] [Google Scholar]
- 16.Tang J, Zhou L, Gao W, et al. Visual DNA microarrays for simultaneous detection of human immunodeficiency virus type-1 and Treponema pallidum coupled with multiplex asymmetric polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;65(4):372–8. doi: 10.1016/j.diagmicrobio.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 17.Tang J, Xu Z, Zhou L, et al. Rapid and simultaneous detection of Ureaplasma parvum and Chlamydia trachomatis antibodies based on visual protein microarray using gold nanoparticles and silver enhancement. Diagn Microbiol Infect Dis. 2010;67(2):122–8. doi: 10.1016/j.diagmicrobio.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Viral hemorrhagic fevers [fact sheet; online]. Available from URL: http://www.cdc.gov/ncidod/dvrd/spb/mnpages/dispages/vhf.htm [Accessed 2011 May 23]
- 19.Maneekan P, Jittmittraphap A, Leaungwutiwong P, et al. Development of single-tube mutiplex RT-PCR for dengue virus typing. Southeast Asian J Trop Med Public Health. 2009;40(6):1254–8. [PubMed] [Google Scholar]
- 20.Trombley AR, Wachter L, Garrison J, et al. Comprehensive panel of real-time TaqMan polymerase chain reaction assays for detection and absolute quantification of filoviruses, arenaviruses, and New World hantaviruses. Am J Trop Med Hyg. 2010;82(5):954–60. doi: 10.4269/ajtmh.2010.09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng Q, Wong C, Rangachari A, et al. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2001;39(8):2937–45. doi: 10.1128/JCM.39.8.2937-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seremba E, Ocama P, Opio CK, et al. Poor performance of hepatitis C antibody tests in hospital patients in Uganda. J Med Virol. 2010;82(8):1371–8. doi: 10.1002/jmv.21817. [DOI] [PubMed] [Google Scholar]
- 23.Tang D, Tang J, Su B, et al. Simultaneous determination of five-type hepatitis virus antigens in 5 min using an integrated automatic electrochemical immunosensor array. Biosens Bioelectron. 2010;25(7):1658–62. doi: 10.1016/j.bios.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Jothikumar N, Sobsey MD, Cromeans TL. Development of an RNA extraction protocol for detection of waterborne viruses by reverse transcriptase quantitative PCR (RT-qPCR) J Virol Methods. 2010;169(1):8–12. doi: 10.1016/j.jviromet.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Lou X, Wang Y, et al. QDs-DNA nanosensor for the detection of hepatitis B virus DNA and the single-base mutants. Biosens Bioelectron. 2010;25(8):1934–40. doi: 10.1016/j.bios.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Wang S, Beutin L, et al. Development of a DNA microarray for detection and serotyping of enterotoxigenic Escherichia coli. J Clin Microbiol. 2010;48(6):2066–74. doi: 10.1128/JCM.02014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Y, Fu C, Zeng X, et al. A novel DNA microarray for rapid diagnosis of enteropathogenic bacteria in stool specimens of patients with diarrhea. J Microbiol Methods. 2008;75(3):566–71. doi: 10.1016/j.mimet.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Jin D, Qi H, Chen S, et al. Simultaneous detection of six human diarrheal pathogens by using DNA microarray combined with tyramide signal amplification. J Microbiol Methods. 2008;75(2):365–8. doi: 10.1016/j.mimet.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 29.Morin NJ, Gong Z, Li XF. Reverse transcription-multiplex PCR assay for simultaneous detection of Escherichia coli O157:H7, Vibrio cholerae O1, and Salmonella typhi. Clin Chem. 2004;50(11):2037–44. doi: 10.1373/clinchem.2004.036814. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Liu D, Cao B, et al. Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J Clin Microbiol. 2006;44(12):4376–83. doi: 10.1128/JCM.01389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hlinomazová Z, Loukotová V, Horáčková M, et al. The treatment of HSV1 ocular infections using quantitative real-time PCR results. Acta Ophthalmol. Epub 2010 Jun 10 [DOI] [PubMed]
- 32.Kandori M, Inoue T, Takamatsu F, et al. Prevalence and features of keratitis with quantitative polymerase chain reaction positive for cytomegalovirus. Ophthalmology. 2010;117(2):216–22. doi: 10.1016/j.ophtha.2009.06.059. [DOI] [PubMed] [Google Scholar]
- 33.Lyamuya E, Bredberg-Rådén U, Massawe A, et al. Performance of a modified HIV-1 p24 antigen assay for early diagnosis of HIV-1 infection in infants and prediction of mother-to-infant transmission of HIV-1 in Dar es Salaam, Tanzania. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12(4):421–6. doi: 10.1097/00042560-199608010-00014. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, Guo Q, He R, et al. A quick and parallel analytical method based on quantum dots labeling for TORCH-related antibodies. Nanoscale Res Lett. 2009;4(12):1469–74. doi: 10.1007/s11671-009-9422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gronlund H, Riber L, Vigre H, et al. Microarray-based genotyping of Salmonella: inter-laboratory evaluation of reproducibility and standardization potential. Int J Food Microbiol. 2011;145(Suppl.1):S79–85. doi: 10.1016/j.ijfoodmicro.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 36.Xiao-Ping K, Yong-Qiang L, Qing-Ge S, et al. Development of a consensus microarray method for identification of some highly pathogenic viruses. J Med Virol. 2009;81(11):1945–50. doi: 10.1002/jmv.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parida M, Posadas G, Inoue S, et al. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–63. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parida M, Horioke K, Ishida H, et al. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43(6):2895–903. doi: 10.1128/JCM.43.6.2895-2903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agrawal A, Tripp RA, Anderson LJ, et al. Real-time detection of virus particles and viral protein expression with two-color nanoparticle probes. J Virol. 2005;79(13):8625–8. doi: 10.1128/JVI.79.13.8625-8628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Kong F, Yang Y, et al. A multiplex PCR-based reverse line blot hybridization (mPCR/RLB) assay for detection of bacterial respiratory pathogens in children with pneumonia. Pediatr Pulmonol. 2008;43(2):150–9. doi: 10.1002/ppul.20749. [DOI] [PubMed] [Google Scholar]
- 41.Chiu CY, Urisman A, Greenhow TL, et al. Utility of DNA microarrays for detection of viruses in acute respiratory tract infections in children. J Pediatr. 2008;153(1):76–83. doi: 10.1016/j.jpeds.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bharaj P, Sullender WM, Kabra SK, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89. doi: 10.1186/1743-422X-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imai M, Ninomiya A, Minekawa H, et al. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J Virol Methods. 2007;141(2):173–80. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Sachithanandham J, Ramamurthy M, Kannangai R, et al. Detection of opportunistic DNA viral infections by multiplex PCR among HIV infected individuals receiving care at a tertiary care hospital in South India. Indian J Med Microbiol. 2009;27(3):210–6. doi: 10.4103/0255-0857.53202. [DOI] [PubMed] [Google Scholar]
- 45.Odenthal M, Koenig S, Farbrother P, et al. Detection of opportunistic infections by low-density microarrays: a diagnostic approach for granulomatous lymphadenitis. Diagn Mol Pathol. 2007;16(1):18–26. doi: 10.1097/PDM.0b013e31802d6916. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Kirby JE, Qian Q. Effective use of JC virus PCR for diagnosis of progressive multifocal leukoencephalopathy. J Med Microbiol. 2009;58:253–5. doi: 10.1099/jmm.0.004432-0. [DOI] [PubMed] [Google Scholar]
- 47.Johnson KM. Hantaviruses: history and overview. Curr Top Microbiol Immunol. 2001;256:1–14. doi: 10.1007/978-3-642-56753-7_1. [DOI] [PubMed] [Google Scholar]
- 48.Krüger DH, Ulrich R, Lundkvist AA. Hantavirus infections and their prevention. Microbes Infect. 2001;3(13):1129–44. doi: 10.1016/S1286-4579(01)01474-5. [DOI] [PubMed] [Google Scholar]
- 49.Meyer BJ, Schmaljohn CS. Persistent hantavirus infections: characteristics and mechanisms. Trends Microbiol. 2000;8(2):61–7. doi: 10.1016/S0966-842X(99)01658-3. [DOI] [PubMed] [Google Scholar]
- 50.Chu YK, Jennings G, Schmaljohn A, et al. Cross-neutralization of hantaviruses with immune sera from experimentally infected animals and from hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome patients. J Infect Dis. 1995;172(6):1581–4. doi: 10.1093/infdis/172.6.1581. [DOI] [PubMed] [Google Scholar]
- 51.Heyman P, Plyusnina A, Berny P, et al. Seoul hantavirus in Europe: first demonstration of the virus genome in wild Rattus norvegicus captured in France. Eur J Clin Microbiol Infect Dis. 2004;23(9):711–7. doi: 10.1007/s10096-004-1196-3. [DOI] [PubMed] [Google Scholar]
- 52.Delfraro A, Clara M, Tomé L, et al. Yellow pigmy rice rat (Oligoryzomys flavescens) and hantavirus pulmonary syndrome in Uruguay. Emerg Infect Dis. 2003;9(7):846–52. doi: 10.3201/eid0907.030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green W, Feddersen R, Yousef O, et al. Tissue distribution of hantavirus antigen in naturally infected humans and deer mice. J Infect Dis. 1998;177:1696–700. doi: 10.1086/515325. [DOI] [PubMed] [Google Scholar]
- 54.Mehla R, Kumar SR, Yadav P, et al. Recent ancestry of Kyasanur Forest disease virus. Emerg Infect Dis. 2009;15(9):1431–7. doi: 10.3201/eid1509.080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaiser PO, Riess T, O’Rourke F, et al. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol. 2011;301(1):7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Salinas LJ, Greenfield RA, Little SE, et al. Tickborne infections in the southern United States. Am J Med Sci. 2010;340(3):194–201. doi: 10.1097/MAJ.0b013e3181e93817. [DOI] [PubMed] [Google Scholar]


