Abstract
Combining standard cytotoxic chemotherapy with BCR-ABL1 tyrosine kinase inhibitors (TKI) has greatly improved the upfront treatment of patients with Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL). However, prognosis remains poor due to the development of drug resistance through both BCR-ABLI-dependent and -independent mechanisms.
The STAT5 transcription factor is activated by BCR-ABL1 and by JAK2-dependent cytokine signaling; therefore, inhibiting its activity could address both mechanisms of resistance in Ph+ ALL.
We show that genetic and pharmacologic inhibition of STAT5 activity suppresses cell growth, induces apoptosis and inhibits leukemogenesis of Ph+ cell lines and patient’s derived newly diagnosed and relapsed/TKI-resistant Ph+ ALL cells ex vivo and in mouse models.
STAT5 silencing leads to decreased expression of the growth-promoting PIM-1 kinase, the apoptosis inhibitor MCL-1 and BCL-2, and to increased expression of pro-apoptotic BIM protein. Mechanistically, we show that changes in the expression of these STAT5-regulated effectors are functionally relevant for the impaired growth of STAT5-silenced Ph+ BV173 cells because the apoptosis of these cells was rescued by BIM silencing and/or restoring BCL-2 expression. Moreover, treatment of Ph+ ALL cells, including samples from relapsed/refractory patients, with the PIM kinase inhibitor AZD1208 and/or the BCL-2 family antagonist Sabutoclax markedly suppressed cell growth and leukemogenesis ex vivo and in mice.
Together, these studies indicate that targeting STAT5 or STAT5-regulated pathways may provide a new approach for therapy development in Ph+ ALL, especially the relapsed/TKI-resistant disease.
Keywords: Acute Lymphoblastic Leukemia, STAT5, Apoptosis
Introduction
Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL) is characterized by the t(9;22) translocation that generates the p190- and, less frequently, the p210-BCR-ABL1 fusion protein, both of which have constitutive tyrosine kinase activity (1,2). The Philadelphia chromosome is the most common cytogenetic abnormality in adult ALL patients, occurring in about 20–30% of the cases (3,4).
The incidence of Ph+ ALL increases with age, with up to 50% being diagnosed in patients older than 60 years (5).
Allogeneic hematopoietic stem cell transplantation (HSCT) is an effective consolidation therapy for patients with Ph+ ALL who have achieved a complete response (CR) after induction of remission chemotherapy (6). However, 10–20% of patients fail to achieve CR and allogeneic HSCT is only available for patients with suitable matched donors. In addition, the odds of treatment-related mortality as well as relapse are high (7).
The outcome of Ph+ ALL patients has improved significantly with the introduction of imatinib and second generation TKI as a first-line therapy, especially when used in combination with chemotherapy (8). However, resistance to TKI develops rapidly in most Ph+ ALL patients and the 5-year overall survival is <50% (9–11).
Thus, inhibiting the BCR-ABL1 kinase with TKI fails to eradicate most Ph+ ALL clones due to BCR-ABL1-dependent and independent mechanisms of resistance (12). Collectively, these findings indicate that additional pathways need to be identified and targeted for an effective treatment of Ph+ ALL.
STAT5 has a pivotal role in promoting cell survival and the subsequent B cell expansion, during early B cell development. CD19-Cre-mediated deletion of STAT5A/B in the B cell compartment impairs IL-7-activated survival pathways, blocking B cell differentiation at the pre-pro-B stage (13–15). Of interest, the defective B-cell development induced by genetic deletion of STAT5 was rescued by restoring expression of STAT5-regulated BCL-2 (16).
In malignant precursor B-cells, deregulated JAK-STAT5 activity may allow survival of leukemic cell independently of stroma-derived cytokine signals (17). The STAT5 pathway is constitutively active in Ph+ ALL and in a subset of B-ALL that contains activating mutations in the JAK1 or JAK2 (18–20). Importantly, STAT5 can be activated in Ph+ leukemia cells either indirectly through JAK2 phosphorylation or directly by BCR-ABL1 since STAT5 is a known substrate of BCR-ABL1 (21), and an intact STAT5 signaling is required for maintenance of BCR-ABL1-driven leukemias (19). Furthermore, STAT5 is a marker of disease progression in Ph+ chronic myeloid leukemia (CML), based on correlation of high STAT5 mRNA levels with TKI resistance and advanced disease stages, irrespective of the presence of tyrosine kinase domain (TKD) BCR-ABL1 mutations (22, 23).
Together, these data suggest that STAT5 itself or STAT5-regulated pathways could serve as rational targets not only to circumvent the BCR-ABL1-dependent TKI resistance of Ph+ ALL but also to suppress growth-promoting STAT5 signals activated through BCR-ABL1-independent mechanisms. In this study, using genetic and pharmacological approaches, we show that STAT5 is critical for the growth of Ph+ ALL cell lines and of newly diagnosed and relapsed/TKI-resistant patient-derived Ph+ ALL cells ex vivo and in mice. Moreover, we found that the growth-promoting effects of STAT5 depend on changes in the expression/activity of PIM-1, BIM, and BCL-2 and that these proteins can serve as therapeutic targets ex vivo and in xenografts of patient’s derived Ph+ ALL cells.
Materials and Methods
Cell lines, Ph+ primary ALL samples, and cell cultures
The SUP-B15 cell line (Ph+ ALL) was purchased from ATCC; the Z181 cell line (Ph+ ALL) was kindly provided by Dr. Z. Estrov, (M.D. Anderson Cancer Center, Houston, TX); the BV173 cell line (Ph+ CML-lymphoid blast crisis) (24) was kindly provided by Dr. N. Donato (NIH, Bethesda, MD). EBV-immortalized B cells GM03798 and GM12878, were purchased from the Coriell Institute (Camden, NJ). All these cell lines and the TKI-resistant T315I-BV173 derivative line (25) were cultured in Iscove’s Modified Dulbecco’s Medium (Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biowest USA), 1% penicillin-streptomycin (Thermo Fisher Scientific) and 1% L-glutamine (Thermo Fisher Scientific) at 37°C, 5% CO2. The Ph-like ALL MUTZ-5 and MHH-CALL-4 cell lines were kindly provided by Dr. M. Carroll (University of Pennsylvania, Philadelphia, PA). These lines were cultured in RPMI supplemented with 20% FBS, 1% penicillin-streptomycin, and 1% L-glutamine at 37°C, 5% CO2.
Cell lines were tested for mycoplasma every 3 months as described (26). Ph+ ALL cell lines were routinely authenticated by monitoring B-cell markers and BCR-ABL1 isoform expression. Ph-like cell lines were authenticated by B-cell immunophenotyping (CD19 and CD10), and by monitoring CRLF2 and phospho-STAT5 expression.
Primary adult human Ph+ ALL cells were kindly provided by: Dr. Alessandro Rambaldi (Hematology and Bone Marrow Transplant Unit, Bergamo, Italy), Dr. Luke F. Peterson (University of Michigan), Dr. Michael Caligiuri (City of Hope Cancer Center, CA), Dr. Pierluigi Porcu (Thomas Jefferson University), and Dr. Martin Carroll (University of Pennsylvania). Main characteristics of the samples are described in Supplementary Table S1. G-CSF-mobilized peripheral blood CD34+ primary cells (J48 and J50) from healthy donors were obtained from the Bone Marrow Transplantation Unit, Thomas Jefferson University. Primary adult human Ph-like cells were provided by Dr. Pierluigi Porcu. Primary cells were maintained in SFEM (Stem Cell Technologies) supplemented with interleukin IL-3 (10 ng/mL), IL-6 (10 ng/mL), IL-7 (10 ng/mL), Flt3-L (20 ng/mL) and Stem Cell Factor (30 ng/mL) (ProSpec).
Relapsed/TKI-resistant Ph+ ALL samples were assessed for the presence of ABL1 TKD mutations by Sanger sequencing; briefly, RNA was extracted with RNAeasy plus mini kit (Qiagen), cDNA was reverse transcribed by using Superscript III (Thermo Fisher Scientific). The ABL1 TKD was PCR-amplified with a forward primer located on BCR exon 1 (p190 FW: 5’-CTCGCAACAGTCCTTCGACA-3’) or with a forward primer located on exon 12 (P210 FW: 5’-CTGCAGATGCTGACCAACTC-3’) and a common reverse primer (TKD REV: 5’-CCTGCAGCAAGGTACTCACA-3’). Gel purified PCR products were sequenced in both directions using TKD REV primer or TKD FW primer (5’-CCCACTGTCTATGGTGTGTCC-3’).
Cell viability
Cell viability was assessed by MTT assay in 96-multiwell plates. Cells were seeded at a concentration of 150,000 or 300,000 cells/ml (depending on length of the treatment and cell line growth rate) and then treated with DMSO (Ctrl) or the specific drug. At the time point of interest, 100 μl of cell cultures were transferred in 96-multiwell plates and incubated with 10 μl of 0.5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, Sigma Aldrich) at 37 °C for three hours. Then, formazan crystals were dissolved with 0.1 M HCl in 2-propanol and absorbance was measured at 570 nm using a μQUANT microplate scanning spectrophotometer (BioTek instruments) and KC4 V.3.4 software.
Apoptosis
Cells were incubated at 100,000/ml with 1 μl of the CellEvent Caspase 3/7 Green Detection Reagent (Thermo Fisher Scientific) for 25 min at 37 °C and analyzed by flow cytometry using a 488 nm excitation laser.
For GFP-positive cell lines, apoptosis was measured by Annexin V staning: 100,000 cells were washed in Annexin V Binding Buffer (10 mM HEPES, 140 mM NaCl, and 2.5 mM CaCl2, pH 7.4), spun and re-suspended in 50 μL of Annexin V Binding Buffer, then incubated with 1.5 μl of Cy 5.5-Annexin V (BD Pharmigen) for 15 min at room temperature (RT). Samples were analyzed by flow cytometry using a 640 nm laser.
Colony formation assays
shRNA lentivirus-transduced or drug-treated SUP-B15, BV173 or Z181 cells were plated in methylcellulose (Stem Cell Technologies) supplemented with Iscove’s Modified Dulbecco’s Medium, 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin—streptomycin and 1% L-glutamine. Colonies were counted 10 days later.
shRNA lentivirus-transduced or drug-treated blast cells from Ph+ ALL, Ph-like patients, or CD34+ cells from healthy donors, were plated in methylcellulose supplemented with SFEM, IL-3 (10 ng/mL), IL-6 (10 ng/mL), IL-7 (10 ng/mL), Flt3-L (20 ng/mL) and Stem Cell Factor (30 ng/mL). Colonies were counted 7–10 days later.
Lentiviral production and cell transduction
STAT5 short hairpin RNA (shSTAT5) designed against the STAT5 mRNA sequence ATCTGGCTTGTTAATGAGTAG (TRCN0000019356) and SCR short hairpin RNA (shSCR) CCTAAGGTTAAGTCGCCCTCG were obtained in the pLKO.1-puro vector (Dharmacon) and subsequently cloned in the Tet-pLKO-puro vector (Addgene, Cambridge, MA, USA) for inducible shRNA expression, using Agel and EcoRI restriction enzyme sites and following Addgene’s protocols.
BIM short hairpin RNA (shBIM) designed against the BIM mRNA sequence CCAGACCACTACTGAATATAA (TRCN0000020155) and cloned into the Tet-pLKO-Neo lentiviral-vector were kindly provided by Dr. Z. Jagani (27).
The PIM1-s cDNA (NCBI: NM_0026483) was generated from total RNA purified from BV173 cells and inserted in the XbaI-SalI restriction enzyme sites of the pUltra, GFP-expressing, lentiviral vector developed by Dr. Malcolm Moore (Addgene, plasmid # 24129). The BCL2 cDNA (isoform α, NM_000633.2) was PCR-amplified from reverse transcribed total RNA of BV173 cells with a forward primer including the XbaI restriction sequence and with a reverse primer including a HA-tag coding sequence (TACCCATACGATGTTCCAGATTACGCT) and an EcoRI restriction site sequence. The product was then inserted into the XbaI-EcoRI sites of the pUltra-hot, mCherry expressing, lentiviral vector (Addgene plasmid # 24130).
The FG12 MCL-1/IRES-GFP lentivirus was kindly provided by Dr. Maria S. Soengas (Spanish National Cancer Centre, Madrid, Spain).
For lentiviral production, 293T cells were transiently transfected with the indicated plasmids, the envelope plasmid pMD2.G and the 2nd generation lentiviral packaging plasmid psPAX2 (Addgene). Infectious supernatant was collected at 24 h, concentrated by ultracentrifugation and used to infect Ph+ ALL cell lines or primary patient’s cells by spinoculation. SUP-B15, BV173 and Z181 cell lines were transduced with the inducible shSTAT5 or shBIM lentivirus and selected in the presence of puromycin (3 μg/ml) or G418 (400 μg/ml) for 5 or 14 days, respectively. shRNAs (shSTAT5 or shBIM) were induced with Doxycycline (2.5 μg/ml). Due to the limited viability of primary Ph+ ALL cells in liquid culture, these cells were transduced with the stable shSTAT5 or shSCR pLKO.1 lentivirus 16h after thawing, and selected in the presence of puromycin. Western blot lysates were obtained after 60 hours of puromycin selection.
For studies of PIM1-s overexpression, shSTAT5-BV173 cells were transduced with the PIM1-s lentivirus and selected by EGFP sorting, using a BD FACSARIAII cell sorter. For studies of MCL-1 or BCL-2 overexpression, shSTAT5-BV173 or shSTAT5-shBIM-BV173 cells were transduced with the MCL-1 or BCL-2 lentivirus and selected by EGFP or mCherry sorting respectively, using a BD ARIAII cell sorter.
Protein analysis
Cells were counted and lysed at a density of 10,000/μL in Laemmli Buffer supplemented with 5% β-Mercaptoethanol. Lysates where run on a 4–20% gradient polyacrylamide gels (Biorad) and transferred onto a nitrocellulose membrane (Fisher Scientific) using a semidry trans-blot transfer cell (Bio-Rad). Membranes were then blocked in 5% non-fat dry milk/TBS-T and incubated with the following primary antibodies: STAT5 (C-17) (1:4000; Santa Cruz Biotechnology # sc-835), p-STAT5 (Y649) (1:1000; BD #611964), STAT3 (1:1000; Transduction lab), β-ACTIN (8H10D10) (1:4000; Cell Signaling #3700S) or β-ACTIN (D6A8) (1:4000; Cell signaling #8457P), BCL-2 (1:1000; BD #610538), MCL-1 (S-19) (1:1000; Santa Cruz # sc-819), BCL-XL (1:1000; Cell Signaling #2762S), PIM-1 (1:250; Cell Signaling #2907S), p-BAD (S112) (40A9) (1:500 Cell Signaling #5284T), BAD (1:1000; Santa Cruz #sc-8044), p-4EBP1 (T37/46) (1:500; Cell Signaling #236B4), 4EBP1 (1:1000; cell Signaling #9644P), p-MYC (phospho-S62) (1:1000; Abcam #ab185656), c-MYC (N-262) (1:1000, Santa Cruz #sc-768), BIM (1:1000; Cell Signaling #2818S), NOXA (1:500; Novus Biological #NB600–1159), PUMA (1:1000; AbCam #ab9643).
After incubation with HRP-conjugated secondary antibodies (Thermo Fisher Scientific), signals were visualized by chemiluminescent reaction using Supersignal West Pico or Dura Chemiluminescent Substrates (Thermo Fisher Scientific). When different antibodies were probed to the same nitrocellulose membrane, previous signals were removed by incubation with 0.5% sodium azide for 10 minutes at RT or by stripping in 62 mM Tris-HCL pH 6.8, 2% SDS, 0.7% β-mercaptoethanol for 20 min at 50 °C.
Quantitative PCR analysis.
RNA was isolated from shSCR-BV173 and shSTAT5-BV173 cells, untreated or treated with Doxycycline at 2.5 μg/ml, using the RNeasy Plus Mini Kit (Qiagen, Limburg, The Netherlands) and reverse-transcribed (2 μg) with the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA). Quantitative PCR was performed with the QuantStudio 12k Flex (Life Technologies) instrument and QuantStudio 12K Flex software, using the following primers: MYC FW 5’-GTCACACCCTTCTCCCTTCG-3’; MYC RV 5’-ATGTCTCCTCCCAGCAGCTC-3’; TRIB3 FW 5’-TGCGTGATCTCCAAGCTGTGT-3’; TRIB3 RV 5’-GCTTGTCCCACAGGGAATCA-3’; ITGα5 FW 5’-GGCTTCAACTTAGACGCGGA-3’; ITGα5 RV 5’-GGCTGGCTGGTATTAGCCTT-3’; ITGβ1 FW 5’-CCGCGCGGAAAAGATGAATTT-3’; ITGβ1 RV 5’-CCACAATTTGGCCCTGCTTG-3’; PIM1 FW 5’-ACACGGACTTCGATGGGACC-3’; PIM1 RV 5’-GATGGTCTCAGGGCCAAGCA-3’; IDH2 FW 5’-GCCGGCACTTTCAAAATGG-3’T; IDH2 RV 5’-TCCTTGACACCACTGCCATC-3’; UBC FW 5’-GTCGCAGTTCTTGTTTGTGGATC-3’; UBC RV 5’-GTCTTACCAGTCAGAGTCTTCACGAAG-3’; GAPDH FW 5’-CCCATCACCATCTTCCAGGAG-3’; GADPH RV 5’-CTTCTCCATGGTGGTGAAGACG-3’.
Animals
Animal experiments were approved by Thomas Jefferson University IACUC under protocol 00012.
For leukemogenesis assays, 106 leukemia cells (shSTAT5-cell lines or primary cells from Ph+ ALL patients) were injected intravenously in 7- to 9-week-old NOD/SCID/IL-2Rγnull mice (The Jackson Laboratory).
To induce STAT5 down-regulation in vivo, mice were continuously treated with Doxycycline (2 g/L) in D(+)-sucrose-supplemented (30 g/L) drinking water starting 72 hours post-cell injection.
IST5–002 was dissolved in 0.2% hydroxypropyl methylcellulose and administered intraperitoneally (50 or 100 mg/kg) daily for a total of 14 days.
Sabutoclax (5 mg/kg; ApexBio) was dissolved in 10% Kolliphor-EL (Sigma-Aldrich)-10% ethanol-80% PBS and administered intraperitoneally every other day for a total of seven doses (14 days).
AZD1208 (100 mg/Kg; SelleckChem) was dissolved in 0.5% hydroxypropyl methylcellulose-0.1% Tween 80 and administrated by oral gavage every day for 14 days. Leukemia formation was monitored by flow cytometry detection of the human CD19 antigen (by antibody #555415 from BD Bioscience) in peripheral blood obtained by retro orbital bleeding.
Statistical analyses
Data, expressed as mean ± s.d. of three experiments, were analyzed for statistical significance by unpaired, two-tailed Student’s t-test. P<0.05 was considered statistically significant. For drugs combination studies, combination indexes (CI) were calculated by the Chou-Talalay method (28) and synergism was defined as CI = 0.1–0.9.
Kaplan-Meier plots for mice survival experiments were generated using the GraphPad Prism 6.0 software. Differences in survival were assessed by log-rank test.
Results
STAT5 silencing decreases cell growth, increases apoptosis, and suppresses leukemogenesis of Ph+ ALL cells
To investigate the role of STAT5 in Ph+ ALL, we transduced human Ph+ leukemia cell lines BV173, SUP-B15 and Z181 with a lentiviral plasmid carrying a doxycycline (Doxy)-inducible shSTAT5 (shSTAT5-Tet-pLKO) and assessed cell growth and survival upon Doxy treatment.
Doxy treatment induced a decrease in STAT5 expression in each cell line (Fig. 1A). However, the effect was more pronounced in BV173 cells and became detectable after a 72 h treatment due to the long half-life of the STAT5 protein (29). As control, levels of STAT3 were not affected by the Doxy-induced shSTAT5 (Fig. 1A).
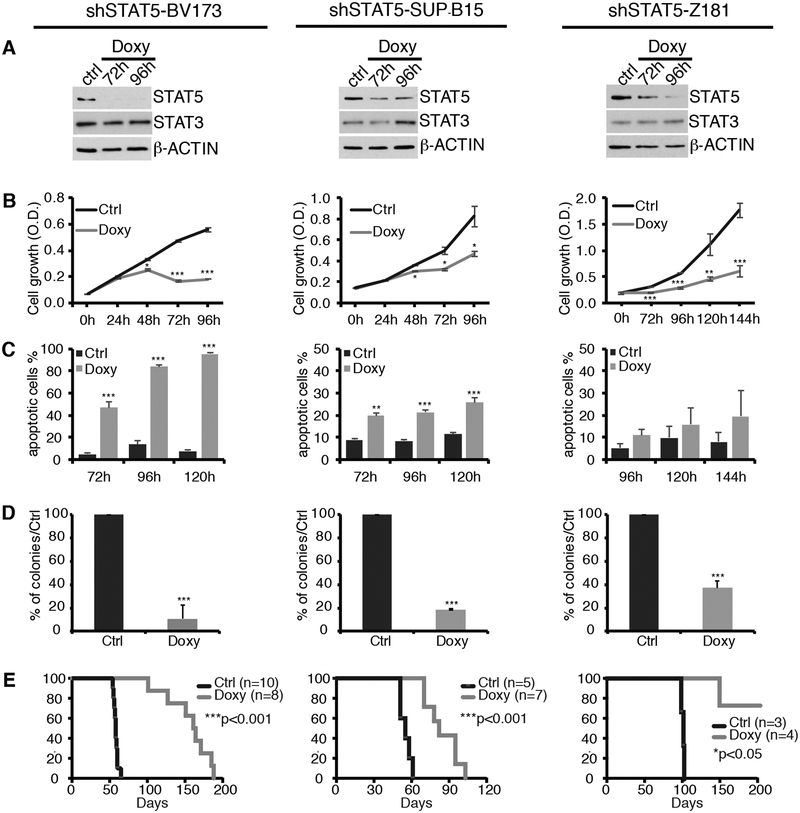
Fig. 1. Effect of STAT5 silencing on cell growth, apoptosis and leukemogenesis of Ph+ leukemia cell lines.
(A-D): Western blot (A), cell growth (B), apoptosis (C), and colony formation (D) of untreated or doxycycline (Doxy)-treated shSTAT5 Ph+ cell lines. Cell growth was determined by MTT assays and data are expressed as optical density (O.D). For Z181 cells, MTT assays were performed until 144 h. Apoptosis was measured as the % of cells expressing activated Caspase 3/7 by flow cytometry analysis. For colony formation assays, cells were seeded in methylcellulose plates (2,500–5,000 cells/dish), with or without Doxy. Colonies were counted 7–10 days after plating. Results are expressed as % inhibition of colony formation in Doxy-treated versus untreated plates. (no asterisk=not significant, *p<0.05, **p<0.01, ***p<0.001). (E) Kaplan-Meier survival plot of untreated or Doxy-treated NSG mice injected with shSTAT5 Ph+ leukemia cell lines. P values indicate statistical significance of the difference in survival between untreated and Doxy-treated mice.
STAT5 silencing markedly reduced cell growth of all three lines, but the effect was more evident in BV173 cells (Fig. 1B), probably reflecting the near complete inhibition of STAT5 expression in these cells (see Fig. 1A).
STAT5 silencing caused a marked increase in the apoptosis of BV173 and SUP-B15 cell lines measured by flow cytometry detection of Caspase 3/7 activation (Fig. 1C). Apoptosis was detected at 72 h and increased after a 96 and 120 h of Doxy treatment, consistent with the kinetics of Doxy-induced STAT5 downregulation. In BV173 cells, apoptosis induced by STAT5 silencing for 72–120 h ranged from 46% to 95%, while in SUP-B15 cells, at the same time points, it ranged from 20% to 26%. Compared to control cells, induction of apoptosis in STAT5-silenced Z181 cells was not statistically significant.
The dependence of Ph+ ALL cell lines on STAT5 expression was also assessed by methylcellulose colony formation assays. These assays revealed that Doxy-treated, STAT5-silenced BV173, SUP-B15 and Z181 cells were markedly less clonogenic than the untreated counterparts (Fig. 1D). Doxy treatment had no effect on STAT5 expression, apoptosis, or colony formation of shScramble-transduced Ph+ ALL cells (Supplementary Fig. 1).
Next, we assessed STAT5 requirement for leukemia development in NOD/SCID-IL-2Rγnull (NSG) mice injected with Ph+ BV173, SUP-B15, or Z181 cells transduced with the Doxy inducible shSTAT5 lentivirus. Mice were injected in the tail vein with 106 cells and, starting at day 7, Doxy was added to the drinking water while control mice were left untreated. Control mice injected with shSTAT5-BV173 or SUP-B15 cells died of leukemia (bone marrow heavily infiltrated by leukemic cells and splenomegaly) within 60 days of leukemic cell injection (median survival = 58 days). Mice injected with shSTAT5-BV173 cells and given Doxy to induce STAT5 silencing survived up to 188 days with a median survival of 162 days; such increase in overall survival was statistically significant (p<0.001) (Fig. 1E). Mice injected with shSTAT5-SUP-B15 cells and treated with Doxy survived up to 105 days with a median survival of 82 days which is significantly longer of that of the untreated mice (median survival = 55 days; p< 0.001) (Fig. 1E). Doxy treatment also induced a statistically significant increase in the survival of NSG mice injected with shSTAT5-Z181 cells (Fig. 1E). Untreated mice injected with shSTAT5-Z181 cells survived significantly longer (median survival = 102 days) than those injected with shSTAT5-BV173 or SUP-B15 cells; however, Doxy-treated mice injected with shSTAT5-Z181 cells survived more than 180 days (Fig. 1E).
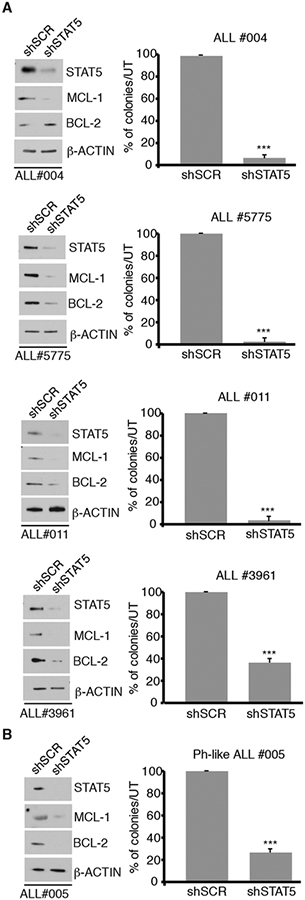
To further evaluate the requirement for STAT5 expression for growth of primary Ph+ ALL cells we utilized colony formation assays of patient-derived primary Ph+ ALL cells. For these assays, peripheral blood blast cells from four Ph+ ALL patients, two newly diagnosed (ALL #004 and ALL #011) and two from relapsed ALL post therapy with TKIs (ALL #5775 and ALL #3961) were transduced with a lentivirus constitutively expressing a STAT5 shRNA used to silence STAT5 expression in K562 cells (30), or a scramble shRNA and plated (105 cells/plate) in methylcellulose in the presence of puromycin. As shown in Fig. 2A, STAT5 expression was downregulated in shSTAT5 lentivirustransduced cells from each patient (left panels). Expression of STAT5-regulated BCL-2 and MCL-1 was also downregulated in STAT5-silenced samples, except for BCL-2 in sample ALL #004. Importantly, STAT5-silenced cells from patient’s samples were markedly less clonogenic than the scramble-transduced counterparts (Fig. 2A, right panels).
Fig. 2. Effect of STAT5 silencing on primary Ph+ or Ph-like ALL cells.
A and B) Western blot and colony formation of Ph+ ALL primary cells (newly diagnosed or relapsed/TKI-resistant) (A) or a newly diagnosed Ph-like ALL sample (B) transduced with a scramble or a STAT5 shRNA lentivirus. 100,000 cells/plate were seeded in the presence of puromycin. Colonies were counted 7–10 days after plating. Results are expressed as % inhibition of colony formation of shSTAT5-transduced versus scramble-transduced Ph+ or Ph-like ALL primary cells (***p<0.001).
Of interest, suppression of colony formation and a marked decrease of BCL-2 and MCL-1 levels compared to the scramble-transduced counterpart was also observed in STAT5-silenced blast cells from a patient with Ph-like ALL (#005, Supplementary Table T2) (Fig. 2B).
Pharmacological inhibition of phospho-STAT5 suppresses Ph+ ALL growth ex vivo and in mice
In silico screening of chemical structure databases has recently identified IST5–002 as a candidate small molecule inhibitor of STAT5 (31). Such activity was confirmed based on its ability to suppress JAK2 and BCR-ABL1 -mediated STAT5 phosphorylation, STAT5A/B dimerization, and to inhibit the transcriptional activity of STAT5A and STAT5B (31). Thus, we assessed if IST5–002 can mimic the ex vivo and in vivo effects of STAT5 silencing in Ph+ ALL cells. Treatment with IST5–002 markedly suppressed the growth of Ph+ cell lines, including the TKI-resistant T315I BV173 derivative (Supplementary Fig. 2). IST5–002-induced growth suppression was associated with markedly decreased STAT5 phosphorylation (Supplementary Fig. 2).
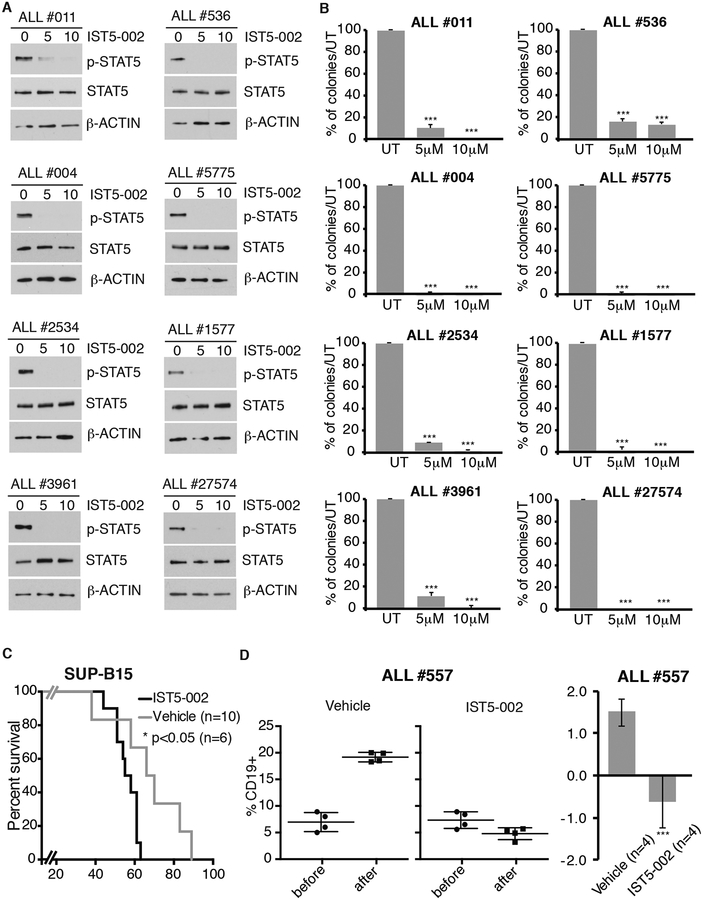
More importantly, treatment with IST5–002 also suppressed STAT5 phosphorylation and colony formation of leukemic blasts from eight Ph+ ALL patients (Fig. 3A and3B), including five samples (ALL #2534, ALL #5775, ALL #1577, ALL#27574 and ALL #3961) from patients with relapsed leukemia post-therapy with TKIs.
Fig. 3. Effect of the STAT5 inhibitor IST5–002 on colony formation and leukemogenesis of Ph+ ALL cells.
Western blot (A) and colony formation (B) of IST5–002-treated primary Ph+ ALL cells. For western blot analysis, cells from Ph+ ALL samples were left untreated (Ctrl) or treated with 5 or 10 μM IST5–002 for 24 h. Whole cell lysates were blotted with anti-p-STAT5, STAT5 and β-actin antibodies. For colony formation assays, cells (100,000/dish) were seeded in methylcellulose plates in presence of medium only or with 5 or 10 μM IST5–002. Colonies were counted after 10 days. Results are expressed as % inhibition of colony formation from IST5–002-treated versus untreated cells (***p<0.001). (C) Kaplan-Meier survival plot of NSG mice injected with SUP-B15 Ph+ ALL cells (106 cells/mouse) and treated with vehicle only or with IST5–002 (50 mg/kg/14 consecutive days) starting 7 days post-cell injection. *p<0.05 denotes statistical significance of the difference in survival between IST5–002-treated versus diluent-treated mice. (D) Peripheral blood leukemia load measured as percentage of CD19+ cells in NSG mice injected with ALL#557 sample before or after treatment with vehicle or IST5–002 (100 mg/kg/14 consecutive days) (right panels); (left panel) fold-change (log2 % CD19 positivity) in leukemia burden based on data shown in right panels (NS=not significant, *p<0.05, **p<0.01, ***p<0.001).
The Ph-like ALL cell lines MUTZ-5 and MHH-CALL-4 exhibit expression of phospho-STAT5 (Supplementary Fig. S3A) driven by the IGH/CRLF2 translocation and JAK2 mutation (32). Treatment with IST5–002 suppressed STAT5 phosphorylation and growth of these cell lines (Supplementary Fig. S3B) as well as STAT5 phosphorylation and colony formation of a primary Ph-like ALL sample (#005) also carrying the IGH/CRLF2 translocation (Supplementary Table 2) (Supplementary Fig. S3C).
To assess whether IST5–002 treatment has detrimental effects on normal hematopoietic cells, we performed colony formation assays of cytokine-treated stem cell-enriched G-CSF-mobilized CD34+ cells, samples J48 and J50, from two healthy donors. Treatment with IST5–002 (5 or 10μM) suppressed cytokine-dependent STAT5 phosphorylation in both samples (Supplementary Fig. S4)t however, IST5–002 had no effect on colony formation at the 5 μM concentration and partially reduced colony formation in sample J50 only at the 10μM concentration (Supplementary Fig. S4).
Then, we tested whether treatment with IST5–002 suppressed leukemogenesis in NSG mice injected with Ph+ ALL cells. For these experiments, we used the Ph+ ALL SUP-B15 cell line and primary cells from a patient with a TKI-resistant Ph+ ALL with the T315I ABL1 kinase domain mutation.
In the SUP-B15 model, mice were treated with vehicle only or with 50 mg/kg of IST5–002 for 14 consecutive days starting 7 days after cell injection. Vehicle-treated mice had a median survival of 56 days; by contrast, mice treated with IST5–002 survived up to 89 days, with a median survival of 68 days (p<0.05) (Fig. 3C).
In the TKI-resistant Ph+ ALL primary cells model, mice (4/group) were injected with 106 cells/mouse and when peripheral blood CD19+ cells were 5–9% (six weeks post cell injection) mice were treated i.p. for 14 days with a daily dose of 100 mg/kg of IST5–002 or with diluent only. Peripheral blood leukemia burden was assessed at the end of the treatment by flow cytometry analysis of CD19+ cells. While vehicle-treated mice showed an increase in the percentage of CD19+ leukemia cells from 5–8% to about 20%, leukemia growth was essentially blocked in the IST5–002-treated mice (Fig. 3D).
These findings support the concept that STAT5 or STAT5-regulated pathways may serve as targets for the treatment of Ph+ ALL.
Mechanisms involved in the growth suppression of STAT-silenced Ph+ ALL cells: role of BCL-2 family members
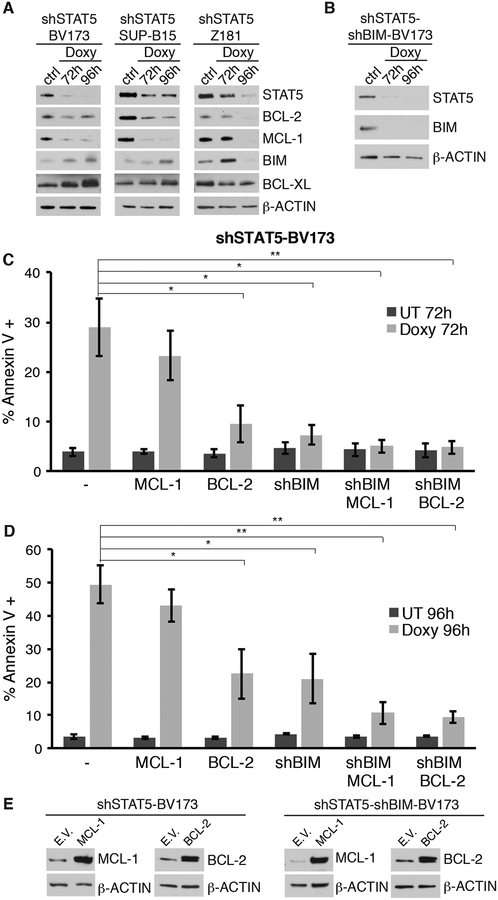
The growth suppression of Ph+ ALL cells induced by STAT5 silencing is largely caused by enhanced apoptosis (Fig. 1C). Thus, we assessed the expression of members of the BCL-2 family, some of which were previously reported to be regulated by STAT5 (33–36); such analysis is a necessary first step to further investigate if there is a functional link between changes in the expression of BCL-2 family proteins and apoptosis induced by STAT5 silencing. Compared to the untreated counterparts, Doxy-treated shSTAT5-BV173, shSTAT5-SUP-B15, and shSTAT5-Z181 cells exhibited a decrease in the expression of anti-apoptotic BCL-2 and MCL-1 proteins (Fig. 4A). Decreased expression of anti-apoptotic BCL-2 and MCL-1 proteins was also observed in STAT5-silenced Ph+ primary ALL cells and the Ph-like primary sample #005 (Fig. 2).
Fig. 4. Effect of BIM silencing or BCL-2/MCL-1 expression on the apoptosis of STAT5-silenced Ph+ ALL cells.
(A, B, E) Western blots show expression of BCL-2, MCL-1 and BIM in the indicated STAT5-silenced Ph+ ALL cell lines (A) and in the shSTAT5-BV173 derivative line transduced with the Doxy-regulated shBIM lentivirus (B), or in shSTAT5-BV173 ectopically expressing MCL-1 or BCL-2 (E, left), or in the shSTAT5-shBIM-BV173 line expressing MCL-1 or BCL-2 (E, right); (C, D) % of apoptotic cells detected by Annexin V staining in shSTAT5-BV173 parental and derivative cell lines, untreated or Doxy treated to silence STAT5 expression.
Doxy-treated shSTAT5-BV173, shSTAT5-SUP-B15, and shSTAT5-Z181 cells also exhibited an increase in the expression of the BH3-only pro-apoptotic BIM protein, although such an increase was detected at 72 but not at 96 h in Z181 cells (Fig. 4A). Expression of anti-apoptotic BCL-XL protein was unchanged (Fig, 4A).
To investigate whether increased BIM expression can explain the enhanced apoptosis of STAT5-silenced Ph+ ALL cells, we generated the shSTAT5-BV173 derivative cell line transduced with a Doxy-regulated BIM shRNA lentivirus (shSTAT5-shBIM-BV173 line) (Fig. 4B) and assessed apoptosis induced by STAT5 silencing. As shown in Fig. 4C–D and Supplementary Fig. S5, downregulation of BIM expression suppressed apoptosis induced by STAT5 silencing; however, a fraction of Doxy-treated shSTAT5-shBIM BV173 cells were apoptotic at 96 h (Fig. 4D), in spite of apparently complete inhibition of BIM expression (Fig. 4B). Next, we generated the shSTAT5-BCL-2-BV173 or the shSTAT5-MCL-1-BV173 derivative line (Fig. 4E, left panel) and assessed whether restoring BCL-2 or MCL-1 expression would also rescue the apoptosis of STAT5-silenced BV173 cells. In these lines, we assessed apoptosis only by Annexin V staining because GFP expression does not allow to measure active Caspase 3/7. As shown in Fig. 4C and 4D, expression of BCL-2 was as effective as BIM silencing in inhibiting apoptosis induced by STAT5 silencing while the effect of MCL-1 expression was negligible. Based on these findings, we generated the shSTAT5-shBIM-BCL-2-BV173 and the shSTAT5-shBIM-MCL-1-BV173 derivative lines (Fig. 4E, right panel) and asked whether combining BIM silencing with BCL-2 or MCL-1 expression would be more effective than BIM silencing or BCL-2/MCL-1 expression alone in blocking apoptosis induced by STAT5 silencing. As shown in Fig. 4C and 4D, combining BIM silencing with BCL-2 or MCL-1 expression rescued almost completely the apoptosis of STAT5-silenced BV173 cells.
Role of the STAT5-regulated PIM-1 gene in the growth of Ph+ ALL cells.
In a previous study (31), we performed oligonucleotide microarray hybridization to generate the gene expression profile of STAT5-silenced Ph+ K562 cells. Thus, we assessed whether the expression of select STAT5-regulated genes identified in K562 cells was also modulated in STAT5-silenced BV173 cells. As shown in Supplementary Fig. S6, expression of MYC, TRIB3, ITGB1, IDH2, ITGA5, PIM1 was markedly reduced in STAT5-silenced BV173 cells, compared to control cells. Then, we focused on STAT5-regulated PIM-1 because of: i) its established oncogenic effects (35); ii) commercially available compounds that inhibit PIM-1 and PIM-2 (37–39); iii) the efficacy of the selective PIM kinase inhibitor AZD1208 in pre-clinical models of acute myeloid leukemia (40).
First, we assessed if PIM-1 protein levels were reduced in STAT5-silenced Ph+ ALL cell lines. Doxy-treated shSTAT5 Ph+ cell lines all showed decreased expression of the short isoform of PIM-1 (PIM1-s) (Supplementary Fig. S7A).
Expression of PIM1-s in blast cells from patients with Ph+ ALL showed sample-to-sample variation; however, it correlated well with levels of total and tyrosine phosphorylated STAT5 (Supplementary Fig. S7B).
Of interest, levels of PIM1 and STAT5A and STAT5B mRNAs were also correlated in microarray dataset from patients with acute B-cell leukemia; in the Ph+ subset only the STAT5A-PIM1 correlation was statistically significant (Supplementary Fig. S7C–F). However, assessing levels of phospho-STAT5 is likely to be more informative than evaluating mRNA levels in correlative studies (41).
Then, we asked whether restoring PIM-1s expression in STAT5 silenced BV173 cells would rescue the growth inhibition of these cells. PIM-1s overexpression did not enhance the overall growth of STAT5-silenced BV173 cells because apoptosis and BIM expression induced by STAT5 silencing was not rescued (Supplementary Fig. S8).
These findings indicate that STAT5 activation downstream of BCR-ABL1 promotes growth and survival of Ph+ ALL cells through different effectors.
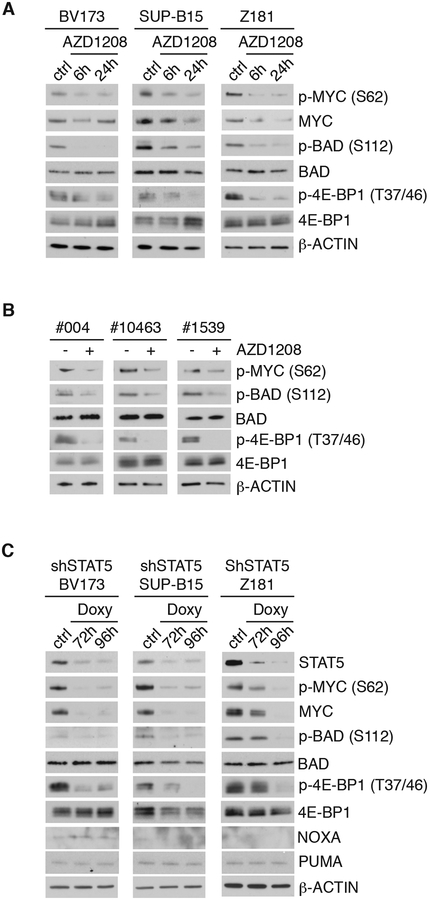
The role of the PIM-1 kinase in Ph+ ALL cells was assessed by pharmacological inhibition with the pan-PIM inhibitor AZD1208 (40).
BV173, SUP-B15, and Z181 cell lines were treated with 3 μM AZD1208 and PIM kinase-regulated pathways were analyzed by western blotting (Fig. 5A).
Fig. 5. Effect of PIM kinase inhibition or STAT5 silencing on signal transduction pathways in Ph+ ALL cells.
Western Blot analysis of PIM-1-regulated proteins in AZD1208-treated Ph+ ALL lines (A) and primary Ph+ ALL samples (B) or in STAT5-silenced Ph+ ALL lines (C).
Previous studies showed that PIM-1 stabilizes c-MYC expression through phosphorylation of Serine 62 (42, 43). Consistent with these findings, AZD1208-treated BV173, SUP-B15 or Z181 cells exhibited reduced levels of phospho (S62) MYC, but total levels of c-MYC were little changed (Fig. 5A).
Phosphorylation of BAD by PIM-1 at Serine 112 is a mechanism through which BAD is sequestered in the cytoplasm in complex with 14-3-3 allowing mitochondrial BCL-2 to suppress BAX-dependent apoptosis (44).
As expected, treatment of Ph+ ALL cell lines with AZD1208 led to a decrease in S112 phosphorylation with no changes in total levels of BAD (Fig. 5A). Expression of NOXA was low or undetectable and the expression of both PUMA and NOXA was not induced by AZD1208 treatment (Fig. 5A)
The translation regulator eukaryotic elongation factor 4E-BP1 is also regulated by PIM kinases, directly at the Threonines 37/46 priming sites and indirectly via mTOR-dependent mechanisms, causing the dissociation from eukaryotic initiation factor 4, and promoting the activation of cap-dependent translation (45–47).
Multiple bands, indicative of hyperphosphorylated 4E-BP1, were detected in untreated BV173, SUP-B15 and Z181 cells (Fig. 5A). Treatment with AZD1208 induced a marked decrease of phospho (T37/46)-4E-BP1 expression in SUP-B15, BV173 and Z181 cells (Fig. 5A) but total 4E-BP1 levels did not change (Fig. 5A).
Consistent with the effect of AZD1208 in Ph+ ALL cell lines, AZD1208-treated blast cells from three Ph+ ALL patients also showed decreased phosphorylation of BAD (S112), c-MYC (S62) and 4E-BP1 (T37/46) (Fig. 5B). Levels of total BAD and 4E-BP1 remained unchanged (Fig. 5B)
Of interest, STAT5 silencing in BV173, SUP-B15, or Z181 cells had effects similar to those of AZD1208 treatment on the phosphorylation of c-MYC (S62), 4E-BP1 (T37/46), and BAD (S112) (Fig. 5C), likely reflecting downregulation of PIM-1 levels. However, total levels of c-MYC were markedly reduced only after STAT5 silencing, suggesting that STAT5 regulates c-MYC expression through multiple pathways, including enhanced transcription (see Supplementary Fig. S6).
Together, these data indicate that PIM-1 might regulate cell growth of Ph+ ALL cells via activation of multiple pathways and suggest that pharmacological inhibition of PIM-1 could suppress Ph+ ALL cell growth.
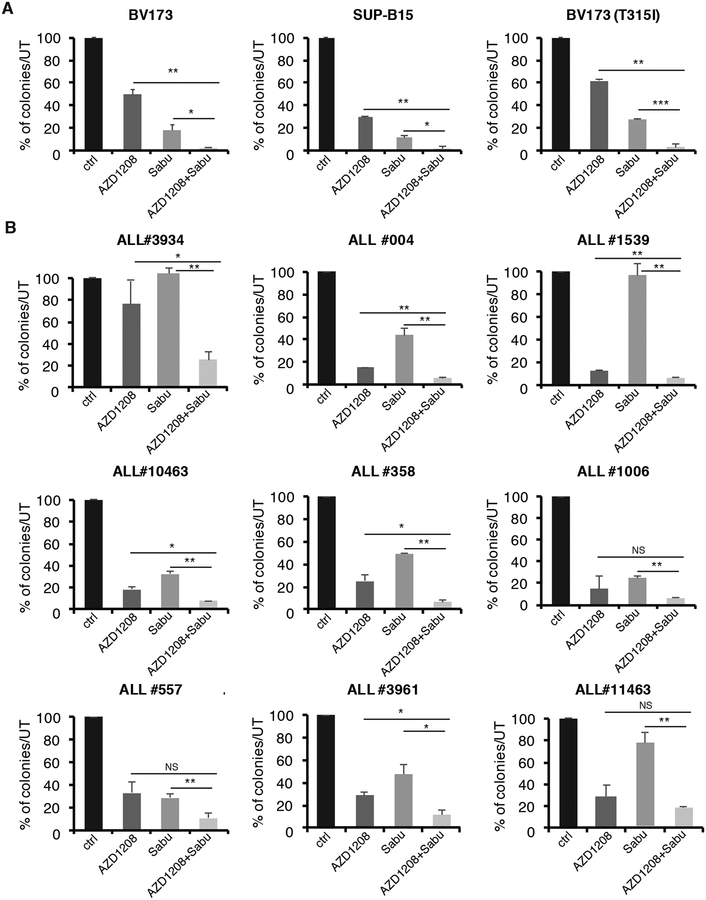
The PIM kinase inhibitor AZD1208 cooperates with the BCL-2 family antagonist Sabutoclax to suppress Ph+ ALL cell growth ex vivo and in NSG mice
Changes in the expression of BCL-2 and BIM appear to be essential for the apoptosis of STAT5-silenced Ph+ ALL cells (Fig. 4) and several growth-promoting pathways are suppressed in these cells by pharmacological inhibition of STAT5-regulated PIM kinase (Fig. 5). Thus, we tested the effects of the PIM kinase inhibitor AZD1208 and the pan-BCL-2 family inhibitor Sabutoclax in Ph+ ALL cells ex vivo and in NSG mice.
Sabutoclax functions as a BH3 mimetic that, like BIM, binds anti-apoptotic members of the BCL-2 family (BCL-2, BCL-XL, MCL-1) blocking their anti-apoptotic activity (48, 49). MTT assays revealed that treatment with AZD1208 or Sabutoclax suppressed the growth of BV173, SUP-B15, and Z181 cells, including the TKI resistant BV173 (T315I) cell line and that the combined treatment was more effective than either drug alone (Supplementary Fig. S9A–D), suggesting that these drugs target non-overlapping pathways.
While treatment with Sabutoclax alone strongly increased apoptosis in each Ph+ ALL cell line, treatment with AZD1208 alone did not. However, the AZD1208/Sabutoclax combination further increased apoptosis in BV173, BV173 (T315I), SUP-B15, and Z181 cells (Supplementary Fig. S9E–H). These findings suggest that suppressing PIM kinase signaling (e.g., phosphorylation of BAD) is insufficient to provide a strong apoptotic signal, but it enhanced apoptosis induced by the BCL-2 family antagonist Sabutoclax.
These findings also suggest that the AZD1208/Sabutoclax combination may inhibit synergistically the growth of Ph+ ALL cells.
Thus, Ph+ ALL cell lines were treated with three doses of AZD1208 and/or Sabutoclax, and the effects on cell growth were analyzed by MTT assay (Supplementary Fig. S10A–D). Combination indexes (C.I.) were calculated using CompuSyn software and plotted in Supplementary Fig. S10E–H.
The combined treatment had synergistic effects at all drug concentrations except one in parental and TKI-resistant BV173 cells and in Z181 cells while the effect was predominantly additive in SUP-B15 cells (Supplementary Fig. S10E–H).
Interestingly, co-treatment with the two drugs also enhanced apoptosis of Ph-like MUTZ-5 and MHH-CALL-4 cell lines compared to treatment with AZD1208 or Sabutoclax alone (Supplementary Fig. S11).
To further analyze the efficacy of the combined AZD1208/Sabutoclax treatment on Ph+ ALL cells, we performed methylcellulose colony assays of drug-treated BV173, BV173 (T315I), SUP-B15 cell lines, and nine primary samples from Ph+ ALL patients. These samples are from six newly diagnosed and three relapsed/TKI-resistant ALL patients (Sample #557 expresses the p210 isoform and carries the T315I ABL1 mutation; sample #3961 expresses the p190 isoform and carries the T315I mutation; and sample #11463 expresses the p190 isoform with no mutations detected in the TKD). Treatment with AZD1208 or Sabutoclax alone suppressed, with varying degrees, colony formation in each case, with the exception of Sabutoclax treatment of ALL samples #1539 and #3934, but the most marked effect was observed when the two drugs were used in combination (Fig. 6A and B).
Fig. 6. Effect of AZD1208 and Sabutoclax on colony formation of Ph+ leukemia cell lines and primary Ph+ ALL cells.
Methylcellulose colony formation of BV173, SUP-B15 and BV173 (T315I) cell lines (A) or primary Ph+ ALL samples (B), untreated or treated with AZD1208 (3 μM), Sabutoclax (80 nM), or a combination of AZD1208 and Sabutoclax. Colonies were counted 7–10 days after plating; results are expressed as % inhibition of colony formation from drug-treated versus untreated cells (*p<0.05, **p<0.01, ***p<0.001).
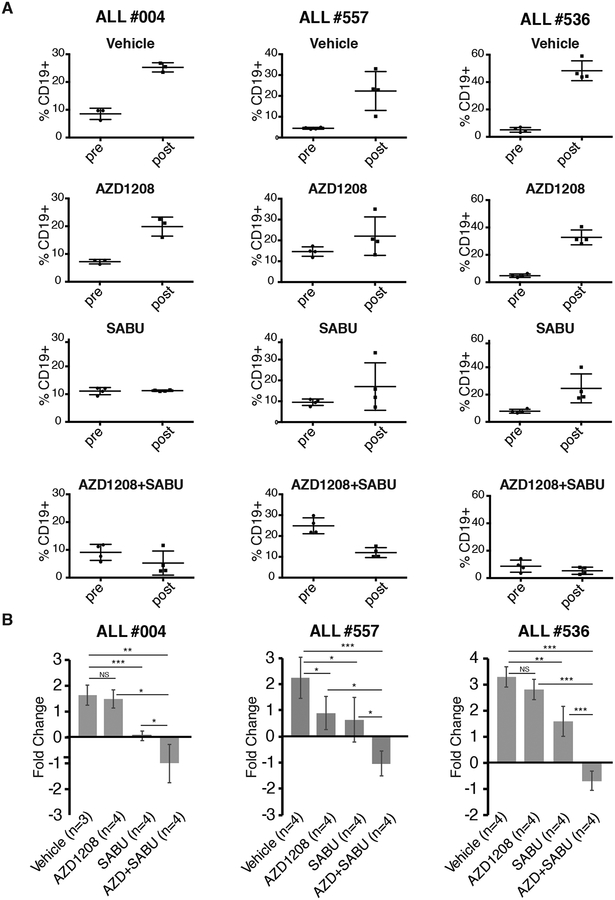
Next, we assessed the effect of the AZD1208/Sabutoclax combination in leukemia progression in vivo. NSG mice were injected with Ph+ ALL #004, #536 or #557 (T315I) cells and, when peripheral blood CD19+ cells were 5–20%, mice were treated for 14 days with AZD1208 alone, Sabutoclax alone, or with both drugs. Then, peripheral blood leukemia burden was assessed; the AZD1208/Sabutoclax combination was more effective than either drug alone in suppressing Ph+ ALL, resulting in leukemia burdens lower than those observed before the treatment, in each Ph+ ALL sample (Fig. 7A and B).
Fig. 7. Effect of AZD1208 and Sabutoclax on Ph+ ALL burden in NSG mice.
(A) Percentage of CD19+ cells in NSG mice injected with ALL#004, ALL#557 or ALL#536 sample before or after treatment with vehicle only, AZD1208, Sabutoclax, or AZD1208 and Sabutoclax; (B) fold-change (log2 % CD19 positivity) in leukemia burden based on data shown in the A panels (NS=not significant, *p<0.05, **p<0.01, ***p<0.001).
Discussion
In this study, we assessed the dependence of Ph+ ALL cells on the expression/activity of STAT5 and its downstream effectors and used such knowledge for targeted therapy of Ph+ ALL in patient-derived xenografts.
The role of STAT5 for in vivo maintenance of oncogenic Abl-driven ALL was previously investigated in p210-BCR-ABL1 or v-Abl-induced leukemia upon conditional deletion of STAT5A/B (18, 19, 50). STAT5 expression was found to be dispensable for p210-BCR-ABL1-driven B-cell leukemia in Balb/c mice while STAT5A/B deletion markedly suppressed v-Abl- and p190-BCR-ABL1-driven B-cell leukemia in C57BL/6 mice (18, 19, 50). Whether such outcomes reflect differences in genetic background of recipient mice and/or biological properties of p210-BCR-ABL1 versus v-Abl-transformed B-cell precursors is, at the moment, unclear.
Despite the potential relevance of STAT5 in BCR-ABL1-driven leukemia, the role of STAT5 expression has not been previously assessed in human Ph+ ALL cells. Using Ph+ ALL cell lines expressing a Doxy-regulated shSTAT5 which inhibits the expression of both STAT5 isoforms, we show here that downregulation of STAT5 expression leads to markedly decreased proliferation and colony formation, induction of apoptosis, and significantly prolonged survival of NSG mice injected with Ph+ ALL cells. Of interest, FACS-sorted CD19+ BV173 isolated from the bone marrow of a mouse which developed leukemia, in spite of being continuously treated with Doxy, expressed STAT5 suggesting that re-expression of STAT5 caused the re-growth of Ph+ ALL.
STAT5-silenced primary Ph+ ALL cells from patients with newly diagnosed or relapsed/TKI-resistant disease were markedly less clonogenic than the empty vector-transduced counterpart.
Of interest, STAT5 silencing also suppressed colony formation from blast cells of a patient with IGH/CRLF2-positive Ph-like ALL.
Based on these findings, we assessed the reliance of Ph+ ALL cells on STAT5 activity by ex vivo and in vivo assays using IST5–002, a small molecule inhibitor of STAT5A/B tyrosine phosphorylation and dimerization, recently identified through structure-based in silico screening of compounds binding to the STAT5 SH2 domain (31).
Such STAT5 inhibitor was highly effective in suppressing colony formation and proliferation of Ph+ and Ph-like ALL cells while sparing a large fraction of normal stem cell-enriched CD34+ hematopoietic cells, in spite of suppressing STAT5 phosphorylation very effectively also in these cells. The in vivo effects of IST5–002 were more modest of those induced by STAT5 silencing; since it is unclear whether any of the commercially available STAT5 inhibitors, including IST5–002, will be ever used in clinical trials, we investigated if Ph+ ALL cells are dependent on STAT5-regulated pathways that may provide alternative targets for the therapy of Ph+ ALL. Targeting such pathways should phenocopy, in part, the effects induced by STAT5 silencing or pharmacological inhibition. We found that the marked apoptosis induced by silencing STAT5 expression in Ph+ BV173 cells was largely dependent on downregulation of BCL-2 or upregulation of pro-apoptotic BIM while modulating MCL-1 expression had negligible effects. While these findings suggest that levels of BCL-2 and BIM are critical for STAT5-regulated survival of BV173 cells, other Ph+ ALL cell lines and patients’-derived primary ALL cells may have different requirements for STAT5-regulated BCL-2 family members. In particular, this would not be surprising based on our findings that levels of BCL-2 family members exhibit wide variations in Ph+ ALL cells (51).
The growth and colony formation of Ph+ ALL cells, including those derived from blast cells of newly diagnosed or relapsed/TKI-resistant Ph+ ALL patients, was also markedly suppressed by pharmacological inhibition of the STAT5-regulated PIM1 kinase. However, restoring PIM1 expression was insufficient to rescue the impaired growth of STAT5-silenced BV173 cells. These findings suggest that the anti-apoptotic signals that may be mediated by the PIM1 kinase through phosphorylation of the BH3-only protein BAD cannot compensate for the increased expression of BIM protein induced by STAT5 silencing.
On the other hand, PIM kinase inhibition suppressed growth-promoting pathways dependent on c-MYC expression and 4EBP-1 phosphorylation (42,45–47), likely explaining the ex vivo growth inhibitory effects of AZD1208 in Ph+ ALL cells.
Given that that Ph+ ALL cells rely for their growth on the expression/activity of STAT5-regulated effectors that can be targeted pharmacologically, we assessed the growth of Ph+ ALL cells ex vivo and in NSG mice, upon treatment with the PIM kinase inhibitor AZD1208 and the pan-BCL-2 inhibitor Sabutoclax.
In most Ph+ ALL samples, including three samples from patients with relapsed/TKI-resistant disease, co-treatment with Sabutoclax and AZD1208 had synergistic or additive growth-suppressive effects compared to the treatment with single agents; these findings are not surprising since AZD1208 and Sabutoclax inhibit non-overlapping pathways regulated, at least in part, by STAT5 in Ph+ ALL cells.
The additive growth-suppressive effect of the AZD1208/Sabutoclax combination was also detected in vivo by leukemogenesis assays in NSG mice injected with three Ph+ ALL primary samples, including sample #557 which carries the TKI-resistant T315I mutation. Although these assays are not directly comparable, it should be noted that in vivo growth suppression by treatment with AZD1208 or Sabutoclax alone did not correlate exactly with data from colony formation assays. In particular, leukemia load was markedly suppressed by treatment with Sabutoclax alone while treatment with AZD1208 alone was effective only in one of three Ph+ ALL samples, in contrast to the statistically significant inhibition of colony formation induced by AZD1208 in eight of nine samples.
While these data point to the need of performing additional studies to assess to extent to which treatment with AZD1208 suppresses Ph+ ALL growth ex vivo, they support the main finding of our in vivo studies which is the marked suppression of leukemia load induced by the AZD1208/Sabutoclax combination.
In summary, our data indicate that targeting STAT5 or STAT5-regulated pathways by pharmacological approaches might provide an alternative treatment for patient with Ph+ ALL, especially those with relapsed or TKI-resistant disease.
Supplementary Material
Acknowledgments
This work was supported, in part, by NCI grant RO1-CA167169 (B. Calabretta) and RO1-CA113580, R21CA178755 and Advancing a Healthier Wisconsin (#5520368) (M. Nevalainen). We thank Dr. C.M. Eischen for critically reviewing the article. We thank Dr. Z. Jagani (Novartis, Cambridge, MA) for kindly providing the Tet-pLKO-Neo shBIM lentiviral vector, Dr. M. Carroll from the Stem Cell and Xenograft Core of the University of Pennsylvania for providing primary Ph+ ALL samples and Dr. N. Flomenberg and the Bone Marrow Transplantation unit at Thomas Jefferson University for providing CD34+ human hematopoietic progenitor cells.
Financial Support: this work was supported, in part, by NCI grant RO1-CA167169 (B. Calabretta) and R01-CA113580, R21CA178755 and Advancing a Healthier Wisconsin (#5520368) (M. Nevalainen).
Footnotes
Conflict of Interest Disclosure: the authors declare no potential conflict of interest.
References
- 1.Faderl S, Kantarjian HM, Talpaz M, Estrov Z. Clinical significance of cytogenetic abnormalities in adult acute lymphoblastic leukemia. Blood. 1998;91:3995–4019. [PubMed] [Google Scholar]
- 2.Lugo TG, Pendergast AM, Muller AJ, and Witte ON. Tyrosine kinase activity and transformation potency by bcr-abl oncogene products. Science. 1990. 247:1079–1082. [DOI] [PubMed] [Google Scholar]
- 3.Wetzler M, Dodge RK, Mrozek K, Carroll AJ, Tantravahi R, Block AW, et al. Prospective karyotype analysis in adult acute lymphoblastic leukemia: the cancer and leukemia Group B experience. Blood. 1999. 93:3983–93. [PubMed] [Google Scholar]
- 4.Gleissner B, Gokbuget N, Bartram CR, Janssen B, Rieder H, Janssen JW, et al. Leading prognostic relevance of the BCR-ABL translocation in adult acute B-lineage lymphoblastic leukemia: a prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002. 99:1536–43. [DOI] [PubMed] [Google Scholar]
- 5.El Fakih R, Jabbour E, Ravandi F, Hassanein M, Anjum F, Ahmed S, and Kantarjian H. Current paradigms in the management of Philadelphia chromosome positive acute lymphoblastic leukemia in adults. American J. Hematol 2017. Am J Hematol. 2013 Oct 3. doi: 10.1002/ajh.24926. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Bassan R, Rossi G, Pogliani EM, Di Bona E, Angelucci E, Cavattoni I, et al. Chemotherapy-phased imatinib pulses improve long-term outcome of adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia: Northern Italy Leukemia Group protocol 09/00. J Clin Oncol. 2010. 28(22):3644–52. [DOI] [PubMed] [Google Scholar]
- 7.Stirewalt DL, Guthrie KA, Beppu L, Bryant EM, Doney K, Gooley T, et al. Predictors of relapse and overall survival in Philadelphia chromosome-positive acute lymphoblastic leukemia after transplantation. Biology of Blood and Marrow Transplantation. 2003. 9:206–12. [DOI] [PubMed] [Google Scholar]
- 8.Yanada M, Takeuchi J, Sugiura I, Akiyama H, Usui N, Yagasaki F, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J. Clin. Oncol 2006. 24:460–6 [DOI] [PubMed] [Google Scholar]
- 9.Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106:3760–7 [DOI] [PubMed] [Google Scholar]
- 10.Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood 2012;119:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pulte D, Jansen L, Gondos A, Katalinic A, Barnes B, Ressing M, et al. Survival of adults with acute lymphoblastic leukemia in Germany and the United States. PloS One. 2014;9:e85554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soverini S, De Benedittis C, Papayannidis C, Paolini S, Venturi C, Iacobucci I, et al. Drug resistance and BCR-ABL kinase domain mutations in Philadelphia chromosome-positive acute lymphoblastic leukemia from the imatinib to the second-generation tyrosine kinase inhibitor era: The main changes are in the type of mutations, but not in the frequency of mutation involvement. Cancer 2014120:1002–9. [DOI] [PubMed] [Google Scholar]
- 13.Goetz CA, Harmon IR, O’Neil JJ, Burchill MA, and Farrar MA. STAT5 activation underlies IL7 receptor-dependent cell development. J. Immunol 2004. 172: 4770–4778. [DOI] [PubMed] [Google Scholar]
- 14.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, et al. Stat5 is essential for early B cell development but not for B cell maturation and function. J. Immunol 2007. 179: 1068–1079. [DOI] [PubMed] [Google Scholar]
- 15.Malin S, McManus S, and Busslinger M. STAT5 in B cell development and leukemia. Curr. Opin. Immunol 2010. 22: 168–176. [DOI] [PubMed] [Google Scholar]
- 16.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol 2010. 11: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012. 22:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol. Med 20102:98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwaller J, Parganas E, Wang D, Cain D, Aster JC, Williams IR, et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell 2000. 6:693–704. [DOI] [PubMed] [Google Scholar]
- 21.Xie S, Wang Y, Liu J, Sun T, Wilson MB, Smithgall TE, et al. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene 200120:6188–95 [DOI] [PubMed] [Google Scholar]
- 22.Nelson EA, Walker SR, Weisberg E, Bar-Natan M, Barrett R, Gashin LB, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 2011;117:3421–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Holbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011117:3409–20. [DOI] [PubMed] [Google Scholar]
- 24.Pegoraro L, Matera L, Ritz J, Levis A, Palumbo A, and Biagini G. Establishment of a Ph1-positive human cell line (BV173). J. Natl. Cacer. Inst 1987. 70: 447–453. [PubMed] [Google Scholar]
- 25.Wu J, Meng F, Ying Y, Peng Z, Daniels L, Bornmann WG, et al. ON012380, a putative BCR-ABL kinase inhibitor with a unique mechanism of action in imatinib-resistant cells. Leukemia. 2010. 24: 869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young L, Sung J, Stacey G, Masters JR. Detection of Mycoplasma in cell cultures. Nature protocols. 2010. 5:929–34 [DOI] [PubMed] [Google Scholar]
- 27.Jagani Z, Wiederschain D, Loo A, He D, Mosher R, Fordjour P, et al. The Polycomb group protein Bmi-1 is essential for the growth of multiple myeloma cells. Cancer Res. 201070:5528–38. [DOI] [PubMed] [Google Scholar]
- 28.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 29.Chia DJ, Subbian E, Buck TM, Hwa V, Rosenfeld RG, Skach WR, et al. Aberrant folding of a mutant Stat5b causes growth hormone insensitivity and proteasomal dysfunction. J. Biol. Chem 2006;281:6552–8. [DOI] [PubMed] [Google Scholar]
- 30.Soliera AR, Mariani SA, Audia A, Lidonnici MR, Addya S, Ferrari-Amorotti G, et al. Gfi-1 inhibits proliferation and colony formation of p210BCR/ABL-expressing cells via transcriptional repression of STAT 5 and Mcl-1. Leukemia. 2012. 26(7):1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Z, Gu L, Vergalli J, Mariani SA, De Dominici M, Lokareddy RK, et al. Structure-based screen Identifies a potent small molecule Inhibitor of Stat5a/b with therapeutic potential for prostate cancer and chronic myeloid leukemia. Mol. Cancer Ther 2015. 14:1777–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tasian SK, Doral MY, Borowitz MJ, Wood BL, Chen IM, Harvey RC, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012. 120: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimoto G, Miyamoto T, Jabbarzadeh-Tabrizi S, Iino T, Rocnik JL, Kikushige Y, et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood. 2009. 114: 5034–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gesbert F and Griffin JD. Bcr/Abl activates transcription of the Bcl-X gene through STAT5. Blood. 2000. 96: 2269–2276. [PubMed] [Google Scholar]
- 35.Lord JD, McIntosh BC, Greenberg PD, Nelson BH. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J Immunol. 2000. 164:2533–41. [DOI] [PubMed] [Google Scholar]
- 36.Nawijn MC, Alendar A, and Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer 2011. 11: 23–34. [DOI] [PubMed] [Google Scholar]
- 37.Chen LS, Redkar S, Bearss WG, Wierda NG, and Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009. 114: 4150–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia Z, Knaak C, Ma J, Beharry ZM, Mciness C, Wang W, et al. Synthesis and evaluation of novel inhibitors of PIM-1 and PIM-2. J. Med. Chem 2009. 52: 74–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curi DA, Beauchamp EM, Blyth GT, Arslan AD, Donato NJ, Giles FJ et al. Pre-clinical evidence of PIM kinase inhibitor activity in BCR-ABL1 unmutated and mutated Philadelphia chromosome-positive (Ph+) leukemias. Oncotarget. 20156:33206–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeton EK, McEachern K, Dillman KS, Palakurthi S, Cao Y, Grondine MR, et al. AZD1208, a potent and selective pan-Pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014;123:905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med. 2011. 208:1135–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–19. [DOI] [PubMed] [Google Scholar]
- 43.Horiuchi D, Camarda R, Zhou AY, Yau C, Momcilovic O, Balakrishnan S, et al. PIM1 kinase inhibition as a targeted therapy against triple-negative breast tumors with elevated MYC expression. Nat. Medicine 2016;22:1321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS letters. 2004;571:43–9. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, Chen LS, Neelapu SS, Miranda RN, Medeiros LJ, and Gandhi V. Transcription and translation are primary targets of Pim kinase inhibitor SGI-1776 in mantle cell lymphoma. Blood. 2012. 120:3491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamburini J, Green AS, Bardet V, Chapuis N, Park S, Willems L, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009. 114:1618–27. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F, Beharry ZM, Harris TE, Lilly MB, Smith CD, Mahajan S, and Kraft AS. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol Ther. 2009. 8:846–53. [DOI] [PubMed] [Google Scholar]
- 48.Wei J, Stebbins JL, Kitada S, Dash R, Placzek W, Rega MF, et al. BI-97C1, an optically pure Apogossypol derivative as pan-active inhibitor of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem. 2010. 53:4166–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goff DJ, Court Recart A, Sadarangani A, Chun HJ, Barrett CL, Krajewska M, et al. A Pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013. 12:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, et al. Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood. 2012119:3550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Dominici M, Porazzi P, Soliera AR, Mariani SA, Addya S, Fortina P, et al. Targeting CDK6 and BCL2 exploits the “MYB addiction” of Ph+ acute lymphoblastic leukemia. Cancer Res. 2018. 78:1097–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.